In addition to recruiting Parkin/autophagy receptors to damaged mitochondria, the authors show that PINK1 triggers PKA displacement from AKAP1 after damage to trigger mitochondrial fission in a Parkin-independent manner, suggesting that PINK1 is a master mitophagy regulator.
Abstract
Mitochondrial fission is essential for the degradation of damaged mitochondria. It is currently unknown how the dynamin-related protein 1 (DRP1)–associated fission machinery is selectively targeted to segregate damaged mitochondria. We show that PTEN-induced putative kinase (PINK1) serves as a pro-fission signal, independently of Parkin. Normally, the scaffold protein AKAP1 recruits protein kinase A (PKA) to the outer mitochondrial membrane to phospho-inhibit DRP1. We reveal that after damage, PINK1 triggers PKA displacement from A-kinase anchoring protein 1. By ejecting PKA, PINK1 ensures the requisite fission of damaged mitochondria for organelle degradation. We propose that PINK1 functions as a master mitophagy regulator by activating Parkin and DRP1 in response to damage. We confirm that PINK1 mutations causing Parkinson disease interfere with the orchestration of selective fission and mitophagy by PINK1.
Introduction
PTEN-induced putative kinase (PINK1), a mitochondrial serine kinase, and Parkin, a cytosolic E3-ubiqutin ligase, function cooperatively to degrade damaged mitochondria by the autophagic machinery (mitophagy). Mutations in PINK1 and Parkin are a cause of early onset familial Parkinson disease (PD; Schapira, 2008), and Parkin deficiency is implicated in sporadic PD (Chung et al., 2004), thus suggesting direct connectivity between mitochondrial homeostasis and PD pathogenesis. Understanding the mechanism of PINK1-Parkin mitophagy may facilitate the development of therapeutic strategies for PD (Schapira et al., 2014).
Mitophagy has three requisite steps: the activation of Parkin E3-ubiquitin ligase activity at the outer mitochondrial membrane (OMM; Narendra et al., 2008, 2010; Tanaka et al., 2010), dynamin-related protein 1 (DRP1)–catalyzed fission of the damaged mitochondria (Twig et al., 2008; Narendra et al., 2010), and the recruitment of the autophagy receptor NDP52 or optineurin to the OMM (Lazarou et al., 2015). The first step is well established: PINK1 normally resides in the inner mitochondrial membrane, but when damage dissipates the mitochondrial membrane potential (mΔΨ), protein import ceases and PINK1 instead accumulates in the OMM (Narendra et al., 2010; Matsuda et al., 2010; Vives-Bauza et al., 2010) via association with TOM7 inthe TOM complex (Hasson et al., 2013). Here, PINK1 phosphorylates Parkin (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012) and ubiquitin (Koyano et al., 2014; Kane et al., 2014) to coactivate Parkin (Wauer et al., 2015), and the ubiquitylation of numerous OMM proteins. OMM ubiquitin tags signal whole-organelle engulfment and degradation by autophagy (Narendra et al., 2010). OMM-bound PINK1 also mediates the recruitment of NDP52 and optineurin, independently of Parkin, which in turn direct the autophagy factors ULK1 and DFCP1 to damaged mitochondria (Lazarou et al., 2015). Fission is required to generate mitochondrial particles sufficiently small to be engulfed by the autophagosome. However, we propose that the established mechanism explaining the fission step is ambiguous.
Normally, OMM-bound A-kinase anchoring protein 1 (AKAP1) binds tetrameric (two regulatory and catalytic subunits) protein kinase A (PKA), via a conserved regulatory subunit binding grove. Here, PKA phospho-inhibits DRP1 at serine 637 to diminish fission (Chang and Blackstone, 2007; Cribbs and Strack, 2007; Cereghetti et al., 2008). PKA is catalytically active, even though the catalytic and regulatory subunits are associated, because local PKA substrates are orientated into a phosphorylatable configuration by AKAP1 (Smith et al., 2013). Thereby mitochondria form elongated fused structures in many cell types as rates of fusion exceed fission. In the majority of studies mitophagy and fission are stimulated by systemically depolarizing mitochondria (using toxins such as carbonyl cyanide m-chlorophenyl hydrazone [CCCP] or antimycin A combined with oligomycin), and fission is driven by the increased cytosolic calcium (released from depolarized mitochondria), which activates DRP1 serine 637 dephosphorylation by calcineurin (Chang and Blackstone, 2007; Cribbs and Strack, 2007; Cereghetti et al., 2008). However, we and others (Schon and Przedborski, 2011) contend that this level of chemically induced mitochondrial damage, including the related cytosolic calcium changes, exceeds physiological limits and is thus probably not mechanistically relevant to the fundamental mitophagy pathway. For instance, it is difficult to rationalize how damaged sections of the mitochondrial reticulum would be selectively distinguished for fission during basal mitophagy; calcium that is released from depolarized/damaged regions of the reticulum will rapidly diffuse throughout the cytosol (according to the diffusion coefficient) and also be strongly buffered by surrounding ER and polarized mitochondria. Thereby calcium cannot function as a local damage signal that is relayed to promote fission. Here, we provide a mechanistic framework to explain “selective fission” and unravel how DRP1 is targeted to recognize damaged mitochondria.
Results and discussion
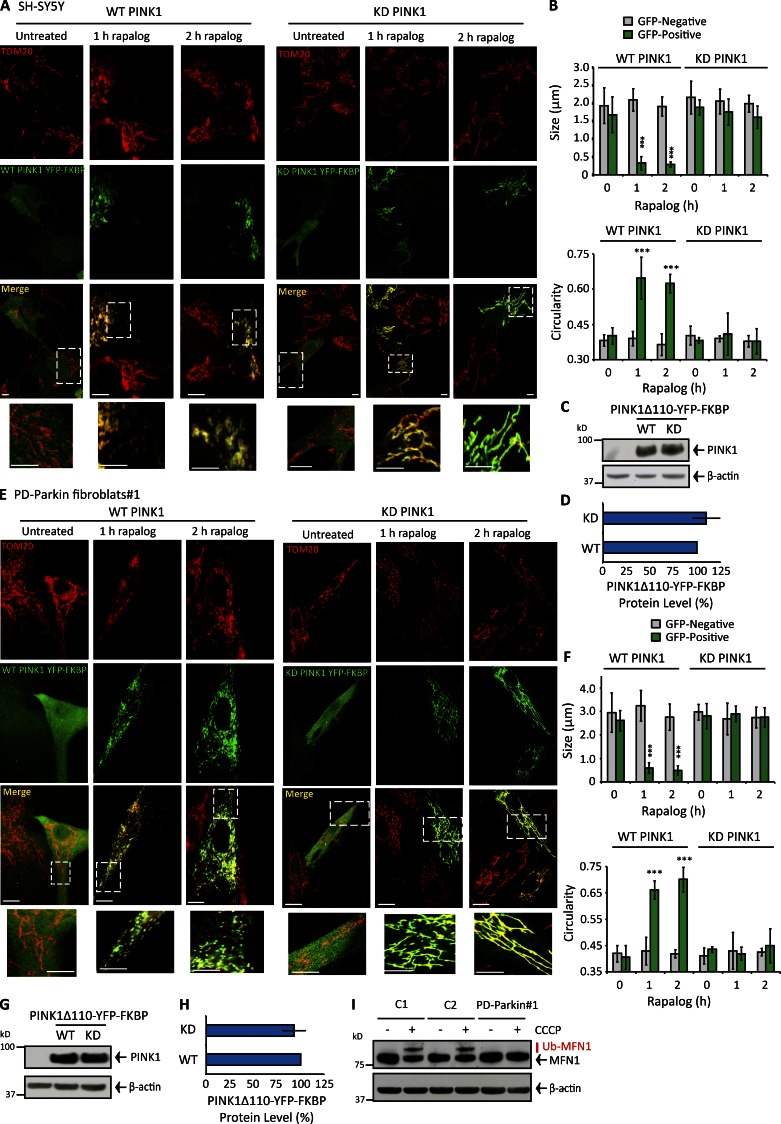
Because PINK1 recruits Parkin and autophagy receptors and factors to damaged mitochondria, we reasoned PINK1 may also promote fission. To clearly and conclusively determine if PINK1 promotes fission independently of calcium-calcineurin, we used a regulated heterodimerization system to target ectopic PINK1 to the OMM while maintaining mΔΨ. Previous studies have used this experimental system to show OMM-localized PINK1 recruits Parkin, optineurin, and NDP52 to the OMM even if mitochondria remain polarized (Lazarou et al., 2012, 2015). Cells were cotransfected with wild-type (WT) or kinase-dead (KD) PINK1Δ110-YFP (lacking the mitochondrial localization sequence and integral domain) and the integral OMM protein Fis1 genetically fused with the FKBP and FRB domains, respectively. FKBP and FRB dimerize in the presence of exogenous rapalog. In transfected SH-SY5Y cells (expressing endogenous PINK1/Parkin; Fig. S1, A–C), rapalog treatment redistributed PINK1 to TOM20-stained mitochondria and triggered kinase-dependent mitochondrial fission (Fig. 1, A–D). Interestingly, WT PINK1Δ110-YFP-FKBP pro-fission activity was conserved in two rapalog-treated PD-Parkin–deficient fibroblast cultures (Fig. 1, E–H; and Fig. S1, N and O) lacking Parkin and associated MFN1 ubiquitylation activity (Fig. 1 I and Fig. S1 D) but expressing PINK1 (Fig. S1 E). Rapalog did not indirectly affect morphology in WT or KD PINK1Δ110-YFP-FKBP–negative cells (Fig. 1, A, B, E, and F; and Fig. S1, N and O), change cytosolic calcium levels (Fig. S1, G–K), depolarize mΔΨ (Fig. S1, L and M), or induce MFN1 ubiquitylation (Fig. S1 F). OMM-bound WT PINK1 is a powerful fission activator independent of Parkin activity and calcium.
Figure 1.
Controlled targeting of PINK1 to the OMM induces robust mitochondrial fission independently of Parkin and calcium-calcineurin. (A and B) Mitochondrial morphology and quantification for TOM20-stained SH-SY5Y cells transfected with WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1 and treated with rapalog for 0–2 h. (C and D) PINK1 protein level (adjusted for β-actin) in SH-SY5Y cells transfected with WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1. (E and F) Mitochondrial morphology and quantification for TOM20-stained PD-Parkin#1 primary human fibroblasts transfected with either WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1, treated with rapalog for 0–2 h. (G and H) PINK1 level (adjusted for β-actin) in primary human PD-Parkin#1 fibroblasts transfected with WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1. (I) MFN1 ubiquitylation in two control primary human fibroblast cultures (C1 and C2) compared with PD-Parkin#1 after 2-h CCCP treatment. Bars: 15 µM; (insets) 5 µM. Each experiment was repeated at least three separate times and the error bars show the SDM. ***, P < 0.001.
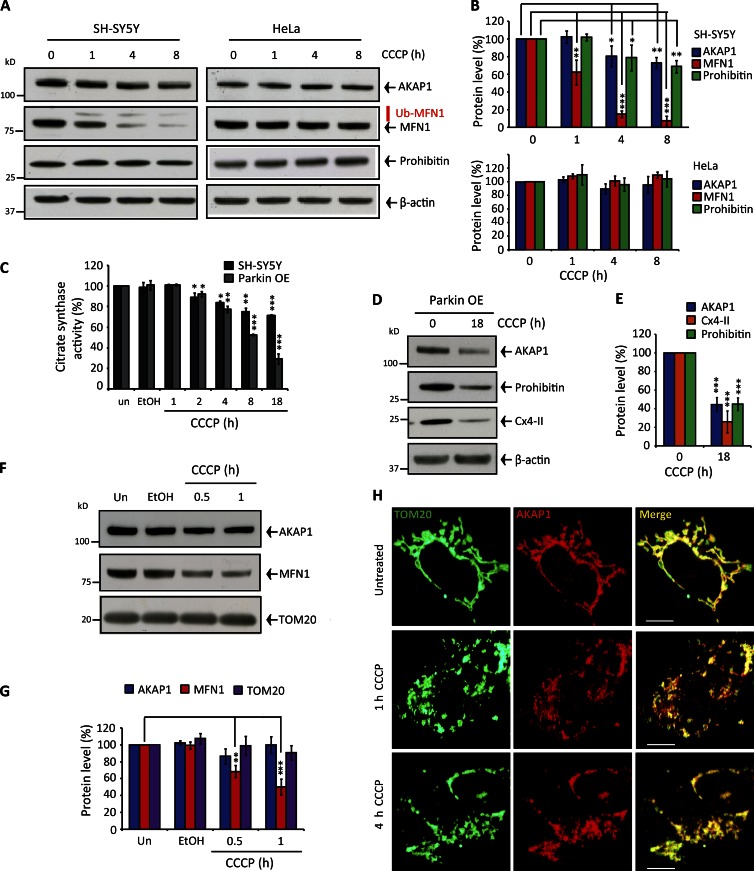
The OMM-associated AKAP1–PKA axis is a core physiological mitochondrial morphology regulator by controlling DRP1 phosphorylation at serine 637 (Newhall et al., 2006; Carlucci et al., 2008; Dickey and Strack, 2011; Kim et al., 2011; Pfluger et al., 2015). Modulating AKAP1-PKA activity is sufficient to hyperfuse or fragment the mitochondrial reticulum without affecting other components of the fission–fusion machinery; DRP1 serine 637 phosphomimic mutagenesis robustly impairs fission, whereas serine-to-alanine mutagenesis confers notable pro-fission activity (Cribbs and Strack, 2007; Cereghetti et al., 2008; Merrill et al., 2011), as does depleting AKAP1 levels (by preventing DRP1 serine 637 phosphorylation; Fig. S2, A–D; Carlucci et al., 2008; Merrill et al., 2011). During hypoxia, AKAP1 is degraded by the E3-ubiquitin ligase Siah2 (Carlucci et al., 2008; Kim et al., 2011). In view of this, we considered that OMM-PINK1 could activate Parkin or another E3-ligase to degrade AKAP1 and drive fission. Instead, AKAP1 levels maintained in CCCP-treated SH-SY5Y cells, whereas the endogenous Parkin substrate MFN1 was markedly degraded, decreasing by 40% and 90% after 1- and 4-h CCCP treatment, respectively (Fig. 2, A and B). Both AKAP1 and MFN1 maintained in CCCP-treated HeLa cells (Fig. 2, A and B) expressing PINK1 but devoid of Parkin/CCCP-induced MFN1 ubiquitylation (Fig. S1, A–C). Eventual AKAP1 loss in SH-SY5Y cells correlated with mitochondrial proteins prohibitin-1, complex IV subunit II (cx4-II), and reduced citrate synthase activity (Fig. 2, A–C), thus reflecting bulk mitophagy. Overexpressing Parkin (Fig. S1 B) amplified CCCP-dependent AKAP1 turnover similarly to other mitophagy marker proteins (Fig. 2, D and E). AKAP1 was not degraded in CCCP-treated primary human fibroblasts used in this study (Fig. S2, E–I). Ejection of AKAP1 from the OMM could explain PINK1-activated fission by displacing PKA and thus preventing DRP1 serine 637 phosphorylation. However, AKAP1 retained in mitochondrial fractions isolated from depolarized SH-SY5Y (Fig. 2, F and G) and HeLa cells (Fig. 3, A and B). AKAP1 also continued to strongly colocalize with TOM20-stained mitochondria after mΔΨ dissipation in SH-SY5Y cells (Fig. 2 H); TOM20-AKAP1 Pearson coefficients were unaffected during 4-h depolarization. Furthermore, the loss of AKAP1 via mitophagy is absolutely consistent with AKAP1 retention in the OMM of damaged mitochondria (Fig. 2, A, B, D, and E); i.e., AKAP1 does not escape bulk organelle turnover. Interestingly, we found that PKA catalytic subunit (Catβ) levels diminished progressively in mitochondrial fractions isolated from CCCP-treated HeLa (Fig. 3, A and B) and SH-SY5Y cells (Fig. S2, J and K), decreasing by 50–60% and 65–80% in both cell types after 1- and 2-h CCCP treatment, respectively. These findings indicate that the AKAP1–PKA axis is somehow disrupted after mitochondrial depolarization and PINK1 activation, regardless of Parkin.
Figure 2.
Mitochondrial depolarization does not trigger AKAP1 degradation or release from the OMM. (A and B) AKAP1, MFN1, and Prohibitin protein levels in 0–8 h depolarized SH-SY5Y or HeLa cells. Quantification was corrected for β-actin. (C) Citrate synthase activity adjusted for protein (BCA) after 0–18-h CCCP treatment in SH-SY5Y cells expressing endogenous and overexpressed Parkin. (D and E) AKAP1, Prohibitin, and Cx4-II protein levels in Parkin-overexpressing SH-SY5Y cells incubated with CCCP for 18 h. Quantification was corrected for β-actin. (F and G) AKAP1, MFN1, and TOM20 protein levels and quantification in mitochondria isolated from CCCP-treated SH-SY5Y cells. un, untreated. (H) Immunocytochemistry demonstrating AKAP1 colocalization with TOM20 in SH-SY5Y cells during 0–4-h CCCP treatment. Pearson coefficients as follows: untreated = 0.84 ± 0.03; 1-h CCCP = 0.85 ± 0.02; 4-h CCCP = 0.83 ± 0.01. Bars, 20 µM. Each experiment was repeated at least three separate times, and error bars indicate SDM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
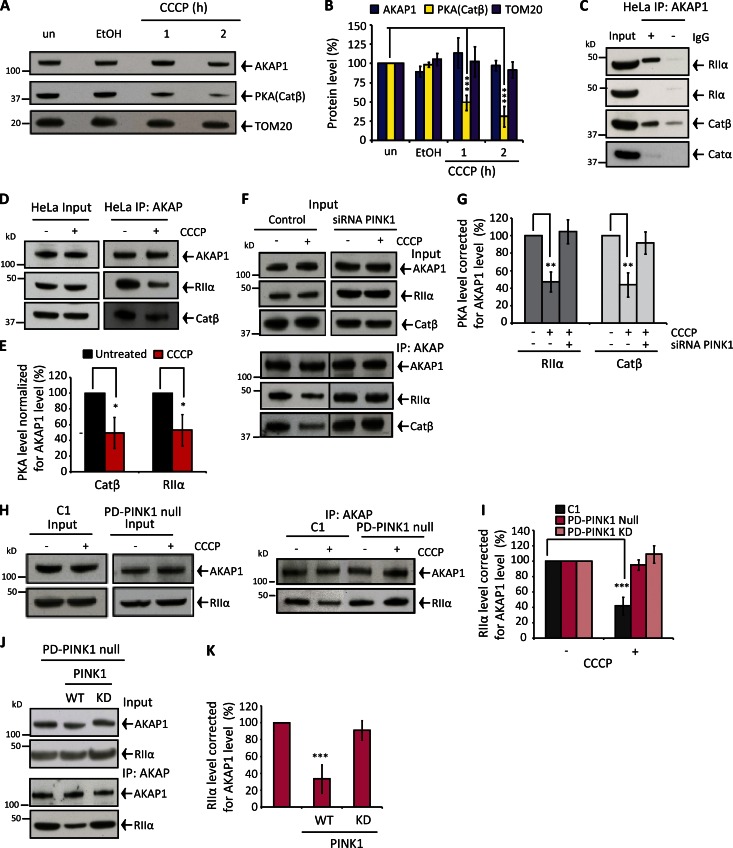
Figure 3.
PINK1 induces PKA displacement from AKAP1. (A and B) Effect of depolarization on AKAP1, PKA catalytic subunit β< and TOM20 protein levels in mitochondria isolated from HeLa cells. (C) AKAP1 IP from untreated HeLa cells and RIα, RIIα, Catβ, Catα, or RIIβ immunoblots. (D and E) AKAP1 CoIP with the PKA subunits RIIα and Catβ in HeLa cells ± 1-h CCCP treatment. RIIα/Catβ abundance corrected for AKAP1 levels and normalized to 100% in untreated samples. (F and G) CoIP between AKAP1 and RIIα/Catβ ± 1-h CCCP in control and pink1-silenced SH-SY5Y cells. RIIα/Catβ levels were corrected for AKAP1 and normalized to untreated samples. (H and I) AKAP1-RIIα CoIP in control (C1), PD-PINK1 null, and PD-PINK1 KD (Fig. S3 Y) fibroblasts after 2-h CCCP treatment. RIIα levels were adjusted for AKAP1 and normalized to untreated samples. (J and K) AKAP1-RIIα CoIP from 2-h CCCP-treated PD-PINK1 null fibroblasts after no transfection or WT or KD (K219M) PINK1 transfection (72 h). RIIα levels are adjusted for AKAP1 and normalized to non-transfected samples. PINK1 expression levels are shown in Fig. S3 O. All experiments were repeated at least three times, and error bars show SDM. **, P < 0.01; ***, P < 0.001.
Next, a series of coimmunoprecipitation (CoIP) experiments revealed the composition of the AKAP1-PKA signaling apparatus and were used to further unravel modulation of AKAP1-PKA after mΔΨ dissipation. AKAP1 exclusively precipitated PKA regulatory subunit II α (RIIα) and Catβ in both HeLa (Fig. 3 C) and SH-SY5Y (Fig. S3 A) cells. AKAP1 was also precipitated in the “reverse” RIIα and Catβ immunoprecipitations (IPs; Fig. S3, B–I). Our findings reveal regulatory and catalytic PKA isoform discrimination and AKAP1–PKA conservation. In contrast, RIIα IP from cytosolic fractions precipitated both Catα and Catβ (Fig. S3, J and K). The mechanism conferring Catβ selectivity with AKAP1 is unclear as is any potential functional significance. Acute CCCP treatment (1 h) diminished both AKAP1-RIIα and AKAP1-Catβ coprecipitation by ∼60–70% in HeLa (Fig. 3, D and E) and SH-SY5Y cells (Fig. 3, F and G), revealing PKA holoenzyme ejection from AKAP1, and Catβ is not selectively released from RIIα. The extent of PKA displacement from AKAP1 is consistent with our findings in depolarized isolated mitochondria (Fig. 3, A and B). Also, progressive PKA loss from AKAP1 is consistent with gradual PINK1 enrichment and activation in the OMM of depolarized mitochondria (not an immediate reaction to mΔΨ dissipation).
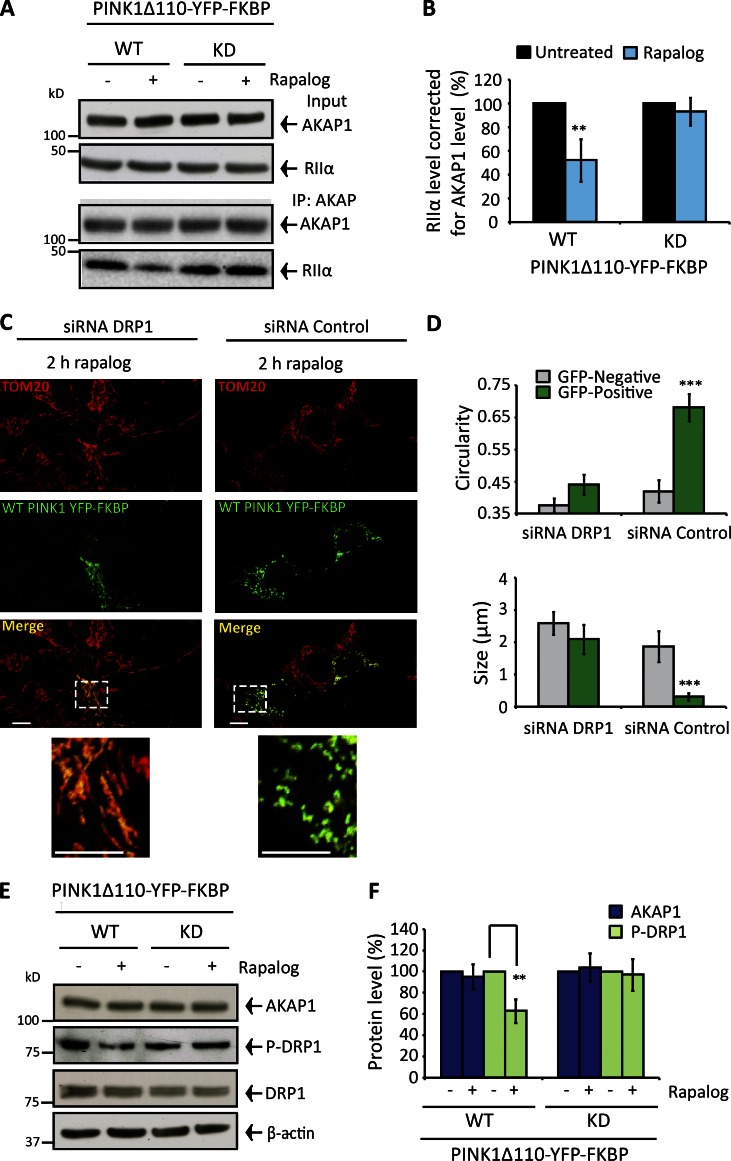
Does PINK1 disrupt the AKAP–PKA axis? To address this question, we silenced pink1 in SH-SY5Y cells, retarding MFN1 ubiquitylation and diminishing PINK1 accumulation after depolarization (Fig. S3, L and M). Complimentary experiments were performed with PD-PINK1 fibroblasts (expressing Parkin) either devoid of PINK1 protein (PD-PINK1 null) or comprising defective kinase activity (PD-PINK1 KD [ILE368ASN]) in conjunction with control fibroblasts (expressing PINK1 and Parkin; Fig. S1, D and E). In pink1-silenced SH-SY5Y cells and both PD-PINK1 fibroblasts, the AKAP1–PKA axis maintained after CCCP treatment but was diminished by 65% and 75% in control silenced SH-SY5Y cells and control fibroblasts, respectively (Fig. 3, F–I; and Fig. S3 N). We also confirmed that the introduction of WT PINK1 into PD-PINK1 null fibroblasts restored depolarization-induced dissociation of RIIα from AKAP1 (Fig. 3, J and K; and Fig. S3 O). Finally, we found WT PINK1Δ110-YFP-FKBP decreased RIIα-AKAP1 coprecipitation by ∼40–45% (transfection efficiency of ∼30–40%) after rapalog treatment (Fig. 4, A and B), and this mode of fission activation was impaired in drp1-silenced cells (Fig. 4 C and D). The abundance of serine 637 phosphorylated DRP1 was diminished in rapalog-treated WT PINK1Δ110-YFP-FKBP–expressing cells, whereas basal DRP1 and AKAP1 levels were unaffected (Fig. 4, E and F). Thereby, PINK1 kinase activity is necessary for PKA displacement from AKAP1 and consequent DRP1-mediated fission following PINK1 accumulation in the OMM.
Figure 4.
Controlled targeting of PINK1 to the OMM disrupts the AKAP1–PKA axis and drives fission via DRP1. (A and B) SH-SY5Y cells transfected with WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1 (48 h) and treated with rapalog for 1 h. (A) AKAP1-RIIα CoIP. (B) RIIα abundance corrected for AKAP1 and normalized to 100% in untreated samples. (C and D) SH-SY5Y cells 72 h drp1- or control-silenced and transfected with WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1 for 48 h. Mitochondrial morphology (TOM20-stained) and quantification after 2-h rapalog treatment. (E and F) AKAP1, DRP1, and phospho–serine 637 DRP1 levels in SH-SY5Y cells expressing WT or KD PINK1Δ110-YFP-FKBP/FRB-Fis1 (48 h) with or without 2-h rapalog treatment. Phospho-DRP1 adjusted for DRP1/β-actin and AKAP1 for β-actin. Experiments were repeated at least three times, and error bars indicate SDM. Bars: 20 µM; (insets) 10 µM. **, P < 0.01; ***, P < 0.001.
In addition to recruiting Parkin/autophagy receptors to damaged mitochondria, we reveal that PINK1 also comprises robust mitochondrial pro-fission activity and thereby can be considered a master mitophagic regulator to ensure efficient mitophagy by coordinating the segregation and ubiquitin/autophagy receptor labeling of damaged organelles. By demonstrating PINK1-dependent dissociation of the AKAP1–PKA axis, we provide a mechanistic rationale to explain how damaged segments of the mitochondrial reticulum (Jin and Youle, 2013) are selectively sensitized for DRP1-mediated fission during basal mitophagy, a fundamental process that occurs in cells and neurons (Bingol et al., 2014). In agreement, a recent study demonstrated the fission and mitophagy of individually damaged organelles in the axons of neurons (Ashrafi et al., 2014) and further emphasizes that damage-induced mitochondrial fragmentation does not require global mitochondrial depolarization. Further investigation is required to establish how PINK1 disrupts the AKAP1–PKA axis. PINK1 might directly phosphorylate AKAP1–PKA or direct other pathways, such as phosphoubiquitin. The notion that OMM-bound PINK1 comprises additional activities/substrates unrelated to Parkin signaling is an emerging concept (Lai et al., 2015), and a recent study has revealed PINK1-dependent phosphorylation of Rab GTPases (Lai et al., 2015).
Interestingly, previous studies have shown fundamental connectivity between PINK1 and PKA activities; overexpressed cytosolic PINK1 robustly augmented PKA-mediated signaling and neuronal morphogenesis via an unknown mechanism (Dagda et al., 2014). Our findings thus further support the notion that PINK1 and PKA activities converge and PINK1 may operate as a PKA regulator. Although we propose that calcineurin is not the fission activator during basal PINK1 mitophagy, because released calcium will rapidly diffuse from damaged mitochondrial sections, the PKA–calcineurin axis exerts key regulatory control by governing the propensity of fission events and can be manipulated to confer either protection or increased susceptibility to apoptotic stimuli (Merrill et al., 2011; Dagda et al., 2011).
Previous studies have shown that the mitochondrial reticulum undergoes random passive fission events (Twig et al., 2008; in the absence of exogenous stimuli/agonists), presumably because DRP1, which repeatedly cycles to the OMM and forms oligomeric spirals (Merrill et al., 2011), is not always phosphorylated by PKA (a function of OMM-PKA Kcat) and/or phosphorylated DRP1 has a decreased propensity, but not complete inability, of forming fission-competent spirals which envelop the OMM (Merrill et al., 2011). We therefore envisage that PINK1-activated fission is not absolutely requisite for mitophagy (because DRP1 will eventually catalyze a successful fission event) but ensures the rapid and efficient segregation of damaged organelles so mitophagy can proceed quickly. Indeed, mitochondrial degradation has been shown to occur within 10–20 min after depolarization (Ashrafi et al., 2014), which presumably limits the possible contagion of damage to surrounding healthy mitochondria. Furthermore, it is also worth noting that rates of passive fission diverge across different cell types and are notably slower in the soma of neurons and in neurons in vivo. Accordingly, the significance of PINK1-activated fission may vary in different tissues with a more prominent role in neurons.
It is interesting to consider Parkin phosphorylation by PINK1 accelerates mitophagy but is also not absolutely required, because phosphorylated ubiquitin forms a secondary mode to fully activate Parkin (Kondapalli et al., 2012; Shiba-Fukushima et al., 2012; Koyano et al., 2014; Kane et al., 2014; Wauer et al., 2015). Therefore phosphoubiquitin and passive fission may serve as a contingency pathway to ensure ubiquitin tagging and segregation of damaged mitochondria if the primary pathways fail. Finally, the reformation of fission proposed here has intriguing implications for the broader understanding of mitophagy and questions the relevance of calcium-driven reactions occurring during systemic depolarization by canonical mitophagy-activating methods; this level of damage occurring in vivo is highly unlikely. It is also interesting to contemplate how calcium may affect and influence other principle mitophagy pathways. Does calcium inadvertently affect PINK1 or OMM-Parkin activity or stability, or even modulate substrate specificity and associated binding proteins?
Materials and methods
Reagents and antibodies
All reagents and chemicals were purchased from Sigma-Aldrich unless otherwise stated. Antibodies were as follows: rabbit polyclonal anti-PKAβ catalytic (Catβ), mouse monoclonal anti–mouse PKAα catalytic (Catα), monoclonal anti-PKAIIα regulatory (RIIα), mouse monoclonal anti-PKAIα regulatory (RIα), mouse monoclonal anti-PKAIIβ regulatory (RIIβ), rabbit monoclonal anti-TOM20, mouse monoclonal anti-TOM20, mouse monoclonal anti-Parkin, (Santa Cruz Biotechnology, Inc.); mouse monoclonal anti-DRP1 (BD); rabbit polyclonal anti-PINK1, rabbit polyclonal anti-AKAP1 (Novus Biologicals); mouse monoclonal anti-PDH E1α (ProteinTech); mouse monoclonal anti-β-actin, mouse monoclonal anti-MFN1, (Abcam); mouse monoclonal anti-HA (Covance); rabbit polyclonal anti-phospho-637 DRP1, rabbit polyclonal anti-Prohibitin, rabbit monoclonal anti-LC3B, rabbit polyclonal anti-MEK1/2 (Cell Signaling Technology); complex IV subunit II was a gift from J.-W. Taanman (University College London, London, England, UK).
Cell culture and treatments
SH-SY5Y cells were cultured in DMEM/F12 (Thermo Fisher Scientific) and HeLa/fibroblasts in DMEM Glutamax (Thermo Fisher Scientific). Media was further supplemented with 10% FBS (Thermo Fisher Scientific), 1 mM pyruvate (Thermo Fisher Scientific), nonessential amino acids (Thermo Fisher Scientific), and penicillin/streptomycin (complete media). Cells were cultured in 37°C 5% CO2 humidified atmosphere.
SH-SY5Y and HeLa cells were incubated with 20 µM CCCP (Sigma-Aldrich) and fibroblasts with 40 µM. Rapalog (ClonTech) was used at 250 nM. All cells were harvested by trypsinization (Thermo Fisher Scientific) followed by the addition of complete media and cell pellets (1,500 g, 5 min, 4°C) were twice washed in ice-cold PBS and frozen as pellets at –80°C until required.
Human primary fibroblasts
Control lines 1 and 2 were cultured from a 52-yr-old male and a 70-yr-old female, respectively. The PD-PINK1 null culture comprises a frameshift deletion in PINK1 (c.261_276del16;p.Try90Leufsx12; provided by J. Hardy, University College London, London, England, UK). PD-Parkin#1 were cultured from a 50-yr-old female and comprise exon 2 and 3 deletion. PD-Parkin#2 were cultured from a 63-yr-old female and comprise exon 4 and 5 depletion. PD-PINK1 KD fibroblasts were acquired from the Coriell Biorepository (ND40066) and comprise a homogeneous ILE368ASN mutation cultured from a 64-yr-old male.
Statistical analysis
All data are expressed as the means ± SD of the mean (SDM). Statistical significance was calculated using the Student’s t test on at least triplicate experiments comparing against control of the equivalent experimental condition. P < 0.05 was considered significant and is denoted with a single asterisk, whereas P < 0.01 and P < 0.001 are denoted with two and three asterisks, respectively.
SDS-PAGE and Western blot analysis
Unless otherwise stated, cell pellets were solubilized in PBS-1% Triton X-100 supplemented with EDTA-free protease/phosphatase inhibitors (Pierce; 15 min, 4°C) and clarified using a table-top centrifuge (13,500 rpm, 10 min, 4°C). DTT-reduced extracts were loaded equally for protein (BCA protein determination assay), separated using SDS-PAGE (precast 4–12% gels, Novex; Thermo Fisher Scientific) and transferred (wet transfer) onto PDVF membranes (Amersham Hybond, 0.45 µm; GE Healthcare). Membranes were blocked using 5% powdered milk-PBS 0.1% Tween 20 solution for at least 1 h (RT) and then incubated with primary antibody overnight (4°C) in blocking buffer, followed by 1:2,000 HRP-conjugated secondary antibodies (Dako) for 1 h (RT) in blocking buffer. ECL chemiluminescent substrate (Thermo Fisher Scientific) was used to detect immunoreactive proteins on x-ray film.
Citrate synthase activity measurements
A total of 20 µg clarified PBS-1% Triton X-100 lysate was added to 200 µM oxaloacetate, acetyl-CoA, and 5,5′-dithio-bis-(2-nitrobenzoic acid (DTNB) in 50 mM Tris, pH 8.0, and measurements were performed in triplicate. The formation of TNB was monitored at 412 nm, and enzymatic rates were calculated during the steady state and corrected for protein amount (using the BCA assay).
Immunocytochemistry
SH-SY5Y cells and fibroblasts were seeded at 3.5 × 106 on 22-mm glass coverslips in six-well plates. Cells were fixed in 4% paraformaldehyde in PBS for 20 min at RT, subsequently washed three times in PBS, and solubilized for 20 min in PBS-0.25% Triton X-100 at RT. Then, SH-SY5Y cells were blocked in PBS-10% goat serum for 1 h at RT and incubated with rabbit or mouse anti-TOM20 (1:1,000) in PBS at 4°C; when required, anti-AKAP1 (1:1,000, 0.25% Triton X-100 included) was included in the overnight incubation. For fibroblasts, the blocking was performed overnight at 4°C in 10% goat serum–PBS. Then, fibroblasts were incubated with rabbit anti-TOM20 (1:300) in PBS for 90 min at RT. Coverslips were washed three times in PBS to terminate primary antibody incubation and then 1:500 anti–rabbit Alexa Fluor 594–conjugated secondary antibody (Thermo Fisher Scientific) was added in PBS for 1 h at RT. When necessary 1:500 anti–mouse Alexa Fluor 488–conjugated secondary antibody was included. Images were acquired using an inverted confocal microscope (LSM 510; ZEISS) and 63× oil-immersion objectives. Alexa Fluor 594 was excited using the 543-nm laser line (HeNe laser) and fluorescence detected with a 560-nm long-pass filter. A 488-nm laser line (argon laser) excited the Alexa Fluor 488 with fluorescence measured between 510 and 550 nm using the appropriate filter.
Mitochondrial morphology analysis
For all experiments, 30–50 cells in at least five different fields of view were analyzed per repeat. Mitochondria morphology (TOM20 immunostained) was quantified as described earlier (Koopman et al., 2008) via ImageJ. In brief, background was subtracted from raw images, which were subsequently linearly contrast optimized, applied with a 7 × 7 “top hat” filter, subjected to a 2 × 3 median filter, and then thresholded to generate binary images. The circularity, mean length and mitochondrial particles per number of cells were used to determine morphological changes.
Transfections and silencing
HiPerFect (QIAGEN) was used to transfect a suspension of SH-SY5Y cells with 30 nM pink1/control siRNA, 10 nM drp1/control siRNA (SMARTpool ON-TARGETplus siRNA; GE Healthcare), or 10 nM akap1/control siRNA (Santa Cruz Biotechnology, Inc.). For drp1 silencing, cells were seeded onto glass coverslips in six-well plates at 3 × 106 for 72 h;after 24 h, cells were transfected with the regulated heterodimerization plasmids (provided by R. Youle, National Institutes of Health, Bethesda, MD). akap1 was silenced in SH-SY5Y cells seeded onto glass coverslips in six-well plates at 2.5 × 106 (for immunofluorescence) or into six-well plates at 4 × 106 (for immunoblot), for 72 h. The Neon system was used to transfect FRB-Fis1 and WT/KD PINK1Δ110-YFP-FKBP or WT/KD (K219M) human PINK1 (as indicated in figures) via electroporation. 2.5 × 107 SH-SY5Y cells or fibroblasts were washed twice in PBS and resuspended in 50 µl buffer R with either 0.5 µg PINK1Δ110-YFP-FKBP and 0.15 µg FRB-Fis1 per 5 × 106 cells, or 1 µg WT/KD PINK1. Transfections were performed according to manufacturers’ instructions. After 48-h expression, the cultures were incubated with 250 nM Rapalog or CCCP treatment for 2 h before immunocytochemistry, IP, or immunoblot analysis.
The following siRNAs were used: AKAP1 (sc-40301; Santa Cruz Biotechnology, Inc.); control (sc-37007; Santa Cruz Biotechnology, Inc.); PINK1 (L-004030-00-0005; GE Healthcare); DRP1 (L-012092-00-0005; GE Healthcare); control (D-001810-10-05; GE Healthcare).
Fluo-4 live-cell imaging
SH-SY5Y cells were seeded onto 22-mm glass coverslips in six-well plates, as described in the Immunocytochemistry section. Cells were loaded with Fluo-4 AM (Thermo Fisher Scientific) for 45 min in Hank’s balanced salt solution (BSS; including 0.0025% Pluronic F-68), at RT. Fluo-4 was excited using the 488-nm laser line (argon laser), using a confocal microscope and 63× oil-immersion objective. For time resolved calcium quantification, fields of view comprising 20–40 SH-SY5Y cells were selected and Z-projection was performed to obtain maximum fluorescent intensities. Each experiment was performed four times in total. Using ImageJ, the mean intensities were determined across the Z-stack and the backgrounds, encompassing extracellular regions were measured and subtracted. Data are presented as the percentage change from the initial measurement or fluorescent intensity (arbitrary units).
SH-SY5Y cells were loaded with 25 nM TMRM+ (Thermo Fisher Scientific). Loading was performed for 30 min in HBSS, at RT. TMRM+ was excited using the 543-nm laser line (HeNe laser) and emission measured using a 560-nm long-pass filter. Fields of view comprising 20–40 cells were selected and Z-projection performed to obtain maximum fluorescence intensities. Experiments were repeated three times. Imaging was performed using a confocal microscope and 63× oil-immersion objectives. Data are presented as fluorescent intensity (arbitrary units).
Rapalog and CCCP were added to 250 nM or 20 µM, respectively, as indicated in figures and in HBSS comprising the equivalent concentration of fluorescent probe.
Mitochondrial isolation and cytosolic fraction preparation
SH-SY5Y or HeLa cells were cultured onto 9 × 10 cm2 plates for each treatment to ∼3.5 × 107 and 2.5 × 107, respectively. After treatment, cells were collected and stored as pellets at −80°C until required. Thawed pellets were resuspended in ice cold 250 mM sucrose, 1 mM EDTA, 10 mM Tris, pH 7.4 (HCl), isolation buffer supplemented with EDTA-free protease-phosphatase inhibitors, ruptured via mechanical homogenization (1,500 rpm, 20 strokes) and then pelleted (1,500 g, 10 min, 4°C). Supernatants (comprising mitochondria) were collected and pellets rehomogenized (1,500 rpm, 20 strokes), subsequently clarified (at 1,500 g, 10 min, 4°C) and supernatants pooled. Mitochondria were pelleted at 11,800 g (12 min, 4°C, with supernatants corresponding to cytosolic fractions) twice washed in isolation buffer, and the final pellets resuspended in 50 µl of isolation buffer for Western blot analysis.
CoIP
1,000 µg clarified PBS-0.5% Triton X-100 lysate (100 µl) from CCCP-treated SH-SY5Y or HeLa cells, 300 µg (100 µl) from CCCP-treated human fibroblasts ± WT or KD PINK1 transfection, 400 µg (100 µl) from rapalog-treated WT/KD PINK1Δ110-YFP-FKBP transfected SH-SY5Y cells, or 700 µg (200 µl) of cytosolic fraction (in mitochondrial preparation buffer) was incubated with either (as indicated in figures) 1 µg anti-AKAP1, 0.2 µg anti-PKARIIα, 0.2 µg anti-PKARIIβ, 0.4 µg anti-PKACα, or 0.4 µg anti-PKACβ for 2 h at 4°C with 30 µl of recombinant protein G agarose (Thermo Fisher Scientific), washed three times in lysis buffer (2,000 g, 2.5 min, 4°C), and eluted in 0.1 M glycine, pH 2.5 (50 µl; Thermo Fisher Scientific) with pH adjusted by 1 M Tris (final volume 60 µl). Samples were reduced (DTT) and heated to 70°C for 10 min before separation by SDS-PAGE.
Online supplemental material
Fig. S1 shows PINK1 and Parkin expression in the cells used in this study as well as data showing mitochondrial polarization and cytosolic calcium levels are not affected by rapalog. Fig. S1 also includes mitochondrial morphology images and analysis for the second PD-Parkin fibroblast cultures that were transfected with the regulated heterodimerization plasmids with or without rapalog. Fig. S2 includes the data showing the relationship between AKAP1 protein level and mitochondrial morphology and shows that AKAP1 is not degraded in the primary human fibroblasts used in this study. Release of PKA from SH-SY5Y depolarized mitochondrial fractions is also included. Fig. S3 shows data mapping the composition of PKA bound to AKAP1. PINK1 silencing efficacy in SH-SY5Y cells is included as is the preservation of AKAP1–PKA in PD-PINK1 KD fibroblasts. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201509003/DC1.
Supplementary Material
Acknowledgments
This study was supported by the Wellcome Trust/Medical Research Council Joint Call in Neurodegeneration award (WT089698), Centre of Excellence in Neurodegeneration award (MR/J009660/1), and National Institute of Health Research grants RCF30AS2012, RCF73TS20145989, and RCF103/AS/2014.
The authors declare no competing financial interests.
Author contributions: K.R. Pryde wrote the manuscript and performed, analyzed, and designed the experiments. H.L. Smith blind-analyzed mitochondrial morphology data, designed experiments, and edited the manuscript. K.-Y. Chau performed and designed experiments and edited the manuscript. A.H.V. Schapira supervised and designed the experiments, wrote the manuscript, and is a National Institute of Healthcare Research Senior Investigator.
Footnotes
Abbreviations used in this paper:
- AKAP1
- A-kinase anchoring protein 1
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- CoIP
- coimmunoprecipitation
- DRP1
- dynamin-related protein 1
- FKBP
- FK506 binding protein 12
- FRB
- 93-amino-acid portion of human FK506 binding protein 12/rapamycin–associated protein
- IP
- immunoprecipitation
- KD
- kinase dead
- mΔΨ
- mitochondrial membrane potential
- OMM
- outer mitochondrial membrane
- PD
- Parkinson disease
- PINK1
- PTEN-induced putative kinase
- PKA
- protein kinase A
- SDM
- SD of the mean
- WT
- wild type
References
- Ashrafi G., Schlehe J.S., LaVoie M.J., and Schwarz T.L.. 2014. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J. Cell Biol. 206:655–670. 10.1083/jcb.201401070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., Foreman O., Kirkpatrick D.S., and Sheng M.. 2014. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 510:370–375. [DOI] [PubMed] [Google Scholar]
- Carlucci A., Adornetto A., Scorziello A., Viggiano D., Foca M., Cuomo O., Annunziato L., Gottesman M., and Feliciello A.. 2008. Proteolysis of AKAP121 regulates mitochondrial activity during cellular hypoxia and brain ischaemia. EMBO J. 27:1073–1084. 10.1038/emboj.2008.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghetti G.M., Stangherlin A., Martins de Brito O., Chang C.R., Blackstone C., Bernardi P., and Scorrano L.. 2008. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. USA. 105:15803–15808. 10.1073/pnas.0808249105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.R., and Blackstone C.. 2007. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 282:21583–21587. 10.1074/jbc.C700083200 [DOI] [PubMed] [Google Scholar]
- Chung K.K., Thomas B., Li X., Pletnikova O., Troncoso J.C., Marsh L., Dawson V.L., and Dawson T.M.. 2004. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 304:1328–1331. 10.1126/science.1093891 [DOI] [PubMed] [Google Scholar]
- Cribbs J.T., and Strack S.. 2007. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8:939–944. 10.1038/sj.embor.7401062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda R.K., Gusdon A.M., Pien I., Strack S., Green S., Li C., Van Houten B., Cherra S.J. III, and Chu C.T.. 2011. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson’s disease. Cell Death Differ. 18:1914–1923. 10.1038/cdd.2011.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda R.K., Pien I., Wang R., Zhu J., Wang K.Z., Callio J., Banerjee T.D., Dagda R.Y., and Chu C.T.. 2014. Beyond the mitochondrion: cytosolic PINK1 remodels dendrites through protein kinase A. J. Neurochem. 128:864–877. 10.1111/jnc.12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey A.S., and Strack S.. 2011. PKA/AKAP1 and PP2A/Bβ2 regulate neuronal morphogenesis via Drp1 phosphorylation and mitochondrial bioenergetics. J. Neurosci. 31:15716–15726. 10.1523/JNEUROSCI.3159-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson S.A., Kane L.A., Yamano K., Huang C.H., Sliter D.A., Buehler E., Wang C., Heman-Ackah S.M., Hessa T., Guha R., et al. . 2013. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 504:291–295. 10.1038/nature12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.M., and Youle R.J.. 2013. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 9:1750–1757. 10.4161/auto.26122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., and Youle R.J.. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205:143–153. 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Scimia M.C., Wilkinson D., Trelles R.D., Wood M.R., Bowtell D., Dillin A., Mercola M., and Ronai Z.A.. 2011. Fine-tuning of Drp1/Fis1 availability by AKAP121/Siah2 regulates mitochondrial adaptation to hypoxia. Mol. Cell. 44:532–544. 10.1016/j.molcel.2011.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C., Kazlauskaite A., Zhang N., Woodroof H.I., Campbell D.G., Gourlay R., Burchell L., Walden H., Macartney T.J., Deak M., et al. . 2012. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2:120080 10.1098/rsob.120080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman W.J., Distelmaier F., Esseling J.J., Smeitink J.A., and Willems P.H.. 2008. Computer-assisted live cell analysis of mitochondrial membrane potential, morphology and calcium handling. Methods. 46:304–311. 10.1016/j.ymeth.2008.09.018 [DOI] [PubMed] [Google Scholar]
- Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. . 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510:162–166. [DOI] [PubMed] [Google Scholar]
- Lai Y.C., Kondapalli C., Lehneck R., Procter J.B., Dill B.D., Woodroof H.I., Gourlay R., Peggie M., Macartney T.J., Corti O., et al. . 2015. Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J. 34:2840–2861. 10.15252/embj.201591593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Jin S.M., Kane L.A., and Youle R.J.. 2012. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell. 22:320–333. 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., and Youle R.J.. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524:309–314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. . 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221. 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill R.A., Dagda R.K., Dickey A.S., Cribbs J.T., Green S.H., Usachev Y.M., and Strack S.. 2011. Mechanism of neuroprotective mitochondrial remodeling by PKA/AKAP1. PLoS Biol. 9:e1000612 10.1371/journal.pbio.1000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.-F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., and Youle R.J.. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhall K.J., Criniti A.R., Cheah C.S., Smith K.C., Kafer K.E., Burkart A.D., and McKnight G.S.. 2006. Dynamic anchoring of PKA is essential during oocyte maturation. Curr. Biol. 16:321–327. 10.1016/j.cub.2005.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger P.T., Kabra D.G., Aichler M., Schriever S.C., Pfuhlmann K., García V.C., Lehti M., Weber J., Kutschke M., Rozman J., et al. . 2015. Calcineurin Links Mitochondrial Elongation with Energy Metabolism. Cell Metab. 22:838–850. 10.1016/j.cmet.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Schapira A.H. 2008. Mitochondria in the aetiology and pathogenesis of Parkinson’s disease. Lancet Neurol. 7:97–109. 10.1016/S1474-4422(07)70327-7 [DOI] [PubMed] [Google Scholar]
- Schapira A.H., Olanow C.W., Greenamyre J.T., and Bezard E.. 2014. Slowing of neurodegeneration in Parkinson’s disease and Huntington’s disease: future therapeutic perspectives. Lancet. 384:545–555. 10.1016/S0140-6736(14)61010-2 [DOI] [PubMed] [Google Scholar]
- Schon E.A., and Przedborski S.. 2011. Mitochondria: the next (neurode)generation. Neuron. 70:1033–1053. 10.1016/j.neuron.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., and Hattori N.. 2012. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep. 2:1002 10.1038/srep01002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F.D., Reichow S.L., Esseltine J.L., Shi D., Langeberg L.K., Scott J.D., and Gonen T.. 2013. Intrinsic disorder within an AKAP-protein kinase A complex guides local substrate phosphorylation. eLife. 2:e01319 10.7554/eLife.01319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., and Youle R.J.. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380. 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. . 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446. 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. . 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA. 107:378–383. 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T., Simicek M., Schubert A., and Komander D.. 2015. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 524:370–374. 10.1038/nature14879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.