Abstract
The stability of β‐catenin is very important for canonical Wnt signaling. A protein complex including Axin/APC/GSK3β phosphorylates β‐catenin to be degraded by ubiquitination with β‐TrCP. In the recent study, we isolated WDR26, a protein that binds to Axin. Here, we found that WDR26 is a negative regulator of the canonical Wnt signaling pathway, and that WDR26 affected β‐catenin levels. In addition, WDR26/Axin binding is involved in the ubiquitination of β‐catenin. These results suggest that WDR26 plays a negative role in β‐catenin degradation in the Wnt signaling pathway.
Keywords: Axin1, ubiquitination, WDR26, Wnt, β‐catenin
Abbreviations
CTLH, C‐terminal to LisH
GID glucose‐induced degradation‐deficient
LisH, lis homology
MO, morpholino oligonucleotides
Wnt signaling plays multiple important roles in disease and embryogenesis 1, 2. β‐Catenin is a key component in the canonical Wnt signaling pathway, and its stability and localization influence Wnt activity. Cytoplasmic levels of β‐catenin are kept low, in the ‘Wnt off’ state, by a degradation complex that includes Axin, APC, and GSK‐3β. CK1α initially phosphorylates β‐catenin at serine 45 (S45) of the N‐terminal region, and GSK‐3β subsequently phosphorylates β‐catenin at threonine 41 (T41), serine 37 (S37), and serine 33 (S33) 3. The phosphorylated β‐catenin is recognized and ubiquitinated at lysine 19 (K19) and lysine 49 (K49) by the ubiquitin E3 ligase, β‐TrCP. The ubiquitinated β‐catenin is degraded by the proteasome 4. In the ‘Wnt on’ state, the Wnt ligand binds to Frizzled receptor and the LRP complex at the cell surface, which leads to the membrane recruitment and activation of scaffold protein, Dishevelled. Activated Dishevelled inactivates the APC/Axin/GSK‐3 complex in the cytoplasm, thus reducing degradation of β‐catenin. The stabilized β‐catenin translocates to the nucleus and associates with TCF/LEF transcription factors to activate Wnt target genes 5.
Axin1 is a scaffold protein including a multidomain that has many functions in biological signaling pathways. Axin1 contains: a regulation of G‐protein signaling (RGS) domain, which binds to APC, at the N‐terminus; a dishevelled and axin (DIX) domain and a protein phosphatase 2 (PP2A)‐binding domain at the C‐terminus; and binding domains of GSK‐3β and β‐catenin in the center region 6, 7. In the canonical Wnt pathway, Axin1 is a component of the β‐catenin destruction complex that negatively controls Wnt signaling. Axin1 down‐regulates the amount of cytoplasmic β‐catenin, to function as a tumor suppressor gene, and several mutations of Axin1 have been identified in tumor cell lines 8, 9. In Xenopus development, Axin1 functions as a ventralizing gene 10. However, in zebrafish development, Axin1 shows dorsalizing activity in the JNK‐mediated signal pathway 11. Thus, Axin1 is a multifunctional gene and its function is dependent on binding partners.
WDR26 contains several protein‐interacting domains: the lis homology domain (LisH); the C‐terminal to LisH motif (CTLH) domain; and a WD40 repeat domain 12. A previous study suggested that WDR26 contributes to the MAPK signaling pathway 13. In a study of chemoattractive migration of leukocytes, WDR26 binds to the Gβ‐gamma complex and positively controls leukocyte migration 14, 15. However, there are no reports that WDR26 is associated with the Wnt signaling pathway. In the yeast Saccharomyces cerevisiae, nine glucose‐induced degradation‐deficient (GID) genes (GID1–GID9) were isolated 16, 17. The GID complex, which comprises GID1–GID9 except for GID6, acts as a polyubiquitination enzyme in yeast. Eight vertebrate homologs that share high similarities of domain architecture to yeast GID complex genes have been identified 12. The following are the yeast GID complex genesand their corresponding vertebrate homologs: GID1/RanBP9; GID2/Rmnd5; GID3/UBE2H; GID4/C17ors39; GID5/ARMc8; GID7/WDR26; GID8/TWA1; and GID9/MAEA 12. Recent studies showed that RMND5 and ARMc8 promote ubiquitination in vertebrates 18, 19, but it is still unknown whether other vertebrate homologs including WDR26 are associated with the ubiquitination pathway.
Previously, WDR26 is identified as an Axin1‐binding protein 20. In the present study, we found that WDR26, with Axin1, controlled β‐catenin levels to negatively regulate the expression of Wnt target genes. We also found that the binding between Axin1 and WDR26 is necessary for the ubiquitination of β‐catenin. These results reveal a function of WDR26 in the canonical Wnt signaling pathway.
Materials and methods
Plasmid construction
The human and Xenopus WDR26 and Axin1 were amplified by RT‐PCR from cDNA templates prepared from HEK 293T cells and Xenopus embryos, respectively, and were subcloned into the pRK5 and modified pCS2+ vectors. Each truncated mutant was constructed by PCR and contained the following amino acid (aa) sequences. xWDR26‐1: 1–283 aa, xWDR26‐2: 1–434 aa, xWDR26‐3: 97–611 aa, xWDR26‐4: 284–611 aa, xWDR26‐5: 435–611 aa, xWDR26‐ΔLisH: 1–63 aa and 97–611 aa, xWDR26‐ΔCTLH: 1–96 aa and 182–611 aa, xWDR26‐ΔLisH‐CTLH: 1–63 aa and 182–611 aa, xAxin1‐1: 1–230 aa, xAxin1‐2: 1–450 aa, xAxin1‐3: 1–610 aa, xAxin1‐4: 231–841 aa, xAxin1‐5: 451–841 aa, xAxin1‐6: 611–841 aa. FLAG tags were fused to the N‐terminus of hWDR26 and xβ‐catenin, and C‐terminus of β‐globin, xWDR26, and its truncated constructs. MYC tags were fused to the N‐terminus of hβ‐catenin and xβ‐catenin, and C‐terminus of hAxin1. HA tag was fused to N‐terminus of ubiquitin.
Embryo handling and morpholino oligonucleotides
Capped mRNA were synthesized from linearized vectors using SP6 (Roche, Basel, Switzerland) and T7 (Toyobo, Osaka, Japan) RNA polymerase. The morpholino oligonucleotides (MO) (Gene Tools, LLC, Philomath, OR, USA) used here were previously reported 21, 22 and are as follows: 5′‐CGCTGCCCCGTTAGCCTGCATGTTA‐3′ (xWDR26‐MO). The specificity of each MO was confirmed by its ability to inhibit the translation of FLAG‐tagged mRNA containing the targeted site with or without five‐mismatched sequences. MO (10 ng) and FLAG‐tagged mRNA (100 pg) were coinjected with β‐globin‐FLAG mRNA (100 pg) as a loading control into the animal poles of four‐cell stage embryos, and the injected animal caps were dissected at stage 10. Lysates from the animal caps were subjected to western blotting with an anti‐FLAG antibody (M2, Sigma, St. Louis, MO, USA).
Morpholino oligonucleotides, mRNA, and plasmids were injected into two animal dorsal blastomeres at the eight‐cell stage for observation of embryo phenotypes and RT‐PCR analysis, into four animal blastomeres for western blot analysis, or into two ventral blastomeres at the four‐cell stage for RT‐PCR analysis. The cytoplasmic and nuclear fractions were prepared as described with modifications 23. Quantitative RT‐PCR analysis: Total RNA was prepared using TRIzol (Invitrogen, Carlsbad, CA, USA) from the injected region at the gastrula or neurula stage. cDNA synthesis was carried out using Moloney murine leukemia virus reverse transcriptase (Invitrogen). The sequences of the primer pairs were previously reported 24, 25, 26 and as follows: xWDR26: Forward 5′‐ATGGCAACCTGCTTGACTCC‐3′; Reverse 5′‐ACAGTACCGTCGTCAGAAGC‐3′. hWDR26: Forward 5′‐CCGGAACTCGCCTGCTTGTC‐3′; Reverse 5′‐TGACATCCTCATCTGACTGG‐3′. Xnr3: Forward 5′‐CTTCTGCACTAGATTCTG‐3′; Reverse 5′‐CAGCTTCTGGCCAAGACT‐3′. Xtwn: Forward 5′‐AACCCAAGAAGGCGACACTATC‐3′; Reverse 5′‐GTGCCGATGGTAGGAAATGATC‐3′. Xenopus embryonic ornithine decarboxylase (xODC) was used for normalization of cDNA samples.
Whole‐mount in situ hybridization
Whole‐mount in situ hybridization was performed as previously described 27. A cDNA fragment of xWDR26 (nucleotides 709–1836; GenBank Accession No. LC066599) was subcloned into pBluescriot vectors, and used as templates to generate the digoxigenin‐labeled antisense and sense RNA probes.
Antibodies, siRNA, and cell lines
The antibodies used here were previously reported 22, 24 and as follows: Peroxidase conjugated anti‐GAPDH (G9295; Wako, Osaka, Japan), horseradish peroxidase‐conjugated anti‐rat IgG (GE, Little Chalfont, UK), anti‐T7 (PM022, MBL, Nagoya, Japan), anti‐HA (3F10, Roche), anti‐WDR26 (NBP1‐78041, Acris Antibodies, Herford, Germany), anti‐β‐catenin (ab32572, Abcam, Cambridge, UK) and anti‐Axin1 (C76H11, Cell Signaling, Danvers, MA, USA). The target sequences of siRNA against human WDR26 were 5′‐TACCAAATTCCGAAATCATGTCA‐3′ (si‐hWDR26‐1), 5′‐GGCCGATTAGCTTTGTTAAATGT‐3′ (si‐hWDR26‐2), and 5′‐ATCCTATTATGTCTTTTACTATT‐3′ (si‐hWDR26‐3). Four‐day Wnt‐3A conditioned medium from L‐Wnt‐3A cells was used for Wnt stimulation of cultured cells. We used the following cell lines: HEK 293T cells, L cells, and L Wnt3A cells. The growth medium for each cell type is described in the American Type Culture Collection 22. Three‐day Wnt‐3A‐conditioned medium from L‐Wnt‐3A cells was used for Wnt stimulation of cultured cells. For western blotting analysis, at least three independent experiments were performed, and representative data were shown in figures.
Results
Interaction between WDR26 and Axin1
To identify novel proteins that bind to human Axin1 (hAxin1), we previously performed a high‐throughput analysis of proteins that coimmunoprecipitated with hAxin1 in HEK 293 cells using direct nanoflow liquid chromatography‐coupled tandem mass spectrometry 20. We identified human WDR26 (hWDR26) as a candidate protein that binds to hAxin1. The interaction of ectopically expressed hWDR26 with hAxin1 was confirmed in HEK 293T cells (Fig. 1A). The endogenous interaction of hWDR26 and hAxin1 was also confirmed in HEK 293T cells (Fig. 1B). Amino acid sequences of both WDR26 and Axin1 are well conserved between vertebrate homologs, and an interaction between ectopically expressed Xenopus WDR26 (xWDR26) with Xenopus Axin1 (xAxin1) was also confirmed in HEK 293T cells (Fig. 1C). To determine the region in xWDR26 responsible for binding to xAxin1, several truncated mutants of xWDR26 were examined using coimmunoprecipitation assays. We found that the N‐terminal region including LisH domain was responsible for binding to xAxin1 (Fig. 1D). Conversely, xWDR26 bound to xAxin1 at the central region including GSK3β‐binding domain (Fig. 1E).
Figure 1.
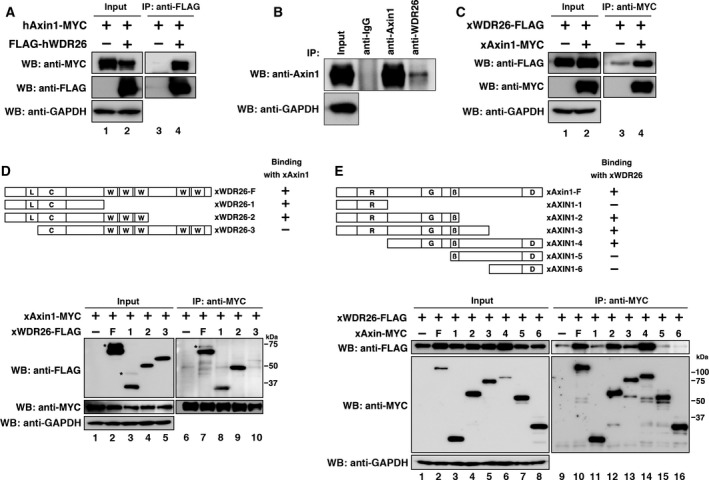
Interaction between WDR26 and Axin1. (A) Interaction between ectopically expressed hWDR26 and hAxin1 in HEK 293T cells. Relative intensity of each western blot was measured by imagej. The ratios of IP/Input of hAxin1 were calculated. Their statistical significances were determined by Mann–Whitney U test. P < 0.01 (between lane 3/lane 1 and lane 4/lane 2). (B) Interaction between endogenous hWDR26 and hAxin1 in HEK 293T cells. Cells were treated with 10 μm MG‐132 for 24 h to improve the detection. The endogenous interaction between hWDR26 and hAxin1was not detected with an anti‐WDR26 antibody (data not shown). (C) Interaction between ectopically expressed xWDR26 and xAxin1 in HEK 293T cells. Relative intensity of each western blot was measured by imagej. The ratios of IP/Input of xWDR26 were calculated. Their statistical significances were determined by Mann–Whitney U test. P < 0.001 (between lane 3/lane 1 and lane 4/lane 2). (D) Interactions among ectopically expressed xAxin1 and truncated xWDR26 constructs. WDR26 protein‐interacting domains are labeled as follows: L: LisH (lis homology domain); C: CTLH (C‐terminal to LisH motif) domain; W: WD40 repeat domain. Asterisks indicate higher molecular weight bands. Constructs were transfected into HEK 293T cells. Relative intensity of each western blot was measured by imagej. The ratios of IP/Input of xWDR26 constructs were calculated. Their statistical significances were determined by Mann–Whitney U test. P > 0.5 (between lane 7/lane 2 and lane 8/lane 3). P > 0.1 (between lane 7/lane 2 and lane 9/lane 4). P < 0.001 (between lane 7/lane 2 and lane 10/lane 5). (E) Interactions among ectopically expressed xWDR26 and truncated xAxin1 constructs. Axin1 protein‐interacting domains are labeled as follows: R: RGS (regulation of G‐protein signaling) domain; G: GSK3‐β binding domain; β: β‐catenin binding domain; D: DIX (Dishevelled and axin) domain. Constructs were transfected into HEK 293T cells. Relative intensity of each western blot was measured by imagej. The ratios of IP/Input of xAxin1 were calculated. Their statistical significances were determined by Mann–Whitney U test. P < 0.01 (between lane 10/lane 2 and lane 11/lane 3). P > 0.5 (between lane 10/lane 2 and lane 12/lane 4). P > 0.05 (between lane 10/lane 2 and lane 13/lane 5). P > 0.1 (between lane 10/lane 2 and lane 14/lane 6). P < 0.01 (between lane 10/lane 2 and lane 15/lane 7). P < 0.01 (between lane 10/lane 2 and lane 16/lane 8).
Expression patterns of xWDR26 during Xenopus embryogenesis
To assess the possible function of xWDR26 in Xenopus embryonic development, we first examined the temporal and spatial expression patterns of xWDR26 by RT‐PCR analysis and whole‐mount in situ hybridization (Fig. 2A,B–I). We found that the expression of xWDR26 remained at a relatively constant level until stage 15, and tended to decrease slightly after stage 18 (Fig. 2A). Whole‐mount in situ hybridization revealed that xWDR26 was expressed broadly until the gastrula stage (data not shown). Expression of xWDR26 was gradually localized to the neural region from the late gastrula (Fig. 2B) to the early neurula stage (Fig. 2C). In the late neurula and tadpole stages, xWDR26 was strongly expressed in the anterior neural region (Fig. 2D,E). These results suggest that xWDR26 might be involved in anterior formation in Xenopus embryos.
Figure 2.
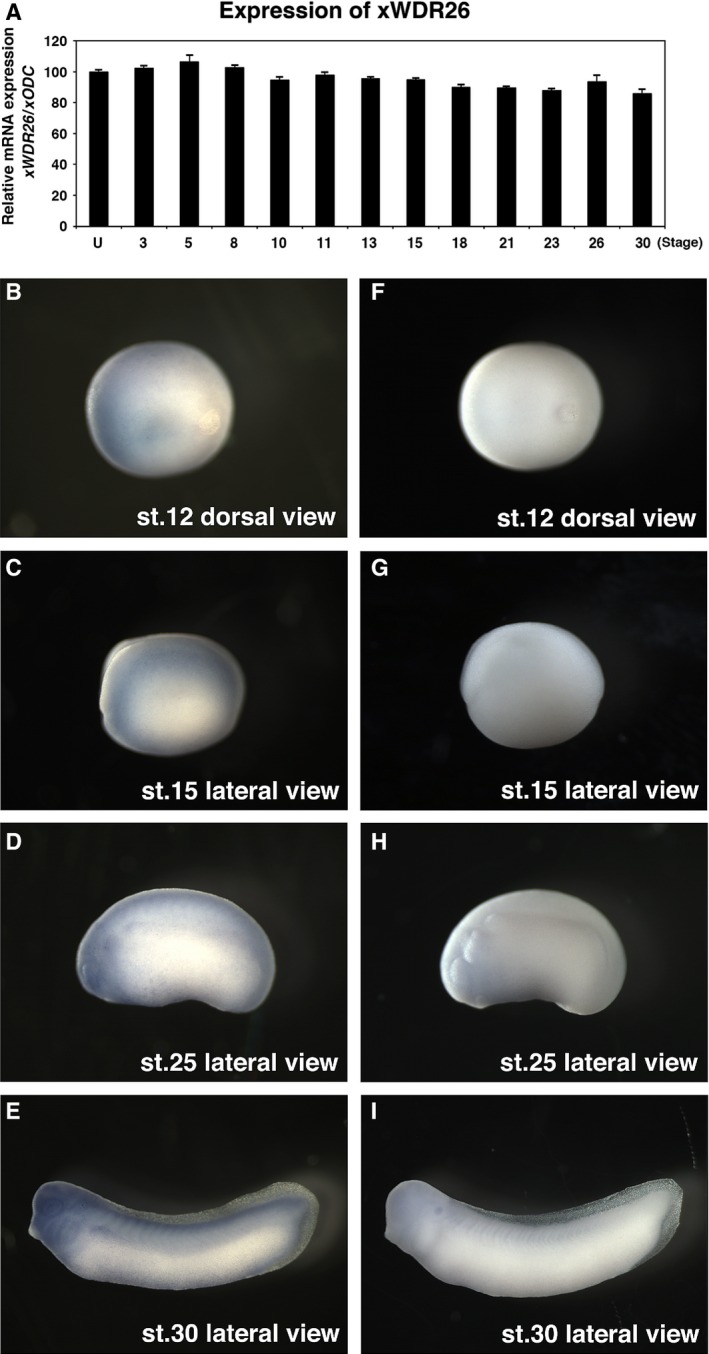
Expression of xWDR26 during Xenopus embryogenesis. (A) Quantitative RT‐PCR revealed temporal expression of xWDR26. Numbers under lanes indicate developmental stages; U, unfertilized eggs. The value obtained for xWDR26 was normalized to the level of xODC (ornithine decarboxylase). The value of unfertilized eggs was set to 100 and other values were computed. Error bars represent standard deviation of the mean in three experiments. Statistical significances of xWDR26/xODC between unfertilized eggs and other stages were determined by Mann–Whitney U test. P > 0.05 (until stage 15, and stage 26), P < 0.01 (after stage 18, except for stage 26). (B–I) Whole‐mount in situ hybridization. (B–E) Anti‐sense RNA probe of xWDR26 cDNA fragment was used. (F–I) Sense RNA probe of xWDR26 cDNA fragment was used. (B and C). Expression of xWDR26 gradually localizes to anterior neural region (B, stage 12; C, stage 15). (D and F) xWDR26 is strongly expressed in the anterior neural region (D, stage 25; E, stage 30).
WDR26 is involved in the canonical Wnt pathway
To determine whether WDR26 is involved in the canonical Wnt pathway, we investigated the effects of knockdown of WDR26 on canonical Wnt activity. We first validated antisense morpholino oligonucleotides for xWDR26 (xWDR26‐MO) in Xenopus embryonic cells and siRNA for hWDR26 in cultured cells (Fig. 3). Although xWDR26‐MO inhibited the translation of FLAG‐tagged mRNA containing the targeted site, the translation of FLAG‐tagged mRNA containing five‐mismatched sequences at the targeted site was not inhibited by xWDR26‐MO (Fig. 3A). This result suggest xWDR26‐MO could be targeted to knockdown specifically xWDR26. The transfection of each si‐hWDR26 RNA reduced hWDR26 mRNA expression (Fig. 3B). Since the knockdown by si‐WDR26‐3 RNA was slightly effective rather than other siRNA, we used si‐WDR26‐3 RNA for the following experiments. A reduction in Wnt activity in the anterior region is necessary for head formation in Xenopus development 28, 29. The injection of xWDR26‐MO into dorso‐animal blastomeres of eight‐cell embryos reduced both head formation at the tadpole stage (Fig. 4A) and the expression of several neural marker genes at the neural stage (Fig. 4B, lane 1 and lane 2). The injection of xWDR26 plasmids containing the MO‐targeted site with five‐mismatched sequences slightly increased the expression of several neural marker genes, except for xRX‐1 (Fig. 4B, lane 1 and lane 3). These reductions of head formation and neural marker gene expression by the knockdown of xWDR26 were partially rescued by coinjection of xWDR26 plasmids (Fig. 4A, B, lane 2 and lane 4). When Xwnt‐8 mRNA is injected into the ventral sides of four‐cell embryos, the target genes of Wnt signaling are induced (Fig. 4C, lane 2 and lane3). Ventral injection of xWDR26‐MO tended to increase the expression of Wnt target genes that were induced by coinjection of Xwnt‐8 mRNA (Fig. 4C, lane 3 and lane 4). Coinjection of xWDR26 mRNA containing the MO‐targeted site with five‐mismatched sequences tended to be restituted with the increasing of Wnt target genes by xWDR26‐MO (Fig. 4C, lane 5 and lane 6). The coinjection of hWDR26 mRNA showed similar results to the coinjection of xWDR26 (Fig. 4D). The knockdown of hWDR26 by siRNA in cultured cells did not affect the expression of a Wnt target gene, Axin2, in the ‘Wnt off’ state (Fig. 4E, lane 1 and lane 2). This suggests that small amounts of WDR26 are enough to keep the ‘Wnt off’ state in the canonical Wnt signaling pathway. However, the exression of Axin2 was increased in the ‘Wnt on’ state (Fig. 4E, lane 3 and lane 4). These results suggest that WDR26 contributes to the negative regulation of the canonical Wnt signaling pathway in the ‘Wnt on’ state.
Figure 3.
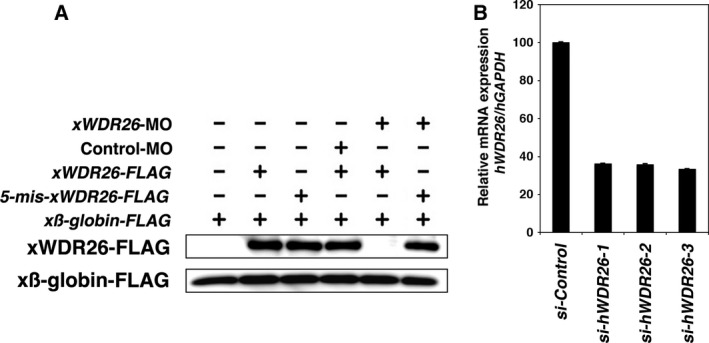
The specificity of antisense morpholino oligonucleotides for xWDR26 and siRNA for hWDR26. (A) Western blotting analysis of injected embryos at stage 10. xWDR26‐MO inhibited the translation of FLAG‐tagged mRNA containing the targeted site. The translation of FLAG‐tagged mRNA containing five‐mismatched sequences at the targeted site was not inhibited by xWDR26‐MO. β‐globin is used as loading control. (B) Quantitative RT‐PCR analysis. Three siRNA against hWDR26 were transfected into HEK 293T cells. GAPDH was used as a loading control. The value obtained for each si‐hWDR26 RNA was normalized to the level of GAPDH. The value of transfection of si‐Control RNA (lane 1) was set to 100 and other values were computed. Error bars represent standard deviation of the mean in three experiments. Statistical significances of hWDR26/hGAPDH between si‐Control RNA transfection and si‐hWDR26 RNA transfection were determined by Mann–Whitney U test. P < 0.01 (between lane 1 and each other lane).
Figure 4.

WDR26 is involved in the canonical Wnt pathway. (A) Phenotypes of xWDR26‐MO injected embryos. Control‐MO (40 ng) or xWDR26‐MO (40 ng) was coinjected with or without 5‐mis‐xWDR26 plasmids (20 pg) into two dorsal animal blastomeres of eight‐cell embryos. Phenotypes are categorized into normal, mild (small eyes), or severe (no eye). The injection of 5‐mis‐xWDR26 plasmids had no effect on head formation. Coinjection of 5‐mis‐xWDR26 plasmids with xWDR26‐MO rescued the phenotypes of the xWDR26‐morphants. The ratio of each phenotype in the injected embryos was indicated in the graph. Lane 1: n = 21, normal (100%). Lane 2: n = 45, normal (8.9%), mild (35.5%), severe (55.6%). Lane 3: n = 16, normal (81.3%), mild (18.7%). Lane 4: n = 14, normal (50.0%), mild (21.4%), severe (28.4%). (B–E) Quantitative RT‐PCR analysis. The value obtained for each marker gene or Wnt target gene was normalized to the level of xODC or hGAPDH. The value of control (lane 1) was set to 100, and other values were computed (other lanes). Error bars represent standard deviation of the mean in three experiments. Statistical significances of values among indicated lanes were determined by Mann–Whitney U test. (B) Quantitative RT‐PCR analysis of anterior neural marker genes, xNCAM, xOTX‐2, xPAX‐6, xSIX‐3, xRX‐1 in the head region of each molpholino‐ or plasmid‐injected embryo at stage 28. The injection was performed with similar methods indicated in (A). The value of neural marker gene/xODC of Control‐MO injected embryos (lane 1) was set to 100 and other values were computed. P < 0.01: all neural marker genes (between lane 1 and lane 2). P < 0.01: neural marker genes except for xRX‐1, P > 0.1: xRX‐1 (between lane 1 and lane 3). P < 0.01: all neural marker genes (between lane 2 and lane 4). (C) Quantitative RT‐PCR analysis of early dorsal Wnt target genes (Xtwn, Siamois, Xnr3) at stage 10. Control‐MO (40 ng), xWDR26‐MO (40 ng), and Xwnt‐8 (0.5 pg) mRNA were ventrally coinjected with xWDR26 mRNA (500 pg). The value of Wnt target gene/xODC of dorsal sectors (lane 1) was set to 100, and other values were computed. The value of uninjected ventral sectors was used as negative control. P < 0.005: all Wnt target genes (between lane 2 and lane 3). P < 0.005: Xnr3 and Siamois, P > 0.1: Xtwn (between lane 3 and lane 4). P < 0.05: all Wnt target genes (between lane 3 and lane 5). P < 0.005: Siamois and Xtwn, P > 0.05: Xnr3 (between lane 5 and lane 6). (D) Quantitative RT‐PCR analysis of early dorsal Wnt target genes (Xtwn, Siamois, Xnr3). Control‐MO (40 ng), xWDR26‐MO (40 ng) and Xwnt‐8 (0.5 pg) mRNA were ventrally coinjected with hWDR26 mRNA (500 pg). The value of Wnt target gene/xODC of ventral sectors (lane1) of Control‐MO injected embryos was set to 100, and other values were computed. P < 0.005: all Wnt target genes (between lane 1 and lane 2). P < 0.005: all Wnt target genes (between lane 1 and lane 3). P < 0.005: all Wnt target genes (between lane 2 and lane 4). (E) Quantitative RT‐PCR analysis of hAxin2 in HEK 293T cells. The siRNA‐transfected cultured cells were stimulated with Wnt3A conditioned medium from L‐Wnt‐3A cells for 6 h. The conditioned medium from L cells was used for unstimulated control. The value of hAxin2/hGAPDH of unstimulated control (lane1) was set to 100, and other values were computed. P > 0.1 (between lane 1 and lane 2). P < 0.005 (between lane 1 and lane 3). P < 0.005 (between lane 3 and lane 4).
WDR26 affects on the stability of β‐catenin
The homolog of WDR26 in yeast, GID7, is a component of the GID complex that mediates polyubiquitination of proteins 16. An increase in β‐catenin protein in the cytoplasm is essential for canonical Wnt signaling 2. We investigated whether WDR26 controls the amount of β‐catenin protein. The expression of either human or Xenopus WDR26 reduced the amount of β‐catenin protein in cultured cells in a dose‐dependent manner (Fig. 5A,B). The reduction of endogenous xβ‐catenin protein in the cytoplasmic fraction was observed in xWDR26 mRNA‐injected Xenopus embryonic cells (Fig. 5C). The injection of hWDR26 mRNA into Xenopus embryonic cells also reduced the amount of endogenous xβ‐catenin protein in the cytoplasmic fraction (Fig. 5D). The knockdown of hWDR26 by siRNA did not affect on the amount of endogenous hβ‐catenin protein in cultured cells (Fig. 5E, lane 1 and lane 2). However, the amount of endogenous hβ‐catenin protein was increased by the knockdown of hWDR26 in Wnt‐stimulated cells (Fig. 5E, lane 3 and lane 4). Moreover, in Xenopus, the endogenous xβ‐catenin protein tended to be up‐regulated by the knockdown of xWDR26 (Fig. 5F). We also confirmed that both knockdown and overexpression of WDR26 did not affect to the expression of β‐catenin mRNA (Fig. S1A, B). Moreover, we investigated β‐catenin stability by the cycloheximide chase assay. In results, the transfection of xWDR26 slightly promoted the degradation of β‐catenin (Fig. 5G). These results suggest that WDR26 controls β‐catenin protein stability in the canonical Wnt signaling pathway.
Figure 5.
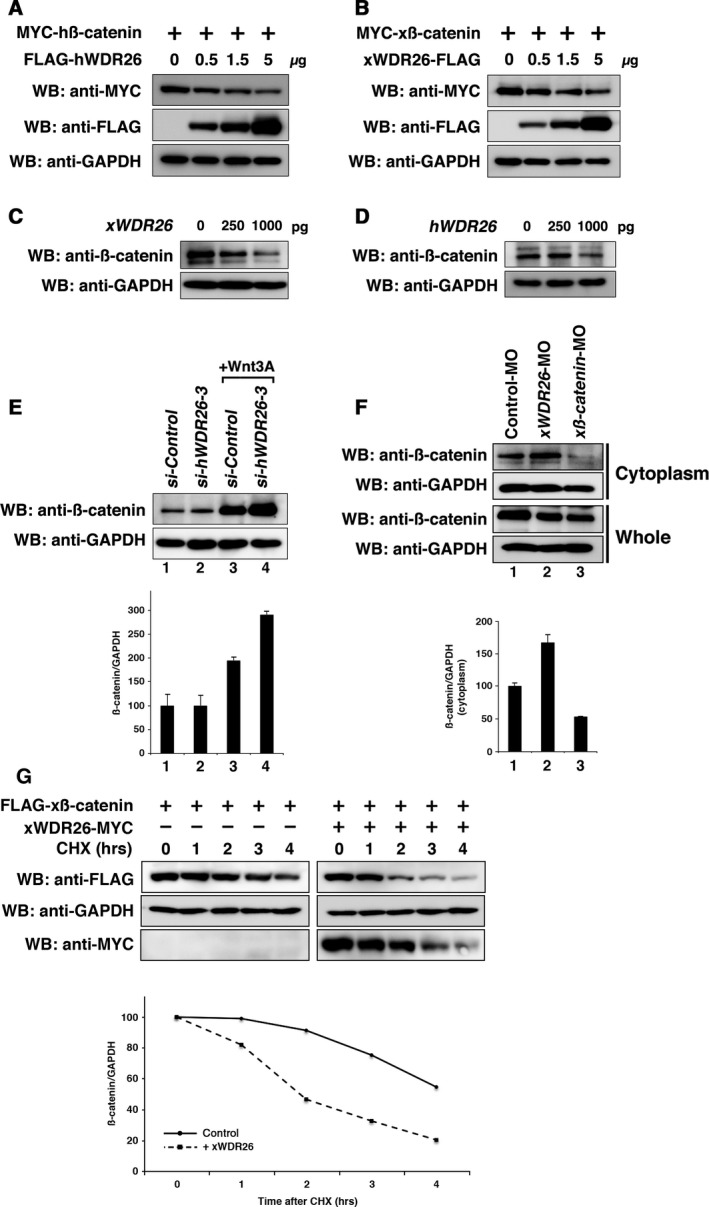
Effects of WDR26 to the stability of β‐catenin. (A–B) Western blotting analysis of ectopically expressed β‐catenin in HEK 293T cells. The amounts of transfected plasmids (hWDR26 and xWDR26) were indicated (0, 0.5, 1.5, 5 μg per dish). GAPDH was used as a loading control. (C–D) Western blotting analysis of endogenous xβ‐catenin in Xenopus embryonic cells. The amounts of injected mRNA (xWDR26 and hWDR26) were indicated (0, 250, 1000 pg per embryo). Cytoplasmic fractions of lysates extracted at the gastrula stage were used here. (E) Western blotting analysis of endogenous hβ‐catenin in HEK 293T cells. si‐hWDR26‐3, an siRNA against hWDR26, was transfected into HEK 293T cells. Twenty‐four hours after transfection, cells were stimulated with Wnt3A for 6 h. Relative intensity of each western blot (upper panel) was measured by imagej. The ratios of IP/Input of hβ‐catenin/hGAPDH were calculated and graphed. The value obtained for each hβ‐catenin signal was normalized to the level of hGAPDH signal. The value of unstimulated control (lane 1) was set to 100, and other values were computed (other lanes). Error bars represent standard deviation of the mean in three experiments. Statistical significances of values among indicated lanes were determined by Mann–Whitney U test. P > 0.1 (between lane 1 and lane 2). P < 0.005 (between lane 1 and lane 3). P < 0.005 (between lane 3 and lane 4). (F) Western blotting analysis of endogenous xβ‐catenin in Xenopus embryonic cells. xWDR26‐MO (40 ng) was injected into animal blastomeres of eight‐cell embryos, and lysates were extracted at the gastrula stage. xβ‐catenin‐MO was used as a positive control. Relative intensity of each western blot (upper panel) was measured by imagej. The ratios of IP/Input of xβ‐catenin/xGAPDH were calculated and graphed (bottom panel). The value for Control‐MO‐injected embryos (lane 1) was set to 100, and other values were computed (other lanes). Error bars represent standard deviation of the mean in three experiments. Statistical significances of values among indicated lanes were determined by Mann–Whitney U test. P < 0.01 (between lane 1 and lane 2). P < 0.01 (between lane 1 and lane 3). (G) Western blotting analysis of the stability of β‐catenin in HEK 293T cells. xWDR26 was transfected at 24 h after transfection of xβ‐catenin. Five hours after the transfection xWDR26, cells were stimulated with cycloheximide (CHX) (20 mg·mL−1) during indicated hours. We measured relative intensity of each western blot (upper panel) by imagej, calculated the ratios of β‐catenin/GAPDH, and graphed (bottom panel). The value for cells untreated with cyclohexmide was set to 100, and other values were computed (other lanes). Solid line indicates ratios of β‐catenin/GAPDH (left panels) as control. Dotted line indicates ratios of β‐catenin/GAPDH with xWDR26 transfection (right panels).
Possible mechanisms of β‐catenin degradation by WDR26
We found that hWDR26 did not bind to hβ‐catenin (Fig. 6A) although hAxin1 binds to both hWDR26 and hβ‐catenin (Figs 1 and 6A and ref. 6). This suggests that WDR26 controls the stability of β‐catenin through binding with Axin1. To examine the binding site of WDR26 with Axin1 in detail, we made deletion constructs of xWDR26, removing either the LisH domain, the CTLH domain, or both domains (see Fig. 6B, upper). The CTLH domain‐deleted construct bound to xAxin1, but the LisH domain‐deleted and the LisH‐CTLH domain‐deleted constructs did not bind (Fig. 6B). The expression of Wnt target genes induced by ventral injection with Xwnt‐8 mRNA was decreased by coinjection with xWDR26 mRNA, but not by mRNA of the LisH domain‐deleted construct (Fig. 6C). Moreover, the expression of the LisH domain‐deleted WDR26 construct hardly reduces the amount of xβ‐catenin protein (Fig. 6D), suggesting that binding between WDR26 and Axin is necessary for β‐catenin degradation. To confirm that WDR26 contributes to the ubiquitination of β‐catenin, we investigated whether the ubiquitination of β‐catenin is altered in the presence of the proteasome inhibitor, MG‐132. First, we confirmed the ubiquitination of xβ‐catenin without the MG‐132 treatment. Although the ubiquitination of xβ‐catenin was not increased by transfection of xWDR26 or xAxin1, both transfection of xWDR26 and xAxin1 slightly increased the ubiquitination of xβ‐catenin (Figs 6E and S2, lane 1 to lane 4). This suggests that the ubiquitinated xβ‐catenin could be immediately degraded via the ubiquitin proteasome pathway in the ‘Wnt off’ state. Under the MG‐132 treatments, the ubiquitination of xβ‐catenin was only slightly increased by cotransfection of xAxin1 alone, but the cotransfection of both xWDR26 and xAxin1 highly increased the ubiquitination of xβ‐catenin (Figs 6F and S2, lane 5 to lane 8). We considered that the amount of β‐catenin was reduced during 24 h before the treatments with MG‐132 (see figure legends). These results suggest that WDR26 regulates the ubiquitination of β‐catenin for its degradation, and that binding of WDR26 and Axin is important for this ubiquitination.
Figure 6.
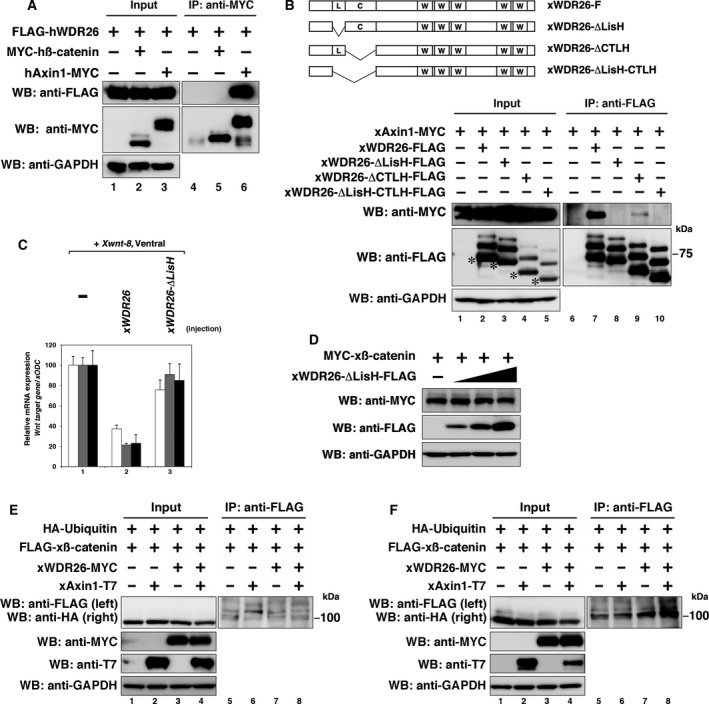
Interaction between WDR26 and Axon1 is critical for β‐catenin degradation. (A) Interaction between ectopically expressed hWDR26 and hβ‐catenin in HEK 293T cells. As a positive control, the binding between hAxin1 and hβ‐catenin was observed. (B) Interactions among ectopically expressed xAxin1 and truncated xWDR26 constructs. A schematic of xWDR26 and truncated constructs is indicated in the upper side. Asterisks indicate original size of each construct, and several bands of high molecular weight were observed. (C) Quantitative RT‐PCR analysis of early dorsal Wnt target genes (Xtwn, Siamois, Xnr3). xWDR26 or xWDR26‐deltaLisH mRNA (500 pg) and Xwnt‐8 (0.5 pg) mRNA were ventrally coinjected. The value obtained for each Wnt target gene was normalized to the level of xODC. The value of Wnt target gene/xODC of only Xwnt‐8‐injected embryos (lane 1) was set to 100, and other values were computed. Statistical significances of values among indicated lanes were determined by Mann–Whitney U test. P < 0.01: all Wnt target genes (between lane 1 and lane 2). P > 0.1: all Wnt target genes (between lane 1 and lane 3). P < 0.01: all Wnt target genes (between lane 2 and lane 3). (D) Western blotting analysis of ectopically expressed β‐catenin in HEK 293T cells. The amounts of transfected plasmid, xWDR26‐deltaLisH, were gradually increased (0, 0.5, 1.5, 5 μg per dish). (E and F) Western blotting analysis of ubiquitinated xβ‐catenin in HEK 293T cells. At 24 h after transfection, cells were untreated (E) or treated with 10 μm MG‐132 for 24 h (F). The HA‐Ubiquitin was cotransfected to detect ubiquitinated xβ‐catenin.
GSK‐3β plays a key role in the β‐catenin degradation, and xWDR26 bound to xAxin1 at the central region including GSK3β‐binding domain (Fig. 1E). Therefore, we investigated whether GSK‐3β affects on the function of WDR26 and the binding between WDR26 and Axin. The lithium chloride and SB216763 are GSK‐3β inhibitors, and increased the stability of β‐catenin (Fig. 7A, B, lane 1 and lane 3). However xWDR26 reduced the stability of β‐catenin under these treatments (Fig. 7A,B, lane 3 and lane 4). This suggests that the function of xGSK‐3β would be not related with the WDR26 function for the β‐catenin stability. Also, xGSK‐3β did not increase and decrease the association between xWDR26 and xAxin1 (Fig. 7C). These suggest that the mechanisms of degradation of β‐catenin by WDR26 would be independent from the canonical degradation mechanisms by the APC/Axin/GSK‐3β complex.
Figure 7.

Effects of GSK‐3β on WDR26 function. (A and B) Western blotting analysis of ectopically expressed β‐catenin in HEK 293T cells with or without lithium chloride treatments (A) and SB216763 treatments (B). GAPDH was used as a loading control. (C) Interaction between ectopically expressed xWDR26 and xAxin1 in HEK 293T cells with or without cotransfection of GSK‐3β. The amounts of transfected plasmids (GSK‐3β) were indicated (0, 0.5, 1.5, 5 μg per dish). GAPDH was used as a loading control. Relative intensity of each western blot was measured by imagej. The ratios of IP/Input of xWDR26 were calculated. Their statistical significances were determined by Mann–Whitney U test. P > 0.1 (between lane 7/lane 2 and lane 8/lane 3). P > 0.1 (between lane 7/lane 2 and lane 9/lane 4). P > 0.1 (between lane 7/lane 2 and lane 10/lane 5).
Discussion
In this study, we showed WDR26 is important for degradation of β‐catenin in the canonical Wnt signaling pathway. In Xenopus development, the inactivation of the Wnt signal by antagonists such as DKK‐1 and Cerberus is necessary for head formation 28, 30. Expression of xWDR26 is strongly localized to the anterior neural region after the neurula stage (Fig. 2D,E). Additionally, knockdown of xWDR26 in the anterior ectodermal region inhibited head formation, including eyes and marker genes (Fig. 4A,B). Moreover, Wnt target genes were increased by the knockdown of xWDR26 (Fig. 4D). These suggest that WDR26 acts as a negative regulator of Wnt signaling during embryogenesis.
The degradation of β‐catenin is regulated by the complex of Axin, APC, and GSK‐3β in the ‘Wnt off’ state. The amount of β‐catenin protein was reduced by the expression of WDR26 (Fig. 5A,B), although we did not confirm direct binding of WDR26 to β‐catenin (Fig. 6A). Interestingly, the deleted construct of xWDR26 that did not bind to xAxin1, did not reduce significantly either the expression of Wnt target genes or β‐catenin levels (Fig. 6C,D). Moreover, the coexpression of Axin1 and WDR26 increased the ubiquitination of β‐catenin (Fig. 6E). These results suggest that the degradation of β‐catenin by WDR26 requires binding between WDR26 and Axin1, and WDR26 is a component of the β‐catenin degradation complex. Axin1 was also expressed in the anterior neural region in Xenopus embryos (data not shown and ref. 31), suggesting that WDR26 and Axin1 regulate anterior formation through their inhibitory effects in Xenopus development. A recent report indicated that oxidative stress induces apoptosis and regulates the expression of WDR26 in cardiomyocytes. Moreover, the expression of WDR26 inhibits apoptosis induced by oxidative stress 32. However, overexpression and activation of β‐catenin induces apoptosis during carcinogenesis and development 33, 34, 35. In melanoma cells, Axin1 acts as a mediator of apoptosis with β‐catenin activation and BRAF inhibition 36. Taken together, these facts suggest that WDR26 and Axin1 play coordinating roles for the inhibitory effects through the degradation of β‐catenin in Wnt signaling pathway.
Vertebrate β‐catenin has 26 lysine residues, and each lysine position is well conserved. Recent studies indicated the presence of several ubiquitination pathways of β‐catenin. β‐TrCP ubiquitinates on lysine 19 and 49 of β‐catenin in a phosphorylation‐dependent manner, while Siah‐1 ubiquitinates β‐catenin on lysine 660 and 671 in the p53‐dependent pathway 37, 38. Jade‐1 ubiquitinates β‐catenin in a similar phosphorylation‐dependent manner to β‐TrCP, but the ubiquitination by Jade‐1 occurs in both ‘Wnt off’ and ‘Wnt on’ states 38, 39. However, it is known that ARMc8/GID5 and RMND5/GID2, predicted vertebrate homologs of the GID complex, contribute to the ubiquitination pathway. ARMc8 binds to α‐catenin to target it for degradation by the proteasome 18, and RMND5 has an E3 ubiquitin‐ligase activity and inhibits forebrain formation in Xenopus, like WDR26 19. Therefore, it is suggested that WDR26 forms a new ubiquitination pathway of β‐catenin with vertebrate GID complex homologs. Further study will be needed to identify how WDR26 controls the ubiquitination of β‐catenin.
Author contributions
HS and TN conceived and supervised the study; TG, JM, and SI performed experiments; TG and HS wrote the manuscript.
Supporting information
Fig. S1. (A) Quantitative RT‐PCR analysis of hβ‐catenin in HEK 293T cells. (B) Quantitative RT‐PCR analysis of xβ‐catenin in Xenopus embryonic cells.
Fig. S2. Semiquantitative analysis of western blots in Fig. 6E,F.
Acknowledgements
This work was supported by Grants‐in‐Aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Edited by Lukas Alfons Huber
*The copyright line for this article was changed on 11 May 2016 after original online publication.
References
- 1. Logan CY and Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20, 781–810. [DOI] [PubMed] [Google Scholar]
- 2. Clevers H (2006) Wnt/beta‐catenin signaling in development and disease. Cell 127, 469–480. [DOI] [PubMed] [Google Scholar]
- 3. Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X and He X (2002) Control of beta‐catenin phosphorylation/degradation by a dual‐kinase mechanism. Cell 108, 837–847. [DOI] [PubMed] [Google Scholar]
- 4. Winer IS, Bommer GT, Gonik N and Fearon ER (2006) Lysine residues Lys‐19 and Lys‐49 of beta‐catenin regulate its levels and function in T cell factor transcriptional activation and neoplastic transformation. J Biol Chem 281, 26181–26187. [DOI] [PubMed] [Google Scholar]
- 5. Bienz M and Clevers H (2000) Linking colorectal cancer to Wnt signaling. Cell 103, 311–320. [DOI] [PubMed] [Google Scholar]
- 6. Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S and Kikuchi A (1998) Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK‐3beta and beta‐catenin and promotes GSK‐3beta‐dependent phosphorylation of beta‐catenin. EMBO J 17, 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S and Kikuchi A (1998) Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of beta‐catenin. J Biol Chem 273, 10823–10826. [DOI] [PubMed] [Google Scholar]
- 8. Hart MJ, de los Santos R, Albert IN, Rubinfeld Band Polakis P (1998) Downregulation of beta‐catenin by human Axin and its association with the APC tumor suppressor, beta‐catenin and GSK3 beta. Curr Biol 8, 573–581. [DOI] [PubMed] [Google Scholar]
- 9. Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T et al (2000) AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus‐mediated transfer of AXIN1. Nat Genet 24, 245–250. [DOI] [PubMed] [Google Scholar]
- 10. Itoh K, Krupnik VE and Sokol SY (1998) Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and beta‐catenin. Curr Biol 8, 591–594. [DOI] [PubMed] [Google Scholar]
- 11. Rui Y, Xu Z, Xiong B, Cao Y, Lin S, Zhang M, Chan SC, Luo W, Han Y, Lu Z et al (2007) A beta‐catenin‐independent dorsalization pathway activated by Axin/JNK signaling and antagonized by aida. Dev Cell 13, 268–282. [DOI] [PubMed] [Google Scholar]
- 12. Francis O, Han F and Adams JC (2013) Molecular phylogeny of a RING E3 ubiquitin ligase, conserved in eukaryotic cells and dominated by homologous components, the muskelin/RanBPM/CTLH complex. PLoS One 8, e75217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu Y, Wang Y, Xia C, Li D, Li Y, Zeng W, Yuan W, Liu H, Zhu C, Wu X et al (2004) WDR26: a novel Gbeta‐like protein, suppresses MAPK signaling pathway. J Cell Biochem 93, 579–587. [DOI] [PubMed] [Google Scholar]
- 14. Sun Z, Tang X, Lin F and Chen S (2011) The WD40 repeat protein WDR26 binds Gβγ and promotes Gβγ‐dependent signal transduction and leukocyte migration. J Biol Chem 286, 43902–43912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Runne C and Chen S (2013) WD40‐repeat proteins control the flow of Gβγ signaling for directional cell migration. Cell Adh Migr 7, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santt O, Pfirrmann T, Braun B, Juretschke J, Kimmig P, Scheel H, Hofmann K, Thumm M and Wolf DH (2008) The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell 19, 3323–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menssen R, Schweiggert J, Schreiner J, Kusevic D, Reuther J, Braun B and Wolf DH (2012) Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite‐induced degradation of gluconeogenic enzymes. J Biol Chem 287, 25602–25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suzuki T, Ueda A, Kobayashi N, Yang J, Tomaru K, Yamamoto M, Takeno M and Ishigatsubo Y (2008) Proteasome‐dependent degradation of alpha‐catenin is regulated by interaction with ARMc8alpha. Biochem J 411, 581–591. [DOI] [PubMed] [Google Scholar]
- 19. Pfirrmann T, Villavicencio‐Lorini P, Subudhi AK, Menssen R, Wolf DH and Hollemann T (2015) RMND5 from Xenopus laevis is an E3 ubiquitin‐ligase and functions in early embryonic forebrain development. PLoS One 10, e0120342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iemura SI and Natsume T (2012) (2012) One‐by‐one sample preparation method for protein network analysis In Protein Interaction (Cai J, ed), pp. 293–310. Intech, Rijeka, Croatia. [Google Scholar]
- 21. Goto T, Fukui A, Shibuya H, Keller R and Asashima M (2010) Xenopus furry contributes to release of microRNA gene silencing. Proc Natl Acad Sci USA 107, 19344–19349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goto T, Sato A, Adachi S, Iemura S, Natsume T and Shibuya H (2013) IQGAP1 protein regulates nuclear localization of β‐catenin via importin‐β5 protein in Wnt signaling. J Biol Chem 288, 36351–36360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shimizu K and Gurdon JB (1999) A quantitative analysis of signal transduction from activin receptor to nucleus and its relevance to morphogen gradient interpretation. Proc Natl Acad Sci USA 96, 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Satoh K, Ohnishi J, Sato A, Takeyama M, Iemura S, Natsume T and Shibuya H (2007) Nemo‐like kinase‐myocyte enhancer factor 2A signaling regulates anterior formation in Xenopus development. Mol Cell Biol 27, 7623–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goto T, Sato A, Shimizu M, Adachi S, Satoh K, Iemura S, Natsume T and Shibuya H (2013) IQGAP1 functions as a modulator of dishevelled nuclear localization in Wnt signaling. PLoS One 8, e60865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shimizu M, Goto T, Sato A and Shibuya H (2013) WNK4 is an essential effector of anterior formation in FGF signaling. Genes Cells 18, 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harland RM (1991) In situ hybridization: an improved whole‐mount method for Xenopus embryos. Methods Cell Biol 36, 685–695. [DOI] [PubMed] [Google Scholar]
- 28. Glinka A, Wu W, Onichtchouk D, Blumenstock C and Niehrs C (1997) Head induction by simultaneous repression of Bmp and Wnt signalling in Xenopus . Nature 389, 517–519. [DOI] [PubMed] [Google Scholar]
- 29. Shibata M, Ono H, Hikasa H, Shinga J and Taira M (2000) Xenopus crescent encoding a Frizzled‐like domain is expressed in the Spemann organizer and pronephros. Mech Dev 96, 243–246. [DOI] [PubMed] [Google Scholar]
- 30. Semënov MV, Tamai K, Brott BK, Kühl M, Sokol S and He X (2001) Head inducer Dickkopf‐1 is a ligand for Wnt coreceptor LRP6. Curr Biol 11, 951–961. [DOI] [PubMed] [Google Scholar]
- 31. Hedgepeth CM, Deardorff MA and Klein PS (1999) Xenopus axin interacts with glycogen synthase kinase‐3 beta and is expressed in the anterior midbrain. Mech Dev 80, 147–151. [DOI] [PubMed] [Google Scholar]
- 32. Feng Y, Zhang C, Luo Q, Wei X, Jiang B, Zhu H, Zhang L, Jiang L, Liu M and Xiao X (2012) A novel WD‐repeat protein, WDR26, inhibits apoptosis of cardiomyocytes induced by oxidative stress. Free Radic Res 46, 777–784. [DOI] [PubMed] [Google Scholar]
- 33. Ahmed Y, Hayashi S, Levine A and Wieschaus E (1998) Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell 93, 1171–1182. [DOI] [PubMed] [Google Scholar]
- 34. Wong MH, Rubinfeld B and Gordon JI (1998) Effects of forced expression of an NH2‐terminal truncated beta‐Catenin on mouse intestinal epithelial homeostasis. J Cell Biol 141, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Damalas A, Ben‐Ze'ev A, Simcha I, Shtutman M, Leal JF, Zhurinsky J, Geiger B and Oren M (1999) Excess beta‐catenin promotes accumulation of transcriptionally active p53. EMBO J 18, 3054–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Biechele TL, Kulikauskas RM, Toroni RA, Lucero OM, Swift RD, James RG, Robin NC, Dawson DW, Moon RT and Chien AJ (2012) Wnt/β‐catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci Signal 5, ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu J, Stevens J, Rote CA, Yost HJ, Hu Y, Neufeld KL, White RL and Matsunami N (2001) Siah‐1 mediates a novel beta‐catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell 7, 927–936. [DOI] [PubMed] [Google Scholar]
- 38. Tauriello DV and Maurice MM (2010) The various roles of ubiquitin in Wnt pathway regulation. Cell Cycle 9, 3700–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I et al (2008) Jade‐1 inhibits Wnt signalling by ubiquitylating beta‐catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 10, 1208–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Quantitative RT‐PCR analysis of hβ‐catenin in HEK 293T cells. (B) Quantitative RT‐PCR analysis of xβ‐catenin in Xenopus embryonic cells.
Fig. S2. Semiquantitative analysis of western blots in Fig. 6E,F.


