Abstract
One major mechanism of phase variable gene expression in prokaryotes is through inversion of the promoter element for a gene or operon. This protocol describes how to detect the promoter orientation of a phase-variable gene by PCR. This protocol, including primer design, is specific to detection of the promoter orientations of hyxR, a LuxR-like response regulator in Extraintestinal Pathogenic Escherichia coli (ExPEC) isolates (Bateman and Seed, 2012); however, this protocol can be generalized to other organisms and genes to discriminate prokaryotic promoter inversions by PCR through size discrimination of the amplification products. Expression of hyxR is regulated through bidirectional phase inversion of the upstream promoter region mediated by a member of the family of site-specific tyrosine recombinases called Fim-like recombinases. The recombinases recognize inverted DNA repeat sequences flanking the promoter and produce a genomic rearrangement, orientating the promoter in favor or disfavor of gene expression.
Materials and Reagents
Escherichia coli (E. coli) isolate UTI893
Sterile distilled, deionized water (diH2O)
Agarose, molecular biology grade, standard sieve
Ethidium Bromide, 1 mg/ml in distilled, deionized water (diH2O) (or other agent to visualize DNA)
Taq polymerase (1 U/µl) with 10× NH4 buffer (APEX Bioresearch Products, catalog number: 42–409)
10 mM dNTP mix, PCR grade (Life Technologies, catalog number: 18427-013)
Tryptone
Yeast extract
NaCl
Tris base
Boric acid
EDTA (pH 8.0)
SDS
Glycerol
Xylene cyanol
Bromophenol blue
- hyxR phase-specific primers, 100 µM stock solution [Integrated DNA Technologies (IDT)]
- 5’ – ACTGATAATAACCAGAGGCTTCTT – 3’
- 5’ – CAGTGATTAACTTTCGAACATATTG – 3’
- 5’ – GCGAAAGTTAATCACTGGTATGACC – 3’
Tris-Borate-EDTA (TBE) (10× stock) (see Recipes)
10× DNA Loading Dye (see Recipes)
Luria-Bertani broth (LB) culture medium (Sigma-Aldrich, catalog number: L3022-250G (see Recipes)
2% TBE agarose gel with EtBr (see Recipes)
20 ml 10× DNA Loading dye6 (xylene cyanol/bromophenol blue) (see Recipes)
Equipment
37 °C Incubator for bacteria with aeration
Thermal cycler (Bio-Rad Laboratories)
Gel electrophoresis system (Owl Separation Systems)
UV Transilluminator with photo documentation (Bio-Rad Laboratories)
Procedure
Start 5 ml overnight culture of E. coli isolate UTI89 (Note 1) in LB at 37 °C with aeration. Culture can be started from a colony on an agar plate or from a −80 °C glycerol stock.
- Setting up the phase-PCR reaction
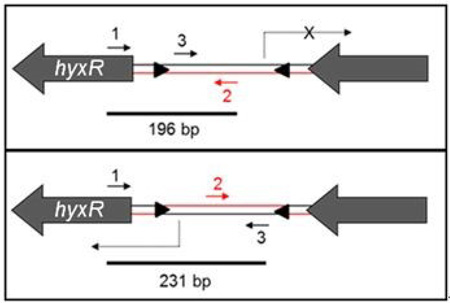
- Phase PCR was performed using a 1:1:1 mixture of 3 phase primers including 1) an anchor primer outside of the invertible region (primer 1); 2) a phase-specific primer for the OFF orientation (primer 2); and 3) a phase-specific primer for the ON orientation (primer 3).
- Reaction mixture (Note 2)
- 1 µl overnight bacterial culture (template)
- 2.5 µl of 10× APEX NH4 buffer (1× final concentration)
- 0.5 µl of 1 unit/µl APEX Taq polymerase (0.5 unit final concentration)
- 1.25 µl of 50 mM MgCl2 (2.5 mM final concentration)
- 0.1 µl of 100 µM stock of each of 3 primers (0.4 µM final concentration of each primer)
- 0.5 µl of 10 mM dNTP mix (0.2 mM final concentration)
- up to 25 µl total volume with sterile diH2O.
- Thermal cycler program:
94°C for 3 min 
× 1 cycle 94°C for 30 sec 
×30 cycles 54°C for 30 sec 72°C for 25 sec 72°C for 6 min 
× 1 cycle Hold at 4°C
Make 50 ml volume of 2% agarose in 1× TBE (see below). Pour gel into casting apparatus.
Cover gel completely with 1× TBE (running buffer).
Mix 5–10 µl of PCR reaction with 0.5–1.0 µl 10× DNA loading dye (approx. 1× final concentration). Load into wells and run samples at 100–120 V for 45 min to 1 h.
Visualize and photograph using a UV gel transilluminator.
Mixed phase populations show two bands by PCR, corresponding to the OFF and ON promoter orientations. The OFF orientation gives a 196 bp amplicon vs. a 231 bp amplicon for the ON orientation.
Recipes
- 1× LB
- Suspend 20 g in 1 L of distilled water
- Autoclave for 15 min at 121 °C
- Final concentration of components (Tryptone 10 g/L, yeast extract 5 g/L, NaCl 5 g/L)
- 1 L 10× TBE buffer stock (Tris-borate-EDTA)
- 108 g Tris base
- 55 g Boric acid
- 40 ml 0.5 M EDTA (pH 8.0)
- diH2O to 1 L
- Stir until completely dissolved.
- 1 L 1× TBE buffer (working) (Note 3)
- 100 ml 10× TBE
- 900 ml diH2O
- Mix well
- 2% TBE agarose gel with EtBr
- 2 g agarose per 100 ml 1×
- TBE Heat in microwave (approx. 2 min) until agarose is dissolved
- Let stand at room temperature for 15–20 min to cool (you do not want to re-solidify)
- Add EtBr to a final concentration of 0.1 µg/ml just prior to pouring gel.
- 20 ml 10× DNA Loading dye (Note 4) (xylene cyanol/bromophenol blue)
- Add 0.025 g each xylene cyanol and bromophenol blue
- 1.25 ml 10% SDS
- 12.5 ml 100% glycerol
- 6.25 ml H2O
- Mix thoroughly and store at room temperature
Acknowledgments
This protocol is adapted from Bateman and Seed (2012).
Footnotes
UTI89 is a prototypic ExPEC cystitis isolate that was obtained from an adult patient with cystitis and has been well described in the literature (Mulvey, 1998). Primers for this protocol were designed to UTI89 genomic sequence, and while ExPEC isolates are similar, their genomic sequence may not be exact. It is recommended that researchers check that the primers listed in this protocol will work with other ExPEC isolates.
All PCR reactions were performed using APEX Taq polymerase and accompanying buffers distributed by Genesse Scientific, Inc (San Diego, California, USA). Thermal cycler parameters were based on the manufacturer’s recommendations. Taq polymerase or related thermostable polymerases would be expected to provide similar performance in this assay with minor modifications of the reactions.
1× TBE is what you will use to make your agarose gel as well as the buffer used to run the gel.
Any commercially available DNA loading dye is appropriate. The recipe given is only a home-made suggestion. Other recipes available use different tracking dyes and/or density agents.
References
- 1.Bateman SL, Seed PC. Epigenetic regulation of the nitrosative stress response and intracellular macrophage survival by extraintestinal pathogenic Escherichia coli. Mol Microbiol. 2012;83(5):908–925. doi: 10.1111/j.1365-2958.2012.07977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282(5393):1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]