Abstract
Eukaryotic mRNAs are monocistronic, and therefore mechanisms exist that coordinate the synthesis of multiprotein complexes in order to obtain proper stoichiometry at the appropriate intracellular locations. RNA‐binding proteins containing low‐complexity sequences are prone to generate liquid droplets via liquid‐liquid phase separation, and in this way create cytoplasmic assemblages of functionally related mRNAs. In a recent iCLIP study, we showed that the Drosophila RNA‐binding protein Imp, which exhibits a C‐terminal low‐complexity sequence, increases the formation of F‐actin by binding to 3′ untranslated regions of mRNAs encoding components participating in F‐actin biogenesis. We hypothesize that phase transition is a mechanism the cell employs to increase the local mRNA concentration considerably, and in this way synchronize protein production in cytoplasmic territories, as discussed in the present review.
Keywords: post‐transcriptional RNA regulon, RNA assemblage, RNA‐binding protein, RNP granule, liquid droplet, low‐complexity sequence
Abbreviations
- ALS
amyotrophic lateral sclerosis
- APC
adenomatous polyposis coli
- CLIP
cross‐linking and immunoprecipitation
- CSD
cold shock domain
- dsRBD
double‐stranded RNA‐binding domain
- FRAP
fluorescence recovery after photobleaching
- FTD
frontotemporal dementia
- iCLIP
individual‐nucleotide resolution cross‐linking and immunoprecipitation
- KH
heterogenous nuclear ribonucleoprotein K homology
- LCS
low‐complexity sequence
- NLS
nuclear localization signal
- RBP
RNA‐binding protein
- RNP
ribonucleoproten
- RRM
RNA recognition motif
Introduction
Since 1961, when Jacob and Monod suggested their operon model, the concept of the polycistronic mRNA encoding functionally related proteins has been a paradigm in bacterial and archaeal gene regulation. It is assumed that this mode of co‐regulation ensures proportional synthesis of components in multiprotein complexes by translational control, and recent ribosome profiling analysis of global gene expression in Escherichia coli by Weissman and co‐workers 1 has shown that this is generally the case. Proportional synthesis avoids dominant‐negative effects of excess components that will have to be eliminated by proteolytic quality control. An added bonus of protein synthesis from polycistronic mRNAs is the proximity of the resulting protein products, thus facilitating expedient macromolecular assembly or catalysis.
The prokaryotic mode of creating multiprotein complexes is in sharp contrast to the way the considerably larger eukaryotic cell, encoding monocistronic mRNAs, achieves proximity and stoichiometry, although classical cases of pyrimidine and purine biosynthesis have evolved multifunctional enzymes and enzyme clustering 2, 3. Faced with this conundrum, Jack Keene 4 suggested that the coordination of gene expression to some extent was delegated from the DNA to the RNA level via the formation of post‐transcriptional RNA regulons. The salient feature behind this hypothesis is the pivotal role of a common RNA‐binding protein (RBP) associating with mRNAs encoding functionally related proteins and in this way dictating a common fate.
In this review, we describe examples of low‐complexity RBPs coordinating RNA metabolism in both mono‐ and multicellular organisms with an emphasis on cytosolic ribonucleoprotein (RNP) granules (Table 1). We present the available evidence regarding their status as liquid droplets, including our own recent study of the participation of the Drosophila RBP Imp in F‐actin formation.
Table 1.
RNA‐binding proteins discussed in this review
| Acronym | Organism | RNA‐binding domains | Low‐complexity sequence | RefSeq |
|---|---|---|---|---|
| hnRNPA1 | human | 2 RRMs | Gly and Ser | NP_002127 |
| hnRNPA2B1 | human | 2 RRMs | Gly | NP_112533 |
| FUS | human | 2 RRMs | Gly, Ser, and Tyr | NP_004951 |
| Whi3p | yeast | 1 RRM | Gln | NP_014202 |
| Puf3p | yeast | 8 pumilio repeats | Asn and Gln | NP_013088 |
| Imp | Drosophila | 4 KH domains | Gln | NP_511111 |
Modular RBPs have a multitude of RNA targets
It is estimated that mammalian cells contain between 800 and 1,600 different RBPs, and about half of these encompasses common RNA‐binding modules such as RRMs, KH domains, CSDs, dsRBDs, RGG motifs, and zinc fingers in various combinations 5, 6, 7. The other half does not contain an easily recognisable module – usually illustrated by the dual behavior of cytosolic aconitase, which switches between high‐affinity RNA‐binding and catalysis depending on the intracellular iron concentration 8. From X‐ray and NMR studies, a good understanding of the structure of the single RNA‐binding module in complex with RNA has emerged 9, 10. Moreover, several structural examples of truncated multi‐domain RBPs in complex with RNA targets are available 11, 12.
There has been a flurry of high‐throughput CLIP data on the targetome of full‐length RBPs and attempts at defining recognition elements. Besides the well‐characterized interaction between Pumilio and its target revealing an 8‐nt recognition “code” 13, 14, many RBPs are promiscuous and exhibit low specificity, although the combination of modules within a single RBP will increase the specificity considerably 15. Because a single RBP may literally have thousands of different interactions with the transcriptome, and hundreds of RBPs may behave in this manner, the complexity is daunting. Nevertheless, the combinatorial binding or competition among RBPs determines the fate of the individual transcript 16, 17.
Are RNP granules membrane‐less liquid droplets?
The cell is compartmentalized, and in recent years the participation of RBPs in the formation of membrane‐less organelles by phase transition has been examined extensively. Seminal studies were carried out with germ granules (termed P granules) in Caenorhabditis elegans, identifying them as liquid droplets formed by phase separation from the cytoplasm 18, but the concept of phase transition of low‐complexity proteins was described earlier for FG‐rich nucleoporins 19. The fortuitous use – by McKnight and co‐workers – of a biotinylated isoxazole derivative to stabilize β‐strand conformations in intrinsically disordered RBPs paved the way for the biochemical isolation of material within these membrane‐less organelles. It turned out that especially low‐complexity RBPs such as FUS (Fused in Sarcoma), hnRNPA1 and hnRNPA2B1 were abundant in the droplets/hydrogels 20, 21 (Fig. 1). Intriguingly, mRNAs in the hydrogels exhibited a preponderance of long 3′UTRs, which provide a platform for RBP interplay, and could mean that mRNAs in liquid droplets are likely to be regulated mRNA species. An emerging theme in mRNA studies is that extensive 3′UTRs act as scaffolds for post‐transcriptional regulatory purposes, and that physiologic responses depend on competing and/or cooperating trans‐acting factors 22, 23, 24.
Figure 1.
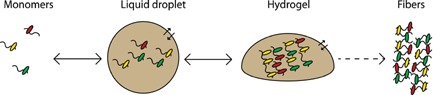
Liquid‐liquid phase separation of RNPs and decreased reversibility. At a critical concentration, monomeric RNPs are partitioned into reversible liquid droplets in a heterotypical manner, illustrated by different colors of low‐complexity RBPs. The presence of RNA (drawn as “snakes”) together with dynamic, multiple weak interactions between intrinsically disordered protein regions (not shown), seems critical for droplet formation. In hydrogels, the interactions between the disordered regions are less dynamic because of cross‐β conformations, and hydrogels appear to exhibit a broad spectrum of reversibility. Formation of amyloid‐like inclusions of low reversibility, associated with disease mutations, is shown at the right. Partitioning of RNPs into droplets/hydrogels increases their local concentration by orders of magnitude 38 and thereby the likelihood of fibrillization.
P bodies and stress granules contain low‐complexity RBPs
P bodies and stress granules are cytoplasmic RNP bodies that exhibit droplet properties. P bodies have been examined in studies of mRNA decay and translational repression, and the marker is usually the decapping enzyme Dcp2 25. Stress granules are formed in response to various cellular stresses such as arsenite and glucose deprivation, and are considered to be a pool of mRNPs stalled during translation initiation with TIA‐1 as a common marker 26, 27. However, a clear distinction between the two types of RNPs, based on the presence of a particular RBP, is not straightforward 28, and neither type seems to be the cause, but rather a consequence, of polysome disassembly 29, 30, 31.
Both types of RNP bodies incorporate low‐complexity RBPs. In particular stress granules are regarded as pivotal in the pathogenesis of the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) via fibrillization of RBPs such as hnRNPA1, hnRNPA2B1, FUS and TDP‐43 32, 33, 34. Although the low‐complexity sequence from hnRNPA1 is sufficient to mediate liquid‐liquid phase separation, the inclusion of the two RRMs in full‐length hnRNPA1, and the presence of RNA, favor reversibility rather than fibrillization 35. This study also showed that fluorescence recovery after photobleaching (FRAP) analysis of hnRNPA1 within liquid droplets exhibited kinetics on the timescale of seconds, both in vitro and in vivo. This is in striking contrast to the near absence of recovery within 15 min in cross‐β fibrillar hydrogels, and suggests a more rigid incorporation of hnRNPA1 in the latter. If a zipper motif within the low‐complexity sequence of hnRNPA1 is removed (termed delta‐hexa mutant), fibrillization is abrogated without affecting liquid droplet formation, thereby uncoupling the two phenomena mechanistically. Moreover, the presence of RNA reduced the necessary critical hnRNPA1 concentration for liquid‐liquid phase separation. Finally, a disease‐causing hnRNPA1 D262V mutation 36 did not appear to alter molecular interactions that drive phase separation, but increased the propensity toward amyloid‐like fibrillization subsequently 35.
Parker and co‐workers 37 also examined the propensity of full‐length hnRNPA1 and the delta‐hexa and D262V mutants to form liquid droplets in vitro, and obtained similar results, namely that a low salt concentration (37.5 mM) and high protein concentration were necessary for phase separation. Regardless of whether the wild‐type or mutants were analyzed by fluorescence microcopy, a physiological salt concentration (150 mM) resulted in a one‐phase system, unless RNA was present together with crowding agents such as polyethylene glycol or Ficoll. However, the kinetics of fibrillization was strikingly different, in that the D262V mutant formed SDS‐resistant fibers, whose rate of formation was increased by prior droplet formation, presumably due to an increased protein concentration within the droplets.
Taken together, the crucial players for forming RNP granules via liquid‐liquid phase separation appear to be RBPs containing low‐complexity sequences and RNA. Multivalency seems to be the key concept behind RNP granule assembly offering synergy between multiple RNA‐binding modules in RBPs, multiple RBP target sites on RNA, and multiple low‐complexity sequences in RBPs 38.
A transition to more solid‐like droplets is accelerated by disease mutations
An emerging theme in studies of liquid droplets is that they evolve into more solid‐like entities, and that this transition is greatly accelerated by disease mutations. A controversial issue has been whether conformations of low‐complexity sequences in liquid droplets are different from those appearing in more fibrous hydrogels. However, a chemical footprinting experiment with N‐acetyl‐imidazole has been unable to identify differences in side‐chain accessibility of the low‐complexity region in hnRNPA2B1, regardless of its presence in liquid droplets, hydrogels – or nuclei for that matter – and suggests cross‐β polymerization as a unifying principle 39. Nevertheless, the physiological relevance of fibrous hydrogels is a pending issue.
Fused in Sarcoma (FUS) is a favorite RBP in studies of liquid‐like compartments both in vitro and in vivo. This is due to several facts: High intracellular concentration, extensive N‐terminal low‐complexity sequence, an ability to shuttle between nucleus and cytoplasm, and importantly, mutations of FUS are seen in relation to the neurodegenerative diseases ALS and FTD. To gain insight into the physiological role of FUS, Alberti and co‐workers 40 generated a FUS transgene that could be expressed at a similar level to the endogenous one at 2 μM in HeLa cells. At steady‐state, FUS is mainly a nuclear protein, but during stress FUS is transported to the cytoplasm, where it is localized in stress granules. Within these compartments, FRAP experiments have shown that exchange with the surrounding cytoplasm is taking place within seconds, and so‐called half‐bleach experiments reveal a dynamic droplet interior 40, so in a physiological setting the propensity to form more solid‐like fibers is low. However, if a patient mutation such as G156E in the low‐complexity QGSY‐rich region or deletion of the C‐terminal nuclear localization signal (NLS) is present, the kinetics of fibrillization is increased. In the latter case, inhibition of nuclear localization increases the cytoplasmic concentration of FUS, thereby facilitating phase separation and the likelihood of subsequent aggregation. This maturation phenomenon is essentially a conversion from a metastable liquid‐like state to a thermodynamically more stable solid‐like state, and takes place within 8 h in vitro. However, it is much slower in vivo, probably because it is efficiently counteracted by chaperones and disaggregases 41, 42; hence cytoplasmic inclusions will first become a severe phenotype with age 40.
A similar conclusion was reached in a C. elegans model of FUS‐dependent neurodegeneration 43. In this study of C‐terminal NLS mutations, the conversion of liquid droplets into fibrillar hydrogels was sufficient to mediate neurotoxicity. An important consequence of the transition from dynamic liquid droplets to more static hydrogels was impairment of on‐site translation in Xenopus retinal neurons, due to entrapped mRNPs. In the ongoing discussion about the physiological significance of hydrogels, a distinction between reversible and irreversible hydrogels may be more fruitful, because the difference in viscosity is slight between the former and liquid droplets 44. Therefore, partial polymerization into a loose yet reversible fibrous mesh could be appropriate for the physiology of longer‐lived RNP granules and the nuclear pore matrix 45.
Whi3p droplet properties depend on RNA identity
In multinucleate large Ashbya cells, the G1 cyclin CLN3 transcript shows a nonrandom spatial clustering due to a glutamine‐rich region in the RBP Whi3p, and this inhomogeneous distribution is crucial for cell‐cycle timing variability and asynchrony of the nuclei 32. An additional mRNA target for Whi3p is the BNI1 transcript encoding formin, which is important for establishing polarity 46. Recombinant Whi3p (28 μM) is able to form protein‐only liquid droplets at 75 mM KCl in vitro, but not at physiological salt concentrations. In contrast, the liquid‐liquid phase separation of protein‐RNA droplets is promoted at physiological salt concentration and 200 nM protein by the presence of RNA in the guise of either CLN3 or BNI1 mRNAs 47. Moreover, the strict dependence on the RRM domain for Whi3p droplet formation reinforces the important role of RNA. Although both mRNAs encompass five putative UGCAU attachment sites for Whi3p, the viscosity of the two types of protein‐RNA droplets, and their propensity to fuse, are different. This implies that the identity of the participating mRNA component could tune the functional specificity and encompass information beyond mere coding ability. Therefore, a combination of low‐complexity sequences and multivalent interactions appears to be critical for intracellular phase transition, although the effect of the spacing pattern of Whi3p attachment sites on mRNA targets is unknown. During droplet maturation, the transition from a liquid to a more fibrillar state of glutamine‐rich RBPs, such as Whi3p, is diminished considerably by the presence of RNA 40, 47. In fact, the negatively charged RNA influences the viscoelastic properties of liquid droplets, as measured by microrheology and FRAP, and gives rise to decreased viscosity within the droplet termed RNA‐induced fluidization 44, 47. The presence of mRNA within these intracellular reactors is therefore crucial for reversibility and, by inference, its subsequent recruitment to the translational apparatus.
RNP granules encompass RNA assemblages
The RNP granule can be regarded as the physical manifestation of the post‐transcriptional RNA regulon, because low‐complexity sequences in intrinsically disordered regions of RBPs would have an ability to interact both homo‐ and heterotypically in a dynamic fashion. The concept of an RNP granule is appealing, because this would allow mRNAs encoding functionally related proteins to be stored/transported in a repressed form and be translated locally in a synchronous manner. The RNP granule concept has been widely accepted among neurobiologists, because mRNAs are transported over considerable distances to obtain local protein synthesis in axons and dendrites 48.
Despite the widespread acclaim of the RNP granule as a mode of harnessing and transporting mRNAs, there are few studies that have identified the molecular motors involved 49, 50, 51. It is generally assumed that long‐range movements of RNP granules take place on microtubules and that short‐range transport will take place on microfilaments, a concept that is based on disturbances following nocodazole and cytochalasin D treatments, respectively. However, because optical resolution has improved, it is not clear whether RNP granules are co‐localizing with microtubules or are tethered via ER/endosomes 52, 53. An additional question related to RNP granules – at least if based on a solid‐like structure – is that of reversibility, i.e. how is the mRNA deployed at the site of translation? The most obvious solution is a signal transduction event leading to a post‐translational modification or proteolysis of the RBP. By breaking protein‐protein, rather than RNA‐protein, interactions, mRNA could be released in an accessible form to the translational apparatus.
Puf3p phosphorylation activates mRNAs encoding mitochondrial proteins
Low‐complexity sequences exhibit enrichment of serine and arginine residues 54, thereby providing ample opportunity for post‐translational modifications such as phosphorylations and methylations, respectively, that could regulate the reversibility of mRNAs within RNP granules 55. For example, chemical inhibition of the dual specificity kinase DYRK3 in HeLa cells affects the dissolution of stress granules by preventing autophosphorylation of its own low‐complexity N‐terminus and phosphorylation of RBPs 56. Moreover, phosphorylation of intrinsically disordered MEG substrates by the C. elegans DYRK3 homologue drives the dissolution of P granules in embryos, whereas condensation is mediated by a PP2A phosphatase 57. In the next section, we describe one of the most clear‐cut examples of a post‐transcriptional RNA regulon that is regulated by environmental cues.
Puf3p associates with cytoplasmic mRNAs encoding mitochondrial proteins in budding yeast 58, 59. The exact mechanism behind the coordination in terms of localization and/or translatability has been unclear, but a recent study suggests that the participation of RNP granules is pivotal for the physiology of the regulon 60. Upon glucose starvation, Puf3p becomes heavily phosphorylated, mainly within its N‐terminal low‐complexity region, and this results in translational activation of bound mRNAs, thereby promoting mitochondrial biogenesis and the capacity for oxidative catabolism of carbon sources 61. From a mechanistic point of view, it should be noted that phosphorylated Puf3p co‐sediments with its target mRNAs in polysomes, suggesting that the translational activation is due to interference with protein‐protein, rather than RNA‐protein, interactions. Moreover, a phosphomutant containing 24 serine/threonine‐to‐alanine mutations exhibits entrapment of target mRNAs in RNP granules and a severe growth arrest in response to glucose depletion. Taken together, this study suggests that signal transduction mediated by glucose deprivation unfurls RNP granules (in this case PUF bodies), rendering monomeric RNPs accessible to the translational apparatus. The coordination in terms of time and space is governed by the RBP Puf3p in a post‐transcriptional RNA regulon 60, 62.
Drosophila Imp coordinates an RNA assemblage involved in growth cone dynamics
In a comparison of the oncofetal RBP IMP family with its Drosophila homologue, we noticed that the latter encompasses a C‐terminal glutamine‐rich low‐complexity sequence (Fig. 2). Moreover, Drosophila Imp has been shown to be part of hydrogels in a biotinylated isoxazole precipitation assay 20. To address the possibility that this particular RBP coordinates the expression of a subset of mRNAs by creating an RNA assemblage within RNP granules 63, we subjected Drosophila S2 cells to either short or long‐wave UV irradiation followed by immunoprecipitation of lysates 64. The iCLIP experimental approach allows the identification of in vivo cross‐linking sites on the entire transcriptome regardless of individual affinities 65, 66, whereas RBP immunoprecipitation on its own preferentially captures stable interactions without pinpointing the precise binding sites within an mRNA. Regardless of the procedure employed, mRNAs encoding components involved in the coordination of F‐actin formation were enriched (Fig. 3). To address the physiological significance of the Imp RNA assemblage, we reduced the level of Imp in Drosophila S2 cells by RNA interference and examined the effect on the steady‐state F‐actin level by staining with phalloidin. S2 cells are spherical without extensive protrusions, but the cellular F‐actin level was nevertheless diminished by 36% upon Imp reduction. By carrying out a global transcriptome analysis, we were able to establish that Imp did not alter the global S2 transcriptome, and that the total cytosolic actin monomer concentration was unchanged. We interpret this as an effect of an RBP on local cytoplasmic events rather than on overall cellular post‐transcriptional regulation. A simple model would be that Imp defines an RNA assemblage ensuring that functionally related mRNAs – i.e. a post‐transcriptional RNA regulon – would be in the vicinity of each other, and produce components at a higher local concentration than possible from dispersed protein synthesis in the cytoplasm.
Figure 2.
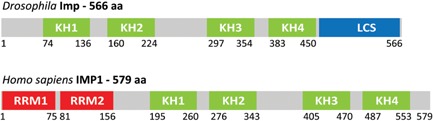
Schematic representation of Drosophila Imp and human IMP1. Numbers refer to amino acid positions bordering various domains. Human IMP1 is composed of 579 amino acids and exhibits six characteristic RNA‐binding modules, namely two N‐terminal RNA recognition motifs (RRM1 and 2), and four C‐terminal hnRNPK homology domains (KH1–4). Whereas two RRMs or four KH domains can be found in other RBPs, the 2 + 4 modular architecture seen in vertebrate IMPs is unique. From both a phylogenetic and experimental standpoint, the four KH domains constitute a functional entity in terms of high‐affinity RNA‐binding, granular RNP assembly, and RNA localization 77. Although Drosophila Imp exhibits rudimentary features of RRMs in minor isoforms, the major isoform of 566 amino acid lacks RRMs. Instead, Drosophila Imp contains a large C‐terminal low‐complexity glutamine‐rich sequence (LCS).
Figure 3.
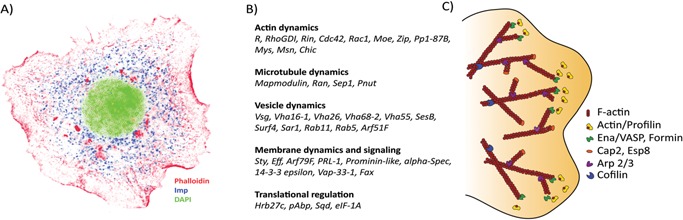
A: Drosophila Imp in cytoplasmic RNP granules. The picture shows a Drosophila S2 cell stained with DAPI (green), anti‐Imp antibody (blue) and phalloidin (red) (all in pseudo‐colors). Imp granules are about 100–200 nm large and located in the perinuclear region and close to the actin mesh. B: Transcripts associated with the Imp‐mediated RNA assemblage. Forty out of the 86 transcripts, identified by three different high‐throughput analyses, participate in growth cone dynamics 64. C: Key proteins involved in the formation and maintenance of F‐actin in lamellipodia. Several effector proteins are needed to nucleate, elongate, depolymerize, bundle and contract the actin filaments in order to reorganize the shape of the leading edge of a cell in response to external guidance cues. Profilin is responsible for the addition of actin monomers to the barbed end of growing filaments by interacting with the barbed‐end protector proteins, the formins and Ena/VASP 78. Formins and Ena/VASP also ensure continuous growth of the filament by inhibiting the binding of capping proteins, Cap2 and Esp8, otherwise capable of blocking the addition of actin monomers 79, 80. In lamellipodia, the mesh‐like form of F‐actin is accomplished by the dendritic nucleator proteins, the Arp2/3 complex, functioning as branch point holders, where they nucleate new filaments that branch off from pre‐existing ones 78. The recycling of actin subunits and actin remodelling is vital for movement of the cell. Cofilin in its unphosphorylated form binds to and twists the actin filament toward the pointed end, and the severing creates additional barbed ends, enhancing the turnover of actin subunits 81.
Imp‐dependent stimulation of F‐actin formation has developmental consequences, especially for processes relying on F‐actin dynamics such a neuronal growth cone steering and synaptogenesis 67. Imp‐deficient flies exhibit a spectrum of neurogenesis defects – almost to the point of being stochastic – and even survivors at the pharate adult stage are unable to eclose, suggesting locomotory failure. The broad spectrum of defects is compatible with what was observed in the cell‐line: namely that all components appear to be formed in the appropriate amount – at least at the transcript level – regardless of Imp. However, the possibility of producing the components in high local concentrations facilitating macromolecular assembly is jeopardized to varying degrees. A study by Besse and co‐workers 68 supports the role of Drosophila Imp in facilitating F‐actin formation during neurogenesis, because defects in mushroom body (somewhat reminiscent of the hippocampus) neurogenesis in Imp‐deficient animals can be partially rescued by Chickadee. The latter is the Drosophila homologue of profilin, which is the main facilitator of F‐actin formation from G‐actin. One would also expect transcripts encoding Rho GTPases, moesin, ena/VASP, and cytosolic actin itself in a post‐transcriptional RNA regulon coordinated by Imp and conveying cues from the membrane to the cytoskeleton, and this was actually observed 64. A recent translational profiling study of the conversion of growth cones into presynaptic terminals in Drosophila photoreceptor R cells has shown that prior to differentiation there is a 40‐fold upregulation of Imp. Moreover, all of the actin regulatory proteins encoded by mRNAs, identified in our iCLIP study, are expressed in R cells during synapse formation 69. Taken together, a picture emerges where one phase transition, namely the partitioning of RNPs into liquid droplets, facilitates a subsequent transition of monomeric actin into F‐actin 70.
The advantages in clustering functionally related mRNAs within an RNA assemblage
Compared with solid‐like cytoplasmic inclusions, liquid droplets offer some intrinsic advantages especially in terms of dynamics, because material can enter and exit the droplet faster but still be concentrated in a small volume 71. The latter is important, because concomitant release of a high local concentration of related mRNAs to the translational apparatus will facilitate a high local protein concentration of newly synthesized interacting species (Fig. 4). Based on RNP body condensation/dissolution behavior following translational inhibition of either initiation or elongation, respectively 29, 72, it is generally assumed that the translational apparatus does not have access to the droplet interior. However, it should be recalled that ribosomal proteins themselves exhibit intrinsically disordered regions termed “waggly tails” 73. A few studies have addressed the presence of translating ribosomes in the vicinity of RNP granules 74, 75, and the study by Mili and co‐workers 76 actually showed translation within cytoplasmic FUS granules generating local protein production from APC‐RNPs in NIH 3T3 protrusions. In general, the understanding of the location and the identity of the ribosomes carrying out the local synthesis is lagging behind, so this should be an important line of enquiry in upcoming studies.
Figure 4.
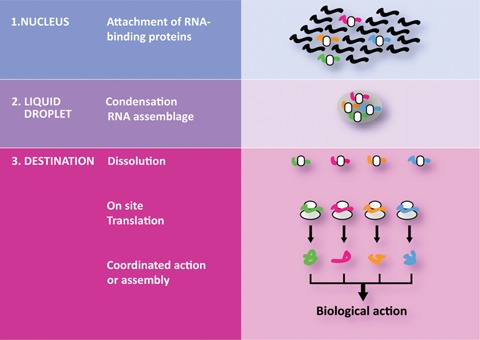
Schematic representation of the role of RNP granules/droplets in the segregation of mRNA assemblages. In the nucleus, transcripts are associated with particular RBPs, that – via low‐complexity sequences – drive the formation of liquid droplets containing mRNAs encoding factors participating in a particular biological process or macromolecular complex. Following cellular trafficking to the final destination, the increase in local mRNA concentration facilitates synchronous on‐site protein production.
Conclusions and prospects
In this essay, we have presented the rationale behind clustering functionally related mRNAs in assemblages, with an emphasis on coordinated and accelerated recruitment to the translational apparatus. An additional bonus of partitioning is fidelity, allowing much less volume and time for aberrant interactions among the resulting protein products. However, this mechanism does not come without a cost, because phase separation of an RBP is a fine balance between physiological assembly and pathological fibrillization.
Recent genome‐wide screenings strongly support the concept of RNA assemblages segregated by particular RBPs. To understand the physiological significance of RNA assemblages, we need a deeper understanding of their dynamic behavior and molecular composition. The crucial role of RNA, salt and temperature for droplet dynamics has been obtained from studies in vitro focusing on the biophysical behavior of tagged low‐complexity sequences. However, visualization of the interplay between RNP granules and the translational apparatus in vivo is strongly needed. Microscopes are now approaching single RNP particle resolution, so studies should be directed toward endogenous components rather than reporters with various tags. Moreover, low‐complexity sequences in homologous RBPs exhibit low levels of conservation, so comparative studies may provide clues to the rationale behind these intrinsically disordered regions in terms of evolutionary rewiring of post‐transcriptional regulation.
A number of granular RBPs have been implicated in neurological diseases. So far we know little about the potential druggability of the conditions, but the concept of modifying particular RNA assemblages is an appealing one, because a complex process might be influenced by a single drug. Blockmirs may be employed to up‐regulate RBPs, whereas another possibility would be to prevent granule formation by modulating post‐translational modifications, such as phosphorylations, or by introducing RNA sponges. Considering the many ongoing human sequencing initiatives, we envisage that RBPs will be implicated in a number of both common and rare diseases, which will stimulate the development of new therapeutic strategies.
References
- 1. Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157: 624–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robitaille AM, Christen S, Shimobayashi M, Cornu M, et al. 2013. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339: 1320–3. [DOI] [PubMed] [Google Scholar]
- 3. French JB, Jones SA, Deng H, Pedley AM, et al. 2016. Spatial colocalization and functional link of purinosomes with mitochondria. Science 351: 733–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keene JD. 2007. RNA regulons: coordination of post‐transcriptional events. Nat Rev Genet 8: 533–43. [DOI] [PubMed] [Google Scholar]
- 5. Castello A, Fischer B, Eichelbaum K, Horos R, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA‐binding proteins. Cell 149: 1393–406. [DOI] [PubMed] [Google Scholar]
- 6. Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA‐binding proteins. Nat Rev Genet 15: 829–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neelamraju Y, Hashemikhabir S, Janga SC. 2015. The human RBPome: from genes and proteins to human disease. J Proteomics 127: 61–70. [DOI] [PubMed] [Google Scholar]
- 8. Zhang DL, Ghosh MC, Rouault TA. 2014. The physiological functions of iron regulatory proteins in iron homeostasis – an update. Front Pharmacol 5: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackay JP, Font J, Segal DJ. 2011. The prospects for designer single‐stranded RNA‐binding proteins. Nat Struct Mol Biol 18: 256–61. [DOI] [PubMed] [Google Scholar]
- 10. Daubner GM, Clery A, Allain FH. 2013. RRM‐RNA recognition: NMR or crystallography and new findings. Curr Opin Struct Biol 23: 100–8. [DOI] [PubMed] [Google Scholar]
- 11. Mackereth CD, Sattler M. 2012. Dynamics in multi‐domain protein recognition of RNA. Curr Opin Struct Biol 22: 287–96. [DOI] [PubMed] [Google Scholar]
- 12. Hennig J, Militti C, Popowicz GM, Wang I, et al. 2014. Structural basis for the assembly of the Sxl‐Unr translation regulatory complex. Nature 515: 287–90. [DOI] [PubMed] [Google Scholar]
- 13. Cheong CG, Hall TM. 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci USA 103: 13635–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hafner M, Landthaler M, Burger L, Khorshid M, et al. 2010. Transcriptome‐wide identification of RNA‐binding protein and microRNA target sites by PAR‐CLIP. Cell 141: 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lunde BM, Moore C, Varani G. 2007. RNA‐binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 8: 479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh G, Pratt G, Yeo GW, Moore MJ. 2015. The clothes make the mRNA: past and present trends in mRNP fashion. Annu Rev Biochem 84: 325–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iadevaia V, Gerber AP. 2015. Combinatorial control of mRNA fates by RNA‐Binding proteins and non‐coding RNAs. Biomolecules 5: 2207–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, et al. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–32. [DOI] [PubMed] [Google Scholar]
- 19. Frey S, Richter RP, Gorlich D. 2006. FG‐rich repeats of nuclear pore proteins form a three‐dimensional meshwork with hydrogel‐like properties. Science 314: 815–7. [DOI] [PubMed] [Google Scholar]
- 20. Kato M, Han TW, Xie S, Shi K, et al. 2012. Cell‐free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han TW, Kato M, Xie S, Wu LC, et al. 2012. Cell‐free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149: 768–79. [DOI] [PubMed] [Google Scholar]
- 22. Jonson L, Christiansen J, Hansen TV, Vikesa J, et al. 2014. IMP3 RNP safe houses prevent miRNA‐directed HMGA2 mRNA decay in cancer and development. Cell Rep 7: 539–51. [DOI] [PubMed] [Google Scholar]
- 23. Bounedjah O, Desforges B, Wu TD, Pioche‐Durieu C, et al. 2014. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res 42: 8678–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berkovits BD, Mayr C. 2015. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522: 363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jain S, Parker R. 2013. The discovery and analysis of P Bodies. Adv Exp Med Biol 768: 23–43. [DOI] [PubMed] [Google Scholar]
- 26. Gilks N, Kedersha N, Ayodele M, Shen L, et al. 2004. Stress granule assembly is mediated by prion‐like aggregation of TIA‐1. Mol Biol Cell 15: 5383–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kedersha N, Anderson P. 2009. Regulation of translation by stress granules and processing bodies. Prog Mol Biol Transl Sci 90: 155–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kedersha N, Stoecklin G, Ayodele M, Yacono P, et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169: 871–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eulalio A, Behm‐Ansmant I, Schweizer D, Izaurralde E. 2007. P‐body formation is a consequence, not the cause, of RNA‐mediated gene silencing. Mol Cell Biol 27: 3970–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu W, Sweet TJ, Chamnongpol S, Baker KE, et al. 2009. Co‐translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee C, Zhang HY, Baker AE, Occhipinti P, et al. 2013. Protein aggregation behavior regulates cyclin transcript localization and cell‐cycle control. Dev Cell 25: 572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ramaswami M, Taylor JP, Parker R. 2013. Altered ribostasis: RNA‐protein granules in degenerative disorders. Cell 154: 727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deng H, Gao K, Jankovic J. 2014. The role of FUS gene variants in neurodegenerative diseases. Nat Rev Neurol 10: 337–48. [DOI] [PubMed] [Google Scholar]
- 35. Molliex A, Temirov J, Lee J, Coughlin M, et al. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim HJ, Kim NC, Wang YD, Scarborough EA, et al. 2013. Mutations in prion‐like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase‐separated liquid droplets by RNA‐binding proteins. Mol Cell 60: 208–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li P, Banjade S, Cheng HC, Kim S, et al. 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483: 336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiang S, Kato M, Wu LC, Lin Y, et al. 2015. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid‐like droplets, and nuclei. Cell 163: 829–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patel A, Lee HO, Jawerth L, Maharana S, et al. 2015. A liquid‐to‐solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162: 1066–77. [DOI] [PubMed] [Google Scholar]
- 41. Cherkasov V, Hofmann S, Druffel‐Augustin S, Mogk A, et al. 2013. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 23: 2452–62. [DOI] [PubMed] [Google Scholar]
- 42. Kroschwald S, Maharana S, Mateju D, Malinovska L, et al. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress‐inducible RNP granules. Elife 4: e06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murakami T, Qamar S, Lin JQ, Schierle GS, et al. 2015. ALS/FTD mutation‐induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 88: 678–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Elbaum‐Garfinkle S, Brangwynne CP. 2015. Liquids, fibers, and gels: the many phases of neurodegeneration. Dev Cell 35: 531–2. [DOI] [PubMed] [Google Scholar]
- 45. Schmidt HB, Gorlich D. 2016. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem Sci 41: 46–61. [DOI] [PubMed] [Google Scholar]
- 46. Lee C, Occhipinti P, Gladfelter AS. 2015. PolyQ‐dependent RNA‐protein assemblies control symmetry breaking. J Cell Biol 208: 533–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang H, Elbaum‐Garfinkle S, Langdon EM, Taylor N, et al. 2015. RNA controls PolyQ protein phase transitions. Mol Cell 60: 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holt CE, Schuman EM. 2013. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 80: 648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanai Y, Dohmae N, Hirokawa N. 2004. Kinesin transports RNA: isolation and characterization of an RNA‐transporting granule. Neuron 43: 513–25. [DOI] [PubMed] [Google Scholar]
- 50. Dienstbier M, Boehl F, Li X, Bullock SL. 2009. Egalitarian is a selective RNA‐binding protein linking mRNA localization signals to the dynein motor. Genes Dev 23: 1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Song T, Zheng Y, Wang Y, Katz Z, et al. 2015. Specific interaction of KIF11 with ZBP1 regulates the transport of beta‐actin mRNA and cell motility. J Cell Sci 128: 1001–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Higuchi Y, Ashwin P, Roger Y, Steinberg G. 2014. Early endosome motility spatially organizes polysome distribution. J Cell Biol 204: 343–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jansen RP, Niessing D, Baumann S, Feldbrugge M. 2014. MRNA transport meets membrane traffic. Trends Genet 30: 408–17. [DOI] [PubMed] [Google Scholar]
- 54. Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. 2015. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett 589: 15–22. [DOI] [PubMed] [Google Scholar]
- 55. Nott TJ, Petsalaki E, Farber P, Jervis D, et al. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 57: 936–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, et al. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152: 791–805. [DOI] [PubMed] [Google Scholar]
- 57. Wang JT, Smith J, Chen BC, Schmidt H, et al. 2014. Regulation of RNA granule dynamics by phosphorylation of serine‐rich, intrinsically disordered proteins in C. elegans. Elife 3: e04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gerber AP, Herschlag D, Brown PO. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA‐binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Blackinton JG, Keene JD. 2014. Post‐transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol 34: 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee CD, Tu BP. 2015. Glucose‐regulated phosphorylation of the PUF protein puf3 regulates the translational fate of its bound mRNAs and association with RNA granules. Cell Rep 11: 1638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gancedo JM. 1998. Yeast carbon catabolite repression. Microbiol Mol Biol Rev 62: 334–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saint‐Georges Y, Garcia M, Delaveau T, Jourdren L, et al. 2008. Yeast mitochondrial biogenesis: a role for the PUF RNA‐binding protein Puf3p in mRNA localization. PLoS ONE 3: e2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Toretsky JA, Wright PE. 2014. Assemblages: functional units formed by cellular phase separation. J Cell Biol 206: 579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hansen HT, Rasmussen SH, Adolph SK, Plass M, et al. 2015. Drosophila Imp iCLIP identifies an RNA assemblage coordinating F‐actin formation. Genome Biol 16: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Darnell RB. 2010. HITS‐CLIP: panoramic views of protein‐RNA regulation in living cells. Wiley Interdiscip Rev RNA 1: 266–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huppertz I, Attig J, D'Ambrogio A, Easton LE, et al. 2014. ICLIP: protein‐RNA interactions at nucleotide resolution. Methods 65: 274–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chia PH, Chen B, Li P, Rosen MK, et al. 2014. Local F‐actin network links synapse formation and axon branching. Cell 156: 208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Medioni C, Ramialison M, Ephrussi A, Besse F. 2014. Imp promotes axonal remodeling by regulating profilin mRNA during brain development. Curr Biol 24: 793–800. [DOI] [PubMed] [Google Scholar]
- 69. Zhang KX, Tan L, Pellegrini M, Zipursky SL, et al. 2016. Rapid changes in the translatome during the conversion of growth cones to synaptic terminals. Cell Rep 14: 1258–71. [DOI] [PubMed] [Google Scholar]
- 70. Weber SC, Brangwynne CP. 2012. Getting RNA and protein in phase. Cell 149: 1188–91. [DOI] [PubMed] [Google Scholar]
- 71. Hyman AA, Weber CA, Julicher F. 2014. Liquid‐liquid phase separation in biology. Annu Rev Cell Dev Biol 30: 39–58. [DOI] [PubMed] [Google Scholar]
- 72. Teixeira D, Sheth U, Valencia‐Sanchez MA, Brengues M, et al. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Peng Z, Oldfield CJ, Xue B, Mizianty MJ, et al. 2014. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell Mol Life Sci 71: 1477–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tatavarty V, Ifrim MF, Levin M, Korza G, et al. 2012. Single‐molecule imaging of translational output from individual RNA granules in neurons. Mol Biol Cell 23: 918–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yasuda K, Zhang H, Loiselle D, Haystead T, et al. 2013. The RNA‐binding protein Fus directs translation of localized mRNAs in APC‐RNP granules. J Cell Biol 203: 737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Shi Z, Barna M. 2015. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA‐Binding proteins. Annu Rev Cell Dev Biol 31: 31–54. [DOI] [PubMed] [Google Scholar]
- 77. Nielsen FC, Nielsen J, Kristensen MA, Koch G, et al. 2002. Cytoplasmic trafficking of IGF‐II mRNA‐binding protein by conserved KH domains. J Cell Sci 115: 2087–97. [DOI] [PubMed] [Google Scholar]
- 78. Dominguez R. 2009. Actin filament nucleation and elongation factors‐structure‐function relationships. Crit Rev Biochem Mol Biol 44: 351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bear JE, Gertler FB. 2009. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci 122: 1947–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu J, Casella JF, Pollard TD. 1999. Effect of capping protein, CapZ, on the length of actin filaments and mechanical properties of actin filament networks. Cell Motil Cytoskeleton 42: 73–81. [DOI] [PubMed] [Google Scholar]
- 81. Carlier MF, Pantaloni D. 1997. Control of actin dynamics in cell motility. J Mol Biol 269: 459–67. [DOI] [PubMed] [Google Scholar]


