Abstract
T follicular helper (Tfh) cells are involved in specific humoral immunity at initial and recall phases. The fact that the transcription repressors B‐cell lymphoma‐6 and Blimp‐1 determine lineages of Tfh cells and other types of effector CD4+ T cells, respectively, suggests that there are unique mechanisms to establish Tfh‐cell identity. In this study, we found that Tfh cells preferentially express the transcriptional coactivator Bob1. Bob1 of Tfh cells was dispensable for the expression of B‐cell lymphoma‐6 and the functional property of the cells for B cell help. However, upon initial immunization of foreign antigens, the percentages of Tfh cells in Bob1−/− mice were much higher than those in wild‐type (WT) mice. In addition, expansion of Tfh cells within Bob1−/−CD4+ T cells transferred into WT mice revealed that the high frequency of Tfh cells was caused by a T‐cell‐intrinsic mechanism. These findings were further supported by the results of in vitro studies demonstrating that Bob1−/− Tfh cells had greater proliferative activity in response to stimuli by CD3/CD28 monoclonal antibody and were also refractory to CD3‐induced cell death in comparison to WT Tfh cells. These results suggest that Tfh cells harbor a Bob1‐related mechanism to restrict numerical frequency against stimulation of TCRs.
Keywords: Apoptosis, Bob1, Cellular proliferation, T‐follicular helper cells
Introduction
Antigen‐specific humoral immune responses are established by the active interaction of B cells and T follicular helper (Tfh) cells, a distinctive subset of effector CD4+ T cells 1, 2. Tfh cell subsets residing in lymphoid tissues and peripheral blood have further advanced our understanding of immune regulation. After the development from naïve CD4+ T cells, Tfh cells start to express B‐cell lymphoma‐6 (Bcl‐6) and cell surface molecules such as CXCR5, PD‐1, and ICOS, which regulate recruitment and activities of Tfh cells. Since other types of effector CD4+ T cells such as Th1 cells, Th2 cells, Treg cells, and Th17 cells are regulated by Blimp‐1, a Bcl‐6 antagonist, it is thought that a unique mechanism specifically operated by Bcl‐6 defines the identity of a Tfh cell lineage 3. Extensive investigations have revealed that the transcription regulators MAF and BATF support the function of Bcl‐6 in Tfh cells to produce IL‐4 and IL‐21 4. Accumulating evidence from studies on circulating Tfh cells have indicated undesirable immune responses by memory Tfh cells in patients with disorders including systemic lupus erythematosus and rheumatoid arthritis 5, 6. However, the mechanisms regulating human Tfh cells have not been fully elucidated.
In this study, we compared the transcriptome of Tfh cells residing in germinal centers (GCs‐Tfh cells) of the tonsils to that of CD4+ T cells of the thymus as a control and dissected unidentified regulators of Tfh cells. The results showed that B‐cell‐specific octamer binding protein 1 (Bob1, OBF1, OCA‐B) was highly expressed in Tfh cells. Bob1 is known as a transcriptional coactivator associated with Oct1 or Oct2 through recognition of POU domains and it assists B‐cell activation to produce antigen‐specific antibodies 7, 8. Analysis of tonsillar lymphocytes and Bob1‐deficient mice revealed that Bob1 regulates proliferation and cell death mediated by CD3, and thereby the numerical frequency of Tfh cells would be maintained. Further investigation focusing on Bob1 would lead to a better understanding of a program of functional resolution of Tfh cells and the pathogenesis of autoimmunity and allergy associated with specific antibody production.
Results
Expression of Bob1 in Tfh cells and Th‐cell subsets
Since the first discovery of Tfh cells as CXCR5+ T cells in GCs of lymphoid follicles, Tfh cells have been shown to comprise subsets residing in lymphoid tissues and peripheral blood 9, 10, 11. To find a molecule(s) regulating human Tfh cells, microarray analysis was initially performed in archetypal GC‐Tfh cells (CD3+CD4+CXCR5hiPD‐1hi) of the tonsils and single‐positive naïve CD4+ T cells (CD3+CD4+CD8−) of the thymus as a control (Fig. 1A and Supporting Information Fig. 1). A total of 611 and 319 genes were extracted as genes preferentially up‐ and downregulated, respectively, in Tfh cells compared to their levels in naïve CD4+ T cells, (Supporting Information Table 1 and 2). In each altered gene set, genes that are known to be expressed in Tfh cells (Bcl‐6, MAF, CXCR5, CXCL13, IL‐21) and in naïve CD4+ T cells (KLF2, IL7R, LEF1, TMSB10, CCR7) were included in the 50 highest upregulated genes. In this study, seven genes of which expression in Tfh cells has not been reported previously were selected, and their transcripts were confirmed by RT‐PCR in purified lymphocytes of the tonsils and thymus (Fig. 1B). Within candidate genes for which expression was validated, we focused on Bob1 of a transcriptional coactivator, because Bob1 regulates GC formation of B cells 7. As assessed by quantitative RT‐PCR analysis, the transcription levels of Bob1 were significantly higher in GC‐Tfh cells than in non‐Tfh cells (CD3+CD4+CXCR5−PD‐1−) and thymic CD4+ T cells in contrast to B‐cell subsets including naïve, GC, and memory B cells of the tonsils (Fig. 1C and Supporting Information Fig. 2). Within CD4+ T cells of tonsils, Th1 cells (CD3+CD4+CXCR5−CCR5+), Th2 cells (CD3+CD4+CXCR5−CCR4+CCR8+), and Th17 cells (CD3+CD4+CXCR5−CCR6+CD161+) exhibited significantly lower expression levels of Bob1 than those of GC‐Tfh cells (Fig. 1D and Supporting Information Fig. 3). Furthermore, when CD4+ T cell subsets of peripheral blood were assessed, the expression levels of Bob1 in Th1, Th2, Th17, and Tfh‐like memory cells (CD3+CD4+CXCR5+) were lower than those in GC‐Tfh cells of tonsils. In accordance with such transcription profiles, Bob1 proteins were clearly detected in GC‐Tfh cells, whereas Bob1 was undetectable in non‐Tfh cells by immunoblot and immunoprecipitation analyses (Fig.1E and Supporting Information Fig. 4). Immunohistochemistry of tonsillar tissues also demonstrated that Bcl‐6+Bob1+CD4+ cells were mainly present within GCs, and they were probably GC‐Tfh cells (Fig. 1F). When GC‐Tfh cells and interfollicular (int)‐Tfh cells (CD3+CD4+CXCR5+PD‐1lo) were investigated, GC‐Tfh cells more abundantly expressed Bob1 than did int‐Tfh cells, implying that GC‐Tfh cells rather than int‐Tfh cells would functionally depend on Bob1 within lymphoid tissues (Fig.1C). Overall, these findings suggest that human Tfh cells primarily express Bob1, while the functional dependency on Bob1 is probably different within Tfh cell subsets.
Figure 1.
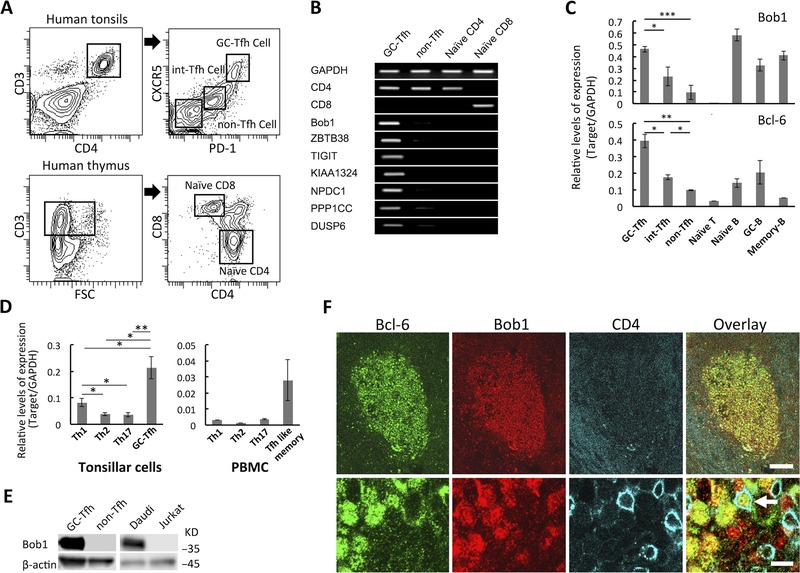
Expression of Bob1 in human Tfh cells. (A) GC‐Tfh cells (CD3+CD4+CXCR5hiPD‐1hi), int‐Tfh cells (CD3+CD4+CXCR5intPD‐1int), and non‐Tfh cells (CD3+CD4+CXCR5−PD‐1−) were purified from human tonsils with a cell sorter (upper panels) (Supporting Information Fig. 1A), and the identity of each population was further examined by ICOS expression (Supporting Information Fig. 1B). Naïve CD4+ T cells (CD3+CD4+CD8−) and CD8+ T cells (CD3+CD4−CD8+) were sorted from the thymus (lower panels). Each gate of isolated lymphocytes is depicted. Data shown are from a single experiment representative of three independent experiments, with one tonsil sample per experiment. (B) Highly expressed genes of Tfh cells as detected by microarray analysis were assessed by RT‐PCR analysis. Data shown are from a single experiment representative of three independent experiments with one tonsil sample per experiment. (C) The gene transcripts of Bob1 and Bcl‐6 in GC‐Tfh, int‐Tfh, and non‐Tfh cells of human tonsils were quantified by quantitative RT‐PCR analysis and compared with those in naïve B cells (CD3−CD19+CD27−CD38−), GC B cells (CD3−CD19+CD27+CD38+), and memory B cells (CD3−CD19+CD27+CD38−). Three B‐cell subsets were sorted from human tonsils (Supporting Information Fig. 2). (D) Quantitative RT‐PCR analysis of Bob1 expression in Th subsets including Th1 cells (CD3+CD4+CXCR5−CCR5+), Th2 cells (CD3+CD4+CXCR5−CCR4+CCR8+), Th17 cells (CD3+CD4+CXCR5−CCR6+CD161+), and Tfh‐like memory cells (CD3+CD4+CXCR5+) isolated from human tonsils or peripheral blood. Each subset was purified by a cell sorter (Supporting Information Fig. 3A and B). (C, D) Data are presented as ratios of target genes and GAPDH mRNA expression. Data are shown as mean ± SD of results pooled from three independent experiments. (E) Bob1 expression in Tfh cells was examined at the protein level. Whole cell extracts from tonsillar Tfh cells and non‐Tfh cells were subjected to Western blot analysis with a specific antibody against Bob1. An anti‐β‐actin antibody was used for the loading control. Daudi B cells and Jurkat T cells were also examined as positive and negative controls, respectively. Data shown are from a single experiment and are representative of three independent experiments with one tonsil sample per experiment. (F) Localization of Bob1+CD4+ T cells in GCs of lymphoid follicles of human tonsils. After staining of frozen tissue sections of tonsils, signals were detected by laser confocal microscopy. Bcl6+ (green), Bob1+ (red), and CD4+ (cyan) cells as indicated by arrows are thought to be GC‐Tfh cells. Upper panels: original magnification, 100× (scale bar, 100 μm). Lower panels: original magnification, 400× (scale bar, 10 μm). Images are from a single experiment representative of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.005; NS, not significant (p > 0.05); unpaired Student's t‐test.
Study of Bob1‐introduced naïve CD4+ T cells
To address the role of Bob1 in Tfh cells, we next introduced Bob1 into naïve CD4+ T cells (CD3+CD4+CD27+CD45RA+) of tonsils and analyzed the expression of a series of molecules characteristic to Tfh cells. After sorting of naïve CD4+ T cells (Fig. 2A), the gene encoding Bob1 was introduced and overexpressed in the cells by using a bicistronic lentivirus vector tagged with ZsGreen. When the transfected cells gated on ZsGreen expression were examined by flow cytometry, the cells did not show cell surface expression of CXCR5 (Fig. 2B and C). We further investigated FACS‐sorted cells gated on ZsGreen expression by employing quantitative RT‐PCR analysis. We failed to find any correlation between the expression levels of Bob1 and those of genes including Bcl‐6, PD‐1, and IL‐21 as well as CXCR5 (Fig. 2D). These findings suggest that Bob1 is not associated with functional expression of these molecules of Tfh cells, although Bob1 may have an unidentified role for Tfh cells.
Figure 2.
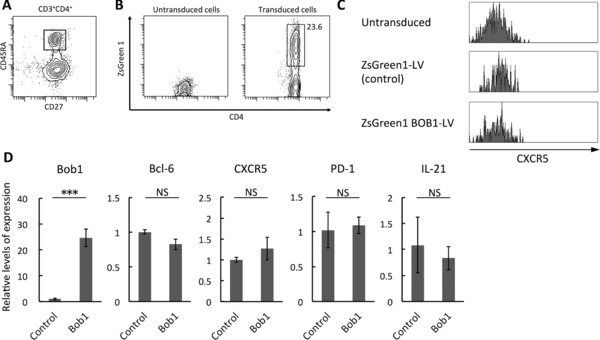
Phenotypes of Bob1‐introduced naïve CD4+ T cells. (A) Naïve CD4+ T cells (CD3+CD4+CD27+CD45RA+) were isolated from human tonsils with a cell sorter. Dot plot is representative of three independent experiments with one tonsil sample per experiment. (B) Naïve CD4+ T cells were transfected with Bob1‐IRES2‐ZsGreen1 lentivirus vector (LV) or control ZsGreen1 LV. Transduced cells were gated on cells positive for ZsGreen1 by FACS analysis (right panel). (C) Histograms showing CXCR5 expression as examined by FACS analysis. Cells gated on cells positive for ZsGreen1 as shown in (B) were investigated. The data are from a single experiment representative of three independent experiments with one tonsil sample per experiment. (D) Quantitative RT‐PCR analysis of Bcl‐6, CXCR5, PD‐1, and IL‐21 expression in Bob1‐introduced naïve CD4+ T cells. ZsGreen1‐positive cells as shown in (B) were isolated with a cell sorter and used for the experiments. The data are relative levels of expression compared with those of control cells after normalizing with GAPDH expression. Data are shown as mean ± SD of results pooled from three independent experiments with one tonsil sample per experiment. ***p < 0.005; NS, not significant (p > 0.05); unpaired Student's t‐test.
High frequency of Bob1−/− Tfh cells in vivo
Following in vitro studies, we used Bob1‐deficient (Bob1−/−) mice to investigate the function of Bob1 in Tfh cells. Even after intraperitoneal immunization of sheep red blood cells (SRBCs) as a foreign antigen, Bob1−/− mice did not have the capacity to form lymphoid follicles with GCs in the spleen due to functional failure of B cells (Fig. 3A) 7. To address the question of whether Bob1−/− mice can induce Tfh cells in response to SRBCs, we analyzed splenic Tfh cells on day 7 after immunization with SRBCs. The results showed the presence of Tfh cells clearly expressing Bcl‐6 in Bob1−/− mice and, unexpectedly, their percentage was much higher than that in Bob1+/+ mice (Fig. 3B and C). In fact, the actual number of Tfh cells in Bob1−/− mice was significantly larger than that in Bob1+/+ mice on day 7 after immunization, while the number of non‐Tfh cells in Bob1−/− mice was smaller than that in Bob1+/+ mice (Fig. 3D). To confirm these results, we further performed adoptive transfer experiments, in which CD45.2+Bob1+/+CD4+ T cells or CD45.2+Bob1−/−CD4+ T cells were introduced into CD45.1+ wild‐type (WT) mice exhibiting normal B‐cell function. When immunized with SRBCs, the percentage of CD45.2+ Tfh cells expressing Bcl‐6 within transferred Bob1−/−CD4+ T cells was significantly higher than that within Bob1+/+CD4+ T cells, being consistent with the results for Bob1−/− mice (Fig. 3E). In these experiments, although slight accumulation of cells expressing Th1‐associated transcription factor T‐bet was observed in transferred Bob1−/−CD4+ T cells, cells positive for Th2‐associated transcription factor GATA3 and Th17‐associated transcription factor RORγt did not show alteration of cellular frequencies (Fig. 3F). Furthermore, we failed to find significant differential mRNA levels of these transcription factors and cytokines characteristic to three Th subsets (IFN‐γ, IL‐4, and IL‐17) between Bob1+/+CD4+ T cells and Bob1−/−CD4+ T cells, suggesting that the functional loss of Bob1 seems not to affect functional balance between Th1 cells, Th2 cell,s and Th17 cells (Fig. 3G).
Figure 3.
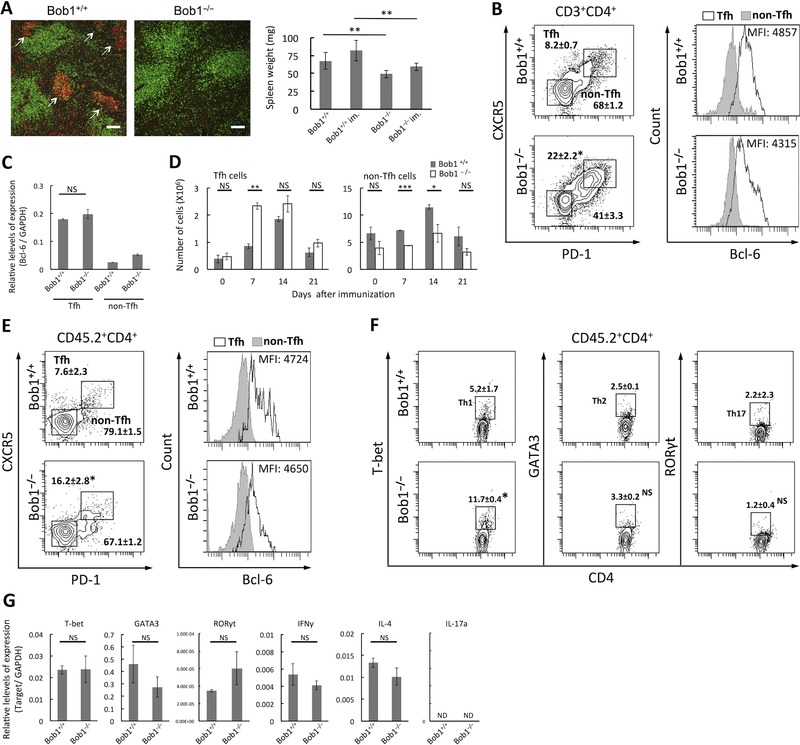
Tfh cells in Bob1‐deficient mice. (A) Formation of GCs in spleens of Bob1+/+ (left panel) and Bob1−/−(middle panel) mice at day 7 after immunization with SRBCs and their weights in immunized (im) and nonimmunized states. After double staining of frozen tissue sections of spleens with PNA (red) and anti‐CD3 (green), signals were detected by an immunofluorescence microscope. Arrows indicate GCs. Original magnification 100× (scale bar: 100 μm). Images are representative of three independent experiments with one mouse per experiment. (B) Percentages of Tfh cells (CD3+CD4+CXCR5+PD‐1+) and non‐Tfh cells (CD3+CD4+CXCR5−PD‐1−) per total CD4+ T cells of spleens of Bob1+/+ and Bob1−/− (left panel). CD4+ T cells gated by positive staining for CD3 and CD4 were examined for CXCR5 and PD‐1 expression (Supporting Information Fig. 5A). The identity of each population was further investigated by FACS analysis to detect intracellular Bcl‐6 expression as depicted in histograms. The values in the plots represent the percent of the population within CD3+CD4+ cells as mean ± SD of samples pooled from three to five independent experiments with one mouse per experiment. (C) Quantitative RT‐PCR analysis of Bcl‐6 in Tfh cells of Bob1+/+ and Bob1−/− mice in (A). The data represent relative levels of expression compared with those of Bob1+/+ Tfh cells after normalizing to GAPDH expression. (D) Absolute numbers of Tfh cells and non‐Tfh cells in spleens of Bob1+/+ and Bob1−/− mice as studied in (A). (E) Percentages of Tfh cells and non‐Tfh cells per total CD4+ T cells of Bob1+/+ and Bob1−/− mice (CD45.2) in primed WT mice (CD45.1). Total CD4+ T cells were isolated from spleen cells of Bob1+/+ and Bob1−/− mice (CD45.2) and adoptively transferred into Ly5.1 WT mice (CD45.1) 24 h before immunization. At day 7 after immunization with SRBCs, donor T cells in recipient spleens were identified by positive staining for both CD45.2 and CD4 (Supporting Information Fig. 5B), and their Tfh‐cell phenotype and Bcl‐6 were analyzed as in (B). The numbers represent the percent of the population within CD45.2+CD4+ cells as mean ± SD. (F) Percentages of Th subsets including Th1, Th2, and Th17 cells per total CD4+ T cells of Bob1+/+ and Bob1−/− mice (CD45.2) in primed WT mice (CD45.1). CD45.2+CD4+ T cells shown in Supporting Information Fig. 5B were subjected to intracellular FACS analysis for detecting T‐bet (Th1 cells), GATA3 (Th2 cells), and RORγt (Th17 cells). The numbers represent the percent of the population within CD45.2+CD4+ cells and are shown as mean and ± SD. (G) Quantitative RT‐PCR analysis of transcription factors and cytokines characteristic of Th1 (T‐bet and IFN‐γ), Th2 (GATA3 and IL‐4), and Th17 cells (RORγt and IL‐17) in total CD4+ T cells of Bob1+/+ and Bob1−/− mice (CD45.2) in primed WT mice (CD45.1). CD45.2+CD4+ cells shown in Supporting Information Fig. 5B were isolated with a cell sorter and used for the experiments. Data are presented as relative levels of expression after normalizing with GAPDH expression. (A, C, D, and G) Data are shown as mean ± SD of results pooled from three independent experiments, each performed with cells pooled from three spleens per experiment. (B, E, and F) Flow cytometry data are representative of three to five independent experiments with one mouse per experiment. *p < 0.05, **p < 0.01, ***p < 0.005; NS, not significant (p > 0.05); unpaired Student's t‐test.
Apoptotic activity and proliferative capacity of Bob1−/− Tfh cells in vitro
CD4+ T cells control their own cell number to ensure effector and memory functions though proliferation and cell death as well after major histocompatibility complex (MHC) class II based stimulation from APCs 12. Thus, we next conducted in vitro studies to evaluate responses of Bob1−/− Tfh cells to TCR stimulation. For culture up to 3 days, stimulation with anti‐CD3 mAb reduced the number of Bob1+/+ Tfh cells due to activation‐induced cell death, whereas Bob1−/− Tfh cells were refractory to the stimulation and maintained their number (Fig. 4A). These cells were further assessed by experiments using annexin V and 7‐amino‐actinomycin D to detect apoptotic cells. The percentages of early (annexin V+7AAD−) and late (annexin V+7AAD+) apoptotic cells were significantly lower in Bob1−/− Tfh cells after CD3 activation (Fig. 4B). We also studied the proliferative capacity of Bob1−/− Tfh cells by FACS analyses for carboxyfluorescein diacetate succinimidyl ester (CFSE)‐labeled cells. After CD3/CD28 stimulation for 3 days, about 40% of the Bob1−/− Tfh cells had undergone proliferation, whereas only about 2% of proliferated cells were detected in Bob1+/+ Tfh cells (Fig. 4C). Taken together, the results indicated that Bob1−/− Tfh cells have a higher proliferative capacity and lower apoptotic susceptibility against TCR‐mediated stimuli than do Bob1+/+ Tfh cells.
Figure 4.
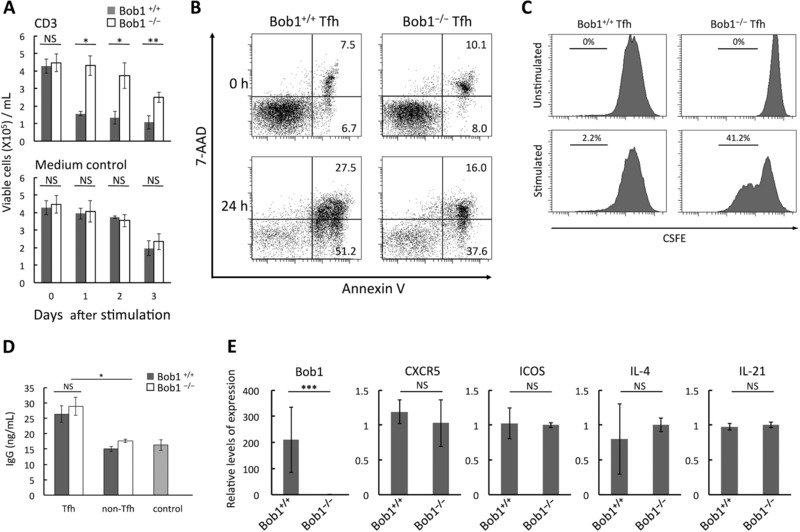
Functional characteristics of Bob1−/− Tfh cells. (A) Viabilities of Bob1+/+ and Bob1−/− Tfh cells against CD3 stimulation. Tfh cells (4 × 105) derived from spleens of Bob1+/+ and Bob1−/− mice after immunization with SRBCs were cultured and stimulated with plate‐bound anti‐mouse CD3 mAb in 200 μL of medium. Viable cells were counted by the trypan blue dye exclusion assay. (B) Reduction of apoptosis of Bob1−/− Tfh cells against CD3 stimulation. Bob1+/+ and Bob1−/− Tfh cells were cultured and stimulated as in (A) and stained with annexin V and 7‐amino‐actinomycin D (7‐AAD) to detect early and late apoptotic cells. The numbers represent the percent of the population. (C) Proliferative capacity of Bob1−/− Tfh cells under a stimulatory condition. Tfh cells obtained from the spleen as in (A) were loaded with CSFE and stimulated by beads coated with anti‐mouse CD3/CD28 mAb. After 3 days, cell proliferation was assessed by FACS analysis. (D) IgG production by Bob1−/− Tfh cells in Tfh‐B cell coculture. Tfh cells of OVA‐primed Bob1+/+OT‐IIor Bob1−/−OT‐II mice were cocultured with B cells of WT mice (CD3− B220+) in a medium containing OVA323‐339 and beads coated with anti‐mouse CD3/CD28 mAb. B cells cultured in the absence of T cells served as control cells. After 4 days, IgG concentrations in culture supernatants were measured by ELISA. (E) Quantitative RT‐PCR analysis of the expression of Tfh‐cell markers in Bob1−/− Tfh cells. The data represent relative levels of expression compared with those of Bob1+/+ Tfh cells after normalizing with GAPDH expression. (A, D, and E) Data are shown as mean ± SD of results pooled from three independent experiments, each performed with three mouse spleens per experiment. (B, C) Flow cytometry data are from a single experiment representative of three independent experiments, each performed with three spleens per experiment. *p < 0.05, **p < 0.01, ***p < 0.005; NS, not significant (p > 0.05); unpaired Student's t‐test.
Capacity of Bob1−/− Tfh cells for B cell help
To investigate the effect of Bob1 deletion on Tfh function to help B cells produce antibodies, we performed mixed cell culture experiments using Tfh cells of OT‐II Bob1+/+ mice or OT‐II Bob1−/− mice with WT B cells. No significant differences were observed between concentrations of IgG in the culture supernatants from Bob1−/− Tfh/WT B‐cell culture and Bob1+/+ Tfh/WT B‐cell culture, indicating that Bob1−/− Tfh cells have the same capacity as that of Bob1+/+ cells to help B cells secrete antibodies (Fig. 4D). We failed to find differential levels of the expression of major functional makers (Bcl‐6, CXCR5, ICOS, IL‐4, and IL‐21) between Bob1+/+ Tfh cells and Bob1−/− Tfh cells (Fig. 4E). Collectively, Bob1 of Tfh cells seems to be dispensable for Tfh cells to fulfill their major role in helping B cells to produce antibodies.
Discussion
Here, we reported a role of Bob1, which was originally identified as a B‐cell regulator to produce high‐affinity antibodies, in the limitation of numbers of Tfh cells. It is known that tumor cells originating from Tfh cells, such as angioimmunoblastic T‐cell lymphoma cells, express Bob1 as well as Bcl‐6, CXCR5, and PD‐1 13. CD4+ T cells can display Bob1 after activation with CD3/CD28 mAb or PMA/ionomycin, further implying the constitutive expression of Bob1 in a certain cell type(s) of effector helper T cells in vivo 14, 15. Our studies using gene and protein analyses revealed that Bob1 is present most abundantly in GC‐Tfh cells and, to a lesser extent, in Th1 cells, Th2 cells, Th17 cells, and Tfh‐like memory cells in human tonsils or peripheral blood. This is not in disagreement with the reported evidence of upregulated genes in Tfh cells, including Bob1 with 2.3‐ to 7.5‐fold higher expression levels than the levels in Th1 cells and Th2 cells 16 (Supporting Information tables). Although the role of Bob1 in Tfh cells has been largely unknown, our exploration of Tfh cells revealed a salient feature of Bob1 in the limitation of cell numbers particularly in association with CD3 stimulation.
Bob1−/− mice lack GC responses against foreign antigens without defects in production of natural antibodies 17. Bob1−/− Tfh cells clearly expressed Bcl‐6, which is a key regulator of Tfh cell differentiation, and other crucial molecules responsible for Tfh cell function, though the numbers of Bob1−/− Tfh cells were significantly increased by themselves. Ectopic expression of Bcl‐6 in human naïve CD4+ T cells can induce a Tfh‐cell phenotype as assessed by the expression of CXCR5 and PD‐1 4. In the same manner, we studied Bob1‐ex‐introduced human naïve CD4+ T cells and obtained results showing that Bob1 is not likely to be associated with the expression of these molecules as found in murine models. It is thought that Tfh cell differentiation is initiated at the time of priming of dendritic cells with the expression of Bcl‐6 18. Our results demonstrate that this early process during Tfh cell differentiation normally occurs in Bob1−/− mice. The fact that Bob1−/− Tfh cells still have the capacity to help B cells produce IgG is further supported by the results of an experiment using mice reconstituted with Bob1‐deficient T cells and WT B cells, which show antigen‐specific IgG responses with the development of GCs 19, 20. Collectively, the results indicate that Bob1 is not necessary for the expression of functional molecules such as Bcl‐6, which exert B‐cell help producing antibodies, even though Bob1 of Tfh cells acts as a controller of cell numbers.
The mechanism by which the number of Tfh cells is controlled remains controversial. Experimental evidence obtained by using mice lacking CD40 or MHC class II on B cells suggests that Tfh cell differentiation is independent of B cells 21. Furthermore, Tfh cell differentiation can occur early after virus infection through processes without B cells 18. According to these findings, it is understandable that Tfh cells were detected even in Bob1−/− mice, which are unable to produce antigen‐specific B cells upon immunization. Several studies have suggested that T–B cell interactions are required for the differentiation of Tfh cells 22, 23, 24. A reduction of Tfh cells in Bob1−/− mice has been reported in a mouse model during influenza infection and in a sanroque mouse model of systemic lupus, which manifests constitutive activation of ICOS signaling by a point mutation of Rouquin1 20, 25. These results seem to be contradictory to our observation of the accumulation of Tfh cells in Bob1−/− mice simply immunized by SRBCs in this study. The conflicting results of our study and the previous study might be due to differences in experimental conditions such as immunization (SRBC injection, autoimmunity or virus infection) and sampling time points. It has been reported that T follicular regulatory cells expressing FoxP3 control the formation of antigen‐specific Tfh cells via suppressive CTLA‐4 function 26. Although further studies are required to clarify a role of Bob1 in the regulation of T follicular regulatory cells, there would be B‐cell‐independent mechanisms for regulating Tfh cell number.
Accumulation of Bob1−/− Tfh cells was observed in WT mice adoptively transferred with Bob1−/−CD4+ T cells. This result indicates that there is a T‐cell‐intrinsic mechanism related to Bob1 for the restriction of Tfh cell numbers. Our in vitro experiments using Bob1−/− Tfh cells under the condition of TCR stimuli by anti‐CD3 mAb demonstrated an uncommon function of Bob1 providing Tfh cell properties. It has been reported that tonsillar Tfh cells have limited proliferative capacity and are prone to apoptosis in vitro, and these characteristics are reflected more in CXCR5hiICOShi GC‐Tfh cells than in CXCR5loICOSint int‐Tfh cells, which express Bob1 at high and low levels, respectively, as found in this study 27. Thus, greater proliferative capacity and lower apoptotic susceptibility of Bob1−/− Tfh cells imply that the level of Bob1 expression is likely to be involved in such functional features. Within GCs of lymphoid follicles, Bob1hi GC‐Tfh cells afford antibody‐producing capacity to GC‐B cells with high expression levels of MHC class II molecules 28. Since appropriate numbers of GC‐Tfh cells are required for optimal differentiation of GC‐B cells, Bob1 may restrict GC‐Tfh cell numbers for efficient differentiation of B cells during the active interaction of GC‐B cells and GC‐Tfh cells 29, 30. In this context, Bob1 of GC‐Tfh cells might play a role in decreasing sensitivities to further activation and proliferation signals through TCR in order to execute their own function. Our RT‐PCR experiments for transcription factors and cytokines characteristic to Th1, Th2, and Th17 cells demonstrated that Bob1 seemed not to be involved in a regulation of functional balance of these T‐cell subsets. On the other hand, the subtle accumulation of T‐bet‐expressing cells within Bob1−/−CD4+ cells was detected by intracellular FACS analysis. It has been reported that Th1 cells share a certain transitional stage and have an overlapped phenotype during their development from naïve CD4+ T cells 31. Further investigations would be needed to clarify whether Bob1 can be involved in regulation of a Th1 population size.
Currently, the precise mechanism by which Bob1 regulates proliferative and apoptotic responses of Tfh cells is not known. Bob1 forms a ternary complex with POU proteins of the ubiquitous factor Oct‐1 or the B‐cell‐specific factor Oct‐2 on conserved octamer motifs, suggesting that a gene(s) regulated by a Bob1/Oct1 complex might be responsible for TCR/CD3 signaling of Tfh cells. The Bob1 protein is able to interact with the cytoplasmic protein of Syk, which is a nonreceptor‐type tyrosine kinase 32. The proximal components of TCR form a multiprotein complex including ZAP‐70/Syk and Lck, and Bob1 in Tfh cells can thus modulate TCR signalosomes including Syk to appropriately adjust cellular responses 33, 34. While it is known that Bob1 and Bcl‐6 can upregulate Syk in B cells, further investigation is needed to determine the role of Bob1 and Bcl‐6 in the control of Syk in Tfh cells 35, 36.
In summary, our study revealed that Bob1 participates in a unique endogenous mechanism to possibly limit TCR‐mediated expansion of Tfh cells. It has been reported that patients with systemic lupus erythematosus or allergic diseases have an increased frequency of ICOS+CD4+ T cells, suggesting a pathological expansion of Tfh cells 37, 38, 39. Because of the functional importance of Tfh cells, a mechanism regulating Tfh cell numbers is a critical issue for the understanding of initial and recall humoral responses and Tfh‐cell related disorders such as autoimmunity, allergy, and cancer 40, 41. Further investigations of Bob1 may shed new light on a mechanism regulating immune homeostasis through the control of Tfh cells.
Materials and methods
Tissues
Surgically resected specimens of tonsils were obtained from patients admitted to Sapporo Medical University Hospital. Thymus tissues were obtained from surgical specimens of patients with cardiovascular disorders who underwent surgery in the Hokkaido Medical Center for Child Health and Rehabilitation. Some of the tissues were stored in OCT compound (Sakura, Tokyo, Japan) at –80°C for frozen tissue sections. All tissues were obtained after receiving informed consent and with the approval of the institutional review boards of Sapporo Medical University and the Hokkaido Medical Center for Child Health and Rehabilitation.
Mice and immunization
All experiments using mice were performed in accordance with the institutional guidelines of Sapporo Medical University for the care and use of animals. Bob1‐deficient mice and OT‐II mice were purchased from Jackson Laboratory. By crossing these strains, we obtained Bob1−/−OT‐II mice. C57BL/6 mice used as control mice and C57BL/6 Ly5.1‐congenic mice expressing CD45.1 were obtained from Sankyo Laboratory (Tokyo, Japan) and RIKEN Bioresource Center (Ibaraki, Japan), respectively. All of the mice were maintained in a specific pathogen‐free animal facility of Sapporo Medical University. Age‐matched (6–12 weeks) and sex‐matched mice from each group, which were simultaneously raised in the same environment at the facility, were used for the experiments. Mice were immunized by intraperitoneal administration of 200 μL of 10% SRBCs in PBS or 100 μg of ovalbumin (OVA) in alum adjuvant.
Microarray analysis
Total cellular RNA was extracted with TRIZOL reagent (Life Technologies) from tonsillar Tfh cells and thymus CD3+CD4+CD8− cells (considered to be naïve CD4+ T cells) and checked with a 2100 Bioanalyzer (Agilent). Then, RNAs of Tfh cells and thymic CD4+ T cells were amplified and labeled with Cy3 and Cy5, respectively. The labeled RNA probes were hybridized to a 3D microarray plate (human 25k oligo chip; Toray, Tokyo, Japan) and signals were analyzed with a 3D gene scanner (Toray). Signals detected for each gene were analyzed by global normalization (Cy3/Cy5 ratio median = 1) and Cy3/Cy5. Data were obtained from a single experiment as summarized in Supporting Information Table 1 and 2.
PCR
Total RNA extracted by TRIZOL reagent was reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The primer pairs used in this study are summarized in Supporting Information Table 3. Quantitative PCR was performed with a Step One Real‐Time PCR System of Assay‐on‐Demand probes according to the instructions of the manufacturer (Applied Biosystems). The levels of expression of target genes were calculated using ΔCT and comparative methods after normalization to glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) expression.
Antibodies
Anti‐human mAbs including APC‐anti‐CD3 (UCHT1), FITC‐anti‐CD3 (UCHT1), APC‐Cy7‐anti‐CD4 (RPA‐T4), PE‐anti‐CD8 (SK1), APC‐anti‐CD8 (SK1), PE‐Cy7‐anti‐CD38 (HIT2), PE‐anti‐PD‐1 (EH12.1), PerCP‐Cy5.5‐anti‐CXCR5 (RF8B2), PE‐anti‐CD27 (M‐T271), FITC‐anti‐CD27 (M‐T271), FITC‐anti‐CD45RA (HI100), APC‐anti‐CCR6 (11A9), PE‐Cy7‐anti‐CXCR3 (1C6), and APC‐Cy7‐anti‐CD19 (SJ25C1) (BD Biosciences) and anti‐mouse mAbs including PE‐Cy7‐anti‐CD3 (145‐2C11), PerCP‐Cy5.5‐anti‐CD4 (RM4‐5), PE‐anti‐B220 (RA3‐682), PE‐anti‐Bcl‐6 (K112‐91), PE‐anti‐T‐bet (4B10), PE‐anti‐GATA3 (L50‐823), PE‐anti‐RORγt (Q31‐378) (BD Bioscience), APC‐anti‐PD‐1 (29.F1A12), FITC‐anti‐CXCR5 (L138D7), and Pacific Blue‐anti‐CD45.2 (104) (Biolegend) were used for flow cytometry. For immunohistochemistry, we used anti‐human Abs including Alexa Fluor 647‐anti‐CD4 (MT310), mouse anti‐Bcl6 (P1F6; Nichirei, Tokyo, Japan), and rabbit anti‐Bob1 pAb (C‐20; Santa Cruz Biotechnology) for staining of human tonsils and we used hamster anti‐mouse CD3 (500A2; Life Technologies) and biotin‐PNA visualized by streptavidin‐PE (Vector Laboratory) for staining of mouse spleens.
Flow cytometry and cell sorting
Human tonsil and thymus tissues were mechanically disrupted and lymphocytes in single cell suspensions were prepared by density gradient centrifugation with Lymphoprep (Axis‐Shield). Mouse total spleen cells were prepared by lysing red blood cells with 0.84% NH4Cl. Subsequently, the cells were subjected to standard staining with specific surface markers for flow cytometry as previously described 42. The cells were analyzed using FACSCanto II and FACSAria II for careful cell sorting (BD Biosciences). In each experiment, samples were analyzed for singlet events with doublet discrimination, and the purity of FACS‐sorted cells reached 95% after validation by reanalysis with FACSCanto II. The cells were also analyzed by intracellular staining for the expression of transcription factors according to the protocol of the Transcription Factor Buffer Set (BD Biosciences). Data were examined by using FACSDiVA software (BD Biosciences).
Immunohistochemical analysis
Immunohistochemistry was conducted as described previously 42. In brief, tissue sections were stained with primary Abs in a moisture box at 4°C overnight and subsequently reacted with secondary goat pAbs conjugated to Alexa Fluor dyes (Invitrogen). Tissues slides were analyzed using an immunofluorescence microscope (IX71; Olympus) or a laser confocal microscope with image examiner software (LSM780; Carl Zeiss).
Western blot analysis
Protein expression analyses were performed using Western blot assay of whole cell extracts according to previously described procedures 42.
Gene transfection
The human Bob1 open reading frame was amplified by PCR from the Bob1 cDNA clone (pF1KB7532; Promega) and ligated into the upstream region of the internal ribosomal entry site of pIRES2‐ZsGreen1 lentivirus vector (Takara, Tokyo, Japan), resulting in Bob1‐IRES2‐ZsGreen1 recombinant vector. Lentiviral particles were prepared in 293T cells with Lenti‐X HTX packaging system and used for transfection (Clontech). Prior to transfection, FACS‐sorted human naïve CD4+ T cells were stimulated with anti‐CD3 (OKT3) and anti‐CD28 mAbs (15E8) obtained from Miltenyi Biotech in serum‐free AIM‐V medium (Invitrogen) containing 50 μg/mL streptomycin and 100 U/mL penicillin. After 48‐h incubation, cells were transfected using RetroNectin (Takara).
Adoptive transfer of lymphocytes
Total CD4+ T cells were isolated from spleen cells of Bob1+/+ and Bob1−/− mice (CD45.2) by using a CD4+ T‐cell isolation kit (Miltenyi Biotech). The cells (5 × 106) were suspended in 200 μL of PBS and transferred intravenously into C57BL/6 Ly5.1‐congenic mice (CD45.1).
T‐cell culture and stimulation
Tfh cells and non‐Tfh cells purified from spleens of mice on day 7 after immunization with SRBCs were cultured on a 96‐well plate‐bound anti‐mouse CD3 mAb (145‐2C11; BD Biosciences) and examined by the trypan blue dye exclusion assay. For examining the capacity of IgG production, Tfh cells (5 × 104) purified from spleens of Bob1+/+OT‐II mice and Bob1−/−OT‐II mice that were primed with OVA were cocultured with B (CD3−B220+) cells (1 × 105) purified from spleens of WT mice in a medium containing OVA323‐339 (1 μg/mL) and beads coated with anti‐mouse CD3/CD28 mAb (Dynabeads Mouse T‐Activator; Life Technologies). After 4 days, the concentration of IgG in the culture supernatant was measured by an IgG mouse ELISA Kit (Abcam). RPMI 1640 medium (Invitrogen) supplemented with 10% FCS, 50 μg/mL streptomycin, and 100 U/mL penicillin was used in all experiments, and all experiments were performed at 37°C in a humidified atmosphere with 5% carbon dioxide.
Detection of apoptosis and cell proliferation
The apoptotic state of cells was assessed by staining with annexin V‐FITC (ImmunoChemistry Technologies) and 7‐amino‐actinomycin D (BD Biosciences) according to the manufacturer's protocols. For evaluating proliferative activities, cells were labeled with 5 μM CSFE (Fluka) in PBS for 15 min at 37°C and examined by flow cytometry.
Statistical analysis
Results are expressed as means and SD. The unpaired Student's t‐test was used to compare experimental groups with p < 0.05 considered significant.
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Abbreviations
- Bcl‐6
B‐cell lymphoma‐6
- Tfh
T follicular helper
- TCR
T cell receptor
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Peer Review Correspondence
Table 1 The 611 genes up‐regulated in Tfh cells of OSAS tonsils. Selected genes had a normalized value of signal intensity of over 300 in Tfh cells and the values were more than twice as high as those in thymic naïve CD4+ T cells.
Table 2 The 319 genes upregulated in the thymic naïve CD4+ T cells rather than Tfh cells of OSAS tonsils. Selected genes had a normalized value of signal intensity of over 300 in thymic naïve CD4+ T cells and the values were more than twice as high as those in Tfh cells.
Table 3 RT‐PCR primers used in this study.
Supporting information Figure 1. Iden‐fica‐on and sor‐ng of human tonsillar T9 cells. (A) Ga#ng strategy for detec#ng T1 cells. Lymphocytes were iden#fied by forward and side sca<er characteris#cs and analyzed for singlet events with doublet discrimina#on using FSC‐W and FSC‐H. CD4+ T cells gated by posi#ve staining for CD3 and CD4 were further characterized by staining for CXCR5 and PD‐1. (B) Expression levels of ICOS in GC‐T1 cells, int‐T1 cells and non‐T1 cells as assessed by flow cytometry. The iden#ty of each popula#on iden#fied in (A) was examined. Data are representa#ve of three independent experiments with one tonsil sample per experiment.
Supporting information Figure 2. Isola‐on of human B cell subsets. B cell subsets (naïve B cells, GC B cells and memory B cells) were isolated from human tonsils with a cell sorter and validated with RT‐PCR for the major marker genes. Data are representa#ve of three independent experiments with one tonsil sample per experiment.
Supporting Information Figure 3. Isola‐on of human CD4+ T cell subsets.(A) Sor#ng of CD4+ T cell subsets (Th1 cells, Th2 cells and Th17 cells) from tonsillar lymphocytes. CD4+ T cells were iden#fied and analyzed as described in Supplementary Figure S1A, and the popula#on showing CXCR5‐ and PD‐1‐ (non‐T1 cells) was further characterized by their expression of CCR5 for Th1 cells, CCR8 and CCR4 for Th2 cells and CD161 and CCR6 for Th17 cells. Data are representa#ve of three independent experiments with one tonsil sample per experiment. (B) Sor#ng of CD4+ T cell subsets (Th1 cells, Th2 cells, Th17 cells and T1‐like memory cells) from human peripheral blood. T1‐like memory cells were iden#fied as CXCR5‐posi#ve cells within CD4+ T cells of PBMCs. The other CD4+ T cell subsets were characterized as shown in (A) within CXCR5‐nega#ve cells. Data are representa#ve of three independent experiments with one blood sample per experiment.
Supporting information Figure 4. Bob1 expression in human T9 cells as inves‐gated by immunoprecipita‐on. Immunoprecipita#on assays were performed as described previously [41]. Briefly, whole cell extracts from the cells were immunoprecipitated with rabbit an#‐Bob1 pAb (C‐20) using GammmaBind G sepharose (GE Healthcare). The whole protein was visualized by silver staining (led panel) as a loading control, and Bob1 was detected by mouse an#‐Bob1 mAb (Wue‐AC5) (right panel). An asterisk (*) indicates a signal of Bob1. Data are representa#ve of three independent experiments with one tonsil sample per experiment.
Supporting information Figure 5. Ga‐ng strategy for iden‐fica‐on of CD4+ T cells in the mouse spleen. (A) Iden#fica#on of CD4+ T cells in Bob1+/+ and Bob1‐/‐ mouse spleens. Cells were iden#fied by forward and side sca<er characteris#cs and analyzed for singlet events with doublet discrimina#on using FSC‐W and FSC‐H. CD4+ T cells gated by posi#ve staining for CD3 and CD4 were further analyzed as shown in Figure 3B. The data are representa#ve of three to five independent experiments with three spleens per experiment. (B) Detec#on of Bob1+/+ and Bob1‐/‐ CD4+ T cells adop#vely transferred into Ly5.1 mice. Cells from recipient spleens were analyzed by forward and side sca<er characteris#cs as shown in (A). Donor cells were iden#fied as both CD4 and CD45.2‐posi#ve cells. The data are representa#ve of three to five independent experiments with one mouse per experiment.
Acknowledgements
This work was supported by Grants‐in‐Aid for Scientific Research from the Japanese Society for the Promotion of Science (JSPS) (to K. Y. [No. 26861398], to R. K. [No. 15K10787], to K. K. [No. 15K20214], to T. N. [No. 26861400], to T. H. [No. 26293370 and No. 26670746], and to S. I. [No. 26670178]) and a grant from GSK Japan (to R. K.).
[The copyright line of this article has been changed since first published on 15 April 2016 from the standard copyright to CC‐BY‐NC.]
References
- 1. Crotty, S. , T follicular helper cell differentiation, function, and roles in disease. Immunity 2014. 41: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takemori, T. , Kaji, T. , Takahashi, Y. , Shimoda, M. and Rajewsky, K. , Generation of memory B cells inside and outside germinal centers. Eur. J. Immunol. 2014. 44: 1258–1264. [DOI] [PubMed] [Google Scholar]
- 3. Hatzi, K. , Nance J. P., Kroenke, M. A. , Bothwell, M. , Haddad, E. K. , Melnick, A. and Crotty, S. , BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 2015. 212: 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kroenke, M. A. , Eto, D. , Locci, M. , Cho, M. , Davidson, T. , Haddad, E. K. and Crotty, S. , Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 2012. 188: 3734–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liarski, V. M. , Kaverina, N. , Chang, A. , Brandt, D. , Yanez, D. , Talasnik, L. , Carlesso, G. et al, Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci. Transl. Med. 2014. 6: 230ra46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang, J. , Shan, Y. , Jiang, Z. , Feng, J. , Li, C. , Ma, L. and Jiang, Y. , High frequencies of activated B cells and T follicular helper cells are correlated with disease activity in patients with new‐onset rheumatoid arthritis. Clin. Exp. Immunol. 2013. 174: 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schubart, D. B. , Rolink, A. , Kosco‐Vilbois, M. H. , Botteri, F. and Matthias, P. , B‐cell‐specific coactivator OBF‐1/OCA‐B/Bob1 required for immune response and germinal centre formation. Nature 1996. 383: 538–542. [DOI] [PubMed] [Google Scholar]
- 8. Lindner, J. M. , Wong, C. S. , Möller, A. and Nielsen, P. J. , A C‐terminal acidic domain regulates degradation of the transcriptional coactivator Bob1. Mol. Cell. Biol. 2013. 33: 4628–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schaerli, P. , Willimann, K. , Lang, A. B. , Lipp, M. , Loetscher, P. and Moser, B. , CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 2000. 192: 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bentebibel, S. E. , Schmitt, N. , Banchereau, J. and Ueno, H. , Human tonsil B‐cell lymphoma 6 (BCL6)‐expressing CD4+ T‐cell subset specialized for B‐cell help outside germinal centers. Proc. Natl. Acad. Sci. USA 2011. 108: E488–E497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morita, R. , Schmitt, N. , Bentebibel, S. E. , Ranganathan, R. , Bourdery, L. , Zurawski, G. , Foucat, E. et al, Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011. 34: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malherbe, L. , Hausl, C. , Teyton, L. and McHeyzer‐Williams, M. G. , Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity 2004. 21: 669–679. [DOI] [PubMed] [Google Scholar]
- 13. Sáez, A. I. , Artiga, M. J. , Sánchez‐Beato, M. , Sánchez‐Verde, L. , García, J. F. , Camacho, F. I. , Franco, R. and Piris, M. A. , Analysis of octamer‐binding transcription factors Oct2 and Oct1 and their coactivator BOB.1/OBF.1 in lymphomas. Mod. Pathol. 2002. 15: 211–220. [DOI] [PubMed] [Google Scholar]
- 14. Moriuchi, M. and Moriuchi, H. , Octamer transcription factors up‐regulate the expression of CCR5, a coreceptor for HIV‐1 entry. J. Biol. Chem. 2001. 276: 8639–8642. [DOI] [PubMed] [Google Scholar]
- 15. Mueller, K. , Quandt, J. , Marienfeld, R. B. , Weihrich, P. , Fielder, K. , Claussnitzer, M. , Laumen, H. et al, Octamer‐dependent transcription in T cells is mediated by NFAT and NF‐κB. Nucleic Acids Res. 2013. 41: 2138–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chatanova, T. , Tangye, S. G. , Newton, R. , Frank, N. , Hodge, M. R. , Rolph, M. S. and Mackey, C. R. , T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non‐Th1/Th2 effector cells that provide help for B cells. J. Immunol. 2004. 173: 68–78. [DOI] [PubMed] [Google Scholar]
- 17. Kim, U. , Qin, X. , Gong, S. , Stevens, S. , Luo, Y. , Nussenzweig, M. and Roeder, R. G. , The B‐cell‐specific transcription coactivator OCA‐B/OBF‐1/Bob‐1 is essential for normal production of immunoglobulin isotypes. Nature 1996. 383: 542–547. [DOI] [PubMed] [Google Scholar]
- 18. Choi, Y. S. , Kageyama, R. , Eto, D. , Escobar, T. C. , Johnston, R. J. , Monticelli, L. , Lao, C. et al, ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 2011. 34: 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin, X. , Reichlin, A. , Luo, Y. , Roeder R. G. and Nussenzweig, M. C. , OCA‐B integrates B cell antigen receptor‐, CD40L‐ and IL 4‐madiated signals for the germinal center pathway of B cell development. EMBO J. 1998. 17: 5066–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karnowski, A. , Chevrier, S. , Belz, G. T. , Mount, A. , Emslie, D. , D'costa, K. , Tarlinton, D. M. et al, B and T cells collaborate in antiviral responses via IL‐6, IL‐21, and transcriptional activator and coactivator, Oct2 and OBF‐1. J. Exp. Med. 2012. 209: 2049–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deenick, E. K. , Chan, A. , Ma, C. S. , Gatto, D. , Schwartzberg, P. L. , Brink, R. and Tangye S. G., Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity 2010. 33: 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haynes, N. M. , Allen, C. D. C. , Lesley, R. , Ansel, K. M. , Killeen, N. and Cyster, J. G. , Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene‐1high germinal center‐associated subpopulation. J. Immunol. 2007. 179: 5099–5108. [DOI] [PubMed] [Google Scholar]
- 23. Zaretsky, A. G. , Taylor, J. J. , King, I. L. , Marshall, F. A. , Mohrs, M. , and Pearce, E. J. , T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J. Exp. Med. 2009. 206: 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnston, R. J. , Poholek A. C., Ditoro, D. , Yusuf, I. , Eto, D. , Barnett, B. , Dent, A. L. et al, BCL6 and Brimp‐1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 2009. 325: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chevrier, S. , Kratina, T. , Emslie, D. , Karnowski, A. and Corcoran, L. M. , Germinal center‐independent, IgM‐mediated autoimmunity in sanroque mice lacking Obf1. Immunol. Cell Biol. 2014. 92: 12–19. [DOI] [PubMed] [Google Scholar]
- 26. Wing, J. B. , Ise, W. , Kurosaki, T. and Sakaguchi, S. Regulatory T cells control antigen‐specific expansion of Tfh cell number and humoral immune responses via the coreceptor CTLA‐4. Immunity 2014. 41: 1013–1025. [DOI] [PubMed] [Google Scholar]
- 27. Rasheed, A. U. , Rahn, H. P. , Sallusto, F. , Lipp, M. and Müller G., Follicular B helper T cell activity is confined to CXCR5hiICOShi CD4 T cells and is independent of CD57 expression. Eur. J. Immunol. 2006. 36: 1892–1903. [DOI] [PubMed] [Google Scholar]
- 28. Weinberg, D. S. , Ault, K. A. , Gurley, M. and Pinkus, G. S. , The human lymph node germinal center cell: characterization and isolation by using two‐color flow cytometry. J. Immunol. 1986. 137: 1486–1494. [PubMed] [Google Scholar]
- 29. Barnett, L. G. , Simkins, H. M. , Barnett, B. E. , Korn, L. L. , Johnson, A. L. , Wherry, E. J. , Wu, G. F. et al, B cell antigen presentation in the initiation of follicular helper T cell and germinal center differentiation. J. Immunol. 2014. 192: 3607–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwickert, T. A. , Victora, G. D. , Fooksman, D. R. , Kamphorst, A. O. , Mugnier, M. R. , Gitlin, A. D. , Dustin, M. L. et al, A dynamic T cell‐limited checkpoint regulates affinity‐dependent B cell entry into the germinal center. J. Exp. Med. 2011. 208: 1243–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nakayamada, S. , Kanno, Y. , Takahashi, H. , Jankovic, D. , Lu, K. T. , Johnson, T. A. , Sun, H. et al, Early Th1 cell differentiation is marked by a Tfh cell‐like transition. Immunity 2011. 35: 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Siegel, R. , Kim, U. , Patke, A. , Yu, X. , Ren, X. , Tarakhovsky, A. and Roeder, R. G. , Nontranscriptional regulation of SYK by the coactivator OCA‐B is required at multiple stages of B cell development. Cell 2006. 125: 761–774. [DOI] [PubMed] [Google Scholar]
- 33. Smith‐Garvin, J. E. , Koretzky, G. A. and Jordan, M. S. , T cell activation. Annu. Rev. Immunol. 2009. 27: 591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krishnan, S. , Warke, V. G. , Nambiar, M. P. , Tsokos, G. C. and Farber, D. L. , The FcR gamma subunit and Syk kinase replace the CD3 zeta‐chain and ZAP‐70 kinase in the TCR signaling complex of human effector CD4 T cells. J. Immunol. 2003. 170: 4189–4195. [DOI] [PubMed] [Google Scholar]
- 35. Corcoran, L. , Emslie, D. , Kratina, T. , Shi, W. , Hirsch, S. , Taubenheim, N. and Chevrier S., Oct2 and Obf1 as facilitators of B:T cell collaboration during a humoral immune response. Front. Immunol. 2014. 5: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juszczynski, P. , Chen, L. , O'Donnell, E. , Polo, J. M. , Ranuncolo, S. M. , Dalla‐Favera, R. , Melnick, A. et al, BCL6 modulates tonic BCR signaling in diffuse large B‐cell lymphomas by repressing the SYK phosphatase, PTPROt. Blood 2009. 114: 5315–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hutloff, A. , Buchner, K. , Reiter, K. , Baelde, H. J. , Odendahl, M. , Jacobi, A. , Dörner, T. et al, Involvement of inducible costimulator in the exaggerated memory B cell and plasma cell generation in systemic lupus erythematosus. Arthritis Rheum. 2004. 50: 3211–3220 [DOI] [PubMed] [Google Scholar]
- 38. Le Coz, C. , Joublin, A. , Pasquali, J. L. , Korganow, A. S. , Dumortier, H. and Monneaux, F. , Circulating TFH subset distribution is strongly affected in lupus patients with an active disease. PLoS One. 2013. 8: e75319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamekura, R. , Shigehara, K. , Miyajima, S. , Jitsukawa, S. , Kawata, K. , Yamashita, K. , Nagaya, T. et al, Alteration of circulating type 2 follicular helper T cells and regulatory B cells underlies the comorbid association of allergic rhinitis with bronchial asthma. Clin. Immunol. 2015. 158: 204–211. [DOI] [PubMed] [Google Scholar]
- 40. Shakya, A. , Goren, A. , Shalek, A. , German, C. N. , Snook, J. , Kuchroo, V. K. , Yosef, N. et al, Oct1 and OCA‐B are selectively required for CD4 memory T cell function. J. Exp. Med. 2015. 212: 2115–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pratama, A. and Vinuesa, C. G. , Control of TFH cell numbers: why and how? Immunol. Cell Biol. 2014. 92: 40–48. [DOI] [PubMed] [Google Scholar]
- 42. Nagashima, T. , Ichimiya, S. , Kikuchi, T. , Saito, Y. , Matsumiya, H. , Ara, S. , Koshiba, S. et al, Arachidonate 5‐lipoxygenase establishes adaptive humoral immunity by controlling primary B cells and their cognate T‐cell help. Am. J. Pathol. 2011. 178: 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Peer Review Correspondence
Table 1 The 611 genes up‐regulated in Tfh cells of OSAS tonsils. Selected genes had a normalized value of signal intensity of over 300 in Tfh cells and the values were more than twice as high as those in thymic naïve CD4+ T cells.
Table 2 The 319 genes upregulated in the thymic naïve CD4+ T cells rather than Tfh cells of OSAS tonsils. Selected genes had a normalized value of signal intensity of over 300 in thymic naïve CD4+ T cells and the values were more than twice as high as those in Tfh cells.
Table 3 RT‐PCR primers used in this study.
Supporting information Figure 1. Iden‐fica‐on and sor‐ng of human tonsillar T9 cells. (A) Ga#ng strategy for detec#ng T1 cells. Lymphocytes were iden#fied by forward and side sca<er characteris#cs and analyzed for singlet events with doublet discrimina#on using FSC‐W and FSC‐H. CD4+ T cells gated by posi#ve staining for CD3 and CD4 were further characterized by staining for CXCR5 and PD‐1. (B) Expression levels of ICOS in GC‐T1 cells, int‐T1 cells and non‐T1 cells as assessed by flow cytometry. The iden#ty of each popula#on iden#fied in (A) was examined. Data are representa#ve of three independent experiments with one tonsil sample per experiment.
Supporting information Figure 2. Isola‐on of human B cell subsets. B cell subsets (naïve B cells, GC B cells and memory B cells) were isolated from human tonsils with a cell sorter and validated with RT‐PCR for the major marker genes. Data are representa#ve of three independent experiments with one tonsil sample per experiment.
Supporting Information Figure 3. Isola‐on of human CD4+ T cell subsets.(A) Sor#ng of CD4+ T cell subsets (Th1 cells, Th2 cells and Th17 cells) from tonsillar lymphocytes. CD4+ T cells were iden#fied and analyzed as described in Supplementary Figure S1A, and the popula#on showing CXCR5‐ and PD‐1‐ (non‐T1 cells) was further characterized by their expression of CCR5 for Th1 cells, CCR8 and CCR4 for Th2 cells and CD161 and CCR6 for Th17 cells. Data are representa#ve of three independent experiments with one tonsil sample per experiment. (B) Sor#ng of CD4+ T cell subsets (Th1 cells, Th2 cells, Th17 cells and T1‐like memory cells) from human peripheral blood. T1‐like memory cells were iden#fied as CXCR5‐posi#ve cells within CD4+ T cells of PBMCs. The other CD4+ T cell subsets were characterized as shown in (A) within CXCR5‐nega#ve cells. Data are representa#ve of three independent experiments with one blood sample per experiment.
Supporting information Figure 4. Bob1 expression in human T9 cells as inves‐gated by immunoprecipita‐on. Immunoprecipita#on assays were performed as described previously [41]. Briefly, whole cell extracts from the cells were immunoprecipitated with rabbit an#‐Bob1 pAb (C‐20) using GammmaBind G sepharose (GE Healthcare). The whole protein was visualized by silver staining (led panel) as a loading control, and Bob1 was detected by mouse an#‐Bob1 mAb (Wue‐AC5) (right panel). An asterisk (*) indicates a signal of Bob1. Data are representa#ve of three independent experiments with one tonsil sample per experiment.
Supporting information Figure 5. Ga‐ng strategy for iden‐fica‐on of CD4+ T cells in the mouse spleen. (A) Iden#fica#on of CD4+ T cells in Bob1+/+ and Bob1‐/‐ mouse spleens. Cells were iden#fied by forward and side sca<er characteris#cs and analyzed for singlet events with doublet discrimina#on using FSC‐W and FSC‐H. CD4+ T cells gated by posi#ve staining for CD3 and CD4 were further analyzed as shown in Figure 3B. The data are representa#ve of three to five independent experiments with three spleens per experiment. (B) Detec#on of Bob1+/+ and Bob1‐/‐ CD4+ T cells adop#vely transferred into Ly5.1 mice. Cells from recipient spleens were analyzed by forward and side sca<er characteris#cs as shown in (A). Donor cells were iden#fied as both CD4 and CD45.2‐posi#ve cells. The data are representa#ve of three to five independent experiments with one mouse per experiment.


