Abstract
RNA silencing is a eukaryote‐specific phenomenon in which microRNAs and small interfering RNAs degrade messenger RNAs containing a complementary sequence. To this end, these small RNAs need to be loaded onto an Argonaute protein (AGO protein) to form the effector complex referred to as RNA‐induced silencing complex (RISC). RISC assembly undergoes multiple and sequential steps with the aid of Hsc70/Hsp90 chaperone machinery. The molecular mechanisms for this assembly process remain unclear, despite their significance for the development of gene silencing techniques and RNA interference‐based therapeutics. This review dissects the currently available structures of AGO proteins and proposes models and hypotheses for RISC assembly, covering the conformation of unloaded AGO proteins, the chaperone‐assisted duplex loading, and the slicer‐dependent and slicer‐independent duplex separation. The differences in the properties of RISC between prokaryotes and eukaryotes will also be clarified. WIREs RNA 2016, 7:637–660. doi: 10.1002/wrna.1356
For further resources related to this article, please visit the WIREs website.
INTRODUCTION
MicroRNAs (miRNAs) are the regulatory small RNAs that control gene expression by inhibition of translation or degradation of messenger RNAs (mRNAs) containing a complementary sequence. To degrade the target mRNAs, miRNAs need to be loaded onto AGO proteins, forming a ribonucleoprotein complex called the RNA‐induced silencing complex (RISC).1, 2, 3 A complex of an AGO and a guide strand alone is referred to as ‘the mature RISC’ or simply ‘the RISC,’ the latter of which will be used throughout this review. The same complex is also called ‘the RISC core’ in the context when the RISC stands for a huge complex including many components required for translational repression and/or deadenylation. The bound guide strand takes the RISC to the target mRNAs, which often possess the sequence complementarity to the guide in the 3′ untranslated region (3′ UTR). For efficient degradation of target mRNAs, the RISC requires GW182 (glycine‐tryptophan protein of 182 kDa). This protein family facilitates the translational repression while triggering mRNA decay by recruiting the CCR4–NOT deadenylase complex4, 5, 6 (Figure 1). This posttranscriptional gene regulation is called miRNA‐mediated gene silencing. In contrast, small interfering RNAs (siRNAs) derived from perfectly complementary RNA duplexes are also incorporated into AGO proteins. The effector complex binds mRNAs containing a fully complementary sequence and endonucleolytically cleaves them. This regulation is referred to as RNA interference (RNAi). RNA silencing with these small RNAs is seen throughout eukaryotes even in some budding yeast species, such as Saccharomyces castellii and Kluyveromyces polysporus, but not in Saccharomyces cerevisiae.7, 8
Figure 1.
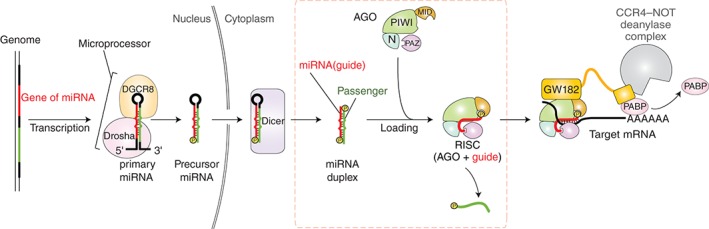
Canonical biogenesis of miRNA in the human system. The gene of a miRNA is transcribed into primary miRNA (pri‐miRNA) and processed by the microprocessor, a complex of an RNase III enzyme Drosha, and its binding partner DGCR8. The product, pre‐miRNA, is transported to the cytoplasm and then cropped at its loop by Dicer. Eventually, the miRNA duplex is loaded onto an AGO protein to form the RISC. The N, PAZ, MID, and PIWI domains of the cartoon of AGO are colored in cyan, pink, wheat, and green, respectively. For clarity, two linker regions L1 and L2 are not shown. After binding to the target mRNAs, the RISC serves as a scaffold for GW182 and CCR4–NOT deadenylase complex that facilitates the mRNA degradation. The nucleotide region serving as the miRNA is colored in red. The generated 5′ monophosphate during the miRNA biogenesis is depicted as a yellow sphere. GW182 interacts with the RISC and poly‐(A) binding proteins (PABP) within its N‐terminal and C‐terminal regions, respectively. The process of the RISC assembly discussed in this review is highlighted with a dotted line.
Recent studies have revealed that loading of siRNA and miRNA duplexes requires heat shock cognate protein 70 (Hsc70), heat shock protein 90 (Hsp90), and their co‐chaperones.9, 10, 11, 12 Using an ATP‐dependent conformational change, these proteins alter guide‐free AGO so as to load bulky RNA duplexes in the nucleic acid‐binding channel. This mechanism is similar to that of the activation of steroid hormone receptors upon the incorporation of the cognate ligand by the Hsp90 chaperone.12, 13, 14 Nowadays, gene knockdown using small RNAs, such as siRNA duplexes, small hairpin RNAs, and miRNA mimics, is a common strategy to repress the expression of a gene of interest, yet little is known about how the regulatory RNAs are loaded onto AGO proteins and how the strands are separated to form the RISC. Further understanding of the molecular mechanism is required for the development of RNAi‐based therapy. This review focuses on the RISC assembly and discusses possible mechanisms of loading and passenger ejection to form the effector complex for gene silencing. (Historically, the process of separating the loaded duplex and discarding one of the strands has been called ‘unwinding.’ This term has been used to describe the process in which helicases separate DNA/RNA duplexes using the energy of ATP hydrolysis. In contrast, the strand separation of the miRNA and siRNA duplexes occurs without ATP hydrolysis. To avoid projecting a false impression, the step of separating duplexes and discarding the passenger strand is rephrased as ‘passenger ejection’ in this review.)
MICRORNA BIOGENESIS
miRNA Processing Machinery
There exist several avenues to generate miRNAs.15 The pathway shown in Figure 1 depicts the long journey of canonical miRNA biogenesis in humans. The genes of miRNA are transcribed by RNA polymerase II in the nucleus16 and folded back on the primary miRNAs (pri‐miRNAs) into a stem‐loop structure. The flanking single‐stranded parts are cleaved by a microprocessor composed of an RNase III enzyme, Drosha, and its binding partner, DGCR8 (DiGeorge syndrome chromosomal region 8) into precursor miRNA (pre‐miRNA).17, 18, 19 The product is exported to the cytoplasm by exportin‐5,20 and the loop region is cropped by another RNase III enzyme, Dicer.21 The resultant guide–passenger duplex (miRNA/miRNA* duplex where miRNA* stands for the passenger strand) that possesses the hallmarks of a 5′ monophosphate and a 2‐nucleotide (nt) 3′ overhang at both termini is loaded onto an AGO protein, followed by passenger strand ejection, to form a RISC.
In contrast, many noncanonical miRNAs that exploit the RNA metabolic activities for maturation have been discovered. For example, bypassing pre‐miRNA processing by Dicer, pre‐miR‐451 with its stem‐loop structure is directly loaded onto Argonaute2 (hAGO2).22, 23 Some transcripts from introns, viral genomes, and tRNAs are processed in a Drosha‐independent manner. Despite structural differences, these canonical and noncanonical miRNAs are eventually incorporated into AGO proteins, indicating the protein's capability to accommodate several different types of structured RNAs. This process is quite important because the AGO protein selects one of the two strands as a guide as well as discriminates the licensed RNAs from others. Given that AGO proteins cannot release the bound guide strand except for a special case,24 the loading step is the single opportunity to choose a guide strand with which the AGO protein will spend the remainder of its life.
Structure of the RISC
In hAGO‐RISCs, the 5′ monophosphate and base moiety at position 1 of the guide strand are recognized between the MID and PIWI domains, while the 3′ end is anchored through its sugar‐phosphate backbone at the PAZ domain (Figure 2(a) and (b)). The seed region of the guide strand (positions 2–8) is thoroughly recognized through the 2′ hydroxyl groups and phosphate backbone in the nucleic acid‐binding channel.25, 26, 27 The base moieties at positions 2–4 are well exposed to the solvent for scanning target mRNAs, which is consistent with the results of the studies of the interaction between the RISC and target strands using single‐molecule fluorescence.28, 29, 30, 31 All of the crystal structures of the RISC, except for one case32 (Figure 2(a): PDB ID: 4W5N), show a similar trajectory of the bound guide RNA with its disordered middle segment25, 26, 27, 32, 33, 34 (Figure 2(b)).
Figure 2.
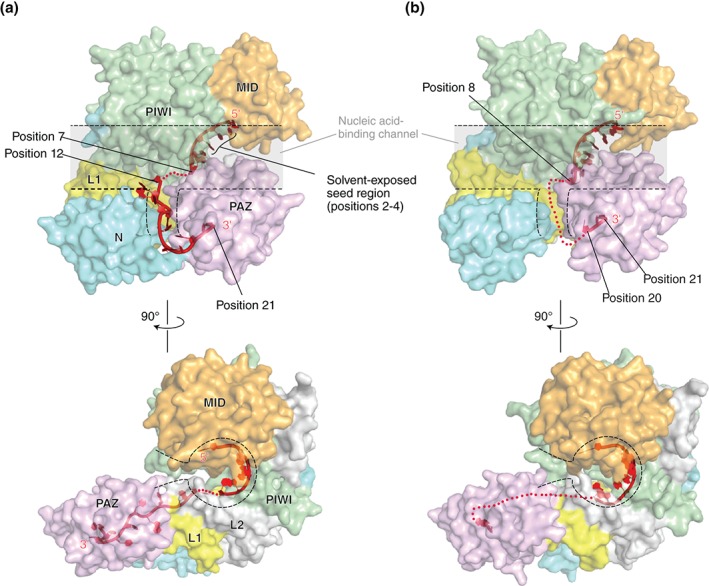
Structures of the RISC of human Argonaute2 (a: PDB ID: 4W5N) and Argonaute1 (b: PDB ID: 4KXT). The transparent surface models of hAGOs are drawn with the same color codes as in Figure 1. The linkers, L1 and L2, are colored in yellow and gray, respectively. The nucleic acid‐binding channels are highlighted with dotted lines. The guide RNA (red) is depicted as a ribbon model. The disordered parts of the guide are shown as dotted lines.
Components Involved in Duplex Loading
The mechanism of duplex loading has been well studied in the fly system. A complex containing Dicer‐2 and R2D2 is essential for the assembly of fly Ago2‐RISC, thereby being called RISC‐loading complex (RLC). R2D2, which contains tandem double‐stranded RNA‐binding domains, recognizes the thermodynamically stable terminus of siRNA duplexes, while Dicer‐2 binds the less stable end.35, 36, 37, 38 The binding manner of Dicer‐2/R2D2 to the duplex reinforces the intrinsic asymmetric guide selection of fly Ago2 by receiving the less stable distal end at the MID domain. R2D2 also plays a role to avoid loading of endogenous siRNAs onto fly Ago1,39, 40, 41, 42 which otherwise may incorporate both siRNA and miRNA duplexes like hAGO proteins that do not have such a small‐RNA sorting system. The significance of Dicer‐2 and R2D2 for duplex loading in flies is evidenced by the deficiency of Ago2‐mediated RNAi in the dcr‐2 null cells.43 In contrast, dicer‐null murine embryonic cells cannot trigger RNAi by long siRNAs, which need to be processed by Dicer, but retains the ability to repress gene expression by siRNAs,44 suggesting that siRNA duplexes are loaded onto the AGO proteins in the absence of Dicer. Moreover, recent studies indicate that Dicer is not necessary for asymmetric duplex loading in mammals.45, 46 These results clearly show that fly Ago2 possesses a different property from that of fly Ago1 and mammalian AGO proteins. Even though Dicer is dispensable for duplex loading in mammals, the existence of a complex containing Dicer‐hAGO‐TRBP (TAR‐RNA‐binding) has been confirmed in vivo and in vitro.47, 48, 49 TRBP contains several double‐stranded RNA‐binding domains like R2D2. The physiological significance of forming the complex in humans still remains unclear. Although this complex was also named RLC, it should be noted that the RLC for fly Ago2 is composed of only Dicer‐2 and R2D2 alone.
Meanwhile, it is known that loading of small RNA duplexes onto AGO proteins requires the ATP‐dependent chaperone activity of Hsc70/Hsp90 in humans as well as in flies and plants,9, 10, 11, 12 possibly by opening the nucleic acid‐binding channel of the AGO protein widely enough to accommodate the bulky duplex. With the aid of the chaperone systems, loading proceeds in an asymmetric manner in which the MID domain preferentially recognizes a thermodynamically unstable terminus of the duplex. As a result, the passenger strand whose 5′ end is not recognized by the MID domain is ejected from the channel, while the remaining guide strand and the AGO form the RISC.
RISC ASSEMBLY COMPLETES THE COMPOSITE STRUCTURE
AGO proteins have been roughly classified into four states based on the types of the bound substrate(s): apo (no substrate), pre‐RISC (with a passenger and a guide), RISC (with a guide), and target complex (with a guide and a target) (Figure 3). The transitions from the apo form to the pre‐RISC, from the pre‐RISC to the RISC, and from the RISC to the target complex correspond to duplex loading, passenger ejection, and target recognition, respectively. In this review, the existence of an intermediate state, hereafter referred to as ‘primary RISC (pri‐RISC),’ between the apo and the pre‐RISC is proposed and explained later (Figure 3). Thus far, most of the crystal structures of eukaryotic AGO proteins were determined in the RISC state25, 26, 27, 33, 34 while the several structures were the target complex of hAGO2.32, 50 The structural comparison of these two states provided insight into the molecular mechanism of the target‐mRNA incorporation. In contrast, no structure of full‐length eukaryotic AGO protein in the apo form or pre‐RISC has been determined. This has hampered the understanding of the molecular bases for loading and passenger ejection. The crystal structure of the isolated MID–PIWI domains from Neurospora crassa QDE2 (Quelling DEfective) is the only structural information about guide‐free AGO protein from eukaryotes51 (Figure 4(a), PDB ID: 2YHA). The comparison of this structure with those of archaeal and bacterial AGO proteins in apo form52, 53 (Figure 4(b) and (c), PDB IDs: 1U04 and 1YVU) indicates differences in the relative position of their MID domain against the PIWI domain (Figure 4(d)). In contrast, the MID and PIWI domains of the Thermus thermophilus AGO‐ (TtAGO), yeast K. polysporus AGO‐ (KpAGO), human Argonaute1‐ (hAGO1), and hAGO2‐RISCs are structurally well aligned across species25, 26, 27, 33, 34, 54 (Figure 4(e), PDB IDs: 3DLH, 4F1N, 4F3T, 4OLA, 4F3T, 4KRE, 4KRF, and 4KXT). This observation raises a hypothesis that the MID domain is hinged to the PIWI domain and free to move in the absence of guide strand. Limited proteolysis with thermolysin digested guide‐free hAGO2 into two fragments corresponding to the N‐PAZ and MID–PIWI lobes. The molecular weight of the MID–PIWI fragment (about 50 kDa)25 indicates the cleavage site in the L2 linker interacting with the MID and PIWI domains. Furthermore, the generated MID–PIWI fragment was rapidly cleaved between the MID and PIWI domains. However, hAGO2 became resistant to proteolysis after incorporation of the guide strand. These results suggest the flexibility between the MID and PIWI domains. The idea of the hinged MID domain is further supported by the structure of TtAGO in complex with a 10‐nt DNA guide (PDB ID: 3DLB).54 As 10 nucleotides are too short to be captured at both termini by the AGO, only the guide 3′ end is recognized at the PAZ domain whereas the 5′ end does not reach the MID domain. As a result, the last 40 residues encompassing the loop L4 to the C‐terminal carboxyl group are completely disordered, and the MID domain occupies the missing space (Figure 4(f) and (g)). This indicates that guide‐free AGO proteins exist in an open conformation where the interface between the MID and PIWI domains is exposed to solvent while the C‐terminal region remains unfolded. This hypothesis could be evidenced by the phosphorylation sites of hAGO2. One of the five sites, Y529 on the MID domain, is located at the interface with the PIWI domain (Figure 4(h)). This tyrosine interacts at its hydroxyl group with the 5′ monophosphate of the bound guide strand.25, 27 A substitution of glutamate for Y529, which mimics a phosphorylated tyrosine, dramatically reduced binding of small RNAs,55 suggesting that the phosphorylated hydroxyl group no longer hydrogen‐bonds with the 5′ monophosphate. Another phosphorylation site, S798 on the loop L4, also interacts at its hydroxyl group with the phosphate backbone of the guide strand at position 525, 27 (Figure 4(h)). Therefore, the phosphorylation of this serine appears to affect binding of small RNAs as well. The crystal structure of hAGO2‐RISC, however, indicates that these phosphorylation sites are not accessible to kinases if the guide‐free hAGO2 retains the same structure as the RISC27 (Figure 4(h)). Thus, phosphorylation of Y529 and S798 strongly supports the idea that guide‐free hAGO2 exposes the interface between the MID and PIWI domains, possibly along with the stretched loop L4, to solvent (Figure 4(i)).
Figure 3.
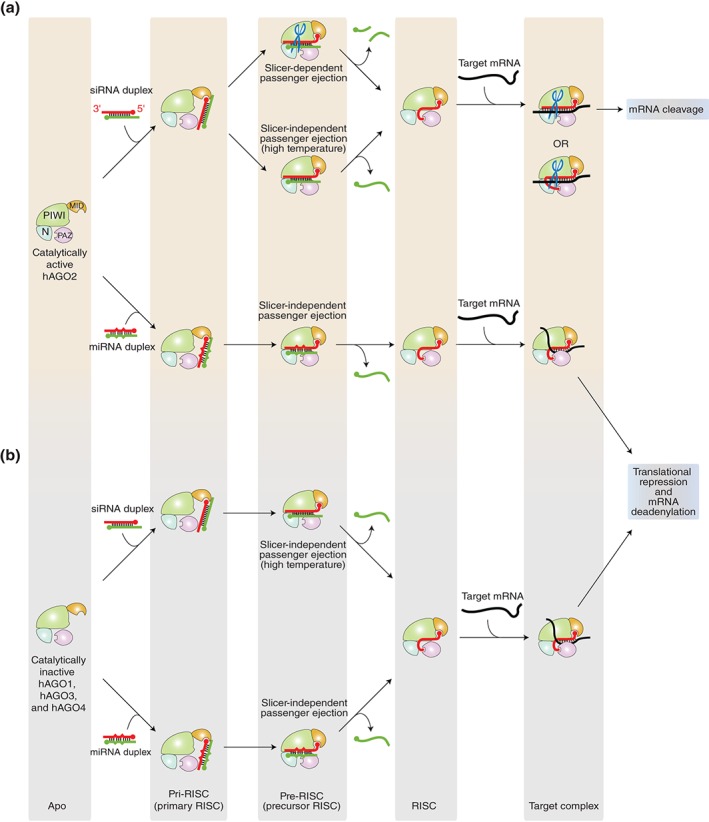
RISC assembly pathways and the downstream events in human system. (a) Catalytically active hAGO2 has two pathways dependent on the types of duplex loaded. The cleavage activity is depicted as scissors. This review proposes the existence of an intermediate state named ‘primary RISC (pri‐RISC)’ between the apo and the pre‐RISC. In this state, only the 5′ monophosphate is captured by the MID domain alone while the rest of the duplex is still exposed to solvent. (b) Catalytically inactive (slicer deficient) AGO proteins, hAGO1, hAGO3, and hAGO4, load siRNA and miRNA duplexes. Their RISC assembly is slicer independent regardless of the types of duplex.
Figure 4.
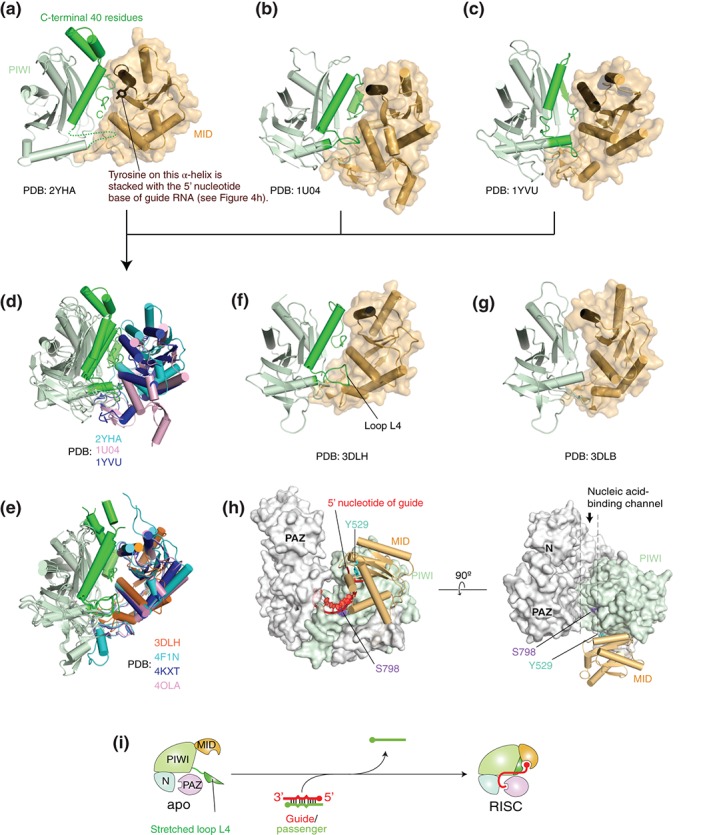
Structural evidence of an open conformation of guide‐free AGO proteins. (a–c) Crystal structures of Neurospora crassa QDE2 MID–PIWI domains (a: PDB ID: 2YHA), Pyrococcus furiosus AGO (b: PDB ID: 1U04), and Aquifex aeolicus AGO (c: PDB ID: 1YVU). For clarity, only the MID (wheat) and PIWI (light green) domains are shown in the ribbon models. The corresponding C‐terminal region that is disordered in TtAGO in complex with a guide of 10 nucleotide (nt) (see g) is colored in green. The MID domain is also drawn as a surface mode. The α‐helix on which the conserved tyrosine is stacked with the first nucleotide base of guide strand is colored in chocolate. (d) Superposed structures of (a)–(c) on their PIWI domain. The MID domains of NcQDE2, PfAGO, and AaAGO are colored in cyan, pink, and blue, respectively. Otherwise, the color code is the same as (a)–(c). (e) Superposed structures of TtAGO‐, KpAGO‐, hAGO1‐, and hAGO2‐RISCs (PDB IDs: 3DLH, 4F1N, 4KXT, and 4OLA, respectively) on their PIWI domain. The MID domains of TtAGO, KpAGO, hAGO1, and hAGO2 are colored in orange, cyan, blue, and pink, respectively. (f–g) Crystal structures of TtAGO in complex with a guide of 21 nt (f: PDB ID: 3DLH) and of 10 nt (g: PDB ID: 3DLB). The color code is the same to (a)–(c). (h) Positions of the phosphorylation sites, Y529 (cyan) and S798 (purple), on hAGO2. The structure of hAGO2‐RISC (PDB ID: 4OLA) is drawn as a surface model except for the MID domain that is shown as a cylinder model. The MID and PIWI domains are colored in wheat and light green, respectively. The bound guide RNA (red) is shown as a ribbon model. The guide is not shown on the right panel for clarity. (i) A hypothetical model of RISC assembly. Guide‐free AGO protein opens the hinged MID‐PIWI domains while the unstructured C‐terminal fragment (green) may be extended to solvent.
PROFOUND CONNECTIONS BETWEEN RISC ASSEMBLY AND CHAPERONES
AGO Protein Was Discovered by Its Interaction With Chaperones
Rat AGO protein was originally discovered as a membrane‐associated protein named GERp95 (Golgi‐Endoplasmic Reticulum Protein 95 kDa).56 Although the physiological function was not identified at that time, it was already known that this protein binds Hsc70, Hsp90, Hsp40 (also known as Hdj‐2: Human DnaJ protein 2), Hop (Hsp70/Hsp90 Organizing Protein homolog), and p23. Hsc70 and Hsp90 are monomeric and dimeric molecular chaperones, respectively, that work together for a vast number of proteins to assist their protein folding and regulate their activation.57, 58 Hsp40, together with Hsc70, helps nascent peptides to be folded properly.59 Hop is a co‐chaperone that binds, via the tetratricopeptide (TRP) domain, to the EEVD motif of Hsc70 and Hsp90.60 Another co‐chaperone, p23, binds to the N‐terminal domain of Hsp90 to stabilize the closed conformation.61 All of them strongly bind to the N domain and the L1 linker of rat AGO2, while only Hsc70 and Hsp40 weakly interact with the MID and PIWI domains56 (Figure 5(a)). Given the susceptibility of unloaded hAGO2 between the MID and PIWI domains,25 the weak interaction with Hsc70/Hsp40 may protect the MID–PIWI domains from its degradation until small RNA duplexes bind to the 5′ nucleotide‐binding pocket on the MID domain (Figure 5(b), top row). Hobman and coworkers reported that inhibition of Hsp90 activity with geldanamycin, which occupies the ATP‐binding site, causes rapid degradation of newly synthesized rat AGO.56 The Hutvagner group observed that the degradation of hAGO1 and hAGO2 by geldanamycin was alleviated by proteasome inhibitor MG132.62 Therefore, Hsp90 protects AGO proteins from proteasome degradation.
Figure 5.
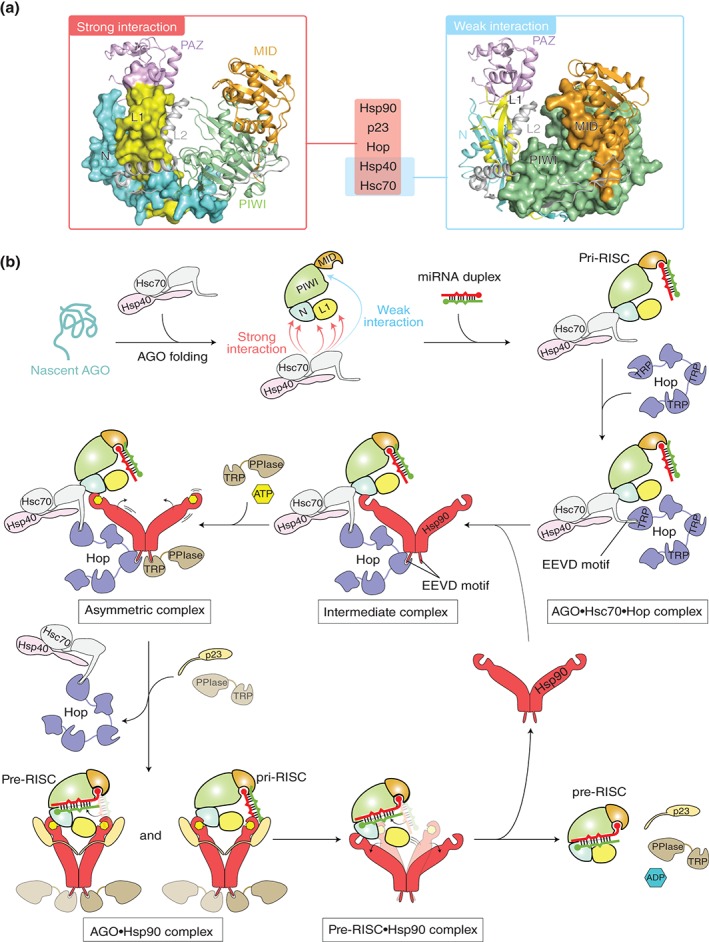
Model of chaperone‐mediated duplex loading. (a) Mapping of the chaperone‐binding sites on the structure of hAGO2 (PDB ID: 4W5N) based on the data of rat AGO256. The regions interacting strongly with Hsp90, p23, Hop, Hsp40, and Hsc70 (left) and weakly with Hsc70 and Hsp40 (right) are shown as a surface model. For clarity, the hAGO2‐bound guide RNA is not shown. (b) Duplex loading by co‐chaperone cycle of Hsp90 in humans. Nascent AGO peptide (aqua) is captured by a complex of Hsc70 (gray) and Hsp40 (pink). The PAZ domain is not shown in order to clarify the interaction between the L1 linker and chaperones. During folding of the AGO protein with the aid of the complex, Hop (slate) uses its TRP motif to recognize at the EEVD tetrapeptide of the Hsc70, which results in an AGO•Hsc70•Hop ternary complex. Hop uses another TRP motif to interact with Hsp90 (red) through the C‐terminal EEVD motif, forming an intermediate complex. Binding of ATP (yellow hexagon) to the N‐terminal domain of the Hsp90 drives the conformational changes, while a Hsp90 co‐chaperone, PPIase, containing a TRP domain (brown) binds to the remaining free EEVD motif of the Hsp90 to form an asymmetric complex. Further binding of PPIase to the C‐terminal EEVD motif of the Hsp90 results in the releases of Hop, which allows Hsp90 to capture the AGO protein. The resultant Hsp90•AGO complex in the closed conformation is stabilized by p23. The widely opened AGO protein accommodates a small RNA duplex. Probably, AGO sorts the 5′ nucleotide of the duplex using the MID domain until the bound duplex is completely accommodated into the channel (see Figure 6). ATP hydrolysis opens the structure of Hsp90 and releases ADP (blue hexagon), PPIase, and p23, along with the pre‐RISC. (Adapted from Ref 13)
Efficient Duplex Loading Requires the Chaperone Machinery
In the absence of ATP, siRNA and miRNA duplexes are loaded onto hAGO2 in cell lysates49 and in in vitro reconstituted systems with immunoprecipitated63 or recombinantly expressed protein.47 However, these loading efficiencies were quite low. Several groups independently reported the significance of chaperone machinery for duplex loading. ATP hydrolysis by Hsc70/Hsp90 is essential for sufficient loading of small RNA duplexes onto fly Ago1 and fly Ago210, 11 as well as onto hAGO2.10 As loading of miRNA duplex onto hAGO2 is promoted in an ATP‐dependent manner,63 the RISC assembly of slicer‐deficient hAGO proteins (hAGO1, hAGO3, and hAGO4) would also need the aid of the chaperone machinery. In Arabidopsis thaliana, ATP binding to Hsp90 promotes loading of siRNA and miRNA duplexes onto AGO1 (AtAGO1)9 and AGO7.64 The ATPase activity of Hsp90 is not, however, required for passenger ejection and target cleavage by fly Ago1 and fly Ago2.10, 11 In addition, fly Ago2 does not need the chaperon machinery to interact with the RLC composed of Dicer 2 and R2D2.10, 11 It is also known that although ATP accelerates multiple turnover target cleavage by fly Ago2‐RISC,65 neither 2‐phenylethynesulfonamide (PES), which blocks the interaction of Hsc70 with its client proteins, nor 17‐allylamino‐17‐demethoxygeldanamycin (17‐AAG), which binds to the ATPase domain of Hsp90, affects turnover.10 On the other hand, hAGO2‐RISC slices the target in multiple rounds in the absence of ATP.66, 67, 68 These results indicate that the chaperone machinery is necessary only for loading of small RNA duplexes and is dispensable for any other steps in the RISC assembly. Therefore, once the passenger strand is discarded, the resultant RISC serves as an autonomous effector complex for RNA silencing.
Analogy of Duplex Loading to the Activation of Steroid Hormone Receptors
A recent study has revealed that fly Ago2 employs Hsc70, Hsp90, Hsp40, Hop, and p23 during loading of siRNA duplexes.12 These are the same proteins co‐purified with rat AGO256 and are also members of the chaperone machinery exploited by steroid hormone receptors when embedding their cognate ligand for activation.14 During loading of small RNA duplexes, Hsc70, Hsp90, and Hop serve as essential core components.12 In contrast, Hsp40 or p23 alone is not able to load siRNA duplexes onto fly Ago2 although either of them promotes the generation of pre‐ and mature‐RISCs in the presence of the three core components,12 suggesting that Hsp40 and p23 facilitate the co‐chaperone cycle of Hsp90 machinery in a manner similar to that of steroid hormone receptor activation (Figure 5(b)). In addition, inhibition of Hsc70 with PES almost completely abolishes the target cleavage activity of fly Ago1, fly Ago2, and hAGO2.10 This result is well explained if the co‐chaperone cycle of Hsp90 is applied to loading of the small RNA duplexes into AGO protein because it begins with the interaction of Hsc70/Hsp40 and the client protein.
Based on these results, a possible chaperone‐dependent loading mechanism is proposed here (Figure 5(b)). Hsc70 and Hsp40, which are general chaperones participating in the folding of nascent peptides,59 bind folded AGO proteins and/or nascent AGO peptides, protecting the susceptible hinge region between the MID and PIWI domains. Using the tetratricopeptide repeat (TPR), Hop recognizes the EEVD motif of Hsc70 and takes the Hsc70/Hsp40/AGO complex to Hsp90. Then, Hop uses another TPR to bind the EEVD motif of Hsp90 in the open conformation where the ATPase activity is inhibited.60 In this intermediate complex (Hsc70/Hsp40/Hsp90/Hop/pri‐RISC), Hsp90 would interact, through its client‐binding amphipathic loop, with hydrophobic patches presumably on the N domain and/or the L1 linker of the AGO protein.56 The remaining EEVD motif of Hsp90 preferentially binds to TRP‐containing large peptidyl‐prolyl cis‐trans isomerases (PPIases) such as FK506‐binding protein 52 (Fkbp52), Fkbp51, and cyclophilin 40 (Cyp40), forming the asymmetric complex.69 Binding of ATP to the center of the N‐terminal domain of Hsp90 triggers the conformational change to the closed state (ATPase‐active form), which weakens the affinity of the EEVD motif to Hop. In addition, p23 binds to the exterior of the N‐terminal domain of Hsp90 to clamp the closed conformation.61 The dissociation of Hop from Hsp90 may be facilitated by binding of another PPIase to the EEVD motif of the Hsp90, thereby promoting the release of Hop. In the transition from the open to closed state, the dimeric Hsp90 drastically moves the two arms, accompanying the bound pri‐RISC. Since nonhydrolyzable ATP analogs do not enhance the formation of pre‐fly Ago1‐RISC efficiently,70 the conformational change upon the ATP binding would not be a main driving force to load duplexes onto the Hsp90‐bound AGO protein. On the other hand, geldanamycin binds to the N‐terminal domain of Hsp90, preventing the interaction with p23, and thus halting the Hsp90 chaperone cycle at the intermediate complex with Hsc70 and Hop.71 After ATP hydrolysis, Hsp90 opens the conformation while releasing p23 and TRP‐containing large PPIase, along with the pre‐RISC. As ATP hydrolysis is essential for sufficient fly Ago1‐RISC loading,70 most of the pri‐RISC loads duplexes mainly by the large conformational transition of Hsp90 from the closed to the open state. Thus, the conformational changes of Hsp90 are regulated by the binding of different co‐chaperones as well as Hsc70, Hsp40, and a client protein.13 Binding of these co‐chaperones to Hsc70/Hsp90 is thought to be a competition with each other against the same motif and is mutually exclusive.72
Co‐immunoprecipitation assays demonstrated that after being washed with 600 mM sodium chloride, Hsc70 and Hsp40 bound to fly Ago1 and fly Ago2, whereas neither Hsp90 nor Hop remained bound.10 This indicates that fly Ago1 or Ago2 form a stable complex with Hsc70 and Hsp40 and constitute a less stable complex with Hsp90 and Hop. These results show that AGO protein always binds to Hsc70 and Hsp40, probably except when it is being captured by Hsp90.
Mochizuki et al. recently reported that in Tetrahymena, a Hsp90 co‐chaperone Coi12p promotes loading of siRNA duplexes onto the Argonaute protein Twi1p73 in an ATP‐dependent manner.74 Coi12p is composed of a TPR and two PPIase domains though it lacks its PPIase activity. Intriguingly, a Coi12p mutant lacking the TRP motif partially restored the loading defect of COI12 knockout cells, suggesting that Coi12p also promotes siRNA loading in an ATP‐independent manner. This is consistent with the fact that most of the large PPIases such as p23, Cyp40, and FKBP52 possess Hsp90‐independent chaperone activity.75, 76 The facilitation of ATP‐dependent siRNA loading requires the TPR domain but not the PPIase domain, which is reminiscent of the typical TPR‐containing long PPIase that competes with Hop for the EEVD motif of Hsp90 and then passes the AGO protein to the Hsp90. However, Hsp70 inhibitor, PES, does not abolish the enhancement of the duplex loading, suggesting that Coi12p promotes the ATP‐dependent siRNA loading in a different manner from that of mammals and flies. In addition, nonhydrolyzable ATP analog enhances the siRNA loading as well, indicating that binding of ATP to Hsp90 rather than its hydrolysis is important, unlike the duplex loading in mammals and flies. These results indicate that for the RISC assembly, different eukaryotes employ the conserved Hsc70/Hsp90 chaperone machinery and co‐chaperones with similar domain structures, yet their mechanisms seem to be quite diverse.
POSSIBLE MECHANISM OF THE GUIDE STRAND SELECTION DURING THE RISC ASSEMBLY
Guide RNA Is the Spine of the RISC
The first crystal structures of apo‐form AGO proteins from thermophilic prokaryotes showed their characteristic crescent shape composed of the N, MID, and PIWI domains with the PAZ on the top52 or two lobes (i.e., the N‐PAZ and MID–PIWI lobes).53 Afterward, several crystal structures that reflect a state of RISC identified the nucleic acid‐binding channel running between the two lobes across all four domains and the L1 and L2 linkers25, 26, 27, 33, 34 (Figure 2(a) and (b)). Thus, the loaded guide strand serves as the spine of the AGO protein. Without the backbone, AGO proteins seem to behave as if they are a soft‐bodied creature literally like Argonauta argo, which is the origin of the name of ‘Argonaute’ based on the appearance of plant Ago mutant leaves. Especially, the MID domain and the PAZ domain are expected to move freely relative to the rest of the molecule mainly comprising the N and PIWI domains. Then, a question comes to light: do apo‐form AGO proteins have any physiological advantage to benefit from being in such an open conformation? This will be discussed here by proposing model mechanisms.
5′ Nucleotide Binding Is a Prerequisite for Loading of the Rest of the Duplex
Tomari and coworkers reported that a Y983E mutant of fly Ago2, whose MID domain binds neither the 5′ monophosphate nor the 5′ base of guide strand,12 failed to load a siRNA duplex even in the presence of a set of Hsc70, Hsp90, Hop, Hsp40, and p23.12 This fact demonstrates that the loading deficiency due to the loss of the 5′ end‐binding activity is not rescued by chaperone activity. Therefore, the loading must be initiated by the recognition of the 5′ end, which is a prerequisite for placing the rest of the duplex into the channel. The crystal structures of the RISC show the bound guide RNA recognized at its sugar‐phosphate backbone by many hydrogen bonds along the nucleic acid‐binding channel.25, 26, 27, 33, 34 This strong interaction indicates that duplexes are unlikely to be unloaded once the stem portion is completely accommodated into the channel. Therefore, the MID domain needs to recognize the 5′ end of the guide strand for the asymmetric loading prior to loading of the whole duplex.
MID Domain Itself Is Capable of Discriminating the 5′ Nucleotide of Duplexes
In mammals, asymmetric selection of guide strand from small RNA duplexes relies on the intrinsic property per se of the AGO proteins.45 The MID domain preferentially recognizes the terminus of the duplex having U or A at the 5′ end and of less thermodynamic stability than the other terminus. The structural and functional study by the Nagar group showed that the hAGO2 MID domain alone preferentially binds UMP and AMP with 10‐fold higher affinity than those of CMP and GMP.77 This result indicates that the MID domain solely is capable of discriminating the 5′ base of small RNA duplexes (Figure 6(a)). The structures of the MID domain in the complex with UMP and AMP identified a nucleotide specificity loop whose main‐chain carbonyl and amino groups are arranged to form hydrogen bonds only with uracil and adenine. This may provide an explanation, from the viewpoint of base specificity, for why AGO‐bound miRNAs and siRNAs show a clear bias for U or A at the 5′ end.78, 79 Nevertheless, the authors succeeded in determining the crystal structures of the MID domain in the CMP‐ or GMP‐bound complex as well.77 Their base moieties were completely disordered whereas the monophosphate and the ribose sugar moieties retained some discernable densities. The observation demonstrates that the pocket possesses strong affinity for the 5′ monophosphate, even in the presence of the repulsion against the cytosine or guanine base moiety. This strong interaction must be reinforced after the MID and PIWI domains form the composite pocket that surrounds the 5′ monophosphate as seen in the structures of RISCs25, 26, 27, 33, 34 (Figure 4(h)).
Figure 6.
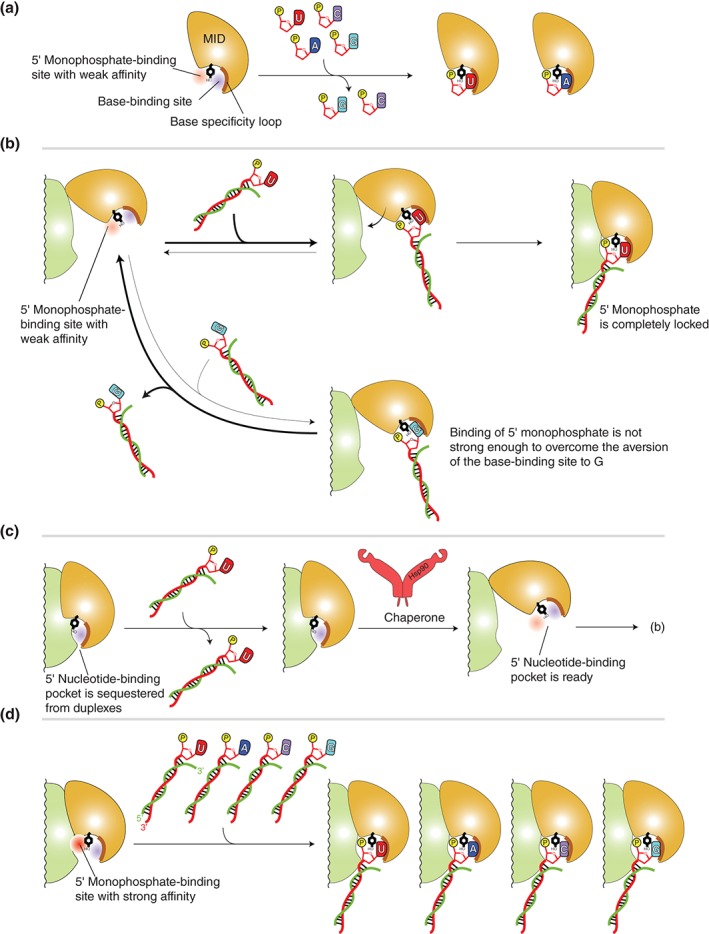
Models of stepwise duplex loading. (a) Interaction between the MID domain alone with nucleoside monophosphates, UMP (red), AMP (blue), CMP (magenta), and GMP (cyan). The MID domain preferentially binds U and A over C and G. The 5′ nucleotide‐binding pocket is divided into the 5′ monophosphate (red)‐ and the base (blue)‐binding sites by a conserved tyrosine residue (black) that is stacked with the 5′ base and its adjacent monophosphate (yellow). The base specificity loop is shown as a thick line (brown). (b) Model of sorting guide strand by the affinity of the MID domain to the 5′ base. Top: The MID domain alone binds to the uracil (or adenine) at the 5′ position of duplexes. The interaction is stable enough to endure until it forms the composite pocket with the PIWI domain. Bottom: The guanine (or cytosine) at the 5′ position of duplexes can bind to the 5′ base‐binding pocket. Owing to the aversion of the nucleotide specificity loop to cytosine and guanine, most of the complexes are dissociated before they form the composite pocket with PIWI domain. Therefore, the duplexes including guanine (or cytosine) at their 5′ position are easily released from the AGO protein. (c) Autoinhibition model. The MID and PIWI domains of unloaded AGO protein interact with each other such that the 5′ nucleotide‐binding pocket is not accessible. Chaperone machinery may pry open the autoinhibited conformation, and the resultant open pocket proceeds to (b). (d) Model of preorganized composite pocket. The MID and PIWI domains of unloaded AGO protein already complete the 5′ monophosphate‐binding site whose affinity is higher than that of the MID domain alone (a). The affinity to the 5′ monophosphate would overwhelm the aversion of the base‐binding site to guanine and cytosine. As a result, the preorganized composite pocket can bind any duplexes, regardless of the types of the 5′ base.
A Model of Two‐Step Sequential Loading
Several reports imply that the duplexes are loaded onto the AGO protein in sequential steps.80, 81 Possible models emerging from the current knowledge will be proposed here. As discussed, recent studies evidenced that the hinged MID domain is opened and exposes the 5′ nucleotide‐binding pocket to solvent, until the AGO protein meets a duplex. Once the MID domain encounters the 5′ monophosphate of any RNAs regardless of shape, they form a transient complex here named ‘primary RISC (pri‐RISC).’ Only if the 5′ base of the bound RNA is U or A, the state of pri‐RISC remains for long enough until the bound 5′ monophosphate is completely locked with the PIWI domain by forming the composite pocket (Figure 6(b), top). Afterward, the pri‐RISC lays the rest of the duplex in the nucleic acid‐binding channel presumably with the help of the chaperone machinery. Alternatively, the complete lock of the 5′ monophosphate may occur concomitantly with the loading of the stem part. Accordingly, only RNA duplexes are allowed to be accommodated into the narrow nucleic acid‐binding channel and form the pre‐RISC. In contrast, when the pri‐RISC is formed with RNAs through G or C at the 5′ end, the interaction does not endure owing to the aversion of the nucleotide specificity loop to guanine and cytosine (Figure 6(b), bottom). Consequently, this sequential mechanism provides AGO proteins with sufficient dwell time to recognize, using the binding site of the MID domain, U or A at the thermodynamically less stable terminus of duplexes. This could explain how AGO proteins select the guide strand when loading duplexes having U or A at one 5′ end and G or C at the other end, such as let‐7, miR‐84, miR‐87, miR‐409, and mir‐424.79, 82, 83, 84, 85
Another possible model is that an interaction between the MID and PIWI domains in the apo form blocks the access of any duplexes to the 5′ nucleotide‐binding pocket (Figure 6(c)). Once such an autoinhibited conformation is pried open, probably by the chaperone machinery, the resultant AGO protein would proceed to the abovementioned two‐step sequential loading mechanism. However, this mode could be denied by the fact that thus far any AGO proteins can be programed with ssRNA bypassing the chaperone pathway to form the RISC. Alternatively, the MID and PIWI domains of guide‐free AGO protein may preorganize the composite pocket similar to that of the RISC. In this case, the affinity of the composite pocket for the 5′ monophosphate would be too strong to discriminate bases. (Figure 6(d)). As a result, AGO may end up with promiscuously loading any duplex regardless of the 5′ base identity. If any, such a composite pocket seen in the RISC seems not to be easily accessible for duplexes25, 26, 27, 33, 34 (Figure 4(h)). Therefore, it is unlikely that the pocket for the 5′ monophosphate is preorganized in the apo form.
UPDATES ON THE PASSENGER EJECTION
siRNAs Can Activate Any hAGO Proteins
After loading of a small RNA duplex into the nucleic acid‐binding channel, the pre‐RISC ejects the passenger strand to form the RISC with the remaining guide strand (Figure 3). There are two types of passenger ejection. The first, ‘slicer‐dependent passenger ejection,’ relies on the RNA cleavage activity of the bound AGO protein.86 Notably, in this pathway, the passenger and guide strands need to have a perfectly complementary stem in order to cleave the passenger strand. The alternative, ‘slicer‐independent passenger ejection,’ heavily depends on the thermodynamic instability of the RNA duplexes.63, 81 A mismatch at the 2–8 steps of the loaded duplex promotes the passenger ejection by slicer‐deficient hAGO proteins, hAGO1, hAGO3, and hAGO4.63 Even perfectly matched duplexes but with low thermodynamic stability due to many G:U pairs in the stem region increased noticeable gene silencing.81 These results indicate that any factors affecting the thermodynamic stability within the duplex, such as mismatches and non‐Watson–Crick base pairings, facilitate the slicer‐independent passenger ejection.
Among four hAGO proteins, only hAGO2 is a catalytically active enzyme capable of cleaving both passenger and target strands.81, 87, 88 Therefore, it has been thought that hAGO2 can load both miRNA and siRNA duplexes, while the slicer‐deficient hAGOs can incorporate only miRNA duplexes to form the RISC. This idea was not, however, consistent with the results of the analyses of the RNAs extracted from immunoprecipitated four hAGO proteins, which showed that all four incorporated both miRNAs and siRNAs,89, 90, 91 proving that even slicer‐deficient AGO proteins can unravel the perfectly matched stem of siRNA duplexes and expel the passenger strand without cleavage. Recently, this discrepancy was explained by the Shin group's in vitro assays which showed that immunoprecipitated hAGO1, hAGO3, and hAGO4 can efficiently separate siRNA duplexes at physiological temperature (37°C) but they fail at 25°C.92 The authors also proposed that even catalytically active hAGO2 ejects the passenger mainly in a slicer‐independent manner.92 The temperature‐dependent passenger ejections of miRNA and siRNA duplexes for each human AGO are summarized in Table 1 and Figure 3.
Table 1.
Passenger Ejection of hAGO Proteins
| Type of Duplex | Passenger Ejection | |||
|---|---|---|---|---|
| hAGO2 | hAGO1, hAGO3, and hAGO4 | |||
| 25°C | 37°C | 25°C | 37°C | |
| siRNA | ✓ | ✓ | ✗ | ✓ |
| miRNA | ✓ | ✓ | ✓ | ✓ |
✓: the passenger is ejected; ✗: the passenger is not ejected.
Models of Two Passenger Ejections
The requirements for each passenger ejection were well characterized, but little is known about their molecular mechanisms due to the lack of structural information of the pre‐RISC. It has been generally believed that the slicer‐independent passenger ejection (the transition from ‘pre‐RISC’ to ‘RISC’ in Figure 3(b)) is the mirror process of target recognition (the transition from ‘RISC’ to ‘target complex’ in Figure 3(b)), regarding the passenger strand as a target of the guide strand.2 This assumption has the consequence that a guide and a passenger in the pre‐RISC are extensively base‐paired and spiral in the channel, with approximately two turns of an A‐form RNA. The AGO protein in such a topology manages to eject the passenger strand in the slicer‐dependent or even the slicer‐independent manner. Possible mechanisms to solve this wire puzzle will be discussed below by taking two scenarios when siRNA duplexes are loaded onto hAGO2 and when miRNA duplexes are incorporated into the slicer‐deficient hAGO proteins, as examples of the slicer‐dependent and the slicer‐independent passenger ejections, respectively.
Slicer‐Dependent Passenger Ejection After Loading of siRNA Duplexes
A cross‐section of the crystal structure of hAGO2‐RISC shows the Y‐shaped nucleic acid‐binding channel (Figure 7(a)). One of the branched ends runs, between the N and L1/L2 domains, toward the PAZ domain (green arrow in Figure 7(a), right). Another one between the N and the PIWI domains is extended from the MID domain (orange arrow in Figure 7(a), right), resulting in the long channel where a guide–passenger duplex is expected to dock.93, 94 The section also shows several spikes in the α7 and α20 projecting from its inner wall of the channel (Figure 7(a), right).
Figure 7.
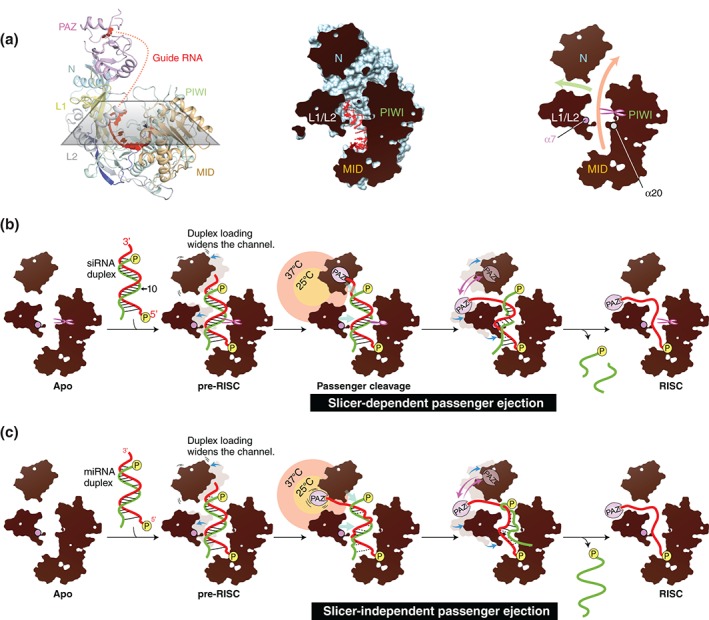
Two model mechanisms of passenger ejection. (a) Y‐shaped nucleic acid‐binding channel of AGO protein. Left: The crystal structure of hAGO2‐RISC (PDB ID: 4OLA) is cut with a section. The color codes of hAGO2 are the same as in Figure 1. Middle: The hAGO2 (light blue) and the bound guide RNA (red) are shown as a surface and a ball‐and‐stick model, respectively. The section area is colored in black. Right: The branched channels. The main channel and the branch are shown by orange and green arrows, respectively. The catalytic site is indicated by scissors. The α7 and α20 are drawn as spheres. (b) Model of the slicer‐dependent passenger ejection. The siRNA duplex composed of a guide (red) and a passenger (green) is loaded onto hAGO2 to form the pre‐RISC. The 5′ monophosphate is shown as a yellow sphere. The N and L1/L2 domains move outward (blue arrow) to expand the width of the nucleic acid‐binding channel. Only the guide RNA is anchored at its 5′ monophosphate and the sugar‐phosphate backbone in the seed region. The loaded siRNA duplex is pushed by the N and L1/L2 domains (cyan arrows) and squeezed out upon the passenger cleavage. The thermal dynamics of the PAZ domain shakes the 3′ end of the guide strand (pink arrows), which facilitates the ejection of the cleaved passenger strand. The territories of the PAZ domain at 25 and 37°C are drawn in a yellow and salmon circles, respectively. The guide 3′ end of siRNA duplex is positioned outside a territory in which the PAZ domain can reach at 25°C. Destabilized base pairs are depicted as dotted line. (c) Model of the slicer‐independent passenger ejection. The guide 3′ end is positioned within a territory that the PAZ domain can reach at 25°C. Because of its thermodynamic instability, the miRNA duplex is heavily distorted by inward pressure.
When a siRNA duplex is loaded onto hAGO2, its 5′ end and sugar‐phosphate backbone of only the guide strand is thoroughly recognized.25, 27 The duplex being loaded pushes the N and L1/L2 domains outward (pre‐RISC in Figure 7(b)). Consequently, the expanded channel generates an inward pressure, which pushes back the loaded duplex from the sides. During passenger ejection, the N domain functions as a wedge to destabilize the base pairings between the 3′ end of the guide and the 5′ end of the passenger.95 On the other hand, the α7 would push the guide–passenger duplex along its minor groove at positions 2–7, as seen in between the α7 and the minor groove of the guide–target duplex bound to hAGO2.32 Therefore, the α7 would serve as another wedge to split the duplex. In this context, hAGO2 cleaves the passenger strand. The nicked duplex no longer withstands the inward pressure between L1/L2 and PIWI domains and is squeezed out. The passenger strand has no specific contacts with the AGO protein, while the thoroughly anchored guide strand remains in the channel to become part of the RISC (Figure 7(b)). As a result, only the passenger strand is ejected from the channel. Thus, hAGO2‐pre‐RISC would separate the siRNA duplex like a nutcracker without energy input.
How do the MID and PAZ domains participate in passenger ejection? In the structures of hAGO2‐RISC and its target complex, the MID domain recognizes the 5′ nucleotide of the guide strand,25, 27, 32 indicating that the MID domain keeps holding the guide 5′ end throughout RISC assembly. Indeed, a mutation of Y529, which is located on the MID domain and essential for recognizing the 5′ nucleotide of guide strand, completely abrogates duplex loading.12, 55 In contrast, a PAZ‐domain truncation mutant of hAGO2 is competent to form the RISC,92, 96 suggesting that the PAZ domain is dispensable for ejecting the passenger strand as long as the AGO protein retains slicer activity. On the other hand, a catalytic mutant of hAGO2 D597A is able to form the RISC at 37°C after loading siRNA duplexes,63, 92 indicating the intrinsic ability of hAGO2 to eject the passenger strand of siRNA duplexes even without cleavage. Intriguingly, the same mutant fails to do so at 25°C.92 In addition, a hAGO2 ΔPAZ mutant fails to form the RISC both at 25 and 37°C when loading a siRNA duplex whose passenger strand is not cleaved due to the existence of a 2′‐O‐methyl modification at position 9.92 These results indicate the significance of the PAZ domain for passenger ejection at high temperature although temperature itself also could affect the stability of the small RNA duplex to some extent. A possible explanation is that when perfectly complementary duplexes are laid along the channel, the 3′ end of their guide strand would be positioned outside of an accessible territory around which the PAZ domain can move at 25°C (yellow circle in Figure 7(b)). In contrast, larger thermal dynamics at 37°C enables the PAZ domain to reach the 3′ end of the guide strand (salmon circle in Figure 7(b)) and vigorously shakes it, thereby dissociating the guide and passenger strands. Capturing of the guide 3′ end by the PAZ domain may be facilitated further at 37°C if the N domain could more efficiently loosen the siRNA duplex.
Passenger Ejection by Slicer‐Deficient AGO Proteins
Slicer‐deficient hAGO1, hAGO3, and hAGO4 are able to eject the passenger strand of siRNA duplexes at 37°C but not at 25°C (Table 1),92 which is consistent with the results of the catalytic mutant of hAGO2 D597A.63, 92 This result suggests that the slicer‐independent passenger ejection basically utilizes the same mechanism as hAGO2, except for the lack of the passenger cleavage (Figure 7(c)). Therefore, the ejection heavily relies on the thermal dynamics of the PAZ domain, as evidenced by the truncation of this domain diminishing passenger release.92, 96 In addition, the slicer‐independent passenger ejection is enhanced if the loaded duplex contains mismatches in the seed region (guide positions 2–8) and/or the 3′ mid region (guide position 12–16).63, 81 The two regions in the duplex seem to contact the spikes protruding from the inner wall of the channel (Figure 7(c)). For example, the α7, one of the spikes, is expected to push the guide–passenger duplex along its minor groove of the seed region as discussed, and would contribute to prying open a bulged stem. Probably, after being loaded into the channel, miRNA duplexes are distorted at the positions of mismatch, which allows the 3′ end of the guide strand to reach the vicinity of the PAZ domain even at 25°C (yellow circle in Figure 7(c)). Thus, the PAZ domain would easily capture the guide 3′ end compared to the case of siRNA duplexes. This model can explain the observation at 25°C that slicer‐deficient AGO proteins fail to form a RISC after loading perfectly complementary duplexes, whereas they can still separate thermodynamically less stable, mismatched stem of miRNA duplexes (Table 1).
DIFFERENCE IN RISC ASSEMBLY BETWEEN EUKARYOTIC AND PROKARYOTIC SYSTEMS
Breaking the Assumption That Eukaryotic and Prokaryotic AGO Proteins Behave the Same Way
In the early studies of the components of the RNAi machinery, structure determination of eukaryotic AGO proteins was hampered by difficulties in sample preparation. Instead, the prokaryotic counterparts from thermophilic species such as Pyrococcus furiosus, Aquifex aeolicus, and T. thermophilus were used for X‐ray crystallography.52, 53, 54, 93, 94 These structures opened the door to the structural basis of AGO protein‐mediated gene silencing. Especially, the series of crystal structures of T. thermophilus AGO (TtAGO) in complexes with a guide,54 a guide–target duplex containing cleavage‐preventing mismatches,93 and an extensively base‐paired guide–target duplex94 provided the first structural insights into the recognition of guide and target strands as well as the target cleavage. Amino acid sequence alignments of eukaryotic and prokaryotic AGO proteins indicated conservation of the N, PAZ, MID, and PIWI domains, and predicted a similarity of their overall structure. Therefore, it seemed reasonable at that time to optimistically assume that understanding of eukaryotic AGO proteins could be reducible to the molecular basis of the prokaryotic counterparts even though they lack the RNAi system and their AGO proteins use DNA as the guide to cleave target DNAs.53, 97, 98
However, structural study of yeast K. polysporus AGO revealed the expansion of the molecular weight of eukaryotic AGO proteins by identifying 11 insertion segments conserved throughout the eukaryotic species and another 8 segments found in only some.26 This structure and others also revealed another difference between eukaryotic and prokaryotic AGO proteins.25, 26, 27 In the eukaryotic RISC, AGO proteins poise a conserved lysine residue over the phosphate backbone of the bound guide RNA at positions 1 and 3 so that its ε‐amino group neutralizes the negative charge.25, 26, 27, 33, 34 In contrast, TtAGO does not have the corresponding lysine residue but instead coordinates a magnesium ion to help the C‐terminal carboxylate group recognize the phosphate backbone of the guide DNA.54 Archaeoglobus fulgidus Piwi protein, which naturally lacks the N and PAZ domains, also positions a magnesium ion close to the same phosphate groups of the bound guide strand.99, 100 These observations prove that eukaryotic and prokaryotic AGO proteins possess different properties though their overall structures look similar, which raises a question of whether they both undergo the same process during the RISC assembly.
Eukaryotic but Not Prokaryotic RISCs Are Ready for Target Cleavage
Even before the structures were available, the amino acid sequence of AGO proteins implied that the PIWI domain adopts an RNase H‐like structure. This fold is found in several enzymes such as reverse transcriptase, integrase, RNase HI, and RNase HII.101 These enzymes recognize DNA–RNA heteroduplexes, and the DNA strand guides the cleavage of the RNA using a conserved catalytic DEDD (Asp‐Glu‐Asp‐Asp) tetrad. The mechanism of RNA cleavage among this group is well investigated in the structure of Bacillus halodurans RNase H1 bound to an RNA–DNA duplex.102 Note that the catalytic glutamate coordinates a magnesium ion as well as directly interacts with the 2′ hydroxyl group of the ribonucleotide adjacent to the scissile phosphate. The corresponding catalytic glutamate was not, however, identified in the first crystal structures of prokaryotic AGO proteins though the structures proved that the PIWI domain displays an RNase H fold.52, 53 Accordingly, it had been thought that AGO proteins are an exceptional family possessing a catalytic DDX (X: Asp or His) triad.101
Meanwhile, the first structural basis of eukaryotic AGO catalytic domain was provided by the crystal structure of guide‐free N. crassa QDE2 (NcQDE2) MID–PIWI domain.51 The catalytic site was shown to be a DDX triad like the prokaryotic counterparts. Afterward, the crystal structure of yeast K. polysporus AGO (KpAGO) was determined, and its comparison to the guide‐free NcQDE2 identified a local conformational change, upon the incorporation of the guide strand, accompanying a rearrangement of a conserved glutamate residue (referred to as a glutamate finger).26 In the KpAGO‐RISC, the glutamate finger moves to the DDX triad and completes the catalytic DEDX tetrad, thereby activating the AGO protein for RNA cleavage. The resultant catalytically active conformation is named the ‘plugged‐in’ conformation because the glutamate finger is inserted into the rest of the catalytic site as when plugging into an outlet turns on a machine (Figure 8(a)). On the other hand, guide‐free NcQDE2 shows the unplugged conformation where the region extended in the plugged‐in conformation of KpAGO is reeled up in the α‐helix (Figure 8(a)).
Figure 8.
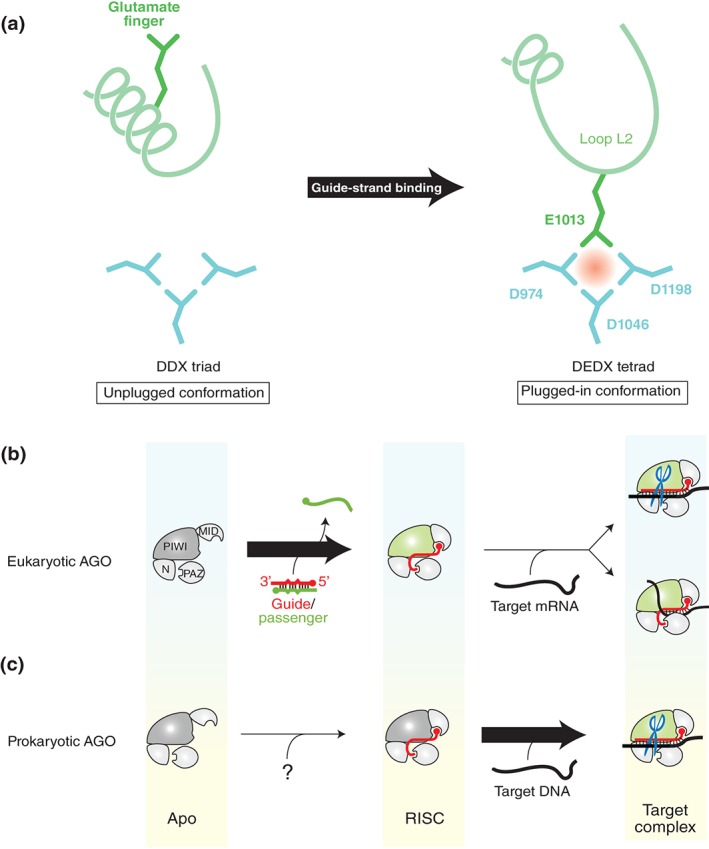
Difference between eukaryotic and prokaryotic AGO proteins. (a) Conformational change activates the AGO protein. In the catalytically inactive state (i.e., unplugged conformation), the region including the glutamate finger (green) folds into an α‐helix (left panel). In the active state (i.e., plugged‐in conformation), the α‐helix is partially unfolded and the glutamate finger is inserted into the DDX triad (aqua) to complete the catalytic DEDX tetrad (right panel). The residue numbers of KpAGO are indicated. (b and c) Requirements for the transition to the plugged‐in conformation. Eukaryotic AGO proteins transition to the plugged‐in state during the RISC assembly (b), whereas the prokaryotic counterparts need to incorporate a target strand to be plugged in (c). The source of guide strand for the prokaryotic AGO proteins remains unknown. The PIWI domain colored in green indicates the plugged‐in conformation.
In addition, the structure of hAGO2‐RISC bound to a complementary 9‐nt target RNA shows that the RISC remains in the plugged‐in conformation even after the seed region is base‐paired with the target strand.32 Therefore, eukaryotic AGO proteins transition, during the RISC assembly, to the plugged‐in conformation that is ready to cleave the target mRNAs without any further conformational change (Figure 8(b)). Given that KpAGO, hAGO2, and NcQDE2 all retain RNA cleavage activity,26, 88, 103 it seems reasonable that only the catalytically active enzymes require the transition to the plugged‐in conformation. However, the crystal structures of hAGO1‐RISC also show the same conformation, despite the lack of cleavage activity.33, 34 This demonstrates that eukaryotic AGO proteins need to have the glutamate finger plugged into the catalytic site during the RISC assembly, regardless of slicer activity. Probably, this transition is an essential step to turn eukaryotic AGO proteins into the effector complex for gene silencing.
The transition to the plugged‐in conformation was rediscovered in TtAGO.104 Like eukaryotic counterparts, TtAGO‐RISC recognizes the 3′ end of guide strand within the PAZ domain. When the TtAGO‐RISC incorporates a target strand, propagation of the guide–target base pairings generates a topological tension toward the 3′ side of the guide strand.94 Once the propagation reaches the guide position 16, the complex structure no longer tolerates the cumulative strain, and the 3′ end of the guide is released from the PAZ domain. This triggers the rearrangement of the glutamate finger. Thus, the transition of prokaryotic AGO proteins to the catalytically active form requires not only the incorporation of target strand but also the zippering between the guide and target. Therefore, prokaryotic RISCs stay in the catalytically inactive conformation (i.e., unplugged), unlike the eukaryotic counterparts (Figure 8(c)).
CONCLUSION
Based on the current knowledge, this review has discussed and proposed several possible models of the RISC assembly: the stepwise loading of small RNA duplexes, the Hsc70/Hsp90‐centered duplex loading, and the passenger ejection by squeezing and thermal dynamics. For a long time, the models of the RISC assembly in eukaryotes have relied heavily on the structural bases of the prokaryotic counterpart.54, 93, 94 As discussed, more recent structural studies revealed several differences in AGO proteins between eukaryotes and prokaryotes.25, 26, 27, 32, 33, 34, 54, 93, 94, 104 The discrepancies warn about the long‐standing consensus that AGO proteins from the two systems undergo the same process during RISC assembly, and also emphasize the significance of the structure determination of eukaryotic system for understanding the mechanism of RNA silencing.
Meanwhile, the appreciation of Hsc70/Hsp90 chaperone and their co‐chaperones during the loading step has been increased.9, 10, 11, 12 The intensive research in flies has revealed that duplex loading into AGO proteins follows the similar Hsp90 chaperone cycle as that of activation of steroid hormone receptors.10, 12 Although the Hsc70/Hsp90 chaperones are broadly used for duplex loading, the participating members of co‐chaperone seem to be diverse in different species.56, 74 The mechanism during the loading step of small RNA duplexes is still almost unexplored, (Box 1) especially in the prokaryotic system. It also remains elusive whether AGO proteins change their affinity to the chaperone machinery depending on the RNA‐free or RNA‐bound form. For example, RISC formation solidifies the structure of the AGO protein with the loaded guide strand as the spine.25 The resultant structure must expose less hydrophobic regions on the surface, which may facilitate the dissociation of some components of the chaperone machinery from the AGO protein. To understand the mechanism of duplex loading, more structural studies of guide‐free AGO protein will be necessary as well.
BOX 1. OUTSTANDING QUESTIONS TO BE ADDRESSED AND POSSIBLE FUNCTIONS TO BE DISCOVERED IN THE FIELD.
What Does the Structure of Unloaded AGO Protein Look Like?
Structural comparison shown in Figure 4 indicates a hinge movement between the MID and PIWI domains in the absence of guide strand. Moreover, disorder of the C‐terminal region is seen in the TtAGO structure whose 5′‐nucleotide‐binding site between the MID and PIWI domains is empty93 (Figure 4(g)). These results indicate a possibility that loading of small RNA duplexes sews each domain of the AGO protein together into the compact structure of RISC. Further study is needed to characterize the structure of apo AGO protein.
Molecular Mechanism of the Temperature Sensitivity of AGO Protein
It has been known that RNAi is a temperature‐dependent biological process. For example, in plants, the level of miRNAs does not change regardless of temperature, while accumulation of siRNAs is temperature dependent.114 Therefore, the levels of virus‐ and transgene‐derived siRNAs are dramatically increased at high temperature. This review has postulated the PAZ domain as the major contributor to promote the RISC assembly at high temperature. The defense system that requires siRNA‐loaded AGO proteins may be activated by the PAZ domain's capability to eject the passenger strand more easily at 37°C than at 25°C. Identification of factors conferring the temperature sensitivity with AGO proteins and elucidation of the molecular mechanisms will be an interesting and significant question to understand any cellular process involving AGO proteins.
Molecular Basis for the Position of Mismatches
The role of mismatch within small RNA duplexes is different depending on its position from the 5′ end of guide strand.115 Given the coevolution of miRNAs and AGO proteins, the significance of the position of mismatches within miRNA duplexes must be explained by the structure of AGO proteins. Indeed, the crystal structures of hAGO2 bound to a 21‐nt guide and 11‐nt targets show the interaction through the minor groove at positions 2–7 but not at 8–9, explaining the tolerance of the protein for mismatches at positions 8–9.32 In the same way, the structure of hAGO bound to a 21‐nt guide–passenger duplex is expected to answer the question of why mismatches at positions 8–11 promote the loading.
Relationship With Chaperones
In humans and flies, duplex loading into AGO proteins heavily relies on the assistance of Hsc70/Hsp90 chaperone machinery and their co‐chaperones.10, 11 In contrast, budding yeast AGO autonomously loads duplexes.26 Comparison of these two groups will reveal the molecular bases of the chaperone‐dependent and chaperone‐independent loading as well as the coevolution of AGO proteins and chaperones. The chaperone dependency also remains an open question in prokaryotic systems.
Do Four hAGO Proteins Function Redundantly?
It is plausible that hAGO1, hAGO3, and hAGO4 are specialized for loading of duplexes containing mismatches at the specific positions because they preferentially incorporate miRNA duplexes over siRNA ones.63, 92 On the other hand, RNA sequencing analyses show that some of the most abundant miRNAs associated with hAGO1, hAGO2, and hAGO3 are different, though the rest of the RNAs are same,89, 116 implying the preference of some miRNA duplexes for being loaded into specific hAGO protein(s). Presumably, AGO proteins recognize not only the shapes of miRNA duplex but also the distribution of base‐pair stability throughout the duplex that is determined by the types of base pair and the positions of mismatches. Comprehensive structural comparison among four hAGO proteins will be able to explain their preference for some specific miRNA duplexes.
In spite of little knowledge about the molecular basis of the RISC assembly, gene knockdown has been a standard technique since the discovery of RNAi to reduce the expression level of a gene of interest. The method is, however, not always controllable due to unknown reasons. mRNA microarray analyses identified very similar sets of transcripts enriched in binding sites for highly expressed endogenous miRNA, suggesting a substantial functional redundancy within four hAGO proteins.88, 89, 105, 106 In contrast, hAGO3 and hAGO4 more efficiently load exogenously expressed small RNAs than hAGO1 and hAGO2,81 though their expression levels are dramatically lower compared with those of hAGO1 and hAGO2.89, 107 A simple explanation is that hAGO3 and hAGO4 preferentially load abundant small RNAs in the cell to maintain the specific pattern of the homeostatic gene expression. On the other hand, hAGO1 and hAGO2 may be susceptible to a subtle change of miRNA members in the cell, thereby taking the helm of the cell development and differentiation stage as well as responding to any intruders carrying foreign RNA. Indeed, hAGO2 loads the circulating RNAs, in human plasma, that originate from bacteria, fungi, and other species.108 Recently, aberrant expression levels of hAGO1 and hAGO3 were observed in the coxsackievirus B3‐induced myocarditis tissue.109 Moreover, the implication of hAGO proteins in the regulation of mRNA splicing110, 111 and of long noncoding RNAs112, 113 implies other aspects of them. Each individual hAGO protein may possess unique roles under specific conditions and/or disease and in different biological processes other than RNA silencing. Given that the loaded guide RNA shapes the character of the AGO protein regardless of the eventual function, a better understanding of how AGO proteins load duplexes would decipher the mechanism of diseases and would provide a solid foundation for new strategies to control the AGO proteins toward next‐generation gene silencing tools and RNAi therapeutics.
ACKNOWLEDGMENTS
The author is grateful to Yukihide Tomari, Shintaro Iwasaki, Hiroshi M. Sasaki, and Edward J. Behrman and the members of the Nakanishi laboratory for critical comments and discussion of the manuscript. I apologize to authors of the relevant primary publications not cited in this review due to space limitation. This work was supported by the PRESTO from the Japan Science and Technology (JST) Agency.
Conflict of interest: The author has declared no conflicts of interest for this article.
REFERENCES
- 1. Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 2013, 14:447–459. [DOI] [PubMed] [Google Scholar]
- 2. Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci 2010, 35:368–376. [DOI] [PubMed] [Google Scholar]
- 3. Jinek M, Doudna JA. A three‐dimensional view of the molecular machinery of RNA interference. Nature 2009, 457:405–412. [DOI] [PubMed] [Google Scholar]
- 4. Jonas S, Izaurralde E. Towards a molecular understanding of microRNA‐mediated gene silencing. Nat Rev Genet 2015, 16:421–433. [DOI] [PubMed] [Google Scholar]
- 5. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011, 12:99–110. [DOI] [PubMed] [Google Scholar]
- 6. Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 2015, 25:651–665. [DOI] [PubMed] [Google Scholar]
- 7. Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science 2009, 326:544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weinberg DE, Nakanishi K, Patel DJ, Bartel DP. The inside‐out mechanism of Dicers from budding yeasts. Cell 2011, 146:262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto‐Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M. In vitro assembly of plant RNA‐induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 2010, 39:282–291. [DOI] [PubMed] [Google Scholar]
- 10. Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP‐dependent RISC loading of small RNA duplexes. Mol Cell 2010, 39:292–299. [DOI] [PubMed] [Google Scholar]
- 11. Miyoshi T, Takeuchi A, Siomi H, Siomi MC. A direct role for Hsp90 in pre‐RISC formation in Drosophila. Nat Struct Mol Biol 2010, 17:1024–1026. [DOI] [PubMed] [Google Scholar]
- 12. Iwasaki S, Sasaki HM, Sakaguchi Y, Suzuki T, Tadakuma H, Tomari Y. Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex. Nature 2015, 521:533–536. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: conformational dynamics and regulation by co‐chaperones. Biochim Biophys Acta 1823, 2012:624–635. [DOI] [PubMed] [Google Scholar]
- 14. Smith DF, Toft DO. Minireview: the intersection of steroid receptors with molecular chaperones: observations and questions. Mol Endocrinol 2008, 22:2229–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okamura K. Diversity of animal small RNA pathways and their biological utility. Wiley Interdiscip Rev RNA 2012, 3:351–368. [DOI] [PubMed] [Google Scholar]
- 16. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004, 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 2004, 14:2162–2167. [DOI] [PubMed] [Google Scholar]
- 18. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432:231–235. [DOI] [PubMed] [Google Scholar]
- 19. Kwon SC, Nguyen TA, Choi YG, Jo MH, Hohng S, Kim VN, Woo JS. Structure of Human DROSHA. Cell 2016, 164:81–90. [DOI] [PubMed] [Google Scholar]
- 20. Yi R, Qin Y, Macara IG, Cullen BR. Exportin‐5 mediates the nuclear export of pre‐microRNAs and short hairpin RNAs. Genes Dev 2003, 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 2004, 11:214–218. [DOI] [PubMed] [Google Scholar]
- 22. Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D, Lai EC. Conserved vertebrate mir‐451 provides a platform for Dicer‐independent, Ago2‐mediated microRNA biogenesis. Proc Natl Acad Sci USA 2010, 107:15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De N, Young L, Lau PW, Meisner NC, Morrissey DV, MacRae IJ. Highly complementary target RNAs promote release of guide RNAs from human Argonaute2. Mol Cell 2013, 50:344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua‐Tor L. The structure of human argonaute‐2 in complex with miR‐20a. Cell 2012, 150:100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature 2012, 486:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science 2012, 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. A dynamic search process underlies microRNA targeting. Cell 2015, 162:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single‐molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell 2015, 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jo MH, Shin S, Jung SR, Kim E, Song JJ, Hohng S. Human Argonaute 2 has diverse reaction pathways on target RNAs. Mol Cell 2015, 59:117–124. [DOI] [PubMed] [Google Scholar]
- 31. Yao C, Sasaki HM, Ueda T, Tomari Y, Tadakuma H. Single‐molecule analysis of the target cleavage reaction by the Drosophila RNAi enzyme complex. Mol Cell 2015, 59:125–132. [DOI] [PubMed] [Google Scholar]
- 32. Schirle NT, Sheu‐Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science 2014, 346:608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua‐Tor L. The making of a slicer: activation of human Argonaute‐1. Cell Rep 2013, 3:1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakanishi K, Ascano M, Gogakos T, Ishibe‐Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote‐specific insertion elements control human ARGONAUTE slicer activity. Cell Rep 2013, 3:1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer‐2‐dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 2004, 117:83–94. [DOI] [PubMed] [Google Scholar]
- 36. Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 2004, 116:831–841. [DOI] [PubMed] [Google Scholar]
- 37. Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 2003, 301:1921–1925. [DOI] [PubMed] [Google Scholar]
- 38. Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science 2004, 306:1377–1380. [DOI] [PubMed] [Google Scholar]
- 39. Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell 2007, 130:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishida KM, Miyoshi K, Ogino A, Miyoshi T, Siomi H, Siomi MC. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell 2013, 49:680–691. [DOI] [PubMed] [Google Scholar]
- 41. Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer‐1. Cell 2007, 130:287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hartig JV, Forstemann K. Loqs‐PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res 2011, 39:3836–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer‐1 and Dicer‐2 in the siRNA/miRNA silencing pathways. Cell 2004, 117:69–81. [DOI] [PubMed] [Google Scholar]
- 44. Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer‐deficient murine embryonic stem cells. Proc Natl Acad Sci USA 2005, 102:12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Suzuki HI, Katsura A, Yasuda T, Ueno T, Mano H, Sugimoto K, Miyazono K. Small‐RNA asymmetry is directly driven by mammalian Argonautes. Nat Struct Mol Biol 2015, 22:512–521. [DOI] [PubMed] [Google Scholar]
- 46. Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA 2012, 18:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC‐loading complex. Proc Natl Acad Sci USA 2008, 105:512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC‐loading complex. Nat Struct Mol Biol 2009, 16:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123:631–640. [DOI] [PubMed] [Google Scholar]
- 50. Schirle NT, Sheu‐Gruttadauria J, Chandradoss SD, Joo C, MacRae IJ. Water‐mediated recognition of t1‐adenosine anchors Argonaute2 to microRNA targets. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID‐PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci USA 2011, 108:10466–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Song JJ, Smith SK, Hannon GJ, Joshua‐Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305:1434–1437. [DOI] [PubMed] [Google Scholar]
- 53. Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site‐specific DNA‐guided endoribonuclease, provides insights into RISC‐mediated mRNA cleavage. Mol Cell 2005, 19:405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide‐strand‐containing argonaute silencing complex. Nature 2008, 456:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, Meister G. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res 2011, 39:2330–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tahbaz N, Carmichael JB, Hobman TC. GERp95 belongs to a family of signal‐transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem 2001, 276:43294–43299. [DOI] [PubMed] [Google Scholar]
- 57. Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 2005, 120:715–727. [DOI] [PubMed] [Google Scholar]
- 58. Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol 2013, 14:630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj‐2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J 1999, 18:1492–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain‐peptide complexes: critical elements in the assembly of the Hsp70‐Hsp90 multichaperone machine. Cell 2000, 101:199–210. [DOI] [PubMed] [Google Scholar]
- 61. Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90‐nucleotide‐p23/Sba1 closed chaperone complex. Nature 2006, 440:1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P‐bodies in human cells. Mol Biol Cell 2010, 21:1462–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y. ATP‐dependent human RISC assembly pathways. Nat Struct Mol Biol 2010, 17:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Endo Y, Iwakawa HO, Tomari Y. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep 2013, 14:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol 2004, 11:599–606. [DOI] [PubMed] [Google Scholar]
- 66. Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua‐Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 2005, 12:340–349. [DOI] [PubMed] [Google Scholar]
- 67. Hutvagner G, Zamore PD. A microRNA in a multiple‐turnover RNAi enzyme complex. Science 2002, 297:2056–2060. [DOI] [PubMed] [Google Scholar]
- 68. Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007, 130:101–112. [DOI] [PubMed] [Google Scholar]
- 69. Pirkl F, Buchner J. Functional analysis of the Hsp90‐associated human peptidyl prolyl cis/trans isomerases FKBP51, FKBP52 and Cyp40. J Mol Biol 2001, 308:795–806. [DOI] [PubMed] [Google Scholar]
- 70. Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer‐independent unwinding. Nat Struct Mol Biol 2009, 16:953–960. [DOI] [PubMed] [Google Scholar]
- 71. Smith DF, Whitesell L, Nair SC, Chen S, Prapapanich V, Rimerman RA. Progesterone receptor structure and function altered by geldanamycin, an hsp90‐binding agent. Mol Cell Biol 1995, 15:6804–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Johnson BD, Schumacher RJ, Ross ED, Toft DO. Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem 1998, 273:3679–3686. [DOI] [PubMed] [Google Scholar]
- 73. Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi‐related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 2002, 110:689–699. [DOI] [PubMed] [Google Scholar]
- 74. Woehrer SL, Aronica L, Suhren JH, Busch CJ, Noto T, Mochizuki K. A Tetrahymena Hsp90 co‐chaperone promotes siRNA loading by ATP‐dependent and ATP‐independent mechanisms. EMBO J 2015, 34:559–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bose S, Weikl T, Bugl H, Buchner J. Chaperone function of Hsp90‐associated proteins. Science 1996, 274:1715–1717. [DOI] [PubMed] [Google Scholar]
- 76. Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp‐40 and the steroid aporeceptor‐associated protein p23. Science 1996, 274:1718–1720. [DOI] [PubMed] [Google Scholar]
- 77. Frank F, Sonenberg N, Nagar B. Structural basis for 5'‐nucleotide base‐specific recognition of guide RNA by human AGO2. Nature 2010, 465:818–822. [DOI] [PubMed] [Google Scholar]
- 78. Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 2008, 320:1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science 2001, 294:858–862. [DOI] [PubMed] [Google Scholar]
- 80. Kobayashi H, Tomari Y. RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta 1859, 2015:71–81. [DOI] [PubMed] [Google Scholar]
- 81. Gu S, Jin L, Zhang F, Huang Y, Grimm D, Rossi JJ, Kay MA. Thermodynamic stability of small hairpin RNAs highly influences the loading process of different mammalian Argonautes. Proc Natl Acad Sci USA 2011, 108:9208–9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21‐nucleotide let‐7 RNA regulates developmental timing in Caenorhabditis elegans . Nature 2000, 403:901–906. [DOI] [PubMed] [Google Scholar]
- 83. Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 2005, 33:2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA‐induced differentiation of HL‐60 cells. Biochem Biophys Res Commun 2004, 322:403–410. [DOI] [PubMed] [Google Scholar]
- 85. Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science 2001, 294:862–864. [DOI] [PubMed] [Google Scholar]
- 86. Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 2006, 7:314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hauptmann J, Dueck A, Harlander S, Pfaff J, Merkl R, Meister G. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat Struct Mol Biol 2013, 20:814–817. [DOI] [PubMed] [Google Scholar]
- 88. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua‐Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 89. Azuma‐Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA 2008, 105:7964–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev 2009, 23:304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 2010, 24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Park JH, Shin C. Slicer‐independent mechanism drives small‐RNA strand separation during human RISC assembly. Nucleic Acids Res 2015, 43:9418–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed‐containing guide DNA and target RNA duplex. Nature 2008, 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 2009, 461:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol 2012, 19:145–151. [DOI] [PubMed] [Google Scholar]
- 96. Gu S, Jin L, Huang Y, Zhang F, Kay MA. Slicing‐independent RISC activation requires the argonaute PAZ domain. Curr Biol 2012, 22:1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol Cell 2013, 51:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, et al. DNA‐guided DNA interference by a prokaryotic Argonaute. Nature 2014, 507:258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'‐end‐specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain‐siRNA guide complex. Nature 2005, 434:663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep 2009, 10:144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal‐dependent catalysis. Cell 2005, 121:1005–1016. [DOI] [PubMed] [Google Scholar]
- 103. Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev 2007, 21:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sheng G, Zhao H, Wang J, Rao Y, Tian W, Swarts DC, van der Oost J, Patel DJ, Wang Y. Structure‐based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand‐mediated DNA target cleavage. Proc Natl Acad Sci USA 2014, 111:652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute‐containing ribonucleoprotein complexes and their bound target mRNAs. RNA 2008, 14:2580–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 2004, 15:185–197. [DOI] [PubMed] [Google Scholar]
- 107. Pfaff J, Hennig J, Herzog F, Aebersold R, Sattler M, Niessing D, Meister G. Structural features of Argonaute‐GW182 protein interactions. Proc Natl Acad Sci USA 2013, 110:E3770–E3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One 2012, 7:e51009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sun S, Ma J, Zhang Q, Wang Q, Zhou L, Bai F, Hu H, Chang P, Yu J, Gao B. Argonaute proteins in cardiac tissue contribute to the heart injury during viral myocarditis. Cardiovasc Pathol 2015, 25:120–126. [DOI] [PubMed] [Google Scholar]
- 110. Ameyar‐Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol 2012, 19:998–1004. [DOI] [PubMed] [Google Scholar]
- 111. Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, Blaustein M, et al. Argonaute‐1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc Natl Acad Sci USA 2014, 111:15622–15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Leucci E, Patella F, Waage J, Holmstrom K, Lindow M, Porse B, Kauppinen S, Lund AH. microRNA‐9 targets the long non‐coding RNA MALAT1 for degradation in the nucleus. Sci Rep 2013, 3:2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Rice AP. Roles of microRNAs and long‐noncoding RNAs in human immunodeficiency virus replication. Wiley Interdiscip Rev RNA 2015, 6:661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Szittya G, Silhavy D, Molnar A, Havelda Z, Lovas A, Lakatos L, Banfalvi Z, Burgyan J. Low temperature inhibits RNA silencing‐mediated defence by the control of siRNA generation. EMBO J 2003, 22:633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Betancur JG, Yoda M, Tomari Y. miRNA‐like duplexes as RNAi triggers with improved specificity. Front Genet 2012, 3:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res 2012, 40:9850–9862. [DOI] [PMC free article] [PubMed] [Google Scholar]


