Abstract
Autoantibodies against galactose-deficient IgA1 drive formation of pathogenic immune complexes in IgA nephropathy. IgG autoantibodies against galactose-deficient IgA1 in patients with IgA nephropathy have a specific amino-acid sequence, Y1CS3, in the complementarity-determining region 3 of the heavy chain variable region compared with a Y1CA3 sequence in similar isotype-matched IgG from healthy controls. We previously found that the S3 residue is critical for binding galactose-deficient IgA1. To determine whether this difference is due to a rare germline sequence, we amplified and sequenced the corresponding germline variable region genes from peripheral blood mononuclear cells of seven patients with IgA nephropathy and six healthy controls from whom we had cloned single-cell lines secreting monoclonal IgG specific for galactose-deficient IgA1. Sanger DNA sequencing revealed that complementarity-determining region 3 in the variable region of the germline genes encoded the Y1C(A/V)3 amino-acid sequence. Thus, the A/V>S substitution in the complementarity-determining region 3 of anti-galactose–deficient-IgA1 autoantibodies of the patients with IgA nephropathy is not a rare germline gene variant. Modeling analyses indicated that the S3 hydroxyl group spans the complementarity-determining region 3 loop stem, stabilizing the adjacent β-sheet and stem structure, important features for effective binding to galactose-deficient IgA1. Understanding processes leading to production of the autoantibodies may offer new approaches to treat IgA nephropathy.
Keywords: IgA nephropathy, immune complexes, immunology
IgA nephropathy (IgAN), the most common primary GN in the world, is characterized by glomerular IgA1-containing immunodeposits.1–3 Up to 40% of patients with IgAN develop ESRD. Until recently, IgAN was not considered an autoimmune disease. New findings revealed that galactose-deficient IgA1 (Gd-IgA1) is recognized by antiglycan antibodies, resulting in formation of circulating pathogenic immune complexes in patients with IgAN.1–3 Levels of such immune complexes, consisting of Gd-IgA1 (autoantigen) and the corresponding autoantibodies, are elevated in the circulation of patients with IgAN.1–3 Glomerular deposition of such complexes has been proposed as the main pathologic mechanism leading to development of this chronic renal disease.1–3
Several studies have shown that IgA1-containing immune complexes activate mesangial cells, resulting in cellular proliferation and overexpression of extracellular matrix components, cytokines, and chemokines that may ultimately lead to ESRD.4 Levels of autoantibodies against Gd-IgA1 of the IgG and IgA1 isotypes are elevated in the circulation of patients with IgAN.5 The serum levels of Gd-IgA1–specific IgG autoantibodies correlate with proteinuria and the amounts of IgA-IgG immune complexes excreted in the urine.6 Moreover, elevated serum levels of Gd-IgA17 and autoantibodies (IgG and IgA) against Gd-IgA18 at the time of renal biopsy correlate with disease severity and predict disease progression. Consequently, serum levels of Gd-IgA1 (autoantigen) and autoantibody may serve as prognostic biomarkers of IgAN.7,9
To better understand the nature of anti–Gd-IgA1 autoantibodies at the molecular level, we have generated single-cell clones that secrete IgG specific for Gd-IgA1. Using single-cell RT-PCR, we have amplified, cloned, and sequenced the transcripts encoding the corresponding heavy-chain (VH) and light-chain variable regions.6 Analysis of the nucleotide sequences encoding the VH segments of IgG antibodies against Gd-IgA1 revealed a consensus sequence in complementarity-determining region 3 (CDR3) encoding Y1CS3 for patients with IgAN, whereas a sequence encoding Y1CA3 was found for healthy controls.6 This finding implied an amino-acid (aa) replacement of A by S in CDR3 of the H chain of IgG autoantibodies from patients with IgAN. Our prior experiments, in which we introduced the S to A substitution into the wild-type sequence of recombinant antibody, confirmed that S3 in CDR3 is important for binding to Gd-IgA1.6 We concluded that the specific VH CDR3 aa sequence with S3 is characteristic for autoantibodies of patients with IgAN and that it determines the ability to bind Gd-IgA1 and form nephritogenic immune complexes in these patients.
In this study, we assessed whether these Gd-IgA1–specific autoantibodies result from utilization of rare alleles of specific VH germline genes in patients with IgAN. We analyzed germline genomic sequences of VH genes that were matched with the VH CDR3 sequences of autoantibodies from seven patients with IgAN and six healthy controls from our previous study.6 Sequence comparisons revealed that the A>S (and in one case, V>S) substitution in the VH CDR3 of the Gd-IgA1–specific IgG autoantibodies of the patients with IgAN did not originate from a rare germline variant of the VH gene. Moreover, we assessed the effect of the presence of S3 versus A3 in CDR3 on the structure of VH CDR3 loop by molecular modeling. Our results indicate that the S3 hydroxyl group spans the CDR3 loop stem, stabilizing the adjacent β-sheet and stem structure. This change in aa sequence is important for effective autoantibody binding to Gd-IgA1.
We used genomic DNA from seven patients with IgAN and six healthy controls from whom we had previously cloned VH regions of IgG antibodies specific for Gd-IgA1.6 The IgG antibodies of the patients used the VH 1–24, 1–46, 3–11, 3–21, 3–23, and 3–30 gene segments and those of the controls used the VH 3–7, 4–4, 4–39, and 4–59 gene segments. PCR amplicons produced with gene-specific primers were cloned and sequenced by the Sanger method in both directions. Germline DNA sequences corresponding to CDR3 were aligned with those obtained by single-cell RT-PCR from the cells secreting the corresponding antibodies (Figure 1A and B).
Figure 1.
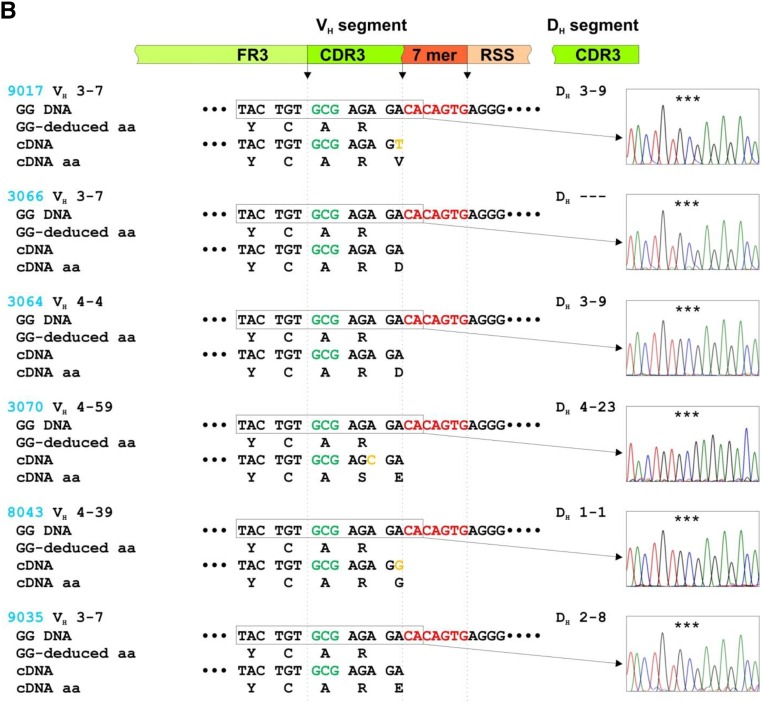
S3 in VH CDR3 of IgG autoantibodies is not encoded in VH germline gene sequences. Alignments of the 3′ termini of VH segments for germline genomic DNA (GG DNA) with reverse-transcribed mRNA from individual IgG-producing clones from seven patients with IgAN (A) (#2047, #1123, #1125, #1139, #1023, #3061, and #3081; red) and six healthy controls (B) (#9017, #3066, #3064, #3070, #8043, and #9035 blue) (cDNA). cDNA sequences were published.6 VH 3′ segment consists of coding 3′ terminus of framework region 3 (FR3) and 5′ part of complementarity-determining region 3 (CDR3), followed by noncoding region sequence composed of conserved 7-mer (CACAGTG; in red) and 5′ terminus of 23-bp spacer nucleotides (recombination signal sequence [RSS]). Chromatograms show GG DNA sequence in rectangles; the triplet-encoding alanine (A) is marked by stars. For cDNA, a coding region of DH segment contributing to CDR3 is shown. In the diagram above the sequences, green marks coding sequences and red marks noncoding sequences. Deduced aa sequences for GG DNA (GG-deduced aa) and cDNA (cDNA aa) are shown to localize the critical A (or V in subject 3061) to serine (S) changes in CDR3 of antibodies from patients with IgAN.
The germline DNA sequences from six of seven patients with IgAN encoded VH CDR3 aa sequence Y1CA3 and in one patient encoded VH CDR3 aa sequence Y1CV3 (Figure 1A). cDNA sequences from the cells producing the antibodies specific for Gd-IgA1 showed that in six of the seven patients with IgAN, the corresponding VH CDR3 aa sequence was Y1CS3(R/K) and in one patient with IgAN was Y1CA3T. This difference between the genomic and cDNA sequences for five of the six patients with IgAN with Y1CS3(R/K) sequence was due to changes in the first and third nucleotides of the codon for A (GCG), thus changing it to a codon for S (TCA or TCC). The sixth patient with Y1CS3R cDNA sequence had all three nucleotides of the codon for V (GTG) in the germline VH gene changed to codon for S (TCC). The one patient with IgAN with Y1CA3T in CDR3 aa cDNA sequence had the codon ACA changed to ACT, but both codons encoded T. In contrast, there was no such codon change in the CDR3 regions in IgH genes versus cDNA sequences in the IgGs from the six healthy controls (Figure 1B). Moreover, computer analysis of VH sequences encoding monoclonal anti–Gd-IgA1 IgG antibodies showed no evidence of VH gene replacement (Z. Zhang, unpublished observations).
These results revealed that S3 in the VH aa sequences of CDR3 region of antibodies against Gd-IgA1 in patients with IgAN did not originate from rare germline-encoded allele(s) of VH gene segments, such as those identified in a murine model of antibody-mediated autoimmune diseases.10 Thus, we propose that the VH aa sequence of CDR3 region with S3 arises from other processes, such as somatic mutations and antigen selection. It was hypothesized that these antibodies may be produced in response to N-acetylgalactosamine (GalNAc)–containing molecules on some viruses or bacteria6 and that the exposed GalNAc on Gd-IgA1 may further drive the autoantibody production and affinity maturation in susceptible individuals.3,11 Some of these processes may be related to infections of the upper respiratory tract, which are frequently associated with flares of disease activity in patients with IgAN, manifested clinically as episodes of visible hematuria.6
The H-chain variable regions of antibodies are formed and modified by multiple processes, including the random VDJ recombination process, junction diversification, VH replacement, and somatic hypermutation combined with clonal selection. These processes increase the diversity of the antibody pool and alter the antigen-binding affinity of the antibody.12 Mutations in VH and VL gene segments usually contribute the most to the changes in the antigen-binding sites,12 although single-point mutations in the D region have also been reported.13,14
Activation-induced cytidine deaminase is essential for somatic hypermutation that initiates cytosine deamination followed by uracil mismatch.15 This process is critical for the immune defense against environmental antigens, as well as other biologic processes.16,17 Multiple factors have been reported to induce activation-induced cytidine deaminase, including CD40L, IL-4, TGF-β, TNF-α, and NF-κB.18,19 Thus, somatic hypermutations of the immunoglobulin VH gene segment may be induced because of changes in local cytokine production associated with a bacterial or viral infection in predisposed individuals.6 Our results support a hypothesis that a process, such as somatic hypermutation, modulates the VH CDR3 regions of the Gd-IgA1–specific antibodies. Future studies are needed to identify genetic and environmental factors that may codetermine these processes in patients with IgAN, leading to production of Gd-IgA1–specific autoantibodies.
Recently, specificities of antibodies have been correlated with protein dynamics. The character of the proposed antibody lock-and-key mechanism is associated with a high level of antibody rigidity.20 One postulated role of somatic mutations of antibodies is the evolution of protein rigidity by introducing hydrogen bonds or packing interactions that crosslink the CDR loops or the strands of the β-sheets, thus facilitating molecular recognition.20 In the antigen-binding site, polar aa residues, particularly R, N, D, S, and T, occur frequently, and these aa residues may provide physicochemical and structural diversity.21
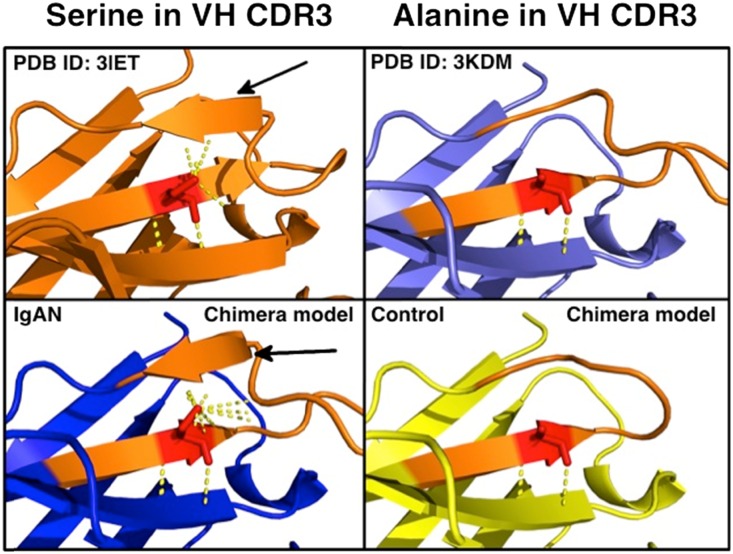
To determine possible structural effects of the S3 in human VH CDR3, we constructed chimeric homology models of the VH CDR3 loops of autoantibodies from patients with IgAN and matching antibodies from healthy controls. As a template and backbone for these models, we used human antitestosterone antibody (PDB ID: 3KDM) because of its high aa sequence similarity to the VH of autoantibodies (Supplemental Figure 1). Notably, CDR3 in these models formed a structured β-sheet for five of six patients with IgAN with the S3 in VH CDR3 (Supplemental Figure 1C, Supplemental Table 1), resembling the CDR3 loop of mouse anti-Tn antibody (PDB ID: 3IET) (Supplemental Figure 1A). In contrast, none of the VH CDR3 from six healthy controls with A3 in CDR3 formed a structured β-sheet, and therefore the VH CDR3 mimicked the 3KDM template (Supplemental Figure 1, B and D; Supplemental Table 1). Upon closer inspection of the models, polar contacts of S3 side-chain hydroxyl group spanned the CDR3 loop stem to stabilize the adjacent β-sheet and stem structure (Figure 2, bottom left panel), similar to polar contacts observed in mouse anti-Tn antibody (PDB ID: 3IET) (Figure 2, top left panel). In contrast, in the VH CDR3 loop of IgG from a healthy control with an A3 residue (Figure 2, bottom right panel), no polar contact was observed between A3 and the adjacent side of the CDR3 loop stem, and the β-sheet structure was not observed; these latter features resemble those of the human antitestosterone template 3KDM (Figure 2, top right panel).
Figure 2.
Serine promotes structural rigidity of VH CDR3 in IgG autoantibodies. Polar contacts of IgG with A>S somatic mutation affect secondary and tertiary structure of variable heavy-chain CDR3. Crystal structure of PDB ID: 3IET (top left), a mouse monoclonal antibody against cancer-associated glycoprotein with terminal GalNAc (also called Tn antigen), demonstrates S residue in CDR3 (red).22 Polar contacts (yellow dotted line) of S hydroxyl R-group span CDR3 loop (orange) and thereby stabilize the adjacent β-sheet and stem structure of CDR3 loop (black arrow). Crystal structure of PDB ID: 3KDM (top right), a human monoclonal antibody against testosterone,29 retains A (red) germline sequence while containing 86% aa sequence identity with CDR3 of IgG of IgAN patient 1123. No polar contact (yellow dotted line) was observed between A (red) and the adjacent side of the CDR3 loop (orange) stem; a β-sheet structure was not observed. Chimeric model of VH of an anti–Gd-IgA1 IgG of IgAN patient 1123 (bottom left) with variable heavy-chain CDR3 loop (orange) with S3 (red) resembles the structure of mouse monoclonal anti-Tn IgG, with increased polar contacts stabilizing the CDR3 loop stem (black arrow). Chimeric model of IgG of a healthy control 3066 (bottom right) with variable heavy-chain CDR3 (orange) with A3 (red) resembles the human monoclonal antitestosterone IgG.
These in silico data are consistent with the hypotheses that the aa S3, as detected in the CDR3 region of autoantibodies against Gd-IgA1 in patients with IgAN, provides a hydrogen bond across CDR3 loop and thereby contributes to stereochemical rigidity that orients neighboring aa residues for interaction with antigen.21 However, these hypotheses, generated by modeling, need to be tested using biochemical and structural biology approaches, such as the crystallographic studies performed for murine monoclonal antibody against cancer-associated Tn antigen that has terminal GalNAc as part of the epitope, similar to the epitope of Gd-IgA1.22
Somatic hypermutation of VH of antibodies induced by environmental antigens is an important mechanism in the immune defense against pathogens, but it may also contribute to production of autoreactive antibodies.23 Recent genomic data suggest that patients with IgAN have compromised mucosal immune responses.24,25 In fact, disease activity in patients with IgAN often coincides with mucosal infections, including those of the upper respiratory tract and/or digestive system.3,11 Thus, an exaggerated somatic hypermutation rate, typically associated with production of high-affinity antibodies, may be a compensatory mechanism in such a situation. Our next-generation sequencing pilot data showed that cDNA sequences corresponding to the VH CDR3 region with YCS3R sequence have a majority of the nucleotides encoding S3 instead of the germline-sequence-encoded A3 in the early clones of EBV-immortalized cells (“first freeze”), as well as in primary cells (PBMCs) of a recalled patient with IgAN (Supplemental Figure 2). Analysis of IgG from serum of this recalled patient by high-resolution mass spectrometry further supported the conclusion that YCS3R occurs in VH CDR3 of the IgG antibodies (data not shown). All these analyses have been done in the context of total observed sequences in VH CDR3 encoding YCA3/S3R aa sequence and not with cells producing autoantibody specific for Gd-IgA1. As expected, most VH CDR3 sequences have not encoded a YCA3/S3R aa sequence; such sequences were excluded from these analyses.
Together, these findings indicate that sequences encoding YCS3R aa sequence occurred naturally in VH CDR3 of IgG. However, future detailed studies are needed to elucidate the mechanism(s) and define extent of this A3 to S3 change in VH CDR3 of IgG in general and Gd-IgA1–specific IgG autoantibodies in patients with IgAN in particular.
Notably, our recent analysis of serum levels of Gd-IgA1 and Gd-IgA1–specific IgG autoantibodies revealed a previously unreported association of the autoantigen and autoantibody in IgAN. Specifically, using serum samples from 135 patients with biopsy-proven IgAN, 79 patients with other renal diseases, and 106 healthy controls, we found that Gd-IgA1 serum levels correlated with levels of Gd-IgA1–specific IgG autoantibodies in patients with IgAN (r=0.4909; P<0.001), but not in healthy controls or disease controls.26 This finding thus further supports the key roles of Gd-IgA1 and the corresponding IgG autoantibodies in the pathogenesis of IgAN.
What do our findings mean for a potential future therapy of patients with IgAN? On the basis of our multihit hypothesis, it is the large immune complexes consisting of Gd-IgA1 autoantigen and anti–Gd-IgA1 autoantibodies that drive the pathogenesis of IgAN.1,9 Thus, suppression of the production of autoantigen or autoantibody would decrease formation of immune complexes. Alternatively, formation of nephritogenic immune complexes could be suppressed through supplementing a competing glycopeptide or a non-crosslinking fragment of anti–Gd-IgA1 antibody; both instances would produce smaller non-nephritogenic immune complexes.1,9
With the new findings from this study that the anti–Gd-IgA1 autoantibodies do not originate from rare alleles of VH gene segments but rather result from somatic hypermutation, one can speculate that modulation of B-cell receptor signaling may reduce the somatic hypermutation process. Alternatively, other means of manipulation of the cells producing the autoantibodies, such as blockade of cytokines, chemokines, or growth factors, may also reduce the load of autoantibodies, as proposed for another B-cell–driven autoimmune disease, systemic lupus erythematosus.27 As serum levels of anti–Gd-IgA1 antibodies are associated with disease activity6 and predict progression of IgAN,8 it is likely that reduced production of anti–Gd-IgA1 antibodies would be of therapeutic benefit.1,4,9
Concise Methods
Genomic DNA
In our previous study, we obtained cDNA VH sequences for monoclonal IgG autoantibodies from seven patients with IgAN and six healthy controls6 (Supplemental Table 1). Here, we analyzed genomic sequence of VH gene segments that were matched with the VH CDR3 sequences of autoantibodies from seven patients with IgAN and six healthy controls. These were the individuals from our previous study for whom we had genomic DNA available.28
Amplification and Sequencing of Germline IgG VH Segments
PCR Primers
Germline genomic VH DNA sequences matching those of monoclonal IgG antibodies specific for Gd-IgA16 were obtained from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov).29 Primers to amplify specific VH genes were designed using Primer-Blast software supplied at the NCBI website (Table 1) and tested in silico for their specificity.
Table 1.
Sequences of primers used to amplify VH genes
| VH Gene | PCR Primers |
|---|---|
| 1–24 | F: ATCCTCTTCTTGGTGGCAGC |
| R: CGCTCTCAGGATGTGGGTTT | |
| 1–46 | F: GGGACACGTCCACGAGCACA |
| R: AGCACAGCTGACTCCTCCCTCA | |
| 3–7 | F: GATGAGCTGGGTCCGCCAGG |
| R: CAGACCGCGACAGGAAGCCA | |
| 3–11 | F: GAGGCTTGGTCAAGCCTGGAGG |
| R: ACAGCCCCGTGCTCAGTGTC | |
| 3–21 | F: TCTGGGGGAGGCCTGGTCAA |
| R: CTGGTGGTCCTGGGGACCCT | |
| 3–23 | F: GGGCTGGAGTGGGTCTCAGC |
| R: ATCTGCACCTGCCTCTGCGG | |
| 3–30 | F: CTGGAGTGGGTGGCAGTTATATCATA |
| R: CACCTGCCTCCAGGGCTGAC | |
| 4–4 | F: CCTCACCTGCACTGTCTCTG |
| R: ACTGAGCTGATGAGGAAGCC | |
| 4–39 | F: AGTATCTATTATAGTGGGAGC |
| R: AGGGGAGGTGAGTGTGAGCCCC | |
| 4–59 | F: CTCACCTGCACTGTCTCTGG |
| R: AGGGAGACCACTGAGCAGAT |
Cloning and Sequencing
PCR amplicons were analyzed by agarose electrophoresis6 and ligated into a plasmid vector (PCR 2.1-TOPO) using a cloning kit (TOPO cloning kit, Invitrogen, Carlsbad, CA) and transformed into TOP10F′-competent cells, according to the manufacturer’s protocol. Positive colonies were expanded in Luria–Bertani medium. Plasmids were purified using QIAprepSpin Miniprep kit (Qiagen, Valencia, CA) and the inserts were sequenced by the Sanger method in both directions from at least three clones using M13 primers with an automated AB3730×1 DNA analyzer (DNA Sequencing Core Facility, University of Alabama at Birmingham, Birmingham, AL). cDNA sequences of VH CDR3 of the IgG were deposited in GeneBank with the following accession numbers: patients with IgAN (2047, FJ746347; 1123, FJ746357; 1125, FJ746355; 1139, FJ746341) and healthy controls (9017, FJ746359; 3066, FJ746353).6 Germline DNA sequences obtained from the corresponding subjects were aligned with human genomic DNA to confirm the identity of the amplified VH genes (chr. 14 IGH locus, sequence ID AC_000146.1).
Statistical Analyses
Data were analyzed using the software supplied by the International Immunogenetic Information System (http://www.imgt.org/) to confirm that the results matched the correct VH genes. Sequences in VH regions were compared with the sequences from cDNA encoding monoclonal IgG antibodies specific for Gd-IgA1.6
Chimera Homology Modeling
PDB ID 3KDM30 was chosen as the homology-modeling template after NCBI BLAST against the PDB databank of full-length aa sequence of IgG 1123 VH indicated 86% identity to that of 3KDM. Chimeric aa sequences for the VH CDR3 sequences from a patient with IgAN and a healthy control were made by using the corresponding VH CDR3 from the patient’s and the control’s IgG VH regions (Supplemental Table 1). PDB ID 3KDM heavy chain was used as a template for homology modeling of chimeric IgG sequences for the patient with IgAN and the healthy control. Modeling was performed using SWISS-MODEL Automatic Modeling Mode online server found at http://www.swissmodel.expasy.org. Models and structures of 3IET and 3KDM were visualized and manipulated using PyMOL v1.6 molecular graphics system (http://www.pymol.org). Additional details are in Supplemental Materials.
Study Approval
The institutional review boards of the University of Alabama and the University of Tennessee Health Sciences Center approved the study. Written informed consent was received from participants (and legal guardian and with the participant’s assent for one pediatric patient) before inclusion in the study.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health DK078244, DK082753, DK106341, GM098539, CA13148, and AI027767 and by a gift from the IGA Nephropathy Foundation of America. M.R. was supported in part by grants CZ.1.07/2.3.00/20.0164 and 15-33686A, Czech Republic.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014101044/-/DCSupplemental.
References
- 1.Suzuki H, Kiryluk K, Novak J, Moldoveanu Z, Herr AB, Renfrow MB, Wyatt RJ, Scolari F, Mestecky J, Gharavi AG, Julian BA: The pathophysiology of IgA nephropathy. J Am Soc Nephrol 22: 1795–1803, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glassock RJ: The pathogenesis of IgA nephropathy. Curr Opin Nephrol Hypertens 20: 153–160, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Wyatt RJ, Julian BA: IgA nephropathy. N Engl J Med 368: 2402–2414, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Novak J, Julian BA, Mestecky J, Renfrow MB: Glycosylation of IgA1 and pathogenesis of IgA nephropathy. Semin Immunopathol 34: 365–382, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J: Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest 104: 73–81, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J: Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao N, Hou P, Lv J, Moldoveanu Z, Li Y, Kiryluk K, Gharavi AG, Novak J, Zhang H: The level of galactose-deficient IgA1 in the sera of patients with IgA nephropathy is associated with disease progression. Kidney Int 82: 790–796, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthoux F, Suzuki H, Thibaudin L, Yanagawa H, Maillard N, Mariat C, Tomino Y, Julian BA, Novak J: Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol 23: 1579–1587, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mestecky J, Raska M, Julian BA, Gharavi AG, Renfrow MB, Moldoveanu Z, Novak L, Matousovic K, Novak J: IgA nephropathy: Molecular mechanisms of the disease. Annu Rev Pathol 8: 217–240, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Raposo B, Dobritzsch D, Ge C, Ekman D, Xu B, Lindh I, Förster M, Uysal H, Nandakumar KS, Schneider G, Holmdahl R: Epitope-specific antibody response is controlled by immunoglobulin V(H) polymorphisms. J Exp Med 211: 405–411, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novak J, Moldoveanu Z, Julian BA, Raska M, Wyatt RJ, Suzuki Y, Tomino Y, Gharavi AG, Mestecky J, Suzuki H: Aberrant glycosylation of IgA1 and anti-glycan antibodies in IgA nephropathy: Role of mucosal immune system. Adv Otorhinolaryngol 72: 60–63, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Prabakaran P, Zhu Z, Chen W, Gong R, Feng Y, Streaker E, Dimitrov DS: Origin, diversity, and maturation of human antiviral antibodies analyzed by high-throughput sequencing. Front Microbiol 3: 277, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan T, Li J, Zhang MY: A single mutation turns a non-binding germline-like predecessor of broadly neutralizing antibody into a binding antibody to HIV-1 envelope glycoproteins. MAbs 3: 402–407, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vale AM, Kapoor P, Skibinski GA, Elgavish A, Mahmoud TI, Zemlin C, Zemlin M, Burrows PD, Nobrega A, Kearney JF, Briles DE, Schroeder HW Jr: The link between antibodies to OxLDL and natural protection against pneumococci depends on D(H) gene conservation. J Exp Med 210: 875–890, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim N, Jinks-Robertson S: Transcription as a source of genome instability. Nat Rev Genet 13: 204–214, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laffleur B, Denis-Lagache N, Péron S, Sirac C, Moreau J, Cogné M: AID-induced remodeling of immunoglobulin genes and B cell fate. Oncotarget 5: 1118–1131, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storb U: Why does somatic hypermutation by AID require transcription of its target genes? Adv Immunol 122: 253–277, 2014 [DOI] [PubMed] [Google Scholar]
- 18.Nagaoka H, Tran TH, Kobayashi M, Aida M, Honjo T: Preventing AID, a physiological mutator, from deleterious activation: regulation of the genomic instability that is associated with antibody diversity. Int Immunol 22: 227–235, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG: Two levels of protection for the B cell genome during somatic hypermutation. Nature 451: 841–845, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Adhikary R, Yu W, Oda M, Zimmermann J, Romesberg FE: Protein dynamics and the diversity of an antibody response. J Biol Chem 287: 27139–27147, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghunathan G, Smart J, Williams J, Almagro JC: Antigen-binding site anatomy and somatic mutations in antibodies that recognize different types of antigens. J Mol Recognit 25: 103–113, 2012 [DOI] [PubMed] [Google Scholar]
- 22.Brooks CL, Schietinger A, Borisova SN, Kufer P, Okon M, Hirama T, Mackenzie CR, Wang LX, Schreiber H, Evans SV: Antibody recognition of a unique tumor-specific glycopeptide antigen. Proc Natl Acad Sci U S A 107: 10056–10061, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foreman AL, Van de Water J, Gougeon ML, Gershwin ME: B cells in autoimmune diseases: insights from analyses of immunoglobulin variable (Ig V) gene usage. Autoimmun Rev 6: 387–401, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiryluk K, Novak J, Gharavi AG: Pathogenesis of immunoglobulin A nephropathy: Recent insight from genetic studies. Annu Rev Med 64: 339–356, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C,, Viola BF, Dallera N, Del. Vecchio L, Barlassina C, Salvi E, Amoroso A, Bertinetto F, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Santoro D, Pani A, Feriozzi S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Maixnerová D, Tesar V, Eitner F, Rauen T, Floege J, Nagy J, Mucha K, Pączek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG: Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Placzek WJ, Makita Y, Renfrow MB, Julian BA, Suzuki Y, Novak J, Suzuki H: Serum levels of galactose-deficient IgA1 in patients with IgA nephropathy correlate with serum levels of IgG autoantibodies. J Am Soc Nephrol 26: 258A, 2015. 25060060 [Google Scholar]
- 27.Kil LP, Hendriks RW: Aberrant B cell selection and activation in systemic lupus erythematosus. Int Rev Immunol 32: 445–470, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP: Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T: The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med 188: 2151–2162, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemi MH, Takkinen K, Amundsen LK, Söderlund H, Rouvinen J, Höyhtyä M: The testosterone binding mechanism of an antibody derived from a naïve human scFv library. J Mol Recognit 24: 209–219, 2011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.