The kinesin-5 Eg5 is essential for mitotic progression, and the lethal effects of Eg5 inhibitors make these inhibitors attractive candidates for chemotherapy drugs. Sturgill et al. show that kinesin-12 and a nonmotile Eg5 mutant form an alternative spindle assembly pathway that provides resistance to Eg5 inhibitors.
Abstract
The microtubule (MT) cytoskeleton bipolarizes at the onset of mitosis to form the spindle. In animal cells, the kinesin-5 Eg5 primarily drives this reorganization by actively sliding MTs apart. Its primacy during spindle assembly renders Eg5 essential for mitotic progression, demonstrated by the lethal effects of kinesin-5/Eg5 inhibitors (K5Is) administered in cell culture. However, cultured cells can acquire resistance to K5Is, indicative of alternative spindle assembly mechanisms and/or pharmacological failure. Through characterization of novel K5I-resistant cell lines, we unveil an Eg5 motility-independent spindle assembly pathway that involves both an Eg5 rigor mutant and the kinesin-12 Kif15. This pathway centers on spindle MT bundling instead of Kif15 overexpression, distinguishing it from those previously described. We further show that large populations (∼107 cells) of HeLa cells require Kif15 to survive K5I treatment. Overall, this study provides insight into the functional plasticity of mitotic kinesins during spindle assembly and has important implications for the development of antimitotic regimens that target this process.
Introduction
During mitosis, microtubules (MTs) organize into a bipolar array termed the spindle that segregates the duplicated genome. Spindle bipolarity is essential for accurate chromosome segregation and is established by separating the duplicated centrosomes in animal cells. Given the importance of spindle bipolarity, the cell deploys a cohort of MT-associated factors to drive centrosome separation. Kinesin-5/Eg5 is the dominant player in mammalian somatic cells (Sawin et al., 1992; Blangy et al., 1995), being optimally tuned for this specific function in the following ways. First, Eg5 organizes into homotetramers with a pair of motor heads on opposing ends of the molecule (Kashina et al., 1996). This enables single molecules to simultaneously engage adjacent MTs (Kapitein et al., 2005). Second, its catalytic cycle is limited by ATP hydrolysis rather than product release, biasing Eg5 to remain attached to MTs (Krzysiak and Gilbert, 2006). With these unique mechanochemical properties, Eg5 harnesses its ATPase activity to slide antiparallel MTs apart, thereby generating centrosome separation forces important for bipolarizing the nascent spindle.
Eg5 also contains unique structural features that dispose the motor to small-molecule inhibitors (Brier et al., 2004; Cox et al., 2005; Maliga and Mitchison, 2006; Lad et al., 2008). We and others have capitalized on Eg5 inhibitors (K5Is) to reveal auxiliary spindle assembly pathways that emerge after chronic exposure K5Is (Raaijmakers et al., 2012; Sturgill and Ohi, 2013; Ma et al., 2014). This approach has improved our understanding of spindle physiology and adaptability, revealing that cytoplasmic dynein and the kinesin-12 Kif15 can drive centrosome separation in K5I-resistant cells (Raaijmakers et al., 2012; Sturgill and Ohi, 2013). Additional studies have shown that Eg5 can become refractory to pharmacological inhibition through the acquisition of mutations that abrogate motor–small molecule interactions (Kasap et al., 2014). Despite this progress, it remains to be tested whether such diverse means of K5I resistance share a commonality that could serve as a focal point for therapeutic intervention.
Here, we identify Kif15 as a molecular linchpin of K5I resistance in HeLa cells. We first describe a novel spindle assembly pathway that involves a spontaneous Eg5 rigor mutant and Kif15. We propose that the Eg5 rigor mutant, which tightly binds MTs regardless of its nucleotide state and/or pharmacological inhibitors, activates Kif15-driven spindle assembly by creating MT bundles, the preferred substrate of Kif15 (Sturgill et al., 2014). Kif15 is not overexpressed in this scenario, contrasting a better-characterized K5I rescue pathway that requires elevated Kif15 levels (Tanenbaum et al., 2009; Vanneste et al., 2009; Sturgill and Ohi, 2013). Given that Kif15 plays a prominent role in the small handful of K5I-resistant cells (KIRCs) characterized thus far, we next test the prevalence of Kif15 in the acquisition of K5I resistance. Using a HeLa cell line largely devoid of Kif15, we find that adaptation to K5Is requires Kif15 under all tested conditions. We conclude that Kif15 is essential for K5I resistance in HeLa cells, even in cases that necessitate additional factors such as the Eg5 rigor mutant discovered here.
Results
KIRC-2 and -3 express a spontaneous Eg5 rigor mutant, Eg5-G268V
We generated K5I-resistant cell lines by treating HeLa cells with a saturating dose of S-trityl-l-cysteine (STLC; DeBonis et al., 2004) and isolating emergent colonies. We assigned the acronym KIRC (K5I-resistant cell) to these cell lines in place of EIC (Eg5-independent cell; Sturgill and Ohi, 2013), because not all adaptation mechanisms may obviate a requirement for Eg5. KIRC-1 was previously published as OL-EIC-1 (Sturgill and Ohi, 2013), whereas KIRC-2 and -3 represent new, uncharacterized cell lines. All three KIRCs were continuously cultured in STLC, whereas the parental HeLa line was maintained in the absence of K5Is unless otherwise noted.
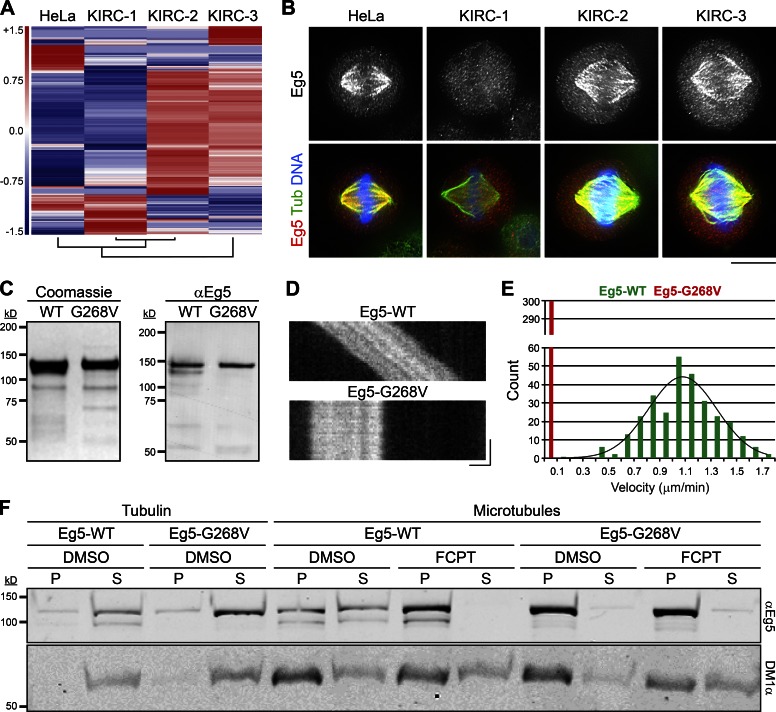
To ensure that the three KIRCs are unrelated, we compared their transcriptomes by RNA sequencing (RNaseq). Hierarchical cluster analysis revealed unique mRNA fingerprints for all three KIRCs (Fig. 1 A), indicating that each cell line is clonally distinct. RNaseq further indicated that none of the KIRCs have significantly altered EG5 expression levels (Table 1). We next monitored the localization of Eg5 in each cell line by immunostaining. Although Eg5 was absent from spindle MTs in KIRC-1 cells, it was readily detectable on spindles in KIRC-2 and -3 cells (Fig. 1 B). This observation raised the possibility that KIRC-2 and -3 cells may express a mutant form of Eg5 that is refractory to STLC.
Figure 1.
KIRC-2 and -3 express a spontaneous Eg5 rigor mutant, Eg5-G268V. (A) Each KIRC line is clonally distinct. Unsupervised cluster and heatmap analysis of indicated cell types. The dendrogram (bottom) shows that KIRC-1 and KIRC-2 are most similar based on gene expression. HeLa is more similar to KIRC-1 and KIRC-2 than KIRC-3. KIRC-3 is the most distinct cell line. (B) Eg5 binds spindle MTs in KIRC-2 and -3 cells despite the presence of STLC. Single optical sections of indicated cell types stained with antibodies targeting Eg5 (grayscale and red) and tubulin (green). DNA, blue. Lookup tables (LUTs) for grayscale and red channel are scaled identically. Bar, 10 µm. See also Fig. S1. (C) Purification of recombinant Eg5 proteins. (left) Protein gel of Eg5-WT and Eg5-G268V preparations stained with Coomassie-blue. (right) Western blot of Eg5-WT and Eg5-G268V preparations probed with antibodies targeting Eg5 (αEg5). (D) Eg5-G268V is immotile. Kymographs of single XR-MTs in gliding assays propelled by Eg5-WT or Eg5-G268V. Bars: (x axis) 1 µm; (y axis) 1 min. See also Videos 1 and 2. (E) Histogram showing gliding velocities from D. Values represent the number of MTs observed to move at indicated velocities in gliding assays powered by Eg5-WT (green) or Eg5-G268V (red). Velocities are reported in micrometers per minute and binned every 0.1 µm/min. n = 300 from three independent experiments. Distributions of Eg5-WT velocities are fit to a single Gaussian (R2 = 0.91). (F) Eg5-G268V does not release from MTs in the presence of ATP. Blot of pellet (P) and supernatant (S) fractions from reactions containing Eg5-WT or Eg5-G268V with tubulin or microtubules in the presence of DMSO or FCPT as indicated, probed with antibodies targeting Eg5 (αEg5) and tubulin (DM1α).
Table 1. EG5and KIF15 expression levels.
| Transcript levels | KIRC-1 | KIRC-2 | KIRC-3 |
|---|---|---|---|
| EG5: fold changea | −0.20 | −0.18 | −0.09 |
| EG5: p-value | 0.51 | 0.54 | 0.76 |
| KIF15: fold changea | 2.38 | 0.06 | 0.11 |
| KIF15: p-value | 5.00 × 10−5 | 0.88 | 0.79 |
Fold change is calculated relative to the parental HeLa line.
RNaseq analysis revealed that KIRC-2 and -3 express a variant EG5 allele alongside the wild-type (WT) form (Table 2). The spontaneous point mutation, guanine 803 to thymine (G803T), encodes a glycine 268 to valine (G268V) substitution within the switch II helix of the motor domain (Rice et al., 1999). G268 is a highly conserved residue among members of the kinesin superfamily, and mutations to the analogous residues within conventional kinesin (G234) and Kip1p (G297) trap the motor heads in a strong MT-binding state (i.e., “rigor”), regardless of the nucleotide cofactor (Rice et al., 1999; Chua et al., 2007). To test whether a G268V substitution similarly affects Eg5, we examined the catalytic and MT-binding properties of recombinant Eg5-WT and Eg5-G268V (Fig. 1 C) in vitro.
Table 2. EG5 expression profile.
| Transcript profile | KIRC-2 | KIRC-3 |
|---|---|---|
| G803T allele frequency | 31.71% | 31.03% |
In conventional MT gliding assays, Eg5-WT moved MTs in an ATP-dependent manner at an expected velocity of 1.1 ± 0.3 µm/min (mean ± SD; Fig. 1, D and E; and Video 1; Kwok et al., 2004). In contrast, Eg5-G268V failed to power MT gliding, despite its ability to capture MTs from solution (Fig. 1, D and E; and Video 2). This result can potentially be explained by an inability of Eg5-G268V to release MTs, even in the presence of ATP. To more rigorously test this idea, we performed MT copelleting assays with Eg5-WT and Eg5-G268V in ATP-containing buffers. Indeed, Eg5-G268V was enriched in the MT pellet compared with Eg5-WT (Fig. 1 F). FCPT, an ATP-competitive K5I that induces tight binding of Eg5 onto MTs (Groen et al., 2008; Rickert et al., 2008), shifted all detectable Eg5-WT into the MT pellet yet did not alter the fractionation of Eg5-G268V (Fig. 1 F).
Coupled with published literature (Rice et al., 1999; Chua et al., 2007), our results demonstrate that Eg5-G268V maintains a high affinity for MTs irrespective of its nucleotide state. This rigor mutation could underlie the ability of Eg5 to bind spindles in KIRC-2 and -3 cells (Fig. 1 B). To test the ability of STLC to displace rigor Eg5 from spindle MTs, we treated KIRC-1 cells with FCPT to induce tight MT binding of WT Eg5 (Table 2) in STLC-treated cells. Although Eg5 was absent from spindles in DMSO-treated KIRC-1 cells, it localized to spindles in FCPT-treated KIRC-1 cells (Fig. S1). This demonstrates that a rigor motor state of Eg5 is dominant to pharmacological inhibition by STLC. Because FCPT and STLC block Eg5 activity through different mechanisms (DeBonis et al., 2004; Rickert et al., 2008), this result cannot be explained by simple competition of the two K5Is for a single drug-binding site. We conclude that Eg5 tetramers in KIRC-2 and -3 containing at least one copy of the rigor mutant Eg5-G268V will stably bind spindle MTs despite the presence of STLC.
Eg5-G268V robustly bundles MTs and is essential for spindle assembly in KIRC-2 and -3
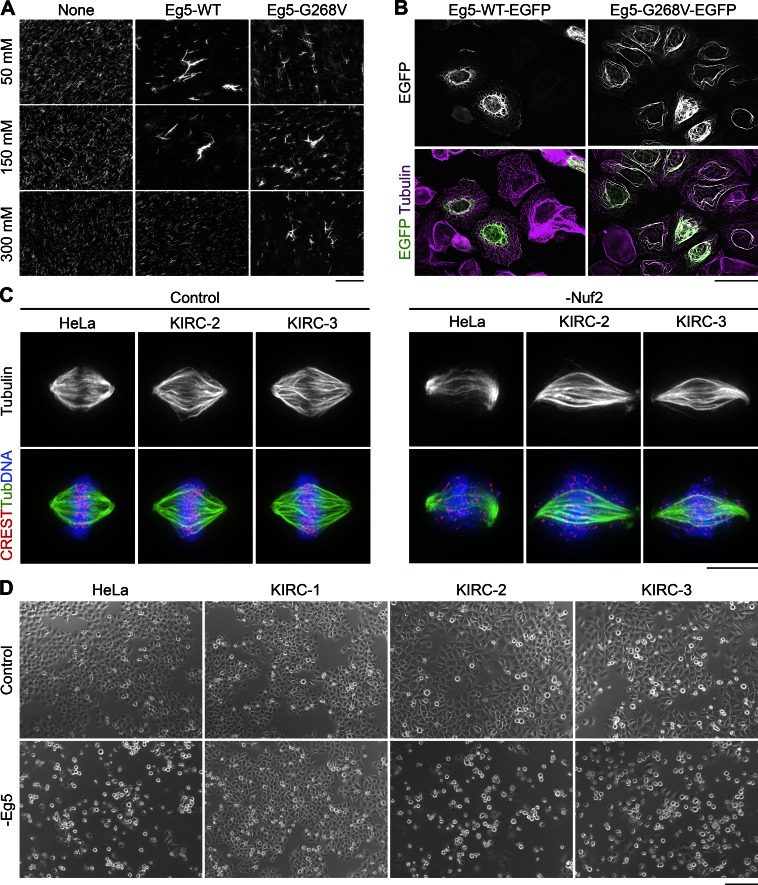
Heterotetramers containing at least two copies of Eg5-G268V would be expected to function as static MT cross-linkers instead of motile MT sliders. We therefore compared the abilities of Eg5-WT and Eg5-G268V to bundle stabilized MTs in vitro. In ATP-containing buffers of near-physiological ionic strength, both Eg5-WT and Eg5-G268V bundled MTs (Fig. 2 A). But as Eg5-WT likely turns over more frequently in these structures, increases in ionic strength may impair its MT bundling capacity more so than that of Eg5-G268V. Indeed, high salt concentrations disrupted the MT bundling effects of Eg5-WT (Cahu and Surrey, 2009) but did not compromise the MT bundling activity of Eg5-G268V (Fig. 2 A). The ability of Eg5-G268V to cross-link MTs in buffers of high ionic strength suggests that it may be well equipped to bundle dynamic MTs in cells.
Figure 2.
Eg5-G268V robustly bundles spindle MTs and is essential for spindle assembly in KIRC-2 and -3 cells. (A) Eg5-G268V bundles MTs in high salt buffers. Squashes from reactions containing XR-MTs (grayscale) alone or in combination with Eg5-WT or Eg5-G268V. Reactions contained 50, 150, or 300 mM KCl as indicated. Bar, 40 µm. (B) Eg5-G268V-EGFP bundles MTs in interphase cells. Single optical sections of HeLa cells expressing Eg5-WT-EGFP or Eg5-G268V-EGFP (grayscale/green) and stained with antibodies targeting tubulin (pseudocolored purple). LUTs for individual channels are scaled identically. Bar, 40 µm. (C) Spindle MTs are unusually bundled in KIRC-2 and -3 cells. Maximum intensity z-projections of indicated cell types 2 d after transfection with nontargeting (Control) or Nuf2-targeting (-Nuf2) siRNAs and stained with antibodies targeting centromeres (CREST, red) and tubulin (grayscale and green). DNA, blue. Bar, 10 µm. (D) Eg5 depletion induces a mitotic arrest in KIRC-2 and -3. Bright-field images of indicated cell types 1 d after transfection with nontargeting (Control) or Eg5-targeting (-Eg5) siRNAs. Bar, 200 µm. See also Fig. S2.
To test the MT bundling capacity of the Eg5 rigor mutant in cells, we expressed Eg5-WT-EGFP or Eg5-G268V-EGFP in HeLa cells. Similar to endogenous Eg5, Eg5-WT-EGFP did not appreciably decorate interphase MTs (Fig. 2 B; Blangy et al., 1995; Sawin and Mitchison, 1995; Goshima and Vale, 2005). In contrast, Eg5-G268V-EGFP strongly decorated and bundled interphase MTs (Fig. 2 B). Mitotic cells expressing Eg5-G268V-EGFP contained spindles with warped poles in comparison to cells expressing Eg5-WT-EGFP (Fig. S2 A), suggesting that the rigor mutant also bundles spindle MTs. To test the MT bundling effects of endogenous Eg5-G268V, we monitored spindle MT architecture in KIRC-2 and -3 cells. Subtle changes in spindle MT bundling are difficult to detect, so we focused on the non–kinetochore–microtubule (non–K-MT) population to increase our ability to detect enhanced MT bundling. We depleted the outer kinetochore component Nuf2 by RNAi to prevent K-MT formation, yielding spindles with reduced end-on K-MT attachments and scattered chromosomes (Fig. 2 C; DeLuca et al., 2002). In comparison to HeLa cells, non–K-MTs in Nuf2-depleted KIRC-2 and -3 cells appeared highly bundled (Fig. 2 C). Altogether, our results indicate that Eg5-G268V converts Eg5 tetramers from motile MT sliders to static MT cross-linkers with the potential to bundle spindle MTs in KIRC-2 and -3.
To test whether Eg5-G268V contributes to spindle assembly in KIRC-2 and -3, we assessed the effects of Eg5 RNAi on the mitotic index (MI) and monopolar index (MPI) of each cell line. As expected, Eg5 depletion increased the parental HeLa line MI from 6.3 ± 1.0 to 80.1 ± 4.2 and MPI from 14.3 ± 1.8 to 100.0 ± 0.0 (mean ± SEM, unless otherwise noted; Fig. 2 D and Fig. S2, B and C). Meanwhile, Eg5 depletion negligibly affected KIRC-1, changing the MI from 6.3 ± 1.1 to 8.2 ± 1.4 and the MPI from 49.7 ± 5.6 to 32.9 ± 1.1 (Fig. 2 D and Fig. S2, B and C). Strikingly, KIRC-2 and -3 responded to Eg5 RNAi similarly to the parental HeLa line. Eg5 RNAi increased the MI of KIRC-2 from 10.6 ± 0.4 to 57.6 ± 1.0 and KIRC-3 from 8.9 ± 0.7 to 51.1 ± 6.7 (Figs. 2 D and S2 B). Furthermore, Eg5 RNAi increased the MPI of KIRC-2 from 52.3 ± 8.4 to 96.3 ± 1.8 and KIRC-3 from 50.0 ± 3.8 to 96.3 ± 0.3 (Fig. S2 C). Because Eg5 depletion, but not pharmacological inhibition, halted spindle assembly and mitotic progression in KIRC-2 and -3, we conclude that Eg5 remains essential in these cell lines independent of its catalytic activity. These results demonstrate that Eg5 remains essential for mitotic progression in some K5I-resistant cells and reveal a potentially novel role for Eg5 as a MT cross-linker during spindle assembly.
Enhanced spindle MT bundling in KIRC-2 and -3 cells affects Kif15 partitioning
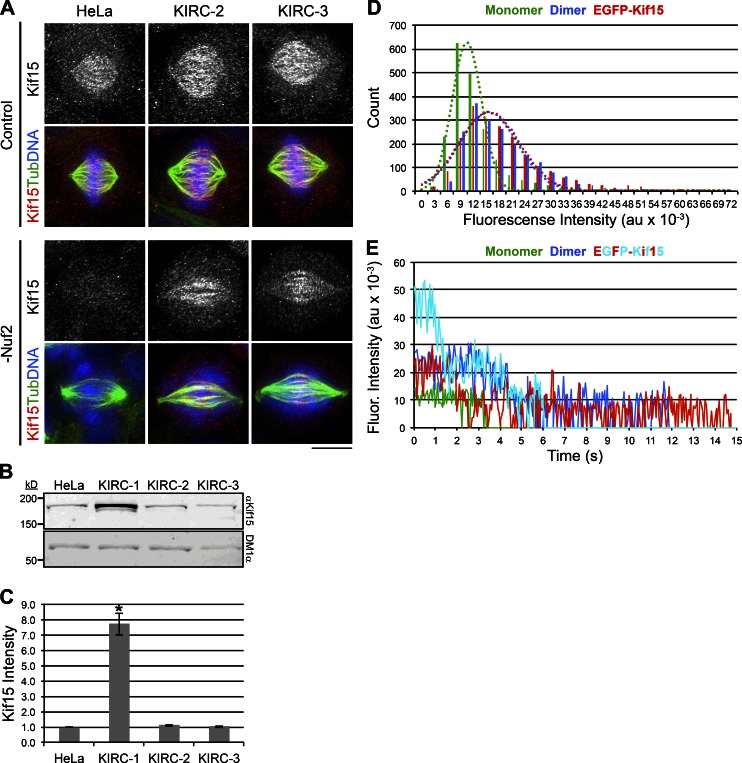
In principle, MT bundling by Eg5-G268V could affect spindle assembly by changing the distribution and activity of other spindle-associated factors. For example, the kinesin-12 Kif15 is normally restricted to K-MTs because of its preferential binding to MT bundles (Sturgill and Ohi, 2013; Sturgill et al., 2014), but excessive MT bundling can distribute Kif15 onto non–K-MTs (Sturgill et al., 2014). Given this, we next monitored Kif15 localization in Nuf2-depleted KIRC-2 and -3 cells. As expected, Kif15 was absent from spindle MTs in Nuf2-depleted HeLa cells (Fig. 3 A; Sturgill and Ohi, 2013). In contrast, Kif15 was readily detectable on spindle MTs in Nuf2-depleted KIRC-2 and -3 cells (Fig. 3 A). These results demonstrate a change in the partitioning of Kif15 within KIRC-2 and -3 cells and reinforce our conclusion that these cell lines exhibit enhanced spindle MT bundling.
Figure 3.
Enhanced spindle MT bundling in KIRC-2 and -3 cells affects Kif15 partitioning. (A) Kif15 mislocalizes to non–K-MTs in KIRC-2 and -3 cells. Maximum intensity z-projections of indicated cell types 2 d after transfection with nontargeting (Control) or Nuf2-targeting (-Nuf2) siRNAs and stained with antibodies targeting Kif15 (grayscale and red) and tubulin (green). DNA, blue. LUTs for grayscale and red channels are scaled identically. Bar, 10 µm. (B) Kif15 protein levels are not elevated in KIRC-2 or -3. Western blot of WCLs prepared from indicated cell types probed with antibodies targeting Kif15 (αKif15) and tubulin (DM1α). (C) Quantitation of B. Values represent the levels of Kif15 protein in indicated cell types normalized to the parental HeLa line. n = 2. Error bars, ± SEM; *, P ≤ 0.1 relative to parental HeLa line. (D) Cellular Kif15 primarily organizes into dimers. Histogram showing initial single-molecule fluorescence intensities of EGFP-Kif15 (red) from HeLa cell extracts. XMAP215-EGFP (green) represents monomer control. EGFP-HSET (blue) represents dimer control. Intensities are indicated in au × 10−3. n ≥ 1945 particles from ≥ 3 fields of view. Single Gaussian fits of fluorescence intensity distributions are overlayed. XMAP215-EGFP (green, R2 = 0.96), EGFP-HSET (blue, R2 = 0.90), and EGFP-Kif15 (red, R2 = 0.93). (E) Example traces showing photobleaching steps of single EGFP-Kif15 molecules from HeLa cell extracts. XMAP215-EGFP (green) represents monomer control. EGFP-HSET (blue) represents dimer control. Two example traces are shown for EGFP-Kif15 (red, cyan) highlighting the variability observed in photobleaching steps. Fluorescence intensities are indicated in au × 10−3, and the baseline is set to the background fluorescence. See also Fig. S3.
We previously demonstrated that Kif15 disperses onto non–K-MTs in KIRC-1 cells and attributed this phenomenon to Kif15 overexpression (Sturgill and Ohi, 2013). To determine whether Kif15 overexpression accounts for its non–K-MT localization in KIRC-2 and -3 cells, we next analyzed Kif15 transcript and protein levels in each cell line. Total Kif15 protein levels were elevated 7.7 ± 0.7-fold in KIRC-1 compared with the parental HeLa line but were unaltered in KIRC-2 and -3 (Fig. 3, B and C). Similarly, KIF15 transcript levels were elevated in KIRC-1 compared with the parental HeLa line but were not significantly altered in KIRC-2 or -3 (Table 1). Because Kif15 is not overexpressed in KIRC-2 and -3, we speculate that enhanced MT bundling by Eg5-G268V is responsible for distributing Kif15 onto non–K-MTs in these cells.
Kif15 targets MT bundles as a consequence of the dimeric motor locking in an active conformation upon simultaneously binding two MTs (Sturgill et al., 2014). Because this was demonstrated with purified Kif15 protein, it remains to be tested whether Kif15 also forms dimers in cells. To examine the oligomerization state of cellular Kif15, we expressed EGFP-labeled Kif15 (EGFP-Kif15) in HeLa cells and subjected the cellular extracts to single-molecule analysis (see Materials and methods). The initial single-molecule fluorescence intensity of EGFP-Kif15 averaged 16,316 ± 398 arbitrary units (au; mean ± the 95% confidence interval [COI]; Figs. 3 D and S3 A). These values resembled those of EGFP-HSET (15,680 ± 400 au, mean ± the 95% COI), a presumed dimer based on the biochemical properties of its Xenopus homolog (Hentrich and Surrey, 2010), and contrasted those of XMAP215-EGFP (10,046 ± 215 au, mean ± the 95% COI; Figs. 3 D and S3 A), a known monomer (Brouhard et al., 2008). Furthermore, these values closely resembled those of recombinant EGFP-Kif15 (Fig. S3 B), which we previously demonstrated to be dimeric (Sturgill et al., 2014). Of note, the cell cycle state of the extracts did not affect the initial fluorescence intensity distributions of EGFP-Kif15 (Fig. S3 B). We primarily observed two-step photobleaching of EGFP-Kif15 molecules; however, in some cases, we observed more than two photobleaching steps (Figs. 3 E and S3 C). We suspect that the latter population corresponds to the small peak around 50,000 au in the initial fluorescence intensity distributions (Fig. 3 D). We conclude that although cellular Kif15 can assemble into higher-order complexes (Drechsler et al., 2014), its primary oligomerization state is a dimer.
Kif15 drives spindle assembly in KIRC-2 and -3
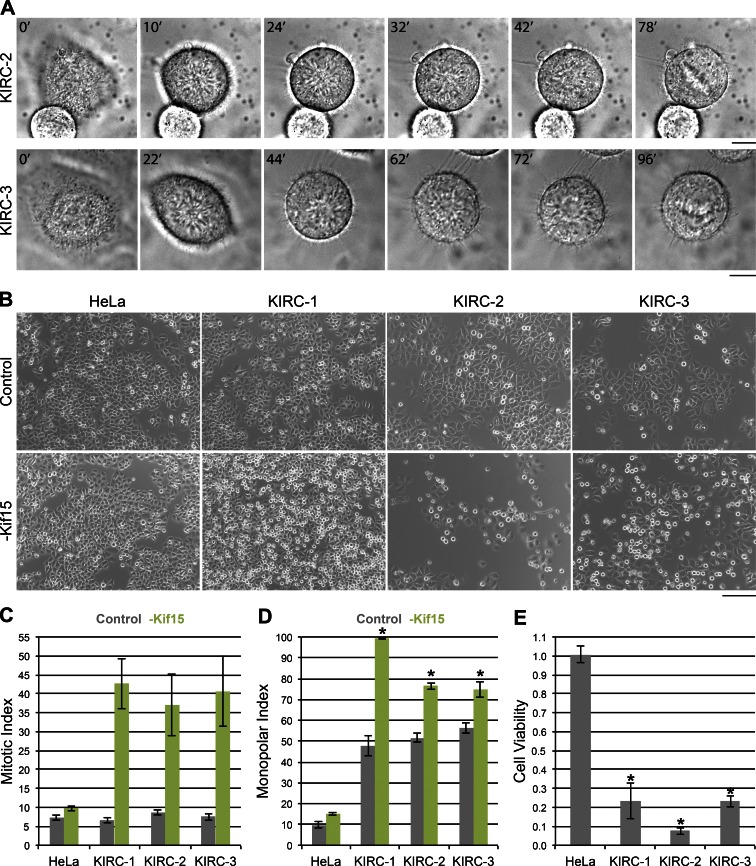
Kif15 drives spindle assembly in KIRC-1 cells through a process termed “reverse jackknifing” (Sturgill and Ohi, 2013), wherein spindles transit through a monopolar geometry before bipolarizing. Unfortunately, overexpression of fluorescently labeled tubulin proved extremely toxic to KIRC-2 and -3 cells, preventing live-cell imaging of tubulin in these cell lines. We therefore used differential interference contrast (DIC) imaging to infer spindle geometry in KIRC-2 and -3 cells as they progressed through mitosis. Chromatin rosettes were apparent in both cell types after mitotic entry and preceding metaphase plate formation (Fig. 4 A). KIRC-2 and -3 also exhibit abnormally high basal MPIs (Figs. 4 D and S2 C), suggesting that spindles assemble through a monopolar intermediate in KIRC-2 and -3. Whether mechanical aspects of spindle assembly in KIRC-2 and -3 cells are similar to KIRC-1 cells remains to be determined.
Figure 4.
Kif15 drives spindle assembly in all three KIRCs. (A) Spindles assemble through a monopolar intermediate in KIRC-2 and -3 cells. Sequential images of a KIRC-2 and -3 cell entering mitosis taken by time-lapse DIC microscopy. Time is indicated in minutes relative to the initial frames. Intermediate frames reveal chromatin rosettes and final frames show metaphase plates. Bar, 10 µm. (B) Kif15 depletion induces a mitotic arrest in all three KIRCs. Bright-field images of indicated cell types 1 d after transfection with nontargeting (Control) or Kif15-targeting (-Kif15) siRNAs. Bar, 200 µm. (C) Quantitation of B. Values represent the mean MI of indicated cell types 1 d after transfection with nontargeting (gray) or Kif15-targeting (green) siRNAs. Error bars, ± SEM; n ≥ 800 cells from three independent experiments. (D) Kif15 depletion prevents spindle assembly in all 3 KIRCs. Values represent the mean MPI of indicated cell types 1 d after transfection with nontargeting (gray) or Kif15-targeting (green) siRNAs. Error bars, ± SEM; n = 300 cells from three independent experiments. *, P ≤ 0.02 relative to control. (E) Kif15 depletion leads to massive cell death in all three KIRCs. Values represent the normalized luminescence of indicated cell types 2 d after transfection averaged over three experiments. Error bars, ± SEM; *, P ≤ 0.01 relative to the parental HeLa line.
To test the requirement of Kif15 during spindle assembly in KIRC-2 and -3 cells, we quantified the effect of Kif15 RNAi on the MI and MPI of each cell line 1 d after siRNA transfection. Consistent with previous work (Tanenbaum et al., 2009; Vanneste et al., 2009), Kif15 depletion had little effect on the parental HeLa line, changing the MI from 7.3 ± 0.8 to 9.8 ± 0.7 (Fig. 4, B and C). In contrast, Kif15 depletion increased the MI of KIRC-1 from 6.6 ± 0.7 to 42.7 ± 6.7, KIRC-2 from 8.7 ± 0.7 to 37.0 ± 8.2, and KIRC-3 from 7.5 ± 0.9 to 40.6 ± 9.2 (Fig. 4, B and C). To test whether this mitotic arrest resulted from failed spindle assembly, we next assessed the effect of Kif15 RNAi on the MPI of each cell line 1 d after siRNA transfection. Again, Kif15 depletion affected the KIRC lines more so than the parental HeLa line, increasing the MPI of the parental HeLa line from 9.7 ± 1.5 to 15.0 ± 0.6, KIRC-1 from 47.7 ± 4.7 to 99.3 ± 0.3, KIRC-2 from 51.7 ± 2.3 to 76.3 ± 1.5, and KIRC-3 from 56.3 ± 2.7 to 74.7 ± 3.8 (Fig. 4 D). Altogether, these results demonstrate that Kif15 contributes to spindle assembly and mitotic progression in all three KIRCs.
We next monitored the effect of Kif15 RNAi on the viability of each cell line 2 d after siRNA transfection. Within each cell line, the ATP-dependent luminescence from a population of Kif15-depleted cells was normalized to that from a population of mock-depleted cells. The normalized luminescence of the parental HeLa line was 1.0 ± 0.0, demonstrating no effect of Kif15 depletion on viability (Fig. 4 E). In contrast, the normalized luminescence averaged 0.2 ± 0.1 in KIRC-1, 0.1 ± 0.0 in KIRC-2, and 0.2 ± 0.0 in KIRC-3, indicative of massive cell death (Fig. 4 E). These data reveal that all three KIRCs use an alternative, Kif15-dependent spindle assembly pathway to survive K5I treatment.
Kif15 is essential for K5I resistance in HeLa cells
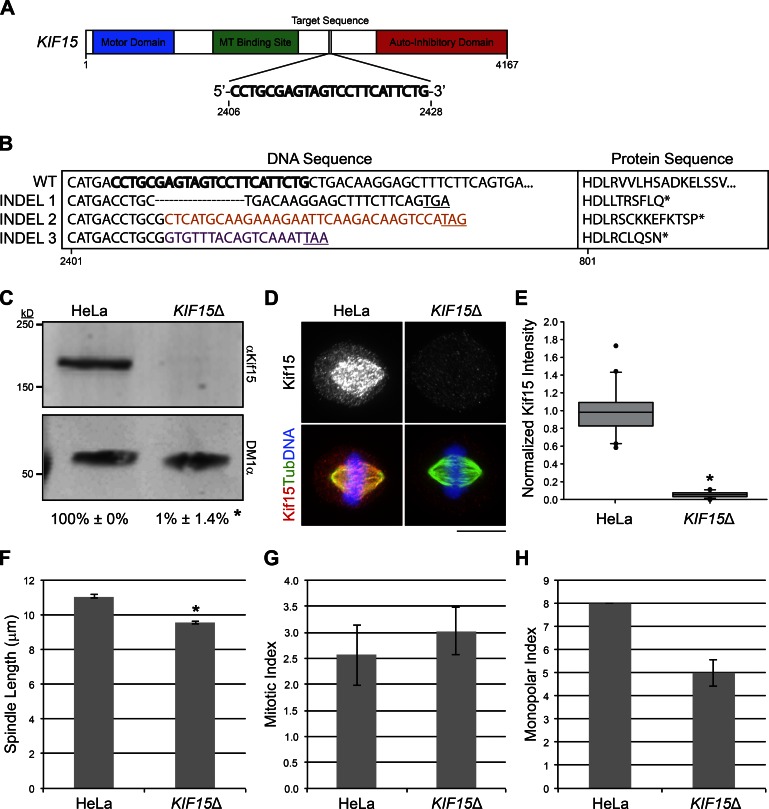
Kif15 is critical for mitotic progression in KIRC-1–3 and also in a limited number of KIRCs analyzed in previous studies (Raaijmakers et al., 2012; Sturgill and Ohi, 2013; Ma et al., 2014). The extent to which Kif15 drives the acquisition of K5I resistance within a cell population, however, has not been systematically tested. To address this issue, we examined the ability of HeLa cells to acquire K5I resistance in the absence of Kif15. To create a HeLa cell line largely devoid of Kif15 protein (KIF15Δ), we used the CRISPR-Cas9 system (Mali et al., 2013) to target nucleotides 2406–2428 of the KIF15 ORF for genomic alteration (see Materials and methods; Fig. 5 A). Within the human genome, the target sequence 5′-CCTGCGAGTAGTCCTTCATTCTG-3′ is unique to exon 20 of KIF15, minimizing the potential for off target effects. Whole-exome sequencing (WES) confirmed disruption of KIF15 in the KIF15Δ line, revealing three distinct insertions/deletions (INDELs; Figs. 5 B and S4). Each INDEL presumably corresponds to one KIF15 allele, consistent with literature suggesting that various HeLa strains possess a relatively stable hypertriploid karyotype (Macville et al., 1999; Adey et al., 2013). The first KIF15Δ INDEL, detected by 26 sequencing reads, is a deletion within exon 20 (corresponding to nucleotides 2,411–2,429 within the ORF), resulting in a frame shift and early stop codon (Figs. 5 B and S4). The second and third KIF15Δ INDELs are nucleotide insertions and also create a premature stop codon (Figs. 5 B and S4). INDELs 2 and 3 were detected in seven and four sequencing reads, respectively.
Figure 5.
Generation of the KIF15Δ cell line. (A) Schematic of KIF15 showing target sequence used for CRISPR-mediated genomic editing. Regions encoding the motor domain (blue), MT binding site (green), and autoinhibitory domain (red) are highlighted. Numbers indicate nucleotide position within the ORF. (B) Three distinct INDELs disrupt KIF15 in the KIF15Δ cell line. DNA sequences (left) and the translated protein sequences (right) for WT and KIF15Δ INDELs 1–3 are shown. Bold letters indicate the guide sequence used for CRISPR. Colored letters indicate inserted nucleotides. Underlined codons and asterisks indicate a premature stop. Numbers indicate nucleotide or amino acid position within the KIF15 ORF. See also Fig. S4. (C) Kif15 protein is undetectable in the KIF15Δ line. Western blot of WCLs prepared from HeLa or KIF15Δ cells probed with antibodies targeting Kif15 (αKif15) and tubulin (DM1α). Numbers represent the levels of Kif15 protein in HeLa or KIF15Δ WCLs normalized to the parental HeLa line, averaged over two experiments. Error bars, ± SD; *, P = 0.006 relative to parental HeLa line. (D) Kif15 spindle localization is undetectable in KIF15Δ cells. Maximum intensity z-projections of HeLa or KIF15Δ cells stained with antibodies targeting Kif15 (grayscale and red) and tubulin (green). DNA, blue. LUTs for individual channels are scaled identically. Bar, 10 µm. (E) Quantitation of D. Values represent the fluorescence intensity of Kif15 on individual spindles in HeLa or KIF15Δ cells normalized to the mean such value from HeLa cells, presented as a box-and-whisker plot highlighting the median, 10th, 25th, 75th, and 90th percentiles. n = 20; *, P = 0.004 × 10−9 relative to parental HeLa line. (F) Spindle length is decreased in the KIF15Δ line. Values represent the mean spindle length in HeLa and KIF15Δ cells. Error bars, ± SEM; n = 150 cells from three independent experiments. *, P ≤ 0.01 × 10−29 relative to the parental HeLa line. (G) Mitotic progression is not significantly altered in the KIF15Δ line. Values represent the mean MI of the HeLa and KIF15Δ cell lines. Error bars, ± SEM; n ≥ 1500 cells from three independent experiments. (H) Values represent the mean MPI of the HeLa and KIF15Δ cell lines. Error bars, ± SEM; n = 300 cells from three independent experiments.
Importantly, no reads covering the target area in exon 20 of KIF15 reported the WT sequence. The absence of WT Kif15 in the KIF15Δ line is further supported by analysis of whole-cell lysate (WCL) by Western blot, wherein Kif15 total protein levels were decreased to 1.0 ± 1.4% (mean ± SD) of that in the parental HeLa line (Fig. 5 C). Furthermore, spindle-bound Kif15 levels were decreased to 5.7 ± 3.1% in the KIF15Δcell line relative to the parental HeLa line (Fig. 5, D and E). These results confirm a near-complete loss of WT Kif15 in KIF15Δ cells.
Messenger RNAs (mRNAs) that contain premature stop codons are typically removed from the cell by nonsense-mediated mRNA decay (Lykke-Andersen and Jensen, 2015). We therefore next examined whether the KIF15Δ line phenocopies the known effects of Kif15 RNAi. The best-characterized effect of Kif15 RNAi is a subtle yet characteristic decrease in spindle length (Tanenbaum et al., 2009; Vanneste et al., 2009). Indeed, spindles in KIF15Δ cells were significantly shorter than those in the parental HeLa cells, measuring 9.5 ± 0.1 µm compared with 11.1 ± 0.1 µm, respectively (Fig. 5 F). Also consistent with Kif15 RNAi, the MI and MPI were unaltered in the KIF15Δ line compared with the parental HeLa line (MI: 3.0 ± 0.5 KIF15Δ, 2.6 ± 0.6 HeLa; MPI: 5.0 ± 0.6 KIF15Δ, 8.0 ± 0.0 HeLa; Fig. 5, G and H). Altogether, these results demonstrate a loss of Kif15 protein and function in KIF15Δ cells, validating this cell line for the study of Kif15 dependency in the acquisition of K5I resistance.
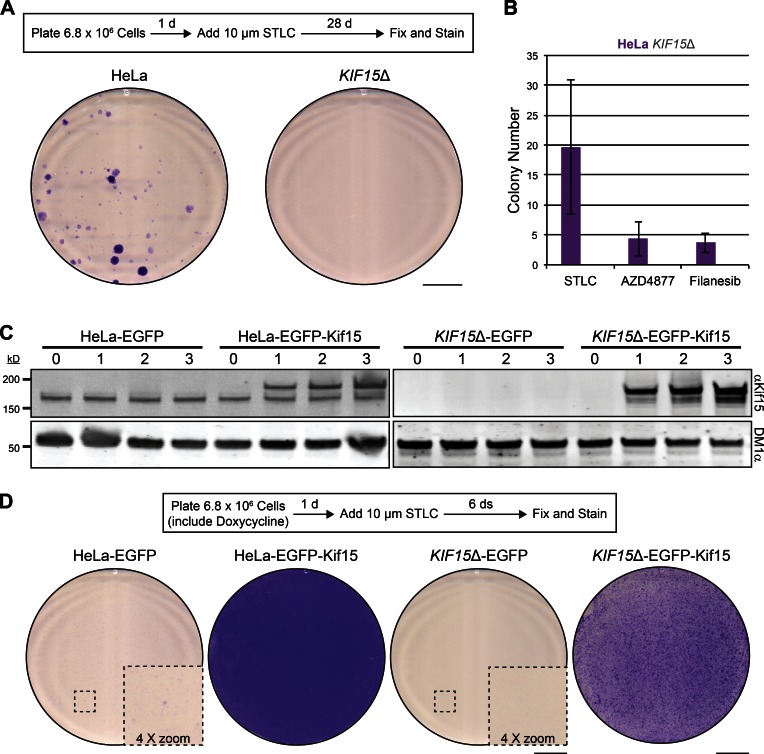
To study the requirement of Kif15 in K5I resistance, we subjected KIF15Δ cells to K5I selection and monitored the emergence of resistant colonies. Both HeLa and KIF15Δ cells were plated at confluency (Fig. S5), treated with STLC for 28 d, and stained with crystal violet (see Materials and methods; Fig. 6 A). Strikingly, no KIF15Δ cells survived this prolonged STLC treatment, whereas STLC-resistant colonies were evident in the HeLa line (Fig. 6 A). We repeated this experiment with two other distinct and clinically relevant K5Is: AZD4877 and Filanesib (Rath and Kozielski, 2012). On average, 20 ± 11 STLC-resistant colonies, 4 ± 3 AZD4877-resistant colonies, and 4 ± 2 Filanesib-resistant colonies emerged from HeLa cell populations, whereas no resistant colonies arose in any case from the KIF15Δ line (Fig. 6 B). These data are the first to indicate that Kif15 is essential for K5I resistance on a cell population level (∼107 cells).
Figure 6.
Kif15 is essential for K5I-resistance in HeLa cells. (A) STLC-resistant colonies emerge from HeLa cell populations, but not from KIF15Δ cell populations. Schematic of experimental design (top). Images of 10-cm dishes seeded with HeLa or KIF15Δ cells, treated with STLC for 28 d, and stained with crystal violet (bottom). Bar, 2 cm. See also Fig. S5. (B) Quantitation of A, with the addition of AZD4877 and Filanesib. Values represent the mean number of colonies detected in 10-cm plates seeded with HeLa (purple) or KIF15Δ (gray) cells and treated with STLC, AZD4877, or Filanesib as indicated for 28 d. Note that no colonies were detected from KIF15Δ cell populations under any scenario. Error bars, ± SEM; n = 3. (C) EGFP-Kif15 levels in transgenic cell lines are under control of doxycycline induction. Western blot of WCLs prepared from indicated cell types probed with antibodies targeting Kif15 (αKif15) and tubulin (DM1α). Numbers indicate days of induction with doxycycline. Note that the lower Kif15 species represents endogenous protein and the higher represents exogenous EGFP-Kif15. (D) EGFP-Kif15 expression rescues K5I resistance in the KIF15Δ cell line. Schematic of experimental design (top). Images of 10-cm dishes seeded with the indicated cell type, induced with doxycycline for 1 d, treated with STLC for 6 d, and stained with crystal violet (bottom). Insets (stippled boxes) are enlarged to show the presence of cell clusters in the HeLa-EGFP line and lack thereof in the KIF15Δ cell line. Bars: 2 cm; (inset) 0.5 cm.
We next used recombination-mediated cassette exchange (RMCE; Khandelia et al., 2011) to restore KIF15 expression in the KIF15Δ line. EGFP or EGFP-Kif15 were placed under control of a doxycycline-inducible promoter and integrated into the genomes of the HeLa and KIF15Δ cell lines. Both the HeLa-EGFP and HeLa-EGFP-Kif15 cell lines expressed endogenous Kif15, whereas KIF15Δ-EGFP and KIF15Δ-EGFP-Kif15 did not (Fig. 6 C). By immunoblotting, we verified that EGFP-Kif15 was expressed in a doxycycline-inducible manner in both the HeLa and KIF15Δ cell lines. EGFP-Kif15 protein levels resembled endogenous Kif15 levels 1 d after doxycycline induction and increased with time (Fig. 6 C). To test the K5I-resistance capacity of these transgenic cell lines, we initiated STLC treatment 1 d after doxycycline induction. Strikingly, the HeLa-EGFP-Kif15 line did not regress in response to STLC and the KIF15Δ-EGFP-Kif15 line initially regressed but rapidly rebounded from STLC treatment (Fig. 6 D). Notably, the HeLa-EGFP-Kif15 cell line grew more vigorously than the KIF15Δ-EGFP-Kif15 cell line in the presence of STLC (Fig. 6 D). This proliferative advantage may be caused by the presence of endogenous Kif15, suggesting that the ability of cells to override the cytotoxic effects of K5Is is highly sensitive to the overall cellular concentration of Kif15 protein. As expected, neither EGFP-expressing transgenic cell line showed robust growth after 6 d of exposure to STLC (Fig. 6 D). Microscopic examination of HeLa-EGFP cultures did, however, reveal small cell clusters (Fig. 6 D) that would likely mature to colonies resembling those in Fig. 6 A after 28 d. Such clusters were undetected in KIF15Δ-EGFP cultures (Fig. 6 D). These data reinforce our conclusion that Kif15 plays a fundamental role in the acquisition of K5I resistance and motivate further experiments to study the timing and extent of KIF15 expression in the rescue of K5I toxicity.
Discussion
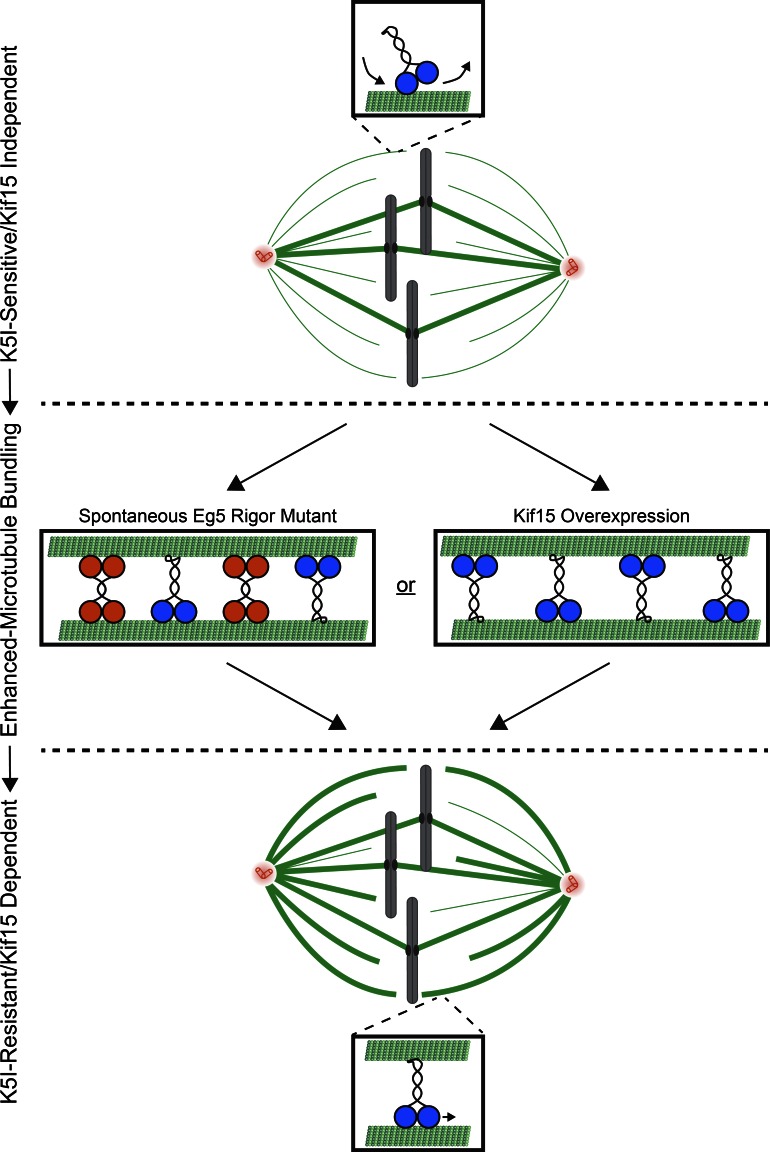
Although previous studies have emphasized overexpression as a means by which Kif15 can confer K5I resistance (Tanenbaum et al., 2009; Vanneste et al., 2009; Raaijmakers et al., 2012; Sturgill and Ohi, 2013; Ma et al., 2014), we demonstrate herein that elevated Kif15 levels are not the only means by which cells can develop Kif15-dependent K5I resistance. Instead, enhanced spindle MT bundling consistently accompanies Kif15-driven spindle assembly. Given that MT bundles serve as the substrate for Kif15 (Sturgill et al., 2014), we propose that MT bundling factors can activate Kif15-driven spindle assembly by altering the bundling architecture of spindle MTs (Fig. 7). Although the MT bundling factor studied here is an Eg5 rigor mutant, we envisage that any MT-associated factor with cross-linking activity can trigger Kif15-dependent spindle assembly.
Figure 7.
Model. Kif15-dependent K5I resistance emerges through enhanced microtubule bundling. Orange, Eg5; blue, Kif15. Thin line, single MT; bold line, bundled MTs. See text for details.
Despite the abundance of MT bundling factors within the cell, the emergence of KIRCs is relatively rare (∼1 in 500,000 cells, or ∼0.0002%). One underlying reason may be that excessive MT bundling interferes with spindle function in somatic cells. This notion is indicated by the lethal consequences of rigor mutations within the kinesin-5 Kip1p of C. albicans (Chua et al., 2007). This effect is particularly noteworthy as Kip1p is nonessential in Candida albicans (Chua et al., 2007), indicating that lethality emerges from the dominant, gain-of-function nature of the mutation. Furthermore, elevated levels of the MT bundling factors TPX2, NuSAP and HURP inhibit normal progression through mitosis in mammalian cells (Gruss et al., 2002; Song and Rape, 2010). We were therefore surprised to find that KIRC-2 and -3 not only tolerate but also require the Eg5 rigor mutant for survival. Given that Eg5-G268V–encoding mRNAs represent ∼1/3 of all EG5 transcripts as determined by RNaseq, a logical prediction holds that the rigor mutant is present at low levels, perhaps just enough to induce non–K-MT binding by Kif15. Indeed, overexpression of EGFP-Eg5-G268V in HeLa cells proved quite toxic (unpublished data). Secondarily, Eg5 tetramers likely contain low copy numbers of the rigor mutant, which may be better tolerated by the cell.
Our work shows that not all K5I-resistant cells are Eg5 independent. Here, the disconnect between K5I resistance and Eg5 dependency stems from the rigor mutant behaving as a static cross-link instead of a motile force generator. But other possibilities are certainly plausible, such as mutations to the drug-binding site (Wacker et al., 2012; Kasap et al., 2014). Because the nature of suppressor mutations are likely to impact disease management strategies, their identification will help in using antimitotic drugs to their maximal potential. Along these lines, this study uncovers a potentially novel spindle assembly function for Eg5. Going forward, it is imperative to determine whether Eg5 contributes to spindle assembly via non–MT-sliding function—e.g., by modulating MT dynamics (Chen and Hancock, 2015) or acting as a mechanical brake (Myers and Baas, 2007; Saunders et al., 2007; Rozelle et al., 2011; Collins et al., 2014; Shimamoto et al., 2015)—to obtain a more comprehensive understanding of the spindle assembly process.
Finally, this study is the first to identify a fundamental requirement for Kif15 in the emergence of K5I-resistant HeLa cells. We speculate that K5Is steer cells into an evolutionary bottleneck that requires Kif15 and that adaptive changes occur during passage through the bottleneck that enable populations to recover and diversify. Further studies are required to test the universality of our finding and identify other features required for life after adaptation to K5Is. Lastly, our findings point to the potential importance of Kif15 small-molecule inhibitors, which may significantly improve the cell death–inducing activity of K5Is in the context of a tumor.
Materials and methods
Cell culture, transfections, and drug treatments
HeLa “Kyoto” (HeLa) cells were cultured in DMEM containing 10% FCS, penicillin, and streptomycin. K5I-resistant cell lines were generated by culturing HeLa cells in 10 µM STLC (Sigma-Aldrich). Individual clonal cell lines were isolated ∼4 wk after the initiation of selection and are continuously cultured in 10 µM STLC.
CRISPR (Mali et al., 2013) was used to generate a HeLa cell line largely lacking the Kif15 protein (KIF15Δ). The sequence 5′-CCTGCGAGTAGTCCTTCATTCTG-3′, which corresponds to nucleotides 2,406–2,428 within the KIF15 ORF, was used to target Cas9 to KIF15 because it is only represented once within the human genome (see WES and analysis), minimizing the chance of off-target genome alterations. A gBlock (IDT) containing the U6 promoter, KIF15 targeting sequence, guide RNA scaffold, and termination signal was synthesized and cloned into pCR-Blunt II-TOPO (Invitrogen). This plasmid was cotransfected with pcDNA3.3-Cas9 (Addgene) twice into HeLa cells using Lipofectamine 2000 according to the manufacturer’s recommendations. Single clones were isolated by limiting dilution, and analyzed for a lack of Kif15 protein by immunostaining.
For transgenesis in HeLa and KIF15Δ cells, we used a high-efficiency and low-background RMCE system (Khandelia et al., 2011). In brief, acceptor cell lines were created by lentiviral transduction of a cassette containing the EF-1α promoter and a blasticidin-resistance gene flanked by two Cre recombinase–specific sites. Cells at 40% confluence were infected with serial dilutions of pEM584 lentivirus stock (Khandelia et al., 2011; a gift from E. Makeyev, King’s College London, London, England, UK) and incubated in DMEM containing 10% FBS, penicillin, and streptomycin, and 10 µg/ml blasticidin S until single colonies emerged. Candidate acceptor clones were isolated and tested for RMCE using pEM784 and pEM791 as previously described (Khandelia et al., 2011; gifts from E. Makeyev). To generate HeLa and KIF15Δ cells that express EGFP or EGFP-Kif15 in a doxycycline-inducible manner, acceptor cell lines were cotransfected in six-well plates with a 1:10 ratio (wt/wt) of pEM784 and either pEM791 or pRO1248 (see Molecular biology and baculovirus construction). 1 d after transfection, cells were cultured in the presence of 1 µg/ml puromycin for 48 h and then incubated in media containing 2 µg/ml puromycin until puromycin-sensitive cells were eliminated. Puromycin-resistant cells were then expanded in media containing 1 µg/ml puromycin and pooled. EGFP or EGFP-Kif15 expression was induced with 2 µg/ml doxycycline in DMEM containing 10% FBS, penicillin, and streptomycin.
siRNA transfections were performed using HiPerfect (QIAGEN) according to the manufacturer’s recommendations. The following siRNAs were used in this study: Kif15, 5′-GGACAUAAAUUGCAAAUAC-3′ (Dharmacon); Nuf2, 5′-AAGCATGCCGTGAAACGTATA-3′ (QIAGEN); and Eg5, 5′-CUGAAGACCUGAAGACAAUdTdT-3′ (QIAGEN; DeLuca et al., 2002; Weil et al., 2002; Tanenbaum et al., 2009). For control depletions, cells were transfected with All Stars siRNA (#1027280; QIAGEN) or Stealth RNAi (#12935111; Invitrogen). Kif15 depletions were analyzed ∼24 h after transfection with the exception of cell viability, which was quantified ∼48 h after transfection. Eg5 depletions were analyzed ∼24 h after transfection, and Nuf2 depletions were analyzed ∼48 h after transfection. Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations and analyzed ∼24 h after transfection. FCPT (a gift from T. Mitchison, Harvard Medical School, Boston, MA) was used at 200 µM and DMSO at 0.1% for 30 min.
RNA sequencing and analysis
Total RNA was prepared from HeLa and KIRCs enriched at the G2/M transition with 9 µM of the CDK1 inhibitor RO-3306 (Vassilev et al., 2006). For each cell line, 107 cells were processed using an RNeasy kit (QIAGEN). Total RNA was subjected to directional mRNA sequencing (single read, 50 nucleotides; 30 million independent reads). RNaseq data were thoroughly quality controlled at multiple stages of data processing as previously described (Guo et al., 2014a). Raw data and alignment quality control were performed using QC3 (Guo et al., 2014b), and expression analysis was performed using MultiRankSeq (Guo et al., 2014c). No quality issue was observed. Raw data were aligned with TopHat 2 (Kim et al., 2013) against human HG19 reference genome. Gene expression of all genes was quantified and normalized into fragments per kilobase of transcript per million reads by Cufflinks (Trapnell et al., 2012). Differential gene expression was performed using Cufflinks. False discovery rate <0.05 was used to correct for multiple testing. Cluster and heatmap were produced using R package Heatmap3 (Zhao et al., 2014).
Whole-exome sequencing and analysis
Genomic DNA was prepared from 5 × 106 each of HeLa and KIF15Δ cells using a Blood and Cell Culture DNA Mini kit (QIAGEN). The genomic DNA samples were subjected to quality control testing and sheared with Covaris. Whole-exome libraries (two-plex captures) were subsequently prepared with a SeqCap EZ Human Exome V3 kit (Nimblegen). Sequencing was performed on a NextSeq 500 (Illumina) at paired-end 150 bp targeting 45 million reads per sample.
WES data quality control was performed at multiple stages: raw and alignment as suggested by (Guo et al., 2014a). QC3 (Guo et al., 2014b) was used to check the quality. The two samples sequenced 91 and 100 million reads each, allowing high coverage in the exome region, including the KIF15 gene. During raw data quality control, we examined base quality by cycle, nucleotide distribution by cycle, base quality score, and GC content and found no quality issues. In alignment quality control, we examined the alignment rate to exome region and mapping quality score and found no quality issues. Alignments were performed using Burrows–Wheeler alignment (Li and Durbin, 2009) against human genome reference HG19. Duplicates were marked using Picard (Daniels et al., 1998) followed by local realignment and local recalibration using the Genome Analysis Toolkit (DePristo et al., 2011) developed by the Broad Institute.
To determine the exact genomic location for CRISPR, a fake read designed using the guide sequence 5′-CCTGCGAGTAGTCCTTCATTCTG-3′ was aligned to the human genome reference HG19 using Burrows–Wheeler alignment. Only one exact alignment was returned at chromosome 3, location 44856425, which is in the 20th exon of KIF15. A clear effect of CRISPR is observable with the Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013) within exon 20 of KIF15.
K5I emergence
To test the emergence of K5I-resistant cells in the HeLa and KIF15Δ cell lines, cells were plated at confluency (6.8 × 106 cells) in 10-cm standard tissue culture dishes (Corning), allowed to adhere for ∼4 h, and then treated with STLC at 10 µM, AZD4877 at 100 nM, or Filanesib at 100 nM for 28 d. AZD4877 was obtained from AstraZeneca. Filanesib is not commercially available and was synthesized at the Chemical Synthesis Core at the Vanderbilt Institute of Chemical Biology. Culturing media was exchanged roughly every other day. On the 28th day, each dish was washed with PBS and fixed with 1% paraformaldehyde in PBS for 15 min at room temperature. Dishes were subsequently washed with PBS and stained with 0.5% crystal violet (Sigma-Aldrich) in H2O for 15 min at room temperature. Excess dye was removed by gentle washing with H2O. Images of dishes were obtained with a CanoScan 8800F (Cannon). For quantifying colony number, colonies were defined as having ≥4 mm perimeter as assessed by analyzing transmitted light images acquired with an EVOS FL digital inverted microscope (AMG) in ImageJ.
To test K5I resistance of the HeLa-EGFP, HeLa-EGFP-Kif15, KIF15Δ-EGFP, and KIF15Δ-EGFP-Kif15 cell lines, cells were plated at confluency (6.8 × 106 cells) in 10-cm standard tissue culture dishes in the presence of 2 µg/ml doxycycline. STLC treatment was initiated ∼24 h after plating and induction. Culturing media containing STLC at 10 µM and 2 µg/ml doxycycline was exchanged roughly every day for 6 d, at which point dishes were fixed with paraformaldehyde, stained with crystal violet, and scanned as described in the preceeding paragraph.
MI, MPI, and cell viability quantitation
MIs (percentage of total cells in mitosis) were determined by cell morphology in transmitted light images acquired with an EVOS FL digital inverted microscope (AMG). Monopolar indices (percentage of preanaphase spindles exhibiting a monopolar geometry) were determined by tubulin staining. Cell viability was determined using CellTiter-Glo (Promega) according to the manufacturer’s recommendations. Cells were grown on coverslips and transfected with siRNA in a 24-well standard tissue culture dish (Corning). 2 d after siRNA transfection, CellTiter-Glo reagent diluted 1:1 in culturing media was added directly onto the coverslip, incubated for 10 min, and transferred to a 384-well flat-bottom white polystyrene plate (Corning). Luminescence was recorded with a Synergy HT microplate reader (BioTek).
Immunostaining, fixed-cell imaging, and live-cell imaging
Cells were fixed with methanol at −20°C for 10 min. The primary antibodies anti–Kif15-Coil2 (αKif15, rabbit; Sturgill and Ohi, 2013), anti–Xenopus laevis Eg5 (αEg5, rabbit; Miyamoto et al., 2004; a gift from T. Mitchison), and CREST (human; Immunovision) were used at 1 µg/ml for 1 h. Generation of αKif15 antibodies was described previously (Sturgill and Ohi, 2013). In brief, the C-terminal 427 amino acids of Kif15 fused to GST (GST-Kif15-Coil2) was used to immunize rabbits (Cocalico). αKif15 antibodies were affinity-purified by passing anti–GST-depleted serum over Affi-Gel 10 coupled to GST-Kif15-Coil2. Antibodies were dialyzed into PBS and frozen in liquid N2. Tubulin was labeled with DM1α (mouse; Sigma-Aldrich) at 1:500 for 30 min. Anti–rabbit, anti–mouse, and anti–human secondary antibodies conjugated to Alexa Fluor 488, 594, or 647 (goat, Invitrogen) were used at 1:1,000 for 45 min. DNA was counterstained with 5 µg/ml Hoechst-33342.
Stained cells were mounted in Prolong (Invitrogen). Images were acquired at 37°C with a DeltaVision Elite image restoration system (GE Healthcare) equipped with a 60× 1.4 numerical aperture lens (Olympus) and a Cool SnapHQ2 charge-coupled device camera (Photometrics). Either single optical slices or z-sections spaced 200 nm apart were acquired and deconvolved (Ratio [conservative], 15 iterations) with SoftWorx (GE Healthcare). Maximum intensity z-projections were generated with SoftWorx (GE Healthcare) where indicated. Images were subsequently processed with ImageJ (adjusting minimum and maximum levels, rotating, and cropping). ImageJ was also used to quantify fluorescence intensities by creating sum intensity z-projections and measuring the raw integrated densities of regions of interest (ROIs). To correct for background fluorescence, the raw integrated density of a region roughly twice the size of the ROI was measured and then scaled to match the size of the ROI based on its area. This background measurement was subsequently subtracted from the raw integrated density of the ROI to determine the final fluorescence intensity. Spindle lengths were measured with the ImageJ line tool based on tubulin staining.
For live-cell imaging of mitotic progression in KIRC-2 and -3 cells, cells were plated onto glass-bottom poly-d-lysine coated dishes (MatTek) ∼24 h before imaging. Cells were imaged in the presence of CO2 at 37°C in L-15 medium without phenol red supplemented with 10% FCS, STLC, antibiotics, and 7 mM K-Hepes, pH 7.7, using the aforementioned DeltaVision Elite system equipped with DIC optics. Images were acquired every 2 min.
Protein gels and immunoblotting
To measure Kif15 protein levels in the various cell lines, cultures grown in a 6-well tissue culture plate (Corning) were trypsinized, washed with PBS, and resuspended in NP-40 buffer (10 mM sodium phosphate, pH 7.2, 150 mM NaCl, 2 mM EDTA, and 1% NP-40) containing protease inhibitors. After 15 min on ice, extracts were clarified at 4°C for 15 min, diluted 1:1 in 2× sample buffer (SB; 100 mM Tris-Cl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, and 200 µg/ml bromophenol blue), and boiled. Samples were loaded at 25 µg total protein onto NuPAGE 10% Bis-Tris polyacrylamide gels (Thermo Fisher Scientific), resolved by SDS-PAGE, and transferred to a nitrocellulose membrane for immunoblotting.
Recombinant Eg5 samples were loaded at 50 ng or 2 µg protein onto NuPAGE 10% Bis-Tris polyacrylamide gels (Thermo Fisher Scientific), resolved by SDS-PAGE, and either transferred to a nitrocellulose membrane for immunoblotting or stained with Coomassie brilliant blue R-250 (Thermo Fisher Scientific).
For immunoblotting, nitrocellulose membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences) diluted 1:1 in PBS for 1 h, and then probed with αKif15 or αEg5 at 1 µg/ml and DM1α (Sigma-Aldrich) at 1:5,000 for 1 h. Secondary antibodies conjugated to Alexa Fluor 700 (Invitrogen) were used at 1:5,000 for 45 min. Bound antibodies were detected using an Odyssey fluorescence detection system (Mandel Scientific). Protein levels were quantified with ImageJ using tubulin as a loading control.
Molecular biology and baculovirus construction
pFASTBAC-HTc-Eg5-WT was constructed by amplifying the Eg5 coding region from pEGFP-C1-Eg5-WT and assembling this fragment into SalI–EcoRI–restricted pFASTBAC-HTc (Thermo Fisher Scientific) by isothermal assembly. pFASTBAC-HTc-Eg5-G268V was generated by introducing a G803T point mutation to pFASTBAC-HTc-Eg5-WT using a QuickChange II site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s recommendations. pFASTBAC-HTc-Eg5-WT and pFASTBAC-HTc-Eg5-G268V were used with the Bac-to-Bac system (Invitrogen) to create baculoviruses that express His6-Eg5-WT and His6-Eg5-G268V, respectively.
pEGFP-N1-Eg5-WT was constructed by inserting an Eg5 ORF-containing a PCR fragment generated from pCR-XL-TOPO-KIF11 (accession no. BC136474) into pEGFP-N1 restricted by EcoRI and SalI. This vector was further modified by site-directed mutagenesis to render the Eg5 mRNA expressed from this plasmid to be resistant to the Eg5 siRNA described in the previous paragraph. For this, we used an oligo pair with a sense sequence of 5′-GTCACAAAAGCAATGTGGAAACCTAACTGAGGATTTAAAAACTATAAAGCAGACCCATTCCCAGGAACTTTGC-3′. pEGFP-N1-Eg5-G268V was generated by site-directed mutagenesis to pEGFP-N1-Eg5-WT as described in the preceeding paragraph.
The KIF15 ORF used in this study is a splice isoform lacking exon 2 (coding sequence for amino acid residues 8–21; ELRSVTNGQSNQPS). Although lacking exon 2, our cDNA is fully functional, as assessed in a previous study (Sturgill et al., 2014). pRO1248 was prepared by isothermal assembly wherein the KIF15 ORF was amplified from a plasmid containing the full-length Kif15 cDNA and assembled into pEM791 restricted with BsrGI–BglII (a gift from E. Makeyev). pEGFP-C1-Kif15 and pEGFP-C1-HSET constructions are previously described (Sturgill et al., 2014).
Protein expression and purification
His6-Eg5-WT and His6-Eg5-G268V were expressed in Sf9 cells for 72 h. Cells were pelleted and resuspended in PNI (50 mM sodium phosphate, 500 mM NaCl, and 20 mM imidazole), 5 mM β-ME, 1% NP-40, 100 µM MgATP, and protease inhibitors. Lysates were incubated on ice for 15 min, sonicated, and clarified by centrifugation at 35,000 rpm for 1 h in a Ti 45 rotor (Beckman). Clarified supernatants were incubated with ∼2 ml Ni2+-NTA agarose (QIAGEN) for 1 h at 4°C and then washed extensively with PNI, 5 mM β-ME, 100 µM MgATP, and 10% glycerol. Protein was eluted with PNI, 5 mM β-ME, 100 µM MgATP, 10% glycerol, and 180 mM imidazole. Peak fractions were combined and dialyzed against 50 mM KH2PO4, 250 mM KCl, 10 mM β-ME, 100 µM MgATP, and 20% glycerol. Protein concentrations were determined using Bradford assays, taking into account that Eg5 exists as a tetramer in solution. Protein was aliquoted, frozen in liquid N2, and stored at −80°C.
His6-EGFP-Kif15 purification was described previously (Sturgill et al., 2014). In brief, pFASTBAC-HTa-EGFP-Kif15 was constructed by amplifying the EGFP-Kif15 coding region from pEGFP-C1-Kif15, and assembling this fragment into EcoRI–XhoI–restricted pFASTBAC-HTa (Thermo Fisher Scientific) by isothermal assembly. pFASTBAC-HTa-EGFP-Kif15 was used with the Bac-to-Bac system (Invitrogen) to create a baculovirus that expresses His6-EGFP-Kif15. His6-EGFP-Kif15 was expressed in Sf9 cells for 72 h and purified using methods described previously (Sturgill et al., 2014), except that it was subjected to size exclusion chromatography on a Hiload 16/60 Superdex 200 preparatory grade column (GE Healthcare). Gel filtration buffer contained 10 mM K-Hepes, pH 7.7, 1 mM DTT, 0.1 mM MgATP, and 300 mM KCl. Peak fractions were combined and concentrated using Amicon centrifugal filter units (EMD Millipore). Protein concentrations were determined using Bradford assays and take into account that Kif15 exists as a dimer in solution. Powdered sucrose was added to 20% wt/vol. Protein was aliquoted, frozen in liquid nitrogen, and stored at −80°C. XMAP215-EGFP-His7 was a kind gift of M. Zanic (Vanderbilt University School of Medicine, Nashville, TN).
Single-molecule imaging and analysis
To prepare cell lysates for single-molecule analysis (Soppina et al., 2014; Norris et al., 2015), HeLa cells cultured in a six-well standard tissue culture dish (Corning) were transfected with plasmids encoding EGFP-Kif15 or EGFP-HSET using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. ∼24 h after transfection, cells were trypsinized and harvested by low-speed centrifugation at 1,500 g at 4°C. The pellet was washed once in DMEM and resuspended in 25 µl lysis buffer (25 mM K-Hepes, 115 mM KOAc, 5 mM NaOAc, 5 mM MgCl2, 0.5 mM EGTA, and 1% Triton X-100, pH 7.4) freshly supplemented with 1 mM ATP, 1 mM benzamidine, 1 mM PMSF, and 10 µg/ml each of leupeptin, pepstatin, and chymostatin. Lysates were clarified by centrifugation at 16,100 g at 4°C, aliquoted, frozen in liquid N2, and stored at −80°C. Mitotic lysates were obtained as above with the following exceptions: 10 µM STLC was added to the cells 12–16 h before lysis to induce a mitotic arrest, cells were harvested by a mitotic shake-off, and the lysis buffer included phosphatase inhibitors (Phosstop; Roche).
All single-molecule photobleaching assays were performed in narrow flow cells (∼10 µl volume) prepared using silanized coverslips as described previously (Soppina et al., 2014; Norris et al., 2015). Flow cells were infused with HeLa cell lysates (∼1:5,000 dilution) or purified XMAP215-EGFP (800 pM) diluted in P12 buffer (12 mM K-Pipes, 1 mM EGTA, and 2 mM MgCl2, pH 6.8) supplemented with 0.5 mg/ml casein for 2 min. Flow cells were then washed with 50 µl P12 buffer supplemented with 0.5 mg/ml casein to remove nonadsorbed fluorescent proteins. The surface-bound EGFP-labeled proteins were imaged at 35°C by total internal reflection fluorescence microscopy on a Ti-E (Nikon) equipped with an H-TIRF module, 100× Apochromat total internal reflection fluorescence 1.49 numerical aperture objective, EMCCD detector (iXon Ultra DU897; Andor Technology), and Perfect Focus. Images were acquired continuously with 50 ms exposures for 15 s using a LuNA solid-state 488 nm laser (∼10 mW power), and image acquisition was controlled by Nikon Elements software. Unlike EGFP-HSET and XMAP215-EGFP, Kif15 was unable to nonspecifically bind silanized coverslips with sufficient affinity for intensity analysis. To increase the number of bound fluorescent particles, GMPCPP-stabilized MTs were adsorbed onto a coverslip via a biotin-streptavidin interaction, and EGFP-Kif15 was locked onto MTs by introducing EGFP-Kif15 (∼1:100 diluted HeLa lysates or 200 pM purified protein) along with 1 mM AMPPNP in P12 buffer supplemented with 0.5 mg/ml casein.
To obtain first-frame intensity values, a 2D Gaussian fitting routine was implemented in ImageJ (http://www.sussex.ac.uk/gdsc/intranet/microscopy/imagej/gdsc_plugins#install), local maxima were detected automatically, and the reported Gaussian intensity values were corrected for local background. For time-lapse intensity traces, background-corrected intensity values were obtained by implementing the 2D Gaussian fitting routine on the same x-y position in time. The intensity was reported as zero when the intensity fell below the required threshold for the fitting routine.
Cumulative distribution functions (CDFs) of first-frame intensity distributions were generated via MATLAB and mean values were obtained as described previously (Norris et al., 2015). CDFs were used for statistical analysis because they are continuous and do not introduce subjective binning. Using MATLAB, intensity CDFs were fit to the hypothetical CDF for a normal distribution using a nonlinear least-squares fit with the free parameters μ (mean) and σ (SD):
Errors were obtained by the bootstrap technique, where each distribution was resampled 200 times and fit to the normal distribution. The SD of the mean parameter μ over the resampled datasets was taken as the standard error for each fitted quantity, and the data were reported as the mean ± the 95% COI, where the 95% COI was defined as two times the standard error. For display (Figs. 3 D and S3 B), Gaussian fits to the binned data were calculated with SigmaPlot (Systat Software).
MT assays
For MT gliding assays, flow cells were constructed with double-stick tape and infused with Eg5-WT or Eg5-G268V at 50 nM for 3 min, and X-rhodamine–labeled GMPCPP MTs (XR-MTs, 1:9 labeled:unlabeled) at 300 nM tubulin in BRB80 for 2 min. Flow cells were washed between each infusion with 3 vol BRB80, 50 mM KCl, 1 mM MgATP, and 500 µg/ml casein. After the final infusion, flow cells were washed with 3 vol BRB80, 50 mM KCl, 1 mM MgATP, 500 µg/ml casein, and oxygen scavenging mix [200 µg/ml glucose oxidase, 35 µg/ml catalase, 25 mM glucose, 70 mM β-ME]. MT gliding was recorded at 5-s intervals by time-lapse microscopy. Images were acquired at ambient temperatures by widefield fluorescence microscopy on a high speed Ti-E (Nikon) equipped with a 100× Apochromat total internal reflection fluorescence 1.49 numerical aperture objective, Andor Neo 5.5 sCMOS camera, and Perfect Focus. Nikon Elements was used for acquisition and ImageJ for subsequent image processing (template matching, kymograph generation, and adjusting minimum and maximum levels) and distance measurements. Gliding velocities were determined by measuring the distance a MT had moved after 2.5 min. Fits of the distributions were calculated with SigmaPlot (Systat Software) and used to determine the mean speed and SD.
For MT bundling assays, XR-MTs at 500 nM tubulin were combined with Eg5-WT or Eg5-G268V at 10 nM in BRB80, 5 mM MgATP, and KCl at 50 mM, 150 mM, or 300 mM as indicated. Reactions were incubated at room temperature for 5 min followed by preparation of a 2-µl squash for imaging. Images were acquired at ambient temperatures by widefield fluorescence microscopy on an Eclipse 90i (Nikon) equipped with a 60× 1.4 numerical aperture (Nikon) or 100× 1.4 numerical aperture (Nikon) objective and a Cool SnapHQ2 CCD camera (Photometrics). Nikon Elements was used for acquisition and ImageJ for subsequent image processing (adjusting minimum and maximum levels, rotating, and cropping).
For MT copelleting assays, tubulin or Taxol-stabilized MTs at 500 nM tubulin were combined with Eg5-WT or Eg5-G268V at 100 nM in 10 mM Hepes, pH 7.7, 50 mM KCl, 1 mM DTT, 5 µM Taxol, and 5 mM MgATP. 50-µl reactions were incubated for 15 min at room temperature and spun over a 100-µl sucrose cushion (10 mM Hepes, pH 7.7, 50 mM KCl, 20 µM Taxol, and 40% sucrose wt/vol) at 60,000 rpm for 20 min. Supernatants were collected and mixed 1:1 with 2X SB. The supernatant/cushion interfaces were washed twice with BRB80, 1 mM DTT, and 20 µM Taxol, followed by complete removal of the cushions. Pellets were resuspended in 100 µl 1X SB. 40 µl of each fraction was boiled, resolved by SDS-PAGE, and transferred to a nitrocellulose membrane for immunoblotting as described in Protein gels and immunoblotting.
Statistical analysis
Statistically relevant differences in experimental data were determined using the T.TEST function in Excel (Microsoft). P values report the two-tailed distribution of a two-sample Student’s t test assuming unequal variance.
Online supplemental material
Fig. S1 shows the Eg5 mechanochemical state induced by rigor is dominant to the effects of STLC (related to Fig. 1). Fig. S2 shows that exogenous Eg5-G268V-EGFP induces spindle MT bundling and reports the MIs and MPIs of Eg5-depleted cell lines (related to Fig. 2). Fig. S3 shows representative fields of view for the single-molecule imaging experiments, analysis of cellular EGFP-Kif15 oligomerization as it relates to cell cycle state, and separation of the representative photobleaching traces (related to Fig. 3). Fig. S4 shows a genetic lesion within exon 20 of KIF15 in the KIF15Δ line (related to Fig. 5). Fig. S5 shows plating cell density for the long-term K5I-resistance experiments (related to Fig. 6). Videos 1 and 2 show representative fields of view during MT gliding assays powered by Eg5-WT and Eg5-G268V, respectively (related to Fig. 2). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201507036/DC1.
Supplementary Material
Acknowledgments
We thank Aaron Groen and Tim Mitchison for FCPT and αEg5, Eugene Makeyev for reagents to perform RMCE, and Marija Zanic for XMAP215-EGFP and use of her total internal reflection fluorescence microscopy system. We also thank AstraZeneca for AZD4877 and the Vanderbilt Institute of Chemical Biology for synthesis of Filanesib. We are grateful to VANTAGE (Vanderbilt Technologies for Advanced Genomics) for sequencing assistance. Members of the Ohi and Zanic laboratories offered critical insight for which we are also grateful.
This work was supported by National Institutes of Health grant R01GM086610 to R. Ohi. R. Ohi is a scholar of the Leukemia and Lymphoma Society.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- au
- arbitrary units
- CDF
- cumulative distribution function
- COI
- confidence interval
- DIC
- differential interference contrast
- FCPT
- 2-[1-(4-fluorophenyl)cyclopropyl]-4-(pyridin-4-yl)thiazole
- INDEL
- insertion/deletion
- K5I
- kinesin-5/Eg5 inhibitor
- KIRC
- kinesin-5– and Eg5 inhibitor–resistant cell
- K-MT
- kinetochore–microtubule
- LUT
- lookup table
- MI
- mitotic index
- MPI
- monopolar index
- MT
- microtubule
- RMCE
- recombination-mediated cassette exchange
- RNaseq
- RNA sequencing
- SB
- sample buffer
- STLC
- S-trityl-l-cysteine
- WCL
- whole-cell lysate
- WES
- whole-exome sequencing
- WT
- wild type
- XR-MT
- X-rhodamine–labeled GMPCPP microtubule
References
- Adey A., Burton J.N., Kitzman J.O., Hiatt J.B., Lewis A.P., Martin B.K., Qiu R., Lee C., and Shendure J.. 2013. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 500:207–211. 10.1038/nature12064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A., Lane H.A., d’Hérin P., Harper M., Kress M., and Nigg E.A.. 1995. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 83:1159–1169. 10.1016/0092-8674(95)90142-6 [DOI] [PubMed] [Google Scholar]
- Brier S., Lemaire D., Debonis S., Forest E., and Kozielski F.. 2004. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry. 43:13072–13082. 10.1021/bi049264e [DOI] [PubMed] [Google Scholar]
- Brouhard G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., and Hyman A.A.. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahu J., and Surrey T.. 2009. Motile microtubule crosslinkers require distinct dynamic properties for correct functioning during spindle organization in Xenopus egg extract. J. Cell Sci. 122:1295–1300. 10.1242/jcs.044248 [DOI] [PubMed] [Google Scholar]
- Chen Y., and Hancock W.O.. 2015. Kinesin-5 is a microtubule polymerase. Nat. Commun. 6:8160 10.1038/ncomms9160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P.R., Roof D.M., Lee Y., Sakowicz R., Clarke D., Pierce D., Stephens T., Hamilton M., Morgan B., Morgans D., et al. . 2007. Effective killing of the human pathogen Candida albicans by a specific inhibitor of non-essential mitotic kinesin Kip1p. Mol. Microbiol. 65:347–362. 10.1111/j.1365-2958.2007.05787.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E., Mann B.J., and Wadsworth P.. 2014. Eg5 restricts anaphase B spindle elongation in mammalian cells. Cytoskeleton (Hoboken). 71:136–144. 10.1002/cm.21158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C.D., Breslin M.J., Mariano B.J., Coleman P.J., Buser C.A., Walsh E.S., Hamilton K., Huber H.E., Kohl N.E., Torrent M., et al. . 2005. Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP. Bioorg. Med. Chem. Lett. 15:2041–2045. 10.1016/j.bmcl.2005.02.055 [DOI] [PubMed] [Google Scholar]
- Daniels J., Holmans P., Williams N., Turic D., McGuffin P., Plomin R., and Owen M.J.. 1998. A simple method for analyzing microsatellite allele image patterns generated from DNA pools and its application to allelic association studies. Am. J. Hum. Genet. 62:1189–1197. 10.1086/301816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBonis S., Skoufias D.A., Lebeau L., Lopez R., Robin G., Margolis R.L., Wade R.H., and Kozielski F.. 2004. In vitro screening for inhibitors of the human mitotic kinesin Eg5 with antimitotic and antitumor activities. Mol. Cancer Ther. 3:1079–1090. [PubMed] [Google Scholar]
- DeLuca J.G., Moree B., Hickey J.M., Kilmartin J.V., and Salmon E.D.. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549–555. 10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. . 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H., McHugh T., Singleton M.R., Carter N.J., and McAinsh A.D.. 2014. The Kinesin-12 Kif15 is a processive track-switching tetramer. eLife. 3:e01724 10.7554/eLife.01724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., and Vale R.D.. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907. 10.1091/mbc.E05-02-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen A.C., Needleman D., Brangwynne C., Gradinaru C., Fowler B., Mazitschek R., and Mitchison T.J.. 2008. A novel small-molecule inhibitor reveals a possible role of kinesin-5 in anastral spindle-pole assembly. J. Cell Sci. 121:2293–2300. 10.1242/jcs.024018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Wittmann M., Yokoyama H., Pepperkok R., Kufer T., Silljé H., Karsenti E., Mattaj I.W., and Vernos I.. 2002. Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4:871–879. 10.1038/ncb870 [DOI] [PubMed] [Google Scholar]
- Guo Y., Ye F., Sheng Q., Clark T., and Samuels D.C.. 2014a Three-stage quality control strategies for DNA re-sequencing data. Brief. Bioinform. 15:879–889. 10.1093/bib/bbt069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhao S., Sheng Q., Ye F., Li J., Lehmann B., Pietenpol J., Samuels D.C., and Shyr Y.. 2014b Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 103:323–328. 10.1016/j.ygeno.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Zhao S., Ye F., Sheng Q., and Shyr Y.. 2014c MultiRankSeq: multiperspective approach for RNAseq differential expression analysis and quality control. BioMed Res. Int. 2014:248090 10.1155/2014/248090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentrich C., and Surrey T.. 2010. Microtubule organization by the antagonistic mitotic motors kinesin-5 and kinesin-14. J. Cell Biol. 189:465–480. 10.1083/jcb.200910125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein L.C., Peterman E.J., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kasap C., Elemento O., and Kapoor T.M.. 2014. DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets. Nat. Chem. Biol. 10:626–628. 10.1038/nchembio.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina A.S., Baskin R.J., Cole D.G., Wedaman K.P., Saxton W.M., and Scholey J.M.. 1996. A bipolar kinesin. Nature. 379:270–272. 10.1038/379270a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia P., Yap K., and Makeyev E.V.. 2011. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc. Natl. Acad. Sci. USA. 108:12799–12804. 10.1073/pnas.1103532108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak T.C., and Gilbert S.P.. 2006. Dimeric Eg5 maintains processivity through alternating-site catalysis with rate-limiting ATP hydrolysis. J. Biol. Chem. 281:39444–39454. 10.1074/jbc.M608056200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok B.H., Yang J.G., and Kapoor T.M.. 2004. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr. Biol. 14:1783–1788. 10.1016/j.cub.2004.09.052 [DOI] [PubMed] [Google Scholar]
- Lad L., Luo L., Carson J.D., Wood K.W., Hartman J.J., Copeland R.A., and Sakowicz R.. 2008. Mechanism of inhibition of human KSP by ispinesib. Biochemistry. 47:3576–3585. 10.1021/bi702061g [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S., and Jensen T.H.. 2015. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16:665–677. 10.1038/nrm4063 [DOI] [PubMed] [Google Scholar]
- Ma H.T., Erdal S., Huang S., and Poon R.Y.. 2014. Synergism between inhibitors of Aurora A and KIF11 overcomes KIF15-dependent drug resistance. Mol. Oncol. 8:1404–1418. 10.1016/j.molonc.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macville M., Schröck E., Padilla-Nash H., Keck C., Ghadimi B.M., Zimonjic D., Popescu N., and Ried T.. 1999. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59:141–150. [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., and Church G.M.. 2013. RNA-guided human genome engineering via Cas9. Science. 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z., and Mitchison T.J.. 2006. Small-molecule and mutational analysis of allosteric Eg5 inhibition by monastrol. BMC Chem. Biol. 6:2 10.1186/1472-6769-6-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto D.T., Perlman Z.E., Burbank K.S., Groen A.C., and Mitchison T.J.. 2004. The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J. Cell Biol. 167:813–818. 10.1083/jcb.200407126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K.A., and Baas P.W.. 2007. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J. Cell Biol. 178:1081–1091. 10.1083/jcb.200702074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Núñez M.F., and Verhey K.J.. 2015. Influence of fluorescent tag on the motility properties of kinesin-1 in single-molecule assays. Biophys. J. 108:1133–1143. 10.1016/j.bpj.2015.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J.A., van Heesbeen R.G., Meaders J.L., Geers E.F., Fernandez-Garcia B., Medema R.H., and Tanenbaum M.E.. 2012. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 31:4179–4190. 10.1038/emboj.2012.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath O., and Kozielski F.. 2012. Kinesins and cancer. Nat. Rev. Cancer. 12:527–539. 10.1038/nrc3310 [DOI] [PubMed] [Google Scholar]
- Rice S., Lin A.W., Safer D., Hart C.L., Naber N., Carragher B.O., Cain S.M., Pechatnikova E., Wilson-Kubalek E.M., Whittaker M., et al. . 1999. A structural change in the kinesin motor protein that drives motility. Nature. 402:778–784. 10.1038/45483 [DOI] [PubMed] [Google Scholar]
- Rickert K.W., Schaber M., Torrent M., Neilson L.A., Tasber E.S., Garbaccio R., Coleman P.J., Harvey D., Zhang Y., Yang Y., et al. . 2008. Discovery and biochemical characterization of selective ATP competitive inhibitors of the human mitotic kinesin KSP. Arch. Biochem. Biophys. 469:220–231. 10.1016/j.abb.2007.10.016 [DOI] [PubMed] [Google Scholar]
- Rozelle D.K., Hansen S.D., and Kaplan K.B.. 2011. Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake. J. Cell Biol. 193:285–294. 10.1083/jcb.201011002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A.M., Powers J., Strome S., and Saxton W.M.. 2007. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr. Biol. 17:R453–R454. 10.1016/j.cub.2007.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., and Mitchison T.J.. 1995. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc. Natl. Acad. Sci. USA. 92:4289–4293. 10.1073/pnas.92.10.4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K.E., LeGuellec K., Philippe M., and Mitchison T.J.. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543. 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Shimamoto Y., Forth S., and Kapoor T.M.. 2015. Measuring Pushing and Braking Forces Generated by Ensembles of Kinesin-5 Crosslinking Two Microtubules. Dev. Cell. 34:669–681. 10.1016/j.devcel.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., and Rape M.. 2010. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol. Cell. 38:369–382. 10.1016/j.molcel.2010.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppina V., Norris S.R., Dizaji A.S., Kortus M., Veatch S., Peckham M., and Verhey K.J.. 2014. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. USA. 111:5562–5567. 10.1073/pnas.1400759111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill E.G., and Ohi R.. 2013. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr. Biol. 23:1280–1290. 10.1016/j.cub.2013.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill E.G., Das D.K., Takizawa Y., Shin Y., Collier S.E., Ohi M.D., Hwang W., Lang M.J., and Ohi R.. 2014. Kinesin-12 Kif15 targets kinetochore fibers through an intrinsic two-step mechanism. Curr. Biol. 24:2307–2313. 10.1016/j.cub.2014.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Macůrek L., Janssen A., Geers E.F., Alvarez-Fernández M., and Medema R.H.. 2009. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr. Biol. 19:1703–1711. 10.1016/j.cub.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., and Pachter L.. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D., Takagi M., Imamoto N., and Vernos I.. 2009. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr. Biol. 19:1712–1717. 10.1016/j.cub.2009.09.019 [DOI] [PubMed] [Google Scholar]
- Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D.C., and Chen L.. 2006. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. USA. 103:10660–10665. 10.1073/pnas.0600447103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker S.A., Houghtaling B.R., Elemento O., and Kapoor T.M.. 2012. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat. Chem. Biol. 8:235–237. 10.1038/nchembio.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D., Garçon L., Harper M., Duménil D., Dautry F., and Kress M.. 2002. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. Biotechniques. 33:1244–1248. [DOI] [PubMed] [Google Scholar]
- Zhao S., Guo Y., Sheng Q., and Shyr Y.. 2014. Advanced heat map and clustering analysis using heatmap3. BioMed Res. Int. 2014:986048 10.1155/2014/986048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.