Abstract
TNF superfamily member 13 (TNFSF13) has been identified as a susceptibility gene for IgA nephropathy in recent genetic studies. However, the role of TNFSF13 in the progression of IgA nephropathy remains unresolved. We evaluated two genetic polymorphisms (rs11552708 and rs3803800) and plasma levels of TNFSF13 in 637 patients with IgA nephropathy, and determined the risk of ESRD according to theses variable. Neither of the examined genetic polymorphisms associated with a clinical outcome of IgA nephropathy. However, high plasma levels of TNFSF13 increased the risk of ESRD. To explore the causal relationship and underlying mechanism, we treated B cells from patients (n=21) with or without recombinant human TNFSF13 (rhTNFSF13) and measured the expression of IgA and galactose-deficient IgA (GdIgA) using ELISA and flow cytometry. Treatment with rhTNFSF13 significantly increased the total IgA level among B cells, and TNFSF13 receptor blockade abrogated this increase. Furthermore, the absolute levels of GdIgA increased with rhTNFSF13 treatment, but the total IgA-normalized levels did not change. Both RNA sequencing and quantitative PCR results showed that rhTNFSF13 did not alter the expression of glycosyltransferase enzymes. These results suggest that high plasma TNFSF13 levels associate with a worse prognosis of IgA nephropathy through the relative increase in GdIgA levels.
Keywords: IgA nephropathy, outcomes, lymphocytes, glomerulonephritis
IgA nephropathy (IgAN) is the most common form of primary glomerulonephritis globally.1 IgAN has certain characteristics with a highly variable clinical course and prognosis: ESRD develops in as many as 30% of patients after 20–30 years, but a substantial proportion of patients have a sustained clinical remission or asymptomatic urinary abnormality alone.2,3 In this regard, a personalized approach to treatment is essential. However, current guidelines for IgAN treatment remain unsatisfactory in the context of personalization4 because there have been few predictive biomarkers identified that are involved in the pathophysiology or useful as disease-specific therapeutic targets.
Substantial progress has been achieved in understanding the pathophysiology of IgAN, which includes the production of galactose-deficient IgA (GdIgA), the formation of an antibody against GdIgA, the deposition of immune complexes, and the activation of intrarenal tissues.5 Recent genome-wide association studies (GWASs) provide scientists with an opportunity to move research vertically from its current state, given the difficulty of knowing the detailed pathophysiology and potential therapeutic targets of IgAN.6–9 With this in mind, GWASs that include Asian populations have discovered new susceptibility loci that are implicated in innate and mucosal immunity.8,9 Among the significant signals that have been identified, tumor necrosis factor superfamily 13 (TNFSF13), which encodes a proliferation-inducing ligand (APRIL), is thought to contribute to the progression of IgAN because of the following issues. TNFSF13 plays a fundamental role in the survival and IgA class switch recombination of B cells,10,11 implying that TNFSF13 may be responsible for pathogenic B cells and thus for the progression of IgAN. Additional research on the potential relationship between mucosal stimulation and the progression of IgAN suggested our hypothesis because both the production of TNFSF13 and the effect of TNFSF13 on B cells are necessary for mucosal homeostasis.9,12,13
In this study, we aimed to address whether TNFSF13 has a relationship with the progression of IgAN. There was a significant and independent relationship between high plasma TNFSF13 and the progression of IgAN. To explore the causal relationship and underlying mechanism (e.g., whether TNFSF13 alters the glycosylation of IgA), we further analyzed kidney tissues and B cells isolated from patients.
Results
Baseline Characteristics
Among the total of 637 IgAN patients recruited, SNPs and plasma levels of TNFSF13 at the time of kidney biopsy were available for 515 and 410 patients, respectively. Table 1 shows the baseline characteristics of the cohorts according to the available data. We designed two healthy cohorts for SNP (n=1068) and plasma (n=163) analyses. All healthy individuals in these groups did not have hypertension, diabetes, kidney dysfunction, or urinary abnormalities. Data on the baseline characteristics of healthy cohorts are available in Supplemental Table 1.
Table 1.
Characteristics of patients with IgA nephropathy at the time of the kidney biopsy
| Parameters | SNP Cohort (n=515) | Plasma Cohort (n=410) | Total (n=637) |
|---|---|---|---|
| Age, yr | 36.6±13.9 | 37.7±13.7 | 36.5±13.7 |
| Male sex, % | 48.7 | 43.4 | 47.4 |
| Current smoker, % | 12.2 | 11.2 | 11.7 |
| Hypertension, % | 27.0 | 28.0 | 26.4 |
| Diabetes mellitus, % | 2.4 | 3.7 | 2.8 |
| Autoimmune disease, % | 4.5 | 5.6 | 5.0 |
| Serum creatinine (mg/dl)a | 1.0 (0.83–1.33) | 1.0 (0.80–1.21) | 1.0 (0.81–1.30) |
| eGFR (ml/min per 1.73 m2) | 78.7±3.1 | 83.9±2.9 | 80.3±3.1 |
| Positive HBs Ag, % | 3.9 | 3.3 | 3.8 |
| Positive HCV Ab,% | 0.4 | 0.5 | 0.5 |
| Proteinuria, % | |||
| – | 12.6 | 19.3 | 15.3 |
| ± | 10.0 | 12.4 | 10.3 |
| 1+ | 16.4 | 18.0 | 16.1 |
| 2+ | 34.3 | 30.0 | 33.5 |
| ≥3+ | 26.7 | 20.2 | 24.8 |
| Hematuria, % | |||
| – | 2.4 | 3.2 | 2.7 |
| ± | 3.6 | 2.7 | 3.4 |
| 1+ | 9.4 | 6.3 | 8.1 |
| 2+ | 25.3 | 22.7 | 25.0 |
| ≥3+ | 59.3 | 65.1 | 60.9 |
| Follow-up period (months)a | 49 (32–85) | 41 (29–57) | 48 (31–79) |
SNP, single-nucleotide polymorphism; HBs Ag, surface antigen of the hepatitis B virus; HCV Ab, anti-hepatitis C virus antibody.
Data are expressed as the median (interquartile range) when the data distribution was skewed.
Genotypic Evaluation of TNFSF13 SNPs and Clinical Outcomes
Two loci in TNFSF13 (i.e., rs11552708 and rs3803800) were selected to test for an association between TNFSF13 with IgAN progression. We selected these loci on the basis of previous studies and minor allele frequencies in Asians.8,9,14,15 The major and minor alleles and genotype frequencies are described in Supplemental Table 2. Neither of the SNPs violated Hardy-Weinberg equilibrium (P>0.05). The AA genotype of rs3803800 displayed a marginal association with IgAN susceptibility (odds ratio for AA [versus others], 1.40 [95% confidence interval, 1.02 to 1.92]; P<0.04), whereas none of the rs11552708 genotypes displayed an association (Supplemental Table 3). We additionally retrieved serum IgA levels of IgAN patients, and the patient group with AA genotype of rs3803800 had higher serum IgA levels than other genotype groups (Supplemental Table 4). These results are consistent with the previous GWAS results.8,9
However, no significant association was observed when the risk of ESRD was assessed between the genotypic groups at the SNPs (Supplemental Figure 1, A and B). Other outcomes, such as the doubling of serum creatinine, were also similar between the genotype groups (data not shown).
Plasma TNFSF13 Levels and Clinical Outcomes
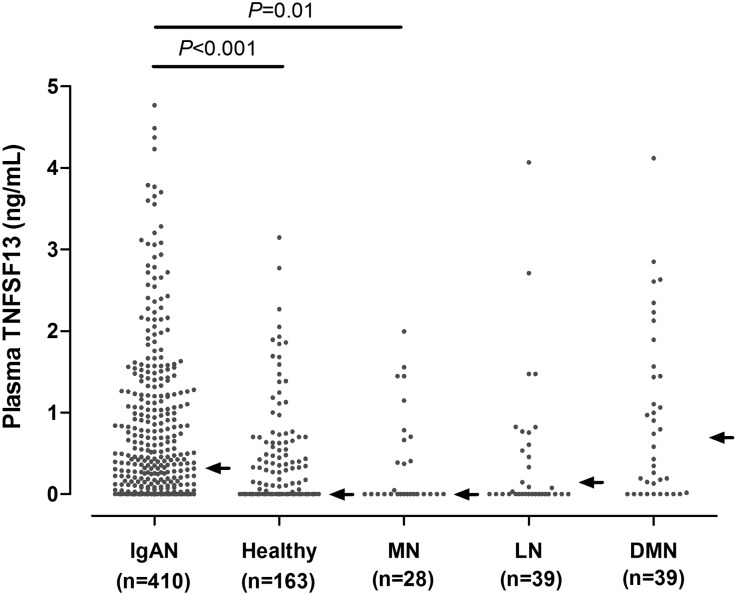
Among the IgAN patients, 31.2% had an undetectable level of TNFSF13. The median value in these patients was 0.32 ng/ml (interquartile range, 0–1.202 ng/ml), and the mean value was 1.86±11.69 ng/ml. The level of TNFSF13 in patients with IgAN was significantly higher than in healthy subjects (median, 0 ng/ml [interquartile range, 0–0.448 ng/ml]; mean, 0.37±0.79 ng/ml) (Figure 1). The discrepancy in TNFSF13 levels between patients and healthy individuals had the same trend with previous study results.16 When comparing the TNFSF13 levels with those of disease controls who had comparable eGFRs (except diabetic nephropathy), the plasma levels of patients with IgAN (median, 0.32 ng/ml; IQR, 0–1.202 ng/ml) were higher than those of patients with membranous nephropathy (median, 0 ng/ml; IQR, 0–0.673 ng/ml) but were not different from those of lupus nephritis patients (median, 0.1 ng/ml; IQR, 0–0.823 ng/ml) and patients with diabetic nephropathy (median, 0.74 ng/ml; IQR, 0.016–1.892 ng/ml). Their TNFSF13 levels were inversely related with baseline eGFR values except for the cases of membranous nephropathy (Supplemental Table 5). When analyzing the correlation between SNPs and plasma TNFSF13 levels, there were no differences in plasma TNFSF13 levels among genotypes (Supplemental Figure 2).
Figure 1.
Comparison of plasma TNFSF13 levels among patients with IgA nephropathy, healthy individuals, and patients with other kidney diseases. Arrows indicate median values. DMN, diabetic nephropathy; IgAN, IgA nephropathy; LN, lupus nephritis; MN, membranous nephropathy.
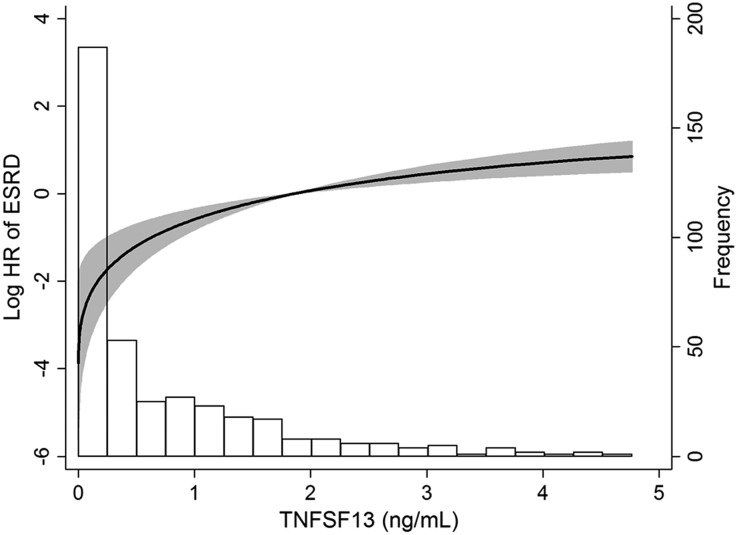
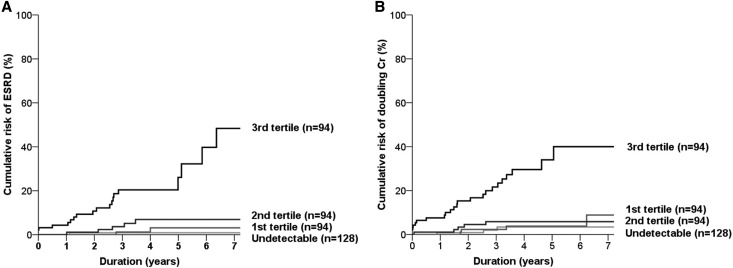
Interestingly, the risk of ESRD varied depending on the plasma TNFSF13 levels (Figure 2). Therefore, we divided patients with IgAN into four groups as follows (Figure 3): the undetectable group (n=128) and three tertile groups, each comprised of a tertile with detectable TNFSF13 levels (each n=94). ESRD risk was significantly different between groups (P<0.001) (Figure 3A). In particular, most ESRD events developed in the third-tertile group. When stepwise multivariate models were conducted to determine the independent relationships, only the third-tertile group was significantly related to high risk of ESRD, regardless of the levels of creatinine and proteinuria (Table 2). Doubling of serum creatinine was also more frequent in the third tertile group than in the undetectable group (Figure 3B and Table 2). The predictive value of TNFSF13 for outcomes remained significant, even when adjusting for eGFR and all other covariates in the analyses.
Figure 2.
Nonlinear increase in ESRD risk according to plasma TNFSF13 levels in 410 patients with IgA nephropathy. Bar represents the histogram according to patient number. HR, hazard ratio.
Figure 3.
Patients with high plasma TNFSF13 levels show worse outcomes of disease. Cumulative risk curves for ESRD (A) and doubling of serum creatinine (B) according to groups determined by plasma levels of TNFSF13. Cr, creatinine.
Table 2.
Risk analyses for ESRD and doubling of serum creatinine according to plasma TNFSF13 levels
| Model One | Model Two | Model Three | |||||
|---|---|---|---|---|---|---|---|
| Group | Outcome | HR (95% CI) | P Value | HR (95% CI) | P Value | HR (95% CI) | P Value |
| Undetectable (n=128) | ESRD | 1 (reference) | 1 (reference) | 1 (reference) | |||
| First tertile (n=94) | 2.86 (0.297 to 27.632) | 0.36 | 3.87 (0.393 to 38.147) | 0.25 | 4.52 (0.395 to 51.635) | 0.23 | |
| Second tertile (n=94) | 5.86 (0.684 to 50.199) | 0.11 | 6.60 (0.751 to 57.933) | 0.09 | 6.974 (0.665 to 73.188) | 0.17 | |
| Third tertile (n=94) | 28.65 (3.835 to 214.076) | 0.001 | 9.47 (1.176 to 76.315) | 0.04 | 10.67 (1.129 to 100.770) | 0.04 | |
| Undetectable (n=128) | DCr | 1 (reference) | 1 (reference) | 1 (reference) | |||
| First tertile (n=94) | 1.42 (0.315 to 6.347) | 0.65 | 1.87 (0.407 to 8.623) | 0.42 | 2.20 (0.465 to 10.350) | 0.32 | |
| Second tertile (n=94) | 2.00 (0.478 to 8.389) | 0.34 | 2.29 (0.531 to 9.886) | 0.27 | 2.38 (0.599 to 13.167) | 0.26 | |
| Third tertile (n=94) | 11.97 (3.591 to 39.871) | <0.001 | 5.21 (1.428 to 19.034) | 0.01 | 5.31 (1.377 to 20.464) | 0.01 | |
Model one: unadjusted for covariates.
Model two: adjusted for age, sex, baseline creatinine, proteinuria, and hematuria.
Model three: adjusted for covariates in model one plus smoking, hypertension, diabetes, autoimmune disease, hepatitis B antigen, anti-hepatitis C antibody, and the use of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers or steroids.
HR, hazard ratio; 95% CI, confidence interval; DCr, doubling of serum creatinine.
For a sensitivity analysis, we reviewed patients’ biopsy slides (n=253) according to the Oxford classification17 and compared pathologic variables among the above four groups (Supplemental Table 6). The third-tertile group showed aggressive pathology, particularly in the context of endocapillary hypercellularity and interstitial fibrosis/tubular atrophy (P<0.05). When additionally comparing the serum IgA levels between TNFSF13 groups, the third-tertile group had higher IgA levels than the undetectable group (Supplemental Table 4).
These results indicated that there was a threshold point of plasma TNFSF13 that predicted outcomes. Accordingly, we calculated the threshold point associated with the increasing risk of 5-year ESRD using a receiver operating characteristic curve. The area under the curve was 0.823 (0.744–0.902) (Supplemental Figure 3). The adjusted threshold point was 0.797 ng/ml, on the basis of the Youden index method.
Ectopic Expression of TNFSF13 in Kidney Tissue
Stromal tissues (e.g., intestinal epithelial cells) expressing TNFSF13 are important in the crosstalk with B cells.12,18 Intriguingly, TNFSF13 was expressed in the glomeruli and tubulointerstitium of patients with lupus nephritis.14 Thus, we investigated whether TNFSF13 is also expressed in kidney tissues of patients with IgAN (n=24) and compared TNFSF13 expression in these patients with healthy individuals (n=6) and patients with lupus nephritis (n=8). As previous results indicated, immunohistochemical staining for TNFSF13 was significantly stronger in both tubulointerstitium and glomeruli isolated from patients with lupus nephritis compared with healthy individuals and patients with IgAN. However, no significant staining differences were observed between healthy individuals and patients with IgAN (Supplemental Figure 4, A and B). Similarly, TNFSF13 mRNA expression in microdissected tubulointerstitium and glomeruli were not different between patients with IgAN and healthy individuals, regardless of the kidney compartment, whereas the tubulointerstitium of lupus nephritis patients expressed higher levels of TNFSF13 mRNA than those from healthy individuals (Supplemental Figure 4C).
B Cell Stimulation with TNFSF13
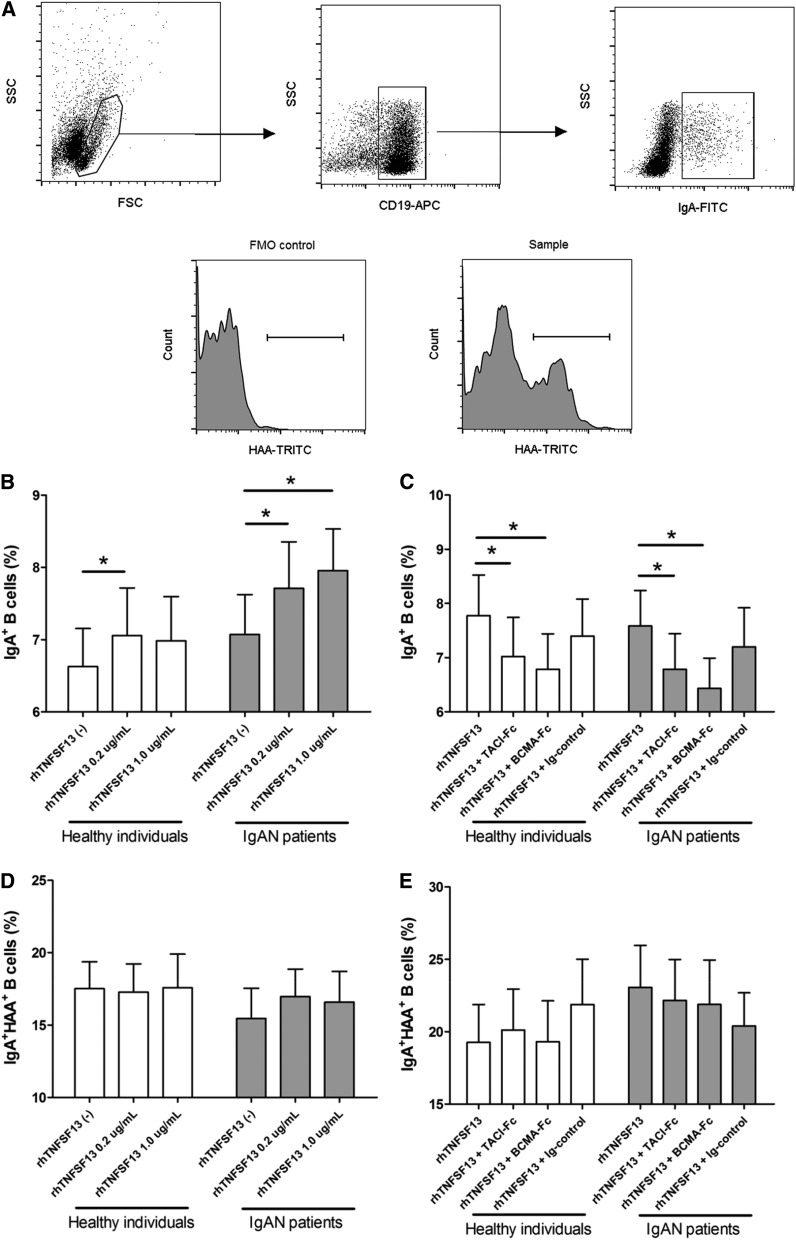
To address the relationship between TNFSF13 and extrarenal pathophysiologies related to IgAN (i.e., the production of GdIgA), we stimulated primary B cells isolated from IgAN patients (n=21) and healthy individuals (n=17) with recombinant human TNFSF13 (rhTNFSF13). GdIgA-producing B cells were identified by treating B cells with GalNAc-specific lectin from Helix aspersa. Before to the stimulation experiments, there was no difference in transmembrane activator and calcium-modulator and cyclophilin ligand interactor (TACI) or B-cell maturation antigen (BCMA) expression between healthy individuals and IgAN patients. However, BCMA expression in the B-cell line was lower than in primary B cells. (Supplemental Figure 5). The flow cytometric analysis results indicated that rhTNFSF13 modestly but significantly increased the proportion of IgA+ cells among B cells from both IgAN patients and healthy individuals, but no dose-dependent effect was observed between 0.2 and 1.0 µg/ml (Figure 4B). Thus, we used 0.2 µg/ml rhTNFSF13 in the following blocker experiments. Blockers for TNFSF13 receptors, such as recombinant human Fc chimeras for TACI (TACI-Fc) and BCMA (BCMA-Fc), significantly abrogated these increases (Figure 4C). In contrast, neither TNFSF13 nor its receptor blockers had an effect on the proportion of IgA+HAA+ B cells in either IgAN patients or healthy individuals (Figure 4, D and E).
Figure 4.
Flow cytometric analyses of B cells activated in culture. (A) Representative gating strategy for IgA+ B cells (upper) and histograms of IgA+HAA+ B cells (lower). (B) IgA-producing B cells according to rhTNFSF13 treatment. (C) IgA-producing B cells according to treatment with receptor blockers. (D) GdIgA-producing B cells among IgA+ B cells according to rhTNFSF13 treatment. (E) GdIgA-producing B cells among IgA+ B cells according to treatment with receptor blockers. In the analysis of receptor blockers, the dose of rhTNFSF13 used was 0.2 µg/ml. The doses of the receptor blockers were 5 µg/ml. Ig (5 µg/ml) indicates the isotype-matched IgG for receptor blockers. The results in the isotype control group did not differ from those of the untreated control group. *P<0.05. FMO, fluorescence minus one.
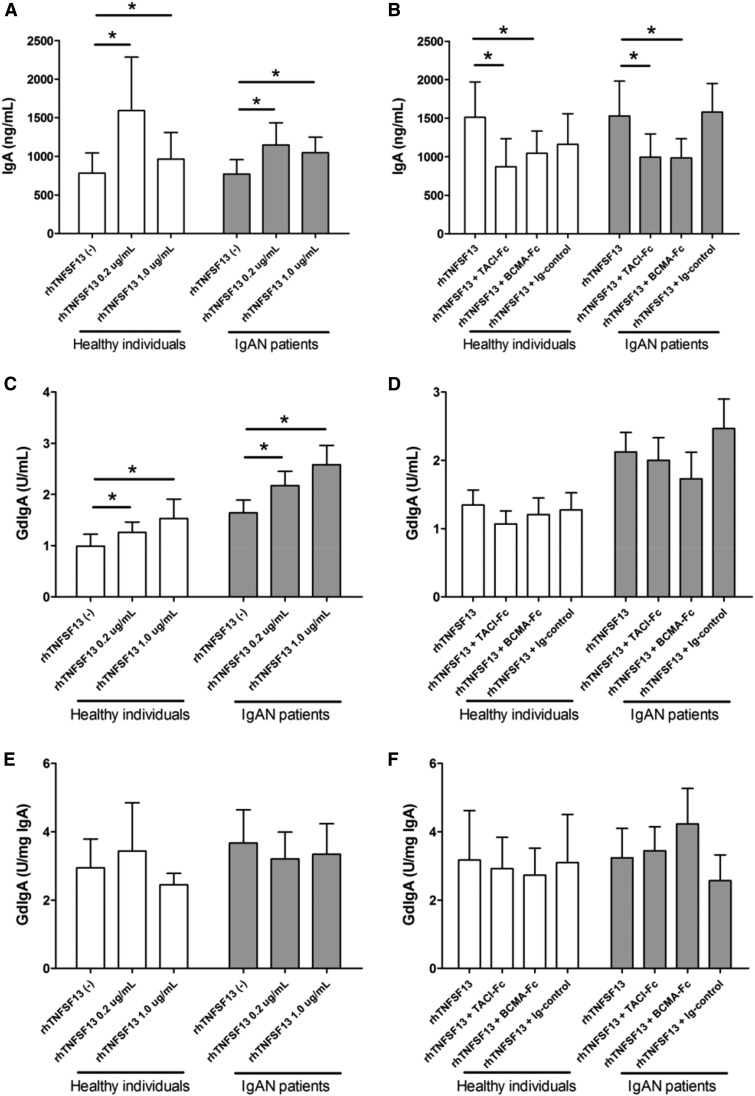
We measured the supernatant levels of IgA and GdIgA to determine the cumulative production of these antibodies in response to rhTNFSF13. Supernatant IgA levels were significantly higher in the rhTNFSF13-treated groups than in the nontreated groups (Figure 5A). These increases were significantly inhibited by the use of receptor blockers (Figure 5B), suggesting that the specific interaction between TNFSF13 and its receptor is involved in IgA production. In the case of GdIgA, the absolute levels were increased by the use of rhTNFSF13, whereas the change was not prominent depending on the use of receptor blockers (Figure 5, C and D). The total IgA-normalized GdIgA levels were not altered after the treatment with rhTNFSF13 or receptor blockers (Figure 5, E and F). These trends were similar between patients with IgAN and healthy individuals.
Figure 5.
Amounts of total IgA and absolute GdIgA, but not relative GdIgA are altered by the rhTNFSF13 treatment. ELISA results for total IgA (A, B), absolute GdIgA (C, D), and total IgA-normalized GdIgA (E, F) according to treatment with rhTNFSF13 (A, C, E) or receptor blockers (B, D, F). In the analysis with receptor blockers, the dose of rhTNFSF13 was 0.2 µg/ml. The doses of receptor blockers and isotype IgG were 5 µg/ml. The results in the isotype control group were not different from the group that was not treated with receptor blockers. *P<0.05.
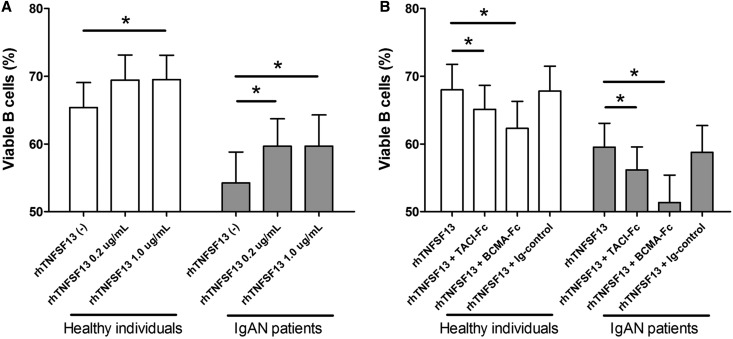
Subsequently, we conducted annexin V staining to examine changes in the proportion of viable B cells induced by treatment with rhTNFSF13 (Figure 6). Baseline B cell viability in patients with IgAN was relatively low compared with healthy individuals. Viable B cells were more prevalent in the rhTNFSF13-treated groups than in the nontreated groups. This effect of rhTNFSF13 was diminished by the use of receptor blockers.
Figure 6.
Prolongation of B cell survival is dependent on the treatment with rhTNFSF13 or its receptor blockers. Proportion of viable B cells according to treatment with TNFSF13 (A) or receptor blockers (B). *P<0.05.
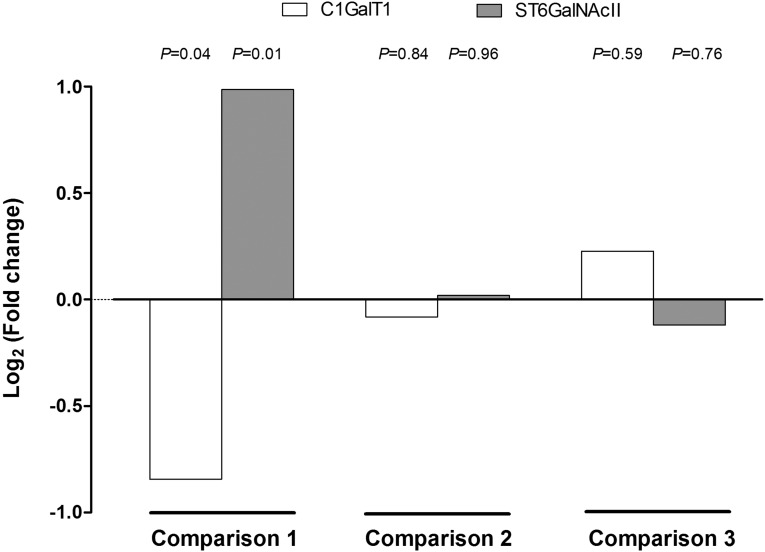
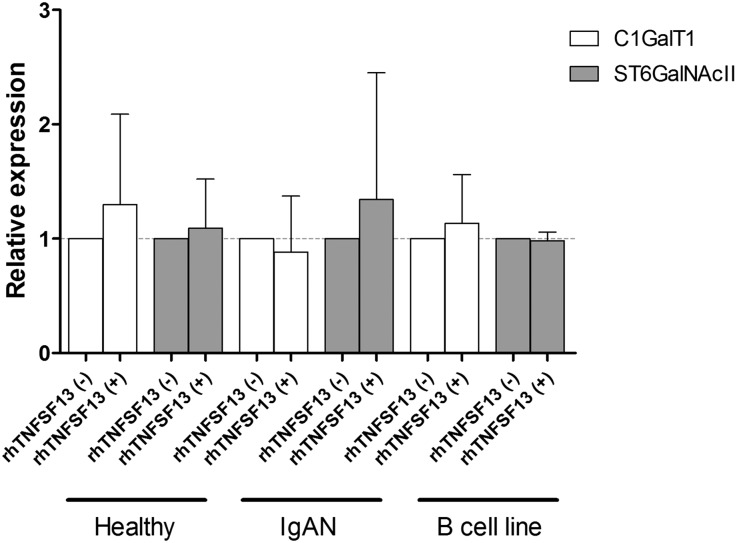
To identify changes in comprehensive gene expression profiles after rhTNFSF13 treatment, RNA sequencing analyses were performed on B cells isolated from patients or healthy individuals. Figure 7, A and B show the genes that were differentially expressed between the rhTNFSF13 treatment and nontreatment groups. In IgAN patients, B cell expression of SFTPB, RPS17L, PTGES, ACE, and MMP9 was increased and the expression of HPX, SLC39A5, PGC, PTGDS, and COL1A1 was decreased after rhTNFSF13 treatment. A list of all of the genes found to be significantly different after rhTNFSF13 treatment is provided in Supplemental Table 7, and its visualization using a volcano plot is shown in Supplemental Figure 6. Using the sequencing data, we explored the gene expression patterns of glycosyltransferase enzymes, such as C1GalT1 and ST6GalNAcII.5 As a result, rhTNFSF13 treatment failed to induce an alteration in the gene expression of glycosyltransferase enzymes in either patients with IgAN or healthy individuals, whereas there was a significant difference in the baseline expression of C1GalT1 and ST6GalNAcII between healthy individuals and patients with IgAN (Figure 7). Additionally, we performed gene set enrichment analysis to assess whether there was a significant overlap within the enriched gene sets.19 However, glycosyltransferase genes such as C1GalT1 and ST6GalNAcII were not included as a significant gene set; significantly intersecting gene sets (false discovery rate <0.1) are shown in Supplemental Table 8. Nevertheless, when the gene set enrichment analysis was conducted after focusing on the glycosyltransferase genes alone (Supplemental Table 9), these genes did not vary depending on TNFSF13 use (P=0.32 in patients with IgAN; P=0.49 in healthy individuals). To validate these data, we conducted real-time quantitative PCR using both primary human B cells and cells from the human lymphoma B-cell line that produces IgA1.20 The expression of C1GALT1 and ST6GalNAcII was not different between the rhTNFSF13-treated and nontreated groups (Figure 8).
Figure 7.
Changes in the expression of glycosyltransferase enzymes (i.e., C1GalT1 and ST6GalNAcII gene) in non-treated patients with IgA nephropathy (versus non-treated healthy individuals) (Comparison 1), rhTNFSF13-treated patients (versus non-treated patients) (Comparison 2), and rhTNFSF13-treated healthy individuals (versus non-treated healthy individuals) (Comparison 3).
Figure 8.
Real-time quantitative PCR for validating the RNA sequencing results, wherein the glycosyltransferase enzyme expression (i.e., C1GalT1 and ST6GalNAcII) was not altered by the rhTNFSF13 treatment in both primary B cells and the IgA-producing B-cell line. The B-cell line results were obtained from four independent experiments.
Discussion
IgAN itself has a multifarious prognosis that ranges from “not problematic throughout the lifetime” to “aggressive progression into ESRD”.2,3 However, there is an inability to use a personalized approach when treating IgAN patients because of insufficient knowledge, particularly with regard to the prognostic biomarkers related to the progression of this disease. With this in mind, this study provides several findings with clinical implications.4 First, TNFSF13 was a successful predictor of ESRD or other outcomes, regardless of the presence of serum creatinine and urinary abnormalities. Second, TNFSF13 was not overexpressed in the kidney, where the TNFSF13-related pathophysiology of kidney damage may be different between IgAN and other autoimmune nephritis diseases, such as lupus nephritis. Third, the aggressive prognosis in the high-plasma-TNFSF13 group may not be attributable to a direct relationship with IgA glycosylation, but it may be related to the relative increase in the GdIgA amount by previously described functions, such as the prolongation of survival in pathogenic B cells.
To achieve personalized therapy for patients with differing clinical outcomes, establishment of a significant biomarker or predictor is essential to clinical practice. Currently, high serum creatinine and proteinuria are the most important parameters used to predict IgAN outcomes and select a treatment regimen.21 However, these parameters actually reflect disease progression, not the origin of the disease itself. Furthermore, a state that lacks these parameters cannot guarantee the benign course of disease.22 In this study, plasma TNFSF13 was associated with baseline kidney functions (i.e., eGFR). In this regard, high plasma TNFSF13 levels may be a mere reflection of decreased kidney function. However, we further found that plasma TNFSF13 was associated with ESRD risk independent of several parameters, including serum creatinine (or eGFR) and proteinuria. Particularly, there was a cut-off value for prediction (0.797 ng/ml in the present cohort); this characteristic provides a further clinical indication that plasma TNFSF13 is a useful and independent biomarker. Future studies will need to address whether high TNFSF13 levels also correlate with subsequent ESRD risk in other kidney diseases, such as lupus nephritis and diabetic nephropathy.
The clinical data in this study did not provide causality for the identified relationship or underlying mechanism. We attempted to explore this issue further using B cells isolated from patients. IgA production or B cell survival varied depending on rhTNFSF13 stimulation, whereas blockers for TNFSF13 receptors yielded the opposite trends. These effects are well documented and fundamental to B-cell biology.10,11 However, the question of whether TNFSF13 alters glycosylation (i.e., under-galactosylation) in the hinge region of IgA has not been studied. Previous studies revealed that cytokines, including interleukin-4 and interleukin-6, increased the production of GdIgA in Epstein-Barr virus-immortalized B cells from IgAN patients; and it is intriguing that interleukin-6 also increased the GdIgA level in healthy individuals.23 In contrast to these findings regarding cytokines, our data, based on flow cytometry, real-time quantitative PCR, and RNA sequencing, do not support the notion that TNFSF13 itself changes the glycosylated structure of IgA. However, when the absolute and relative values of GdIgA in ELISA and the flow cytometric measurements of the viability of B cells were observed, TNFSF13 may be involved in the worst outcome of IgAN through the relative increase of GdIgA by prolongation of B-cell survival. TNFSF13 and its receptors (data not shown) were similarly expressed in the kidney tissues of IgAN patients and healthy individuals, which suggests that the role of TNFSF13 may not be part of intrarenal processes.
The current therapy for IgAN patients includes the use of blockers of the renin-angiotensinogen system and steroids,24,25 which can result in several side-effects, particularly after long-term use (e.g., decreased kidney function, anemia, hypotension, osteoporosis, and steroid dependency).26,27 This issue hampers the general utilization of these therapies. Other immunomodulatory agents and tonsillectomy are not used as a standard regimen, although these therapies may provide a benefit to patients in certain disease states.28,29 The lack of understanding of the wide variability in prognosis in IgAN patients is a real problem because it prevents potential pathways that may be targets for IgAN intervention from being discovered. Recently, TACI-Fc, a receptor blocker for TNFSF13, has been developed and investigated for its effect on autoimmune disease, although both optimistic and negative aspects related to this treatment have coexisted until now.30,31 Although the results of our work do not provide a definitive reason to use a TNFSF13 blocker in patients with IgAN, these data may be a foundation for future strategies related with a drug intervention for TNFSF13 in progressive IgAN cases.
In addition to the hypothesis-driven approach to evaluate the role of TNFSF13 on IgAN, we used RNA sequencing to produce unbiased data showing gene expression after treatment with TNFSF13. A total of 29 genes have been identified to be differentially expressed in B cells from IgAN patients after TNFSF13 treatment. Although most of these genes have not yet been linked to IgAN, several of them, including PTGES, MMP9, and ACE, have been associated with innate immunity or inflammation and may thereby potentially affect the progression of IgAN.32–34 This aspect is one of the strengths of these data that may be helpful in future studies of IgAN or B-cell biology.
Weaknesses of the clinical results in this study include a lack of causality and inevitable missing values in certain covariates. This experiment focused primarily on GdIgA in B cells, but not on other pathophysiological steps, anti-GdIgA autoantibody production, and intrarenal processes. The conclusions of the experiments were drawn primarily on the basis of results from a mixed B-cell population rather than GdIgA B cells, which may affect the interpretation of the sequencing as well as other experimental data. Additionally, power limitation due to current sample size might underlie our negative results.
This study has implications with regard to the clinical and experimental annotation of a susceptibility locus identified by genome-wide association studies. Although replication of the data in an independent cohort is needed, TNFSF13 may be a successful biomarker for IgAN progression, and its association with a poor outcome may be attributable to the relative increase of GdIgA by its known role in B cell survival rather than the alteration of glycosylation structure. The ability to specifically identify and potentially target biomarkers related to aggressive outcomes in IgAN will provide an important basis for developing personalized therapies with improved efficacy and long-term safety. To this end, the data from this study support the future investigation of targeted therapies to facilitate the use of immune regulation to attenuate the progression of IgAN.
Concise Methods
The study protocol complies with the Declaration of Helsinki and received full approval from the institutional review boards at Seoul National University Hospital (no. H-1306–108–500). Risks of clinical outcomes (e.g., ESRD and doubling of serum creatinine) were compared between the genetic polymorphisms (rs11552708 and rs3803800) or by the plasma levels of TNFSF13 among 637 IgAN patients. To evaluate the expression of TNFSF13 in kidney tissue, we used immunohistochemistry and real-time quantitative PCR to compare kidney tissues from patients with IgAN (n=24) with tissues from healthy individuals (n=6) and patients with lupus nephritis (n=8). B cells from patients (n=21) and healthy individuals (n=17) were stimulated with or without rhTNFSF13. Standard protocols were used for cell culture experiments, immunohistochemistry, real-time quantitative PCR, flow cytometry, ELISA, RNA sequencing, and statistical analyses. Detailed methods are provided in the Supplemental Material (Supplemental Table 10).
Disclosures
None.
Supplementary Material
Acknowledgments
This study was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, Ministry of Health and Welfare, Republic of Korea (HI10C1268). The biospecimens for IgAN patients were provided by the Seoul National University Hospital Human Biobank, a member of the National Biobank of Korea, which is supported by the Ministry of Health and Welfare, Republic of Korea. Clinical and SNP data of the Illumina cohort was provided by the Korean Genome Analysis Project (4845-301), the Korean Genome and Epidemiology Study (4851-302), and the Korea Biobank Project (KBP-2013-35), which were supported by the Korea Center for Disease Control, Republic of Korea.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015060677/-/DCSupplemental
References
- 1.Donadio JV, Grande JP: IgA nephropathy. N Engl J Med 347: 738–748, 2002 [DOI] [PubMed] [Google Scholar]
- 2.D’Amico G: Natural history of idiopathic IgA nephropathy: role of clinical and histological prognostic factors. Am J Kidney Dis 36: 227–237, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Reich HN, Troyanov S, Scholey JW, Cattran DC Toronto Glomerulonephritis Registry : Remission of proteinuria improves prognosis in IgA nephropathy. J Am Soc Nephrol 18: 3177–3183, 2007 [DOI] [PubMed] [Google Scholar]
- 4.Kidney Disease. Improving Global Outcomes (KDIGO) Glomerulonephritis Work Group : KDIGO Clinical Practice Guideline for Glomerulonephritis. Kidney Int Suppl 2: 139–274, 2012 [Google Scholar]
- 5.Suzuki H, Fan R, Zhang Z, Brown R, Hall S, Julian BA, Chatham WW, Suzuki Y, Wyatt RJ, Moldoveanu Z, Lee JY, Robinson J, Tomana M, Tomino Y, Mestecky J, Novak J: Aberrantly glycosylated IgA1 in IgA nephropathy patients is recognized by IgG antibodies with restricted heterogeneity. J Clin Invest 119: 1668–1677, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, Kumar A, Peden JF, Maxwell PH, Morris DL, Padmanabhan S, Vyse TJ, Zawadzka A, Rees AJ, Lathrop M, Ratcliffe PJ: HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol 21: 1791–1797, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiryluk K, Li Y, Sanna-Cherchi S, Rohanizadegan M, Suzuki H, Eitner F, Snyder HJ, Choi M, Hou P, Scolari F, Izzi C, Gigante M, Gesualdo L, Savoldi S, Amoroso A, Cusi D, Zamboli P, Julian BA, Novak J, Wyatt RJ, Mucha K, Perola M, Kristiansson K, Viktorin A, Magnusson PK, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boland A, Metzger M, Thibaudin L, Wanner C, Jager KJ, Goto S, Maixnerova D, Karnib HH, Nagy J, Panzer U, Xie J, Chen N, Tesar V, Narita I, Berthoux F, Floege J, Stengel B, Zhang H, Lifton RP, Gharavi AG: Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet 8: e1002765, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ, Sun LD, Sim KS, Li Y, Foo JN, Wang W, Li ZJ, Yin XY, Tang XQ, Fan L, Chen J, Li RS, Wan JX, Liu ZS, Lou TQ, Zhu L, Huang XJ, Zhang XJ, Liu ZH, Liu JJ: A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 44: 178–182, 2012 [DOI] [PubMed] [Google Scholar]
- 9.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerová D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Pączek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG: Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackay F, Schneider P, Rennert P, Browning J: BAFF AND APRIL: a tutorial on B cell survival. Annu Rev Immunol 21: 231–264, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Castigli E, Scott S, Dedeoglu F, Bryce P, Jabara H, Bhan AK, Mizoguchi E, Geha RS: Impaired IgA class switching in APRIL-deficient mice. Proc Natl Acad Sci U S A 101: 3903–3908, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, Knowles DM, Rescigno M, Cerutti A: Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity 26: 812–826, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Kiryluk K, Novak J: The genetics and immunobiology of IgA nephropathy. J Clin Invest 124: 2325–2332, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neusser MA, Lindenmeyer MT, Edenhofer I, Gaiser S, Kretzler M, Regele H, Segerer S, Cohen CD: Intrarenal production of B-cell survival factors in human lupus nephritis. Mod Pathol 24: 98–107, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Kawasaki A, Tsuchiya N, Ohashi J, Murakami Y, Fukazawa T, Kusaoi M, Morimoto S, Matsuta K, Hashimoto H, Takasaki Y, Tokunaga K: Role of APRIL (TNFSF13) polymorphisms in the susceptibility to systemic lupus erythematosus in Japanese. Rheumatology (Oxford) 46: 776–782, 2007 [DOI] [PubMed] [Google Scholar]
- 16.McCarthy DD, Kujawa J, Wilson C, Papandile A, Poreci U, Porfilio EA, Ward L, Lawson MA, Macpherson AJ, McCoy KD, Pei Y, Novak L, Lee JY, Julian BA, Novak J, Ranger A, Gommerman JL, Browning JL: Mice overexpressing BAFF develop a commensal flora-dependent, IgA-associated nephropathy. J Clin Invest 121: 3991–4002, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts IS, Cook HT, Troyanov S, Alpers CE, Amore A, Barratt J, Berthoux F, Bonsib S, Bruijn JA, Cattran DC, Coppo R, D’Agati V, D’Amico G, Emancipator S, Emma F, Feehally J, Ferrario F, Fervenza FC, Florquin S, Fogo A, Geddes CC, Groene HJ, Haas M, Herzenberg AM, Hill PA, Hogg RJ, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Li LS, Li PK, Liu ZH, Mackinnon B, Mezzano S, Schena FP, Tomino Y, Walker PD, Wang H, Weening JJ, Yoshikawa N, Zhang H Working Group of the International IgA Nephropathy Network and the Renal Pathology Society : The Oxford classification of IgA nephropathy: pathology definitions, correlations, and reproducibility. Kidney Int 76: 546–556, 2009 [DOI] [PubMed] [Google Scholar]
- 18.Wells JM, Rossi O, Meijerink M, van Baarlen P: Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A 108[Suppl 1]: 4607–4614, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC: PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273, 2003 [DOI] [PubMed] [Google Scholar]
- 20.Serino G, Sallustio F, Cox SN, Pesce F, Schena FP: Abnormal miR-148b expression promotes aberrant glycosylation of IgA1 in IgA nephropathy. J Am Soc Nephrol 23: 814–824, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Floege J, Eitner F: Current therapy for IgA nephropathy. J Am Soc Nephrol 22: 1785–1794, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Vivante A, Afek A, Frenkel-Nir Y, Tzur D, Farfel A, Golan E, Chaiter Y, Shohat T, Skorecki K, Calderon-Margalit R: Persistent asymptomatic isolated microscopic hematuria in Israeli adolescents and young adults and risk for end-stage renal disease. JAMA 306: 729–736, 2011 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Raska M, Yamada K, Moldoveanu Z, Julian BA, Wyatt RJ, Tomino Y, Gharavi AG, Novak J: Cytokines alter IgA1 O-glycosylation by dysregulating C1GalT1 and ST6GalNAc-II enzymes. J Biol Chem 289: 5330–5339, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coppo R, Peruzzi L, Amore A, Piccoli A, Cochat P, Stone R, Kirschstein M, Linné T: IgACE: a placebo-controlled, randomized trial of angiotensin-converting enzyme inhibitors in children and young people with IgA nephropathy and moderate proteinuria. J Am Soc Nephrol 18: 1880–1888, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Pozzi C, Bolasco PG, Fogazzi GB, Andrulli S, Altieri P, Ponticelli C, Locatelli F: Corticosteroids in IgA nephropathy: a randomised controlled trial. Lancet 353: 883–887, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Schoolwerth AC, Sica DA, Ballermann BJ, Wilcox CS Council on the Kidney in Cardiovascular Disease and the Council for High Blood Pressure Research of the American Heart Association : Renal considerations in angiotensin converting enzyme inhibitor therapy: a statement for healthcare professionals from the Council on the Kidney in Cardiovascular Disease and the Council for High Blood Pressure Research of the American Heart Association. Circulation 104: 1985–1991, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Lv J, Xu D, Perkovic V, Ma X, Johnson DW, Woodward M, Levin A, Zhang H, Wang H TESTING Study Group : Corticosteroid therapy in IgA nephropathy. J Am Soc Nephrol 23: 1108–1116, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maes BD, Oyen R, Claes K, Evenepoel P, Kuypers D, Vanwalleghem J, Van Damme B, Vanrenterghem YF: Mycophenolate mofetil in IgA nephropathy: results of a 3-year prospective placebo-controlled randomized study. Kidney Int 65: 1842–1849, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Liu LL, Wang LN, Jiang Y, Yao L, Dong LP, Li ZL, Li XL: Tonsillectomy for IgA nephropathy: a meta-analysis. Am J Kidney Dis 65: 80–87, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Isenberg D, Gordon C, Licu D, Copt S, Rossi CP, Wofsy D: Efficacy and safety of atacicept for prevention of flares in patients with moderate-to-severe systemic lupus erythematosus (SLE): 52-week data (APRIL-SLE randomised trial). Ann Rheum Dis 74: 2006–2015, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kappos L, Hartung HP, Freedman MS, Boyko A, Radü EW, Mikol DD, Lamarine M, Hyvert Y, Freudensprung U, Plitz T, van Beek J ATAMS Study Group : Atacicept in multiple sclerosis (ATAMS): a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Neurol 13: 353–363, 2014 [DOI] [PubMed] [Google Scholar]
- 32.Remuzzi G, Perico N, Macia M, Ruggenenti P: The role of renin-angiotensin-aldosterone system in the progression of chronic kidney disease. Kidney Int Suppl 68: S57–S65, 2005 [DOI] [PubMed] [Google Scholar]
- 33.Parks WC, Wilson CL, López-Boado YS: Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol 4: 617–629, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Zhou A, Scoggin S, Gaynor RB, Williams NS: Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene 22: 2054–2064, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.