Inclusion bodies containing aggregated disease-associated proteins and polyubiquitin conjugates are universal hallmarks of neurodegenerative diseases. Here, Bersuker et al. examine the necessity and sufficiency of ubiquitin conjugation for targeting proteins to inclusion bodies.
Abstract
Inclusion bodies (IBs) containing aggregated disease-associated proteins and polyubiquitin (poly-Ub) conjugates are universal histopathological features of neurodegenerative diseases. Ub has been proposed to target proteins to IBs for degradation via autophagy, but the mechanisms that govern recruitment of ubiquitylated proteins to IBs are not well understood. In this paper, we use conditionally destabilized reporters that undergo misfolding and ubiquitylation upon removal of a stabilizing ligand to examine the role of Ub conjugation in targeting proteins to IBs that are composed of an N-terminal fragment of mutant huntingtin, the causative protein of Huntington’s disease. We show that reporters are excluded from IBs in the presence of the stabilizing ligand but are recruited to IBs after ligand washout. However, we find that Ub conjugation is not necessary to target reporters to IBs. We also report that forced Ub conjugation by the Ub fusion degradation pathway is not sufficient for recruitment to IBs. Finally, we find that reporters and Ub conjugates are stable at IBs. These data indicate that compromised folding states, rather than conjugation to Ub, can specify recruitment to IBs.
Introduction
Inclusion bodies (IBs) composed of aggregated forms of normally soluble proteins are a universal pathognomonic hallmark of neurodegenerative diseases (Ross and Poirier, 2004). Whether IB formation contributes to the demise of neurons in which they occur or serves as part of a cytoprotective response is controversial (Arrasate et al., 2004; Ross and Poirier, 2005). The universal presence of 26S proteasome subunits (Ii et al., 1997) and conjugated ubiquitin (Ub; Lowe et al., 1988; DiFiglia et al., 1997) within these disease-associated, intracellular aggregates has long suggested a role for disrupted protein (Hipp et al., 2014) and Ub homeostasis (Bennett et al., 2005, 2007; Dantuma and Bott, 2014) as a common mechanism underlying these otherwise clinically diverse disorders. In spite of this correlation between Ub accumulation and disease pathology, a mechanistic understanding of why components of the Ub–proteasome system (UPS) machinery are sequestered at IBs and its relationship to disease etiology is still lacking.
Dominantly inherited neurodegenerative disorders, most commonly caused by mutations that result in the production of proteins with altered folding and solubility properties, have been highly informative in investigating the link between protein aggregation and Ub homeostasis (Bennett et al., 2007; Atkin and Paulson, 2014). Genetic expansion of a CAG tract in the first exon of the huntingtin (htt) gene beyond a critical threshold of ∼35 causes Huntington’s disease (HD) and results in the synthesis of a mutant htt protein containing an expanded polyglutamine (polyQ) tract (MacDonald et al., 1993). Expression of exon 1 htt fragments or fusion proteins containing polyQ tracts longer than 35–40 residues leads to the formation of insoluble amyloid aggregates of htt in vitro (Scherzinger et al., 1999) and in cell culture (Waelter et al., 2001) and causes neurodegeneration and formation of intraneuronal IBs when expressed in the brains of transgenic (Mangiarini et al., 1996) or knock-in (Menalled, 2005) mice. Like IBs in other neurodegenerative disorders, htt IBs are robustly immunoreactive with antibodies to Ub conjugates (Davies et al., 1997), and poly-Ub chains are significantly enriched in the brains of human HD patients and HD transgenic mice (Bennett et al., 2007), confirming that mutant htt expression is a suitable system with which to study the relationship between protein aggregation and disrupted Ub homeostasis.
Several nonmutually exclusive models have been posited to explain why Ub conjugates accumulate in IBs found in patients and models of HD and other neurodegenerative diseases. One prominent model holds that cellular “quality control” machinery recognizes mutant htt as a terminally misfolded protein (Steffan et al., 2004; Bhat et al., 2014), thereby directing the protein for destruction by the 26S proteasome. However, htt fusions with long or short polyQ tracts exhibit similarly long half-lives (>30 h; Tsvetkov et al., 2013; unpublished data) that are atypical of Ub-dependent proteasome system substrates. Additionally, both short and long polyQ sequences are intrinsically disordered before aggregation (Wetzel, 2012) and are thus unlikely to adopt a conformation that is recognized as misfolded by quality control Ub ligases. Moreover, mutant htt is recruited to IBs before and independently of Ub, and only a small fraction of soluble htt is conjugated to Ub (Hipp et al., 2012). It is therefore unlikely that the majority of Ub present in IBs is conjugated to htt.
A second model posits that mutant htt interferes with the function of the 26S proteasome, causing ubiquitylated forms of many different (non-htt) proteins to accumulate and, eventually, to become sequestered in IBs. Although this model is consistent with data showing a strong correlation between mutant htt aggregation and accumulation of synthetic short-lived substrates of the UPS in cell culture (Bence et al., 2001; Bennett et al., 2005), these UPS substrate reporters do not accumulate (Bett et al., 2009; Maynard et al., 2009) or only accumulate transiently (Ortega et al., 2010) in mouse models of HD. In cell culture, mutant htt aggregation and recruitment of Ub conjugates to nascent IBs is not temporally correlated with accumulation of UPS reporters (Hipp et al., 2012). In vitro, purified soluble or aggregated forms of mutant htt inhibit 26S proteasome activity only when they are ubiquitylated, suggesting that they could compete with other ubiquitylated proteins for binding to Ub receptors on 26S proteasomes (Bennett et al., 2005; Hipp et al., 2012). Because htt conjugates comprise only a small fraction of soluble cellular Ub conjugates (Riley et al., 2010), it is highly unlikely that ubiquitylated htt is present at sufficient concentrations in vivo to effectively outcompete other UPS substrates. These findings are inconsistent with a model in which mutant htt directly impairs the function of 26S proteasomes. As such, the mechanism underlying the accumulation of Ub conjugates at IBs in cells challenged with mutant htt remains unclear.
A third model to explain the accumulation of Ub conjugates in IBs after htt aggregation holds that expression of mutant, aggregation-prone proteins like htt impairs global protein homeostasis (Hartl et al., 2011; Margulis and Finkbeiner, 2014), possibly by sequestering one or more molecular chaperones required for the folding of chaperone-dependent clients (Park et al., 2013; Yu et al., 2014). This model is supported by studies showing that expression of mutant htt causes the manifestation of multiple mutant phenotypes at permissive temperatures in Caenorhabditis elegans harboring temperature-sensitive alleles of unrelated genes (Gidalevitz et al., 2006) and induces the misfolding of engineered, metastable luciferase-based “proteostasis sensors” in cultured mammalian cells (Gupta et al., 2011). The cumulative deficit caused by the challenge of mutant htt to global protein folding capacity could interfere with the folding of otherwise normal proteins, diverting misfolded proteins to the UPS and eventually exceeding the capacity of the 26S proteasome (Hipp et al., 2012). This model provides an explanation for why unrelated ubiquitylated proteins accumulate in cells challenged with aggregation-prone proteins like mutant htt, but does not explain why these Ub conjugates associate with IBs or why there is an ∼20-h delay between the onset of htt aggregation and the accumulation of synthetic fluorescent reporters of UPS activity (Hipp et al., 2012).
In this work, we have used conditionally destabilized mutants of proteins that undergo ligand-regulated misfolding and Ub-dependent degradation together with pharmacological inhibition of Ub activation to directly assess the role of Ub conjugation in recruitment of proteins to IBs. Surprisingly, our results indicate that Ub is neither necessary nor sufficient to target proteins to IBs. Instead, we find that recruitment of proteins to IBs is specified exclusively by adoption of nonnative folding states.
Results
Conditionally destabilized UPS substrates are recruited to IBs
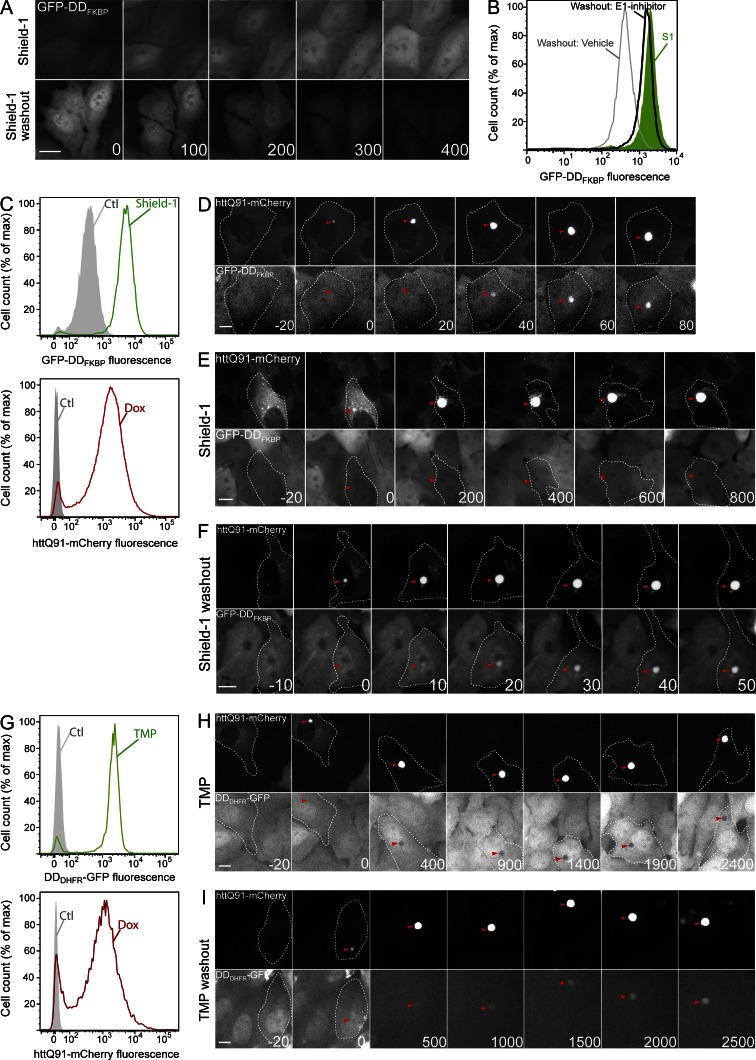
To assess the role of Ub conjugation in recruitment of proteins to htt IBs, we used fluorescence time-lapse microscopy to monitor the effect of mutant htt aggregation on the intracellular localization of fluorescent reporters engineered to become reversibly and conditionally misfolded by the removal of a small ligand (Fig. S1 A). One of these reporters, GFP-DDFKBP, is composed of GFP fused to a variant FK506 binding domain from FKBP12. This domain is tightly folded in the presence of the synthetic ligand shield-1 but is misfolded and efficiently targeted for Ub-dependent degradation in the absence of this ligand (Banaszynski et al., 2006; Egeler et al., 2011). For these experiments, we generated a stable U2OS cell line (U2-DDFKQ91) that expresses GFP-DDFKBP constitutively and mutant htt exon 1 fused to mCherry fluorescent protein (httQ91-mCherry) under the control of a tetracycline-inducible (TET-ON) promoter. Addition of shield-1 to these cells in the absence of doxycycline (dox) caused a uniform ∼10-fold increase in the mean GFP fluorescence of the population (Fig. 1, A and C, top). This fluorescence was distributed diffusely in the nuclear and cytoplasmic compartments (Fig. 1 A). After washout of shield-1, GFP-DDFKBP fluorescence levels decayed to baseline (Fig. 1, A and B; and Fig. S1 B). This decay (t1/2 of ∼90 min) was abrogated by addition of an inhibitor of the Ub-activating enzyme E1 (E1 inhibitor; Fig. 1 B) at concentrations sufficient to inhibit Ub conjugation, as assessed by the rapid depletion of ubiquitylated proteins (Fig. S1 C; Chen et al., 2011). These results confirm that in U2-DDFKQ91 cells, GFP-DDFKBP is rapidly degraded in a Ub-dependent manner upon withdrawal of shield-1 (Egeler et al., 2011).
Figure 1.
Conditionally destabilized reporters are recruited to IBs. (A) GFP-DDFKBP is stabilized by shield-1. U2OS cells stably expressing GFP-DDFKBP and tetracycline-inducible httQ91-mCherry (U2-DDFKQ91) were treated with 1 µM shield-1 or subjected to shield-1 washout and imaged by time-lapse microscopy. (B) GFP-DDFKBP undergoes Ub-dependent degradation. GFP-DDFKBP fluorescence in U2-DDFKQ91 cells was determined by flow cytometry analysis in the presence of 1 µM shield-1 (S1) or after washout of shield-1 for 6 h in the presence or absence of 10 µM E1 inhibitor. (C) U2-DDFKQ91 cells were treated with 1 µM shield-1 (top) or 1 µg/ml dox (bottom) for 72 h and analyzed by flow cytometry. (D–F) Misfolded but not folded GFP-DDFKBP is recruited to httQ91-mCherry IBs. U2-DDFKQ91 cells were treated with 1 µg/ml dox for 48 h and imaged by time-lapse microscopy in the absence of shield-1 (D), in the presence of 1 µM shield-1 (E), or after shield-1 washout (F). (G) Cells stably expressing DDDHFR-GFP and tetracycline-inducible httQ91-mCherry (U2-DDDHQ91) were treated with 10 µM TMP (top) or 1 µg/ml dox (bottom) for 72 h and analyzed by flow cytometry. (H and I) Misfolded but not folded DDDHFR-GFP is recruited to httQ91-mCherry IBs. U2-DDDHQ91 cells were treated with 1 µg/ml dox for 48 h and imaged by time-lapse microscopy in the presence of 10 µM TMP (H) or after TMP washout (I). Time stamps indicate elapsed time in minutes. For montages of httQ91-mCherry, t = 0 min indicates the frame in which IBs were first observed, and negative values correspond to time in minutes preceding this frame. (A, D–F, H, and I) Microscopy panels are representative of n > 50 (A and D–F) and n > 10 (H and I) independent observations. Arrowheads indicate IBs. (B, C, and G) For flow cytometry, n > 10,000 cells were analyzed. Ctl, control. Bars, 10 µm.
To assess the effect of IB formation on GFP-DDFKBP, we induced expression of httQ91-mCherry in U2-DDFKQ91 cells. Addition of dox resulted in increased httQ91-mCherry fluorescence intensity (Fig. 1 C, bottom; and Fig. S1 D) and a time-dependent increase in the fraction of cells with IBs, which reached a plateau by 72 h (Fig. S1 E). Time-lapse video microscopy of these cells revealed that, as previously observed (Hipp et al., 2012), httQ91-mCherry redistributed from an initially diffuse cytoplasmic localization to a single, centrally located IB once its levels exceeded a critical concentration (Fig. 1 D). Although there was some cell-to-cell variability in the time course, aggregation of httQ91-mCherry occurred rapidly, with httQ91-mCherry fluorescence at IBs reaching a plateau within 100 min (Fig. S1 F). This behavior is consistent with previous studies demonstrating that mutant htt IBs nucleate the efficient and rapid recruitment of soluble htt monomers into amyloid aggregates (Scherzinger et al., 1997, 1999). GFP-DDFKBP fluorescence was excluded from IBs when httQ91-mCherry expression was induced in the presence of shield-1–stabilized GFP-DDFKBP (Fig. 1 E). In contrast, GFP-DDFKBP fluorescence accumulated at nascent IBs in the absence of shield-1 (Fig. 1 D and Fig. S1 F) or after washout of shield-1 (Fig. 1 F). GFP-DDFKBP maintained a cytoplasmic/nuclear distribution in cells expressing diffuse httQ91-mCherry (Fig. 1, D and F). Thus, recruitment of GFP-DDFKBP is exclusively a property of misfolded, UPS-targeted conformers of GFP-DDFKBP.
The recruitment of misfolded conformers of GFP-DDFKBP to IBs led us to ask whether this property is common to misfolded conformers of other conditionally destabilized proteins. One such protein, DDDHFR-GFP, is a fluorescent reporter consisting of bacterial dihydrofolate reductase engineered to undergo degradation by the UPS when expressed in the absence of the folate analogue trimethoprim (TMP; Iwamoto et al., 2010). We generated a stable U2OS line (U2-DDDHQ91) that expresses DDDHFR-GFP constitutively and TET-ON httQ91-mCherry. In the absence of TMP and dox, basal GFP fluorescence in these cells was not detectable above background (Fig. 1 G), consistent with the very short half-life (t1/2 of ∼30 min) of this reporter (Fig. S1 H). DDDHFR-GFP fluorescence increased dramatically after exposure of U2-DDDHQ91 cells to TMP (Fig. 1 G) and decayed rapidly after TMP washout (Fig. S1 B). This decay was blocked by the E1 inhibitor (Fig. S1 G), indicating that, like GFP-DDFKBP, turnover of DDDHFR-GFP is dependent on Ub conjugation. Time-lapse imaging revealed that, as with GFP-DDFKBP, folded DDDHFR-GFP (i.e., in the presence of TMP) was excluded from IBs (Fig. 1 H), whereas washout of TMP led to sequestration of DDDHFR-GFP in mCherry-positive IBs (Fig. 1 I). The kinetics of DDDHFR-GFP recruitment to IBs were considerably slower than those exhibited by GFP-DDFKBP, likely reflecting the intrinsically faster turnover of DDDHFR-GFP (Fig. S1 H). These data show that the ability to be recruited to htt IBs in the absence of ligands is a common property of two structurally unrelated protein reporters containing unrelated conditional degrons. In the absence of the ligand, both proteins adopt nonnative (misfolded) states that are targeted for Ub-dependent conjugation and destruction by the 26S proteasome. Thus, recruitment to IBs could be a consequence of the (mis)folding state, Ub conjugation, or both.
Ub conjugation is neither necessary nor sufficient for recruitment of misfolded GFP-DDFKBP to IBs
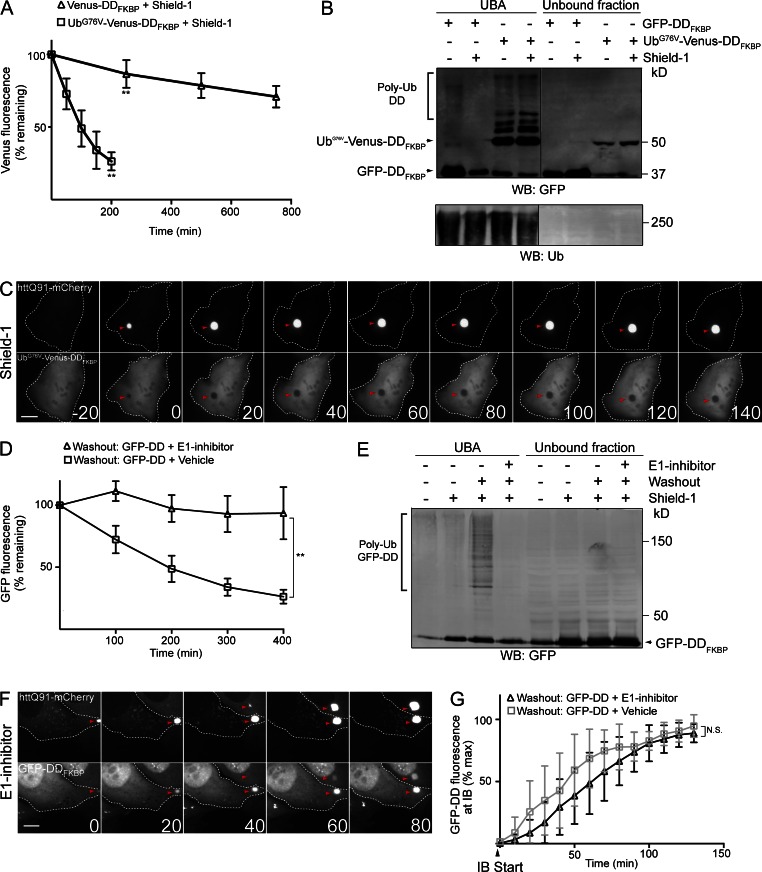
To determine whether conjugation to Ub is sufficient to recruit GFP-DDFKBP to IBs, we developed a reporter, UbG76V-Venus-DDFKBP, containing a noncleavable Ub (UbG76V) fused to the N terminus of Venus-DDFKBP that, despite being tightly folded in the presence of shield-1, is constitutively ubiquitylated and degraded by the proteasome. N-terminal UbG76V cannot be removed by Ub-specific proteases and functions as a dominant degron that can target even correctly folded, long-lived proteins for polyubiquitylation and degradation by the UPS (Johnson et al., 1992; Dantuma et al., 2000). Fluorescence time-lapse imaging of cells expressing UbG76V-Venus-DDFKBP after translation shutoff showed that this reporter is indeed rapidly degraded (t1/2 of ∼90 min), even in the presence of shield-1 (Fig. 2 A and Fig. S2, A and B). This degradation was completely blocked by the E1 inhibitor (Fig. S2 B). In contrast, an identical construct lacking the N-terminal UbG76V was extremely stable in the presence of shield-1 (t1/2 > 12 h; Fig. 2 A) and was degraded more slowly than UbG76V-Venus-DDFKBP in the absence of shield-1 (Fig. S2 B), indicating that UbG76V fusion functions as a dominant Ub-dependent degron, even in the presence of a stabilizing ligand. Consistent with its rapid Ub-dependent turnover, ubiquitylated forms of UbG76V-Venus-DDFKBP were detected (Fig. 2 B) after affinity capture of Ub conjugates by the Ub-associated (UBA) domain of hPLIC-2 (UBAhPLIC-2; Raasi et al., 2005). Thus, the UbG76V fusion promotes constitutive ubiquitylation and degradation of folded, shield-1–bound UbG76V-Venus-DDFKBP.
Figure 2.
Ub conjugation is not necessary or sufficient for recruitment of GFP-DDFKBP to IBs. (A) Fusion to UbG76V decreases the half-life of folded Venus-DDFKBP. Cells were transiently transfected with UbG76V-Venus-DDFKBP or Venus-DDFKBP for 48 h in the presence of 1 µM shield-1, and mean Venus fluorescence of cells (n > 10) was determined by time-lapse microscopy after translation shutoff with 25 µM emetine. (B) Folded UbG76V-Venus-DDFKBP is constitutively ubiquitylated. Cells were transiently transfected with GFP-DDFKBP or UbG76V-Venus-DDFKBP in the presence of 1 µM shield-1 or vehicle for 72 h. Subsequently, Ub conjugates were affinity captured by incubating cell lysates with UBAhplic-2-conjugated beads, and eluate and unbound fractions were analyzed by SDS-PAGE and immunoblotting with anti-GFP and anti-Ub antibodies. (C) Folded and ubiquitylated UbG76V-Venus-DDFKBP is not recruited to IBs. Cells transiently transfected with httQ91-mCherry and UbG76V-Venus-DDFKBP for 48 h in the presence of 1 µM shield-1 were imaged by time-lapse microscopy. (D) E1 inhibitor blocks degradation of misfolded GFP-DDFKBP. U2-DDFKQ91 cells were treated with 1 µg/ml dox and 1 µM shield-1 for 48 h. Mean GFP-DDFKBP fluorescence of cells (n > 10) was determined by time-lapse microscopy after shield-1 washout in the presence or absence of 10 µM E1 inhibitor. (E) E1 inhibitor blocks ubiquitylation of misfolded GFP-DDFKBP. U2-DDFKQ91 cells incubated with 1 µM shield-1 were treated for 3 h with 10 µM E1 inhibitor or vehicle. After a 15-min shield-1 washout, Ub conjugates were affinity captured from cell lysates with UBAhplic-2 beads and eluate, and unbound fractions were analyzed by SDS-PAGE and immunoblotting with anti-GFP antibody. (F) Misfolded, nonubiquitylated GFP-DDFKBP is recruited to IBs. U2-DDFKQ91 cells were treated for 48 h with 1 µM shield-1 and 1 µg/ml dox. Cells were subsequently treated for 3 h with 10 µM E1 inhibitor, subjected to shield-1 washout, and imaged by time-lapse microscopy. (G) E1 inhibitor does not block recruitment of GFP-DDFKBP to IBs. GFP-DDFKBP fluorescence at IBs was measured in cells (n > 10) after shield-1 washout in the presence or absence of 10 µM E1 inhibitor. IB start indicates the frame in which httQ91-mCherry IBs were first detected. Time stamps indicate elapsed time in minutes. For C and F, t = 0 indicates the frame in which IBs were first observed, and negative values correspond to time in minutes preceding this frame. Data points indicate mean ± SD. Microscopy panels C and F are representative of n > 20 independent observations. Bars, 10 µm. Arrowheads indicate IBs. Western blots (WBs) are representative experiments from at least two independent repeats. Statistically relevant differences (α = 0.05): **, P < 0.0001.
Time-lapse microscopy imaging revealed that UbG76V-Venus-DDFKBP was recruited to IBs in the absence of shield-1 (Fig. S2 C), indicating that the UbG76V fusion does not interfere with the sequestration of misfolded UbG76V-Venus-DDFKBP. In contrast, addition of shield-1 completely prevented UbG76V-Venus-DDFKBP recruitment to IBs (Fig. 2 C). These results demonstrate that Ub conjugation is not sufficient to recruit folded UPS substrates such as UbG76V-Venus-DDFKBP to IBs but do not rule out the possibility that Ub functions as an essential IB targeting signal for misfolded proteins.
To determine whether Ub conjugation is necessary for IB recruitment, we used the E1 inhibitor to block conjugation of Ub to misfolded GFP-DDFKBP. Before adding E1 inhibitor, cells expressing this reporter were incubated with shield-1 overnight to allow folded GFP-DDFKBP to accumulate. Upon washout of shield-1, GFP-DDFKBP fluorescence declined rapidly (Fig. 2 D), and high molecular weight forms of GFP-DDFKBP were detected after affinity capture by UBAhPLIC-2 beads (Fig. 2 E). In contrast, when shield-1 was washed out in the presence of E1 inhibitor, GFP-DDFKBP was markedly stabilized (Fig. 2 D and Fig. S2 D), and ubiquitylated forms of the reporter were not captured by UBAhPLIC-2 beads (Fig. 2 E). Therefore, the combination of shield-1 washout and E1 inhibitor can be used to acutely generate a cohort of misfolded, nonubiquitylated GFP-DDFKBP molecules. These conditions (shield-1 washout with E1 inhibitor) did not impede recruitment of httQ91-mCherry to IBs (Fig. S2 E), confirming that Ub conjugation is not required for IB formation. Strikingly, recruitment of misfolded GFP-DDFKBP to IBs after shield-1 washout occurred with similar kinetics irrespective of the presence of the E1 inhibitor (Fig. 2, F and G). Thus, Ub conjugation is neither necessary nor sufficient for recruitment of GFP-DDFKBP to IBs. Together, these data suggest that the folding state, but not the ubiquitylation state, is the primary determinant of recruitment of proteins to IBs.
Misfolded GFP-DDFKBP and Ub conjugates are stabilized at IBs
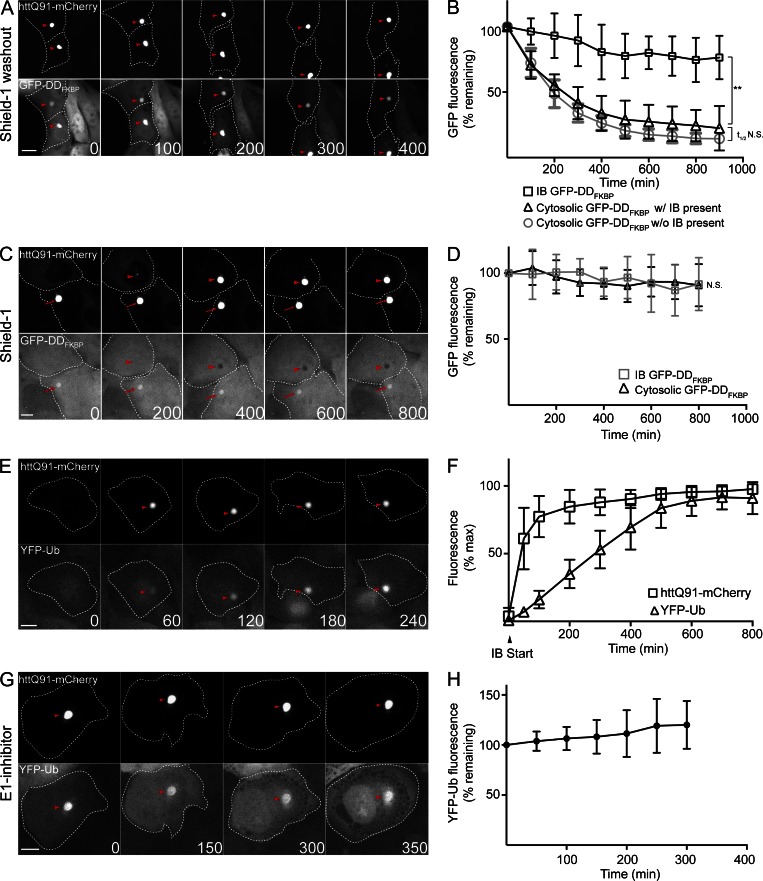
It has previously been proposed that targeting of Ub conjugates to IBs facilitates degradation of misfolded, ubiquitylated proteins via autophagy (Kawaguchi et al., 2003; Nagaoka et al., 2004; Lamark and Johansen, 2012). However, we found that GFP-DDFKBP fluorescence at IBs after shield-1 washout was extremely stable (t1/2 > 16 h; Fig. 3, A and B). In contrast, GFP-DDFKBP fluorescence in cytosolic regions adjacent to IBs, or in the cytosol of cells that have only diffuse httQ91-mCherry fluorescence, decayed rapidly (t1/2 of ∼150 min) upon shield-1 washout (Fig. 3 B). The rate of decay was similar to the decay kinetics of this reporter in the absence of shield-1 in cells not expressing htt (Fig. S1 B). These data suggest that stabilization of GFP-DDFKBP fluorescence at IBs is not a consequence of global impairment of protein turnover by htt IBs. Acute addition of shield-1 to dox-induced U2-DDFKQ91 cells led to stabilization of cytoplasmic GFP-DDFKBP but did not lead to decreased GFP-DDFKBP fluorescence within IBs that were present before addition of the stabilizing ligand (Fig. 3, C and D; and Fig. S3 A). Thus, misfolded GFP-DDFKBP at IBs is stable and refractory to folding by shield-1.
Figure 3.
Misfolded GFP-DDFKBP and Ub conjugates are stable at IBs. (A) Misfolded GFP-DDFKBP is stable at IBs. U2-DDFKQ91 cells were treated with 1 µg/ml dox and 1 µM shield-1 for 48 h and imaged by time-lapse microscopy after washout of shield-1. (B) Quantification of mean GFP-DDFKBP fluorescence at IBs in the cytosol/nucleus of cells with IBs present and in the cytosol/nucleus of cells without IBs in cells (n > 10) imaged in A. (C) Misfolded GFP-DDFKBP is sequestered at IBs. U2-DDFKQ91 cells were treated with 1 µg/ml dox for 48 h. Cells were subsequently imaged by time-lapse microscopy after addition of 1 µM shield-1 and 25 µM emetine to prevent accumulation of cytosolic and nuclear GFP-DDFKBP fluorescence (for addition of emetine only; Fig. S3 A). Arrowheads indicate IBs that formed after addition of shield-1, and arrows indicate IBs that were present before addition of shield-1. (D) Quantification of mean GFP-DDFKBP fluorescence at IBs and in the cytosol/nucleus in cells (n > 10) imaged in C. (E) YFP-Ub accumulates at IBs. U2-Ub-Q91 cells were treated with 1 µg/ml dox for 48 h and imaged by time-lapse microscopy. (F) Quantification of YFP-Ub and httQ91-mCherry fluorescence at IBs in cells (n > 10) imaged in E. IB start indicates the frame in which httQ91-mCherry IBs were first detected. (G) Ub conjugates are stable at IBs. U2-Ub-Q91 cells were treated with 1 µg/ml dox for 48 h. Cells were subsequently treated for 3 h with 10 µM E1 inhibitor and imaged by time-lapse microscopy. (H) Quantification of YFP-Ub fluorescence at IBs in cells (n > 15) imaged in G. Arrowheads indicate IBs in A, E, and G. Time stamps indicate elapsed time in minutes. Bars, 10 µm. Data points indicate mean ± SD. Statistically relevant differences (α = 0.05): **, P < 0.0001.
To test whether stabilization at IBs is unique to misfolded GFP-fusion reporters like GFP-DDFKBP or DDDHFR-GFP or is a general property of Ub conjugates recruited to these sites, we monitored partitioning of conjugated Ub to IBs with Ub fused to YFP (YFP-Ub; Qian et al., 2002) in stable U2OS cells that express YFP-Ub constitutively and httQ91-mCherry under the control of a TET-ON promoter (U2-Ub-Q91; Fig. S3 B). YFP-Ub was recruited to IBs in these cells after a variable (∼10–50 min) time lag (Fig. 3, E and F; and Fig. S3, C and D), consistent with our previous observation in transiently transfected U2OS cells (Hipp et al., 2012). A conjugation-defective YFP-Ub variant containing a glycine to alanine mutation at the C terminus (YFP-UbG76A) was not recruited to IBs (Fig. S3 E), indicating that recruitment of Ub to IBs requires its covalent conjugation to proteins.
We assessed the stability of Ub conjugates at IBs under conditions where Ub conjugation was globally inhibited by the E1 inhibitor. Despite a significant reduction in levels of total Ub conjugates observed after a 3-h treatment with E1 inhibitor (Fig. S1 C), YFP-Ub fluorescence at IBs remained stable for >5 h (Fig. 3, G and H). Moreover, analysis of YFP-Ub mobility at IBs by FRAP indicated that YFP-Ub did not exchange between IBs and the cytosol (Fig. S3 F). These data indicate that Ub conjugates, like misfolded DD reporters, are stably associated with IBs.
Misfolded firefly luciferase (Fluc)–GFP is recruited to IBs
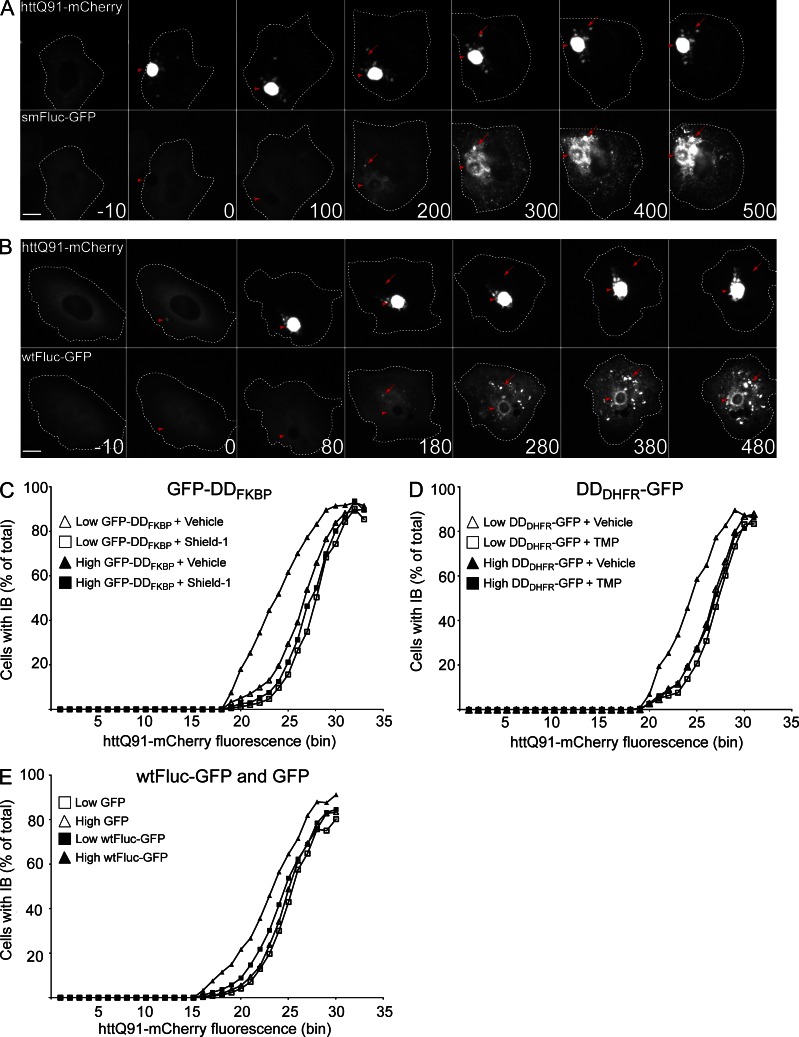
Unlike engineered degradation domains (DDs), which can be switched between homogenous folded and misfolded states, most proteins exist in a dynamic equilibrium between a single, folded native conformation and an ensemble of partially folded conformers, reflecting either the kinetics of folding to or partial unfolding from the native state (Hipp et al., 2014). To study the partitioning of metastable proteins between the cytosol and IBs, we exploited a conformationally destabilized variant of Fluc (smFluc-GFP), a metastable protein engineered to have a steep dependence on cytoplasmic chaperones to remain folded, active, and soluble (Gupta et al., 2011). smFluc-GFP has been previously shown to aggregate and colocalize with IBs in HEK293 cells expressing mutant htt (Gupta et al., 2011). We found that in cells coexpressing httQ91-mCherry, smFluc-GFP became associated with the IB after a variable delay of 1–2 h (Fig. 4 A, arrowheads). It is probable that this delay underlies the more peripheral association of smFluc-GFP compared with GFP-DDFKBP, which was recruited to httQ91-mCherry foci more rapidly (Fig. S1 F). In contrast to the DD reporters, recruitment of smFluc-GFP to IBs was temporally accompanied by the appearance of multiple GFP foci (Fig. 4 A, arrows) that were distinct from and not associated with IBs or mCherry puncta. The appearance of these non-IB–associated smFluc-GFP foci usually followed the emergence of httQ91-mCherry IBs (Fig. S4 A). We also observed that wild-type (wt) Fluc-GFP (wtFluc-GFP) was recruited to both IB-associated and peripheral foci with kinetics similar to those observed for smFluc-GFP (Fig. 4 B), suggesting that the equilibrium between folded and partially folded states (i.e., the intracellular protein folding environment) can be altered by a process temporally associated with the solubility transition of httQ91-mCherry. httQ91-mCherry may induce wtFluc-GFP misfolding/aggregation by sequestering chaperones essential for wtFluc-GFP folding (Frydman et al., 1994), some of which could function to suppress mutant htt aggregation. If true, we reasoned that expression of wtFluc-GFP or misfolded chaperone clients such as DD reporters should increase the propensity of httQ91-mCherry to aggregate by competing with htt for this limited pool of chaperones. To test this model, we used pulse-shape analysis (PulSA; Ramdzan et al., 2012) to generate IB dose–response profiles (Bersuker et al., 2013; Ormsby et al., 2013) that report the critical concentration at which httQ91-mCherry forms IBs (Fig. S4 B) in cells expressing low or high levels of GFP-DDFKBP and DDDHFR-GFP reporters (Fig. S4 E). The httQ91-mCherry IB dose–response profiles for cells expressing GFP-DDFKBP or DDDHFR-GFP shifted to the left when the ligands shield-1 or TMP, respectively, were absent, indicating that expression of these misfolded proteins caused IBs to form at a lower total httQ91-mCherry concentration (Fig. 4, C and D). This shift was more pronounced in cells expressing high levels of the reporters (Fig. 4, C and D). Addition of shield-1 (Fig. S4 C) or TMP (Fig. S4 D) did not alter the dose–response profile of httQ91-mCherry aggregation in the absence of the ligand-binding reporters. Thus, expression of misfolded proteins decreases the concentration at which httQ91-mCherry transitions into IBs in a dose-dependent manner. These data support a model whereby misfolded proteins and httQ91-mCherry compete with each other for access to cellular factors that maintain their respective solubility. A similar effect on httQ91-mCherry IB formation was observed in cells expressing high levels of wtFluc-GFP, a metastable protein, but not stably folded GFP (Fig. 4 E). Together, these data suggest that metastable and conformationally challenged proteins alter the cytoplasmic milieu to reduce the critical concentration at which httQ91-mCherry aggregates.
Figure 4.
Misfolded Fluc-GFP is recruited to IBs. (A and B) smFluc-GFP and wtFluc-GFP are recruited to IBs and distinct foci. Cells were transiently transfected with htt91-mCherry and smFluc-GFP (A) or wtFluc-GFP (B) for 48 h and imaged by time-lapse microscopy. Arrowheads indicate IBs, and arrows indicate Fluc-GFP foci that do not colocalize with IBs. Time stamps indicate elapsed time in minutes. t = 0 min indicates the frame in which IBs were first observed, and negative values correspond to time in minutes preceding this frame. Bars, 10 µm. (C) GFP-DDFKBP decreases the concentration at which httQ91-mCherry forms IBs. Cells stably expressing tetracycline-inducible httQ91-mCherry (U2-Q91) were transiently transfected with GFP-DDFKBP in the presence of vehicle or 1 µM shield-1 for 24 h. After addition of 1 µg/ml dox for 72 h, cells expressing low or high levels of GFP-DDFKBP were analyzed by PulSA. (D) DDDHFR-GFP decreases the concentration at which httQ91-mCherry forms IBs. U2-Q91 were transiently transfected with DDDHFR-GFP in the presence of vehicle or 10 µM TMP for 24 h. After addition of 1 µg/ml dox for 72 h, cells expressing low or high levels of DDDHFR-GFP were analyzed by PulSA. (E) wtFluc-GFP decreases the concentration at which httQ91-mCherry forms IBs. U2-Q91 were transiently transfected with GFP or wtFluc-GFP for 24 h. After addition of 1 µg/ml dox for 72 h, cells expressing low or high levels of wtFluc-GFP or GFP were analyzed by PulSA. Microscopy in A is representative of n > 10, and B is representative of n > 20 independent observations. Flow cytometry in C–E are representative experiments of at least two independent repeats.
Discussion
The ubiquity of Ub conjugates at intracellular IBs has long suggested a link between protein aggregation and disrupted protein homeostasis, but an understanding of their biological significance and the specific mechanisms by which these Ub conjugates are formed and recruited to IBs has remained obscure. The prevailing model—that most Ub is conjugated to the principal aggregated components of IBs, e.g., α-synuclein in Lewy bodies (Tofaris et al., 2003) or htt in HD IBs (Steffan et al., 2004), and contributes to sequestration of these proteins (Lee and Lee, 2002; Kawaguchi et al., 2003; Iwata et al., 2005)—is inconsistent with evidence that these proteins, which are typically mostly or entirely intrinsically disordered, are poor substrates for Ub conjugation and turnover (Eliezer et al., 2001; Wetzel, 2012). This conclusion is also supported by time-lapse studies in human cells showing that Ub arrives at IBs after deposition of mutant htt (Hipp et al., 2012), histological studies of IBs in human and mouse brains showing that not all IBs contain Ub (Sampathu et al., 2003; Gong et al., 2012), Ub-independent targeting of quality control substrates to nuclear IBs in yeast (Miller et al., 2015), and recruitment of a nonubiquitylated, misfolded reporter GFP-250 to IBs (García-Mata et al., 1999). The data reported here establish that Ub is neither necessary nor sufficient to direct proteins to IBs. However, because UbG76V fusions predominantly undergo ubiquitylation by K48- and K29-linked chains (Koegl et al., 1999), we do not exclude the possibility that other Ub chain linkages are sufficient to target proteins to IBs. K63-linked ubiquitylation by the E3 Ub ligase parkin has been reported to target the Parkinson’s disease–associated protein DJ-1 to IBs (Olzmann et al., 2007), and K63-linked chains have been reported to promote IB formation by mutant tau and SOD1 (Tan et al., 2008).
Here, we use two different models of ligand-dependent, conditional misfolding to show that folding state is the key determinant of protein recruitment to cytoplasmic, polyQ IBs. We find that two structurally unrelated proteins, mutant DHFR and FK506-binding domain, are recruited to htt IBs only when they are conformationally destabilized by removal of high affinity small molecule ligands. Although these proteins in their unliganded state are ubiquitylated and normally targeted for destruction by the 26S proteasome, our data show that they become diverted to htt IBs where they are protected from degradation. Surprisingly, we find that Ub conjugation is neither necessary nor sufficient for this recruitment. We propose a model in which protein aggregation disrupts intracellular protein homeostasis by competing for limiting chaperones and protein folding machinery and whereby protein aggregates divert misfolded or partially folded proteins away from the proteasome to IBs. The stabilization of Ub conjugates at IBs may explain why Ub chains accumulate in brain tissues from HD patients (Bennett et al., 2007).
The finding that IB-associated Ub conjugates are refractory to proteasomal degradation is surprising given evidence that proteasomes remain proteolytically active at IBs (Wigley et al., 1999; Schipper-Krom et al., 2014). It is likely that 26S proteasomes are recruited to IBs by the poly-Ub chains on the IB surface. One possibility is that misfolded, polyubiquitylated proteins at IBs are bound by the 19S cap of the proteasome but are not efficiently unfolded and therefore not subject to proteolysis. This may be caused by incorporation of ubiquitylated proteins into highly stable polyQ aggregates, as was previously demonstrated for short polyQ sequences bearing degradation signals recruited to IBs composed of ataxin-1 containing an expanded polyQ stretch (Verhoef et al., 2002). Previous studies have suggested that IBs can be cleared by basal (Iwata et al., 2005; Bové et al., 2011) or pharmacologically induced (Bové et al., 2011) macroautophagy and that Ub conjugation may signal autophagic clearance of proteins at IBs (Yao, 2010). We did not observe clearance of IBs or Ub conjugates at IBs in time-lapse microcopy experiments, even after transcriptional shutoff of httQ91-mCherry expression (unpublished data). IB turnover was also not observed in a study in which individual striatal neurons expressing mutant htt were imaged over several days, despite evidence that these neurons up-regulated autophagy pathways (Tsvetkov et al., 2013). Our data suggest that htt IBs do not function as sites of proteolysis but leave open the possibility that IBs composed of other aggregation-prone proteins are cleared by autophagy.
Our data supports a model in which mutant htt expression increases the likelihood that proteins adopt misfolded conformations by disrupting the protein folding environment. Indeed, we observed that recruitment of wtFluc-GFP and smFluc-GFP to IBs coincided with the appearance of Fluc-GFP aggregates, suggesting that the Fluc-GFP folding equilibrium is compromised by httQ91 aggregation. Titration of chaperones by IBs is one mechanism by which mutant htt may disrupt Fluc-GFP folding. A recent study suggested that sequestration of Hsc70 by IBs leads to defects in endocytosis by preventing Hsc70-mediated uncoating of clathrin-coated vesicles (Yu et al., 2014). Another study in a yeast polyQ model proposed that sequestration of the chaperone Sis1p at IBs inhibits shuttling of a proteasome substrate to the nucleus for degradation (Park et al., 2013). Although these studies suggest that mutant htt in IBs is the “toxic species” that disrupts protein homeostasis, we find that expression of wtFluc-GFP decreases the concentration at which diffuse httQ91 transitions into IBs, suggesting that soluble htt competes with wtFluc-GFP, and potentially other chaperone clients, for folding machinery. This conclusion is consistent with studies showing that worms harboring temperature-sensitive mutations exhibit an earlier appearance of polyQ aggregates (Gidalevitz et al., 2006).
Our results establish that a misfolded conformation is sufficient to target proteins to polyQ IBs. Disruption of microtubules with nocodazole did not block recruitment of GFP-DDFKBP to IBs (unpublished data), indicating that this reporter, unlike some aggregation-prone proteins (Johnston et al., 1998), is not recruited to IBs through microtubule-directed transport. Interestingly, misfolded conformers of GFP-DDFKBP and DDDHFR-GFP were recruited to IBs with very different kinetics. These results may be explained by the more rapid turnover of DDDHFR-GFP. Another possibility is that GFP-DDFKBP and DDDHFR-GFP sample different unfolded or misfolded conformations or contain partially folded domains that differentially promote IB recruitment. The glutamine/asparagine (Q/N)-rich domains of the RNA-binding proteins TDP-43 (Fuentealba et al., 2010) and TIA-1 (Furukawa et al., 2009) are sufficient to recruit these proteins to IBs, and htt aggregates can seed the aggregation of the Q/N-rich domain of TIA-1 in vitro (Furukawa et al., 2009; Kayatekin et al., 2014). Our data suggest that although inhibition of protein quality control with the E1 inhibitor affects GFP-DDFKBP solubility and clearance (Fig. S2 D), mutant htt IBs do not inhibit degradation or promote the aggregation of GFP-DDFKBP not associated with IBs. These findings suggest that, in addition to titrating one or more limiting components of the protein folding machinery, IBs could recruit constitutively misfolded GFP-DDFKBP independently of disrupting protein homeostasis. Although considerably less efficient than homotypic seeding, heterotypic seeding has been shown to occur in vitro and in vivo and has been proposed to contribute to cooperative interactions between protein aggregates in diseases of protein misfolding (Morales et al., 2013). Cross-seeding may trap GFP-DDFKBP within IBs and may explain why GFP-DDFKBP is an integral component of IBs and why it is refractory to proteasome-mediated degradation. In contrast, Fluc-GFP is only recruited to the IB periphery after a significant lag time, perhaps reflecting the time required for misfolded forms of the protein to accumulate. However, we consider it equally possible that interaction of GFP-DDFKBP with IBs could be bridged by as yet unidentified proteins or chaperones. Additional studies are needed in order to elucidate the mechanism by which proteins that adopt misfolded conformations are targeted to IBs.
Materials and methods
Plasmids
httQ91-mCherry encodes mCherry fused to the N terminus of htt exon 1 as described previously (Hipp et al., 2012). Tetracycline-inducible httQ91-mCherry was generated by inserting httQ91-mCherry into the KpnI site in pTRE-Tight (Takara Bio Inc.). GFP-DDFKBP and Venus-DDFKBP encode GFP or Venus, respectively, that were fused to the N terminus of mutant FKBP12 (F36V, E31G, R71G, and K105E; a gift from T. Wandless, Stanford University, Stanford, CA) and inserted into BamHI and XhoI sites in pcDNA3.1. DDDHFR-GFP encodes GFP that was fused to the C terminus of mutant Escherichia coli dihydrofolate reductase (R12Y, G67S, and Y100I; a gift from T. Wandless) and inserted into BamHI and NotI sites in pcDNA3.1. UbG76V-Venus-DDFKBP was created by inserting UbG76V between NheI and KpnI sites in Venus-DDFKBP. wtFluc-GFP and smFluc-GFP (a gift from U. Hartl, Max Planck Institute of Biochemistry, Munich, Germany) were described previously (Gupta et al., 2011).
Cell lines
U2OS cells expressing tetracycline-inducible httQ91-mCherry (U2-Q91) were generated by cotransfection of U2OS TET-ON (Takara Bio Inc.) cells with pTRE-Tight httQ91-mCherry and linear puromycin marker (Takara Bio Inc.) in a 20:1 weight/weight ratio for 72 h, selection in media containing 1 µg/ml puromycin, and cloning by limited dilution. The clonal line exhibiting the highest httQ91-mCherry fluorescence in the presence of dox was cotransfected with pTRE-Tight and pcDNA3.1/Hygro in a 20:1 weight/weight ratio for 72 h and selected in media containing 500 µg/ml hygromycin, and clonal lines exhibiting the highest httQ91-mCherry fluorescence were isolated by limited dilution cloning. GFP-DDFKBP and DDDHFR-GFP lines U2-DDFKQ91 and U2-DDDHQ91, respectively, were generated by transfection of U2-Q91 cells with these plasmids and selection in 400 µg/ml Zeocin followed by cloning by limited dilution.
Microscopy
For live cell time-lapse microscopy, U2OS cells were seeded in glass-bottom 4-well imaging chambers (Lab-Tek II; Thermo Fisher Scientific) in DMEM with 10% FBS. Cells were transfected the following day with FuGENE 6 (Roche) according to the manufacturer’s instructions and imaged after 48 h. For induction of expression in TET-ON cells stably expressing httQ91-mCherry, cells were treated with 1 µg/ml dox for 48 h before imaging. Imaging was performed on an inverted microscope (Axiovert 200M; ZEISS) encased in a chamber (Perspex) heated to 37°C and continuously perfused with humidified 5% CO2. Digital 12-bit images were acquired at multiple stage positions using a cooled charge-coupled device camera (CoolSNAP HQ; Photometrics) with an exacte light source (X-Cite; Lumen Dynamics), filter cubes for visualizing mCherry, YFP, or GFP (Chroma Technology Corp.), and a 40× air objective. MetaMorph software (Molecular Devices) was used to control the microscope, and ImageJ (National Institutes of Health) was used for image processing and quantification of fluorescence intensity.
For shield-1 and TMP washout experiments, cells were washed three times with PBS, and 5 µM recombinant FKBP (F36V) or DHFR was added to the media before imaging. For half-life experiments, cells were treated with 25 µM emetine. For E1 inhibitor experiments, cells were treated with 10 µM E1 inhibitor (Chen et al., 2011) for 3 h before shield-1 washout.
FRAP experiments were performed using a confocal microscope (FV-1000; Olympus) containing a CO2/temperature incubator (Life Imaging Services). The microscope was equipped with a sim scanner (FV1200; Olympus), which enables simultaneous photobleaching with a 405-nm laser while scanning with a 488-nm laser. All experiments were performed using a 60×/1.35 oil objective. FRAP experiments were performed by exposing a 5-µm region in single cells to 100% laser intensity for 0.45 ms. Imaging was typically performed at 1% laser intensity using the 488-nm laser. The interval between image scans varied depending on the duration of recovery and was either 1 s or 250 ms for slow and fast recovery, respectively. 10 prebleach and 180 postbleach frames were recorded for each series. As cells tended to move during the imaging time, the ImageJ StackReg plugin was used for quantitative analysis. The mean fluorescence intensities of the bleached region for each time point were normalized to the mean of the last five prebleach values. These values were divided by the total cell fluorescence in order to correct for changes in total fluorescence.
Poly-Ub affinity capture
Stably or transiently transfected cells were harvested with trypsin and PBS, washed twice with PBS, lysed in cold buffer containing 50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mM EDTA, pH 7.5, and supplemented with protease inhibitors (Roche) and 5 mg/ml N-ethylmaleimide. Lysates were rotated at 4°C for 15 min and cleared by centrifugation at 20,000 g for 20 min. 0.5 mg of cleared lysate was diluted fivefold with cold binding buffer containing 10 mM Hepes, 20 mM imidazole, 150 mM NaCl, and 0.5% Triton X-100, pH 7.4, and added to 25 µl Affigel-10 beads (Bio-Rad Laboratories) covalently conjugated to purified hplic2UBA prepared as a 50% slurry in binding buffer. Lysate and beads were incubated overnight at 4°C and washed three times with binding buffer, and bound proteins were eluted by incubating at 65°C for 10 min in 2× SDS loading buffer. Eluted proteins and 2% of unbound fraction were resolved by SDS-PAGE, transferred for 100 min at 95 V to a polyvinylidene difluoride membrane, and immunoblotted with antibodies against Ub (P4G7; Abcam) or GFP (JL-8; Takara Bio Inc.). The membrane was immunoblotted with a secondary goat anti–mouse antibody (IR dye 800; LI-COR Biosciences) and scanned using an imaging system (Odyssey; LI-COR Biosciences).
Flow cytometry
For PulSA flow cytometric analysis, cells were analyzed on a flow cytometer (LSR II; BD) equipped with 488- and 535-nm lasers. U2-Q91 cells were transiently transfected with the indicated GFP constructs and treated with stabilizing ligands or vehicle. The following day, expression of httQ91-mCherry was induced with 1 µg/ml dox, and cells were incubated for an additional 72 h before harvesting and analysis. 20,000–30,000 cells were analyzed in the high GFP gate (Fig. S4 E) for each sample, and measurements for mCherry and GFP peak width, peak height, and total intensity were collected. Flow cytometry analysis software (Flow Jo; Tree Star) was used to subdivide the GFP axis into low and high GFP fluorescence gates (Fig. S4 E). The total mCherry intensity of cells in each GFP gate was subdivided into 41 bins of equal intensity. For analysis of the fraction of cells with IBs by PulSA, cells with IBs were identified using an mCherry peak width versus peak height scatter plot, and a lasso gate was drawn around the IB population. The fraction of cells with IBs was plotted against binned total mCherry fluorescence intensity (Fig. S4 B).
Statistical analysis
P-values were generated by applying a two-tailed t test to pooled data from individual cells for given time points. P-values >0.05 were reported as nonsignificant. To compare half-life measurements (t1/2), decay data for individual cells were fitted to a one-phase exponential decay equation using Prism (GraphPad Software), and p-values for pooled half-life data were generated using a t test.
Online supplemental material
Fig. S1 shows additional characterization of reporter cell lines and effects of E1 inhibitor on levels of poly-Ub conjugates. Fig. S2 shows half-life of the UbG76V-Venus-DD reporter and the effect of E1 inhibitor on GFP-DDFKBP localization and recruitment of httQ91-mCherry to IBs. Fig. S3 shows additional characterization of U2-Ub-Q91 cells and YFP-Ub FRAP data. Fig. S4 explains the PulSA method and shows control experiments for the PulSA experiments in Fig. 4. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201511024/DC1.
Supplementary Material
Acknowledgments
The authors are grateful to T. Wandless and J. Olzmann for helpful discussions and C. Gottlieb, M. To, and J. Stevenson for critical reading of the manuscript.
This work was funded by a grant from the National Institute of Neurological Disorders and Stroke (NS42842) to R.R. Kopito. K. Bersuker is the recipient of a National Institutes of Health predoctoral grant (2T32HG000044-16). M. Brandeis was supported by the Israel Science Foundation (1759/14) and the Israel Ministry of Health (3-100-39).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- DD
- degradation domain
- Fluc
- firefly luciferase
- HD
- Huntington’s disease
- htt
- huntingtin
- IB
- inclusion body
- PulSA
- pulse-shape analysis
- TMP
- trimethoprim
- Ub
- ubiquitin
- UPS
- Ub–proteasome system
- wt
- wild type
References
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., and Finkbeiner S.. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810. 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Atkin G., and Paulson H.. 2014. Ubiquitin pathways in neurodegenerative disease. Front. Mol. Neurosci. 7:63 10.3389/fnmol.2014.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L.A., Chen L.C., Maynard-Smith L.A., Ooi A.G., and Wandless T.J.. 2006. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 126:995–1004. 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., and Kopito R.R.. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555. 10.1126/science.292.5521.1552 [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Bence N.F., Jayakumar R., and Kopito R.R.. 2005. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol. Cell. 17:351–365. 10.1016/j.molcel.2004.12.021 [DOI] [PubMed] [Google Scholar]
- Bennett E.J., Shaler T.A., Woodman B., Ryu K.-Y., Zaitseva T.S., Becker C.H., Bates G.P., Schulman H., and Kopito R.R.. 2007. Global changes to the ubiquitin system in Huntington’s disease. Nature. 448:704–708. 10.1038/nature06022 [DOI] [PubMed] [Google Scholar]
- Bersuker K., Hipp M.S., Calamini B., Morimoto R.I., and Kopito R.R.. 2013. Heat shock response activation exacerbates inclusion body formation in a cellular model of Huntington disease. J. Biol. Chem. 288:23633–23638. 10.1074/jbc.C113.481945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bett J.S., Cook C., Petrucelli L., and Bates G.P.. 2009. The ubiquitin-proteasome reporter GFPu does not accumulate in neurons of the R6/2 transgenic mouse model of Huntington’s disease. PLoS One. 4:e5128 10.1371/journal.pone.0005128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K.P., Yan S., Wang C.-E., Li S., and Li X.-J.. 2014. Differential ubiquitination and degradation of huntingtin fragments modulated by ubiquitin-protein ligase E3A. Proc. Natl. Acad. Sci. USA. 111:5706–5711. 10.1073/pnas.1402215111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bové J., Martínez-Vicente M., and Vila M.. 2011. Fighting neurodegeneration with rapamycin: mechanistic insights. Nat. Rev. Neurosci. 12:437–452. 10.1038/nrn3068 [DOI] [PubMed] [Google Scholar]
- Chen J.J., Tsu C.A., Gavin J.M., Milhollen M.A., Bruzzese F.J., Mallender W.D., Sintchak M.D., Bump N.J., Yang X., Ma J., et al. 2011. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. J. Biol. Chem. 286:40867–40877. 10.1074/jbc.M111.279984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N.P., and Bott L.C.. 2014. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front. Mol. Neurosci. 7:70 10.3389/fnmol.2014.00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N.P., Lindsten K., Glas R., Jellne M., and Masucci M.G.. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538–543. 10.1038/75406 [DOI] [PubMed] [Google Scholar]
- Davies S.W., Turmaine M., Cozens B.A., DiFiglia M., Sharp A.H., Ross C.A., Scherzinger E., Wanker E.E., Mangiarini L., and Bates G.P.. 1997. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 90:537–548. 10.1016/S0092-8674(00)80513-9 [DOI] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., and Aronin N.. 1997. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 277:1990–1993. 10.1126/science.277.5334.1990 [DOI] [PubMed] [Google Scholar]
- Egeler E.L., Urner L.M., Rakhit R., Liu C.W., and Wandless T.J.. 2011. Ligand-switchable substrates for a ubiquitin-proteasome system. J. Biol. Chem. 286:31328–31336. 10.1074/jbc.M111.264101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer D., Kutluay E., Bussell R. Jr., and Browne G.. 2001. Conformational properties of α-synuclein in its free and lipid-associated states. J. Mol. Biol. 307:1061–1073. 10.1006/jmbi.2001.4538 [DOI] [PubMed] [Google Scholar]
- Frydman J., Nimmesgern E., Ohtsuka K., and Hartl F.U.. 1994. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 370:111–117. 10.1038/370111a0 [DOI] [PubMed] [Google Scholar]
- Fuentealba R.A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M.I., Weihl C.C., and Baloh R.H.. 2010. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 285:26304–26314. 10.1074/jbc.M110.125039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., and Nukina N.. 2009. Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 29:5153–5162. 10.1523/JNEUROSCI.0783-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata R., Bebök Z., Sorscher E.J., and Sztul E.S.. 1999. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146:1239–1254. 10.1083/jcb.146.6.1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., and Morimoto R.I.. 2006. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 311:1471–1474. 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- Gong B., Kielar C., and Morton A.J.. 2012. Temporal separation of aggregation and ubiquitination during early inclusion formation in transgenic mice carrying the Huntington’s disease mutation. PLoS One. 7:e41450 10.1371/journal.pone.0041450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R., Kasturi P., Bracher A., Loew C., Zheng M., Villella A., Garza D., Hartl F.U., and Raychaudhuri S.. 2011. Firefly luciferase mutants as sensors of proteome stress. Nat. Methods. 8:879–884. 10.1038/nmeth.1697 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., and Hayer-Hartl M.. 2011. Molecular chaperones in protein folding and proteostasis. Nature. 475:324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hipp M.S., Patel C.N., Bersuker K., Riley B.E., Kaiser S.E., Shaler T.A., Brandeis M., and Kopito R.R.. 2012. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. J. Cell Biol. 196:573–587. 10.1083/jcb.201110093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp M.S., Park S.-H., and Hartl F.U.. 2014. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24:506–514. 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Ii K., Ito H., Tanaka K., and Hirano A.. 1997. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J. Neuropathol. Exp. Neurol. 56:125–131. 10.1097/00005072-199702000-00002 [DOI] [PubMed] [Google Scholar]
- Iwamoto M., Björklund T., Lundberg C., Kirik D., and Wandless T.J.. 2010. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 17:981–988. 10.1016/j.chembiol.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A., Riley B.E., Johnston J.A., and Kopito R.R.. 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280:40282–40292. 10.1074/jbc.M508786200 [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Bartel B., Seufert W., and Varshavsky A.. 1992. Ubiquitin as a degradation signal. EMBO J. 11:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J.A., Ward C.L., and Kopito R.R.. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883–1898. 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., and Yao T.P.. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 115:727–738. 10.1016/S0092-8674(03)00939-5 [DOI] [PubMed] [Google Scholar]
- Kayatekin C., Matlack K.E.S., Hesse W.R., Guan Y., Chakrabortee S., Russ J., Wanker E.E., Shah J.V., and Lindquist S.. 2014. Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc. Natl. Acad. Sci. USA. 111:12085–12090. 10.1073/pnas.1412504111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M., Hoppe T., Schlenker S., Ulrich H.D., Mayer T.U., and Jentsch S.. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 96:635–644. 10.1016/S0092-8674(00)80574-7 [DOI] [PubMed] [Google Scholar]
- Lamark T., and Johansen T.. 2012. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012 10.1155/2012/736905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-J., and Lee S.-J.. 2002. Characterization of cytoplasmic α-synuclein aggregates. Fibril formation is tightly linked to the inclusion-forming process in cells. J. Biol. Chem. 277:48976–48983. 10.1074/jbc.M208192200 [DOI] [PubMed] [Google Scholar]
- Lowe J., Blanchard A., Morrell K., Lennox G., Reynolds L., Billett M., Landon M., and Mayer R.J.. 1988. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson’s disease, Pick’s disease, and Alzheimer’s disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J. Pathol. 155:9–15. 10.1002/path.1711550105 [DOI] [PubMed] [Google Scholar]
- MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N., et al. The Huntington’s Disease Collaborative Research Group . 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 72:971–983. 10.1016/0092-8674(93)90585-E [DOI] [PubMed] [Google Scholar]
- Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., and Bates G.P.. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 87:493–506. 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- Margulis J., and Finkbeiner S.. 2014. Proteostasis in striatal cells and selective neurodegeneration in Huntington’s disease. Front. Cell. Neurosci. 8:218 10.3389/fncel.2014.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard C.J., Böttcher C., Ortega Z., Smith R., Florea B.I., Díaz-Hernández M., Brundin P., Overkleeft H.S., Li J.-Y., Lucas J.J., and Dantuma N.P.. 2009. Accumulation of ubiquitin conjugates in a polyglutamine disease model occurs without global ubiquitin/proteasome system impairment. Proc. Natl. Acad. Sci. USA. 106:13986–13991. 10.1073/pnas.0906463106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled L.B. 2005. Knock-in mouse models of Huntington’s disease. NeuroRx. 2:465–470. 10.1602/neurorx.2.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.B., Ho C.-T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., et al. 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34:778–797. 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales R., Moreno-Gonzalez I., and Soto C.. 2013. Cross-seeding of misfolded proteins: implications for etiology and pathogenesis of protein misfolding diseases. PLoS Pathog. 9:e1003537 10.1371/journal.ppat.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka U., Kim K., Jana N.R., Doi H., Maruyama M., Mitsui K., Oyama F., and Nukina N.. 2004. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J. Neurochem. 91:57–68. 10.1111/j.1471-4159.2004.02692.x [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Li L., Chudaev M.V., Chen J., Perez F.A., Palmiter R.D., and Chin L.-S.. 2007. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J. Cell Biol. 178:1025–1038. 10.1083/jcb.200611128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormsby A.R., Ramdzan Y.M., Mok Y.-F., Jovanoski K.D., and Hatters D.M.. 2013. A platform to view huntingtin exon 1 aggregation flux in the cell reveals divergent influences from chaperones hsp40 and hsp70. J. Biol. Chem. 288:37192–37203. 10.1074/jbc.M113.486944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Z., Díaz-Hernández M., Maynard C.J., Hernández F., Dantuma N.P., and Lucas J.J.. 2010. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J. Neurosci. 30:3675–3688. 10.1523/JNEUROSCI.5673-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., Hayer-Hartl M., and Hartl F.U.. 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 154:134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Qian S.-B., Ott D.E., Schubert U., Bennink J.R., and Yewdell J.W.. 2002. Fusion proteins with COOH-terminal ubiquitin are stable and maintain dual functionality in vivo. J. Biol. Chem. 277:38818–38826. 10.1074/jbc.M205547200 [DOI] [PubMed] [Google Scholar]
- Raasi S., Varadan R., Fushman D., and Pickart C.M.. 2005. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 12:708–714. 10.1038/nsmb962 [DOI] [PubMed] [Google Scholar]
- Ramdzan Y.M., Polling S., Chia C.P.Z., Ng I.H.W., Ormsby A.R., Croft N.P., Purcell A.W., Bogoyevitch M.A., Ng D.C.H., Gleeson P.A., and Hatters D.M.. 2012. Tracking protein aggregation and mislocalization in cells with flow cytometry. Nat. Methods. 9:467–470. 10.1038/nmeth.1930 [DOI] [PubMed] [Google Scholar]
- Riley B.E., Kaiser S.E., Shaler T.A., Ng A.C.Y., Hara T., Hipp M.S., Lage K., Xavier R.J., Ryu K.-Y., Taguchi K., et al. 2010. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J. Cell Biol. 191:537–552. 10.1083/jcb.201005012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C.A., and Poirier M.A.. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(7, Suppl):S10–S17. 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- Ross C.A., and Poirier M.A.. 2005. Opinion: What is the role of protein aggregation in neurodegeneration? Nat. Rev. Mol. Cell Biol. 6:891–898. 10.1038/nrm1742 [DOI] [PubMed] [Google Scholar]
- Sampathu D.M., Giasson B.I., Pawlyk A.C., Trojanowski J.Q., and Lee V.M.-Y.. 2003. Ubiquitination of α-synuclein is not required for formation of pathological inclusions in α-synucleinopathies. Am. J. Pathol. 163:91–100. 10.1016/S0002-9440(10)63633-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G.P., Davies S.W., Lehrach H., and Wanker E.E.. 1997. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 90:549–558. 10.1016/S0092-8674(00)80514-0 [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G.P., Lehrach H., and Wanker E.E.. 1999. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington’s disease pathology. Proc. Natl. Acad. Sci. USA. 96:4604–4609. 10.1073/pnas.96.8.4604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper-Krom S., Juenemann K., Jansen A.H., Wiemhoefer A., van den Nieuwendijk R., Smith D.L., Hink M.A., Bates G.P., Overkleeft H., Ovaa H., and Reits E.. 2014. Dynamic recruitment of active proteasomes into polyglutamine initiated inclusion bodies. FEBS Lett. 588:151–159. 10.1016/j.febslet.2013.11.023 [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.-Z., Cattaneo E., et al. 2004. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 304:100–104. 10.1126/science.1092194 [DOI] [PubMed] [Google Scholar]
- Tan J.M.M., Wong E.S.P., Kirkpatrick D.S., Pletnikova O., Ko H.S., Tay S.-P., Ho M.W.L., Troncoso J., Gygi S.P., Lee M.K., et al. 2008. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 17:431–439. 10.1093/hmg/ddm320 [DOI] [PubMed] [Google Scholar]
- Tofaris G.K., Razzaq A., Ghetti B., Lilley K.S., and Spillantini M.G.. 2003. Ubiquitination of α-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. J. Biol. Chem. 278:44405–44411. 10.1074/jbc.M308041200 [DOI] [PubMed] [Google Scholar]
- Tsvetkov A.S., Arrasate M., Barmada S., Ando D.M., Sharma P., Shaby B.A., and Finkbeiner S.. 2013. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat. Chem. Biol. 9:586–592. 10.1038/nchembio.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef L.G.G.C., Lindsten K., Masucci M.G., and Dantuma N.P.. 2002. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum. Mol. Genet. 11:2689–2700. 10.1093/hmg/11.22.2689 [DOI] [PubMed] [Google Scholar]
- Waelter S., Boeddrich A., Lurz R., Scherzinger E., Lueder G., Lehrach H., and Wanker E.E.. 2001. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol. Biol. Cell. 12:1393–1407. 10.1091/mbc.12.5.1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel R. 2012. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. J. Mol. Biol. 421:466–490. 10.1016/j.jmb.2012.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley W.C., Fabunmi R.P., Lee M.G., Marino C.R., Muallem S., DeMartino G.N., and Thomas P.J.. 1999. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145:481–490. 10.1083/jcb.145.3.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T.-P. 2010. The role of ubiquitin in autophagy-dependent protein aggregate processing. Genes Cancer. 1:779–786. 10.1177/1947601910383277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Shibata Y., Shah B., Calamini B., Lo D.C., and Morimoto R.I.. 2014. Protein aggregation can inhibit clathrin-mediated endocytosis by chaperone competition. Proc. Natl. Acad. Sci. USA. 111:E1481–E1490. 10.1073/pnas.1321811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.