ABSTRACT
Objectives
North Africa has a complex demographic history of migrations from within Africa, Europe, and the Middle East. However, population genetic studies, especially for autosomal genetic markers, are few relative to other world regions. We examined autosomal markers for eight Tunisian and Libyan populations in order to place them in a global context.
Materials and Methods
Data were collected by TaqMan on 399 autosomal single nucleotide polymorphisms on 331 individuals from Tunisia and Libya. These data were combined with data on the same SNPs previously typed on 2585 individuals from 57 populations from around the world. Where meaningful, close by SNPs were combined into multiallelic haplotypes. Data were evaluated by clustering, principal components, and population tree analyses. For a subset of 102 SNPs, data from the literature on seven additional North African populations were included in analyses.
Results
Average heterozygosity of the North African populations is high relative to our global samples, consistent with a complex demographic history. The Tunisian and Libyan samples form a discrete cluster in the global and regional views and can be separated from sub‐Sahara, Middle East, and Europe. Within Tunisia the Nebeur and Smar are outlier groups. Across North Africa, pervasive East‐West geographical patterns were not found.
Discussion
Known historical migrations and invasions did not displace or homogenize the genetic variation in the region but rather enriched it. Even a small region like Tunisia contains considerable genetic diversity. Future studies across North Africa have the potential to increase our understanding of the historical demographic factors influencing the region. Am J Phys Anthropol 161:62–71, 2016. © 2016 The Authors American Journal of Physical Anthropology Published by Wiley Periodicals, Inc.
Keywords: SNP, haplotype, population genetics, North Africa
Abbreviations
- ALFRED
allele frequency database
- PHYLIP
phylogeny inference package
- PCA
principal component analysis
- UID
unique identifier
Tunisia is a country located in the extreme north of Africa. It is bounded by the Sahara desert to the South, by the Mediterranean Sea on the North and East, and by Algeria to the West. Mitochondrial DNA evidence supports the idea that Tunisia was occupied more than 20,000 YBP by a group originating from sub‐Saharan Africa (Frigi et al., 2010; Soares et al., 2012). By 15,000 YBP the Mechtoïdes, also known as Ibero‐Maurisans, appear (Bedoui, 2002). This group had anatomical similarities with the European Cro‐Magnons who expanded in Iberia during the same period. Before 9,000 years ago, the Sahara went through a wet period (Aumassip et al., 1988) which allowed several mesolithic cultures to flourish. The local population in Tunisia at that time may have coexisted with and mixed with sub‐Saharan migrants (Dutour et al., 1988). Around 8,000 YBP a proto‐Mediterranean community known as Capsian (Camps, 1968, 1975; Camps‐Fabrer, 1989; Hachid, 2000) arrived and spread widely in what is now Tunisia. Many relics of this group are found in Gafsa, a town in southern Tunisia. The Capsians could have undergone admixture with pre‐existing populations or else replaced them. Since 4,000 YBP, Berbers have expanded through all of North Africa. The term Berber refers to a heterogeneous group of indigenous peoples of North Africa who vary ethnically and culturally (Collignon, 1886). In present day Tunisia, two main Berber tribes are distinguished–the Zenata and the Ketama–although in some areas other groups are more commonly found. For instance, the Accaras tribes, originally from the Western Sahara, live in southern Tunisia near Smar.
In the historical period, the Tunisian region saw a great range of invaders, migrants, and colonists that included Phoenicians, Greeks, Romans, Vandals, Byzantines, Arabs, Spanish, Ottomans, Andalousians, and French. Most of these groups left some imprint upon the modern Berbers. However, the most important change in recent centuries arose from the influx of Arabs and Bedouins leading to the major part of the original population being converted to Islam and Arabized. The eleventh century, for instance, was marked by the arrival of about 400,000 Bedouins belonging to Eastern tribes of Beni Hilal and Beni Souleim (Abdel Waheb, 2004). The arrival of Morisco Andalousians, expelled from Iberia in the early 1600s, is considered the second most important demographic event in Tunisia's recent history. More than 80,000 Andalousians settled in the northernmost part of Tunisia (Abdel Waheb, 1917). During the colonial period (about 1881‐1921), about 116,000 Europeans (from Italy, Spain, Malta, Greece, Austria, Russia, and Belgium) were living in Tunisia, about 99,000 of whom were in the capital, Tunis. In the same time frame, Tunisia was a refuge for people from Morocco, Algeria, and Libya (Belhedi, 1992; Ben Hamida, 2002).
The southern part of Tunisia's population is genetically similar to the Libyan population since the two areas contain the same tribes. The Libyan population is primarily of Berber origin; the name is taken from a particular Berber tribe—the “Libou” which means free man. More than 20% of the population speaks an Amazigh language.
Even with such a rich and complex demographic history the human populations living north of the Sahara desert in Africa have received little attention in published population genetics studies. More specifically, numerous studies targeting particular genes, especially those suspected to be of clinical interest, have accumulated in the scientific literature on particular North African populations, but as yet few studies of North African populations incorporating large sets of DNA polymorphisms exist. One of two recent exceptions is the report by Henn et al. (2012) which does sample a large number of autosomal SNPs from seven locations in North Africa with about 18 individuals from each site; however, the study has a limited number of comparative population samples from nearby geographical regions. Their work supports the presence of indigenous genomic variation extending back 12 to 40 thousand years ago with ancient gene flow in different periods from south of the Sahara, Southwest Asia, and Europe. The study of Bekada et al. (2015) studied mtDNA, Y‐chromosome, and autosomal DNA markers on several hundred individuals from four locations in Algeria. They compared their results to previously studied groups (Berber and Arab) in other Algerian communities along with a small number of populations in adjacent regions (Yoruba, Palestinians, French Basque). The authors describe a complex genetic landscape with genetic diversity present that does not readily correlate with geography or linguistics. Their analyses also indicate that gene flow from beyond North Africa has more often been mediated by females (mitochondrial evidence) than by males (Y‐chromosome evidence) while the more indigenous ancient genome patterns are more often carried by males. They caution against generalizing from limited population sampling or from small numbers of polymorphisms whether uniparental or autosomal.
A small sample of Mozabites from Algeria has been part of the Human Genome Diversity Panel (Cann et al., 2002) for some years giving rise to many publications and data available on various public electronic resources including, for example, the ALFRED (allele frequency database), the HGDP/CEPH database (accumulating results from many labs around the world), and the HGDP selection browser. But, while the Mozabite sample results are valuable and interesting, the Mozabites still represent only a small window into a large geographical region. Other recent studies have somewhat broader comparisons with populations elsewhere in the world but they have relatively small sets of polymorphisms; for instance Fadhlaoui‐Zid et al. (2015) report on the diversity of Y‐chromosome markers in Sousse Tunisia and Khodjet‐el‐Khil et al. (2011) present results on Tunisian and Libyan populations for an autosomal set of SNPs (SNPforID 34‐plex panel) selected for ancestry informativeness. Comparisons with a broad sampling of other populations from around the globe are still lacking and a richer sampling of populations across North Africa is desirable. Such studies are needed in order to make it possible to appreciate better how genetic variation in North Africa fits into the rich mosaic of global human genetic diversity. New population studies focusing on various world regions have been appearing in the last decade sampling large numbers of autosomal DNA markers and providing some answers as well as new questions about the origins and development of modern human populations (e.g., review by Pugach and Stoneking, 2015). Practical applications would also benefit in many domains ranging from medical studies searching for disease‐susceptibility genes to forensic work focused on the identification of the victims and perpetrators of natural and human‐made disasters (Phillips et al., 2009). The reported studies suggest that a great deal of the diversity present in the region as a whole is uniquely related to North Africa and distinctly different from other major zones such as Europe, sub‐Saharan Africa, and Southwest Asia.
In this report, we present the results of a population genetics study on the DNA markers from eight population samples (331 individuals) from different regions of Tunisia and Libya in North Africa. Our study examines these eight North African groups for a set of 399 autosomal SNPs also studied in 57 other populations. Some combined analyses included the 7 North African groups and a Spanish Basque group studied by Henn et al. (2012) for the subset of 102 SNPs that are shared in common. The results support the view that this part of North Africa is a genetically unique region of the world worthy of more studies of autosomal genetic markers.
MATERIAL AND METHODS
Populations studied
The 65 populations (a total of 2,914 individuals) in the present study are listed in Supporting Information Table S1A where they are organized by geographical region of the world. The table also includes the three character abbreviations used in various tables and figures as well as the sample unique identifier (UID) employed in ALFRED (Allele Frequency Database; http://alfred.med.yale.edu) where additional information can be found about the populations sampled. Except for the new groups from Tunisia and Libya (331 individuals), these same population samples have been used in many previous studies (e.g. three recent studies: Murdoch et al., 2013; Heffelfinger et al., 2014; Kidd et al., 2014). The names of all seven newly collected populations from Tunisia correspond to the cities and towns in which individuals were sampled. The “Libyans” include individuals from six cities in Libya. The sampling locations of the new populations are shown on the map in Figure 1.
Figure 1.
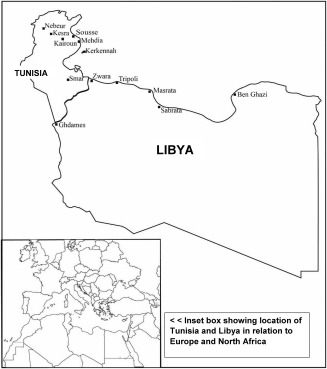
Map showing sampling locations of Tunisian and Libyan populations.
A recent study by Henn et al. (2012) made available genotypes for 145 individuals from seven populations in North Africa (Western Sahara, northern and southern Morocco, Algeria, Tunisia, Libya, and Egypt) along with a sample of Spanish Basque. Supporting Information Table S1B lists the populations and the number of individuals in each group from the Henn et al. (2012) study.
DNA and SNP typing
The DNA of the 57 non‐Tunisian populations was extracted from lymphoblastoid cell lines. The DNA for the Tunisians and Libyans was provided by Dr. Lotfi Cherni and purified from blood. All individuals were typed with TaqMan® assays (Life Technologies, California, USA) in 3 µl reactions in 384‐well plates using the manufacturer's protocol. Following PCR the plates were read by an AB7900 using SDS software. In general, data were very complete for most individuals studied for each of the 399 SNPs (on average 99.2% complete).
The 399 SNPs employed in this study are identified in Supporting Information Table S2 by their dbSNP rs‐number. The SNPs are listed by chromosome and nucleotide position. The distance between adjacent SNPs is shown as the number of basepairs. The table also identifies which SNPs are organized as multi‐SNP haplotypes for the analyses and which SNPs were utilized as single SNPs. A total of 299 genetic markers are in the 65 population dataset; 159 SNPs were organized into 59 multi‐allelic haplotypes while the remaining 240 SNPs were used as simple diallelic markers.
The DNA markers included in this study were those identified across a number of past projects. They are not random markers but had been selected largely because they display substantial genetic variation among populations from most of the world's major geographical regions or in particular regions. Some of the SNPs have been identified previously as being useful in helping to characterize ancestry for major continental regions and/or for distinguishing among populations within particular geographical regions for studies of anthropological or forensic interest. Some of the other SNPs emerged from studies seeking to characterize genes of special interest (e.g. in studies of alcohol or lactose metabolism) or in surveys of linkage disequilibrium on the autosomal genome. No SNPs were chosen because they were thought to distinguish North African groups from other populations.
The analysis of 73 populations including eight populations from the study by Henn et al. (2012), used the 102 SNPs common to that study and the present report. Supporting Information Table S2 identifies these 102 SNPs. In the combined analysis the 102 SNPs were organized as 90 genetic markers consisting of 81 individual diallelic SNPs and 21 SNPs defining nine multi‐allelic haplotypes.
Statistical analyses
Data on all SNPs studied were tested for deviations from Hardy‐Weinberg ratios in all population samples using simple chi‐square tests and/or simulation. SNP frequencies were estimated by gene counting. Haplotypes were inferred by the PHASE software–version 2.1.1 (Stephens et al., 2001; Stephens and Scheet, 2005). F st, a statistic measuring the extent of genetic variation between populations, was calculated from the allele frequencies using the formula of Wright (1969).
Tau genetic distances (Kidd and Cavalli‐Sforza, 1974) were the input for the principal components analyses and for the tree analyses except that the bootstrap analyses used the Reynolds distance (Reynolds et al., 1983), which is virtually identical numerically.
Principal Component Analysis (PCA) of the populations was based on the population‐specific SNP and multi‐SNP haplotype allele frequencies. Tau genetic distances were computed by the DISTANCE program and the resulting pairwise matrix was used as input for the PCOMPNTS program.
The STRUCTURE program (version 2.3.4; Pritchard et al., 2000; Falush et al., 2007) clusters individuals into a pre‐specified number of population clusters. A total of 10,000 burn‐in iterations were followed by 10,000 Markov Chain Monte Carlo iterations employing the admixture model assuming correlated allele frequencies. A total of 20 independent replicates were run at each cluster level (K). The similarity of the runs at each K level was evaluated by the CLUMPP software (Jakobsson and Rosenberg, 2007) as implemented at the online resource CLUMPAK (Kopelman et al., 2015). The DISTRUCT utility at the CLUMPAK server was utilized to align and coordinate the color scheme for estimated cluster membership across the best runs at each K level.
Phylogenetic population trees were generated. As an initial step the neighbor joining method was employed using tau genetic distances as input. We then applied a search algorithm to improve the fit of an additive tree structure to the genetic distances (Kidd and Sgaramella‐Zonta, 1971). A more detailed description of the logic underlying this approach can be found in Kidd et al. (2011). Bootstrap values for each tree were obtained by software programs that are part of the Phylogeny Inference Package (PHYLIP) software (Felsenstein 1989; Felsenstein, 2009; PHYLIP version 3.61), using the gene frequencies for each population and SEQBOOT, which uses the GENDIST, NEIGHBOR, and CONSENSE programs; 1,000 trees were examined.
RESULTS
No significant deviations from Hardy‐Weinberg ratios were found beyond those nominal results expected by chance given the large number of tests. The allele frequencies for each SNP in each of the 65 populations studied have been added to the ALFRED database. Supporting Information Figure S1 in the supplemental material displays an example bar plot of allele frequencies for one of the 59 multi‐SNP haplotypes that form part of the data set analyzed.
The average heterozygosity across the 65 populations studied for the 299 DNA polymorphisms is 0.34. The average heterozygosity of the 59 multi‐SNP haplotypes is 0.51 and the 240 single SNPs is 0.29. Supporting Information Figure S2 compares the average heterozygosity of each population for the 299 and 90 marker datasets. For 62 of the 65 populations we studied, the average heterozygosity is higher for the 299 marker dataset than for the 90 marker subset available in the 73 population analysis (avg. difference 0.015 for 65 groups). In general, the North African populations have a higher average heterozygosity compared with the other major world regions (except for the South Central Asians which are comparable) for both the 299 and 90 marker datasets. The average F st of the SNPs is 0.28 for the 65 population analysis. This value is large and emphasizes the extent to which these allele frequencies vary across the populations since in a more random selection of autosomal SNPs the expected average Fst on populations from the major continental regions of the world would be ∼0.14—where SNPs with very low global heterozygosity and those known to have been under selection are excluded (Kidd et al., 2014).
Figure 2 and Supporting Information Figure S3 show the strong geographical clustering of the 65 populations studied based on the results of the PCA analysis. The first two principal components (Fig. 2) account for 87.4% of the genetic variation provided by the polymorphisms in the dataset. The eight Tunisian and Libyan population samples cluster together between the populations of sub‐Saharan Africa and Southwest Asia in each of the two‐dimensional views summarized by the PCA figures but they are closer to the Southwest Asians and the populations of the southern or Mediterranean part of Europe. The North Africans are closest to that part of the sub‐Saharan cluster containing the Ethiopian Jews, African Americans, and some of the populations of East Africa and farthest from the West and Central African populations in the dataset.
Figure 2.
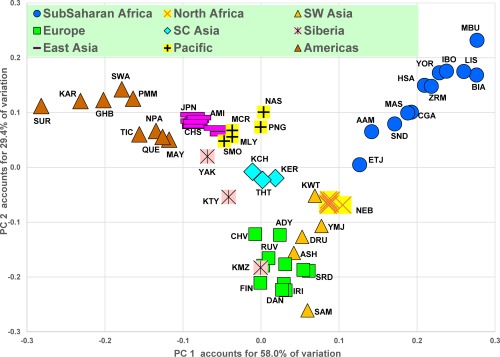
Principal components analysis. Plot of results for first two principal components. PCA is based on pairwise Tau genetic distances for 65 populations.
Supporting Information Figures S7 to S17 display SNP and haplotype allele frequency bar plots across the 65 populations for nine SNPs (Supporting Information Table S3) contributing to functional variation; these include variation for phenotypes related to alcohol metabolism, the flushing response to alcohol, hair form, tooth size and shape, pigmentation (eye, hair, skin), classic PTC‐tasting, and other taste‐receptor genes. Along with Supporting Information Figure S1 these figures illustrate various examples of clinal changes in SNP or haplotype allele frequencies across and sometimes within geographical regions.
In order to show more detail of how the North African populations are related to one another in the PCA space, separate PCA analyses focused on the North African groups (for both the 299 and 90 marker datasets) were plotted and the results can be found in the Supporting Information Figures S18–S23.
In one of the additional focused PCA analyses (Supporting Information Fig. S23), four of the Southwest Asian populations are included as outliers with the 15 North African groups. These additional PCA plots, limited to North African and SW Asian populations, do not indicate any simple East‐West set of relationships for the 15 North African populations among themselves for the markers studied. The Egyptians, Saharawi, Algerians, and South Moroccan groups tend to be consistent outliers with the Tunisian and Libyan groups being more centrally located in the PCA space. Among the Tunisian and Libyan samples the Nebeur (NW Tunisia) and Smar (SE Tunisia) do tend to be outliers. The Tunisian population sample from the Henn et al. (2012) study also tends to be somewhat of an outlier and clusters closest to the Nebeur from NW Tunisia. Henn et al. indicate their Tunisians (sampled in southern Tunisia from Chenini and Douiret) were notably more inbred than the individuals from their other locations across North Africa demonstrating much longer runs of homozygosity in their high density SNP study. This is consistent with that group (Supporting Information Fig. S2) having the lowest average heterozygosity among the 15 North African populations in both the 90 and 299 marker datasets but still higher than the heterozygosities for many other groups (e.g. Europeans; see Supporting Information Fig. S2). The Smar, which were sampled only about 40 kilometers from Chenini/Douiret in southern Tunisia, have higher average heterozygosity and do not cluster very closely with the other Tunisians sampled.
In the STRUCTURE analyses of the 65 population dataset, the North African population samples clearly emerge as a separate geographical grouping at K = 8 clusters and remain so through all the other K values tested. The specified number of clusters ranged from K = 2 through 14. Figure 3 presents the individual bar plots for the runs with the highest likelihood values (out of the 20 runs at each K level) for levels K = 5 through 8 and provides a visual rendering of the strong and persistent regional presence of North Africa. The CLUMPP results indicate that highest likelihood runs represent the predominant (most common) patterns across the 20 different runs at K levels 2 through 6. At K = 7 and higher the runs with the highest likelihood did not usually represent the most common patterns. The number of alternative solutions/patterns observed increases as K increased above 7. Inspection of the alternative patterns revealed population subclusters in many other world regions but not for the 8 populations representing North Africa. Usually, North Africa remained a distinct region among the alternative solutions; when this was not the case, it was always because the North Africans resembled to some extent some populations in Southwest Asia. A bar plot (Fig. 4) showing the average estimates of cluster membership by population highlight the results at K = 8. The genetic markers provide clear evidence of strong, predominant clusters in the sub‐Sahara, North Africa, northern Europe, East Asia, Pacific, and Americas. At K = 8 Southwest Asia is a transitional zone sharing similarity with neighboring regions.
Figure 3.
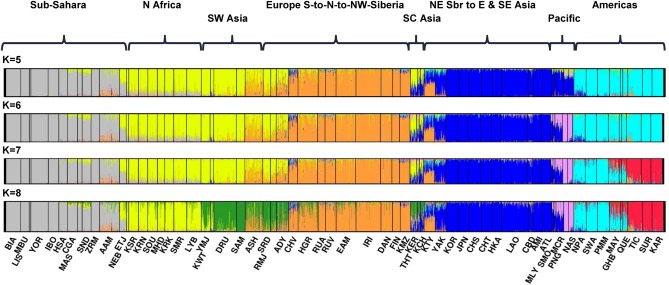
STRUCTURE individual bar plots—65 populations, 299 markers study—displaying results for runs with highest likelihood out of 20 runs in each cluster K = 5 to 8. Each individual has a separate column in the bar plot and the individuals in a population are clustered together in the display but the STRUCTURE analysis was unsupervised, i.e. not informed about an individual's population membership. Black vertical lines identify the population boundaries. The height extent of each color within an individual's color bar corresponds to the estimated membership of the individual in one of the clusters; each cluster is assigned a separate color. The bars with multiple colors can be interpreted as genetic admixture or as relative probabilities of belonging to the different clusters. Since there is a separate color bar for each of the more than 2900 individuals, the interval width covered by a population varies and corresponds to the number of individuals in the population.
Figure 4.
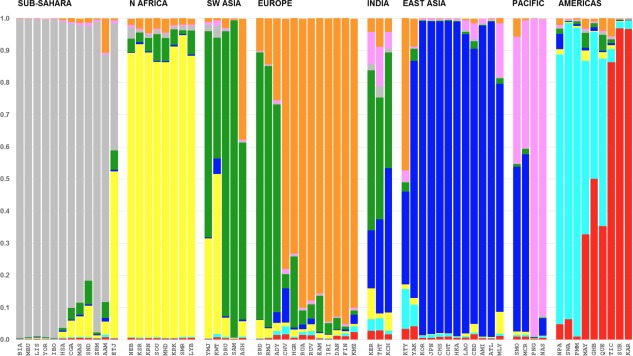
STRUCTURE population bar plot showing average estimated cluster membership in each of 65 populations for the runs with highest likelihood out of 20 runs for cluster K = 8. There are 65 population bars and each population bar has the same width. The height extent of each color within a population bar corresponds to the average estimated cluster membership for all the individuals in the population.
A visual overview of the STRUCTURE results on the combined dataset of 73 populations can be found in Supporting Information Figure S6. The results are similar to the results in Figure 3 but not as clear cut since the 73 population analysis is based on less information–a subset of 102 SNPs compared with the 399 SNPs contributing to the 65 population dataset. The seven North African populations from the Henn et al. (2012) study cluster with the 8 different North African population samples of the present study. (Supporting Information Tables S1A and S1B provide details on the ordering of the populations in the STRUCTURE individual bar plots.) At first glance the fifteen North African population samples appear distinct from the other major regions of the world as in the 65 population analysis; however, the transition from North Africa to Southwest Asia is not as clear cut as it is in Figure 3. The Yemenites and the Kuwaiti resemble the North Africans more than they do the other Southwest Asian populations. Some other geographical regions do not emerge as consistently or distinctly (e.g., North and South America as separate regions; northern Europe is not very distinct from Southwest Asian and southern Europe). However, broad regions like sub‐Saharan Africa, North Africa, East/Southeast Asia, the Pacific Islands, and the Americas as a whole emerge clearly and persist across the K levels displayed. The Spanish Basque population from the Henn et al. (2012) study is placed among the populations of southern Europe and in these STRUCTURE results blends in rather well there.
A more focused STRUCTURE analysis was also carried out for the 299 polymorphism data set by omitting the 25 populations from East Asia, Pacific, and Americas but was otherwise similar to the analysis on 65 populations. This allows us to see if the STRUCTURE program can identify any additional interesting cluster patterns within North Africa and the adjacent regions remaining in the analysis that did not emerge in the previous 65 population analysis because of the distraction provided by the strong clusters in East Asia, Pacific, and Americas. Figure 5 presents the individual bar plots for the most commonly occurring cluster patterns at each K level from 2 to 6. The most common cluster pattern typically includes the highest likelihood run in this more focused STRUCTURE analysis. The cluster pattern observed for sub‐Saharan, North African, and Southwest Asian populations at K = 5 (Fig. 5) in this analysis focused on 40 populations is very similar to what is seen at K = 8 (Fig. 3) for the 65 populations.
Figure 5.
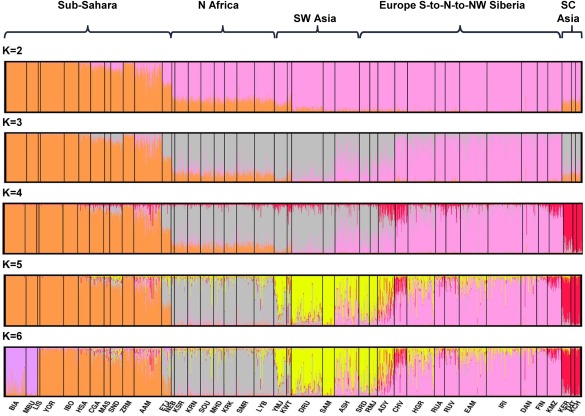
STRUCTURE individual bar plots—40 populations, 299 markers study—displaying results for the most common cluster pattern out of 20 runs in each cluster for K = 2–6. This analysis omits the 25 populations from East Asia, the Pacific, and the Americas. The highest likelihood run in this more focused analysis is usually found among the runs of the most common cluster pattern.
Figure 6 displays the best of the population trees identified in the least squares search for the 65 population dataset. Many of the junctures with the highest bootstrap values (≥90%) are indicated on the image. The pairwise genetic distances between the populations in the least squares method are expected to be related to time in generations since the populations separated divided by twice the effective population size. A key assumption is that allele frequency differences between the populations are due to random genetic drift and little or no gene flow since the populations separated. The specific tree presented is the best fit found after examining a large sampling of the huge number of theoretically possible trees using a heuristic search algorithm but a better tree may exist. The details of Figure 6 are in general similar to the results of the PCA and STRUCTURE but the details are a little more complex. While the North Africans are again between sub‐Saharan, Southwest Asian, and European groupings, six of the eight North African groups cluster on a branch that contains two of the S.W.Asians (Samaritans, Yemenite Jews) and two groups of Mediterranean Europe (Sardinians, Roman Jews). The remaining two North Africans (Nebeur, Smar) are on the long sub‐Saharan branch but very close to the juncture leading to the S.W.Asians and Europeans and very distant from the sub‐Saharans. The colored balloons and text labels in Figure 6 surrounding the major geographical regions highlight the distinctiveness of the major tree branches which correspond to major geographical regions.
Figure 6.
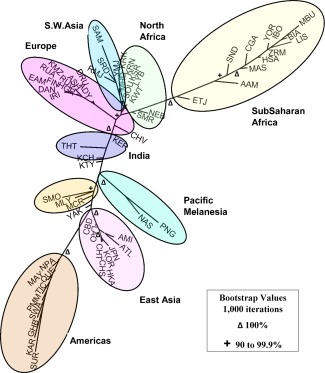
Best least squares population tree for the 65 populations based on the pairwise genetic distances between the populations. Some of the highest bootstrap values (based on 1,000 iterations) are indicated on the image by symbols Δ (100%) and + (90 to 99.9%).
The bootstrap simulations for 1,000 trees for this 65 population dataset show that the eight North African groups separate in 100% of the trees from all the Sub‐Saharan African populations studied and from the African‐Americans. For 91.7% of the trees, the eight North African populations also separated from the Southwest Asian populations along with all of the other populations studied to the North (Europe) and the East (Asia, Pacific, and the Americas). In general, the population samples from Tunisia and Libya clustered together. The Nebeur population, located in northwestern Tunisia, separated from the other North African locales sampled (98.5% of the trees) but is still tightly clustered with them from the perspective of the 65 populations studied as can be seen in the PCA plots (Fig. 1, Supporting Information Fig. S4). There was a weak trend (60% of the trees) for the Smar, the population farthest to the South within Tunisia among the seven Tunisian locales sampled, to separate from the six other North African populations. The reference populations in continental regions like the Americas and East/Southeast Asia also separated from the rest of the world in 100% of the trees generated. Among the Pacific island populations the Nasioi and the Papuans separate consistently in all the trees from the rest of the world while the Micronesians and the Samoans separated in 97.9% of the trees from the Nasioi, Papuans, and the rest of the world (as also can be seen in the best population tree displayed in Fig. 6). The Southwest Asian and the European populations do not separate very strongly from one another for the set of DNA markers studied; there is a more gradual transition (which is also evident in the PCA and STRUCTURE results) with particular groupings showing stronger differences, for instance, northern Europeans from southern Europe and Southwest Asia.
DISCUSSION
The PCA (Fig. 2), STRUCTURE (Fig. 3) and population tree (Fig. 6) results on 65 populations clearly show that the Tunisian and Libyan populations in North Africa represent a distinctive regional pattern of human genetic variation in the context of a broad sampling of human populations worldwide. Similar PCA and STRUCTURE analyses on a subset of 90 polymorphisms that add an additional seven populations from elsewhere in North Africa (Morocco to Egypt) generate comparable results (Supporting Information Figs. S3–S6). The more focused STRUCTURE results on 40 populations (Fig. 5) studied on 299 polymorphisms do not reveal any additional subclustering within the North African area for the genetic variation sampled by the 399 autosomal SNPs in this study. In the context of worldwide variation the Tunisian and Libyan groups appear to be very homogeneous. However, PCA analyses limited to North Africa and also North Africa with some SW Asian groups as outliers (Supporting Information Figs. S18–S21) reveal that even within the relatively small zone represented by Tunisia the local populations are different from one another. Including the seven North African populations from Henn et al. (2012) in similar restricted PCA analyses (based on 90 polymorphisms) also provides evidence of the diversity of the populations within the North African region (Supporting Information Figs. S22, S23), although no simple geographical patterns emerge.
A reasonable ancient settlement scenario for North Africa–based on various studies with different types of markers could be proposed as follows. Berbers arose from ancient events across North Africa with various sub‐regional differences. The autosomal SNPs analyzed here are not able to give a clear picture of what those pre‐historic events were. The Berbers constitute the main genetic background of the North African population as a whole even though in historical times several substantial migrations occurred into the region from Middle Eastern populations and elsewhere. Migrations during the historical period enriched the North African populations rather than replaced them. For many markers analyzed, North African populations display intermediate frequencies between European and African populations, possibly reflecting ancient as well as known historical admixture along with genetic drift. During the thousands of years of development a variety of populations contributed at different levels not only to allelic diversity of North Africa but also to different amounts of linkage disequilibrium between markers that are physically close to one another. Recombination events over many human generations should have generated new haplotypes that should be specific to the North African region; see Supporting Information Figures S14, S15 for some examples. Along with recombination, some specific and founder mutations, also described in the published literature, are in agreement with a common and ancient North African genetic background. All these demographicevents lead to the findings in the present study that North Africa constitutes a distinct population genetic entity.
Population migrations within and between the geographical regions during pre‐history and historical times have certainly made North Africa a cross‐road of change—culturally and genetically. The positioning of the North African groups studied near the core of the tree is consistent with this. The heterozygosity evidence is supportive of a complex history for North African populations. Divergent theories on the peopling of North Africa exist to explain the accumulated evidence from different research areas. There are those who say that a civilization radiated to North Africa 40,000 years ago built by the Aterian, after which North Africa was depopulated (see the introduction to Rando et al., 1998). Indeed, uniparental genetic data support the arrival of sub‐Saharans around 20,000 years ago according to Frigi et al. (2010). Other studies show that the introduction of sub‐Saharan mtDNA lineages in North Africa is older than 30,000 YBP (Soares et al., 2012). The mixture between Iberian and sub‐Saharan Saharan populations was described in papers such as Periera et al. (2010), but more studies are needed to substantiate that mixture.
The set of 399 autosomal SNPs studied here on 65 populations were not specifically selected to differentiate the Tunisian and Libyan populations representing North Africa from populations in other regions of the world. The DNA markers genotyped on the eight populations from North Africa had already been typed on the 57 reference populations. They are a subset of markers that had accumulated across a number of research projects and were originally selected for study most often because they had been shown to be highly heterozygous on average in most regions of the world or sometimes in a particular geographical region. The STRUCTURE analyses (Supporting Information Fig. S6) on 73 populations (with 15 populations sampled across North Africa) reinforce the idea that North Africa is not only genetically diverse but also a distinctive world region for human variation. More systematic studies—better, denser sampling of populations across North Africa as well as larger DNA marker sets—will likely be interesting and they will also make it easier to identify subsets of markers that differentiate North Africa from other world regions as well as markers that may show distinctive patterns within North Africa. Small, efficient sets of SNPs and other classes of DNA markers emerging from such efforts would be of benefit in a variety of areas such as anthropological research of normal human variation and ancestry, medical studies searching for common disease‐related mutations, and forensic applications identifying human remains in natural and man‐made disasters such as airline crashes, battle field casualties, and the victims and perpetrators of terrorist bombing events. Such work can also help fine tune studies that throw more light on the evolution of modern human populations. Recent studies (Henn et al., 2012; Bekada et al., 2015) have already supported the role of migrations during pre‐history and more recent eras affecting the development of human populations in North Africa. Botigué et al. (2013) explored the evidence for gene flow from North Africa and its possible effect on genetic variation in southern Europe. If, as some recent papers have suggested (Osborne et al., 2008; Balter, 2011), early human waves of migration out of Africa could have originated in part from North Africa before the formation of the Sahara desert, then the only way to help validate this is via empirical evidence that characterizes the autochthonous genome patterns still discernible in current day populations from across North Africa so that they can be compared to the descendant groups in other world regions. Broad‐based evidence is needed. Simple characterizations of groups and regions based on limited initial information is understandable, but will need to be updated and new perspectives obtained by additional analyses. For example, Sanchez‐Quinto et al. (2012) characterize the Tunisian population as homogeneous and inbred based on a small relatively inbred Tunisian sample reported by Henn et al. (2012); but that broad characterization is now untenable in the context of our more extensive sampling of populations in Tunisia which shows that the Henn et al. Tunisian sample is something of an outlier for Tunisia as a whole.
North Africa is certainly not the only world region deserving more intensive study by population genetics. The present study and other recent studies by Henn et al. (2012) and Fadhlaoui‐Zid et al. (2015) indicate that the effort has the potential to yield important new understanding of the genetic history of North Africa and adjacent regions.
ELECTRONIC RESOURCES CITED
ALFRED: http://alfred.med.yale.edu
HGDP‐CEPH Database: http://www.cephb.fr/en/index.php
HGDP selection browser: http://hgdp.uchicago.edu/cgi-bin/gbrowse/HGDP/
CLUMPAK (Cluster Markov Packager Across K): http://clumpak.tau.ac.il/
Supporting information
Supporting Information
ACKNOWLEDGMENTS
Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Department of Justice. The authors want to thank all the collaborators who helped to collect the reference population samples. Sources for the populations previously studied can be found in earlier publications (e.g., Murdoch et al., 2013; Heffelfinger et al., 2014). Special thanks go to the thousands of individuals around the world who volunteered to give blood and saliva samples for studies of gene frequency variation. Some of the software calculations for this project were carried out on the Louise cpu cluster at the Yale University Biomedical High Performance Computing Center.
Grant sponsor: National Institute of Justice, U.S. Department of Justice; Grant numbers: 2010‐DN‐BX‐K225 and 2013‐DN‐BX‐K023; Grant sponsor: Tunisian Ministry of Higher Education and Scientific Research; Grant sponsor: University of Monastir; Grant sponsor: University of Tunis El Manar; Grant sponsor: NIH; Grant numbers: RR19895 and RR029676‐01.
LITERATURE CITED
- Abdel Waheb HH. 1917. Coup d'œil général sur les apports ethniques étrangers en Tunisie. Revue Tunisienne 24:305–316. [Google Scholar]
- Abdel Waheb HH. 2004. Resumé de l'histoire de la tunisie. Maison d'édition Eljanoub [Google Scholar]
- Aumassip G, Ferhat N, Heddouche A, Vernet R. 1988. Le milieu saharien aux temps préhistoriques In Aumassip G, Ferhat N, Heddouche A, Vernet R, Thinon M, Dutour O, Onrubia‐Pintado J, Grebenart D, Ould Khattar M, Tauveron M, Striedter KH, Dupuy C, Amblard S, Quechon G, Gaussen J, Bedeaux R, Bathily M, Mori F, editors. Milieux, hommes et techniques du Sahara préhistorique: Problèmes actuels. Paris: Editions L'Harmattan, pp 9–29. [Google Scholar]
- Balter M. 2011. Was North Africa the launch pad for Modern Human migrations? Science 331:20–23. [DOI] [PubMed] [Google Scholar]
- Bedoui C. 2002. La cuvette de meknassy: recherches sur les formes et les dépots quaternaires. Thèse de doctorat; Université de Tunis I: 230 pages. [Google Scholar]
- Bekada A, Arauna LR, Deba T, Calafell F, Benhamamouch S, Comas D. 2015. Genetic heterogeneity in Algerian human populations. PloS One 10:e0138453. doi:10.1371/journal.pone.0138453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhedi A. 1992. Société espace et développement en Tunisie, Publication de la faculté des sciences humaines et sociales de Tunis; p. 26. [Google Scholar]
- Ben Hamida A. 2002. Cosmopolitisme et colonialisme. Le cas de Tunis, Cahiers de l'Urmis N°8/En ligne http://urmis.revues.org/index4.html
- Botigué LR, Henn BM, Gravel S, Maples BK, Gignoux CR, Corona E, Atzmon G, Burns E, Ostrer H, Flores C, Bertranpetit J, Comas D, Bustamante CD. 2013. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci USA 110:11791–11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps G. 1968. Tableau chronologique de la Préhistoire récente du Nord de l'Afrique. Bulletin de la Société préhistorique Française 65:609–622. [Google Scholar]
- Camps G. 1975. Les civilisations préhistoriques de l'Afrique du Nord et du Sahara. Revue de l'occident musulman et de la Méditerranée 20:179–181. [Google Scholar]
- Camps‐Fabrer H. 1989. Capsien et Natoufien au Proche‐Orient. Colloque « L'homme maghrébin et son environnement depuis 100000 ans'» Maghnia (Algérie) et Trav. du LAPMO:71‐104.
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne‐Tamir B, Cambon‐Thomsen A, Chen Z, Chu J, Carcassi C, Contu L, Du R, Excoffier L, Ferrara GB, Friedlaender JS, Groot H, Gurwitz D, Jenkins T, Herrera RJ, Huang X, Kidd J, Kidd KK, Langaney A, Lin AA, Mehdi SQ, Parham P, Piazza A, Pistillo MP, Qian Y, Shu Q, Xu J, Zhu S, Weber JL, Greely HT, Feldman MW, Thomas G, Dausset J, Cavalli‐Sforza LL. 2002. A human genome diversity cell line panel. Science 296:261–262. [DOI] [PubMed] [Google Scholar]
- Collignon R. 1886. Ethnologie de la Tunisie. Bulletins de la Société d'anthropologie de Paris 9:620–622. [Google Scholar]
- Dutour O, Vernet R, Aumassip G. 1988. Le peuplement prehistorique du Sahara. In Aumassip G, Ferhat N, Heddouche A, Vernet R, Thinon M, Dutour O, Onrubia‐Pintado J, Grebenart D, Ould Khattar M, Tauveron M, Striedter KH, Dupuy C, Amblard S, Quechon G, Gaussen J, Bedeaux R, Bathily M, Mori F, editors. Milieux, hommes et techniques du Sahara prehistorique Problemes actuels. Paris: Editions L'Harmattan, pp 39–52. [Google Scholar]
- Fadhlaoui‐Zid K, Garcia‐Bertrand R, Alfonso‐Sánchez MA, Zemni R, Benammar‐Elgaaied A, Herrera RJ. 2015. Sousse: extreme genetic heterogeneity in North Africa. J Hum Genetics 60:41–49. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2007. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP‐Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166. [Google Scholar]
- Felsenstein J. 2009. PHYLIP (Phylogeny Inference Package) version 3.7a. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle. [Google Scholar]
- Frigi S, Cherni L, Fadhlaoui‐Zid K, Benammar‐Elgaaied A. 2010. Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations. Hum Biol 82:367–384. [DOI] [PubMed] [Google Scholar]
- Hachid M. 2000. Les premiers Berbères: Entre Méditerranée, Tassili et Nil. Edusud.
- Heffelfinger C, Pakstis AJ, Speed WC, Clark AP, Haigh E, Fang R, Furtado MR, Kidd KK, Snyder MP. 2014. Positive selection and haplotype structure at TLR1. Europ J Hum Genetics 22:551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Gravel S, Wang W, Brisbin A, Byrnes JK, Fadhlaoui‐Zid K, Zalloua PA, Moreno‐Estrada A, Bertranpetit J, Bustamante CD, Comas D. 2012. Genomic ancestry of North Africans supports back‐to‐Africa migrations. PloS Genetics 8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing withlabel switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Cavalli‐Sforza LL. 1974. The role of genetic drift in the differentiation of Icelandic and Norwegian cattle. Evolution 28:381–395. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Sgaramella‐Zonta LA. 1971. Phylogenetic Analysis: concepts and methods. Am J Hum Genetics 23:235–252. [PMC free article] [PubMed] [Google Scholar]
- Kidd KK, Speed WC, Pakstis AJ, Furtado MR, Fang R, Madbouly A, Maiers M, Middha M, Friedlaender FR, Kidd JR. 2014. Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci Intl Genetics 10:23–32. [DOI] [PubMed] [Google Scholar]
- Kidd JR, Friedlaender F, Pakstis AJ, Furtado M, Fang R, Wang X, Nievergelt CM, Kidd KK. 2011. SNPs and haplotypes in Native American populations. Am J Phys Anthropol 146:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjet‐el‐Khil H, Fadhlaoui‐Zid K, Cherni L, Phillips C, Fondevila M, Carracedo A, Ben Ammar‐Elgaaied A. 2011. Genetic analysis of the SNPforID 34‐plex ancestry informative SNP panel in Tunisian and Libyan populations. Forensic Sci Intl Genetics 5:e45–47. doi:10.1016/j.fsigen.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 115:1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JD, Speed WC, Pakstis AJ, Heffelfinger CE, Kidd KK. 2013. Worldwide population variation and haplotype analysis at the serotonin transporter gene SLC6A4 and implications for association studies. Biol Psychiatry 74:879–889. [DOI] [PubMed] [Google Scholar]
- Osborne AH, Vance D, Rohling E, Barton N, Rogerson M, Fello N. 2008. A humid corridor across the Sahara for the migration “Out of Africa” of early modern humans 120,000 years ago. Proc Natl Acad Sci USA 105: 16444–16447. [DOI] [PMC free article] [PubMed]
- Phillips C, Prieto L, Fondevila M, Salas A, Gomez‐Tato, A , Alvarez‐Dios J, Alonso A, Blanco‐Verea A, Brion M, Montesino M, Carracedo A, Lareu MV. 2009. Ancestry analysis in the 11‐M Madrid bomb attack investigation. PloS One 4:e6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Cerny V, Cerezo M, Silva NM, Hajek M, Vasikova A, Kujanova M, Brdicka R, Salas A. 2010. Linking the sub‐Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel. Eur J Hum Genetics 18:915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach I, Stoneking M. 2015. Genome‐wide insights into the genetic history of human populations. Invest Genet 6:6. doi:10.1186/s13323-015-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. 1983. Estimation of the co‐ancestry coefficient: basis for a short‐term genetic distance. Genetics 105:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando JC, Pinto F, Gonzalez AM, Hernandez M, Larruga JM, Cabrera VM, Bandelt H‐J. 1998. Mitochondrial DNA analysis of Northwest African populations reveals genetic exchanges with European, Near‐Eastern, and sub‐Saharan populations. Ann Hum Genet 62:531–550. [DOI] [PubMed] [Google Scholar]
- Sanchez‐Quinto F, Botigue LR, Civit S, Arenas C, Avila‐Arcos MC, Bustamante CD, Comas D, Lalueza‐Fox C. 2012. North African populations carry the signature of admixture with Neandertals. PloS One 7:e47765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Alshamali F, Pereira JB, Fernandes V, Silva NM, Afonso C, Costa MD, Musilova E, Macaulay V, Richards MB, Cerny V, Pereira L. 2012. The expansion of mtDNA Haplogroup L3 within and out of Africa. Mol Biol Evol 29:915–927. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing‐data imputation. Am J Hum Genetics 76:449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1969. Evolution and the genetics of populations Volume 2: The Theory of Gene Frequencies. Chicago: University of Chicago Press, p. 511. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


