Abstract
Ovarian cancer is the leading cause of death among women with gynecologic malignancies. The development and progression of ovarian cancer are complex and a multiple-step process. New biomarker molecules for diagnostic and prognostic are essential for novel therapeutic targets and to extend the survival time of patients with ovarian cancer. Long noncoding RNAs (lncRNAs) are non–protein-coding transcripts longer than 200 nucleotides that have recently been found as key regulators of various biological processes and to be involved in the development and progression of many diseases including cancers. In this review, we summarized the expression pattern of several dysregulated lncRNAs (HOTAIR, H19, XIST, and HOST2) and the functional molecular mechanism of these lncRNAs on the initiation and progression of ovarian cancer. The lncRNAs as biomarkers may be used for current and future clinical diagnosis, therapeutics, and prognosis.
Key Words/Abbreviations: lncRNA, Ovarian cancer, HOTAIR, H19, XIST, HOST2, OC - ovarian cancer, lncRNA - long noncoding RNA, EOC - epithelial ovarian cancer, SOC - serous ovarian cancer, ORF - open reading frame, ncRNAs - noncoding RNAs, HOTAIR - HOX transcript antisense intergenic RNA, XIST - X inactive-specific transcript, Xi - X chromosome inactivation, MALAT1 - metastasis-associated lung adenocarcinoma transcript 1, MEG3 - maternally expressed gene 3, HOST2 - human ovarian cancer-specific transcript 2, ICR - imprinting control region, PRC2 - polycomb-repressive complex 2, EMT - epithelial-mesenchymal transition, IGFII - insulin-like growth factor II, UCA1 - urothelial cancer associated 1, MMP2 - matrix metallopeptidase 2, MMP9 - matrix metallopeptidase 9, NEAT - 1-nuclear paraspeckle assembly transcript 1, PSPC1 - protein-coding gene paraspeckle component 1
Ovarian cancer (OC) is one of the gynecologic malignancies with the highest mortality rate. The incidence of OC accounts for 3% of the gynecological cancer total incidence, but the number of OC deaths accounts for 5% of all gynecologic cancer number of deaths.1 Every year, about 200,000 women are diagnosed with ovarian carcinoma, and almost 125,000 die of it. Although 85% of OC is epithelial OC (EOC), it is the most serious histological subtype and threat for women’s health.2 Currently, OC is generally treated by a combination of surgery and chemotherapy with cisplatin and paclitaxel, and appropriate radiotherapy, or other treatments. Although patients with OC have short-term relief, the recurrence and metastasis rate are still very high. Moreover, the 5-year survival rate of patients who have surgical treatment followed by adjuvant chemotherapy has remained less than 40%.3,4 Because of lack of thorough understanding of the resistance and molecular mechanisms of progression in OC, conventional treatments are limited. Increased efforts to get a better understanding of the specific molecular mechanisms in OC are required, and thus assist in the development of new diagnostic and therapeutic strategies.
RNAs longer than 200 nucleotides that lack the ability to encode proteins are classified as long noncoding RNAs (lncRNAs).5–8 About 76% of the human genome are transcribed to generate lncRNAs,6,8 and they reside in the nucleus and/or in the cytoplasm.9 With the help of high-throughput sequencing technologies and other advanced research techniques, the biofunctions of lncRNAs are increasingly defined.10 Increasing evidence suggests that lncRNAs play a distinctly important role in many different cellular processes, including cell migration, growth, invasion, apoptosis, and differentiation.11,12 However, the biofunctions and molecular mechanisms of lncRNAs in human diseases including cancers have remained a mystery.13
In this review, we describe the functions and molecular mechanisms of lncRNAs in the initiation and progression of OC. The current and prospective applications of lncRNAs in clinical OC research are also explored, with an emphasis on lncRNA-based diagnosis, prognosis, and therapeutics.
LncRNA
LncRNAs are noncoding RNA, which are longer than 200 nucleotides in length. In addition, they are very conservative and do not function as templates for protein synthesis. According to the location of the lnRNA gene on the genome, lncRNAs can be divided into 5 categories: (1) sense lncRNA, (2) antisense lncRNA, (3) bidirectional lncRNA, (4) intronic lncRNA, and (5) intergenic lncRNA.10,14–16
In various aspects of research on lnRNAs, the functions and mechanisms of lncRNAs are the most difficult and least understood. Now, new research indicates that lncRNAs have been implicated in various aspects of gene regulation, including imprinting, epigenetic regulation, trafficking of nucleus and cytoplasm, transcription, mRNA splicing, and so on.10–12 lncRNAs have also been related to many diverse biological processes, such as cell cycle, cell proliferation, cell apoptosis, and cell differentiation.17–19 On the basis of the pattern of their functions, lncRNAs have been divided into 4 subtypes, namely, signals (X-inactive specific transcript [XIST]20 and PANDA21), decoys (Gas521), guides (HOX transcript antisense intergenic RNA [HOTAIR]22 and lincRNA-p2123), and scaffolds (Anril24). Previous studies have found that the abnormal expression or dysfunction of lnRNA is closely related to the initiation and progression of human diseases, including cancer. LncRNAs are closely involved in tumor development, for example, in gastric cancer, the maternally expressed gene 3 (MEG3) expression is clearly reduced, and MEG3 overexpression can inhibit cell proliferation and induce the cell apoptosis of gastric cancer.20 Therefore, in some types of cancer, specifically expressed lncRNAs may be used as tumor biomarkers, and they can be served as therapeutic targets as well to conquer cancer.
To summarize, lncRNAs can play a role through a variety of mechanisms in cancer to influence its development. Despite the increasing knowledge about the functions of lncRNA in cancer, the modes of action of most lncRNAs in most cancers remain unclear. Widely understanding the mechanism of action, regulatory pathways, hierarchical structure, and operation of the network of lncRNAs, we will greatly improve our recognitions of their function in cancer accordingly and find new way of treatment through regulating their function in OC.
DYSREGULATED lncRNAs IN OC
HOST2
In 2003, Rangel et al21 found a new transcript, human OC-specific transcript 2 (HOST2), specific expression in human OC tissue. The HOST2 gene is located on chromosome 10 with a length of 2.9 kb, and it does not have an open reading frame, does not encode proteins, and belongs to lncRNA.21 LncRNA HOST2 have a highly specific expression in EOC, but the expression is very low or none in a variety of other common malignancies. After inhibiting the expression of HOST2, the migration, invasion, and proliferation of OVCAR-3 cells were significantly reduced.21
Recently, the mechanisms of lncRNA HOST2 regulating the biological behavior in EOC cells by binding to microRNA let-7b was reported.22 The study first indicated that the lncRNA HOST2, which is highly expressed in EOC, functions as a molecular sponge which allows direct binding to let-7b and then inhibits the function of let-7b in the EOC. Moreover, they found that given the overexpression of HOST2 in EOC, HOST2/let-7b may contribute to the regulation of growth and development in EOC. Thus, we can easily infer that the expression of let-7–targeting genes HMGA2, c-myc, Dicer, and Imp3 increases, thus promoting the initiation and development of EOC. In addition, they observed that suppression of the activity of let-7b can increase both the mRNA and protein levels of HMGA2, c-Myc, Dicer, and Imp3. The schematic diagram of HOST2 promoting the endogenous expression of metastasis-promoting–related genes, which were targeted by let-7b, is shown in (Fig. 1). The results showed that the functions of let-7b can inhibit the expression of the oncogenes that negatively regulate cell growth and proliferation. In addition, the regulation between HOST2 and let-7b can provide innovative strategies for better diagnosis and treatment of this deadly malignant tumor.
FIGURE 1.
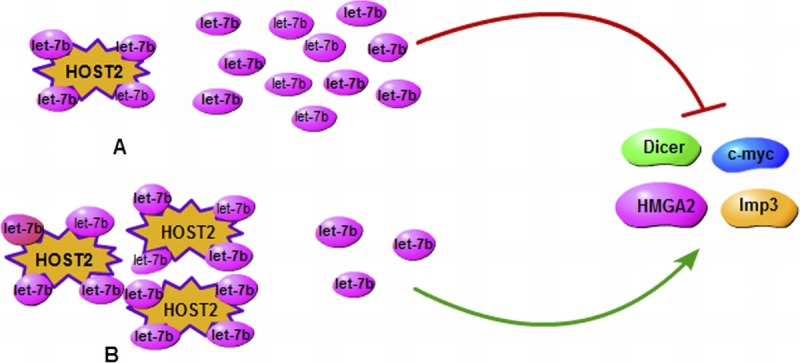
A schematic diagram of HOST2 enhances the endogenous expression of metastasis-promoting genes that are targeted by let-7b. A, Down-regulation of HOST2 may increase let-7b activity, which would inhibit expression of its target genes (HMGA2, c-myc, Dicer, and Imp3). B, High regulation of HOST2 binds to more let-7b and increases expression of its target genes (HMGA2, c-myc, Dicer, and Imp3).
HOTAIR
HOX transcription antisense RNA (HOTAIR) is the first trans-acting LncRNA to be found, which is located in human chromosome 12q13.13 with a length of 6232 bp.23,24 The human HOX gene family is divided into 4 subfamilies: HOXA, HOXB, HOXC, and HOXD, whereas LncRNA HOTAIR is identified from a custom tilling array of the HOXC locus.25,26 HOTAIR is located in the antisense strand of the HOXC gene cluster, but it could regulate the HOXD gene cluster.26 The most familiar regulatory mechanism of HOTAIR is that the 5′ or 3′ ends of HOTAIR can be respectively combined with PRC2 (composed of EZH2, SUZ12, and EED)25 and histone demethylase complex (LSD1, Co REST, and REST),27 and it mediates these 2 complexes binding to specific genomic loci, then respectively makes histone H3 trimethylated at lysine 27(H3K27me3) and histone H3 dimethyl Lys4(H3K4me2), and then causes the chromosome in a closed state and silences the expression of the target gene25 (Fig. 2). Thus, HOTAIR can regulate the expression of HOXD epigenetically, which was sited at human chromosome 2q.26
FIGURE 2.
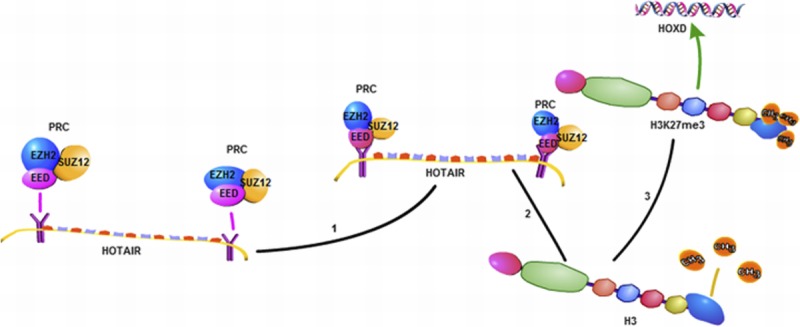
A schematic diagram of the mechanisms of HOTAIR. HOTAIR when combined with PRC2 (composed of EZH2, SUZ12, and EED) can trimethylate histone H3 at lysine 27 (H3K27me3) and inhibit HOXD gene expression.
The expression of HOTAIR is very high in the EOC tissue compared with normal ovarian tissues. In addition, high expression of HOTAIR is closely associated with the occurrence and development, invasion, metastasis, and prognosis in EOC.28 These researches emphasized the great clinical significance of HOTAIR in patients with EOC, and HOTAIR expression may act as a biomarker in predicting the progression of EOC. They also pointed out that the knockdown HOTAIR can inhibit cellular epithelial-mesenchymal transition (EMT) and reduce potential of tumorigenesis and migration ability via regulating the EMT-related gene. That is, HOTAIR can be used as a potential therapeutic target to treat patients with EOC.
Recently, studies have reported that HOTAIR can play a role by regulating the cell cycle and apoptosis pathway in serous OC (SOC).28 The study showed that silencing of HOTAIR can delay the progression of cell cycle and promote the apoptosis of the A2780 and OVCA429 SOC cells, evidencing that HOTAIR may participate in the regulation of cell cycle and apoptosis to promote cell proliferation in SOC. In addition to these studies, they also disclosed that the mechanism of HOTAIR in SOC is partly through regulating the expression of cell cycle– and apoptosis-related genes, such as cyclin E, BCL-2 and caspase-9, caspase-3, and BRCA1. These research can be extended to our existing cognition about the downstream genes of HOTAIR, including the cell cycle–related and apoptosis-related proteins. In addition, these results in the present studies give us a stronger appreciation of the significance of the abnormal expression of lncRNAs in the SOC, and offer us a theoretical basis for the targeted therapy of SOC by lncRNA-based targeted strategies.
H19
The H19 gene is located at human chromosome 11p with a full length of 2.3 kb, which is the first lncRNA to be found related with cancer.29 Numerous studies have shown that H19 is differentially expressed in many cancers, and for its function in cancer, it plays a dual role: promote the tumor or suppress the cancer.30 Several groups reported that H19/IGF2 gene imprinting is associated with differences in methylation, chromatin conformation, and transcription cycle of the allelic of H19.31,32
Mizrahi et al33 reported that the expression of H19 was detected in 90% of ascites cells of patients with OC by means of situ hybridization. By decreasing the expression of H19 in a subcutaneous nude mice model for OC, it could be observed that 40% of the tumor growth was inhibited.33 This indicates that overexpression H19 promotes the growth and the knockdown of the expression of H19 can inhibit the growth of OC.
Recently, Medrzycki et al’s31 research reported that histone H1.3 can inhibit the expression of H19 and suppress the growth of OC cells. H1.3, a variant of H1, is highly expressed in the OVCAR-3 cell line, which is an EOC cell line. In addition, excessive expression of H1.3 can reduce the growth rate and colony formation of OVCAR-3 cells, indicating that H1.3 may play a suppressive role in cancer cells. Moreover, in multiple different OC cell lines, overexpression of H1.3 can inhibit the expression of H19, whereas down-regulation of H1.3 can increase the expression of H19, suggesting histone H1.3 as a specific inhibitor of H19. Histone H1.3 can regulate H19 expression mainly because overexpression of histone H1.3 can make the occupancy of H1.3 better at the imprinting control region (ICR), which encompasses the H19 regulator region, and also accompanied by the increase of DNA methylation and the decrease in the insulator protein CCCTC binding factor occupancy at the ICR (Fig. 3). Most of all, the synergies of increased expression of H1.3 and reduced expression of H19 can better inhibit the growth of ovarian epithelial cancer. These studies provide some new theoretical basis for better targeting treatment of EOC.
FIGURE 3.
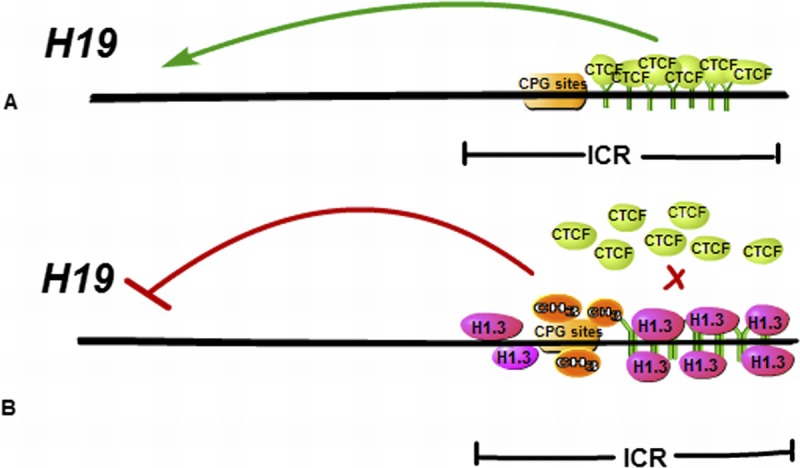
A schematic diagram of epigenetic mechanisms of H19 repression mediated by H1.3 Histone H1.3 overexpression leads to increased occupancy of H1.3 at the H19 regulator region encompassing the ICR, concomitant with increased DNA methylation and reduced occupancy of the insulator protein CTCF at the ICR, which can inhibit the expression of H19.
XIST
X-inactive specific transcript (XIST) is a lnRNA that is located at the X chromosome inactivation (Xi) center, which is important in the initial phase of X chromosome inactivation.34 The precise molecular mechanism of XIST in the initial phase of X chromosome inactivation remains elusive, but there are plenty of evidence suggesting that XIST plays a stronger effect in the process of cell differentiation and proliferation and the maintenance of genomic stability.35 As XIST is located at the X chromosome, it is involved in gynecological diseases. Studies have shown that the expression of XIST is detected in almost all female cells, except in gynecological malignancies such as cervical cancer, breast cancer, and OC.32
Benoit et al36 showed that XIST was not detected in more than half of the OC cells with Barr body staining compared with normal ovarian cells. In addition, other studies indicated that reduction of the expression of XIST can decrease the sensitivity of OC cells to paclitaxel, suggesting that XIST can serve as a biomarker of the prognosis of OC. This mechanisms could be interpreted by the XIST toxic effects of paclitaxel and might also be considered relevant in the reactivation of X-“specific resistance gene” and increased expression of these genes on the X chromosome.37 Another explanation is that loss of XIST is caused by genetic instability, thereby promoting resistance to chemotherapy in OC.37
Others
LncRNA UCA1 is cloned from BLZ-211, which is a human bladder carcinoma cell line, and the full length of UCA1 cDNA is 1442 bp.38 Early studies have shown that UCA1 is associated with the development of embryonic and bladder cancer, and it is also observed as highly expressed in OC and other tumors.39 In addition, UCA1 plays a key role in promoting invasion and metastasis of tumor cells by up-regulating the expression of matrix metallopeptidase 2 (MMP2) and MMP9 in OC.38 Recently, Yang et al40 indicated that UCA1 could bind to miR-485-5p directly in EOC as an endogenous sponge. Low expression of UCA1 could down-regulate the expression of MMP14, and MMP14 is one of the target genes of miR-485-5p. In addition, UCA1 could improve EOC metastasis, and it is also associated with poor prognosis. Therefore, UCA1 can be a new prognostic biomarker and may serve as a potential target for the treatment of EOC.
LSINCT5 is an lncRNA with a length of 2.6 kb and is located in the cell nucleus.40 Studies have shown that the expression of LSINCT5 is very high in both OC cell lines and tissues.41,42 It also shows that knockdown of LSINCT5 can reduce cell proliferation and induce changes in the expression of some genes, such as NEAT-1 and PSPC1.41 Although the function mechanism of LSINCT5 in OC is still unclear, it plays a significant role in the initiation and development of OC.43 So, LSINCT5 may be used as an appropriate target for the treatment of OC.
LncRNA MEG3 is located at the human chromosome 14q32.3, which is also called maternally expressed gene 3.44 Studies have found that MEG3 is expressed in many normal tissues, including ovarian tissue45; however, the level of MEG3 is very low or rarely detected in some tumor tissues and tumor cell lines, including breast cancer, cervix cancer, hepatocellular cancer, colon cancer, and so on.46–48 Studies have demonstrated that MEG3 showed low or no expression in EOC tissues and OC cells compared with normal ovarian tissues,49 and overexpression of MEG3 could inhibit proliferation and promote apoptosis in the OC cells; therefore, MEG3 may be a tumor suppressor gene in OC.50 The low expression of MEG3 in OC is caused by multiple mechanisms, and the increased hypermethylation of the MEG3 promoter can reduce the expression of MEG3 RNA in EOC.49 In addition, the study also noted that MEG3 can control the proliferation of OC cells via targeting p53 and/or RB1.50 So, through further research, MEG3 may be used as a potential therapeutic target and may provide new treatment strategies for EOC.
BC200 is a lncRNA with a length of 200 nucleotides and expressed specifically in the human nervous system.10,51 The expression of BC200 was significantly decreased in OC tissue compared with that in normal ovarian epithelium.52 This suggests that the low expression of BC200 may be associated with the development of OC. The low expression of BC200 may be used as a potential diagnostic marker for patients with OC. In addition, down-regulation of BC200 can lead to the proliferation of OC cells and inhibit the sensitivity of OC cells to carboplatin.52 However, the current research also shows that BC200 has no obvious effect on the invasion and migration of OC cells. In a summary, the dysregulation of BC200 was detected in OC, and this evidence provide some clinical implications of BC200 for the diagnosis and targeted therapy of OC.
PVT1 is an lncRNA with a length of 1.9 kb, originally identified as a transcriptional unit from a human homologous sequence to PVT1.53 The amplification and overexpression of PVT1 can increase cell proliferation and inhibit cell apoptosis, suggesting that it is an antiapoptotic gene.54 Recently, it was reported that PVT1 is associated with cisplatin resistance in OC.55 It also indicated that overexpression of PVT1 can inhibit the expression of transforming growth factor-β1, p-smad and caspase-3 in OC, which were associated with cell apoptosis.55 In short, PVT1 expression is closely related with carboplatin resistance in OC by regulating cell apoptotic pathways.
CONCLUSIONS AND PROSPECTIVE
To summarize, lncRNAs play a major role in the control of OC cell growth, division, metastasis, invasion, proliferation, and drug resistance22,28,31,33,37,38,43,50,52,55 (Fig. 4). The abnormal expression of some lncRNAs can contribute to the development and progression of OC. In addition, aberrant expression of some lncRNAs in OC can provide new information for disease detection, diagnosis, and treatment to patients with EOC. LncRNA is associated with the sensitivity of chemotherapy drug in OC, so the detection of lncRNA level within tumor cells can predict patients’ sensitivity to chemotherapy. In aspects of molecular therapy, lncRNA as an emerging target can provide new information for targeting therapy of OC. In addition, lnRNA as a tumor marker has a higher sensitivity and specificity for the diagnosis and prognosis of OC.56 However, compared with microRNA, the research of lncRNA is still in the fledgling stage. Only a small amount of lncRNAs are identified and found to be associated with OC, and the molecular mechanisms of their role in the development and progression of OC are still not yet well defined. Accordingly, further work on lncRNAs are required to understand the molecular mechanism and to serve for the clinical prognosis and therapy of diseases.
FIGURE 4.
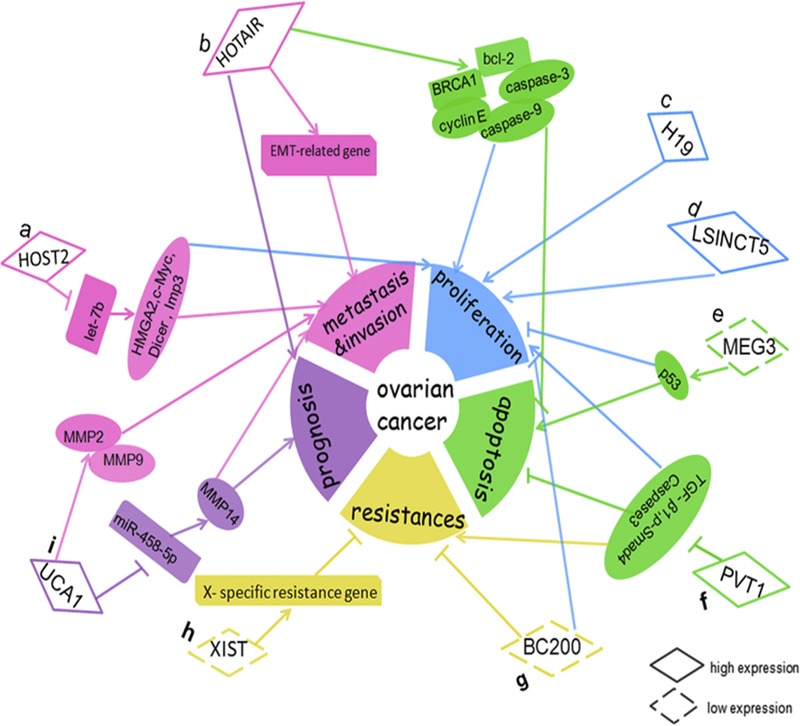
Dysregulation and functional roles of lncRNAs in OC. A, HOST2 could increase cell metastasis, invasion, and proliferation via binding to let-7b and inhibiting the function of let-7b, and then increase expression of its target genes (HMGA2, c-myc, Dicer, and Imp3). B, HOTAIR represented a prognostic marker and increased cell metastasis and invasion by regulating EMT-related genes; HOTAIR promoted SOC cell proliferation and decreased cell apoptosis by regulating certain cell cycle– and apoptotic-related genes (cyclin E, BCL-2, caspase-9, caspase-3, and BRCA1). C, H19 increased cell proliferation. D, LSINCT5 increased cell proliferation. E, MEG3 promoted cell proliferation and decreased cell apoptosis via p53. F, PVT1 inhibited cell apoptosis and increased cell proliferation and carboplatin resistance in OC by knockdown the expression of TGF-β1, p-smad4, and Caspase-3. G, BC200 inhibited OC cell proliferation and increased the sensitivity of OC cells to carboplatin. H, XIST could inhibit OC cell resistance to paclitaxel via reactivating the X-specific resistance gene. I, UCA1 promoted invasion and metastasis by up-regulating MMP2 and MMP9 expression in OC cells; UCA1 represented a prognostic marker and enhanced EOC metastasis through the UCA1-miR-485-5p-MMP14 axis.
Footnotes
This work was supported by Natural Science Foundation of China (81572577, 81372366, 81672993), Overseas, Hong Kong & Macao Scholars Collaborated Researching Fund of National Natural Science Foundation of China (81428018), 111 project (111-2-12), The Hunan Province Natural Science Foundation of China (2016JJ1027, 2016JC2035), the Project of Innovation-driven Plan of Central South University (2016CX023), the Open-End Fund for the Valuable and Precision Instruments of Central South University, and the Fundamental Research Funds for the Central Universities of Central South University.
The authors declare no conflicts of interest.
REFERENCES
- 1.Sopik V, Iqbal J, Rosen B, et al. Why have ovarian cancer mortality rates declined? Part II. Case-fatality. Gynecol Oncol. 2015;138:750–756. [DOI] [PubMed] [Google Scholar]
- 2.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aletti GD, Gallenberg MM, Cliby WA, et al. Current management strategies for ovarian cancer. Mayo Clin Proc. 2007;82:751–770. [DOI] [PubMed] [Google Scholar]
- 4.Lage H, Denkert C. Resistance to chemotherapy in ovarian carcinoma. Recent Results Cancer Res. 2007;176:51–60. [DOI] [PubMed] [Google Scholar]
- 5.Ma MZ, Chu BF, Zhang Y, et al. Long non-coding RNA CCAT1 promotes gallbladder cancer development via negative modulation of miRNA-218-5p. Cell Death Dis. 2015;6:e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Y, Wang J, Chen X, et al. Large-scale study of long non-coding RNA functions based on structure and expression features. Sci China Life Sci. 2013;56:953–959. [DOI] [PubMed] [Google Scholar]
- 8.Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ. 2012;54:44–54. [DOI] [PubMed] [Google Scholar]
- 9.Ye N, Wang B, Quan ZF, et al. Functional roles of long non-coding RNA in human breast cancer. Asian Pac J Cancer Prev. 2014;15:5993–5997. [DOI] [PubMed] [Google Scholar]
- 10.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. [DOI] [PubMed] [Google Scholar]
- 11.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976–10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Luo H, Liao Q, et al. Systematic study of human long intergenic non-coding RNAs and their impact on cancer. Sci China Life Sci. 2013;56:324–334. [DOI] [PubMed] [Google Scholar]
- 13.Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–165. [DOI] [PubMed] [Google Scholar]
- 16.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:6391–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parasramka MA, Maji S, Matsuda A, et al. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther. 2016;161:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–376. [DOI] [PubMed] [Google Scholar]
- 19.Fang XY, Pan HF, Leng RX, et al. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356:357–366. [DOI] [PubMed] [Google Scholar]
- 20.Sun M, Xia R, Jin F, et al. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. [DOI] [PubMed] [Google Scholar]
- 21.Rangel LB, Sherman-Baust CA, Wernyj RP, et al. Characterization of novel human ovarian cancer-specific transcripts (HOSTs) identified by serial analysis of gene expression. Oncogene. 2003;22:7225–7232. [DOI] [PubMed] [Google Scholar]
- 22.Gao Y, Meng H, Liu S, et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet. 2015;24:841–852. [DOI] [PubMed] [Google Scholar]
- 23.He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan Y, Chang HY. HOTAIR: flight of noncoding RNAs in cancer metastasis. Cell Cycle. 2010;9:3391–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grier DG, Thompson A, Kwasniewska A, et al. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154–171. [DOI] [PubMed] [Google Scholar]
- 27.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jun-jun Qiu YJH, Gong Yang K. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res. 2015:238–248. [DOI] [PubMed] [Google Scholar]
- 29.Leighton PA, Ingram RS, Eggenschwiler J, et al. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. [DOI] [PubMed] [Google Scholar]
- 30.Hibi K, Nakamura H, Hirai A, et al. Loss of H19 imprinting in esophageal cancer. Cancer Res. 1996;56:480–482. [PubMed] [Google Scholar]
- 31.Medrzycki M, Zhang Y, Zhang W, et al. Histone h1.3 suppresses h19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014;74:6463–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami T, Zhang C, Taniguchi T, et al. Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene. 2004;23:6163–6169. [DOI] [PubMed] [Google Scholar]
- 33.Mizrahi A, Czerniak A, Levy T, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med. 2009;7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weakley SM, Wang H, Yao Q, et al. Expression and function of a large non-coding RNA gene XIST in human cancer. World J Surg. 2011;35:1751–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaligne R, Heard E. X-chromosome inactivation in development and cancer. FEBS Lett. 2014;588:2514–2522. [DOI] [PubMed] [Google Scholar]
- 36.Benoit MH, Hudson TJ, Maire G, et al. Global analysis of chromosome X gene expression in primary cultures of normal ovarian surface epithelial cells and epithelial ovarian cancer cell lines. Int J Oncol. 2007;30:5–17. [PubMed] [Google Scholar]
- 37.Huang KC, Rao PH, Lau CC, et al. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol Cancer Ther. 2002;1:769–776. [PubMed] [Google Scholar]
- 38.Wang F, Li X, Xie X, et al. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582:1919–1927. [DOI] [PubMed] [Google Scholar]
- 39.Wang Fan ZJXX. Invasion and migration effect and mechanism of long non -coding RNA UCA1 in ovarian cancer cells. Modern Oncology. 2015. [Google Scholar]
- 40.Yang Y, Jiang Y, Wan Y, et al. UCA1 functions as a competing endogenous RNA to suppress epithelial ovarian cancer metastasis. Tumour Biol. 2016. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 41.Hecht SS, Hatsukami DK, Bonilla LE, et al. Quantitation of 4-oxo-4-(3-pyridyl)butanoic acid and enantiomers of 4-hydroxy-4-(3-pyridyl)butanoic acid in human urine: a substantial pathway of nicotine metabolism. Chem Res Toxicol. 1999;12:172–179. [DOI] [PubMed] [Google Scholar]
- 42.Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. [DOI] [PubMed] [Google Scholar]
- 43.Silva JM, Boczek NJ, Berres MW, et al. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505. [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi N, Wagatsuma H, Wakana S, et al. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. [DOI] [PubMed] [Google Scholar]
- 45.Zhou Y. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;83:R45–R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Zhou Y, Mehta KR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–5126. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y, Zhong Y, Wang Y, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. [DOI] [PubMed] [Google Scholar]
- 48.Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng X, Li J, Yang L, et al. Promoter hypermethylation influences the suppressive role of maternally expressed 3, a long non-coding RNA, in the development of epithelial ovarian cancer. Oncol Rep. 2014;32:277–285. [DOI] [PubMed] [Google Scholar]
- 50.Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129:773–779. [DOI] [PubMed] [Google Scholar]
- 51.Tiedge H, Chen W, Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13:2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu DI, Wang T, Ren C, et al. Downregulation of BC200 in ovarian cancer contributes to cancer cell proliferation and chemoresistance to carboplatin. Oncol Lett. 2016;11:1189–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beck-Engeser GB, Lum AM, Huppi K, et al. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guan Y, Kuo WL, Stilwell JL, et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. [DOI] [PubMed] [Google Scholar]
- 55.Liu E, Liu Z, Zhou Y, et al. Overexpression of long non-coding RNA PVT1 in ovarian cancer cells promotes cisplatin resistance by regulating apoptotic pathways. Int J Clin Exp Med. 2015;8:20565–20572. [PMC free article] [PubMed] [Google Scholar]
- 56.Xiong XD, Ren X, Cai MY, et al. Long non-coding RNAs: An emerging powerhouse in the battle between life and death of tumor cells. Drug Resist Updat. 2016;26:28–42. [DOI] [PubMed] [Google Scholar]


