Zhu et al. show that CHCHD2, a mitochondrial protein, primes the differentiation potential of human induced pluripotent stem cells (hiPSCs) to neuroectodermal lineages through binding and sequestering SMAD4 to the mitochondria and suppressing the activity of the TGFβ signaling pathway. CHCHD2 expression could be a useful tool to assay hiPSC differentiation potential.
Abstract
Human induced pluripotent stem cell (hiPSC) utility is limited by variations in the ability of these cells to undergo lineage-specific differentiation. We have undertaken a transcriptional comparison of human embryonic stem cell (hESC) lines and hiPSC lines and have shown that hiPSCs are inferior in their ability to undergo neuroectodermal differentiation. Among the differentially expressed candidates between hESCs and hiPSCs, we identified a mitochondrial protein, CHCHD2, whose expression seems to correlate with neuroectodermal differentiation potential of pluripotent stem cells. We provide evidence that hiPSC variability with respect to CHCHD2 expression and differentiation potential is caused by clonal variation during the reprogramming process and that CHCHD2 primes neuroectodermal differentiation of hESCs and hiPSCs by binding and sequestering SMAD4 to the mitochondria, resulting in suppression of the activity of the TGFβ signaling pathway. Using CHCHD2 as a marker for assessing and comparing the hiPSC clonal and/or line differentiation potential provides a tool for large scale differentiation and hiPSC banking studies.
Introduction
Induced pluripotent stem cells (iPSCs), derived by transduction of somatic cells with OCT4, SOX2, KLF4, and c-MYC are defined as pluripotent in view of their ability to self-renew and differentiate into cell types representative of three embryonic germ layers (Takahashi et al., 2007; Park and Daley, 2009); however, several studies have shown considerable variation in their differentiation potential (Narsinh et al., 2011; Tobin and Kim, 2012). The mechanistic basis of this variation is poorly understood, but several hypotheses to account for these differences have been proposed, such as incomplete epigenetic reprogramming (Ma et al., 2014), microRNA expression (Vitaloni et al., 2014), donor cell type (Kim et al., 2010), reprogramming factor selection (Buganim et al., 2014), differential activity of endogenous TGFβ signaling pathways (Zhou et al., 2010; Pauklin and Vallier, 2013), and genetic variation between individual donors of the somatic cells used to generate iPSCs (Rouhani et al., 2014).
Human embryonic stem cell (hESC) lines vary in their propensity for differentiation (Osafune et al., 2008), but growing evidence suggests that even greater variability may be present in human iPSCs (hiPSCs; Narsinh et al., 2011; Buganim et al., 2014; Ma et al., 2014), even though the genetic background of hiPSCs is likely to be more variable given their greater availability compared with hESC lines. Detailed comparisons of the ability of both hESC and hiPSC to generate specific types of somatic cells indicate that despite using identical transcriptional networks to generate cells such as those of the neuroepithelium, some hiPSC lines respond to such developmental programs with significantly reduced efficiency (Hu et al., 2010).
Parameters such as methylome analysis, expression of transcript regulators, and analyses of aneuploidy cannot be used to distinguish high- and low-quality hiPSC lines (Buganim et al., 2014). γH2A.X deposition patterns may distinguish the differentiation potential of hiPSCs (Wu et al., 2014); however, it would be helpful to have a rapid assay to assess the differentiation potential of hiPSCs. In this study, we identified CHCHD2, whose expression is often low or absent in hiPSCs when compared with hESCs, which is an efficient correlate of the potential of such hiPSCs to give rise to neuroectodermal lineages on differentiation.
Results
Identification of differentially expressed transcripts between hESCs and hiPSCs
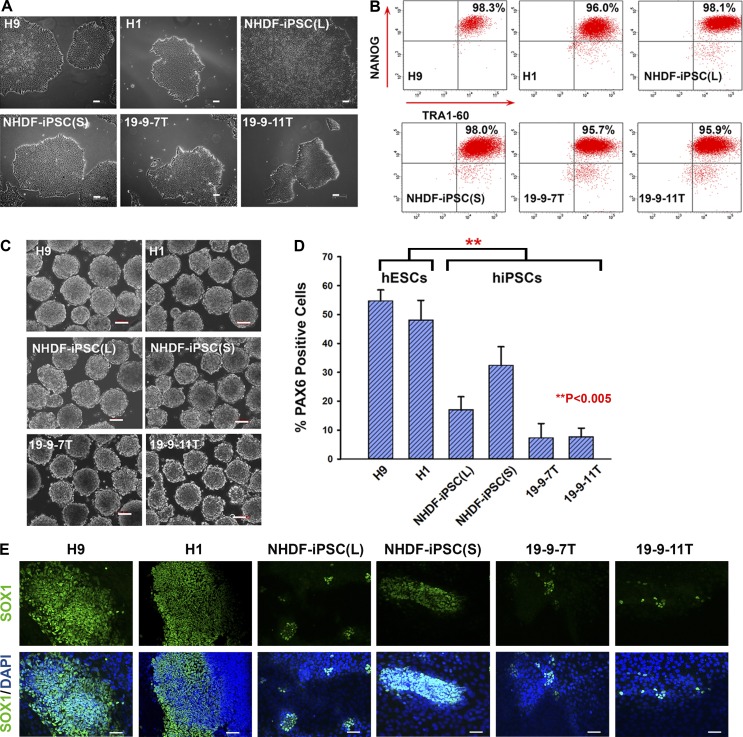
Six independently derived pluripotent stem cells lines were used, including two human embryonic stem cell lines (H9 and H1; WiCell Inc.) and four hiPSC lines generated using the lentiviral, nonintegrating Sendai virus and episomal vectors (NHDF-iPSC(L), NHDF-iPSC(S), 19-9-7T, and 19–9-11T; Table 1 and Fig. 1 A). The lentiviral- and Sendai-derived hiPSC lines were generated and characterized in our laboratory (Jiang et al., 2014; Chichagova et al., 2016) and fulfilled all pluripotency criteria, whereas the episomal-derived lines (19-9-7T and 19-9-11T) were purchased from WiCell Inc. (Yu et al., 2009). These pluripotent stem cells, cultured under identical feeder-free conditions, were differentiated into neural stem cells (NSCs) as outlined in Materials and methods. During pluripotent culture, all hESC and hiPSC lines demonstrated similar expression of the key pluripotency markers NANOG and TRA-1-60 (Fig. 1 B) in addition to the maintenance of pluripotent stem cell morphology (Fig. 1 A). We subjected all hESC and hiPSC lines to neuroectodermal differentiation using an embryoid body (EB)–based differentiation method (Fig. 1 C) and observed that all hiPSC lines showed a significant reduction in their differentiation ability as indicated by a reduction in the number of PAX6-positive cells (Fig. 1 D) and reduced SOX1 expression when compared with hESCs (Fig. 1 E), corroborating previously published data (Hu et al., 2010).
Table 1. Schematic summary of hESCs and hiPSCs used in this study.
| Cell type | Cell line | Generated method | Provider |
|---|---|---|---|
| hESCs | H9 | — | WiCell |
| H1 | — | WiCell | |
| hiPSCs | NHDF-iPSC(L) | Lentivirus | Our laboratory |
| NHDF-iPSC(S) | Sendai virus | Our laboratory | |
| 19-9-7T | Nonintegrating vector | WiCell | |
| 19-9-11T | Nonintegrating vector | WiCell |
Figure 1.
Variations in the ability of hiPSCs to undergo neuroectodermal differentiation. (A) Phase-contrast images of hESCs and hiPSCs used in this study. Bars, 100 µm. (B) Representative flow cytometric analysis indicating a high expression level of the pluripotency markers TRA1-60 and NANOG. (C) All human pluripotent stem cells formed EBs in suspension culture. Bars, 100 µm. (D) Graph representation of flow cytometric analysis for PAX6 expression at day 8 of neural induction process. Data are shown as mean ± SEM (n = 3). **, P < 0.005. (E) Immunofluorescence with SOX1 antibody at day 15 of neural induction process (nuclei were labeled with blue-fluorescent DAPI). Bars, 100 µm.
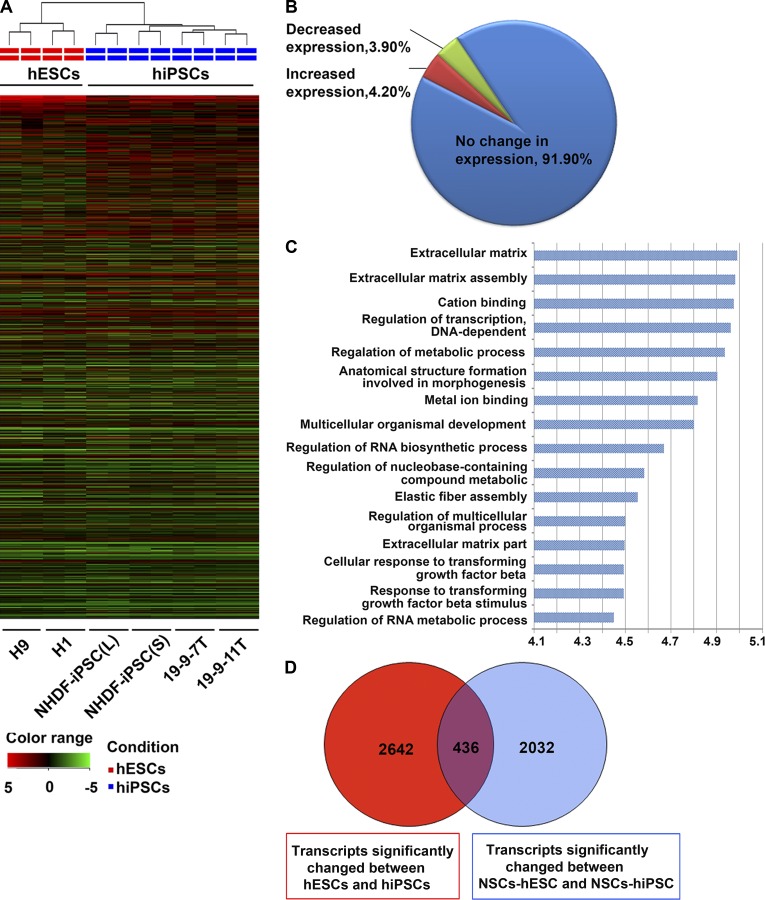
The possibility of a hiPSC-specific defect leading to this observation prompted us to perform transcriptomic analysis of the pluripotent stem cell lines used in this work. Total RNA was extracted from undifferentiated hiPSCs and hESCs and also from NSCs obtained using the monolayer differentiation protocol (Fig. S1, A–D; this protocol was selected because it generates homogenous populations of NSCs) and hybridized to the Agilent SurePrint G3 Human Gene Expression 8 × 60K v2 as described in Materials and methods. We used a cutoff fold change of >1.5 and P < 0.05 to identify differentially expressed genes between hESCs and hiPSCs. 4.2% of transcripts displayed decreased expression in hESCs compared with hiPSCs, and 3.9% showed decreased expression in hiPSCs compared with hESCs (Fig. 2, A and B; Fig. S2 A; and Table S2). Gene Ontology analysis (Genespring software) suggested that the majority of genes differentially expressed between hESCs and hiPSCs were likely to be involved in the synthesis and assembly of components of the extracellular matrix, regulation of transcription, metabolic processes, and embryonic morphogenesis (Fig. 2 C and Table S3).
Figure 2.
Transcriptional profiling demonstrating differences between hiPSCs and hESCs and NSCs derived therefrom. (A) Unsupervised hierarchical clustering of global gene expression data in hESCs and hiPSCs. (B) Global view of gene expression comparison between hESCs and hiPSCs. The array data were filtered using P < 0.05 and fold change >1.5. (C) Gene Ontology analysis of genes with different expression levels in hESCs and hiPSCs. The Gene Ontology terms are noted on the y axis and the log10(p-value) on the x axis. (D) Venn diagram analysis visualizing the overlap between the genes differently expressed in hESCs versus hiPSCs as well as NSCs derived from both sources.
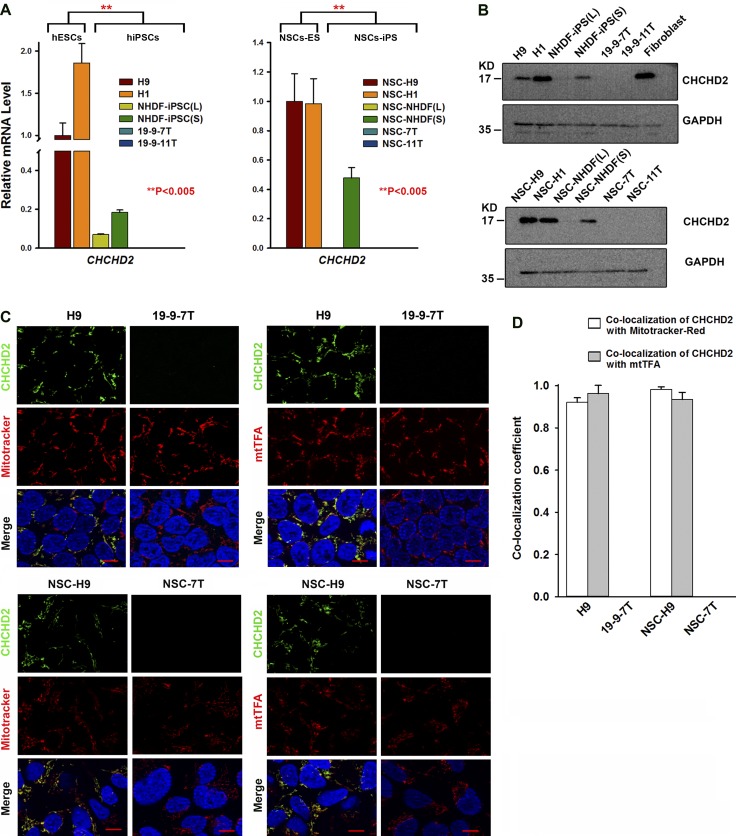
Transcriptomic data obtained from NSCs generated from all pluripotent stem cell lines (Fig. S1 and Fig. S2, B and C) was analyzed similarly. We found that 3.1% of transcripts were significantly increased and 3.4% of transcripts were significantly decreased in NSCs derived from hiPSCs when compared with NSCs derived from hESCs (Table S4). Venn diagram analysis identified 436 common transcripts whose expression significantly differed between hESCs and hiPSCs at both the pluripotent and the NSC stages (Fig. 2 D and Table S5). Several of those highly changed transcripts between hESCs and hiPSCs (DPP6, FAM15A5, TCERG1L, and CTSF) were identified in other recent studies (Ruiz et al., 2012; Huang et al., 2013), and their expression differences in hESCs and hiPSCs were confirmed by quantitative RT-PCR analysis (Fig. S2, D–G). However, one of the significantly changed transcripts identified in our study is CHCHD2, which was previously shown to be expressed in hiPSC-derived neuronal cells in a time-dependent manner (Shimojima et al., 2015). Quantitative RT-PCR analysis of CHCHD2 expression in all hESC and hiPSC lines at the pluripotent and NSC stages (Fig. 3 A) confirmed the array data and indicated that hESC lines express significantly higher levels of this gene compared with the four hiPSC lines included in this study, and this persists in the NSCs obtained from those lines. These findings were further confirmed at the protein level by Western immunoblotting (Fig. 3 B). The coding sequence of CHCHD2 is highly conserved between Homo sapiens, Mus musculus, and Rattus norvegicus (Fig. S2 H); therefore, the possibility that this gene plays a significant and hitherto unknown role in maintenance of pluripotency and differentiation into neuroectodermal cell types was worthy of further investigation.
Figure 3.
CHCHD2 expression in human pluripotent stem cells and NSCs derived therefrom. (A) Quantitative RT-PCR results indicate that CHCHD2 expression is significantly lower in hiPSCs than in hESCs, and this difference is maintained in NSCs. Data are shown as mean ± SEM (n = 3). **, P < 0.005. (B) Western blot analysis of CHCHD2 in human pluripotent stem cells and NSCs derived therefrom. (C) Immunofluorescence with CHCHD2 antibody and MitoTracker red or CHCHD2 and mtTFA antibodies showing CHCHD2 expression in H9 and NSC-H9, but not in 19-9-7T or NSC-7T. Please note that CHCHD2 expression is only localized to the mitochondria of a subset of hESCs and all NSCS derived therefrom (nuclei were labeled with blue-fluorescent DAPI). Bars, 10 µm. (D) Schematic chart of colocalization coefficients between CHCHD2 and MitoTracker red or CHCHD2 and mtTFA in hiPSCs and NSCs derived therefrom. Data are shown as mean ± SEM (n ≥ 4).
We investigated CHCHD2 localization by immunofluorescence in hESC/hiPSC and NSCs derived therefrom in combination with MitoTracker red and mitochondrial transcription factor A (mtTFA; TFAM) to investigate if CHCHD2 expression does indeed localize to mitochondria as suggested (Aras et al., 2015; Fig. 3 C). This indicated that not all hESCs expressed CHCHD2 (Fig. 3 C); however, in cells that did, its localization corresponded perfectly with MitoTracker and mtTFA antibody staining, suggesting a mitochondrial colocalization pattern (Fig. 3, C and D). The hiPSC line, 19-9-7T showed no CHCHD2 expression, corroborating the quantitative RT-PCR and Western immunoblotting data (Fig. 3, A–C). Similarly, all NSCs derived from hESCs (H9) expressed CHCHD2 at the mitochondria (Fig. 3, C and D), whereas a complete lack of CHCHD2 expression was observed in NSCs derived from the 19–7-7T hiPSC line (Fig. 3, C and D).
CHCHD2 and its likely involvement in mitochondrial metabolism, apoptosis, and cell migration
Recent publications have attributed several functions to CHCHD2, including regulation of mitochondrial metabolism (Seo et al., 2010; Zubovych et al., 2010; Aras et al., 2015; Liu et al., 2015). Our group and others have provided evidence that hESCs and hiPSCs rely on the glycolytic pathway and have an underdeveloped mitochondrial network and low mitochondrial activity (Armstrong et al., 2010; Prigione et al., 2010). Given the potential involvement of CHCHD2 in regulation of mitochondrial metabolism and its mitochondrial localization, as well as the differences in expression patterns between hESC and hiPSC lines, we went on to examine several parameters of mitochondrial function as shown in Fig. S3 (A–F). Very few significant differences in mitochondrial complex IV quantity and activity could be detected between hESCs and hiPSCs and the respective NSCs derived therefrom (Fig. S3, A and B), apart from a slight decrease in the rate of oxygen consumption observed in NSCs derived from hiPSCs (Fig. S3, C and D) and a corresponding decrease in the relative levels of ATP (Fig. S3, E and F), very likely because of the absence (or, in some hiPSC lines, such as NHDF-iPSC(S), very low expression) of CHCHD2 in the mitochondria. Similarly, we could detect no significant differences between hESCs and hiPSCs in the expression levels of genes encoded by the mitochondrial genome. The mitochondria of pluripotent stem cells are few and show an immature morphology (Armstrong et al., 2010; Prigione et al., 2010); this was confirmed by transmission electron microscope (TEM) imaging of all pluripotent stem cell lines used in this work (Fig. S3 G). The same analysis also indicated greater numbers of mitochondria in NSCs derived from all pluripotent stem cell lines consistent with the increase in mitochondrial biogenesis during differentiation (Fig. S3 G). Moreover, the mitochondrial morphology NSCs shows more prominent cristae, indicating some degree of maturation but no obvious differences between hESC and hiPSC derived NSCs could be determined (Fig. S3 G). Together, these data suggest that despite showing significant differences in expression of CHCHD2, hESCs and hiPSCs do not show notable differences in mitochondrial activity. Hence, it is unlikely that CHCHD2 function in pluripotent stem cells is linked to a possible function within mitochondria, which is consistent with their glycolytic nature. However, some differences are observed in NSCs derived from pluripotent stem cells with different levels of CHCHD2 expression, which suggests that with the onset of differentiation, a mitochondrial CHCHD2-related function may become more prevalent.
Ectopic expression of CHCHD2 in NIH3T3 fibroblasts has been linked to increased motility after mechanical damage of cell monolayers (Seo et al., 2010). Accordingly, wound-healing studies in NSCs derived from all the hESCs and hiPSCs (Fig. S4 A) indicated reduced NSC migration capacity (Fig. S4 B) in hiPSC-derived NSCs compared with hESC-derived NSCs. Down-regulation of CHCHD2 by RNAi (Fig. S4, C and D) in hESC-derived NSCs resulted in reduced NSC migration capacity (Fig. S4 E). In contrast, transient up-regulation of CHCHD2 in NSCs derived from the NHDF-iPSC(L) hiPSC line (Fig. S4, F and G) resulted in increased migration capacity (Fig. S4 H). Together, these data suggest that reduced CHCHD2 expression in hiPSCs and NSCs derived therefrom may impact cell migration, which could lead to differentiation-related impairments, as cell migration and differentiation are two highly coordinated events during early embryonic development (Friedl and Gilmour, 2009).
Mitochondria-localized CHCHD2 protein can regulate Bax oligomerization and apoptosis by binding to mitochondrial Bcl-xL (Liu et al., 2015). In response to apoptotic stimuli, CHCHD2 expression decreases and loses its mitochondrial localization, which is accompanied by decreased Bcl-xL–Bax interaction and increased Bax homo-oligomerization and Bax–Bak hetero-oligomerization, thus negatively regulating the apoptotic cascade upstream of Bax oligomerization. Accordingly, our results indicate that there are no significant differences in the rate of cell death among all six pluripotent stem cell lines despite variable expression of CHCHD2 (Fig. S4 I). However, upon exposure to cisplatin, which can induce cell death in hESCs and hiPSCs, pluripotent stem cell lines (19-9-7T) lacking CHCHD2 expression showed a higher rates of apoptosis (Fig. S4 J). Down-regulation of CHCHD2 in hESCs by RNAi (Fig. S4, K and L) enhanced cell death upon cisplatin exposure (Fig. S4 M), whereas transient overexpression of CHCHD2 in the hiPSC line 19-9-7T (which lacks CHCHD2 expression) reduced cisplatin-induced cell death (Fig. S4, N–P). Together, these data suggest that CHCHD2 can regulate stress-induced apoptosis in pluripotent stem cell lines.
CHCHD2 expression primes pluripotent stem cells to differentiate toward neuroectodermal lineages
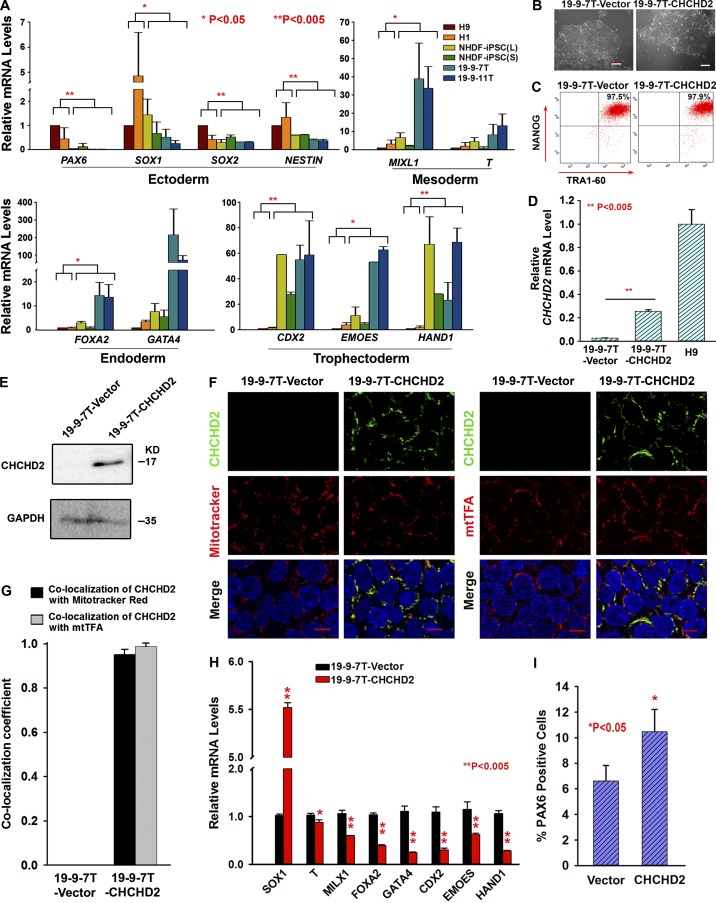
Because NHDF-iPSC(L), NHDF-iPSC(S), 19-9-7T, and 19-9-11T hiPSC lines underexpress CHCHD2 and show reduced capacity to differentiate to NSCs, we determined their potential to generate cells representative of the three embryonic germ layers during EB-mediated differentiation. This relied on quantitative RT-PCR analysis of genes specific to meso-endoderm and mesoderm (MIXL1 and T), endoderm (FOXA2 and GATA4), ectoderm (SOX1, SOX2, NESTIN, and PAX6) and trophectoderm (CDX2, EOMES, and HAND1) and enabled us to assess whether inferior differentiation of hiPSC lines lacking CHCHD2 expression was related to a general differentiation defect or specific to the neuroectodermal lineage only. The morphology of EBs generated from NHDF-iPSC(L), NHDF-iPSC(S), 19-9-7T, and 19-9-11T lines was similar (unpublished data), but significant differences were observed in the relative expression levels of embryonic germ layer markers. Expression of ectodermal markers is higher in EBs derived from hESC lines (Fig. 4 A) correlating with higher expression of CHCHD2 (Fig. 3, A and B). In contrast, hESC-derived EBs show a lower expression of all the other markers (with the exception of T only) typical of mesodermal, endodermal, and trophoectodermal differentiation (Fig. 4 A) when compared with hiPSC-derived EBs. Higher expression of endodermal and mesodermal markers during differentiation of hiPSC with very low or absent expression of CHCHD2 indicates that their differentiation to other lineages is not affected, corroborating previously published data (Jiang et al., 2014). Together, these data suggest that lower expression of CHCHD2 in hiPSCs may be associated with a reduced differentiation capacity of pluripotent stem cell lines to neuroectodermal lineages, which is in turn compensated by differentiation to other embryonic and primitive lineages.
Figure 4.
CHCHD2 and its impact on the spontaneous differentiation of human pluripotent stem cells. (A) Quantitative RT-PCR results with markers of neuroectoderm (PAX6, SOX1, SOX2, and NESTIN), mesoderm (MIXL1 and T), endoderm (FOXA2 and GATA4), and trophectoderm (CDX2, EMOES, and HAND1) at day 6 of differentiation. Data are shown as mean ± SEM (n = 3). *, P < 0.05; **, P < 0.005. (B) Phase-contrast images of 19-9-7T cells stably transfected with vector or CHCHD2 construct. Bars, 200 µm. (C) Representative flow cytometric analysis indicating a high percentage of TRA1-60 and NANOG expression. (D) Quantitative RT-PCR analysis to assess the overexpression of CHCHD2 in CHCHD2 stable overexpression 19-9-7T. Data are shown as mean ± SEM (n = 3). ** P < 0.005. (E) Western blot analysis to assess the overexpression of CHCHD2 in CHCHD2 stable overexpression 19-9-7T. (F) Double staining of CHCHD2 and MitoTracker red or CHCHD2 and mtTFA indicates that CHCHD2 expression is localized to the mitochondria in 19-9-7T-CHCHD2 cells (nuclei were labeled with blue-fluorescent DAPI). Bars, 10 µm. (G) Schematic chart of colocalization coefficient between CHCHD2 and MitoTracker red or CHCHD2 and mtTFA. Data are shown as mean ± SEM (n ≥ 4). (H) Quantitative RT-PCR analysis indicating increased expression of neuroectodermal markers at the expense of other germ lineage markers upon stable overexpression of CHCHD2 in 19-9-7T. Data are shown as mean ± SEM (n = 3). **, P < 0.005. (I) Flow cytometric analysis at day 8 of neural induction indicating increased commitment to neuroectodermal lineages upon overexpression of CHCHD2 in 19-9-7T. Data are shown as mean ± SEM (n = 3). *, P < 0.05.
We investigated the impact of CHCHD2 expression levels on the pluripotent phenotype in more detail by stably transfecting a CHCHD2 expression construct into the hiPSC line with the lowest levels of CHCHD2 expression (19-9-7T; see Materials and methods for more details). This gave several morphologically similar hiPSC clones (Fig. 4, B and C) that expressed significantly more CHCHD2 (assessed by quantitative RT-PCR, Western blot, and immunofluorescence analysis) than control transfected cells (Fig. 4, D–G). Although the level of CHCHD2 expression in the overexpressing hiPSC clone (19-9-7T–CHCHD2) was not as high as that seen in hESCs (Fig. 4 D), this was still able to increase the expression of neuroectodermal markers at the expense of markers delineating embryonic and primitive germ layers (Fig. 4 H) and to enhance differentiation to neuroectodermal lineages (Fig. 4 I).
To address the question of whether defined media conditions could bypass the preferential lineage determination imposed by CHCHD2 expression, hESCs and hiPSCs with high and low levels of CHCHD2 expression were subjected to a monolayer differentiation protocol that gives rise to definitive endodermal progenitors (Rashid et al., 2010; Hannan et al., 2013) characterized by the expression of SOX17 at day 3 as well as a defined EB-based differentiation that gives rise to primitive (CD34+CD43+) and definitive (CD34+CD43−) hematopoietic progenitors at day 6 of differentiation (Kennedy et al., 2012). Both of these defined differentiations indicated that the hiPSC lines without CHCHD2 expression show higher SOX17 expression and a higher percentage of hematopoietic progenitors upon endodermal and hematopoietic differentiation, respectively (Fig. S5, A and B). CHCHD2 overexpression reversed these trends in hiPSC lines with low endogenous CHCHD2 levels. Together with the lower propensity to differentiate toward neuroectodermal lineages (refer to Fig. 1, D and E), these data suggest that expression of CHCHD2 primes pluripotent stem cell differentiation to neuroectodermal lineages at the expense of mesoendodermal and primitive lineages.
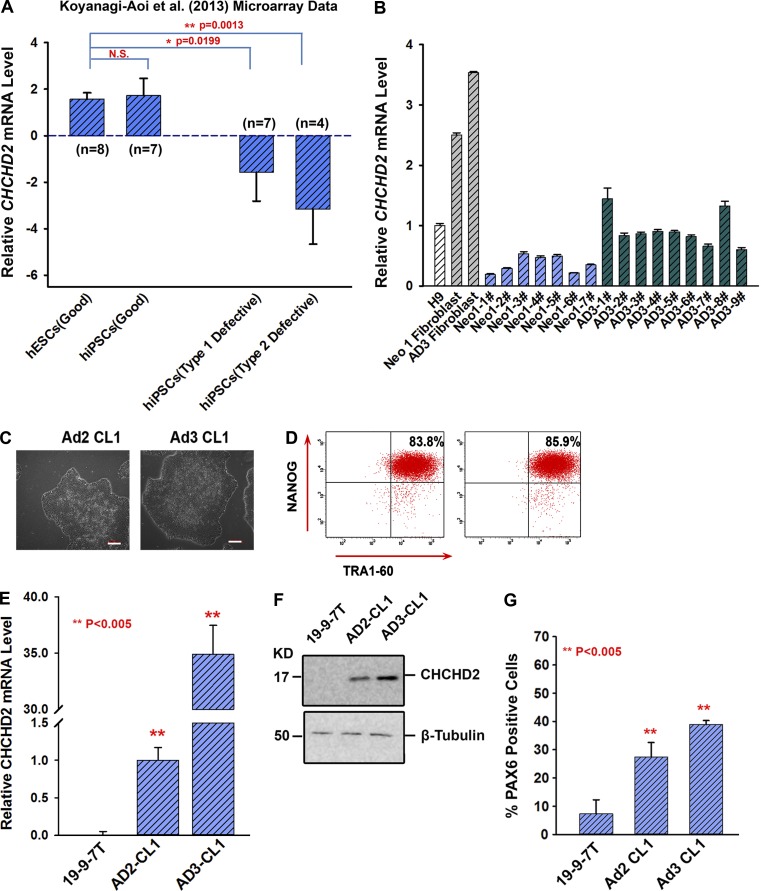
To confirm that our observations were not a laboratory-specific phenomenon, we analyzed CHCHD2 expression in a set of seven hiPSC lines shown to differentiate efficiently to neural lineages and 11 hiPSC lines that were classified either as type I defective (unable to differentiate efficiently to neural lineages and containing OCT4-positive undifferentiated cells in pretransplantation samples) or type II defective (unable to differentiate efficiently to neural lineages, but not containing OCT4-positive cells in pretransplantation samples; Koyanagi-Aoi et al., 2013). Eight hESC lines deemed free of this neural differentiation inability from this study were also included in our analysis (Koyanagi-Aoi et al., 2013). As can be seen in Fig. 5 A, significant differences were found in CHCHD2 expression between hESC and hiPSC lines with good neural differentiation potential and defective hiPSCs that show inefficient neural differentiation. These data suggest that our findings are applicable to hiPSC lines derived, characterized, and differentiated in other laboratories and are not a laboratory-specific phenomenon.
Figure 5.
CHCHD2 expression correlates with the neural differentiation potential of hiPSC derived and characterized in our laboratory as well as others. (A) Large-scale transcriptional data from eight hESCs and 18 hiPSCs reported by Koyanagi-Aoi et al. (2013) to have different potentials to differentiate into neural lineages were analyzed for CHCHD2 expression. Data are shown as mean ± SEM (n = 3). (B) Relative CHCHD2 mRNA levels in H9, neonatal (Neo-1) fibroblast, adult (AD3) fibroblast, and reprogrammed hiPSC clones derived from them (Neo1-1# to 7# and AD3-1# to 9#). Data are shown as mean ± SEM (n = 3). (C) Phase contrast images of hiPSC lines Ad2 CL1 and Ad3 CL1. Bars, 100 µm. (D) Representative FACS analyses showing a high expression of TRA1-60 and NANOG in Ad2 CL1 and Ad3 CL1. (E) Relative expression of CHCHD2 in 19-9-7T, Ad2 CL1, and Ad3 CL1. Data are shown as mean ± SEM (n = 3). **, P < 0.005. (F) Western blot analysis of CHCHD2 in 19-9-7T, Ad2 CL1, and Ad3 CL1 lines. (G) Representative FACS analysis at day 8 of neural induction showing variable proportion of PAX6-positive cells from hiPSCs. Data are shown as mean ± SEM (n = 3). **, P < 0.005.
All four hiPSC lines included in this study generated with three different methods displayed lower CHCHD2 expression, indicating that this is unlikely to be related to the hiPSC derivation method (Fig. 3, A and B). We investigated the hypothesis that low CHCHD2 expression is caused by clonal variations associated with the reprogramming process. To investigate this further, we reprogrammed a human neonatal fibroblast line (Neo 1 fibroblasts; Fig. 5 B) and two human adult dermal fibroblasts (AD2 and AD3; Fig. 5 B). To investigate if the CHCHD2 differential expression pattern was correlated with differential ability to undergo neuroectodermal differentiation, we selected two established hiPSC clones (Ad2 CL1 and Ad3 CL1; Fig. 5 C) derived from two different adult fibroblast samples. After confirming their pluripotent phenotype (Fig. 5 D) and CHCHD2 expression in relation to our lowest-expressing hiPSC line (19-9-7T; Fig. 5, E and F), we subjected them to the same differentiation protocol used for the neural lineages shown in Fig. 1 D and observed that hiPSCs with higher levels of CHCHD2 expression differentiated to neuroectodermal lineages more efficiently (Fig. 5 G). Together, our data suggest that CHCHD2 expression can vary in hiPSC clones; however, expression above a certain threshold seems to correlate with an enhanced ability to undergo neuroectodermal differentiation. Furthermore, differential expression of CHCHD2 within clones derived from the same fibroblast sample reinforces the concept that this is an intraclone variation issue and is not related to genetic heterogeneity of fibroblasts samples from which hiPSCs are derived.
Regulation of CHCHD2 expression during reprogramming
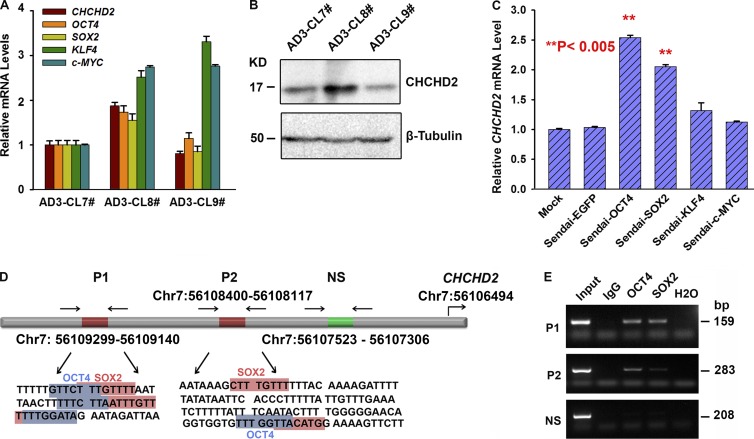
To address the question of how such intraclonal variability is generated with respect to CHCHD2 expression, we analyzed the expression of OCT4, SOX2, KLF4, and c-MYC in three hiPSC clones (AD3: CL7, CL8, and CL9) derived from one adult fibroblast sample (AD3; Fig. 6 A). We observed that higher CHCHD2 expression correlated with higher OCT4 and SOX2 expression (Fig. 6, A and B, note AD3-CL8), suggesting that these two master pluripotency factors may regulate CHCHD2 expression. Furthermore, we performed Sendai virus–based transduction of OCT4, SOX2, KLF4, and c-MYC in the same adult fibroblast sample (AD3) and monitored the expression of CHCHD2 3 d after transduction. Again, we observed increased CHCHD2 expression in samples transduced with OCT4 and SOX2 but no change in KLF4 and c-MYC transduced fibroblasts (Fig. 6 C). Bioinformatic screening found two OCT4 and two SOX2 binding sites in the 3-kb predicted CHCHD2 promoter region (Fig. 6 D). Chromatin immunoprecipitation (ChIP) assays using the H9 hESC line indicated binding of OCT4 to P1 and P2 regions of CHCHD2 promoter (Fig. 6 E). SOX2 showed stronger binding to the P1 region of the CHCHD2 promoter and weaker binding to the P2 region (Fig. 6 E). Control experiments with KLF4 and c-MYC antibodies showed no binding in either of the two selected CHCHD2 promoter regions. Together, these data indicate that OCT4 and SOX2 bind to the CHCHD2 promoter region and regulate its expression. We also provide evidence to support the argument that higher OCT4 and SOX2 expression in selected hiPSC clones results in higher CHCHD2 expression, which further indicates intraclonal variation with respect to their differentiation to neuroectodermal lineages. The high expression of CHCHD2 in fibroblasts compared with clonal hiPSC lines suggests that other factors in addition to OCT4 and SOX2 are likely to regulate CHCHD2 expression at the pluripotent stem cell stage and during their differentiation. This work is currently ongoing in our group with the aim of better understanding the role of this gene in hiPSC biology and cell lineages derived therefrom.
Figure 6.
Variable expression of CHCHD2 in hiPSC clones and regulation of its expression during the reprogramming process. (A) Relative CHCHD2, OCT4, SOX2, KLF4, and c-MYC mRNA levels in AD3-CL7#, AD3-CL8#, AD3-CL9#. Data are shown as mean ± SEM (n = 3). (B) Western blot analysis of CHCHD2 in AD3-CL7#, AD3-CL8#, and AD3-CL9#. (C) Relative CHCHD2 mRNA levels in AD3 fibroblasts and AD3 fibroblasts transduced with Sendai-EGFP, Sendai-OCT4, Sendai-SOX2, Sendai-KLF4, and Sendai-c-MYC for 3 d. Data are shown as mean ± SEM (n = 3). **, P < 0.005. (D) Schematic presentation of CHCHD2 promoter showing the location of the putative OCT4 and SOX2 binding sites, as well as the location of primers used for ChIP. OCT4 and SOX2 binding sites are highlighted in blue and red. NS, nonspecific primers for region without potential OCT4, SOX2 binding sites; P1, P2 specific primers for OCT4, SOX2 potential binding sites. (E) ChIP assays demonstrating the capacity of OCT4 and SOX2 to bind to the CHCHD2 upstream fragment in H9 cells.
CHCHD2 influences the differentiation and survival of human pluripotent stem cells through the TGFβ signaling pathway
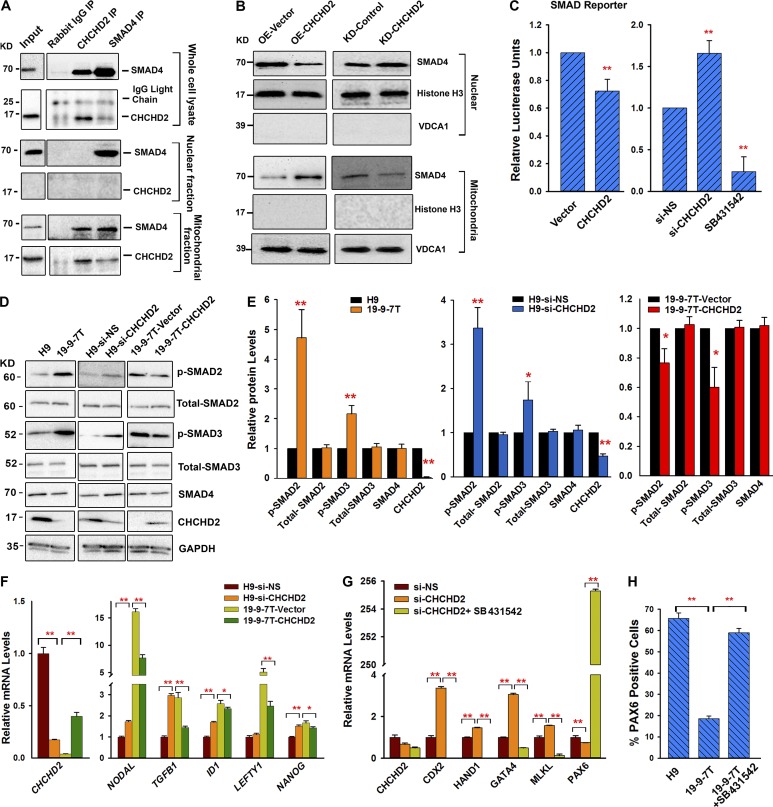
High TGFβ signaling activity has been linked to poor neuroectodermal differentiation (Zhou et al., 2010; Morizane et al., 2011; Pauklin and Vallier, 2013). Given the correlation between lack of CHCHD2 expression and the lower ability to give rise to neuroectodermal derivatives described in this paper, we investigated possible interactions between CHCHD2 and the TGFβ signaling pathway. Previous studies have shown that CHCHD2 and SIRT1 as well as SIRT1 and SMAD 4 (an essential protein involved in signal transduction of the TGFβ signaling pathway) can interact with each other (Law et al., 2009; Chen et al., 2014). We performed coimmunoprecipitation experiments with CHCHD2 and SMAD4 antibodies in lysates of whole cells and nuclear and mitochondrial fractions. These experiments indicated that SMAD4 interacts with CHCHD2 in the mitochondria (Fig. 7 A). Given the interaction between CHCHD2 and SMAD4, we went on to investigate SMAD4 expression using Western immunoblotting of nuclear and mitochondrial extracts of control and respective CHCHD2-overexpressing hiPSC lines (19-9-7T) as well as control and respective knockdown CHCHD2 cell lines. This analysis indicated that SMAD4 is predominantly expressed in the nucleus; however, upon CHCHD2 overexpression, less SMAD4 is found in the nucleus and more in mitochondria. This is further corroborated by CHCHD2 down-regulation experiments, which indicate higher SMAD4 expression in the nucleus and lower expression in the mitochondria, suggesting a shuffling mechanism between the two (Fig. 7 B).
Figure 7.
CHCHD2 represses TGFβ signaling activity. (A) CHCHD2 interacts with SMAD4. CHCHD2 and SMAD4 were immunoprecipitated (IP) and analyzed for the presence of SMAD4 and CHCHD2 by Western blot from the protein lysates of whole cells, as well as nuclear and mitochondrial fractions. (B) CHCHD2 regulates Smad4 intracellular localization. SMAD4 is predominantly expressed in the nucleus; however, upon CHCHD2 overexpression, less SMAD4 is found in the nucleus and more in mitochondria. Upon CHCHD2 knockdown, more SMAD4 is found in the nucleus and less in mitochondria. Nuclear and mitochondrial fractions were isolated from 19-9-7T overexpressed with vector (OE-Vector) or CHCHD2 (OE-CHCHD2) and H9 transfected with si-NS (KD-Control) or si-CHCHD2 (KD-CHCHD2). Histone H3 and VDCA1 were used as nuclear and mitochondrial markers, respectively. (C) SMADs transcriptional activity is repressed by CHCHD2. Cells were cotransfected with the reporter plasmid with the vector or CHCHD2 construct (left) or cotransfected with the reporter plasmid with si-NS or si-CHCHD2 (right). Cells that were transfected with the reporter and incubated with 10 µM SB431542 were used as controls. Data are shown as mean ± SEM (n = 3). **, P < 0.005. (D) Western blot analysis of phosphorylated-SMAD2 (p-SMAD2), total-SMAD2, phosphorylated-SMAD3 (p-SMAD3), total-SMAD3, SMAD4, and CHCHD2 in the cells as indicated. (E) Relative protein expression levels in indicated cells. Data are shown as mean ± SEM (n = 3). *, P < 0.05; **, P < 0.005. (F) Quantitative RT-PCR results of TGFβ-related genes in the cells as indicated. Data are shown as mean ± SEM (n = 3). *, P < 0.05; **, P < 0.005. (G) Quantitative RT-PCR results of markers of neuroectoderm (PAX6), mesoderm (MIXL1), endoderm (GATA4), and trophectoderm (CDX2, HAND1) at day 6 of spontaneous differentiation of H9 cells transfected with si-NS or si-CHCHD2 or si-CHCHD2 with TGFβ inhibitors. Data are shown as mean ± SEM (n = 3). *, P < 0.05; **, P < 0.005. (H) Graph representation of flow cytometric analysis for PAX6 expression at day 8 of the monolayer differentiation process. Data are shown as mean ± SEM (n = 3). **, P < 0.005.
To further prove the interaction between CHCHD2 and TGFβ activity, the Cignal SMAD reporter, which contains tandem repeats of the SMAD transcriptional response element, was transfected into HEK293 cells in which CHCHD2 was transiently overexpressed or down-regulated by RNAi (Fig. 7 C). The specificity of this reporter was tested by adding a TGFβ inhibitor (SB 431542) to the culture media, which resulted in a significant down-regulation of reporter activity as expected. Down-regulation and overexpression of CHCHD2 in these cells caused a respective increase and decrease in the activity of the SMAD reporter (Fig. 7 C), suggesting an inverse link between CHCHD2 expression and TGFβ signaling activity. Accordingly, we also found that the expression of two key phosphorylated receptor-regulated SMADs (pSMAD2 and pSMAD3) was increased in cell lines lacking CHCHD2 expression (19-9-7T) or human ESC lines where CHCHD2 expression was down-regulated by RNAi (Fig. 7, D and E). In contrast, overexpression of CHCHD2 in the 19-9-7T hiPSC line, which lacks endogenous CHCHD2 expression, resulted in a decrease in expression of the phosphorylated SMADs without notable changes in total SMAD2, SMAD3, or SMAD4 expression (Fig. 7, D and E), corroborating findings reported in Fig. 7 C via the TGFβ reporter. Furthermore, expression of five targets of the TGFβ signaling pathway (NODAL, TGFβ1, ID1, LEFTY, and NANOG) showed reciprocal correlation with CHCHD2 expression (up-regulated in response to CHCHD2 down-regulation and down-regulated when CHCHD2 was up-regulated; Fig. 7 F), strongly suggesting a direct link between CHCHD2 expression and the TGFβ pathway activity.
To investigate if we could modulate the impact of CHCHD2 on human pluripotent stem cell differentiation via modulation of TGFβ pathway activity, we repeated the spontaneous differentiation experiment with hESCs in which CHCHD2 expression was down-regulated by RNAi in addition to down-regulation of both CHCHD2 and inhibition of the TGFβ signaling pathway. Although down-regulation of CHCHD2 suppressed differentiation to neuroectodermal lineages and enhanced differentiation to trophoectodermal, endodermal, and mesodermal lineages (Fig. 7 G), combined inhibition of CHCHD2 with TGFβ signaling pathway reversed these impacts (Fig. 7 G), suggesting that the TGFβ pathway acts downstream of CHCHD2. Similar results were obtained when TGFβ inhibition was applied during directed differentiation of human iPSC that lack CHCHD2 expression (Fig. 7 H), previously shown to be deficient at differentiating toward neuroectodermal lineages (Fig. 1, D and E). Furthermore, the reduction in cell survival caused by the application of cisplatin in hiPSC lines lacking CHCHD2 expression was rescued by inhibition of the TGFβ signaling pathway (Fig. S5 C), suggesting that the interaction between TGFβ signaling and CHCHD2 is important not only for the early differentiation of human pluripotent stem cells but also for their survival in culture.
Discussion
Through transcriptional comparison of two hESC and four hiPSC lines generated using three different methods, we have shown that hiPSCs are inferior in their ability to differentiate to neuroectodermal lineages, and we have identified a new marker, CHCHD2, whose expression differs significantly between hESC and hiPSC lines. Low CHCHD2 expression in hiPSC seems unrelated to the method of derivation (lentiviral, Sendai, or episomal vectors) and is not a laboratory-specific phenomenon because the data of other groups confirm our findings (Koyanagi-Aoi et al., 2013). Our data suggest that differential expression of CHCHD2 is caused by clonal variations in the expression of OCT4 and SOX2, which bind to the CHCHD2 promoter and activate its expression in a clone-dependent manner. CHCHD2 is able to interact directly with the TGFβ pathway by binding Smad4 in the mitochondria. This causes a reduction in nuclear location of SMAD4, which results in suppression of TGFβ activity and enhanced differentiation toward neuroectodermal lineages.
To date, there is paucity of information about the functions of CHCHD2 (in particular, its possible contribution to the maintenance of pluripotency), yet it is highly conserved among human, mouse, and rat, suggesting an important role for CHCHD2 (Aras et al., 2015). Most published studies suggest that CHCHD2 is a regulator of mitochondrial metabolism active in both the mitochondria and the nucleus. In the former, it regulates the activity of cytochrome c oxidase, whereas in the nucleus, it stimulates transcription of a subset of genes that includes COX4I2 (and itself under stress conditions). CHCHD2 knockdown results in reduced mitochondrial complex IV activity and increased reactive oxygen species production (Aras et al., 2013, 2015). Our studies indicate that although CHCHD2 localizes to the mitochondria, nuclear localization is not observed even when mitochondrial entry of CHCHD2 is prevented by application of inhibitors such as Mito Block 6 (Aras et al., 2015) in both hiPSCs and NSCs derived therefrom (unpublished data). What CHCHD2 does in the mitochondria is less clear. Although mutations in nonmammalian CHCHD2 homologues, such as HAR-1 in Caenorhabditis elegans, disrupt mitochondrial network formation and impair ATP production (Zubovych et al., 2010), we might anticipate a similar response to the lower levels of CHCHD2 expression in hiPSCs, but despite differential expression of CHCHD2 between the hESC and hiPSC lines used in this study, we observed no significant differences in mitochondrial complex IV quantity and activity or relative ATP levels and oxygen consumption rates (Fig. S3, A–F). This is not unexpected, because hESCs and hiPSCs have immature mitochondria and rely heavily upon glycolysis for ATP generation (Armstrong et al., 2010; Bukowiecki et al., 2014); however, differentiation of pluripotent stem cells into cell types with higher ATP demand is accompanied by an increase in mitochondrial numbers and an increase in ATP synthesis by oxidative phosphorylation (Facucho-Oliveira et al., 2007; Yanes et al., 2010). In accordance with this, we found that NSCs derived from hiPSC lines (with lower CHCHD2 expression) showed a reduced oxygen respiration rate and reduced relative ATP levels, but again, this was modest when compared with hESC-derived NSCs (Fig. S3, A–F). Such NSCs have higher mitochondrial mass (Fig. S3 G) but lower complex IV activity (Fig. S3 A). The higher mitochondrial mass relative to the amount of complex IV protein may reflect the need for other mitochondrial functions. Neural progenitor cells tend to still rely on glycolysis (Gershon et al., 2013), because their normal niche in the brain is quite hypoxic, so there is little need for mitochondrial respiratory complexes. NSCs might need the larger numbers of mitochondria because they use a lot more fatty acid metabolism such as mitochondrial fatty acid β oxidation (Knobloch et al., 2013). A similar requirement for fatty acid oxidation might be common to many types of adult stem cells, particularly during quiescence (Ito et al., 2012); however, we cannot be absolutely sure the same applies to our hiPSC-derived NSCs. It has already been shown that hESC-derived NSCs have a lower ATP demand and reduced mitochondrial activity when compared with hESCs (Birket et al., 2011), which indicates that the presence of a higher number of mitochondria does not guarantee a higher complex IV quantity and activity. Nevertheless, this prompted us to investigate mitochondrial parameters that could lead to reduced ATP synthesis, such as mitochondrial membrane potential, but we did not observe significant differences in membrane potential between hESCs and hiPSCs and NSCs derived therefrom, nor did we observe increased production of reactive oxygen species (a hallmark of defective oxidative phosphorylation) when hESCs were compared with hiPSCs or hESC-derived NSCs were compared with hiPSC-derived NSCs (unpublished data). In view of this, it is unlikely that subtle differences in mitochondrial respiration function observed between hESC- and hiPSC-derived NSCs are the cause of the impaired neuroectodermal differentiation observed as consequence of lower CHCHD2 expression in hiPSCs.
The precise mechanism by which variable levels of CHCHD2 expression dictate the corresponding variability in neuroectodermal differentiation capacity prompted us to investigate the impact of this gene upon the TGFβ signaling pathway. High TGFβ activity negatively impacts the ability of hESCs and hiPSCs to differentiate toward neuroectoderm (Zhou et al., 2010; Morizane et al., 2011; Pauklin and Vallier, 2013), and many defined differentiation protocols for this lineage involve dual SMAD inhibition, which enables the pluripotent stem cells to progress with differentiation (Chambers et al., 2009; Zhou et al., 2010; Pauklin and Vallier, 2013). Interestingly, we discovered that CHCHD2 interacts directly with SMAD4, a co-Smad, capable of subsequent binding to the receptor regulated Smads, resulting in a complex that enters the nucleus, where it acts as a transcription factor for various target genes of the TGFβ signaling pathway. Smad4 has been shown to translocate to mitochondria, where it associates with mitochondrial protein cytochrome c oxidase subunit II to regulate the apoptotic response (Pang et al., 2011). Furthermore, CHCHD2 also has been shown to bind and regulate cytochrome c oxidase activity in 293 cells (Aras et al., 2015); nevertheless, a direct interaction between CHCHD2 and SMAD4 as reported herein has not been reported previously. As shown here, some hESC and hiPSC lines are able to undergo differentiation to neuroectoderm without the additional need for TGFβ inhibition, suggesting different levels of activity for this pathway across hESC and hiPSC lines. This concurs with differential endogenous expression of CHCHD2 and the ability to give rise to neuroectoderm across human pluripotent stem cell lines as reported herein and leads us to hypothesize that hESC and hiPSC cell lines with endogenous CHCHD2 expression are able to self-regulate TGFβ signaling activity via the CHCHD2–SMAD4 interaction. This is backed up by our experimental data, which indicate that in the presence of CHCHD2, SMAD4 nuclear expression is lower, whereas its mitochondrial expression is higher. Together, these data suggest that CHCHD2 sequesters SMAD4 to the mitochondria via direct protein binding and makes it less available to enter the nucleus to activate the TGFβ signaling targets. Accordingly, we have shown that hiPSC lines with absent or reduced expression of CHCHD2 have higher expression of phosphorylated receptor-regulated Smads, nuclear Smad4, and TGFβ signaling targets and higher TGFβ activity, whereas the opposite is true for pluripotent stem cell lines with endogenous CHCHD2 expression, strongly suggesting a direct inverse relationship between CHCHD2 and TGFβ pathway activity in pluripotent stem cells. This is reinforced by inhibition of TGFβ pathway activity, which is able to rescue the neuroectodermal differentiation ability of cell lines with absent CHCHD2 expression and their response to apoptotic stimuli, providing further evidence that CHCHD2 most likely controls the neuroectodermal differentiation of hESCs and hiPSCs via regulation of TGFβ pathway activity.
Although we report here a novel interaction between CHCHD2 and SMAD4, is still possible that CHCHD2 regulates pluripotent stem cell differentiation through other interacting factors. CHCHD2 has been suggested to interact with YBX1 (Wei et al., 2015), which has recently been shown to play a role in positively affecting the expression of Nanog and other pluripotency-related genes (Guo et al., 2016). Particularly, down-regulation of YBX1 results in high expression of mesoderm markers (Guo et al., 2016). In addition, YBX-1 binds to the SOX2 promoter and down-regulates its expression in MCF7 and ZR751 (Jung et al., 2014). In addition, YBX-1 can interact physically and functionally with CTCF, which is a pluripotency factor in hESCs (Chernukhin et al., 2000; Balakrishnan et al., 2012). Although the CHCHD2 and YBX1 interaction needs to be confirmed in human pluripotent stem cells, these findings raise the tantalizing possibilities that CHCHD2 impacts differentiation and that differentiation could also be dependent on YBX1 or novel, yet-unidentified interacting proteins.
In conclusion, our study underlines the incomplete understanding of the mechanisms by which somatic cell–induced reprogramming affects the ability of hiPSCs to undergo neuroectodermal differentiation. Although a few papers have reported differences in the ability of pluripotent stem cells to undergo directed differentiation to various lineages, this is a relatively unexplored territory and very important for comparing hiPSC lines and selecting the most appropriate hiPSC lines for future cell banking and regenerative purposes (Hu et al., 2010; Zimmermann et al., 2012; Solomon et al., 2015). Multilineage differentiation (including differentiation to ectodermal cells) is generally accepted as an indicator of hiPSC line “quality,” but markers of such quality and methods to improve this characteristic are incompletely met needs. Much progress has been made toward the establishment of “naive” or “ground state” pluripotency, but the data highlighted in this current study suggest the involvement of the recently described gene CHCHD2 in several molecular processes that contribute to induced pluripotency and the ability of hiPSC clones to undergo neuroectodermal differentiation with similar efficiency to hESCs. In view of this, we propose that CHCHD2 expression is a valuable indicator of hiPSC quality that can be performed readily as a measure of how successful the expanded hiPSC clones are in their ability to undergo neuroectodermal differentiation. It is clear that future work needs to address the possibly multiple mechanisms through which CHCHD2 may interact with other systems to contribute to the differentiation potential of pluripotent stem cells.
Materials and methods
hPSC culture
All hESC and hiPSC lines were maintained on plates coated with Matrigel (growth factor reduced; BD) with mTeSR1 (STEMCELL Technologies) at 37°C, 5% CO2, and 21% O2 according to WiCell Inc. protocols. Cells were passaged every 4–5 d at ∼80% confluence by using 0.02% EDTA (Versene). Colonies containing clearly visible differentiated cells were manually removed before further passaging.
NSCs culture
NSCs were maintained on flasks coated with poly-l-ornithine/laminin (Sigma-Aldrich) with N2B27 medium containing DMEM/F12, N2 supplement (1:100, Invitrogen), B27 supplement (1:50, Invitrogen), 100 mM nonessential amino acids (Invitrogen), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) plus 10 ng/ml FGF2 (R&D Systems) and 10 ng/ml EGF (R&D Systems). The cells were passaged every 4–6 d at ∼90% confluence by using Accutase. All NSCs used in this study were between passages 6 and 10.
EB-based neural differentiation from hESCs and hiPSCs
The procedure used for neural induction of hPSCs was based on a previously described protocol with a minor modification (Hu et al., 2010). In brief, on day 0, hPSCs were enzymatically detached by collagenase IV and dispase treatment and then dissociated into small clumps and cultured in suspension with mTeSR1 in a low-attachment plate for 24 h. From day 1, the aggregates were transferred into differentiation medium (KO-DMEM; Invitrogen), 2 mM l-glutamine (Invitrogen), 100 mM nonessential amino acids (Invitrogen), and 20% serum replacement (Invitrogen) for 6 d. The aggregates were then adhered to plates coated with Matrigel (growth factor reduced; BD) in neural induction medium consisting of DMEF/F12, N2 supplement, and 2 µg/ml heparin as detailed previously (Hu et al., 2010).
Gene expression analysis
The SurePrint G3 Human Gene Expression 8 × 60K v2 Microarray kit from Agilent were used for the gene expression assay of each cell line with two biological replicates. RNA samples were prepared from by ReliaPrep RNA Cell Minprep System (Promega). Data were analyzed using Genespring software.
Accession numbers
Microarray data have been submitted to the Gene Expression Omnibus and are available under accession number GSE67325.
Flow cytometry analysis
Whole cultures (adherent cells or suspension aggregates) were dissociated by treatment with Accutase (Invitrogen) for 3 min and analyzed for expression of various neural and pluripotency markers by flow cytometric analysis, which was performed with a FACSCaliber (BD). Data were analyzed with CellQuest Pro (BD) as described in the manufacturer's instructions. At least 10,000 events were analyzed in each replicate. Antibodies used for FACS were FITC-conjugated TRA-1-60 (1:200, FCMAB115F; EMD Millipore), NANOG conjugated with Alexa Fluor 647 (D73G4, 1:200; Cell Signaling Technology), PAX6 (PRB-278P, 1:100; Covance), NESTIN (MAB5326, 1:200; EMD Millipore), anti–rabbit IgG-FITC (Sigma-Aldrich), and anti–mouse IgG-FITC (Sigma-Aldrich).
RNA extraction, reverse transcription, and quantitative RT-PCR
Total RNA isolation was performed using the ReliaPrep RNA Cell Minprep System (Promega). RNA quality was evaluated using the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and reverse transcription was performed using the GoScript Reverse Transcription System. Quantitative RT-PCR was performed using the QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific) and the GoTaq qPCR Master (Promega) according to the manufacturer’s instructions. Primer sequences used for quantitative RT-PCR are provided in Table S1.
Western blotting
30 µg protein from whole-cell extracts or cellular fraction extracts was used for Western blotting analysis. Antibodies used for Western blotting were CHCHD2 (HPA027407, 1:200; Sigma-Aldrich) and GAPDH (G9545, 1:2,000; Sigma-Aldrich), SIRT1 (2493, 1:1,000; Cell Signaling Technology), SMAD4 (9515, 1:1,000; Cell Signaling Technology), p-SMAD2 (3108, 1:1,000; Cell Signaling Technology), SMAD2 (5339, 1:1,000; Cell Signaling Technology), p-SMAD3 (9520, 1:1,000; Cell Signaling Technology), SMAD3 (9523, 1:1,000; Cell Signaling Technology), histone H3 (ab1791, 1:3,000; Abcam), VDAC1 (ab15895, 1:1,000; Abcam), and β-tubulin (T4026, 1:800; Sigma-Aldrich). Quantification of Western blotting results was performed using ImageJ software.
Immunofluorescence staining
In brief, cells were fixed in 4% PFA (Sigma-Aldrich) for 15 min, followed by a 10-min permeabilization step (0.1% Triton X-100 in PBS for internal cell markers). The blocking step was performed by incubation in 2% BSA for 30 min. Cells were incubated with primary antibodies at 4°C overnight and further incubated with secondary antibodies for 1 h. For MitoTracker dye staining, cells were cultured in the normal media with 150 nM MitoTracker red (Invitrogen) for 30 min before further fixation and immunostaining. Antibodies against the following proteins were used at the indicated dilutions: CHCHD2 (HPA027407, 1:200; Sigma-Aldrich), SOX1 (4194S, 1:200; Cell Signaling Technology), NESTIN (MAB5326, 1:200; EMD Millipore), TUJ1 (MRB-435P, 1:500; Covance), mtTFA (ab119684, 1:500; Abcam), anti–mouse IgG-FITC (1:800; Sigma-Aldrich), and anti–rabbit IgG-Cy3 (1:1,000; Jackson ImmunoResearch Laboratories, Inc.). Nuclei were labeled with DAPI (Thermo Fisher Scientific). Colocalization coefficient studies were performed using ImageJ software by calculating Manders’ colocalization coefficient, which describes the amount of colocalizing pixels of GFP using pixels generated by RFP.
Mitochondrial complex IV quantity and activity
Mitoprofile Human Complex IV Activity and Quantity from Mitosciences (Invitrogen) was used according to the manufacturer’s instructions for the determination of complex IV activity and levels using 40 µg protein per well in at least two independent measurements in quadruplicate. A Multiskan Ascent 96 microplate instrument from Thermo Fisher Scientific was used for analysis.
Measurement of respiration rates using a Seahorse XFe-96 analyser
Respiration rates were measured on adherent cells using a Seahorse XFe-96 analyzer (Seahorse Bioscience) according to the manufacturer’s instructions. The assay plates were coated with Matrigel (growth factor reduced; BD). Cells were seeded at ∼36 h (for hPSCs) or 24 h (for NSCs) before measurement in their normal growth medium. hESCs and hiPSCs were seeded at 4 × 104 cells per well as single cells (with ROCK inhibitor for the first 6 h and then washed out twice by PBS), and NSCs were seeded as single cells totaling 5 × 104 cells per well. The assay was performed in bicarbonate-free DMEM (Sigma-Aldrich) with 15 mM glucose, 2 mM l-glutamine, and 0.5 mM sodium pyruvate; media was warmed to 37°C and the pH set to 7.4. Cells were washed twice and preincubated in the medium for 1 h before measurement. Oligomycin was used at 1.5 µM, FCCP was added in two injections of 0.3 µM to 0.5 µM. Rotenone and antimycin A was added at 1 µM and 2 µM. Oxygen consumption values were measured three times after each injection. After the assay, a standard protein assay was performed. The oxygen consumption rate was normalized to protein levels.
ATP measurement
ATP levels were measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions. In brief, 7 × 104 hPSCs (with 10 µM ROCK inhibitor for the first 6 h, and then washed out twice by PBS) or NSCs as single cells were seeded 18 h (for hPSCs) or 12 h (for NSCs) in their culture media before the measurement. Cells were then washed twice with warm PBS and incubated for 6 h in the measurement medium with 5 mM glucose plus 2 mM pyruvate, 5 mM glucose plus 2 mM pyruvate plus 1.5 µM oligomycin (glycolytic ATP generation), 5 mM 2-deoxy-d-glucose plus 2 mM pyruvate (oxidative ATP production), or 5 mM 2-deoxy-d-glucose plus 2 mM pyruvate plus 1.5 µM oligomycin. After incubation, cells were lysed, and lysates were incubated with the luciferin/luciferase reagents. Samples were measured using a NovoStar MBG Labtech microplate luminometer and the results referred to the cell viability by neutral red measurement (Sigma-Aldrich).
Wound-healing study
For wound-healing migration assays, NSCs were seeded onto poly-l-ornithine/laminin–coated six-well plates at a density of 7 × 105 cells per well. 16 h after seeding, confluent cells were scratched by a fine pipette tip, washed with warmed PBS, and incubated in their normal growth media. After 0 and 24 h, pictures were taken under a phase microscope. Cell migration distance was determined by measuring the wound width. The relative recovery rate was calculated as: [(initial wound width − wound width at the time of measurement)/initial wound width] × 100%.
Nondirected EB differentiation
In brief, on day 0, hPSCs were enzymatically detached by collagenase IV and dispase treatment, dissociated into small clumps, and cultured in suspension with mTeSR1 in a low-attachment plate for 24 h. Aggregates (EBs) were then transferred into the differentiation media (KO-DMEM, 2 mM l-glutamine, 100 mM nonessential amino acids, and 20% knockout serum replacement; Invitrogen). The medium was changed every 2 d.
Stable cell lines
The CHCHD2 full coding sequences were isolated from cDNA generated from the H9 hESC line using the following oligonucleotides: CHCHD2 forward, 5′-GTCGCTTAGCTCTTCGGTGG-3′; CHCHD2 reverse, 5′-TACAGAGTAGGGACACCCCC-3′. The full-length fragment was ligated into the pCAG-IP vector. hiPSCs were transfected with control empty vector or CHCHD2 construct by Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. 2 d after transfection, stable clones were selected by using puromycin (0.5–1.0 µg/ml) for 7 d. Positive clones were expanded in mTeSR1 with 0.5 µg/ml puromycin and labeled as 19-9-7T vector or 19-9-7T-CHCHD2. The expression level of CHCHD2 in stable cell lines was assessed by quantitative RT-PCR, Western blotting, and immunostaining.
Fibroblast reprogramming
Fibroblast cells were reprogrammed using the CytoTune-iPS 2.0 Sendai Reprogramming kit (Invitrogen) according to the manufacturer’s instructions. In brief, fibroblast cells were seeded at 105 cells per well. 24 h later, cells were then transfected with Sendai virus from the Cytotune 1 kit (Thermo Fisher Scientific). 7 d after transfection, cells were dissociated into single cells by trypsinization (0.05% trypsin; Gibco) and then plated onto Matrigel-coated plates with mTeSR1 media at a density of 2 × 104 ∼ 1 × 105 cells per well of a six-well plate. The media was changed every day until day 28, and pluripotent colonies were mechanically transferred to a four-well plate for further expansion and characterization.
ChIP analysis
ChIP analysis was performed using the ChIP-IT Express Chromatin Immunoprecipitation kit (Active Motif) according to the manufacturer’s instructions. Five percent of the total genomic DNA from the nuclear extract was used as the input. The primers used for this procedure are provided in Table S1. The antibodies used for ChIP were OCT4 (39811; Active Motif), SOX2 (39843; Active Motif), and rabbit IgG (AB-105-C; R&D Systems).
Monolayer neural differentiation
The monolayer protocol was modified from previous published protocols with minor modifications. In brief, we used a combination of three inhibitors (1 µM compound C; Sigma-Aldrich), together with 500 ng/ml rmNoggin and 10 μM SB431542 (Sigma-Aldrich) to induce neural differentiation At day 14, the cells were picked as clumps, and plated onto a poly-l-ornithine/laminin–coated plate with N2 medium. When the polarized cells were visible, the cells were dissociated into single cells using Accutase and expanded in the N2B27 medium plus 10 ng/ml basic FGF and 10 ng/ml EGF.
Neurosphere-forming assay
NSCs were dissociated to single cells using 0.05% trypsin and cultured in suspension conditions. Neurosphere formation was observed from day 2 or day 3 in NSC culture media supplemented with basic FGF and EGF. When basic FGF and EGF were withdrawn, NSCs spontaneously differentiated into neuronal cells. Differentiated spheres were attached onto coverslips coated with Matrigel for immunofluorescence staining.
Transmission electron microscopy
Cells were collected and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, for 20 min at room temperature. After being rinsed for 10 min in the same buffer, cells were postfixed with 1% OsO4 in 0.04 M phosphate buffer containing 0.14 M sucrose for 10 min at 4°C. After dehydration with a standard ethanol series and infiltration with epoxy resin, cells were transferred to beam capsules for polymerization in the oven. The capsules were separated from the polymerized resin with a razor blade, and embedded cells in hardened blocks were viewed with an optical microscope so that the appropriate area was chosen for ultrathin sectioning. Subsequently, ultrathin sections were obtained using an ultramicrotome (Sorvall MT-6000; DuPont) with a diamond knife. Heavy metal staining was done with 4% uranyl acetate and lead citrate, and the samples were examined through the electron microscope (H-7100; Hitachi) at 50 kV.
Cell transfection
hPSCs or NSCs were transfected with vector or CHCHD2 construct using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. hPSCs or NSCs were transfected with si-NS or si-CHCHD2 using Lipofectamine RNAiMAX Transfection Reagent according to the manufacturer’s instructions.
Apoptosis assay
Apoptosis was quantified by Annexin V/FITC and phosphoinositide double staining by using the FITC Annexin V Apoptosis Detection kit I (BD, 556547) according to the manufacturer’s instructions. To induce apoptosis, cells were cultured in the normal medium with 1 µg/ml cisplatin for 18 h.
Endoderm differentiation
The procedure for definitive endoderm differentiation of hPSCs was based on a previously described protocol (Rashid et al., 2010; Hannan et al., 2013). In brief, for initiation of definitive endoderm differentiation, hPSCs were detached with EDTA (15575–020; Thermo Fisher Scientific), filtered through a 70-µm filter (352350; Corning) and seeded as small clumps in E8 media with 10 µM Y27632 (Y0503; Sigma-Aldrich) on gelatin-coated plates preincubated overnight with mouse embryonic fibroblast media. Differentiation was started 48 h later by changing media to day 1 media, consisting of CDM-PVA media + 100 ng/ml Activin A + 80 ng/ml FGF2 + 3 µM CHIR99021 (CT99021; Stratech Scientific) + 10 µM LY (V1201; Promega) + 10 ng/ml BMP4 (R&D Systems), followed by day 2 media, consisting of CDM-PVA media + 100 ng/ml Activin A + 80 ng/ml FGF2 + 10 µM LY + 10 ng/ml BMP4, and day 3 media, consisting of RPMI + 2% B27 + 100 ng/ml ActiviNa + 80 ng/ml FGF2.
Mesoderm differentiation
Before differentiation, cell lines were cultured on recombinant human vitronectin (A14700; Thermo Fisher Scientific) in StemPro hESC SFM medium (A1000701; Thermo Fisher Scientific). To generate uniform in shape and size EBs, hPSCs colonies were manually cut and scraped by using the StemPro EZPassage Stem Cell Tool (23181–010; Thermo Fisher Scientific). EBs were cultured in media supplemented with recombinant factors as previously described (Kennedy et al., 2012). All recombinant factors are human and purchased from PeproTech. Analysis of expression of KDR, CD34, and CD43 at the indicated time points was performed by flow cytometry using KDR-PE (560494; BD), CD34-APC (555824; BD), and CD43-FITC (MHCD4301; Thermo Fisher Scientific) antibodies. Analyses were performed by gating on single cells using forward scatter height versus area and followed by gating on live cells and lack of DAPI uptake. Stained cells were analyzed using LSR II (BD), and data analysis was performed using BD FACSDiva software (BD).
Coimmunoprecipitation
For coimmunoprecipitation, cells were washed with cold PBS three times. Cell lysates were prepared in coimmunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 2 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail; Roche), and protein concentration was measured using Bradford reagent (Bio-Rad Laboratories). Protein A/G PLUS agarose (sc-2003; Santa Cruz Biotechnology, Inc.) beads were washed three times with the coimmunoprecipitation buffer. 800 µg protein recovered from cell supernatants was precleared with 20 µl bead slurry and 2 µg rabbit IgG for at least 2 h on a rotor at 4°C. The beads were removed by centrifugation at 2,500 g for 5 min at 4°C. Immunoprecipitation was performed by overnight incubation and rotation with the precleared cell lysate, 20 µl beads, 2 µg of rabbit anti-CHCHD2 antibody (HPA027407; Sigma-Aldrich), rabbit anti-SMAD4 antibody (ab208804; Abcam), rabbit anti-SIRT1 antibody (2493; Cell Signaling Technology), or rabbit IgG. The beads with bound protein complexes were recovered by centrifugation at 2,500 g for 5 min and washed three times with coimmunoprecipitation buffer. The samples from immunoprecipitation were examined using Western blot analysis.
Cellular subfractionation
Mitochondria were isolated from cells with the Mitochondrial Isolation kit (89874; Thermo Fisher Scientific) according to the manufacturer’s protocol. The nuclear fraction was obtained by low-speed centrifugation, and the mitochondrial fraction was obtained after high-speed centrifugation of the nuclear supernatant.
Luciferase assays
Cells were seeded at a density of 105 per well into 24-well plates 24 h before transfection. To examine the effect of CHCHD2 on the SMAD reporter, control vector, CHCHD2 expression plasmids, si-NS, or si-CHCHD2 was cotransfected with the reporter (Cignal SMAD Reporter, CCS-017L; QIAGEN) by Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions. Cell extracts were prepared 48 h after transfection. Luciferase activity was evaluated with a Dual-Luciferase Assay System (Promega) according to the manufacturer’s recommendations.
Microscopy
Cell morphology was visualized using a microscope (Diaphot 300; Nikon) with the following objectives: 10× NA 0.25 and 20× NA 0.40 at room temperature. Digital images were recorded using a digital camera (DXM1200; Nikon). Fluorescence sections were visualized using a microscope (Axio Imager Z1; Nikon) with the following objectives: 10× NA 0.25, 20× NA 0.40, and 40× NA 1.3 at room temperature. Digital images were recorded using a digital camera (ApotomeCam; Nikon).
Statistical analysis
All values are shown as means ± SEM (n = 3). Statistical significance was assessed using Student's t test. For all statistical tests, the 0.05 confidence level was considered statistically significant. In all figures, * denotes P < 0.05, ** denotes P < 0.005, and *** denotes P < 0.001 in an unpaired Student's t test.
Online supplemental material
Fig. S1 shows NSCs generated from hESCs and hiPSCs using a monolayer protocol. Fig. S2 shows the gene expression study in hESCs and hiPSCs and NSCs derived therefrom. Fig. S3 shows CHCHD2 and its likely involvement in mitochondrial metabolism. Fig. S4 shows CHCHD2 and its likely involvement in cell migration and apoptosis. Fig. S5 shows the mesodermal and endodermal differentiation of human pluripotent stem cells; CHCHD2 protects hESC and hiPSC from apoptosis upon application of proapoptotic stimuli by regulating TGFβ. Table S1 lists the oligonucleotides used for quantitative RT-PCR analysis and ChIP. Table S2 lists the genes showing differential expression between the hESCs and hiPSCs included in this study. Table S3 shows Gene Ontology analysis of transcripts showing significant differences in expression levels between the hESCs and hiPSCs included in this study. Table S4 lists the genes showing differential expression between hESC- and hiPSC-derived NSCs included in this study. Table S5 lists common transcripts whose expression significantly differed in hESCs and hiPSCs at both the pluripotent and NSC stages. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201601061.dv.
Supplementary Material
Acknowledgments
The authors thank Dr. Kathryn White for help with the transmission electron microscopy analysis.
This work was funded by grants from the Biotechnology and Biological Sciences Research Council UK (BB/I020209/1) and European Research Council (614620).
The authors declare no competing financial interests.
Author contributions: L. Zhu designed and performed research, analyzed data, and contributed to manuscript writing and final approval. A. Gomez-Duran, G. Saretzki, S. Jin, K. Tilgner, D. Melguzo-Sanchis, G. Anyfantis, L. Vallier, J. Al-Aama performed some of the research and data analysis and gave final approval of the manuscript. P. Chinnery designed research, gave final approval of the manuscript, and contributed to fundraising. M. Lako and L. Armstrong designed and performed research and data analysis, wrote the paper, gave final approval of the manuscript, and contributed to fundraising.
Footnotes
Abbreviations used:
- EB
- embryoid body
- hESC
- human embryonic stem cell
- hiPSC
- human induced pluripotent stem cell
- iPSC
- induced pluripotent stem cell
- mtTFA
- mitochondrial transcription factor A
- NSC
- neural stem cell
References
- Aras S., Pak O., Sommer N., Finley R. Jr., Hüttemann M., Weissmann N., and Grossman L.I.. 2013. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res. 41:2255–2266. 10.1093/nar/gks1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aras S., Bai M., Lee I., Springett R., Hüttemann M., and Grossman L.I.. 2015. MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion. 20:43–51. 10.1016/j.mito.2014.10.003 [DOI] [PubMed] [Google Scholar]
- Armstrong L., Tilgner K., Saretzki G., Atkinson S.P., Stojkovic M., Moreno R., Przyborski S., and Lako M.. 2010. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 28:661–673. 10.1002/stem.307 [DOI] [PubMed] [Google Scholar]
- Balakrishnan S.K., Witcher M., Berggren T.W., and Emerson B.M.. 2012. Functional and molecular characterization of the role of CTCF in human embryonic stem cell biology. PLoS One. 7:e42424 10.1371/journal.pone.0042424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M.J., Orr A.L., Gerencser A.A., Madden D.T., Vitelli C., Swistowski A., Brand M.D., and Zeng X.. 2011. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J. Cell Sci. 124:348–358. 10.1242/jcs.072272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Markoulaki S., van Wietmarschen N., Hoke H., Wu T., Ganz K., Akhtar-Zaidi B., He Y., Abraham B.J., Porubsky D., et al. 2014. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 15:295–309. 10.1016/j.stem.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowiecki R., Adjaye J., and Prigione A.. 2014. Mitochondrial function in pluripotent stem cells and cellular reprogramming. Gerontology. 60:174–182. 10.1159/000355050 [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., and Studer L.. 2009. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27:275–280. 10.1038/nbt.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.C., Chiang W.F., Huang H.H., Chen P.F., Shen Y.Y., and Chiang H.C.. 2014. Role of SIRT1 in regulation of epithelial-to-mesenchymal transition in oral squamous cell carcinoma metastasis. Mol. Cancer. 13:254 10.1186/1476-4598-13-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernukhin I.V., Shamsuddin S., Robinson A.F., Carne A.F., Paul A., El-Kady A.I., Lobanenkov V.V., and Klenova E.M.. 2000. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 275:29915–29921. 10.1074/jbc.M001538200 [DOI] [PubMed] [Google Scholar]
- Chichagova V., Sanchez-Vera I., Armstrong L., Steel D., and Lako M.. 2016. Generation of human induced pluripotent stem cells using RNA-based Sendai virus system and pluripotency validation of the resulting cell population. Methods Mol. Biol. 1353:285–307. 10.1007/7651_2015_205 [DOI] [PubMed] [Google Scholar]
- Facucho-Oliveira J.M., Alderson J., Spikings E.C., Egginton S., and St John J.C.. 2007. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J. Cell Sci. 120:4025–4034. 10.1242/jcs.016972 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Gilmour D.. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10:445–457. 10.1038/nrm2720 [DOI] [PubMed] [Google Scholar]
- Gershon T.R., Crowther A.J., Tikunov A., Garcia I., Annis R., Yuan H., Miller C.R., Macdonald J., Olson J., and Deshmukh M.. 2013. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer Metab. 1:2 10.1186/2049-3002-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Xue Y., Yang G., Yin S., Shi W., Cheng Y., Yan X., Fan S., Zhang H., and Zeng F.. 2016. Nanog RNA-binding proteins YBX1 and ILF3 affect pluripotency of embryonic stem cells. Cell Biol. Int. 40:847–860. 10.1002/cbin.10539 [DOI] [PubMed] [Google Scholar]
- Hannan N.R., Fordham R.P., Syed Y.A., Moignard V., Berry A., Bautista R., Hanley N.A., Jensen K.B., and Vallier L.. 2013. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Rep. 1:293–306. 10.1016/j.stemcr.2013.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., and Zhang S.C.. 2010. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 107:4335–4340. 10.1073/pnas.0910012107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Shen Y., Xue Z., Bibikova M., April C., Liu Z., Cheng L., Nagy A., Pellegrini M., Fan J.B., and Fan G.. 2013. A panel of CpG methylation sites distinguishes human embryonic stem cells and induced pluripotent stem cells. Stem Cell Rep. 2:36–43. 10.1016/j.stemcr.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D.E., Schafer Z.T., Evans R.M., Suda T., Lee C.H., and Pandolfi P.P.. 2012. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 18:1350–1358. 10.1038/nm.2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Habibollah S., Tilgner K., Collin J., Barta T., Al-Aama J.Y., Tesarov L., Hussain R., Trafford A.W., Kirkwood G., et al. 2014. An induced pluripotent stem cell model of hypoplastic left heart syndrome (HLHS) reveals multiple expression and functional differences in HLHS-derived cardiac myocytes. Stem Cells Transl. Med. 3:416–423. 10.5966/sctm.2013-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wu F., Wang P., Ye X., Abdulkarim B.S., and Lai R.. 2014. YB-1 regulates Sox2 to coordinately sustain stemness and tumorigenic properties in a phenotypically distinct subset of breast cancer cells. BMC Cancer. 14:328 10.1186/1471-2407-14-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M., Awong G., Sturgeon C.M., Ditadi A., LaMotte-Mohs R., Zúñiga-Pflücker J.C., and Keller G.. 2012. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Reports. 2:1722–1735. 10.1016/j.celrep.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I., et al. 2010. Epigenetic memory in induced pluripotent stem cells. Nature. 467:285–290. 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobloch M., Braun S.M., Zurkirchen L., von Schoultz C., Zamboni N., Araúzo-Bravo M.J., Kovacs W.J., Karalay O., Suter U., Machado R.A., et al. 2013. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 493:226–230. 10.1038/nature11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi-Aoi M., Ohnuki M., Takahashi K., Okita K., Noma H., Sawamura Y., Teramoto I., Narita M., Sato Y., Ichisaka T., et al. 2013. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 110:20569–20574. 10.1073/pnas.1319061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law I.K., Liu L., Xu A., Lam K.S., Vanhoutte P.M., Che C.M., Leung P.T., and Wang Y.. 2009. Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics. 9:2444–2456. 10.1002/pmic.200800738 [DOI] [PubMed] [Google Scholar]
- Liu Y., Clegg H.V., Leslie P.L., Di J., Tollini L.A., He Y., Kim T.H., Jin A., Graves L.M., Zheng J., and Zhang Y.. 2015. CHCHD2 inhibits apoptosis by interacting with Bcl-x L to regulate Bax activation. Cell Death Differ. 22:1035–1046. 10.1038/cdd.2014.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Morey R., O’Neil R.C., He Y., Daughtry B., Schultz M.D., Hariharan M., Nery J.R., Castanon R., Sabatini K., et al. 2014. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 511:177–183. 10.1038/nature13551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane A., Doi D., Kikuchi T., Nishimura K., and Takahashi J.. 2011. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J. Neurosci. Res. 89:117–126. 10.1002/jnr.22547 [DOI] [PubMed] [Google Scholar]
- Narsinh K.H., Sun N., Sanchez-Freire V., Lee A.S., Almeida P., Hu S., Jan T., Wilson K.D., Leong D., Rosenberg J., et al. 2011. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 121:1217–1221. 10.1172/JCI44635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-Gerald C.S., Sato Y., Cowan C.A., Chien K.R., and Melton D.A.. 2008. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26:313–315. 10.1038/nbt1383 [DOI] [PubMed] [Google Scholar]
- Pang L., Qiu T., Cao X., and Wan M.. 2011. Apoptotic role of TGF-β mediated by Smad4 mitochondria translocation and cytochrome c oxidase subunit II interaction. Exp. Cell Res. 317:1608–1620. 10.1016/j.yexcr.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Park I.H., and Daley G.Q.. 2009. Human iPS cell derivation/reprogramming. Curr Protoc Stem Cell Biol Chapter 4:Unit 4A.1. 10.1002/9780470151808.sc04a01s8 [DOI] [PubMed] [Google Scholar]
- Pauklin S., and Vallier L.. 2013. The cell-cycle state of stem cells determines cell fate propensity. Cell. 155:135–147. (published erraturm appears in Cell. 2014. 156:1338) 10.1016/j.cell.2013.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A., Fauler B., Lurz R., Lehrach H., and Adjaye J.. 2010. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 28:721–733. 10.1002/stem.404 [DOI] [PubMed] [Google Scholar]
- Rashid S.T., Corbineau S., Hannan N., Marciniak S.J., Miranda E., Alexander G., Huang-Doran I., Griffin J., Ahrlund-Richter L., Skepper J., et al. 2010. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 120:3127–3136. 10.1172/JCI43122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani F., Kumasaka N., de Brito M.C., Bradley A., Vallier L., and Gaffney D.. 2014. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 10:e1004432 10.1371/journal.pgen.1004432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Diep D., Gore A., Panopoulos A.D., Montserrat N., Plongthongkum N., Kumar S., Fung H.L., Giorgetti A., Bilic J., et al. 2012. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 109:16196–16201. 10.1073/pnas.1202352109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Lee W.H., and Suk K.. 2010. Identification of novel cell migration-promoting genes by a functional genetic screen. FASEB J. 24:464–478. 10.1096/fj.09-137562 [DOI] [PubMed] [Google Scholar]
- Shimojima K., Okumura A., Hayashi M., Kondo T., Inoue H., and Yamamoto T.. 2015. CHCHD2 is down-regulated in neuronal cells differentiated from iPS cells derived from patients with lissencephaly. Genomics. 106:196–203. 10.1016/j.ygeno.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Solomon S., Pitossi F., and Rao M.S.. 2015. Banking on iPSC--is it doable and is it worthwhile. Stem Cell Rev. 11:1–10. 10.1007/s12015-014-9574-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Okita K., Nakagawa M., and Yamanaka S.. 2007. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2:3081–3089. 10.1038/nprot.2007.418 [DOI] [PubMed] [Google Scholar]
- Tobin S.C., and Kim K.. 2012. Generating pluripotent stem cells: differential epigenetic changes during cellular reprogramming. FEBS Lett. 586:2874–2881. 10.1016/j.febslet.2012.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaloni M., Pulecio J., Bilic J., Kuebler B., Laricchia-Robbio L., and Izpisua Belmonte J.C.. 2014. MicroRNAs contribute to induced pluripotent stem cell somatic donor memory. J. Biol. Chem. 289:2084–2098. 10.1074/jbc.M113.538702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Vellanki R.N., Coyaud É., Ignatchenko V., Li L., Krieger J.R., Taylor P., Tong J., Pham N.A., Liu G., et al. 2015. CHCHD2 Is Coamplified with EGFR in NSCLC and Regulates Mitochondrial Function and Cell Migration. Mol. Cancer Res. 13:1119–1129. 10.1158/1541-7786.MCR-14-0165-T [DOI] [PubMed] [Google Scholar]
- Wu T., Liu Y., Wen D., Tseng Z., Tahmasian M., Zhong M., Rafii S., Stadtfeld M., Hochedlinger K., and Xiao A.. 2014. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell. 15:281–294. 10.1016/j.stem.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Yanes O., Clark J., Wong D.M., Patti G.J., Sánchez-Ruiz A., Benton H.P., Trauger S.A., Desponts C., Ding S., and Siuzdak G.. 2010. Metabolic oxidation regulates embryonic stem cell differentiation. Nat. Chem. Biol. 6:411–417. 10.1038/nchembio.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., and Thomson J.A.. 2009. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 324:797–801. 10.1126/science.1172482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Su P., Li D., Tsang S., Duan E., and Wang F.. 2010. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 28:1741–1750. 10.1002/stem.504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A., Preynat-Seauve O., Tiercy J.M., Krause K.H., and Villard J.. 2012. Haplotype-based banking of human pluripotent stem cells for transplantation: potential and limitations. Stem Cells Dev. 21:2364–2373. 10.1089/scd.2012.0088 [DOI] [PubMed] [Google Scholar]
- Zubovych I.O., Straud S., and Roth M.G.. 2010. Mitochondrial dysfunction confers resistance to multiple drugs in Caenorhabditis elegans. Mol. Biol. Cell. 21:956–969. 10.1091/mbc.E09-08-0673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.