Monaco et al. demonstrate that calcium signals in activated human T cells consist of a cytoplasmic and a nuclear component, which are both required for the immune response. Blockade of nuclear calcium signaling inhibits T cell activation and induces an anergy-like state.
Abstract
Calcium signals in stimulated T cells are generally considered single entities that merely trigger immune responses, whereas costimulatory events specify the type of reaction. Here we show that the “T cell calcium signal” is a composite signal harboring two distinct components that antagonistically control genomic programs underlying the immune response. Using human T cells from healthy individuals, we establish nuclear calcium as a key signal in human T cell adaptogenomics that drives T cell activation and is required for signaling to cyclic adenosine monophosphate response element–binding protein and the induction of CD25, CD69, interleukin-2, and γ-interferon. In the absence of nuclear calcium signaling, cytosolic calcium activating nuclear factor of activated T cells translocation directed the genomic response toward enhanced expression of genes that negatively modulate T cell activation and are associated with a hyporesponsive state. Thus, nuclear calcium controls the T cell fate decision between a proliferative immune response and tolerance. Modulators of nuclear calcium–driven transcription may be used to develop a new type of pro-tolerance immunosuppressive therapy.
Introduction
Upon stimulation from the environment, many cell types use calcium signals for intracellular processing of information and the induction of appropriate biological responses through activating specific gene expression programs (Berridge et al., 2000; Clapham, 2007). To generate diversity in signal transduction using a single second messenger, cells exploit the spatial and temporal profiles of calcium transients (Rizzuto and Pozzan, 2006; Bading, 2013). This process is well documented in the nervous system, where the partitioning of calcium signaling events in subcellular compartments and microdomains enables neurons to build a repertoire of stimulus-specific responses. For example, the genomic events that specify the expression patterns of target genes in synaptically stimulated neurons are differentially controlled by nuclear versus cytoplasmic calcium signals (Hardingham et al., 1997; Chawla et al., 1998; Mauceri et al., 2011). In particular, calcium signals in the cell nucleus function as key regulators of plasticity-related gene expression in neurons and are needed for the long-term implementation of different neuroadaptations including memory formation, acquired neuroprotection, and the development of chronic pain (Limbäck-Stokin et al., 2004; Papadia et al., 2005; Zhang et al., 2009; Bading, 2013; Simonetti et al., 2013; Weislogel et al., 2013).
Calcium regulates many cellular functions by forming a complex with calmodulin (CaM), a ubiquitously expressed calcium-binding protein. Upon binding of calcium, CaM increases its affinity for its target proteins, which include the cytoplasmic serine/threonine phosphatase calcineurin (CaN) and the nuclear calcium/CaM-dependent protein kinase IV (CaMKIV; Crabtree, 1999; Hook and Means, 2001; Hogan et al., 2003). The instructive role of calcium signals in mounting adaptive responses in other tissues such as the heart or the immune system is generally appreciated (Feske et al., 2001; Oh-hora and Rao, 2008; Higazi et al., 2009). In nonneuronal cells, however, the complexity of calcium transients and possible functional diversity of spatially distinct signals is less well explored.
In antigen-stimulated T lymphocytes, increases in intracellular calcium levels are critical for the immune response (Dolmetsch et al., 1998; Lewis, 2001; Feske, 2007). Both local signals in the immunological synapse (Lioudyno et al., 2008; Quintana et al., 2011) and cytoplasmic calcium microdomains have gene transcription–regulating functions (Di Capite et al., 2009; Kar et al., 2011). In contrast, the role of nuclear calcium signaling is virtually unexplored in T cells. In particular, it has not been considered that calcium signals in the cytosol and the nucleus may serve distinct functions in T cells that could explain differences in the responses to antigen challenge. T cells can undergo two very different types of physiological responses: activation, leading to a productive immune response, or anergy, leading to tolerance. Anergy is characterized by functional unresponsiveness and is induced when T cell receptor (TCR) stimulation is not accompanied by a costimulatory event (Macián et al., 2004). The costimulatory signal involves phosphatidylinositol-3-kinase and PKC signaling cascades; it is initiated physiologically by the binding of CD80/CD86 receptor on the antigen-presenting cell to the CD28 receptor and can be induced in vitro by the exposure of T cells to either CD28 antibodies or chemical inducers of PKC such as PMA. At the genomic level, the decision between activation and anergy depends on whether nuclear factor of activated T cells (NFAT), upon its stimulus-induced translocation to the nucleus, forms a transcription factor complex with AP1 (Macián et al., 2001). The transcriptional program induced by NFAT/AP1, which includes interleukin (IL)-2 and IFNγ, initiates a productive immune response, whereas genes induced by NFAT lead only to T cell tolerance (Macián et al., 2000). One of the hallmarks of anergic T cells is their reduced ability to produce IL-2 (Bandyopadhyay et al., 2007). The uncoupling of the activation of NFAT and AP1 is one reason for the lack of IL-2 production after TCR stimulation. In addition, in anergic T cells, active mechanisms of transcriptional repression of IL-2 are induced. This involves in particular the activation of the repressor complex cyclic adenosine monophosphate (cAMP) response element binding protein (CREB)–cAMP responsive element modulator (CREM) that antagonizes both CREB- and AP1-mediated transcription (Masquilier and Sassone-Corsi, 1992; Powell et al., 1999). The expression levels of CREM tightly correlate with the suppression of IL-2 transcription (Bodor and Habener, 1998; Juang et al., 2005). Indeed, CREM is part of a set of genes referred to as “anergy inducing genes”; they include other transcriptional repressors and NFAT targets such as Egr2 and Egr3 (Wells, 2009), but also phosphatases, proteases, and inhibitory receptors that act as negative regulators of TCR-induced signaling and transcription (Baine et al., 2009). Collectively, the expression of these genes defines a genetic signature of reduced responsiveness (Lechner et al., 2001; Macián et al., 2002; Safford et al., 2005). In this study, we tested the hypothesis that the T cell activation process is differentially controlled by spatially distinct calcium signals. We used recombinant calcium sensors containing specific targeting sequences to monitor calcium transients in the cytosol and the nucleus during human T cell activation. A functional analysis revealed a key role for nuclear calcium in T cell activation. The stimulation of human T cells in the absence of nuclear calcium signaling redirects the transcriptional response toward a gene expression profile characteristic of a hyporesponsive state.
Results
Activation of TCR induces nuclear and cytoplasmic calcium transients
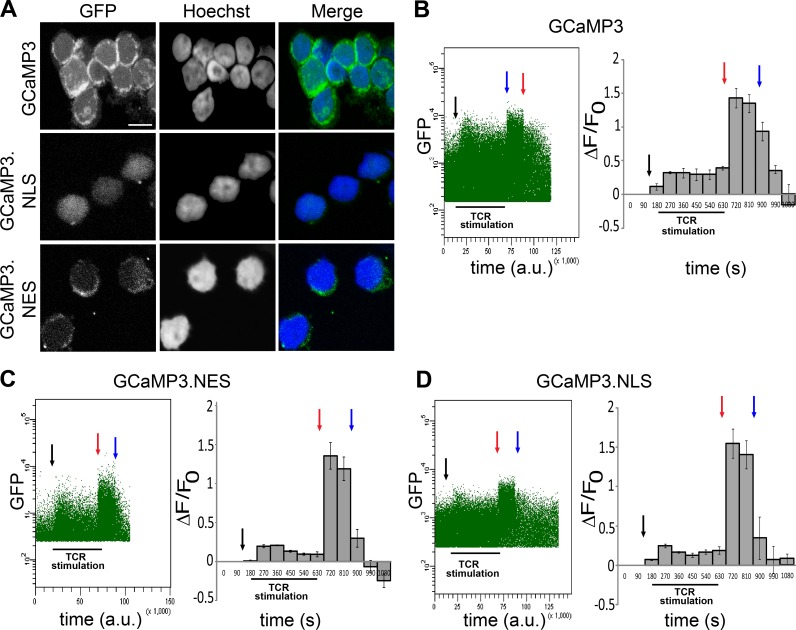
We used compartment-specific recombinant calcium sensors to determine the spatial features of calcium signals in primary human T cells after activation. Plasmids containing expression vectors for GCaMP3 or GCaMP3 fused to amino acid sequences encoding for either a nuclear localization signal (GCaMP3.NLS) or a nuclear export signal (GCaMP3.NES) were transfected into primary human T cells isolated from healthy donors. GCaMP3 consists of a circularly permuted GFP fused to CaM and the CaM target peptide, M13 (Tian et al., 2009; Bengtson et al., 2010). Upon binding of calcium, GCaMP3 undergoes a conformational change that increases its fluorescence.
Using confocal microscopy, we confirmed that GCaMP3 is present throughout the cell, whereas expression of GCaMP3.NLS and GCaMP3.NES is restricted to the nuclear and cytosolic compartments, respectively (Fig. 1 A). Changes in intracellular calcium concentrations were monitored using flow cytometry for a period of 9 min after TCR stimulation using CD3 antibodies. Stimulus-induced increases in intracellular calcium levels were readily detectable in both the cytoplasm and the nucleus and remained at an elevated plateau for the duration of the experiment (Fig. 1, B–D). The stimulation of T cells with a combination of CD3 and CD28 antibodies induced similar nuclear calcium transients (Fig. S1). These results indicate that TCR stimulation generates a composite calcium signal that consists of a nuclear and a cytoplasmic component.
Figure 1.
TCR activation induces calcium transients in the cytoplasm and the nucleus in primary human T cells. (A) Confocal images showing the subcellular localization of the genetically encoded calcium indicators GCaMP3 (cytoplasm and nucleus), GCaMP3.NLS (nucleus), and GCaMP3.NES (cytoplasm). Bar, 5 µm. (B–D) Representative experiments showing calcium transients in human T cells transfected with the indicated calcium sensors and stimulated with CD3 antibodies. Anti–mouse IgG (black arrows) was added for antibody cross-linking, and calcium signals were measured for 9 min. Ionomycin (red arrows) and EGTA (blue arrows) were added to obtain the maximum (Fmax) and minimum (Fmin) signal, respectively. Calcium signals were analyzed by monitoring changes in GFP mean fluorescence as a function of time. The dot plots (left) are representative examples of at least four independent experiments (a.u., arbitrary units). Histograms (right) show mean GFP fluorescence (ΔF/F0) from seven (GCaMP3), four (GCaMP3.NLS), and four (GCaMP3.NES) independent experiments. Error bars represent SEM.
Nuclear calcium induces the expression of activation markers and cytokines
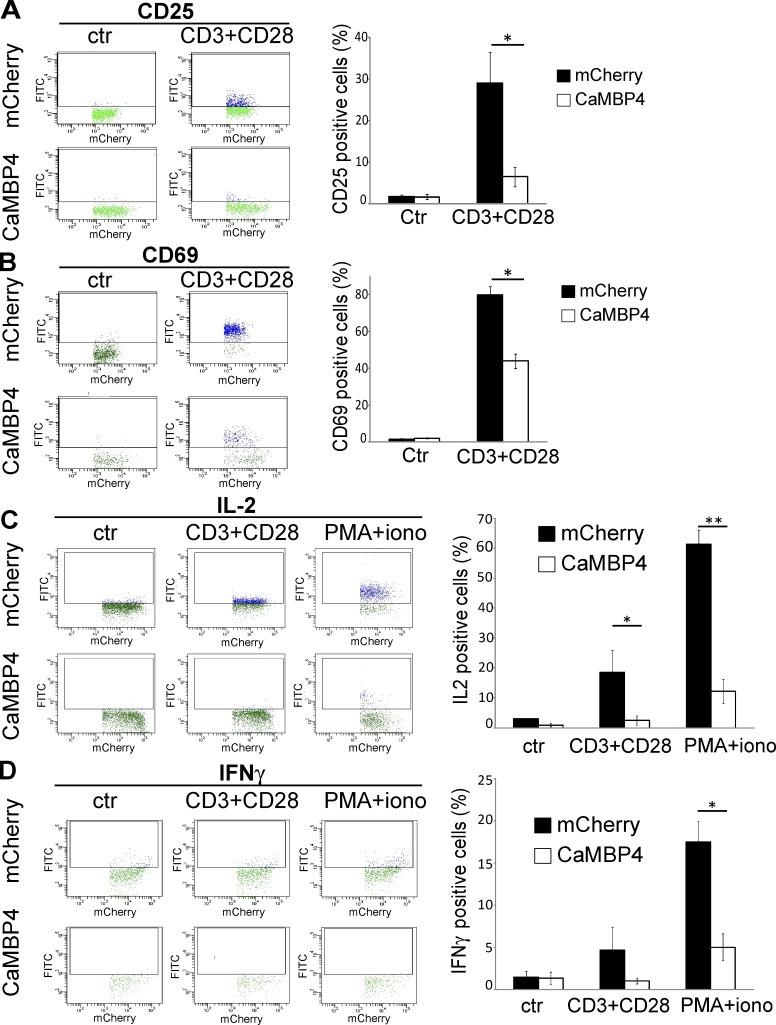
To investigate possible functional differences of nuclear and cytoplasmic calcium signals, we transfected primary human pan-T cells with expression vectors for either CaMBP4 fused to mCherry (CaMBP4.mCherry) or mCherry as a control. CaMBP4.mCherry is a nuclear protein that contains four repeats of the M13 binding peptide that binds to and thereby inactivates the nuclear calcium/CaM complex (Wang et al., 1995). It is not a calcium buffer and therefore does not reduce intranuclear calcium concentrations; instead, it acts as a recombinant competitive inhibitor of calcium/CaM-dependent processes in the cell nucleus. Calcium imaging experiments confirmed that CaMBP4.mCherry does not alter calcium transients in response to TCR stimulation (Fig. S2 A). This inhibitor has been used in several studies to investigate the role of nuclear calcium in signal-regulated gene expression and adaptive processes in the nervous system of mice, rats, and Drosophila melanogaster (Limbäck-Stokin et al., 2004; Papadia et al., 2005; Zhang et al., 2009; Bading, 2013; Simonetti et al., 2013; Weislogel et al., 2013). To trigger activation, human pan-T cells were either exposed to antibodies to CD3 and CD28 or were chemically activated with a mixture of PMA and ionomycin. In control cells transfected with an expression vector for mCherry, these stimulation protocols induced the typical profile of activation markers, which include the IL-2 receptor α chain (CD25) and CD69 (Fig. 2, A and B) and the key regulatory cytokines, IL-2 and IFNγ (Fig. 2, C and D). The presence of CaMBP4.mCherry in T cell nuclei led to a far smaller induction of CD69 and CD25 expression (Fig. 2, A and B) and virtually completely blocked the stimulus-induced increase in the production of IL-2 and IFNγ (Fig. 2, C and D). CaMBP4.mCherry expression had the same inhibitory effect on IL-2 and CD25 production in naive CD4+ and CD8+ human T cells obtained by negative selection and stimulated with either a combination of CD3 and CD28 antibodies or with PMA and ionomycin (Fig. S3). Expression of an inactive form of CaMBP4 (mut-CaMBP4.mCherry; Lau et al., 2015) did not block IL-2 production in T cells (Fig. S2 B).
Figure 2.
Nuclear calcium controls the expression of activation markers and cytokines in primary human T cells. T cells transfected with expression vectors for CaMBP4.mCherry (CaMBP4) or mCherry either were left unstimulated (ctr) or were stimulated with a combination of antibodies to CD3 and CD28 or with a combination of PMA and ionomycin. Expression of CD25 (A), CD69 (B), IL-2 (C), and IFNγ (D) in T cells was analyzed by flow cytometry with FITC-conjugated antibodies. Dot plots (left) are representative examples of three independent experiments; histograms (right) show the mean percentage of FITC-positive cells (blue dots) from three independent experiments. Error bars represent SEM. Statistically significant differences are indicated with asterisks (*, P < 0.05; **, P < 0.01).
A productive immune response culminates in T cell proliferation and clonal expansion. To determine the role of nuclear calcium signaling in T cell proliferation, we performed a mixed lymphocyte reaction (MLR). The proliferative response measured in the MLR serves as an indicator of T cell activation in response to exposure to peripheral blood mononuclear cells (PBMCs) from a different donor (Chen et al., 2003). We performed a nonradioactive, fluorescence-based MLR assay using the cell-permeable dye carboxyfluorescein diacetate succinimidyl ester (CFSE; Popma et al., 2000; Fig. S4). Responder T cells transfected with mCherry or CaMBP4.mCherry were labeled with CFSE and cultured with PBMCs for 3 d to induce a proliferative response. On days 1–3 after stimulation, we observed a decrease in the fluorescence intensity of the mCherry-expressing control cells, which is caused by the dilution of CFSE after cell division (Fig. S4 A). Quantitative analysis revealed that compared with mCherry control, CaMBP4.mCherry expression dramatically decreased the responders’ proliferative response (Fig. S4 B). Thus, in the absence of nuclear calcium signaling, activated human T cells are unable to proliferate, supporting the hypothesis that nuclear calcium signaling is necessary for a productive T cell response.
Nuclear calcium regulates CREB and CREM but not NFAT nuclear translocation
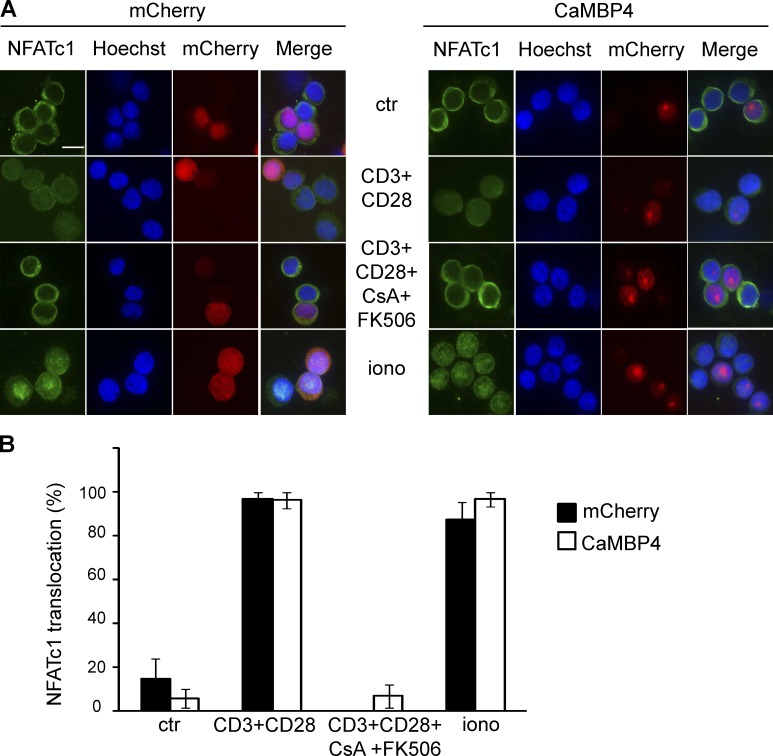
To investigate a possible role for nuclear calcium in the T cell fate decision process, we determined the effect of CaMBP4.mCherry expression on the nuclear translocation of NFAT. Immunocytochemical analysis of the subcellular localization of endogenous NFATc1 (NFAT2) in control cells expressing mCherry revealed that the typical stimuli used to induce activation (i.e., treatment with combination of CD3 and CD28 antibodies) or anergy (i.e., ionomycin treatment) promoted translocation of NFAT2 to the nucleus, and that this translocation was blocked by a mixture of the CaN inhibitors, cyclosporine A and FK506 (Fig. 3, A [left] and B). Expression of CaMBP4.mCherry did not affect this nuclear translocation (Fig. 3, A [right] and B). A similar result was obtained for NFAT1, whose translocation to the nucleus upon stimulation of the cells with a combination of CD3 and CD28 antibodies was not affected by the expression of CaMBP4.mCherry (Fig. S5). These results are not unexpected because the calcium-dependent dephosphorylation of NFATs by CaN that triggers their nuclear translocation is taking place in the cytosol (Macián et al., 2002) and should function even when nuclear calcium signaling is blocked.
Figure 3.
Nuclear calcium signaling does not regulate nuclear translocation of NFATc1 in primary human T cells. (A) T cells transfected with expression vectors for CaMBP4.mCherry (CaMBP4) or mCherry were left unstimulated (ctr), were stimulated with a combination of antibodies to CD3 and CD28 in the presence or absence of cyclosporine A (CsA) and FK506, or were stimulated with ionomycin (iono) for 30 min. Cells were stained with antibodies against NFATc1. Nuclei were stained with Hoechst. Bar, 5 µm. (B) Quantitative analysis of NFATc1 nuclear localization (n = 3). Error bars represent SEM.
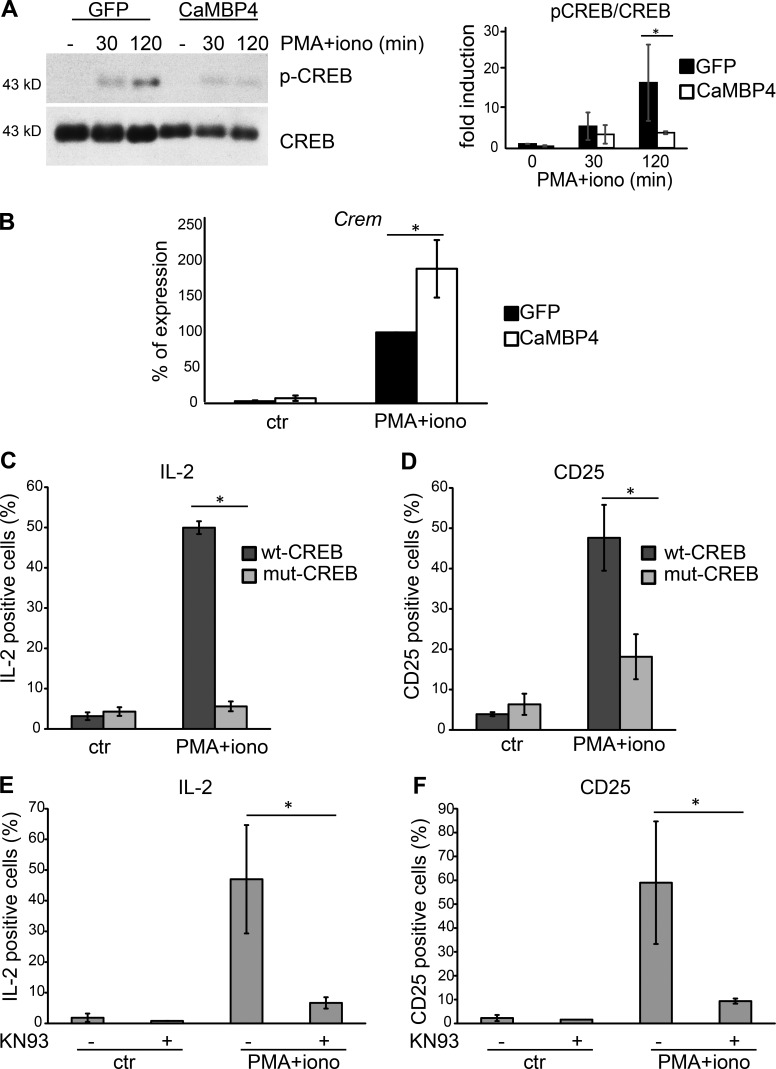
We next investigated the phosphorylation of CREB on its activator site, serine 133, and the stimulus-induced expression of CREM. In the nervous system, CREB is considered the prototypical nuclear calcium–regulated transcription factor that controls diverse forms of neuroadaptations (Hardingham et al., 2001; Bading, 2013). Its activity is primarily regulated by CaMKIV, a serine/threonine kinase located in the cell nucleus (Hook and Means, 2001; Racioppi and Means, 2008). CREB also has a major role in the immune system (Wen et al., 2010). In T cells, CREB activity is induced by CD3 and CD28 costimulation, and it promotes activation, proliferation, and cytokine production (Yu et al., 2001). In contrast, CREM, an isoform of CREB that lacks the transactivation domain, binds to the distal AP1 site of the IL-2 promoter (Powell et al., 1999) and suppresses IL-2 transcription in anergic cells. Thus, whereas CREB activation is required for T cell activation, the increased expression of CREM is associated with tolerance (Wells, 2009). Given the observation that the blockade of nuclear calcium signaling abolished IL-2 expression but left the signal-regulated nuclear translocation of NFATs unaffected (Figs. 2 and 3), we predicted that nuclear calcium differentially regulates CREB and CREM function. Indeed, we found that the PMA + ionomycin–induced phosphorylation of CREB on serine 133 was blocked by the inhibition of nuclear calcium signaling, whereas the expression of CREM mRNA increased under those conditions (Fig. 4, A and B). To investigate whether CREB and CaMKIV are involved in the molecular mechanism by which nuclear calcium signaling controls T cell activation, we determined the effects of inhibition of CREB and CaMKIV on the production of activation markers, IL-2 and CD25. The expression of mut-CREB.GFP, a mutant, transcriptionally inactive version of CREB (containing a serine-to-alanine mutation at position aa 133) fused to GFP but not overexpression of wt-CREB.GFP (wild-type CREB-GFP fusion protein) strongly reduced IL-2 and CD25 expression after 6 h of PMA + ionomycin treatment (Fig. 4, C and D). A similar reduction of IL-2 and CD25 expression in PMA + ionomycin–treated human T cells was observed using KN93, a general inhibitor of CaM kinases (Fig. 4, E and F). Thus, inhibition of either CaMKIV or inhibition of its target transcription factor CREB produced effects on T cell activation that are similar to the ones observed after blockade of nuclear calcium signaling, suggesting that similar to adaptive changes in the nervous system (Bading, 2013), a nuclear calcium–CaMKIV-CREB signaling module controls T cell activation. In the absence of nuclear calcium signaling, a cytoplasmic calcium signal causing nuclear translocation of NFATs may direct the immune response toward an anergy-like state.
Figure 4.
Nuclear calcium signaling differentially regulates CREB activation and CREM expression in primary human T cells. (A, left) Immunoblot analysis of CREB and the serine 133–phosphorylated form of CREB (pCREB) in T cells transfected with expression vectors for CaMBP4.GFP (CaMBP4) or GFP and stimulated with a combination of PMA and ionomycin for 30 or 120 min. A representative example is shown. (right) Quantitative analysis of three experiments; fold induction is relative to the pCREB/CREB ratio in unstimulated GFP-expressing T cells. (B) QRT-PCR analysis of Crem mRNA in T cells transfected with expression vectors for CaMBP4.GFP (CaMBP4) or GFP. Cells were stimulated with a combination of PMA and ionomycin for 6 h or were left unstimulated (ctr). mRNA levels are expressed relative to the mRNA level in GFP-expressing T cells stimulated with a combination of PMA and ionomycin, which was set to 100%. n = 3. Statistically significant differences are indicated with an asterisk (*, P < 0.05). (C and D) FACS analysis of the percentage of IL-2– and CD25-expressing T cells transfected with GFP-tagged expression vectors for either wild-type CREB (wt-CREB) or mutant CREB containing a serine-to-alanine mutation at aa 133 (mut-CREB). Cells were stimulated with a combination of PMA and ionomycin for 6 h or were left unstimulated (ctr). n = 3. Statistically significant differences are indicated with asterisks (*, P < 0.05). (E and F) FACS analysis of the percentage of IL-2– and CD25-expressing T cells. Cells were stimulated with a combination of PMA and ionomycin for 6 h or were left unstimulated (ctr). n = 3. Pretreatment with KN93 was done as indicated. Statistically significant differences are indicated with asterisks (*, P < 0.05). Error bars represent SEM.
Gene expression profiling
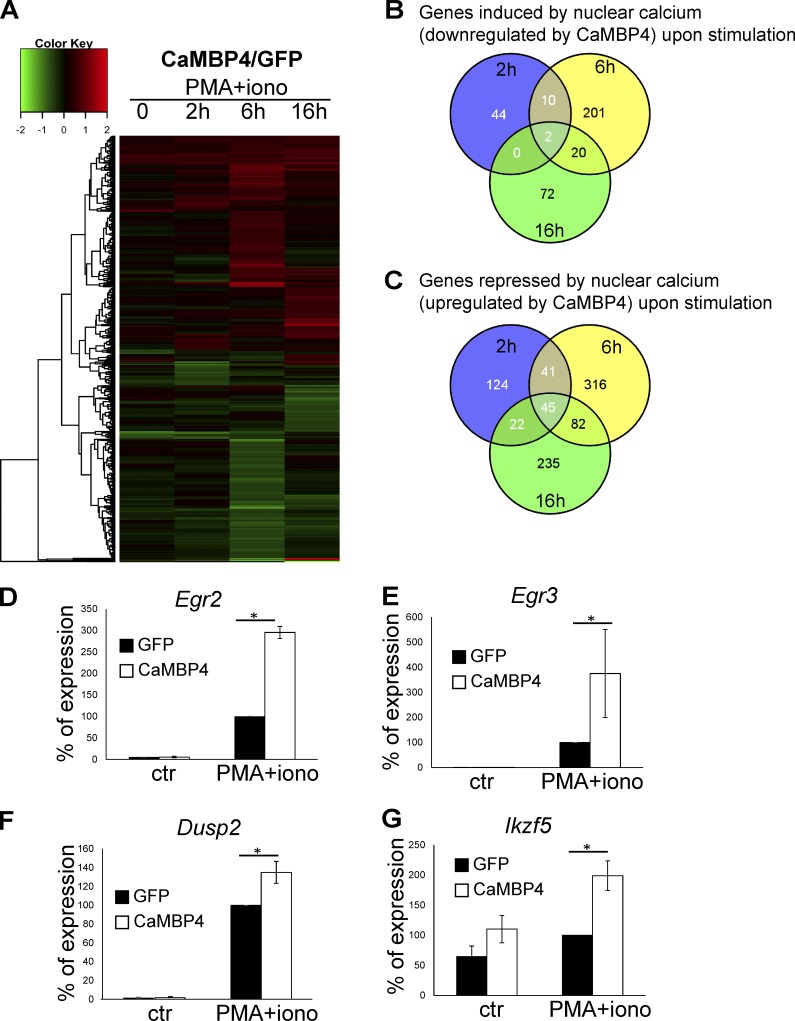
To further investigate the possible role of nuclear calcium as a decision maker in the immune response, we determined the human T cells’ gene expression signatures using microarray analysis. The transcriptome profiles of T cells transfected with expression vectors for CaMBP4.GFP or GFP were compared. The fold increase or decrease in gene expression in CaMBP4.GFP-expressing cells compared with GFP-expressing controls was determined 2, 6, and 16 h after stimulation with PMA + ionomycin. Genes up-regulated or down-regulated with a p-value <0.05 in CaMBP4.GFP versus control (GFP) samples were considered differentially regulated. The cutoff was set to 1.3- and 0.7-fold for up-regulation and down-regulation, respectively. Using these criteria, we identified 29 genes up-regulated and 12 genes down-regulated in CaMBP4.GFP-expressing unstimulated T cells (Fig. 5 A). In PMA + ionomycin–treated T cells, the increase of a total of 349 genes was reduced by CaMBP4.GFP (Fig. 5, A and B), whereas the same condition led to an up-regulation of 865 genes (Fig. 5, A and C). The regulation by nuclear calcium signaling of many genes has been verified by quantitative RT-PCR (QRT-PCR) analyses; Fig. 5, D–G illustrates several examples. The transcriptome data also confirmed the protein expression analyses shown in Fig. 2.
Figure 5.
Analysis of nuclear calcium–regulated human T cell adaptogenomics. Representation of gene profiling data of human T cells transfected with expression vectors for CaMBP4.GFP (CaMBP4) or GFP and stimulated with a combination of PMA and ionomycin for 2, 6, and 16 h. (A) Heatmap with cluster dendrogram of 939 genes differentially expressed (log2 fold change) in at least one condition. Maximum value is set to 2.0. Fold ratio values greater than or equal to 2.0 are depicted with saturated red; values less than or equal to −2.0 are depicted with saturated green. (B and C) Number of genes induced by nuclear calcium signaling (i.e., down-regulated by CaMBP4.GFP; B) and repressed by nuclear calcium signaling (up-regulated by CaMBP4; C) at different time points on the basis of Illumina Human Sentrix-12 BeadChip arrays. (D–G) Validation of gene regulatory events by QRT-PCR. Gene expression was analyzed in T cells transfected with expression vectors for CaMBP4.GFP (CaMBP4) or GFP. Cells were stimulated with a combination of PMA and ionomycin for 2 h (Egr2 and Egr3) and 6 h (DUSP2 and Ikzf5) or were left unstimulated (ctr). mRNA levels are expressed relative to those in GFP-expressing T cells stimulated with PMA and ionomycin, which was set to 100% (n = 3). Statistically significant differences are indicated with an asterisk (*, P < 0.05). Error bars represent SEM.
Blockade of nuclear calcium signaling induces an anergy-like gene program
A bioinformatics assessment of our microarray data revealed that human T cells stimulated under conditions of nuclear calcium signaling blockade fail to induce expression of numerous genes necessary for activation. Instead, they acquire a transcription profile with characteristic features of a hyporesponsive state. Tables 1 and 2 provide comprehensive lists of the nuclear calcium–regulated genes with known functions in T cell activation and the induction or maintenance of adaptive tolerance, respectively (Macián et al., 2000, 2002; Lechner et al., 2001; Safford et al., 2005; Bandyopadhyay et al., 2007; Baine et al., 2009; Wells, 2009).
Table 1. Genes induced by nuclear calcium signaling (i.e., down-regulated by CaMBP4) with a role in T cell activation.
| Signaling | MAPKs pathway | Transcription |
|---|---|---|
| PRKCQ (PKCθ) | MAP4K2 | STAT2 |
| PLCG2 (PLCγ2) | RASGRP3 | SMAD3 |
| Receptors | IFN signaling | SMAD4 |
| CD27 | IFITM | SMAD7 |
| CD8A | IRF8 | FOXO1 |
| IL-2R | IFI27 | EOMES |
| TGFβ R | IRFI35 | |
| Cytokines | Inflammation and cytotoxic activity | Adhesion molecules |
| IFNγ | KLRD1 (CD94) | ITGB2 |
| IL-2 | KLRB1 | ITGAL |
| IL-3 | GZMA (granzyme) | ICAM3 |
| IL-4 | GZMK (granzyme) | ITGB7 |
| GM-CSF (CSF2) | GZMH (granzyme) | Rho GTPase signaling |
| TNFSF13B (BAFF) | PTGER2 | RAP-GEF |
| TNFSF10 | ARHGAP4 |
Table 2. Genes repressed by nuclear calcium signaling (i.e., up-regulated by CaMBP4) with a function in promoting anergy.
| Signaling | Inhibitors of MAPKs pathway | E3 ubiquitin ligases | Proteasome |
|---|---|---|---|
| Fas | PTPN7 | RNF19B | PSMA6 |
| CD97 | PTPN22 | CBL | PSMA1 |
| GDI1 | PTPN6 | Transcription | PSMC6 |
| RGS16 | DUSP6 | NFATC1 | PSMD12 |
| GEM | DUSP5 | EGR1 | PSMD6 |
| Sh2D2A | DUSP2 | EGR2 | PSMB2 |
| Receptors | DUSP8 | EGR3 | PSMC1 |
| FasL | DUSP1 | STAT4 | PSMC4 |
| CD44 | DUSP4 | STAT6 | PSMA3 |
| IL7R | DGKα | Jarid2 | Other functions |
| TNFRSF4 | Inhibitors of proliferation | Nr4a2 | Actn4 |
| ICOS-LG | BTG2 | Nr4a3 | CCRN4L |
| Inhibitory receptors | IER5 | IRF4 | Hif1a |
| PDCD1 (PD1) | CDKN1A (P21) | Zfp36 | S100a |
| CTLA4 | GADD45B | Stx11 | |
| PFN1 | |||
| CDKN1B (p27 kip) |
Among the prominent promoters of a productive immune response whose expression is down-regulated by CaMBP4.GFP are the cytokines IL-2, IL-3, IL-4, and IFNγ (see also Fig. 2), as well as cytokine receptors and genes involved in the regulation of cytotoxic activity and inflammation, such as granzyme A and granzyme K (Table 1). This impairment in signaling mediated by secreted factors is accompanied by alterations in the expression of genes involved in the control of cytoskeletal dynamics and cell–cell interaction. This includes components of the Rho GTPase signaling pathway and different classes of adhesion molecules (Table 1), which could structurally impair the immunological synapse and further compromise the immune response (Pernis, 2009).
The inhibition of nuclear calcium signaling also led to changes in the expression of key components and regulators of PKC and MAPK signaling, whose activation upon TCR stimulation is crucial for T cell activation (Kwon et al., 2010; Kortum et al., 2013). We observed a reduction in the expression of PKCθ and in the levels of phospholipase C γ2 (Table 1), which catalyzes the generation of the PKC activator, DAG. Inhibition of nuclear calcium signaling further impaired PKC signaling through an increase in expression of DAG kinase α (Table 2), which inactivates DAG. The MAPK cascades are major targets of genomic alterations caused by nuclear calcium signaling blockade. The expression of several enzymes involved in the activation of these pathways and in signal processing are altered in CaMBP4.GFP-expressing T cells. This included down-regulation of expression of the Ras guanyl nucleotide-releasing protein (RasGRP3), which is involved in TCR proximal signaling to Ras, and MAP4K2, an upstream activator of both the stress-activated protein kinase/c-Jun N-terminal kinase signaling pathway and the p38 MAPK signaling pathway (Zha et al., 2006; Cook et al., 2007; Table 1). Moreover, the blockade of nuclear calcium signaling led to a striking increase in the expression of inhibitors of the MAPK signaling pathway, which includes many dual-specificity phosphatases (DUSPs) and protein tyrosine phosphatases (PTPs) that dephosphorylate and thereby inactivate several components of both the MAPKs and the TCR signaling cascades (Table 2). These results, summarized in Tables 1 and 2, illustrate at the genomic level that the inhibition of nuclear calcium signaling prevented T cell activation. They also indicate that the T cells appear to acquire a transcriptional profile characteristic of an anergy-like state (Macián et al., 2000, 2002; Lechner et al., 2001; Safford et al., 2005; Bandyopadhyay et al., 2007; Baine et al., 2009). Indeed, we uncovered an entire network of anergy-related genes that were induced in activated T cells in which nuclear calcium signaling was blocked (Table 2). This includes the NFAT-induced early growth response gene 2 and 3 (Egr2 and Egr3; Harris et al., 2004), which act as transcriptional repressors and impair IL-2 transcription in anergy (Safford et al., 2005), and many other transcription factors with known functions in anergy (Macián et al., 2002; Safford et al., 2005; Wells, 2009). The concerted action of anergy-promoting transcriptional regulators is complemented by an increased expression of inhibitory cell-surface molecules, PDCD1 (PD-1) and CTLA4 (Greenwald et al., 2001; Chikuma et al., 2009; Table 2). Finally, anergic T cells typically show an increase in the components of the ubiquitin protein degradation pathway (Heissmeyer et al., 2004). We observed a similar increase in expression of nine proteasome subunits and some E3 ubiquitin ligases in T cells activated under conditions of nuclear calcium signaling blockade (Table 2). Collectively, these findings revealed that nuclear calcium is a key signal in the T cell activation process and that, in its absence, the genomic response is redirected toward an anergy-like gene program.
Nuclear calcium blockade induces human T cell unresponsiveness
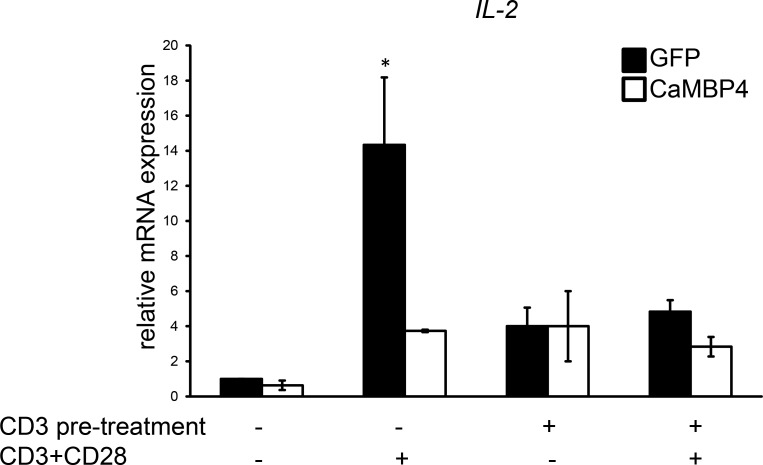
T cells exposed to CD3 antibodies acquire a state of anergy that is characterized by a lack of IL-2 synthesis upon subsequent stimulation using a combination of CD3 and CD28 antibodies (Heissmeyer et al., 2004). We next investigated whether the blockade of nuclear calcium signaling induces a comparable hyporesponsive state. We found that the efficacy of the classic anergy-inducing protocol (i.e., exposure to CD3 antibodies) to block the induction of IL-2 expression upon stimulation with CD3 and CD28 antibodies is very similar to that obtained with CaMBP4.GFP expression (Fig. 6). These findings indicate that treatment with CD3 antibodies and blockade of nuclear calcium signaling were equally effective in shifting human T cells toward a hyporesponsive, anergy-like state.
Figure 6.
Blockade of nuclear calcium signaling induces an anergy-like state in activated human T cells. QRT-PCR analysis of IL-2 expression in human T cells transfected with expression vectors for CaMBP4.GFP (CaMBP4) or GFP. Cells were stimulated as indicated with CD3 antibodies alone or with a combination of CD3 and CD28 antibodies (for 2 h) with or without overnight pretreatment with CD3 antibodies (n = 3). The results are expressed relative to the IL-2 mRNA levels of unstimulated GFP-expressing T cells. Statistically significant differences are indicated with an asterisk (*, P < 0.05). Error bars represent SEM.
Discussion
In this study, we have investigated the role of spatially distinct calcium signals in the control of transcriptional responses associated with the human T cell immune response. The use of compartment-specific calcium sensors revealed that the T cell calcium signal induced by antigen challenge is a composite signal consisting of a cytosolic and a nuclear component that serve different functions in the immune response. The role of cytosolic calcium is to trigger the translocation of NFATs to the nucleus, whereas the generation of nuclear calcium transients is critical for the activation of CREB and the induction of NFAT/AP1-dependent genomic responses. A schematic illustration of the distinct roles of cytosolic and nuclear calcium signals in T cell activation is shown in Fig. 7. The identification of nuclear calcium as a key signal for T cell activation fits well into an emerging concept that places nuclear calcium signaling at the center of adaptive responses induced by stimuli from the environment. In the nervous system, nuclear calcium functions as an evolutionarily conserved signaling end point in synapse-to-nucleus communication that promotes gene expression programs needed for the long-term implementation of several different forms of adaptations including memory, acquired neuroprotection, and chronic pain (Limbäck-Stokin et al., 2004; Papadia et al., 2005; Zhang et al., 2009; Bading, 2013; Simonetti et al., 2013; Weislogel et al., 2013). In the heart, nuclear calcium signaling is involved in the development of hypertrophy (Higazi et al., 2009), and outside the animal kingdom, nuclear calcium regulates symbiotic gene programs in plants (Oldroyd and Downie, 2006).
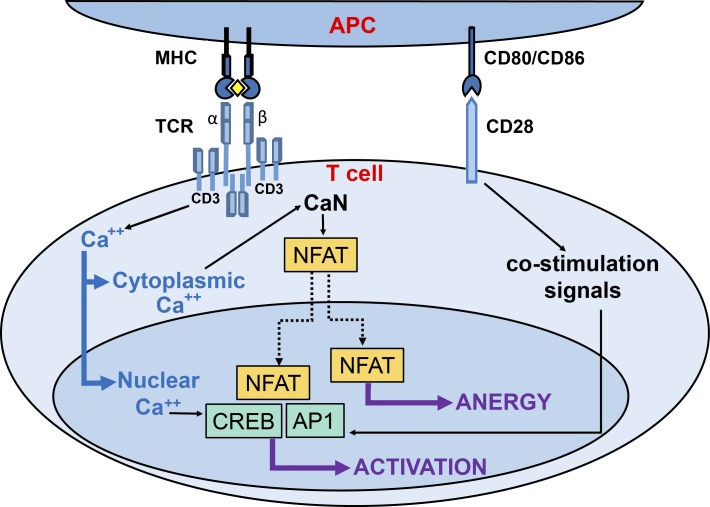
Figure 7.
Differential role of spatially distinct calcium signals in the human T cell immune response. Schematic illustration of the roles of nuclear and cytoplasmic calcium signals in the decision of stimulated T cells to mount a proliferative immune response or develop tolerance. In the immunological synapse, the stimulation of TCR and CD28 by the corresponding receptors on the antigen-presenting cell (APC) trigger the activation of different signaling pathways that lead to the induction of the immune response. After being activated by binding to the major histocompatibility complex (MHC)/antigen complex (the yellow rhombus represents the antigen), CD3 subunit of TCR induces an intracellular calcium increase, which activates CaN and NFAT in the cytoplasm and CREB and AP1 in the nucleus. The selective blockade of nuclear calcium signaling inhibits CREB activation but not NFAT translocation to the cell nucleus. As a consequence, T cell activation is blocked, and the immune response is redirected toward an anergy-like state.
Our study revealed that inhibition of calcium signaling in the cell nucleus shifts the T cell immune response from activation toward a hyporesponsive state characterized by a gene expression profile similar to that of anergic T cells. This observation has far-reaching clinical implications, as it identifies a novel signal in the T cell fate decision process that provides an unexpected new path for modulating the genomic responses underlying immune reactions. A means of preventing a proliferative T cell response that at the same time redirects the immune reaction toward tolerance may improve the acceptance of foreign cells and tissue after transplantations. Attempts to selectively block T cell costimulation through targeting CD40L or CD28 have been considered one possible therapeutic route to prevent allograft rejection but spare the side effects of a generalized immune suppression (Larsen et al., 1996; Laskowski et al., 2002; Sho et al., 2002; Vincenti et al., 2010; Zhang et al., 2011b). The results of our study suggest a novel strategy to accomplish “pro-tolerance immunosuppression,” in which blockade of nuclear calcium signaling may enable foreign cells to be recognized as self. So far, drugs that block nuclear calcium signaling have not been developed. However, the approach used in this study bears the potential for clinical application. Targeting T cells in humans with viral vectors carrying expression cassettes for CaMBP4 may represent a feasible route for establishing pro-tolerance immunosuppression.
Our findings also provide a conceptual framework for developing novel immune-enhancement treatments. Increasing the duration and/or amplitude of nuclear calcium signals is likely to enhance the transcriptional responses associated with T cell activation. In healthy individuals, this may result in a superinduction of cytokine and cytokine receptor expression and more robust immune responses. In conditions of immunodeficiency, augmenting nuclear calcium signaling may help restore functionality. The development of drugs that target the nuclear calcium clearance machinery and thus prolong stimulus-induced nuclear calcium transients represents one possible strategy in the design of novel treatments to boost T cell function.
Materials and methods
Cell culture and transfections
Human PBMCs were obtained by Ficoll-Hypaque (Linaris) density gradient centrifugation of heparinized blood from healthy volunteers. Human pan-T cells were purified with negative magnetic bead selection using the Pan T cell isolation kit (Miltenyi Biotec). Naive human T cells were isolated using the Naive T cells isolation kit (Miltenyi Biotec). Transfections were done using the Human T cells Nucleofector kit (Amaxa Biosystems) according to the manufacturer’s instructions. For MLR, responder cells (T cells) and stimulator cells (PBMCs) were isolated from blood of different healthy human donors.
Expression plasmids
The GCaMP3 plasmid has been described (Tian et al., 2009). To direct the expression of GCaMP3 to different cellular compartments, three copies of a nuclear localization signal or a nuclear export signal were inserted at the C terminus of GCaMP3 to generate GCaMP3.NLS and GCaMP3.NES, respectively (Bengtson et al., 2010). We generated two different plasmids containing an expression cassette for CaMBP4 fused to either mCherry or GFP. The CaMBP4.mCherry expression vector was used in FACS and immunocytochemistry, in which an mCherry expression vector served as a control. The CaMBP4.GFP expression vector was used for FACS, followed by QRT-PCR, Western blot, and transcriptome analysis experiments, in which an EGFP expression vector (pEGFP-N1; Takara Bio Inc.) served as a control. The mutated inactive version of CaMBP4 (mut-CaMBP4.mCherry) has been described (Lau et al., 2015). Wt-CREB.GFP and mut-CREB.GFP were generated by insertion of an expression cassette for wt-CREB and mut-CREB (Tan et al., 2012) into pEGFP-N1.
Cell stimulation
For stimulation with CD3 + CD28 antibodies, 24-well plates were precoated with 6 µg/ml goat anti–mouse antibodies in PBS (Dianova) overnight. After 30-min blocking with RPMI containing 10% FBS, the wells were incubated with 1 µg/ml CD3 antibodies (eBioscence) and 10 µg/ml CD28 antibodies (BD) diluted in RPMI containing 10% FBS for 1 h. The plates were then washed with RPMI, and the cells were plated for the indicated time (Wabnitz et al., 2007). Alternatively, cells were incubated with 1 nM PMA (Sigma-Aldrich) and 1 µM ionomycin (MP Biomedicals) for the indicated time. For anergy induction in vitro, T cells were pretreated overnight with plate-bound CD3 antibodies. The cells were then washed and left for 3 h. The cells were restimulated with plate-bound CD3 and CD28 antibodies for 2 h. Where indicated, T cells were treated with the CaM kinases inhibitor, KN93 (10 µM), for 30 min before stimulation.
Calcium measurements
To measure calcium signals in different intracellular compartments, T cells were transfected with expression vectors containing GCaMP3, GCaMP3.NLS, and GCaMP3.NES. The cells were preincubated on ice with CD3 antibodies or a combination of CD3 and CD28 antibodies (eBioscience) for 15 min. The cells were washed twice and resuspended in RPMI containing 2% FBS. The intracellular calcium concentrations were analyzed by flow cytometry (FACSAria; BD) by monitoring changes in the mean fluorescence of the GCaMP3-positive population over time. Samples were run for 90 s to measure the baseline and then treated with 0.6 µg/ml goat anti–mouse antibodies to enable antibody cross-linking. Samples were run for an additional 9 min. At the end of the analysis, 1 µM ionomycin (MP Biomedicals) was added to obtain the maximum signal. After 2 min, 50 mM EGTA (Sigma-Aldrich) was added to eliminate the signal. Analysis was done using Diva software (BD). The data are presented as ΔF/F0 = (F − F0)/F0, where F represents the mean of fluorescence in a region of interest (a certain interval of time) from which the mean background fluorescence intensity was subtracted; F0 represents baseline F (measured before stimulation with the secondary antibody).
For measuring total intracellular calcium levels in cells transfected with CaMBP4.mCherry and mCherry transfected, the cells were loaded with 0.2 µg/ml indo-1 for 45 min, resuspended in RPMI, and preincubated with CD3 antibodies. Baseline fluorescence signal was recorded for 10 min, and goat anti-mouse antibodies were added to induce cross-linking. The signal was measured for 9 min, and ionomycin was added to obtain the maximal signal. The ratios of fluorescence signal intensity at a wavelength of 390 versus 496 nm (bound/free indo-1) indicated the intracellular calcium levels.
Flow cytometry
For surface staining of membrane receptors, transfected cells were fixed for 10 min with 1.5% PFA at RT, washed in FACS buffer (PBS containing 0.5% BSA and 0.07% NaN3), and incubated with FITC- or phycoerythrin (PE)-labeled antibodies against CD25 and CD69 (BD) diluted in FACS buffer for 1 h at RT. For intracellular staining of cytokines, 10 µg/ml brefeldin A (Invitrogen) was added during the last 4 h of stimulation to obtain intracellular accumulation of cytokines. The cells were subsequently fixed in 1.5% PFA and permeabilized in FACS buffer containing 0.05% saponin. Cells were washed and incubated for 30 min with FITC- or PE-conjugated antibodies against IL-2 or IFNγ (BD), washed again, and analyzed by FACS. FITC-conjugated antibodies were used in FACS experiments with mCherry-transfected cells, and PE-conjugated antibodies were used in FACS experiments with GFP-transfected cells. For all FACS experiments, the analysis was restricted to the transfected cells (i.e., the mCherry- or GFP-expressing population). Data acquisition was done using an LSRII (BD), and data were analyzed using Diva software. For isolation of total RNA (for QRT-PCR and microarray analysis) or proteins (for immunoblot analysis), the GFP-expressing cells were sorted on a FACSAria cytometer (BD) after having been resuspended in ice-cold RPMI containing 10% FCS, 1 µg/ml propidium iodide, and 20 U/ml DNase (Ciccolini et al., 2005).
Immunocytochemistry
For analysis of the subcellular localization of NFAT-c1 (also known as NFAT2) and NFAT1, cells were stimulated with CD3 + CD28 antibodies for 6 h or 1 µM ionomycin for 30 min or were left unstimulated. Nuclear translocation of NFAT was inhibited by incubation of the cells with a combination of 0.1 µM cyclosporine A (Fluka) and 0.1 µM FK506 (Axxora) for 30 min before stimulation. The cells were centrifuged in a cytospin for 5 min at 500 rpm onto poly-l-lysine–coated coverslips (Sigma-Aldrich), fixed in 4% PFA for 20 min at RT, and permeabilized by washing three times in wash buffer (PBS containing 0.5% NP-40 and 0.01% NaN3). Nonspecific binding was blocked by incubation with wash buffer containing 10% FBS for 30 min at RT. The antibody incubation was done for 1 h at RT using antibodies against NFATC1 (Enzo Life Sciences) to detect NFAT-c1 or antibodies against NFAT1 (Merck Millipore) followed by incubation with Alexa Fluor 488–conjugated goat anti–rabbit antibodies (Invitrogen) for 1 h at RT (Feske et al., 2000). Counterstain of nuclei was done with Hoechst 33258 (2 µg/ml; Serva). Samples were mounted in Mowiol. Stained samples were analyzed using a DM IRBE microscope (Leica Biosystems) and a CCD camera (Spot insight2; Visitron Systems) with 100× (numerical aperture 1.3) or 40× (numerical aperture 1.25–0.75) oil-immersion objectives. Data were acquired with Spot Advance software. Nuclear NFAT-c1 and NFAT1 immunoreactivity was quantified using ImageJ. Results are expressed as percentage of NFAT-c1 and NFAT1 nuclear immunoreactivity obtained in stimulated control cells expressing mCherry. Images of GCaMP3-transfected cells were acquired using a confocal laser-scanning microscope (TCS SP2; Leica Biosystems) equipped with an inverted fluorescence microscope (DM IRE2; Leica Biosystems) and Leica confocal scan software.
QRT-PCR
FACS was used to isolate GFP-expressing transfected cells. mRNA expression levels of Crem, Egr2, Egr3, Dusp2, Ikzf5, IL-2, and B2M were measured by QRT-PCR using real-time TaqMan technology with a sequence detection model 7300 PCR system (Applied Biosystems). Total RNA was isolated with RNeasy Mini kit (QIAGEN); DNaseI treatment was for 15 min according to the manufacturer’s instructions (QIAGEN). For the generation of first-strand cDNA, 1 µg total RNA was reverse transcribed by extension of oligo(dT) primers using SuperScript III reverse transcription (Invitrogen) according to the manufacturer’s instructions. QRT-PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems) with Assays-on-Demand Gene Expression Products. The human probes were Assays-on-Demand Gene Expression Products with TaqMan MGB probes, FAM dye labeled. The thermal cycling conditions were 10 min at 95°C, 55 cycles of 15 s for denaturation at 95°C, and 60 s for annealing and extension at 60°C. The expression levels of the target mRNA were normalized to the relative ratio of the expression of B2M mRNA using the ΔΔCt cycle threshold method. Each QRT-PCR assay was performed at least three times, and the results are expressed as means ± SEM (Zhang et al., 2011a).
Immunoblot analysis
GFP-expressing cells were sorted, stimulated with PMA + ionomycin for the indicated time, and subjected to standard immunoblot analysis. Antibodies against CREB (Cell Signaling Technology) and the serine-133 phosphorylated form of CREB (EMD Millipore) were used.
Mixed lymphocyte reaction
Responder cells (T cells) and stimulator cells (PBMCs) were isolated from different healthy human donors. T cells were transfected with expression vectors for mCherry or CaMBP4.mCherry. PBMC stimulator cells were irradiated with 3,000 cGy from a 137Cs source. T cells and PBMCs were co-cultured at a ratio of 1:3. T cells (4 × 105) were incubated with 1 µM CFSE in serum-free RPMI for 15 min and washed twice with RPMI supplemented with 10% FBS. T cells were cultured with 1.2 × 106 PBMCs in 2 ml RPMI medium, and aliquots of the cell suspension were harvested every day. For FACS analysis, cells were fixed in 1.5% PFA; analysis of CFSE fluorescence was restricted to mCherry-expressing cells (Tzachanis et al., 2002). Proliferation was calculated based on the percentage of cells in which CFSE fluorescence decreased over time. The proliferation gate was set to the basal CFSE fluorescence immediately after CFSE loading.
Gene profiling and bioinformatics analysis
The quality of total RNA from GFP-expressing human T cells was determined by gel analysis using the total RNA Nano chip assay on an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA concentrations were determined using the NanoDrop spectrophotometer (NanoDrop Technologies). Microarray analysis was performed in the Genomics and Proteomics Core Facility at the German Cancer Research Center (Heidelberg, Germany). Biotin-labeled cRNA samples for hybridization on Illumina Human Sentrix-12 BeadChip arrays (Illumina) were prepared according to Illumina’s recommended sample-labeling procedure based on the modified Eberwine protocol (Eberwine et al., 1992). We selected the genes up- or down-regulated with a p-value <0.05 in CaMBP4.GFP versus control (GFP) samples (t test with Benjamini–Hochberg correction for multiple comparisons). A cutoff of 1.3 was applied for up-regulated genes and 0.7 for down-regulated genes. To represent the data, we created a heatmap with cluster dendrogram of 939 differentially expressed genes (log2 fold change) at a false discovery rate of 5% in at least one of the following conditions: CaMBP4.GFP/GFP at 0, 2, 6, or 24 h. The heatmap was depicted by color-coding genewise log2 fold change with the heatmap.2 function from the gplots programming tool CRAN (https://cran.r-project.org/package=gplots). Default maximum value is 2.0. A ratio greater than or equal to 2.0 is depicted with saturated red; less than or equal to −2.0 is depicted with saturated green.
Statistics
Data represent means ± SEM. Analysis of variance with Tukey’s post hoc test was used for statistical analysis.
Study approval
The blood samples were taken from healthy human donors. Written informed consent was received from the donors before inclusion in the study.
Online Supplemental Material
Fig. S1 shows that the stimulation of T cells with CD3 antibodies or with a combination of CD3 and CD28 antibodies induced similar nuclear calcium transients. Fig. S2 shows that CaMBP4.mCherry does not alter calcium transients in response to TCR stimulation (A) and that the expression of an inactive form of CaMBP4 (mut-CaMBP4.mCherry) does not block IL-2 production (B). Fig. S3 shows that CaMBP4.mCherry expression had the same inhibitory effect on IL-2 and CD25 production in human pan-T cells and human naive T cells. Fig. S4 shows that CaMBP4.mCherry inhibits T cell proliferation in an MLR. Fig. S5 shows that CaMBP4.mCherry does not inhibit NFAT1 nuclear translocation upon stimulation with a combination of CD3 and CD28 antibodies.
Supplementary Material
Acknowledgments
We are grateful to Yen Wei Tan for helping with the bioinformatics analysis. We thank Gabriele Hölzl-Wenig and Sven Rüffer for technical assistance in flow cytometry, Ursula Weiss for immunoblot analyses, and Pegah Jabbari Lak for her help with some gene expression analyses. We thank Eckehard H. Freitag, Anna M. Hagenston, and David Lau for providing constructs and C. Peter Bengtson, David Lau, Daniela Mauceri, and Henning Kirchgessner for discussions.
This work was supported by a European Research Council Advanced Grant (H. Bading) and the Sonderforschungsbereich 938 of the Deutsche Forschungsgemeinschaft (Y. Samstag).
The authors declare no competing financial interests.
Author contributions: H. Bading conceived the project. H. Bading and S. Monaco wrote the manuscript. S. Monaco performed all experiments. B. Jahraus purified human T cells and contributed to the experiment design. H. Bading, S. Monaco, and Y. Samstag supervised project planning. All authors discussed the results and commented on the manuscript.
Footnotes
Abbreviations used:
- CaMKIV
- calcium/CaM-dependent protein kinase IV
- cAMP
- cyclic adenosine monophosphate
- CaN
- calcineurin
- CFSE
- carboxyfluorescein diacetate succinimidyl ester
- CREB
- cAMP response element–binding protein
- CREM
- cAMP-responsive element modulator
- DUSP
- dual-specificity phosphatase
- IL
- interleukin
- MLR
- mixed lymphocyte reaction
- NES
- nuclear export signal
- NFAT
- nuclear factor of activated T cells
- PBMC
- peripheral blood mononuclear cell
- PE
- phycoerythrin
- PTP
- protein tyrosine phosphatase
- QRT-PCR
- quantitative RT-PCR
- TCR
- T cell receptor
References
- Bading H. 2013. Nuclear calcium signalling in the regulation of brain function. Nat. Rev. Neurosci. 14:593–608. 10.1038/nrn3531 [DOI] [PubMed] [Google Scholar]
- Baine I., Abe B.T., and Macian F.. 2009. Regulation of T-cell tolerance by calcium/NFAT signaling. Immunol. Rev. 231:225–240. 10.1111/j.1600-065X.2009.00817.x [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S., Soto-Nieves N., and Macián F.. 2007. Transcriptional regulation of T cell tolerance. Semin. Immunol. 19:180–187. 10.1016/j.smim.2007.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson C.P., Freitag H.E., Weislogel J.M., and Bading H.. 2010. Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys. J. 99:4066–4077. 10.1016/j.bpj.2010.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M.J., Lipp P., and Bootman M.D.. 2000. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1:11–21. 10.1038/35036035 [DOI] [PubMed] [Google Scholar]
- Bodor J., and Habener J.F.. 1998. Role of transcriptional repressor ICER in cyclic AMP-mediated attenuation of cytokine gene expression in human thymocytes. J. Biol. Chem. 273:9544–9551. 10.1074/jbc.273.16.9544 [DOI] [PubMed] [Google Scholar]
- Chawla S., Hardingham G.E., Quinn D.R., and Bading H.. 1998. CBP: A signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 281:1505–1509. 10.1126/science.281.5382.1505 [DOI] [PubMed] [Google Scholar]
- Chen J.C., Chang M.L., and Muench M.O.. 2003. A kinetic study of the murine mixed lymphocyte reaction by 5,6-carboxyfluorescein diacetate succinimidyl ester labeling. J. Immunol. Methods. 279:123–133. 10.1016/S0022-1759(03)00236-9 [DOI] [PubMed] [Google Scholar]
- Chikuma S., Terawaki S., Hayashi T., Nabeshima R., Yoshida T., Shibayama S., Okazaki T., and Honjo T.. 2009. PD-1-mediated suppression of IL-2 production induces CD8+ T cell anergy in vivo. J. Immunol. 182:6682–6689. 10.4049/jimmunol.0900080 [DOI] [PubMed] [Google Scholar]
- Ciccolini F., Mandl C., Hölzl-Wenig G., Kehlenbach A., and Hellwig A.. 2005. Prospective isolation of late development multipotent precursors whose migration is promoted by EGFR. Dev. Biol. 284:112–125. 10.1016/j.ydbio.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Clapham D.E. 2007. Calcium signaling. Cell. 131:1047–1058. 10.1016/j.cell.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Cook R., Wu C.C., Kang Y.J., and Han J.. 2007. The role of the p38 pathway in adaptive immunity. Cell. Mol. Immunol. 4:253–259. [PubMed] [Google Scholar]
- Crabtree G.R. 1999. Generic signals and specific outcomes: Signaling through Ca2+, calcineurin, and NF-AT. Cell. 96:611–614. 10.1016/S0092-8674(00)80571-1 [DOI] [PubMed] [Google Scholar]
- Di Capite J., Ng S.W., and Parekh A.B.. 2009. Decoding of cytoplasmic Ca(2+) oscillations through the spatial signature drives gene expression. Curr. Biol. 19:853–858. 10.1016/j.cub.2009.03.063 [DOI] [PubMed] [Google Scholar]
- Dolmetsch R.E., Xu K., and Lewis R.S.. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 392:933–936. 10.1038/31960 [DOI] [PubMed] [Google Scholar]
- Eberwine J., Yeh H., Miyashiro K., Cao Y., Nair S., Finnell R., Zettel M., and Coleman P.. 1992. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA. 89:3010–3014. 10.1073/pnas.89.7.3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S. 2007. Calcium signalling in lymphocyte activation and disease. Nat. Rev. Immunol. 7:690–702. 10.1038/nri2152 [DOI] [PubMed] [Google Scholar]
- Feske S., Draeger R., Peter H.H., Eichmann K., and Rao A.. 2000. The duration of nuclear residence of NFAT determines the pattern of cytokine expression in human SCID T cells. J. Immunol. 165:297–305. 10.4049/jimmunol.165.1.297 [DOI] [PubMed] [Google Scholar]
- Feske S., Giltnane J., Dolmetsch R., Staudt L.M., and Rao A.. 2001. Gene regulation mediated by calcium signals in T lymphocytes. Nat. Immunol. 2:316–324. 10.1038/86318 [DOI] [PubMed] [Google Scholar]
- Greenwald R.J., Boussiotis V.A., Lorsbach R.B., Abbas A.K., and Sharpe A.H.. 2001. CTLA-4 regulates induction of anergy in vivo. Immunity. 14:145–155. 10.1016/S1074-7613(01)00097-8 [DOI] [PubMed] [Google Scholar]
- Hardingham G.E., Chawla S., Johnson C.M., and Bading H.. 1997. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 385:260–265. 10.1038/385260a0 [DOI] [PubMed] [Google Scholar]
- Hardingham G.E., Arnold F.J., and Bading H.. 2001. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat. Neurosci. 4:261–267. 10.1038/85109 [DOI] [PubMed] [Google Scholar]
- Harris J.E., Bishop K.D., Phillips N.E., Mordes J.P., Greiner D.L., Rossini A.A., and Czech M.P.. 2004. Early growth response gene-2, a zinc-finger transcription factor, is required for full induction of clonal anergy in CD4+ T cells. J. Immunol. 173:7331–7338. 10.4049/jimmunol.173.12.7331 [DOI] [PubMed] [Google Scholar]
- Heissmeyer V., Macián F., Im S.H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y.C., Dustin M.L., and Rao A.. 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat. Immunol. 5:255–265. 10.1038/ni1047 [DOI] [PubMed] [Google Scholar]
- Higazi D.R., Fearnley C.J., Drawnel F.M., Talasila A., Corps E.M., Ritter O., McDonald F., Mikoshiba K., Bootman M.D., and Roderick H.L.. 2009. Endothelin-1-stimulated InsP3-induced Ca2+ release is a nexus for hypertrophic signaling in cardiac myocytes. Mol. Cell. 33:472–482. 10.1016/j.molcel.2009.02.005 [DOI] [PubMed] [Google Scholar]
- Hogan P.G., Chen L., Nardone J., and Rao A.. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205–2232. 10.1101/gad.1102703 [DOI] [PubMed] [Google Scholar]
- Hook S.S., and Means A.R.. 2001. Ca(2+)/CaM-dependent kinases: From activation to function. Annu. Rev. Pharmacol. Toxicol. 41:471–505. 10.1146/annurev.pharmtox.41.1.471 [DOI] [PubMed] [Google Scholar]
- Juang Y.T., Wang Y., Solomou E.E., Li Y., Mawrin C., Tenbrock K., Kyttaris V.C., and Tsokos G.C.. 2005. Systemic lupus erythematosus serum IgG increases CREM binding to the IL-2 promoter and suppresses IL-2 production through CaMKIV. J. Clin. Invest. 115:996–1005. 10.1172/JCI22854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar P., Nelson C., and Parekh A.B.. 2011. Selective activation of the transcription factor NFAT1 by calcium microdomains near Ca2+ release-activated Ca2+ (CRAC) channels. J. Biol. Chem. 286:14795–14803. 10.1074/jbc.M111.220582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortum R.L., Rouquette-Jazdanian A.K., and Samelson L.E.. 2013. Ras and extracellular signal-regulated kinase signaling in thymocytes and T cells. Trends Immunol. 34:259–268. 10.1016/j.it.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.J., Wang R., Ma J., and Sun Z.. 2010. PKC-θ is a drug target for prevention of T cell-mediated autoimmunity and allograft rejection. Endocr. Metab. Immune Disord. Drug Targets. 10:367–372. 10.2174/1871530311006040367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen C.P., Elwood E.T., Alexander D.Z., Ritchie S.C., Hendrix R., Tucker-Burden C., Cho H.R., Aruffo A., Hollenbaugh D., Linsley P.S., et al. . 1996. Long-term acceptance of skin and cardiac allografts after blocking CD40 and CD28 pathways. Nature. 381:434–438. 10.1038/381434a0 [DOI] [PubMed] [Google Scholar]
- Laskowski I.A., Pratschke J., Wilhelm M.J., Dong V.M., Beato F., Taal M., Gasser M., Hancock W.W., Sayegh M.H., and Tilney N.L.. 2002. Anti-CD28 monoclonal antibody therapy prevents chronic rejection of renal allografts in rats. J. Am. Soc. Nephrol. 13:519–527. [DOI] [PubMed] [Google Scholar]
- Lau D., Bengtson C.P., Buchthal B., and Bading H.. 2015. BDNF reduces toxic extrasynaptic NMDA receptor signaling via synaptic NMDA receptors and nuclear-calcium-induced transcription of inhba/activin A. Cell Reports. 12:1353–1366. 10.1016/j.celrep.2015.07.038 [DOI] [PubMed] [Google Scholar]
- Lechner O., Lauber J., Franzke A., Sarukhan A., von Boehmer H., and Buer J.. 2001. Fingerprints of anergic T cells. Curr. Biol. 11:587–595. 10.1016/S0960-9822(01)00160-9 [DOI] [PubMed] [Google Scholar]
- Lewis R.S. 2001. Calcium signaling mechanisms in T lymphocytes. Annu. Rev. Immunol. 19:497–521. 10.1146/annurev.immunol.19.1.497 [DOI] [PubMed] [Google Scholar]
- Limbäck-Stokin K., Korzus E., Nagaoka-Yasuda R., and Mayford M.. 2004. Nuclear calcium/calmodulin regulates memory consolidation. J. Neurosci. 24:10858–10867. 10.1523/JNEUROSCI.1022-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioudyno M.I., Kozak J.A., Penna A., Safrina O., Zhang S.L., Sen D., Roos J., Stauderman K.A., and Cahalan M.D.. 2008. Orai1 and STIM1 move to the immunological synapse and are up-regulated during T cell activation. Proc. Natl. Acad. Sci. USA. 105:2011–2016. 10.1073/pnas.0706122105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián F., García-Rodríguez C., and Rao A.. 2000. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19:4783–4795. 10.1093/emboj/19.17.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián F., López-Rodríguez C., and Rao A.. 2001. Partners in transcription: NFAT and AP-1. Oncogene. 20:2476–2489. 10.1038/sj.onc.1204386 [DOI] [PubMed] [Google Scholar]
- Macián F., García-Cózar F., Im S.H., Horton H.F., Byrne M.C., and Rao A.. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 109:719–731. 10.1016/S0092-8674(02)00767-5 [DOI] [PubMed] [Google Scholar]
- Macián F., Im S.H., García-Cózar F.J., and Rao A.. 2004. T-cell anergy. Curr. Opin. Immunol. 16:209–216. 10.1016/j.coi.2004.01.013 [DOI] [PubMed] [Google Scholar]
- Masquilier D., and Sassone-Corsi P.. 1992. Transcriptional cross-talk: Nuclear factors CREM and CREB bind to AP-1 sites and inhibit activation by Jun. J. Biol. Chem. 267:22460–22466. [PubMed] [Google Scholar]
- Mauceri D., Freitag H.E., Oliveira A.M., Bengtson C.P., and Bading H.. 2011. Nuclear calcium-VEGFD signaling controls maintenance of dendrite arborization necessary for memory formation. Neuron. 71:117–130. 10.1016/j.neuron.2011.04.022 [DOI] [PubMed] [Google Scholar]
- Oh-hora M., and Rao A.. 2008. Calcium signaling in lymphocytes. Curr. Opin. Immunol. 20:250–258. 10.1016/j.coi.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E., and Downie J.A.. 2006. Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9:351–357. 10.1016/j.pbi.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Papadia S., Stevenson P., Hardingham N.R., Bading H., and Hardingham G.E.. 2005. Nuclear Ca2+ and the cAMP response element-binding protein family mediate a late phase of activity-dependent neuroprotection. J. Neurosci. 25:4279–4287. 10.1523/JNEUROSCI.5019-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernis A.B. 2009. Rho GTPase-mediated pathways in mature CD4+ T cells. Autoimmun. Rev. 8:199–203. 10.1016/j.autrev.2008.07.044 [DOI] [PubMed] [Google Scholar]
- Popma S.H., Krasinskas A.M., McLean A.D., Szeto W.Y., Kreisel D., Moore J.S., and Rosengard B.R.. 2000. Immune monitoring in xenotransplantation: The multiparameter flow cytometric mixed lymphocyte culture assay. Cytometry. 42:277–283. [DOI] [PubMed] [Google Scholar]
- Powell J.D., Lerner C.G., Ewoldt G.R., and Schwartz R.H.. 1999. The -180 site of the IL-2 promoter is the target of CREB/CREM binding in T cell anergy. J. Immunol. 163:6631–6639. [PubMed] [Google Scholar]
- Quintana A., Pasche M., Junker C., Al-Ansary D., Rieger H., Kummerow C., Nuñez L., Villalobos C., Meraner P., Becherer U., et al. . 2011. Calcium microdomains at the immunological synapse: how ORAI channels, mitochondria and calcium pumps generate local calcium signals for efficient T-cell activation. EMBO J. 30:3895–3912. 10.1038/emboj.2011.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racioppi L., and Means A.R.. 2008. Calcium/calmodulin-dependent kinase IV in immune and inflammatory responses: Novel routes for an ancient traveller. Trends Immunol. 29:600–607. 10.1016/j.it.2008.08.005 [DOI] [PubMed] [Google Scholar]
- Rizzuto R., and Pozzan T.. 2006. Microdomains of intracellular Ca2+: Molecular determinants and functional consequences. Physiol. Rev. 86:369–408. 10.1152/physrev.00004.2005 [DOI] [PubMed] [Google Scholar]
- Safford M., Collins S., Lutz M.A., Allen A., Huang C.T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., and Powell J.D.. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472–480. 10.1038/ni1193 [DOI] [PubMed] [Google Scholar]
- Sho M., Sandner S.E., Najafian N., Salama A.D., Dong V., Yamada A., Kishimoto K., Harada H., Schmitt I., and Sayegh M.H.. 2002. New insights into the interactions between T-cell costimulatory blockade and conventional immunosuppressive drugs. Ann. Surg. 236:667–675. 10.1097/00000658-200211000-00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti M., Hagenston A.M., Vardeh D., Freitag H.E., Mauceri D., Lu J., Satagopam V.P., Schneider R., Costigan M., Bading H., and Kuner R.. 2013. Nuclear calcium signaling in spinal neurons drives a genomic program required for persistent inflammatory pain. Neuron. 77:43–57. 10.1016/j.neuron.2012.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y.W., Zhang S.J., Hoffmann T., and Bading H.. 2012. Increasing levels of wild-type CREB up-regulates several activity-regulated inhibitor of death (AID) genes and promotes neuronal survival. BMC Neurosci. 13:48 10.1186/1471-2202-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Hires S.A., Mao T., Huber D., Chiappe M.E., Chalasani S.H., Petreanu L., Akerboom J., McKinney S.A., Schreiter E.R., et al. . 2009. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 6:875–881. 10.1038/nmeth.1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzachanis D., Berezovskaya A., Nadler L.M., and Boussiotis V.A.. 2002. Blockade of B7/CD28 in mixed lymphocyte reaction cultures results in the generation of alternatively activated macrophages, which suppress T-cell responses. Blood. 99:1465–1473. 10.1182/blood.V99.4.1465 [DOI] [PubMed] [Google Scholar]
- Vincenti F., Charpentier B., Vanrenterghem Y., Rostaing L., Bresnahan B., Darji P., Massari P., Mondragon-Ramirez G.A., Agarwal M., Di Russo G., et al. . 2010. A phase III study of belatacept-based immunosuppression regimens versus cyclosporine in renal transplant recipients (BENEFIT study). Am. J. Transplant. 10:535–546. 10.1111/j.1600-6143.2009.03005.x [DOI] [PubMed] [Google Scholar]
- Wabnitz G.H., Köcher T., Lohneis P., Stober C., Konstandin M.H., Funk B., Sester U., Wilm M., Klemke M., and Samstag Y.. 2007. Costimulation induced phosphorylation of L-plastin facilitates surface transport of the T cell activation molecules CD69 and CD25. Eur. J. Immunol. 37:649–662. 10.1002/eji.200636320 [DOI] [PubMed] [Google Scholar]
- Wang J., Campos B., Jamieson G.A. Jr., Kaetzel M.A., and Dedman J.R.. 1995. Functional elimination of calmodulin within the nucleus by targeted expression of an inhibitor peptide. J. Biol. Chem. 270:30245–30248. 10.1074/jbc.270.51.30245 [DOI] [PubMed] [Google Scholar]
- Weislogel J.M., Bengtson C.P., Müller M.K., Hörtzsch J.N., Bujard M., Schuster C.M., and Bading H.. 2013. Requirement for nuclear calcium signaling in Drosophila long-term memory. Sci. Signal. 6:ra33 10.1126/scisignal.2003598 [DOI] [PubMed] [Google Scholar]
- Wells A.D. 2009. New insights into the molecular basis of T cell anergy: Anergy factors, avoidance sensors, and epigenetic imprinting. J. Immunol. 182:7331–7341. 10.4049/jimmunol.0803917 [DOI] [PubMed] [Google Scholar]
- Wen A.Y., Sakamoto K.M., and Miller L.S.. 2010. The role of the transcription factor CREB in immune function. J. Immunol. 185:6413–6419. 10.4049/jimmunol.1001829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.T., Shih H.M., and Lai M.Z.. 2001. Multiple signals required for cyclic AMP-responsive element binding protein (CREB) binding protein interaction induced by CD3/CD28 costimulation. J. Immunol. 166:284–292. 10.4049/jimmunol.166.1.284 [DOI] [PubMed] [Google Scholar]
- Zha Y., Marks R., Ho A.W., Peterson A.C., Janardhan S., Brown I., Praveen K., Stang S., Stone J.C., and Gajewski T.F.. 2006. T cell anergy is reversed by active Ras and is regulated by diacylglycerol kinase-alpha. Nat. Immunol. 7:1166–1173. 10.1038/ni1394 [DOI] [PubMed] [Google Scholar]
- Zhang S.J., Zou M., Lu L., Lau D., Ditzel D.A., Delucinge-Vivier C., Aso Y., Descombes P., and Bading H.. 2009. Nuclear calcium signaling controls expression of a large gene pool: Identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 5:e1000604 10.1371/journal.pgen.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.J., Buchthal B., Lau D., Hayer S., Dick O., Schwaninger M., Veltkamp R., Zou M., Weiss U., and Bading H.. 2011a A signaling cascade of nuclear calcium-CREB-ATF3 activated by synaptic NMDA receptors defines a gene repression module that protects against extrasynaptic NMDA receptor-induced neuronal cell death and ischemic brain damage. J. Neurosci. 31:4978–4990. 10.1523/JNEUROSCI.2672-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Fresnay S., Welty E., Sangrampurkar N., Rybak E., Zhou H., Cheng X.F., Feng Q., Avon C., Laaris A., et al. . 2011b Selective CD28 blockade attenuates acute and chronic rejection of murine cardiac allografts in a CTLA-4-dependent manner. Am. J. Transplant. 11:1599–1609. 10.1111/j.1600-6143.2011.03624.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.