Abstract
Extracellular vesicles (EVs), including exosomes and microvesicles, have emerged as promising drug delivery vehicles for small RNAs (siRNA and miRNA) due to their natural role in intercellular RNA transport. However, the application of EVs for therapeutic RNA delivery may be limited by loading approaches that can induce cargo aggregation or degradation. Here, we report the use of sonication as a means to actively load functional small RNAs into EVs. Conditions under which EVs could be loaded with small RNAs with minimal detectable aggregation were identified, and EVs loaded with therapeutic siRNA via sonication were observed to be taken up by recipient cells and capable of target mRNA knockdown leading to reduced protein expression. This system was ultimately applied to reduce expression of HER2, an oncogenic receptor tyrosine kinase that critically mediates breast cancer development and progression, and could be extended to other therapeutic targets. These results define important parameters informing the application of sonication as a small RNA loading method for EVs and demonstrate the potential utility of this approach for versatile cancer therapy.
KEY TERMS: Exosomes, siRNA delivery, microRNA, HER2, cancer nanoparticle, nanobiotechnology, biotherapeutic, biopharmaceutical
INTRODUCTION
Small RNAs such as small interfering RNA (siRNA) and microRNA (miRNA) have promise as anticancer therapeutics due to their ability to post-transcriptionally control oncogene expression levels 5, 14, 23. However, the inability of naked small RNAs to cross cell membranes and avoid degradation by extracellular nucleases as well as rapid filtration from the bloodstream requires protective delivery approaches for prospective small RNA therapeutics 6, 15, 27, 32, 35. Amongst many exciting developments in the field, lipid nanoparticle-based delivery of siRNA for RNA interference (RNAi) therapy has been successfully demonstrated in clinical trials 8, and additional approaches are currently under investigation 35. Yet, RNAi approaches are presently predominantly limited to the liver owing to the biodistribution of their delivery vehicles 35. Thus, new delivery approaches that offer the possibility of controllable biodistribution are needed to enable the full potential small RNA therapeutics.
One such approach involves the use of extracellular vesicles (EVs), which have emerged as promising potential delivery vehicles for therapeutic RNAs due to their status as natural carriers of small RNAs 31. EVs, including exosomes, microvesicles and other subtypes, are cell-derived nanovesicles that are produced by an abundance of different cell types 2, 16, 22. EV biodistribution can be controlled via peptide conjugation strategies 1, 9, 34 as well as through producer cell source selection 28, 34, and EVs have previously been loaded exogenously with various nucleic acids for genetic therapy approaches 1, 9, 11, 21, 25, 26. However, loading of functional small RNAs into EVs remains a limiting factor. Electroporation has been employed successfully as a loading method for small RNAs in some studies 1, 9 but not in others 26, and this technique can induce siRNA aggregation 19, which can limit siRNA functional efficacy. Therefore, alternative approaches for loading EVs with small RNAs for therapeutic applications are needed.
In this study, we explored the potential of sonication as a method for loading small RNAs in EVs. Sonication has previously been employed to generate nanoparticles for siRNA delivery 12, 20 and to load EVs with small molecules 18 and protein cargo 17, yet its utility as a small RNA loading technique for EVs is unknown. Parameters of sonication and its effects on EV and nucleic acid integrity as well as siRNA aggregation were assessed. Functionality and potency of EVs loaded with siRNA by sonication were also evaluated with regard to knockdown of the oncogene HER2, a therapeutic target that is overexpressed in numerous cancers leading to enhanced tumorigenesis and malignancy 3, 4, 24.
MATERIALS AND METHODS
EV Isolation and Characterization
EVs were isolated from two different cell lines, HEK293T and MCF-7, using differential centrifugation as described previously 21. Briefly, media that was previously depleted of EVs by ultracentrifugation was collected from cells grown in T150 flasks at ~90% confluency and centrifuged at 300 × g for 10min. Supernatant was then centrifuged at 2000 × g for 20min, 10,000 × g for 30min, and 2h at 100,000 × g to pellet EVs using Optiseal tubes (Beckman Coulter) and a T70i ultracentrifuge rotor (Beckman Coulter). All centrifugation steps were carried out at 4ºC. Pelleted EVs were resuspended in 1X PBS and subsequently washed with PBS buffer using 300kDa MWCO filters and protein content was measured by BCA assay. EV size and concentration were assessed by nanoparticle tracking analysis (NTA) using a NanoSight LM10. Previously isolated EVs were diluted twenty-fold in 400μl of PBS before loading into the NanoSight analysis chamber and data were collected from three different fields of view per sample (1min each; at least 100 particle tracks) and analyzed by NTA2.1 software. All experiments were independently replicated two additional times (n=3).
EV identity was also assessed via immunoblotting using standard approaches. RIPA buffer was used for EV lysis and a 1:1 ratio of PBS to Odyssey Blocking Buffer reagent was employed as the blocking buffer. Antibodies were obtained from Cell Signaling Technologies unless indicated; primary antibodies anti-Alix (2171) and anti-TSG101 (sc-7964; Santa Cruz Biotechnology) were used at a 1:500 dilution; anti-GAPDH (2118) was used at a 1:2000 dilution;. The secondary antibody was goat anti-rabbit IRDye 800CW (956579-01-4; LI-COR), used at a dilution of 1:10,000. Bands were detected with a LI-COR Odyssey CLX Imager and the data were quantified using its associated software.
Sonication Parameter Optimization and EV Loading
Loading of EVs with siRNA, miRNA or single stranded DNA (ssDNA) oligonucleotides was carried out in identical fashion. Parameter optimization experiments utilized the following molecules:
siRNA (sequence: 5′-GGUGCCAGUUCUCCAAGAUUdTdT-3′ (Dharmacon GE Life Sciences CTM-120916))
miRNA (sequence: 5′-CAA AGU GCU GUU CGU GCA GGU AG-3′ (Dharmacon GE Life Sciences CTM-197815))
ssDNA (sequence: 5′-TGCTAGCTATCTAGTAGCCTAGTTA-3′ (Integrated DNA Technologies))
For each experiment, 1000pmol of nucleic acids were incubated with 100μg of EVs in 100μl total volume a 1.5ml tube at room temperature for 30min followed by sonication in a water bath sonicator, (VWR® symphony™; cat# 97043-964, 2.8 L capacity, dimensions 24L × 14W × 10D cm) at 35kHz for 30s unless otherwise indicated. Tubes were then placed on ice for 1min and sonicated again for the same time period as in the first sonication step. Nucleic acids were pre-labeled for detection by mixing 1000pmol of nucleic acids with 10μl dye reagent at room temperature for 5min according to manufacturer’s instructions for Quant-iT™ PicoGreen Assay Kits (ThermoFisher Scientific, cat# P11496) or Quant-iT™ microRNA Assay Kits (ThermoFisher Scientific, cat# Q32882). The described ratio of nucleic acids to EVs was chosen based on preliminary experiments that established detection limits for labeled nucleic acids.
Nucleic acid loading was quantified using Quant-iT™ kits following extensive washing using a 300kDa MWCO filter to remove excess unincorporated nucleic acids as previously described 21. Experimental controls included sonicated siRNA only (no EVs) and siRNA mixed with EVs without sonication. Measured amounts of labeled nucleic acids were used to create a standard curve in order to quantify EV-associated cargo. In the case of DNA oligo encapsulation, samples were digested with DNase I prior to extensive washing and inactivation as described in our previous work 21. However, this step was unable to be carried out for RNA encapsulation due to inability to successfully inactivate RNase A, potentially due to a high binding affinity of this enzyme to EV lipids.
Electroporation
Electroporation of EVs was carried out as previously described 21. Briefly, EVs were mixed with nucleic acids in electroporation buffer (1.15mM potassium phosphate, pH=7.2, 25mM potassium chloride, 21% Optiprep) and electroporation was carried out at 400V and 125μF with two pulses using Gene Pulser/Micropulser Cuvettes (Bio-Rad #165-2089) in a GenePulser Xcell electroporator (Bio-Rad). All samples were then filtered through Nanosep centrifugal devices with Omega membranes (300kDa MWCO; Pall #OD300C33) to remove free cargo and buffer components, transferred into 0.5ml tubes, and 1mM EDTA was added to alleviate nucleic acid aggregation based on the results of Kooijmans et al 19. Samples were then incubated at room temperature for 15min and transferred to Nanosep tubes for centrifugation at 5000 × g at 4°C for 5min to remove buffer and unincorporated cargo.
Nucleic Acid Aggregation
To assess aggregation of nucleic acid cargo induced by the loading process, equal amounts of PicoGreen-labeled ssDNA (5′-TGCTAGCTATCTAGTAGCCTAGTTA-3′) or siRNA (5′-GGUGCCAGUUCUCCAAGAUUdTdT-3′) were subjected to sonication or electroporation under the conditions described above. Samples were transferred into 300kDa MWCO filters and washed with 1X PBS three times to remove any soluble, unincorporated nucleic acids from the filter, leaving aggregates of >300kDa within the filter. This remnant was then resuspended in 1X PBS and quantified using a SpectraMax Gemini plate reader with excitation wavelength = 480nm and emission wavelength = 520nm.
EV Uptake Analysis by Flow Cytometry
To assess EV uptake, HEK293T-derived EVs were labeled with the green fluorescent membrane dye PKH67 (Sigma-Aldrich). Briefly, EVs suspended in PBS were filtered using Nanosep 300K spin columns (Pall) at 8000 × g and resuspended in diluent C solution (Sigma-Aldrich). PKH67, diluted to 4μM in diluent C, and EVs (800μg/mL) were combined at 1:1 ratio for 5min with frequent mixing, followed by the addition of EV-depleted FBS at a 1:2 ratio of FBS to PKH67-EV mixture for 1 min. Labeled EVs were filtered again using Nanosep 300K spin columns and resuspended in PBS. As a control, the same concentration of EVs was incubated in diluent C for 5min followed by EV-depleted FBS for 1min and filtered as described. Loading of Cy3-labeled siRNA in PKH67-labeled EVs was carried out as described under sonication method.
PKH67-labeled EVs (25μg) were then added to 100,000 HEK293T cells in a 12 well plate in 1mL DMEM supplemented with 10% EV-depleted FBS for 22h at 37°C. Unlabeled EVs and siRNA alone were used as negative controls for PKH67 and Cy3 labeling, respectively. Unloaded PKH67-labeled EVs and transfected Cy3-labeled siRNA were used as positive controls. HEK293T cells incubated with Cy3-siRNA loaded PKH67-labeled EVs or appropriate controls were collected from the wells using 500μL of 5mM EDTA for 2min and collected in 1.5mL centrifuge tubes. Cells were washed twice with FACS buffer (0.5% BSA in 1X PBS) at 200 × g for 2min each and resuspended in FACS buffer. The samples were run on a FACSCanto II Cell Analyzer (BD Biosciences) and analyzed by FlowJo Single Cell Analysis Software. Gating was done first on live cells in forward light scatter/side scatter. PKH67+ and Cy3+ populations were gated from the live cell population.
Gene Knockdown via siRNA-Loaded EVs
Functionality of loaded siRNA was assessed using siRNAs targeting GAPDH and HER2, respectively:
GAPDH: Dharmacon, ON-TARGETplus GAPDH Control siRNA, Catalog # D-001830-01-20
HER2: Cell Signaling, SignalSilence® HER2/ErbB2 siRNA II, Catalog #6283
GAPDH knockdown experiments were performed with EVs harvested from HEK293T cells, while HER2 knockdown experiments utilized MCF-7 derived EVs. The corresponding cell types were also used as the target cells in these experiments to avoid any potential cross-cell type uptake complications. EVs (100μg/ml) were added to cell cultures (12 well plates) at volumes appropriate to achieve the indicated final concentrations of siRNA based on the calculated loaded amounts. Negative controls for these experiments included untreated cells and cells incubated with EVs that were not loaded with siRNA at a concentration equivalent to that used to deliver the highest amount of siRNA (no significant difference), while siRNA transfection with the HiPerFect reagent (Qiagen) following the manufacturer’s recommended protocol was employed as a positive control. Scrambled siRNA (Sigma Aldrich, MISSION® siRNA Universal Negative Control #1; SIC001) at 20nM final concentration was also used as an additional control. At least three replicates were carried out for each experiment, and at least three independent experiments were performed for each study.
Using the iScript cDNA Synthesis Kit (Bio-Rad), cDNA was created from cellular RNA (isolated via Qiagen RNeasy Mini Kits). Quantitative PCR (ABI 7900 Fast HT) using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used to detect RNA levels, with settings as recommended for the Supermix reagent. The real time PCR data were analyzed using the ΔΔCt method. Target genes GAPDH and HER2 were assessed using gene specific primers, with β-actin and p53 mRNAs selected as controls that were not expected to be affected by the siRNA treatment. The sequences of the primers used for this study are listed below:
GAPDH F: 5′-TGTTGCCATCAATGACCCCTT-3′
GAPDH R: 5′-CTCCACGACGTACTCAGCG-3′
p53 F: 5′-GTC CCA AGC AAT GGA TGA TTT GAT GC-3′
p53 R: 5′-GGC ATT CTG GGA GCT TCA TCT GG-3′
HER2 F: 5′-CAA CTG CAC CCA CTC CTG TGT GGA CC-3′
HER2 R: 5′-CGC TTG ATG AGG ATC CCA AAG ACC ACC C-3′
β-actin F: 5′-CTG TGG CAT CCA CGA AAC TA-3′
β-actin R: 5′-CGC TCA GGA GGA GCA ATG-3′
Protein isolation and immunoblotting were carried out using standard methods as described previously; additional antibodies for these studies included anti-β-actin (8457) (1:2000 dilution) and anti-HER2 (2165) (1:500 dilution). Densitometry was performed using LI-COR Odyssey software.
Statistical Analysis
Parametric statistical tests (one-way analysis of variance (ANOVA) with Bonferroni post-hoc test, 2-sample t-test) were used as appropriate and statistical significance level is indicated for each figure where it was calculated. Data were plotted as mean +/− standard deviation.
RESULTS
Effects of Sonication on EVs and Nucleic Acids
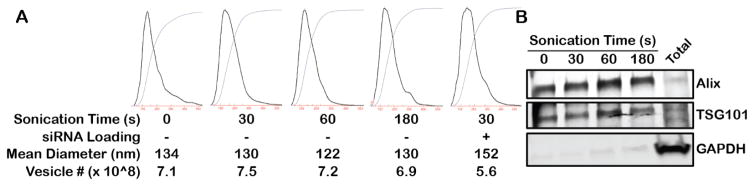
To assess the viability of sonication as a loading method for small nucleic acids into EVs, the impact of this process on each component of the potential end product was investigated. An immediate concern was the possibility for sonication to irreparably disrupt the integrity of lipid-based EVs, as sonication is often employed as a means to disrupt lipids or cell membranes. HEK293T-derived EVs were sonicated for varying times from 0–180s in the presence or absence of siRNA and EV size and number were quantified via NTA. The size distribution and total number of EVs in all experiments was similar, with essentially no difference in mean diameter or EV count over increasing sonication time up to 180s, suggesting minimal EV agglomeration (Figure 1A). Further, immunoblot analysis showed no degradation of exosomal protein markers 16 in the sonicated samples (Figure 1B).
Figure 1. Impact of sonication on EV integrity.
(A) HEK293T-derived EV size and concentration were assessed by NTA after the indicated sonication times in the presence or absence of siRNA for loading. Data are representative of 3 independent trials (n=3); no statistical difference in mean diameter or vesicle number was calculated for any group using one-way ANOVA with Bonferroni post-hoc test (P>0.05). (B) Immunoblot analysis of exosomal markers Alix and TSG101 and cellular protein marker GAPDH was conducted on HEK293T EVs sonicated for the indicated times. Total = total cellular protein under control conditions (no sonication).
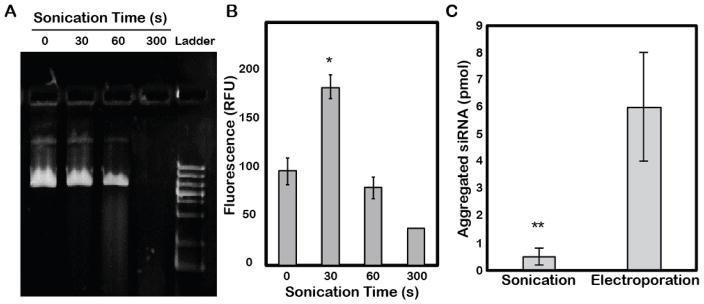
The effects of sonication on the potential nucleic acid cargo of EVs were also tested, as the labile nature of these molecules might make them susceptible to degradation. Indeed, exposure to sonication for increasing times induced degradation of plasmid DNA as indicated on an agarose gel (Figure 2A), although minimal degradation was observed for a 30s sonication time. Additionally, siRNA incorporation into HEK293T-derived EVs via sonication was found to decrease for times greater than 30s (Figure 2B), potentially indicative of siRNA degradation at longer times. Based on these results, further experiments were conducted with a 30s sonication time. In addition, potential sonication-induced nucleic acid aggregation was investigated based on prior reports of electroporation-induced siRNA aggregation 19. Large aggregates captured by a 300kDa MWCO filter were quantified as an indicator of overall aggregation following sonication of siRNA, and the amount of these aggregates was ~12 fold less than those induced by electroporation using previously optimized parameters 1, 11, 19, 21 (Figure 2C).
Figure 2. Effect of sonication on nucleic acid integrity.
(A) Plasmid DNA was sonicated for the indicated times and samples were analyzed by agarose gel electrophoresis (0.6% agarose). DNA was visualized by staining with syto-60 dye. This gel is representative of 3 independent experiments. (B) PicoGreen dye-labeled siRNA was sonicated for the indicated times for loading into HEK293T-derived EVs and quantified (ex=480nm; em=520nm) after extensive washing with 1X PBS to remove unbound excess siRNA (*P<0.05 relative to 0s control by t-test; n=3). (C) In the absence of EVs, equal amounts of siRNA were subjected to electroporation (400V and 125μF with two pulses) or sonication (30s, 35kHz) and subsequently washed with 1X PBS using a 300kDa MWCO filter. siRNA remaining in the filter was quantified as aggregated siRNA. **P<0.01 by t-test; n=3.
Quantification of Cargo Loading and Function in Sonicated EVs
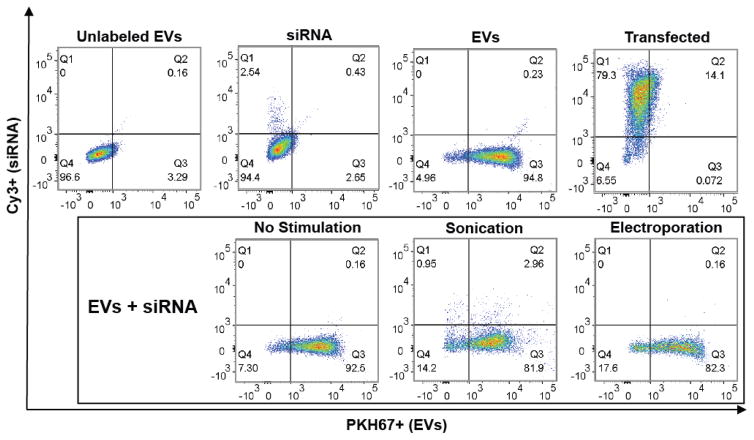
Upon validation of nucleic acid and EV integrity under sonication for 30s, levels of various small nucleic acid cargoes were loaded by sonication and quantified. Active loading via sonication enabled siRNA incorporation into HEK293T EVs at an average of 325% of the passive loading (control) level measured, while increased loading was also observed for miRNA (267% of control level) and ssDNA (225% of control level) (Figure 3). Next, cellular uptake of EVs loaded with siRNA was assessed to ensure that sonication did not negatively impact EV cargo delivery through potential disruption of surface proteins or lipids that might prevent intracellular cargo delivery. Cy3-labeled GAPDH siRNA was loaded into PKH67-labeled HEK293T EVs via sonication and, after removal of excess unincorporated siRNA, these EVs were incubated with HEK293T cells for 22h. Flow cytometry analysis revealed diminished uptake of labeled EVs associated with sonication (Figure 4; 84.86% for sonicated EVs with siRNA loaded vs. 95.03% for EVs without sonication or siRNA and 92.66% for EVs mixed with siRNA without sonication). However, siRNA uptake was significantly increased in cells exposed to EVs loaded with siRNA by sonication (Figure 4; 2.96% for sonicated EVs with siRNA loaded vs. 0.16% for EVs mixed with siRNA without sonication). Additionally, siRNA content in cells exposed to EVs loaded by sonication (2.96%) was higher than that in cells exposed to EVs loaded by electroporation using previously optimized parameters 1, 11, 19, 21 (0.16%) (Figure 4).
Figure 3. Validation of active loading of small nucleic acids into EVs by sonication.
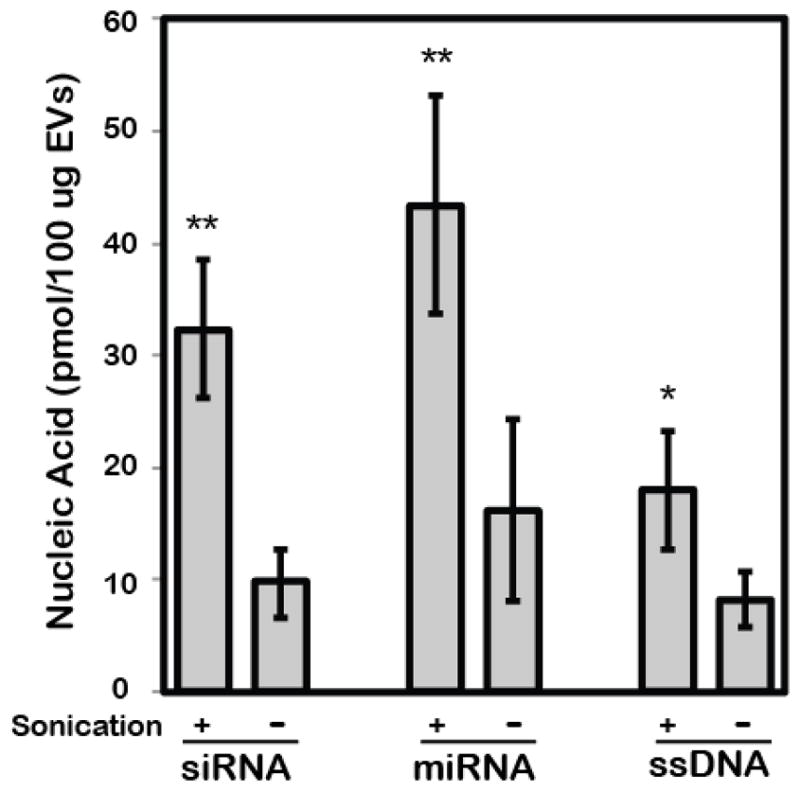
Loading of siRNA, miRNA and ssDNA was assessed with or without sonication after extensive washing to remove unincorporated nucleic acids. In each experiment, the starting materials were 1000pmol of nucleic acids and 100μg of HEK293T-derived EVs. **P<0.01, *P<0.05 by t-test; n=3.
Figure 4. Assessment of EV and siRNA uptake following sonication.
To assess EV uptake, flow cytometry analysis of PKH67-labeled HEK293T-derived EVs and Cy3-labeled siRNA was conducted. PKH67+ and Cy3+ populations were gated from live cells, with the upper right quadrant indicative of cells containing both labeled EVs and labeled siRNA. The upper four panels show control conditions: cells incubated with unlabeled EVs alone (Unlabeled EVs), siRNA alone (siRNA), labeled EVs alone (EVs), or transfected with labeled siRNA (Transfected). The lower three panels represent cells incubated with EVs loaded with siRNA by different methods: passively by mixing (No Stimulation), sonication as described in Materials and Methods (Sonication), or electroporation as described in Materials and Methods (Electroporation).
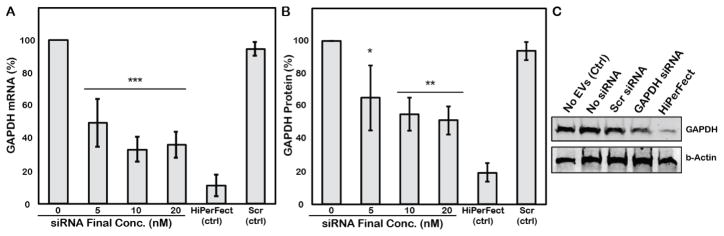
While cell uptake of siRNA may be enhanced by loading into EVs by sonication, overall cellular siRNA levels were low and the data collected do not indicate whether the siRNA is delivered to the cytoplasm, which is necessary for functional activity. Thus, functional evaluation of gene knockdown was assessed in HEK293T and MCF-7 cells. Initial proof-of-concept experiments were performed in HEK293T cells. HEK293T-derived EVs loaded with siRNA targeted to GAPDH, a gene that codes for an ~37kDa enzyme involved in glycolysis, were exposed at different concentrations to HEK293T cells for 24h. Dose-dependent knockdown of GAPDH mRNA was observed for EVs loaded by siRNA via sonication, with levels as low as 33.3±7.6% of the gene expression levels observed in control cells achieved (Figure 5A). This result corresponded with induced downregulation of GAPDH protein expression to a 51.4±8.6% of control cell protein levels as assessed by immunoblot in cells exposed to a 20nM final siRNA concentration via HEK293T EVs (Figure 5B). EVs loaded with scrambled siRNA induced expression of GAPDH mRNA and protein levels that were >94% of the control cell expression levels, while GAPDH levels in cells treated with EVs without siRNA were indistinguishable from those in cells without any treatment.
Figure 5. Functional evaluation of EVs loaded with GAPDH-targeted siRNA by sonication.
HEK293T-derived EVs loaded with siRNA targeted to GAPDH were exposed to HEK293T cells at concentrations sufficient to create the indicated final concentrations of siRNA. (A) GAPDH mRNA levels were assessed by qPCR using p53 as a housekeeping control and quantified by the ΔΔCt method. (B) GAPDH protein levels were assessed by immunoblotting using β-actin as a housekeeping control. In both panels, the same GAPDH-targeted siRNA (5nM final concentration) was transfected into cells using the HiPerFect reagent as a positive control. Also in both panels, EVs loaded by sonication with a scrambled siRNA sequence (Scr (ctrl)) were included (20nM final siRNA concentration) as a control. ***P<0.001, **P<0.01, *P<0.05 relative to 0nM siRNA control by one-way ANOVA with Bonferroni post-hoc test; n=3. (C) Representative immunoblot used to generate data in B. Scr siRNA and GAPDH siRNA were loaded into EVs by sonication, final concentration 20nM.
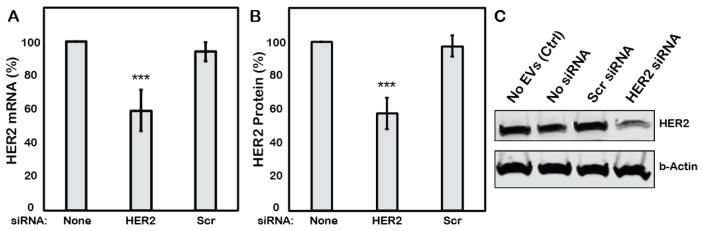
Having validated the activity of EVs loaded with siRNA via sonication, further studies were conducted in the MCF-7 breast cancer cell line with siRNA targeted to oncogene HER2 to assess the potential of the overall approach on a therapeutically relevant target. EVs isolated from MCF-7 cells were used in these studies to avoid potential issues of cell uptake specificity with regard to EVs, which is not well characterized. MCF-7-derived EVs loaded with HER2-targeted siRNA by sonication were administered to MCF-7 cells at a concentration (100μg/ml) resulting in a final siRNA concentration of 20nM. HER2 mRNA knockdown to 59.2±12% of control levels was observed (Figure 6A), corresponding to downregulation of HER2 protein expression to 51.8±10.5% of control levels (Figure 6B). EVs loaded with scrambled siRNA induced expression of HER2 mRNA and protein levels that were >94% of the control cell expression levels as determined by qPCR and immunoblot analysis, respectively. Thus, EVs loaded with siRNA via sonication demonstrated significant bioactivity suggesting potential therapeutic efficacy against a critical target oncogene.
Figure 6. Functional evaluation of EVs loaded with HER2-targeted siRNA by sonication.
MCF-7-derived EVs loaded with siRNA targeted to HER2 were exposed to MCF-7 cells at concentrations sufficient to create a 20nM final concentration of siRNA. (A) HER2 mRNA levels were assessed by qPCR using p53 as a housekeeping control and quantified by the ΔΔCt method. (B) HER2 protein levels were assessed by immunoblotting using β-actin as a housekeeping control. In both panels, EVs loaded by sonication with a scrambled siRNA sequence (Scr (ctrl)) were included (20nM final siRNA concentration) as a control. **P<0.01, *P<0.05 by t-test; n=3. (C) Representative immunoblot used to generate data in B. Scr siRNA and HER2 siRNA were loaded into EVs by sonication, final concentration 20nM.
DISCUSSION
Sonication has been identified as a potential loading method for the creation of therapeutic EVs 17, 18, however, the compatibility of this process with a crucial class of potential EV cargo, therapeutic nucleic acids and especially small RNAs, has not been previously defined. Here, we have demonstrated that sonication can increase the loading of a variety of small nucleic acids into EVs. We further showed that loaded EVs retain their integrity and ability to deliver small RNA cargo after sonication. Finally, these studies showed that EVs loaded with siRNA via sonication can mediate knockdown of target genes leading to cognate protein downregulation.
Overall, our data suggest that sonication may be a suitable alternative to electroporation for small nucleic acid loading in EVs. To be clear, the studies presented here did not attempt to optimize parameters for electroporation-mediated EV loading; such work has been reported by our group and others previously 1, 11, 19, 21. However, it was observed that less >300kDa siRNA aggregates were formed under sonication as compared to electroporation (Figure 2C). Of course, this does not rule out the possibility of smaller aggregate formation by sonication, and a complete assessment of aggregation induced by these methods would require further study. Additionally, increased cell uptake of EV-associated siRNA was observed for sonicated EVs compared to electroporated EVs (Figure 4). Yet, overall delivery of labeled siRNA to cells via sonicated EVs was still very low (2.96%), especially given that cell uptake of labeled EVs was >80%. One potential explanation for this is degradation of siRNA associated with the EV surface. The levels of siRNA associated with EVs (Figure 3) correspond to several thousand copies per EV, however interior encapsulation of this amount of siRNA is unlikely based on the volume fraction of the interior compartments of EVs compared to the total solution volume for sonication. Thus, loading levels based purely on diffusion-mediated incorporation following sonication would be expected to be lower than those observed, and substantial amounts of siRNA may be associated with the EV surface. Any potential degradation would further limit loading efficiency, which is already limited based on the same volume fraction between EVs and total solution as well as the high siRNA:EV ratio needed to ensure labeled siRNA detection in these studies. Improving overall loading efficiency, such as by potentially reusing a loading solution for multiple batches of EVs, should be explored to promote scalability and practicality of this approach for future applications. Further, the ratio of siRNA:EV uptake must be improved to enable eventual clinical utility of this approach and should be a major focus of future work in this area.
In the context of the relatively low siRNA delivery measured by flow cytometry, the knockdown levels observed (Figures 6,7) are more encouraging. Functional knockdown of target genes by sonication-mediated siRNA-loaded EVs was observed to be specific, as scrambled siRNA sequences delivered in the same manner induced minimal changes in target gene and protein expression. The high uptake of sonicated EVs overall (Figure 4) suggests that improved siRNA delivery and increased knockdown may be possible with continued development of this approach. Further, these data indicate that despite any potential limitations associated with cargo degradation, EVs loaded with siRNA via sonication have therapeutic potential.
The utility of siRNA-loaded EVs for HER2 knockdown has potential clinical relevance for cancer therapy. HER2 knockdown via siRNA is an established method of tumor cell growth inhibition, including in the MCF-7 breast cancer cells used in these studies 7, 13. Critically, siRNA can be successfully employed to reduce the growth rate of HER2 antibody (trastuzumab)-resistant cancer cells by 50–65% 33. In addition to siRNA, miRNAs are also potential vectors for HER2 silencing. For example, miR-331-3p has recently been shown to reduce proliferation and increase apoptosis in colorectal cancer cells via HER2 silencing 36. Small RNA loading into EVs via sonication enables increased options for delivery of both siRNA and miRNA against HER2 and other oncogenes, and thus may be useful in a variety of cancer therapeutic approaches.
Broadening the utility of EVs for small RNA delivery is crucial due to the overall potential of EVs as alternatives to current cancer therapeutic approaches. Several elegant approaches, including aptamer-based strategies and multiple nanoparticle formulations, have been employed towards therapeutic RNAi generally 8, 27, 32, 35 and for HER2 silencing specifically 29, 30. Yet, the vast majority of these approaches are ultimately synthetic in nature and face common obstacles as delivery vehicles to tumors, including induction of immune or inflammatory responses and toxicological limitations 10. The use of EVs as natural, cell-derived drug carriers, may avoid many of these limitations and has the additional potential of being integrated into a personalized medicine approach, where a patient’s own cells could be used to generate drug carriers for treatment of their disease 22. In this last case, the continued development of exogenous loading methods such as sonication will be critical in enabling widespread utility.
Overall, these studies validate sonication as a method for loading functional small RNAs into EVs for targeted oncogene knockdown. The proof-of-concept experiments conducted here are expected to be applicable to a wide variety of EVs, enabling potential selection of an optimized cell source towards control of EV biodistribution upon delivery 34. Further, these methods should allow for incorporation of many different small RNA cargos targeting numerous genes mediating cancer and other diseases.
Acknowledgments
This work was supported by NIH R00 grant HL112905, by an ORAU Ralph E. Powe Junior Faculty Enhancement Award, and by two University of Maryland Tier 1 seed grants (all to SMJ).
Biography
Steven M. Jay is an Assistant Professor in the Fischell Department of Bioengineering at the University of Maryland. Through generous support from the University and the National Institutes of Health (NIH), he has established a laboratory focused on biotherapeutic development and delivery, with specific interests in cancer therapy and vascular regeneration. Originally from the great state of Kentucky, Dr. Jay received a B.S.B.E. (Biological Engineering) from the University of Georgia before completing his Ph.D. in Biomedical Engineering under Dr. Mark Saltzman at Yale University. He then received postdoctoral training jointly from Dr. Richard T. Lee at the Brigham and Women’s Hospital and Dr. Linda Griffith at the Massachusetts Institute of Technology, and has also trained with Drs. Brian Rymond and Russ Mumper (University of Kentucky), Drs. Karen Burg and Thomas Jenkins (Clemson University), Dr. William Kisaalita (University of Georgia) and Drs. Themis Kyriakides and Jordan Pober (Yale University) among others. In addition to the NIH and the University of Maryland, Dr. Jay’s work has been supported by the National Science Foundation and the Oak Ridge Associated Universities. Dr. Jay’s research is enabled by the fortitude, diligence and intelligence of his trainees and by inspiration from his colleagues, friends and family.
Footnotes
AUTHOR CONTRIBUTIONS
TNL, AJ, DBP, BP, NKL, NA and JSS performed the research and analyzed data. TNL, AJ, DBP and SMJ contributed to conception and design of experiments and wrote the manuscript. All authors reviewed, edited and approved of the final manuscript.
CONFLICT OF INTEREST
Authors Tek N. Lamichhane, Anjana Jeyaram, Divya B. Patel, Babita Parajuli, Natalie K. Livingston, Navein Arumugasaamy, John S. Schardt and Steven M. Jay declare that they have no conflicts of interest.
ETHICS AND INFORMED CONSENT
No human studies were carried out by the authors for this article. No animal studies were carried out by the authors for this article.
References
- 1.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 2.Andaloussi EL, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 3.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 4.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 5.Blenkiron C, Miska EA. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16(Spec No 1):R106–113. doi: 10.1093/hmg/ddm056. [DOI] [PubMed] [Google Scholar]
- 6.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudhury A, Charo J, Parapuram SK, Hunt RC, Hunt DM, Seliger B, Kiessling R. Small interfering RNA (siRNA) inhibits the expression of the Her2/neu gene, upregulates HLA class I and induces apoptosis of Her2/neu positive tumor cell lines. Int J Cancer. 2004;108:71–77. doi: 10.1002/ijc.11497. [DOI] [PubMed] [Google Scholar]
- 8.Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M, Falzone R, Harrop J, Cehelsky J, Bettencourt BR, Geissler M, Butler JS, Sehgal A, Meyers RE, Chen Q, Borland T, Hutabarat RM, Clausen VA, Alvarez R, Fitzgerald K, Gamba-Vitalo C, Nochur SV, Vaishnaw AK, Sah DW, Gollob JA, Suhr OB. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 9.Cooper JM, Wiklander PB, Nordin JZ, Al-Shawi R, Wood MJ, Vithlani M, Schapira AH, Simons JP, El-Andaloussi S, Alvarez-Erviti L. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov Disord. 2014;29:1476–1485. doi: 10.1002/mds.25978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jong WH, Borm PJ. Drug delivery and nanoparticles:applications and hazards. Int J Nanomedicine. 2008;3:133–149. doi: 10.2147/ijn.s596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li J, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJ. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat Protoc. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 12.Esmaeilzadeh Gharehdaghi E, Amani A, Khoshayand MR, Banan M, Esmaeilzadeh Gharehdaghi E, Amini MA, Faramarzi MA. Chitosan nanoparticles for siRNA delivery: optimization of processing/formulation parameters. Nucleic Acid Ther. 2014;24:420–427. doi: 10.1089/nat.2014.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faltus T, Yuan J, Zimmer B, Kramer A, Loibl S, Kaufmann M, Strebhardt K. Silencing of the HER2/neu gene by siRNA inhibits proliferation and induces apoptosis in HER2/neu-overexpressing breast cancer cells. Neoplasia. 2004;6:786–795. doi: 10.1593/neo.04313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farooqi AA, Rehman ZU, Muntane J. Antisense therapeutics in oncology: current status. Onco Targets Ther. 2014;7:2035–2042. doi: 10.2147/OTT.S49652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavrilov K, Saltzman WM. Therapeutic siRNA: principles, challenges, and strategies. Yale J Biol Med. 2012;85:187–200. [PMC free article] [PubMed] [Google Scholar]
- 16.Gyorgy B, Hung ME, Breakefield XO, Leonard JN. Therapeutic applications of extracellular vesicles: clinical promise and open questions. Annu Rev Pharmacol Toxicol. 2015;55:439–464. doi: 10.1146/annurev-pharmtox-010814-124630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV, Batrakova EV. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2015 doi: 10.1016/j.nano.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kooijmans SA, Stremersch S, Braeckmans K, de Smedt SC, Hendrix A, Wood MJ, Schiffelers RM, Raemdonck K, Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J Control Release. 2013;172:229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Kundu AK, Chandra PK, Hazari S, Pramar YV, Dash S, Mandal TK. Development and optimization of nanosomal formulations for siRNA delivery to the liver. Eur J Pharm Biopharm. 2012;80:257–267. doi: 10.1016/j.ejpb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamichhane TN, Raiker RS, Jay SM. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamichhane TN, Sokic S, Schardt JS, Raiker RS, Lin JW, Jay SM. Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine. Tissue Eng Part B Rev. 2015;21:45–54. doi: 10.1089/ten.teb.2014.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClorey G, Wood MJ. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–6487. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peer D, Lieberman J. Special delivery: targeted therapy with small RNAs. Gene Ther. 2011;18:1127–1133. doi: 10.1038/gt.2011.56. [DOI] [PubMed] [Google Scholar]
- 28.Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2015;199:145–155. doi: 10.1016/j.jconrel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan WB, Jiang S, Zhang Y. Quantum-dot based nanoparticles for targeted silencing of HER2/neu gene via RNA interference. Biomaterials. 2007;28:1565–1571. doi: 10.1016/j.biomaterials.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, 2nd, Giangrande PH. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res. 2012;40:6319–6337. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 32.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wichmann H, Guttler A, Bache M, Taubert H, Rot S, Kessler J, Eckert AW, Kappler M, Vordermark D. Targeting of EGFR and HER2 with therapeutic antibodies and siRNA: a comparative study in glioblastoma cells. Strahlenther Onkol. 2015;191:180–191. doi: 10.1007/s00066-014-0743-9. [DOI] [PubMed] [Google Scholar]
- 34.Wiklander OP, Nordin JZ, O’Loughlin A, Gustafsson Y, Corso G, Mager I, Vader P, Lee Y, Sork H, Seow Y, Heldring N, Alvarez-Erviti L, Smith CE, Le Blanc K, Macchiarini P, Jungebluth P, Wood MJ, Andaloussi SE. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316. doi: 10.3402/jev.v4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wittrup A, Lieberman J. Knocking down disease: a progress report on siRNA therapeutics. Nat Rev Genet. 2015;16:543–552. doi: 10.1038/nrg3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao D, Sui Y, Zheng X. miR-331-3p inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt and ERK1/2 pathways in colorectal cancer. Oncol Rep. 2016;35:1075–1082. doi: 10.3892/or.2015.4450. [DOI] [PubMed] [Google Scholar]