Abstract
Successful clearance of a microbial infection depends on the concerted action of both the innate and adaptive arms of the immune system. Accurate recognition of an invading pathogen is the first and most crucial step in eliciting effective antimicrobial defense mechanisms. In recent years, remarkable progress has been made towards understanding the molecular details of how the innate immune system recognizes microbial signatures, commonly called pathogen-associated molecular patterns (PAMPs). For viral pathogens, nucleic acids — both viral genomes and viral replication products — represent a major class of PAMPs that trigger antiviral host responses via activation of germline-encoded innate immune receptors. Here we summarize recent advances in intracellular innate sensing mechanisms of viral RNA and DNA.
Introduction
Virtually all cells of a mammalian host organism have the capacity to detect the presence of an invading pathogen by recognizing ‘non-self’ structural components through germline-encoded innate immune sensors, called pattern recognition receptors (PRRs). Over the past ten years, significant progress has been made in identifying the precise viral pathogen signatures (or PAMPs) recognized by PRRs, such as specific modifications (e.g. a 5′-triphosphate moiety) of viral RNA (vRNA), or mislocalized cytoplasmic viral DNA (vDNA) [1,2]. Mammalian cells have evolved a large repertoire of PRRs, which can be grouped with respect to their subcellular localization. While Toll-like receptors (TLRs) and C-type lectin receptors (CLRs) recognize virion components in endosomes and on cell membranes (reviewed in [3]), the detection of incoming and actively replicating viruses is mediated by PRRs that are localized inside the cell. Most intracellular PRRs recognize viral nucleic acids and have the remarkable ability to distinguish ‘non-self’ RNA or DNA from the large pool of cellular RNAs and DNAs. At least three major classes of intracellular sensors of viral infection have been identified: (1) RIG-I-like receptors (RLRs) which sense vRNA species in the cytoplasm and play important roles in the detection of RNA viruses; (2) a structurally unrelated group of vDNA receptors (e.g. cGAS and IFI16) localized in the host cytoplasm and/or nucleus; and (3) members of the NOD-like receptor (NLR) family which, besides their established roles in sensing bacterial infections, have also been implicated in detecting viral pathogens. In addition, several other proteins have been implicated in vRNA or vDNA sensing, although their physiological roles have yet to be fully established (as discussed below).
Following ligand recognition, PRRs activate antiviral signaling cascades that converge on a group of well-characterized kinases, namely TANK-binding kinase 1 (TBK1), mitogen-activated protein kinases (MAPKs), and IκB kinase α (IKKα) and IKKβ. Through phosphorylation events, these kinases subsequently activate the interferon (IFN)-regulatory factors 3 and 7 (IRF3/7), AP-1, and NF-κB, respectively. These proteins transcriptionally induce the gene expression of type-I IFNs (mainly IFN-α subtypes and IFN-β), type-III IFN (IFN-λ), and other pro-inflammatory cytokines such as members of the interleukin (IL) protein family [1,2]. Furthermore, some PRRs activate inflammasomes, which are caspase-1-activating multi-protein complexes that cleave pro-IL-1β and pro-IL-18 to generate their mature forms [4]. Secreted IFNs bind to their respective surface receptors on both infected and uninfected neighboring cells, inducing signal transduction that leads to the expression of numerous IFN-stimulated genes (ISGs) [5,6]. ISGs encode for proteins that exert distinct antiviral effector functions such as cleavage of vRNA or induction of apoptosis. In addition, some ISGs encode for PRRs or for proteins involved in PRR signal transduction, leading to positive feedback amplification of the antiviral response in infected cells, and also sensitizing uninfected cells to fight off the viral attack. Induction of IFNs and other pro-inflammatory cytokines not only limits the spread of the viral pathogen to surrounding cells, but also facilitates viral clearance by recruiting and stimulating cells of the adaptive immune system.
In this opinion article, we summarize recent findings on the molecular mechanisms of how intracellular innate immune receptors detect vRNA and vDNA, and further outline unresolved questions in this rapidly progressing field.
Cytosolic sensing of vRNA
Detection of vRNA by RLRs
Cytosolic vRNA is predominantly recognized by DExD/H-box RNA helicases of the RLR family (Figure 1). This family consists of retinoic acid-inducible gene-I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2), all of which are able to directly bind RNA through their helicase and C-terminal domains (CTD) [1,2]. In addition, both RIG-I and MDA5 possess a pair of caspase activation and recruitment domains (CARDs), which mediate downstream signaling and thereby cytokine induction. In contrast, LGP2 lacks the CARD signaling module. Following vRNA binding, RIG-I switches from an auto-inhibited ‘closed’ conformation into its active tetrameric form, in which the CARDs are exposed [7–9]. In contrast to RIG-I, MDA5 is thought to adopt an extended conformation under normal conditions; upon vRNA binding, MDA5 then forms filaments along the viral dsRNA strand [10••]. The exposed CARDs of RIG-I and MDA5 associate with the CARD of the mitochondrial transmembrane adaptor protein MAVS/IPS-1/VISA/Cardif, inducing prion-like filament structures that represent signaling-active RLR-MAVS complexes [11,12]. MAVS serves as a scaffolding protein to assemble a multi-protein complex consisting of several tumor necrosis factor receptor-associated factors (TRAFs), TBK1 and IKKs, which together mediate IRF3/7 and NF-κB activation.
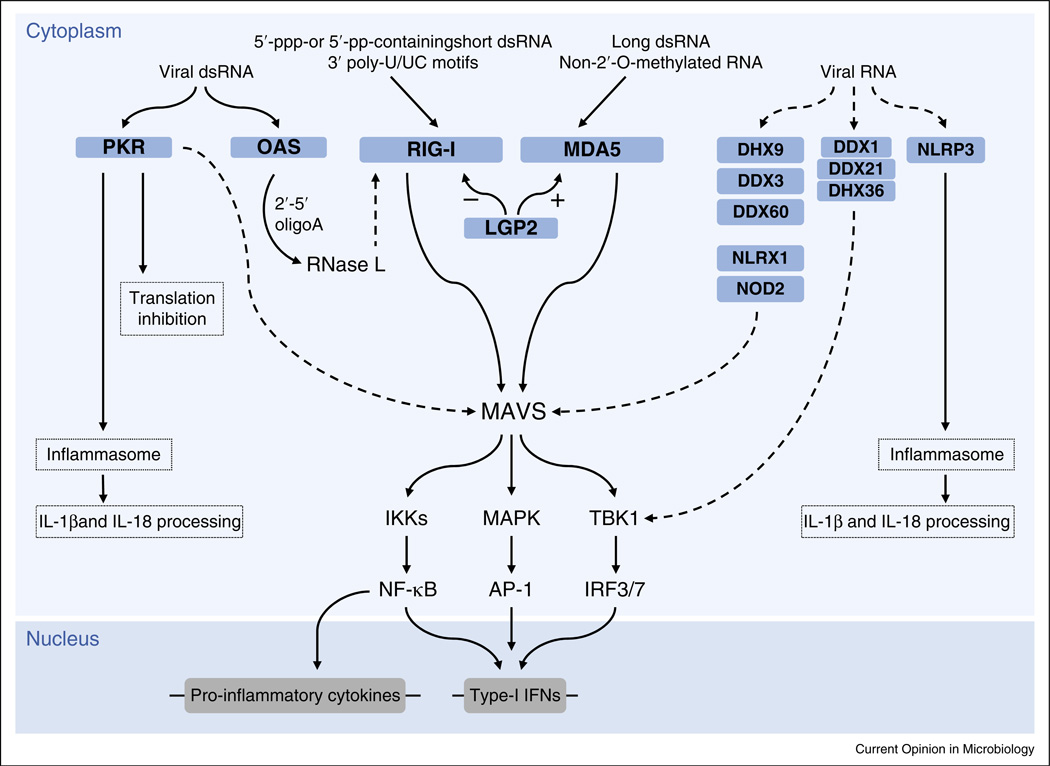
Figure 1.
Detection of cytosolic vRNA by RLRs and other proteins. Viral 5′-triphosphate (5′-ppp)-containing or 5′-diphosphate (5′-pp)-containing short dsRNA as well as poly-U/UC motifs are recognized by RIG-I, whereas MDA5 binds to long dsRNA or non-2′-O-methylated vRNA. The RNA binding and/or signaling activities of RIG-I and MDA5 are negatively and positively regulated by LGP2, respectively. Signaling induced by RIG-I and MDA5 converges on MAVS, which serves as a scaffolding protein to activate the key transcription factors NF-κB, AP-1 and IRF3/7 via several kinases (IKKs, MAPK, TBK1). NF-κB, AP-1 and IRF3/7 then act in concert to induce the gene expression of type-I IFNs and other pro-inflammatory cytokines. PKR and OAS both recognize viral dsRNA. Upon activation, PKR leads to inhibition of cellular translation. Furthermore, PKR activates the inflammasome, resulting in IL-1β and IL-18 processing and release. Upon dsRNA binding, OAS produces 2′–5′ oligoA, which activates RNase L. RNA fragments generated by RNase L can serve as RIG-I ligands, amplifying RIG-I-mediated antiviral signaling. Viral RNA is also sensed (directly or indirectly) by DHX9, DDX3, DDX60, NLRX1 and NOD2, leading to activation of MAVS-dependent signaling. The DDX1–DDX21–DHX36 complex signals downstream via the adaptor protein TRIF (not depicted), leading to TBK1 and IRF activation. In response to RNA virus infection, NLRP3 and inflammasomes are activated, which leads to maturation of IL-1β and IL-18. Solid arrows indicate well-established signaling events. Dashed arrows indicate signaling events that are indirect or that have not yet been fully elucidated.
RLR activation is tightly regulated by several posttranscriptional modifications (PTMs) and interacting proteins to avoid accidental ‘misfiring’ [2]. Both RIG-I and MDA5 require activation via dephosphorylation of specific Ser/Thr residues in their CARDs by the phosphatase PP1α or PP1γ [13••,14]. Dephosphorylation of the RIG-I and MDA5 CARDs allows for effective MAVS interaction and downstream signaling, likely mediated by charge-dependent rearrangement of the tandem CARDs. Additional PTMs that are required specifically for RIG-I activation include Lys63-linked ubiquitination marks in its CARDs and CTD mediated by the E3 ligases TRIM25 and RIPLET, respectively [15–17]. Mechanistically, Lys63-polyubiquitin chains induce RIG-I oligomerization and RIG-I-MAVS binding [15,18,19]. Conversely, Ser/Thr phosphorylation (induced by protein kinase C and casein kinase II), Lys48-linked polyubiquitination, and removal of K63-linked polyubiquitin by several deubiquitinating enzymes (CYLD, USP3 and USP21) inhibit RIG-I activation (reviewed in detail in [2]). Furthermore, it was recently reported that the RNA-binding activity of LGP2 is regulated by Pumilio proteins (PUM1/2) [20].
The functional relevance of RIG-I and MDA5 in cell-intrinsic antiviral responses was demonstrated by infection studies in RIG-I and MDA5 knockout cells and mice. RIG-I was shown to be critical for detecting many negative-sense RNA viruses (influenza virus, vesicular stomatitis virus, arenaviruses), as well as hepatitis C virus (HCV) and Japanese encephalitis virus, which are both positive-sense RNA viruses. In contrast, MDA5 plays a predominant role in Picornaviridae detection. Moreover, both RIG-I and MDA5 detect Flavivirus family members (dengue virus and West Nile virus), reoviruses and paramyxoviruses (measles virus [MV], Sendai virus [SeV], and respiratory syncytial virus) (reviewed in [1,2]). The functional relevance of LGP2 in antiviral IFN responses remains incompletely understood. A role for LGP2 in promoting ligand binding and signaling of MDA5 has been described [21,22,23•]. On the other hand, RIG-I-dependent signaling appears to be repressed by LGP2 [24,25], supporting an inhibitory role for LGP2 specifically in RIG-I signaling.
The nature of RNA ligands stimulating RIG-I activity has been well characterized. Short 5′-triphosphorylated vRNAs (~10–20 nucleotides) with a dsRNA stretch near the 5′ end were shown to activate RIG-I [26,27]. Recently, it was shown that a 5′-diphosphate moiety in the vRNA can also stimulate RIG-I [28•]. In addition, poly(U/UC)-rich motifs, found in the HCV genomic RNA, can be sensed by RIG-I [29]. In contrast to those of RIG-I, MDA5 ligands are less well defined. They are thought to be aggregated or long dsRNA, as occuring during the replication of positive-strand RNA viruses [30–32]. In addition, 2′-O-methylation of RNAs has been shown to serve as a motif that allows MDA5 to distinguish between ‘non-self’ and ‘self’ RNA [33] (Figure 1).
While these studies provided important insights into the structures and motifs of vRNA ligands for RIG-I, and to some extent also for MDA5, research on physiological PAMPs stimulating RLR activation during an authentic viral infection has just begun. Recent studies showed that in vivo RLR ligands can be either viral replication products or incoming viral genomes [34–36,37•]. During IAV and SeV infection, RIG-I recognizes genomic RNAs as well as the RNA of defective interfering (DI) particles [34,35]. A novel RNA–protein crosslinking technique combined with next-generation sequencing revealed that, during MV infection, RIG-I associates with the 5′-triphosphorylated leader transcript and trailer RNAs. In addition, DI RNAs of MV and internal regions within the MV genome are recognized by RIG-I [37•]. Recently, RIG-I was shown to sense the 5′-ε region of the pregenomic RNA of hepatitis B virus (HBV) [38•]. In MV-infected cells, MDA5 binds to positive-strand AT-rich vRNA, which probably originates from viral mRNA [37•]. To date, the only physiological PAMP described for LGP2 is the antisense RNA of the L region of encephalomyocarditis virus (EMCV) [23•]. More investigation is clearly required to identify the physiological PAMPs of RLRs during other viral infections.
Non-RLR proteins involved in the detection of cytosolic vRNA
While the role of RLRs in cytosolic vRNA sensing is well established, other cytoplasmic proteins have been shown to bind vRNA and to engage in antiviral signaling as well (Figure 1). Notably, many of these molecules regulate RLR signaling, and also have other functions in RNA metabolism and gene transcription. DEAD-box protein 3 (DDX3) was first proposed to function as vRNA receptor, activating MAVS-dependent signaling and IFN induction [39]; however recent research indicates that DDX3 exerts a regulatory role in innate signaling by augmenting the activation of TBK1 or IKKε via direct interaction [40]. Moreover, it has been recently reported that DDX3 functions as an antiviral effector protein that restricts HBV transcription [41]. DDX60 was shown to bind both vRNA and RLRs, promoting RLR-vRNA interaction and thereby induction of antiviral cytokines [42]. Another study indicated that DDX60 acts as an antiviral restriction factor for HCV; however, the precise mechanism of how DDX60 functions remains elusive [43]. DEAH-box protein 9 (DHX9) and a triple complex of DDX1, DDX21 and DHX36 were reported to mediate vRNA-mediated host responses specifically in myeloid dendritic cells (reviewed in [1]). It has been proposed that DDX helicases, most of which are constitutively expressed (in contrast to RLRs that are IFN-inducible genes), may play an important role in vRNA sensing specifically early in infection, when RLR abundance is low. However, future studies are necessary to assess the contribution of individual DDX helicases to vRNA sensing and to identify their roles in other RNA-related processes.
Protein kinase R (PKR) and 2′,5′-oligoadenylate synthetase (OAS), two of the most well-studied dsRNA-binding proteins, play also critical roles in host responses to vRNA [44,45]. PKR exerts numerous antiviral functions, including inhibition of cap-dependent translation via phosphorylation of eIF2α, induction of autophagy, and activation of the NLRP3-dependent inflammasome [44]. Recent studies indicate that PKR acts in concert with RLRs to trigger IFN-mediated antiviral innate immunity. Upon viral infection, PKR induces and localizes to distinct cytoplasmic bodies called stress granules (SG), which are thought to serve as platforms for the interaction of RLRs (and also other antiviral proteins) with vRNAs [46,47]. A recent study implicated DHX36 in dsRNA-dependent PKR activation and PKR-mediated SG formation, facilitating RLR-dependent vRNA detection and antiviral signaling [48]. While these studies indicated that PKR may act as an upstream regulator of RLR-mediated innate sensing and signaling, the relevance of SG as a critical subcellular compartment for IFN-mediated antiviral defenses remains to be fully established. OAS produces, upon its binding to cytoplasmic dsRNA, 2′–5′-linked oligoadenylate (2′–5′ oligoA), a second messenger that is subsequently recognized by RNase L [49]. Following activation, RNase L degrades both cellular RNA and vRNA, ultimately limiting virus replication. Interestingly, RNA fragments generated by RNase L have been reported to serve as endogenous ligands for RIG-I, amplifying the IFN response [50,51].
Several members of the NLR superfamily have also been implicated in vRNA-induced innate immune responses [52]. It should be pointed out that for many of these molecules, it remains to be determined if they can directly sense vRNA. The current view is that most of these molecules are activated indirectly through mechanisms triggered by vRNA or other ‘danger signals’ of viral infection. Specifically, NLRP3 is thought to be indirectly activated in response to RNA virus infection via receptor-interacting proteins 1 and 3 (RIP1/3), leading to activation of the inflammasome [53]. NOD2 (nucleotide-binding oligomerization domain 2), known for its role in conferring responsiveness to intracellular bacterial peptidoglycan, is also activated during RNA virus infection, inducing signaling via MAVS [54]; however, the precise details of what triggers NOD2 activation in this context are unknown. Similarly, NLRX1 is activated during RNA virus infection, inducing MAVS activation; however, in contrast to NOD2 and NLRP3, NLRX1 appears to have direct vRNA-binding capacity [52,55]. While these studies strengthened the existence of RLR-independent vRNA sensing mechanisms, more detailed investigation is needed to define the relevance of these molecules in ‘non-self’ RNA recognition and antiviral innate immunity.
Intracellular sensing of vDNA
It has long been recognized that the presence of foreign DNA in the host cytoplasm — either arising from infection with DNA viruses or intracellular bacteria, or artificially introduced by transfection — can trigger innate immune activation. In addition, recent studies have indicated that host cells can also sense vDNA in the nucleus. Remarkable progress has been made in the past few years that has led to the identification of multiple intracellular vDNA receptors and a critical endoplasmic reticulum (ER)-resident adaptor protein called STING (also called MITA, MPYS, or ERIS) that bridges most vDNA receptors to downstream signaling events (Figure 2) [56].
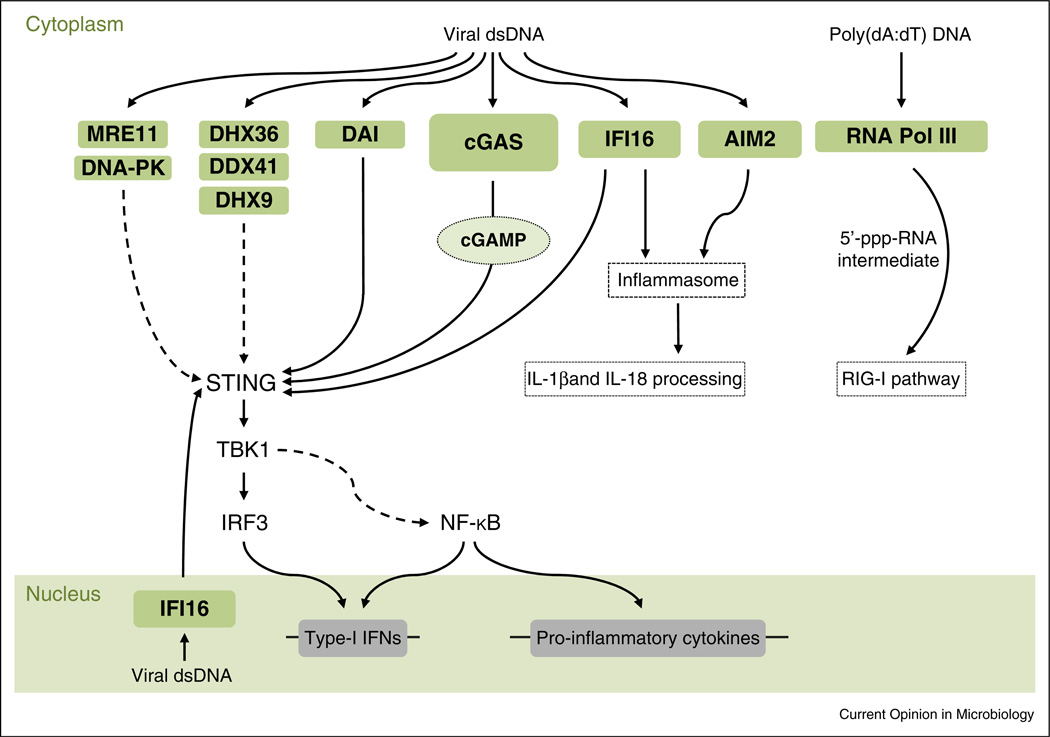
Figure 2.
Proteins involved in the detection of intracellular vDNA. Cytoplasmic vDNA triggers activation of a number of different innate immune receptors. Following vDNA binding, cGAS produces the cyclic dinucleotide cGAMP, which serves as a second messenger leading to the activation of the adaptor protein STING. Activation of STING induces the gene expression of type-I IFNs and other pro-inflammatory cytokines via the TBK1-IRF3 axis. Proteins involved in DNA repair (MRE11 and DNA-PK), DDX/DHX proteins (DHX36, DDX41, DHX9), and DAI have also been reported to sense vDNA and to induce antiviral signaling via STING. AIM2 senses cytoplasmic vDNA and subsequently activates inflammasomes, leading to IL-1β and IL-18 maturation. IFI16 is another cytosolic vDNA sensor that triggers inflammasome activation. In addition, IFI16 induces IFN gene expression via STING. In many cell types, IFI16 also localizes to the nucleus, where it senses herpesviral DNA. RNA Pol III specifically detects poly(dA:dT) DNA and subsequently transcribes it into 5′-triphosphate (5′-ppp)-containing short dsRNA that serves as a PAMP for RIG-I. Solid arrows indicate well-established signaling events. Dashed arrows indicate signaling events that are indirect or that have not yet been fully elucidated.
Detailed functional studies in STING-deficient cells demonstrated that STING is essential for IFN-α/β-mediated responses to various DNA stimuli including viral, bacterial, parasitic and synthetic dsDNA. Furthermore, studies in STING-knockout mice demonstrated that STING is critical for in vivo protection against herpes simplex virus-1 (HSV-1) [57]. Following activation, STING dimerizes and translocates from the ER to the Golgi network and perinuclear structures where it engages in TBK1 binding. STING then facilitates the recruitment of IRF3 to TBK1 and subsequent IRF3 phosphorylation. Moreover, STING has been reported to promote NF-κB activation via TBK1 [58]. In addition to its role in vDNA-dependent immune signaling, there is increasing evidence suggesting a role for STING in IFN-mediated antiviral responses to RNA viruses (reviewed in [59]); the precise mechanism by which STING mediates this response, however, is not well understood.
The first cytosolic vDNA receptor mediating antiviral immunity described was the IFN-inducible DAI (DNA-dependent activator of IRFs) [60]. DAI was suggested to signal via STING, triggering IRF3-mediated and NF-κB-mediated cytokine production. However, mice and several human cell types lacking DAI elicit normal vDNA-dependent antiviral responses [61], suggesting a very restricted or cell type-specific role for DAI, or redundancy with other vDNA sensors. In 2009, the cellular RNA polymerase III (Pol III) was described as a receptor for AT-rich dsDNA, transcribing it into 5′-triphosphate-containing small dsRNA, a ligand subsequently recognized by RIG-I [62,63]. While some studies support a role of the Pol III-RIG-I axis in sensing vDNA [64,65], others argue against an involvement of Pol III in IFN-mediated antiviral responses [66,67]. Thus, the relevance of Pol III in vDNA sensing remains to be fully established.
IFN-inducible protein 16 (IFI16), a member of the Pyrin and HIN domain (PYHIN) protein family, is another recently identified cytosolic vDNA receptor [67]. IFI16 was shown to cooperatively assemble into filaments on ‘non-self’ dsDNA [68], which leads to STING activation and IRF3-dependent and NF-κB-dependent responses. Furthermore, IFI16 can interact with the adaptor protein ASC (apoptosis-associated speck-like protein containing CARD) to activate the inflammasome [69]. Depletion of human IFI16 and murine p204, the apparent IFI16 funcional ortholog in mice, indicated that IFI16 is critical for type-I IFN-mediated innate immunity to HSV-1. Additionally, IFI16 was recently shown to play a role in the host intrinsic defense to human immunodeficiency virus-1 (HIV-1), and it is postulated that ssDNA with secondary structures that mimic dsDNA can be recognized by IFI16 [70]. Interestingly, recent studies showed that IFI16 can also be found in the nucleus of many cell types, suggesting a role for IFI16 in nuclear vDNA sensing (reviewed in detail in [71]). In fact, most DNA viruses (e.g. herpesviruses, papillomaviruses, polyomaviruses) replicate in the nucleus, which would suggest that host cells may have evolved the ability to discriminate between ‘self’ and ‘non-self’ DNA in the nucleus. Further studies are required to determine the precise mechanisms that allow IFI16 to sense specifically vDNA in the nucleus. It also remains to be elucidated how IFI16 signals from the nucleus to STING and inflammasomes in the cytoplasm. In addition to its ability to detect vDNA, a recent study indicates that IFI16 also regulates the type-I IFN response to RNA virus infection [72].
AIM2 (absent in melanoma 2), another PYHIN protein, is also implicated in vDNA recognition and inflammasome activation [73]. Following viral infection (vaccinia virus and murine cytomegalovirus), AIM2-deficient cells are impaired in pro-inflammatory cytokine production, supporting the importance of AIM2 in vDNA-dependent innate immune defense [74]. Several other proteins involved in DNA repair pathways, such as DNA-PK (DNA-dependent protein kinase) and MRE11, as well as DDX41, DHX9 and DHX36 have been proposed to mediate vDNA-dependent antiviral responses (Figure 2), although further research is clearly needed to determine the physiological relevance of these putative vDNA recognition receptors (reviewed in [71]).
In 2013, a fascinating new enzyme — the cyclic GMP-AMP synthase (cGAS) — was identified as a major cytoplasmic DNA sensor [75••]. Upon binding to cytoplasmic dsDNA, cGAS produces the cyclic dinucleotide cGAMP (cyclic GMP-AMP) [76,77], which is characterized by an unusual 2′–5′ phosphodiester bond similar to OAS-generated 2′–5′ oligoA [78]. The second messenger cGAMP is subsequently recognized by STING via direct binding, leading to type-I IFN induction. Intriguingly, cGAMP can spread from infected cells to neighboring cells via gap junctions, mediating IFN-independent activation of uninfected bystander cells to block the viral attack [79••]. Biochemical and structural analyses showed that cGAS binds to the sugar-phosphate backbone of DNA, indicating a sequence-independent mechanism of cGAS activation [45]. This also strengthens the overall concept that mislocalized vDNA in the cytoplasm represents a ‘danger signal’ that provokes an innate immune response. Unusual DNA:RNA hybrids were also reported to be sensed by cGAS [80]. Functional studies in cGAS-deficient cells and mice showed that cGAS is crucial for an IFN-mediated antiviral response following infection with DNA viruses, bacteria, and even RNA viruses [81,82]. Interestingly, infection with HIV and other retroviruses also triggers cGAMP production, an activity which is dependent on the ability of cGAS to recognize retroviral cDNA upon reverse transcription [83•,84].
Conclusions and perspectives
Although recent work has shed light on the molecules and pathways involved in vRNA and vDNA sensing, the exact nature of the physiological ligands for most receptors is still unknown. While several in vivo PAMPs have been identified for RIG-I using next-generation sequencing, physiological ligands of MDA5 and LGP2 during viral infection are largely unknown. Given that LGP2’s function is still enigmatic, the identification of vRNA species recognized by LGP2 may clarify its role in antiviral innate immunity. More broadly, identification of the physiological properties of immunostimulatory molecules will certainly advance our understanding of how the innate immune system discriminates between ‘self’ and ‘non-self’ nucleic acids, which may have important implications for many infectious and inflammatory diseases.
One of the biggest questions that arises from recent studies is the question of why such a multiplicity of sensors exists. It is unlikely that all these sensors function redundantly; instead, it is possible that specific sensors act in a cell-type specific manner. Alternatively, some of these receptors may work together in the infected cell, either in a temporal or site-specific manner, to effectively detect virus infection. The latter point is particular important for RNA virus infections which are known to produce a variety of different PAMPs (different genomic RNAs, DI RNAs, various replication intermediates), likely triggering the activation of several distinct PRRs. More detailed studies are needed to identify the dynamic role of individual sensors in the context of an authentic viral infection, and to determine the crosstalk between different sensing pathways. Lastly, it is possible that some of the described sensor molecules are not all true sensors, but instead have regulatory roles in innate sensing pathways, or act antivirally through completely different mechanisms. Indeed, several non-RLR helicases and some candidate vDNA sensors also have fundamental roles in gene induction and nucleic acid biogenesis and metabolism.
In contrast to the vRNA signatures, cytosolic DNA receptors are thought to detect mislocalized ‘non-self’ DNA in the cytoplasm. Interestingly, the sensor IFI16 has been found to also localize to the nucleus, opening up the field of nuclear vDNA sensing. As vDNA recognition by intracellular receptors is currently believed to be sequence-independent, it has yet to be resolved how IFI16 distinguishes between host DNA and vDNA in the nucleus. Intriguing hypotheses are that IFI16 recognizes underchromatization or a loose chromatin structure of vDNA, which is in stark contrast to the tight chromatin structure of host DNA.
Finally, an important ongoing topic in this field is the investigation of how aberrant signaling induced by vRNA and vDNA sensors can be controlled to avoid excessive or prolonged immune activation. This is particularly important in light of recent evidence demonstrating that aberrant activation of intracellular nucleic acid sensors can lead to autoimmune diseases such as systemic lupus erythematosus, type-I diabetes and Aicardi–Goutières syndrome (reviewed in [85]).
Acknowledgments
We apologize to all researchers whose critical contributions to the field could not be cited due to space constraints. Current research in the Gack laboratory is supported by National Institutes of Health Grants (AI087846, AI097699, and AI104415), a John and Virginia Kaneb Fellowship, and the Alexander and Margaret Stewart Trust Foundation.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Goubau D, Deddouche S, Reis e Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang JJ, Davis ME, Gack MU. Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev. 2014;25:491–505. doi: 10.1016/j.cytogfr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Chen I, Ichinohe T. Response of host inflammasomes to viral infection. Trends Microbiol. 2015;23:55–63. doi: 10.1016/j.tim.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng J, Yong HY, Panutdaporn N, Liu C, Tang K, Luo D. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res. 2014;43:1216–1230. doi: 10.1093/nar/gku1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 10. Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152:276–289. doi: 10.1016/j.cell.2012.11.048. The authors solve the crystal structure of MDA5 bound to dsRNA, which reveals that MDA5 binds to internal regions of the vRNA and then assembles into large filaments for antiviral downstream signaling.
- 11.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146:448–461. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. Elife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, Finberg RW, Gack MU. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity. 2013;38:437–449. doi: 10.1016/j.immuni.2012.11.018. This study uncovers that serine/threonine dephosphorylation of the N-terminal CARDs of RIG-I and MDA5 by PP1α/γ is essential for RLR-MAVS interaction and antiviral IFN responses.
- 14.Davis ME, Wang MK, Rennick LJ, Full F, Gableske S, Mesman AW, Gringhuis SI, Geijtenbeek TB, Duprex WP, Gack MU. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe. 2014;16:19–30. doi: 10.1016/j.chom.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 16.Oshiumi H, Miyashita M, Matsumoto M, Seya T. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013;9:e1003533. doi: 10.1371/journal.ppat.1003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, Gack MU. The ubiquitin-specific protease USP15 promotes RIG-I-mediated antiviral signaling by deubiquitylating TRIM25. Sci Signal. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature. 2014;509:110–114. doi: 10.1038/nature13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narita R, Takahasi K, Murakami E, Hirano E, Yamamoto SP, Yoneyama M, Kato H, Fujita T. A novel function of human Pumilio proteins in cytoplasmic sensing of viral infection. PLoS Pathog. 2014;10:e1004417. doi: 10.1371/journal.ppat.1004417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Childs KS, Randall RE, Goodbourn S. LGP2 plays a critical role in sensitizing mda-5 to activation by double-stranded RNA. PLoS One. 2013;8:e64202. doi: 10.1371/journal.pone.0064202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJ, et al. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. Elife. 2014;3:e01535. doi: 10.7554/eLife.01535. By using next-generation sequencing, the authors identify that during EMCV infection LGP2 recognizes the anti-sense RNA of the L region of EMCV, stimulating MDA5 activation.
- 24.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M., Jr Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkataraman T, Valdes M, Elsby R, Kakuta S, Caceres G, Saijo S, Iwakura Y, Barber GN. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol. 2007;178:6444–6455. doi: 10.4049/jimmunol.178.10.6444. [DOI] [PubMed] [Google Scholar]
- 26.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 27.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 28. Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514:372–375. doi: 10.1038/nature13590. Previously it had been demonstrated that a 5′triphosphate moiety in the vRNA potently triggers RIG-I activation. This paper shows that a diphosphate group at the 5′-end of vRNA (e.g. Reovirus genomes) can bind and activate RIG-I.
- 29.Schnell G, Loo YM, Marcotrigiano J, Gale M., Jr Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog. 2012;8:e1002839. doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Q, Hato SV, Langereis MA, Zoll J, Virgen-Slane R, Peisley A, Hur S, Semler BL, van Rij RP, van Kuppeveld FJ. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012;2:1187–1196. doi: 10.1016/j.celrep.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao X, Ding Q, Lu J, Tao W, Huang B, Zhao Y, Niu J, Liu YJ, Zhong J. MDA5 plays a critical role in interferon response during hepatitis C virus infection. J Hepatol. 2014 doi: 10.1016/j.jhep.2014.11.007. http://dx.doi.org/10.1016/j.jhep.2014.11.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 33.Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baum A, Sachidanandam R, Garcia-Sastre A. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 2010;107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rehwinkel J, Tan CP, Goubau D, Schulz O, Pichlmair A, Bier K, Robb N, Vreede F, Barclay W, Fodor E, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Weber M, Gawanbacht A, Habjan M, Rang A, Borner C, Schmidt AM, Veitinger S, Jacob R, Devignot S, Kochs G, et al. Incoming RNA virus nucleocapsids containing a 5′-triphosphorylated genome activate RIG-I and antiviral signaling. Cell Host Microbe. 2013;13:336–346. doi: 10.1016/j.chom.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Runge S, Sparrer KM, Lassig C, Hembach K, Baum A, Garcia-Sastre A, Soding J, Conzelmann KK, Hopfner KP. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog. 2014;10:e1004081. doi: 10.1371/journal.ppat.1004081. The physiological ligands of RIG-I and MDA5 during infection with measles virus are identified using next-generation sequencing.
- 38. Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, Watanabe T, Iijima S, Sakurai Y, Watashi K, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity. 2015;42:123–132. doi: 10.1016/j.immuni.2014.12.016. This paper uncovers that RIG-I functions as a sensor of HBV infection by recognizing the 5′-ε region of the pregenomic RNA of HBV. In addition, RIG-I directly inhibits the HBV polymerase.
- 39.Oshiumi H, Sakai K, Matsumoto M, Seya T. DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur J Immunol. 2010;40:940–948. doi: 10.1002/eji.200940203. [DOI] [PubMed] [Google Scholar]
- 40.Gu L, Fullam A, Brennan R, Schroder M. Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol Cell Biol. 2013;33:2004–2015. doi: 10.1128/MCB.01603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko C, Lee S, Windisch MP, Ryu WS. DDX3 DEAD-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J Virol. 2014;88:13689–13698. doi: 10.1128/JVI.02035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Mol Cell Biol. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaller CK, Li Z, George CX, Samuel CE. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol. 2011;23:573–582. doi: 10.1016/j.coi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hornung V, Hartmann R, Ablasser A, Hopfner KP. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onomoto K, Jogi M, Yoo JS, Narita R, Morimoto S, Takemura A, Sambhara S, Kawaguchi A, Osari S, Nagata K, et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reineke LC, Lloyd RE. The stress granule protein G3BP1 recruits PKR to promote multiple innate immune antiviral responses. J Virol. 2014;89:2575–2589. doi: 10.1128/JVI.02791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoo JS, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M, Lai JC, Lattmann S, Nagamine Y, Matsui T, et al. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLoS Pathog. 2014;10:e1004012. doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013;110:1652–1657. doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banerjee S, Chakrabarti A, Jha BK, Weiss SR, Silverman RH. Cell-type-specific effects of RNase L on viral induction of beta interferon. MBio. 2014;5:e00856–e00814. doi: 10.1128/mBio.00856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lupfer C, Kanneganti TD. The expanding role of NLRs in antiviral immunity. Immunol Rev. 2013;255:13–24. doi: 10.1111/imr.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Jiang W, Yan Y, Gong T, Han J, Tian Z, Zhou R. RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat Immunol. 2014;15:1126–1133. doi: 10.1038/ni.3015. [DOI] [PubMed] [Google Scholar]
- 54.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong M, Yoon SI, Wilson IA. Structure and functional characterization of the RNA-binding element of the NLRX1 innate immune modulator. Immunity. 2012;36:337–347. doi: 10.1016/j.immuni.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konno H, Barber GN. The STING controlled cytosolic-DNA activated innate immune pathway and microbial disease. Microbes Infect. 2014;16:998–1001. doi: 10.1016/j.micinf.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol. 2014;88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maringer K, Fernandez-Sesma A. Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection. Cytokine Growth Factor Rev. 2014;25:669–679. doi: 10.1016/j.cytogfr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 61.Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 62.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Minamitani T, Iwakiri D, Takada K. Adenovirus virus-associated RNAs induce type I interferon expression through a RIG-I-mediated pathway. J Virol. 2011;85:4035–4040. doi: 10.1128/JVI.02160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melchjorsen J, Rintahaka J, Soby S, Horan KA, Poltajainen A, Ostergaard L, Paludan SR, Matikainen S. Early innate recognition of herpes simplex virus in human primary macrophages is mediated via the MDA5/MAVS-dependent and MDA5/MAVS/RNA polymerase III-independent pathways. J Virol. 2010;84:11350–11358. doi: 10.1128/JVI.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A. 2014;111:E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, Laustsen A, Hansen K, Ostergaard L, Fitzgerald KA, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Orzalli MH, Knipe DM. Cellular sensing of viral DNA and viral evasion mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, Fitzgerald KA, Kurt-Jones EA. Interferon gamma-inducible protein (IFI) 16 transcriptionally regulates type i interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem. 2014;289:23568–23581. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. This study identifies cGAS as a cytosolic DNA receptor that produces the cyclic dinucleotide cGAMP, which triggers type-I IFN induction.
- 76.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′–5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. The authors show that the second messenger cGAMP is able to spread via gap junctions from infected cells to neighboring uninfected cells, activating an antiviral innate immune response.
- 80.Mankan AK, Schmidt T, Chauhan D, Goldeck M, Honing K, Gaidt M, Kubarenko AV, Andreeva L, Hopfner KP, Hornung V. Cytosolic RNA:DNA hybrids activate the cGAS-STING axis. EMBO J. 2014;33:2937–2946. doi: 10.15252/embj.201488726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. This paper identifies that the reverse-transcribed DNAs of HIV and other retroviruses trigger cGAS activation, which leads to antiviral cytokine induction.
- 84.Lahaye X, Satoh T, Gentili M, Cerboni S, Conrad C, Hurbain I, El Marjou A, Lacabaratz C, Lelievre JD, Manel N. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity. 2013;39:1132–1142. doi: 10.1016/j.immuni.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 85.Kato H, Fujita T. Autoimmunity caused by constitutive activation of cytoplasmic viral RNA sensors. Cytokine Growth Factor Rev. 2014;25:739–743. doi: 10.1016/j.cytogfr.2014.08.003. [DOI] [PubMed] [Google Scholar]