Abstract
The current study reports the successful application of a fast and efficient genetic screening system for common hearing loss (HL) genes based on SNPscan genotyping technology. Genetic analysis of 115 variants in common genes related to HL, GJB2, SLC26A4 and MT-RNR, was performed on 695 subjects with non-syndromic hearing loss (NSHL) from the Northern China. The results found that 38.7% (269/695) of cases carried bi-allelic pathogenic variants in GJB2 and SLC26A4 and 0.7% (5/695) of cases carried homoplasmic MT-RNR1 variants. The variant allele frequency of GJB2, SLC26A4 and MT-RNR1 was 19.8% (275/1390), 21.9% (304/1390), and 0.86% (6/695), respectively. This approach can explain ~40% of NSHL cases and thus is a useful tool for establishing primary molecular diagnosis of NSHL in clinical genetics.
Introduction
Hearing loss (HL) is one of the most common sensorineural disorders, affecting 1–2 neonates in every 1000 and the prevalence increases with age [1]. In fact, 50% to 60% of childhood HL can be attributed to genetic defects[1]. With at least 100 genes identified to date (http://heretidaryhearingloss.org/, updated in Sep. 2016), non-syndromic HL (NSHL, with no symptoms in other organs) is an extraordinarily heterogeneous trait. Autosomal recessive (AR) NSHL is typically pre-lingual and is present in ~80% of HL cases. GJB2 is (OMIM*121011) the most common gene involved in ARNSHL. In some populations, pathogenic GJB2 variants account for up to 50% of all ARNSHL cases [2]. SLC26A4 (OMIM*605646), the second most frequent gene involved in ARNSHL and is present in 4–10% of NSHL cases among different populations [3–5]. A defect in the mitochondrial gene, MT-RNR1, is associated with extreme sensitivity to aminoglycoside ototoxicity and accounts for maternally inherited NSHL [6, 7]. The traditional method for diagnosis of NSHL relied on Sanger sequencing. Although direct sequencing is the gold standard approach for genetic testing, it is expensive, time consuming and has low throughput. An SNPscan technique has been developed, using a patented SNP genotyping technology based on a double ligation reaction and multiplex fluorescence PCR, which is able to perform multiplex (48–192) genotyping for up to 192 kinds of SNPs[8]. This technique is uniquely suited to genetic screening for NSHL due to the extremely strong genetic heterogeneity of NSHL. In the current study, the majority of pathogenic variants of GJB2, SLC26A4 and MT-RNR1 were obtained and a primary genetic test for NSHL was designed using an SNPscan genotyping technique with the ability to simultaneously detect 115 different variants in GJB2 (n = 36), SLC26A4 (n = 77), and MT-RNR1 (n = 2), the common genes related to HL.
Materials and Methods
Subjects and Auditory Evaluation
A total of 695 affected subjects were recruited in 2014. Patients from the Department of Otolaryngology (n = 132), 3rd Hospital of Peking University (n = 132) and from 12 different rehabilitation centers for the deaf around the area of Beijing, China (n = 563) were included. This study was approved by the Ethics Committee of 3rd Hospital of Peking University (Approval number 20141212). Written informed consent was obtained from adult participants and from guardians on behalf of minors/children prior to their participation in the study. A comprehensive medical history was obtained from each subject via a questionnaire regarding onset age, evolution, degree and symmetry of hearing loss, hearing aids, cochlear implants, presence of tinnitus, pressure in the ears, vertigo, history of noise exposure and other relevant clinical manifestations. Each participant received a physical examination to exclude any additional distinguishing physical findings. A comprehensive auditory evaluation, including otoscopy, pure-tone audiometry (PTA, at frequencies from 500 to 4000Hz) and immittance testing was performed. The severity of HL was defined according to PTA as mild (26-40dBHL), moderate (41–70dBHL), severe (71-90dBHL) or profound (> 90dBHL). Middle ear pressure, ear canal volume and tympanic membrane mobility was evaluated according to immittance testing.
Screening of GJB2, SLC26A4 and MT-RNR1
Peripheral blood samples (2 ml) were drawn from all subjects for DNA extraction and genetic analysis. Genomic DNA was isolated from whole blood using a DNA extraction kit (Axygen Scientific Inc., Union City, CA, USA). A total of 115 variants of GJB2 (NM_004004, n = 36), SLC26A4 (NM_000441, n = 77) and MT-RNR1(NR_137294, n = 2) were included. Of these, 97 pathogenic variants (GJB2 = 18, SLC26A4 = 77, MT-RNR1 = 2) were pathogenic and 18 GJB2 variants were unclassified. All the 97 pathogenic variants of GJB2, SLC26A4 and MT-RNR1 genes included in this primary screening system were taken from deafness variant database (http://deafnessvariationdatabase.org) where clear indication of the pathogenicity associated with HL were given. Screening of these pathogenic variants in common HL genes was carried out to establish molecular diagnosis for HL cases. Genetic analysis was performed using a custom-built 48-Plex SNPscanTM Kit (Genesky Biotechnologies Inc., Shanghai, China).
Results
Based on the questionnaires, HL in 568 subjects was pre-lingual, HL in 47 subjects was post-lingual, and the age of HL onset in 80 subjects was uncertain. According to audiograms, the severity of sensorineural hearing loss was mild (n = 1), moderate (n = 15), severe (n = 69) and profound (n = 610). Thirteen subjects had a history of noise exposure and 60 subjects had a history of using aminoglycoside antibiotics (dosage uncertain), such as gentamicin and/or streptomycin. The mothers of 65 subjects suffered from possible viral infections, such as colds or urticaria, during the first 3 months of pregnancy. Comprehensive family medical histories and clinical examinations found no additional clinical abnormalities, such as cardiovascular disease, kidney disease, visual problems or neurological disorders, in any subject (Table 1).
Table 1. Clinical Information of 695 Chinese HL Subjects.
| Onset age | Hearing loss | Environmental factors | ||||||
|---|---|---|---|---|---|---|---|---|
| Pre-lingual | Post-lingual | Unknown | Mild | Moderate | Severe | Profound | Noise-exposure | Ototoxic drug |
| 568 | 47 | 80 | 1 | 15 | 69 | 610 | 13 | 60 |
GJB2
Pathogenic variants
Genetic analysis revealed that 17 subjects carried only one GJB2 mutant allele and 129 (18.6%, 129/695) subjects carried bi-allelic pathogenic variants in GJB2, including 57 homozygotes and 72 compound heterozygotes (Fig 1). Therefore, the allele frequency of pathogenic variants in GJB2 gene was 19.8% (275/1390). The allele frequency of c.235delC, the most prevalent pathogenic variant of GJB2, was 13.0% (181/1390), followed by c.299_300 del AT, c.257C > G, c.34_35insG and c.511_512insAACG with allele frequencies of 4.5% (63/1390), 0.58% (8/1390), 0.43% (6/1390) and 0.43% (6/1390), respectively. These 5 pathogenic variants account for 96.0% (264/275) of all GJB2 mutant alleles. In addition, c.235delC was a compound heterozygote, with 9 different pathogenic variants, in 73 patients and was present as a mono-allelic variant in 2 patients who carried bi-allelic pathogenic variants of SLC26A4. Overall, 11 pathogenic variants of GJB2 (Fig 2) constituted 14 different genotypes (Table 2).
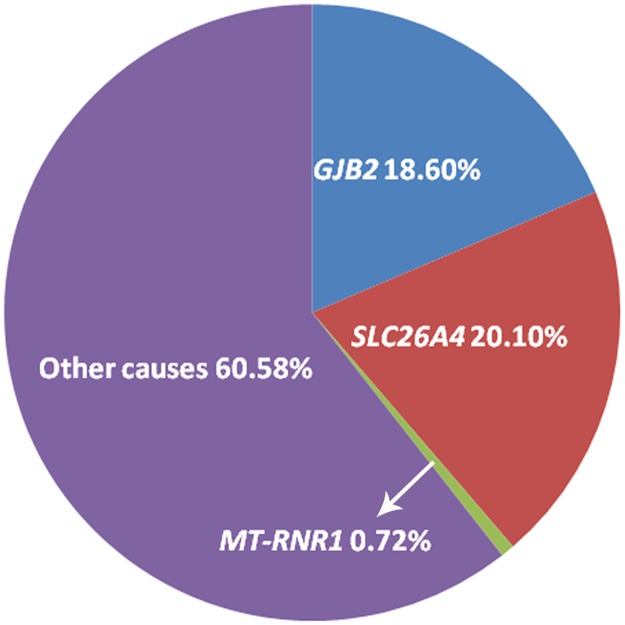
Fig 1. Molecular etiology of 695 Chinese patients with NSHL.
The percentages of biallelic GJB2, SLC26A4 and homoplasmic MT-RNR1 pathogenic variants were 18.6%, 20.1% and 0.72%, respectively. Screening of 97 different pathogenic variants in 3 genes revealed that 129 (18.6%, 129/695) cases carried biallelic GJB2 variants and 140 patients (20.1%,140/695) carried biallelic SLC26A4 variants. Homoplasmic MT-RNR1 m.1555A > G occurred in 5 cases (0.72%,5/695), however, MT-RNR1 variants are not necessarily causative for HL.
Fig 2. Location of pathogenic variants of GJB2 and SLC26A4 in the 695 subjects with NSHL.
(a)Overall, 11 different GJB2 pathogenic variants were identified in 695 subjects, out of which, one subject carried a heterozygous c.IVS1+1G > A mutant allele. The number of the mutant alleles of the other 10 different pathogenic variants, c.9G>A, c.34_35insG, c.139G>T, c.176_191del16, c.230G>A, c.235delC, c.257C>G, c.299_300delAT, c.427C>T and c.511_512insAACG are shown here. (b) In addition, 41 SLC26A4 pathogenic variants were identified in 695 subjects, the number of the mutant alleles of the most frequent 9 variants, c.414delT, c.919-2A>G, c.1174A>T, c.1226G>A, c.1229C>T, c.1707+5G>A, c.1975G>C, c.2027T>A and c.2168A>G are shown here.
Table 2. Chinese Patients with Bi-allelic Pathogenic variants in GJB2.
| Genotypes | Number (%) of patients | Inheritance mode | Clinical information | |
|---|---|---|---|---|
| HL | Onset age (Years) | |||
| Homozygous | ||||
| c.235delC/ c.235delC | 52 (7.48%) | AR * | Severe, profound | Did not pass NBHS~ 13 |
| c.299_300delAT/ c.299_300delAT | 5 (0.72%) | AR | Profound | 3 months~3.5 |
| Compound heterozygous | ||||
| c.235delC/c.299_300delAT | 40 (5.76%) | AR | Severe, profound | 6 months~3.5 |
| c.235delC/c.34_35insG | 6 (0.86%) | AR | Severe, profound | 1~5 |
| c.235delC/c.511_512insAACG | 6 (0.86%) | AR | Severe, profound | 9 months~5 |
| c.235delC/c.257C>G | 4 (0.58%) | AR | Severe, profound | 6months~3 |
| c.235delC/c.427C>T | 4 (0.58%) | AR | Severe, profound | 11 months~4 |
| c.235delC/c.176_191del16 | 3 (0.43%) | AR | Profound | 1~2.5 |
| c.299_300delAT/c.257C>G | 3 (0.43%) | AR | Severe, profound | Did not pass NBHS ~2 |
| c.235delC/c.9G>A | 2 (0.29%) | AR | Severe, profound | 1~3 |
| c.176_191del16/c.230G>A | 1 (0.14%) | AR | Profound | 1.5 |
| c.176_191del16/c.139G>T | 1 (0.14%) | AR | Profound | 4 |
| c.235delC/c.IVS1+1G>A | 1 (0.14%) | AR | Severe | 1 |
| c.299_300delAT/c.427C>T | 1 (0.14%) | AR | Severe | 6 months |
| Total | 129 (18.6%) | |||
*AR: Autosomal recessive
Unclassified variants
Genetic analysis of 18 unclassified GJB2 variants revealed that 7 variants in 50 cases. Specifically, 3 cases carried bi-allelic variants (one c. 95G > A homozygote and 2 c.109G > A homozygotes) and 47 cases carried 1 GJB2 variant. The allele frequency of c.109G > A was 3.2% (44/1390) and c.95G > A, c.283G > A, and c.571T > C shared the same allele frequency of 0.14% (2/1390). Moreover, c.23C > T, c.408C > A and c.416G > A occurred in one case each as a heterozygous variant, sharing an allele frequency of 0.07% (1/1390) (Tables 3 and 4).
Table 3. Unclassified GJB2 variants in Patients with NSHL.
| Variants | Number of patients | Clinical information | |
|---|---|---|---|
| HL | Onset age (Years) | ||
| Homozygous | |||
| c.95G>A/ c.95G>A | 1 | Profound | Unclear |
| c.109G>A /c.109G>A | 2 | Mild | 3 |
| Profound | 6 months | ||
| Heterozygous | |||
| c.23C>T | 1 | Severe | 1 |
| c.109G>A | 40 | Moderate-profound | Did not pass NBHS*~4 |
| c.283G>A | 2 | Severe-profound | 6 months~5 |
| c.408C>A | 1 | Profound | 5 |
| c.416G>A | 1 | Severe | 6 |
| c.571T>C | 2 | Severe, profound | 6 months ~2 |
| Total | 50 | ||
*NBHS: Newborn hearing screening
Table 4. The allele frequency of Unclassified GJB2 variants in Patients with NSHL.
| Variants | Number (%)of mutant alleles | Allele frequency in Database | ||
|---|---|---|---|---|
| ExAC(East Asian) | 1000 genomes(Han Chinese in Beijing) | ClinVar | ||
| c.23C>T | 1(0.07%) | 0% | 0% | - |
| c.95G>A | 2(0.14%) | 0% | - | - |
| c.109G>A | 44(3.2%) | 7.2% | 3.9% | GO-ESP 0.13%; GMAF 1.5% |
| c.283G>A | 2(0.14%) | 0% | 0% | GMAF 0.02% |
| c.408C>A | 1(0.07%) | 0% | - | - |
| c.416G>A | 1(0.07%) | 0.01% | 0.49% | GO-ESP 0.054%; GMAF 0.04% |
| c.571T>C | 2(0.14%) | 0.2% | 0.49% | GMAF 0.02% |
-: no record
SLC26A4
A total of 164 patients (23.6%, 164/695) had molecular defects in the SLC26A4 gene and 140 patients (20.1%, 140/695) carried bi-allelic pathogenic variants in SLC26A4 (Fig 2), including 54 homozygotes and 86 compound heterozygotes. Twenty-four cases carried a heterozygous SLC26A4 mutant allele and 3 of these also carried bi-allelic pathogenic variants of GJB2. Thus, the mutant allele frequency of SLC26A4 was 21.9% (304 /1390). Genetic analysis of SLC26A4 revealed 52 different genotypes (Table 5) comprised by 41 SLC26A4 variants (Table 6). The 5 most prevalent pathogenic variants in SLC26A4 were c.919-2A > G (12.2%, 169/1390), c.2168A > G (2.5%, 35/1390), c.1975G > C (1.0%,14/1390), c.1174A > T (0.65%,9/1390) and c.1707+5G > A(0.5%, 7/1390), and accounted for 77.0% (234/304) of all mutant SLC26A4 alleles(Fig 2). The most prevalent pathogenic variant, c.919-2A > G, accounting for 55.6% (169/304) of all SLC26A4 mutant alleles, was in compound heterozygosity with 24 different SLC26A4 variants in 60 cases.
Table 5. Chinese Patients with Bi-allelic Pathogenic variants in SLC26A4.
| Genotypes | Number (%) of patients | Inheritance mode | Clinical information | |
|---|---|---|---|---|
| HL | Onset age(Years) | |||
| Homozygous | ||||
| c.919-2A>G/c.919-2A>G | 50 (7. 19%) | AR * | Moderate, severe, profound | Did not pass NBHS~ 8 |
| c.414delT/c.414delT | 2 (0. 29%) | AR | Profound | 5 months, 1 |
| c.1707+5G>A/c.1707+5G>A | 1 (0. 14%) | AR | Profound | 2 |
| c.2168A>G/c.2168A>G | 1 (0. 14%) | AR | Profound | 2 |
| Compound heterozygous | ||||
| c.919-2A>G/c.2168A>G | 17 (2. 44%) | AR | Moderate, severe, profound | 6 months ~11 |
| c.919-2A>G/c.1975G>C | 9 (1. 29%) | AR | Severe, profound | 2 ~3. 5 |
| c.919-2A>G/c.1174A>T | 3 (0. 43%) | AR | Severe, profound | 2~3 |
| c.919-2A>G/c.1226G>A | 3 (0. 43%) | AR | Profound | 2~3 |
| c.919-2A>G/c.1707+5G>A | 3 (0. 43%) | AR | Severe, profound | 8months~2 |
| c.919-2A>G/c.279T>A | 2 (0. 29%) | AR | Profound | 2, 4 |
| c.919-2A>G/c.754T>C | 2 (0. 29%) | AR | Profound | 2, 7 |
| c.919-2A>G/c.916_917insG | 2 (0. 29%) | AR | Profound | 1. 5, 3 |
| c.919-2A>G/c.1225C>T | 2 (0. 29%) | AR | Severe, profound | 1, 2 |
| c.919-2A>G/c.1229C>T | 2 (0. 29%) | AR | Profound | 2, 4 |
| c.2168A>G/c.1174A>T | 2 (0. 29%) | AR | Severe, profound | 2, 3. 5 |
| c.919-2A>G/c.1343C>T | 2 (0. 29%) | AR | Severe, profound | 2~3 |
| c.2168A>G/c.1927G>T | 2 (0. 29%) | AR | Moderate, severe | 1, 1 |
| c.2168A>G/c.227C>T | 1 (0. 14%) | AR | Severe | 1 |
| c.235C>T/c.1594A>C | 1 (0. 14%) | AR | Profound | 2 |
| c.249G>A/c.1707+5G>A | 1 (0. 14%) | AR | Severe | 2. 5 |
| c.269C>T/c.1229C>T | 1 (0. 14%) | AR | Severe | 4 |
| c.281C>T/c.919-2A>G | 1 (0. 14%) | AR | Severe | 2 |
| c.281C>T/c.1174A>T | 1 (0. 14%) | AR | Profound | 2 |
| c.414delT/c.919-2A>G | 1 (0. 14%) | AR | Profound | 1 |
| c.439A>G/c.2027T>A | 1 (0. 14%) | AR | Profound | 3 |
| c.563T>C/c.919-2A>G | 1 (0. 14%) | AR | Profound | 2 |
| c.563T>C/c.2168A>G | 1 (0. 14%) | AR | Profound | 12 |
| c.589G>A/c.2168A>G | 1 (0. 14%) | AR | Profound | 1. 5 |
| c.754T>C/c.1226G>A | 1 (0. 14%) | AR | Profound | 6 |
| c.916_917insG/c.1975G>C | 1 (0. 14%) | AR | Severe | 1 |
| c.919-2A>G/c.946G>T | 1 (0. 14%) | AR | Profound | 2 |
| c.919-2A>G/c.1079C>T | 1 (0. 14%) | AR | Profound | 2 |
| c.919-2A>G/c.1173C>A | 1 (0. 14%) | AR | Severe | 3 |
| c.919-2A>G/c.1340delA | 1 (0. 14%) | AR | Profound | 4 |
| c.919-2A>G/c.1343C>A | 1(0. 14%) | AR | Severe | 2 |
| c.919-2A>G/c.1371C>A | 1 (0. 14%) | AR | Profound | 1 |
| c.919-2A>G/c.1489G>A | 1 (0. 14%) | AR | Profound | 3 |
| c.919-2A>G/c.1547_1548InsC | 1 (0. 14%) | AR | Severe | 1 |
| c.919-2A>G/c.1586T>G | 1 (0. 14%) | AR | Moderate | 10 |
| c.919-2A>G/c.1673A>T | 1 (0. 14%) | AR | Severe | 3 |
| c.1174A>T/c.1229C>T | 1 (0. 14%) | AR | Severe | 1 |
| c.1225C>T/c.1594A>C | 1 (0. 14%) | AR | Profound | 1 |
| c.1226G>A/c.2168A>G | 1 (0. 14%) | AR | Profound | 1. 5 |
| c.1229C>T/c.2027T>A | 1 (0. 14%) | AR | Profound | 6 months |
| c.1327G>C/c.2168A>G | 1 (0. 14%) | AR | Profound | 1 |
| c.1334T>G/c.1547_1548InsC | 1 (0. 14%) | AR | Profound | 9 months |
| c.1340delA/c.2027T>A | 1 (0. 14%) | AR | Profound | 2 |
| c.1343C>T/c.1975G>C | 1 (0. 14%) | AR | Severe | 3 |
| c.1686_1687insA/c.2168A>G | 1 (0. 14%) | AR | Profound | 1 |
| c.1686_1687insA/c.2027T>A | 1 (0. 14%) | AR | Severe | 2. 5 |
| c.1707+5G>A/c.1975G>C | 1 (0. 14%) | AR | Profound | 3 |
| c.2027T>A/c.2168A>G | 1 (0. 14%) | AR | Severe | 4 |
| Total | 140 (20. 1%) | |||
*AR: Autosomal recessive
Table 6. Recessive Pathogenic Variants in SLC26A4 in Patients with NSHL.
| Base pair change | Effect or Amino acid change | TM* domain | Number of subjects | Number of mutant alleles |
|---|---|---|---|---|
| c.147C>G | Ser49Arg | NH2 | 2 | 2 |
| c.227C>T | Pro76Leu | NH2 | 1 | 1 |
| c.235C>T | Arg79Ter | NH2 | 1 | 1 |
| c.249G>A | Trp83 Ter | NH2 | 1 | 1 |
| c.269C>T | Ser90Leu | Exon 1 | 1 | 1 |
| c.279T>A | Ser93Arg | Exon 1 | 2 | 2 |
| c.281C>T | Thr94Ile | Exon 1 | 3 | 3 |
| c.414delT | Frame-shift | Exon 3 | 3 | 5 |
| c.439A>G | Met147Val | Exon 3 | 1 | 1 |
| c.563T>C | Ile188Thr | Exon 4 | 2 | 2 |
| c.589G>A | Gly197Arg | Exon 4 | 1 | 1 |
| c.754T>C | Ser252Pro | EC LOOP3 | 3 | 3 |
| c.916_917insG | Frame-shift | Exon 7 | 3 | 3 |
| c.919-2A>G | Splice site | None | 119 | 169 |
| c.946G>T | Gly316 Ter | Exon 7 | 1 | 1 |
| c.1079C>T | Ala360Val | Exon 8 | 1 | 1 |
| c.1173C>A | Cys391 Ter | Exon 9 | 2 | 2 |
| c.1174A>T | Asn392Thr | Exon 9 | 9 | 9 |
| c.1225C>T | Arg409Cys | EC LOOP5 | 3 | 3 |
| c.1226G>A | Arg409His | EC LOOP5 | 5 | 5 |
| c.1229C>T | Thr410Met | EC LOOP5 | 5 | 5 |
| c.1327G>C | Glu443Gln | IC LOOP5 | 1 | 1 |
| c.1334T>G | Leu445Trp | IC LOOP5 | 1 | 1 |
| c.1336C>T | Gln446 Ter | IC LOOP5 | 1 | 1 |
| c.1340delA | Frame-shift | IC LOOP5 | 2 | 2 |
| c.1343C>A | Ser448 Ter | IC LOOP5 | 1 | 1 |
| c.1343C>T | Ser448Leu | IC LOOP5 | 3 | 3 |
| c.1371C>A | Asn457Lys | Exon 11 | 1 | 1 |
| c.1489G>A | Gly497Ser | Exon12 | 1 | 1 |
| c.1547_1548InsC | Frame-shift | COOH | 2 | 2 |
| c.1586T>G | Ser529Ala | COOH | 1 | 1 |
| c.1594A>C | Ser532Arg | COOH | 2 | 2 |
| c.1673A>T | Asn558Ile | COOH | 1 | 1 |
| c.1686_1687insA | Frame-shift | COOH | 2 | 2 |
| c.1707+5G>A | Splice site | COOH | 6 | 7 |
| c.1927G>T | Glu643 Ter | COOH | 2 | 2 |
| c.1975G>C | Val659Leu | COOH | 14 | 14 |
| c.1991C>T | Ala664Val | COOH | 1 | 1 |
| c.2027T>A | Leu676Gln | COOH | 5 | 5 |
| c.2167C>G | His723Asp | COOH | 1 | 1 |
| c.2168A>G | His723Arg | COOH | 34 | 35 |
| Total | 164 | 304 |
*TM: Transmembrane domains
MT-RNR1 pathogenic variants
MT-RNR1 m.1555A > G occurred in 6 patients (5 homoplasmons and 1 heteroplasmon). The mutant allele frequency of MT-RNR1 was 0.86% (6/695). The percentage of homoplasmic MT-RNR1 pathogenic variants was 0.72% (5/695) (Table 7). Of the 5 cases with homoplasmic variants, 1 case had normal hearing, and then was found to be hard of hearing at 19 months, 2 weeks after the use of amikacin and 1 case showed characteristic down-sloping high-frequency hearing loss (Fig 3). MT-RNR1 m.1494C > T occurred in no subjects.
Table 7. HL cases with MT-RNR1 m.1555A>G.
| Number | Age (years) | Onset age(years) | Hearing loss | History of aa* | m. 1555A>G |
|---|---|---|---|---|---|
| 1 | 5 | 19 months | All frequencies, profound | Amikacin | Homo |
| 2 | 3 | 1 | All frequencies, profound | Unclear | Homo |
| 3 | 22 | Unclear | All frequencies, profound | Unclear | Homo |
| 4 | 3 | 2 | All frequencies, profound | Unclear | Homo |
| 5 | 3 | 2 | High frequencies. severe | Unclear | Homo |
| 6 | 7 | 3 days | All frequencies, profound | Unclear | Hetero |
*Aa: aminoglycoside antibiotics
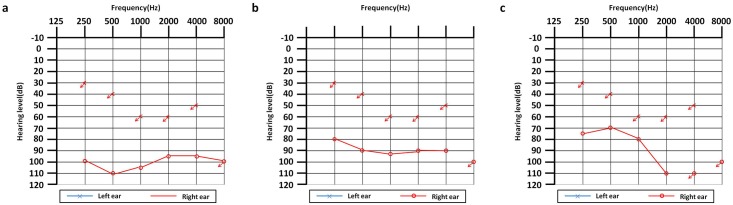
Fig 3. Patients with characteristic audiograms.
(a) One case (female, 4 years old, onset at birth) with bi-allelic pathogenic variants in GJB2 had a cochlear implant in the left ear and had profound hearing loss in the right ear. (b) One case (male, 3 years old, onset at 1 year old) with bi-allelic pathogenic variants in SLC26A4 had a cochlear implant in the left ear and had severe hearing loss in the right ear. (c)One case (male, 3 years old, onset at 2 yrs) with homoplasmic MT-RNR1 m.1555A > G had a cochlear implant in the left ear and had severe hearing loss in the right ear with a down-sloping shaped audiogram, which is characteristic of aminoglycoside-induced HL.
Discussion
GJB2, SLC26A4
The GJB2 gene, encoding gap junction protein Connexin 26(CX26), is the most frequent causative gene for NSHL. In this study, bi-allelic GJB2 pathogenic variants accounted for 18.6% of HL among the 695 subjects in this study. This is comparable with the previously reported frequency of 17.9% in 2063 Chinese HL patients [9]. According to previous reports, pathogenic variants of c.35delG, c.167delT, and c.235delC are the 3 most frequent pathogenic variants in Caucasian, Ashkenazi Jewish, and Asian populations [10–14]. The frequency of c.235delC varies from 4% to 30.4% among different regions in China [9]. In this study, GJB2 c.235delC was the most common pathogenic variant with a frequency of 13.0% (181/1390). Moreover, the panel in this study contained 18 unclassified GJB2 variants with undetermined pathogenicity and 7 of these variants were detected in 50 of the 695 cases examined. The allele frequencies of c.109G > A, c.95G > A, c.283G > A, c.571T > C, c.23C > T, c.408C > A and c.416G > A were 3.2%, 0.14%, 0.14%, 0.14%, 0.07%, 0.07%, and 0.07%, respectively. It’s important to know that the pathogenic association between these variants and HL is uncertain and therefore the 3 subjects (one c.95G > A and 2 c.109G > A homozygotes) cannot assumed to be deaf or hard of hearing on the basis of having variants of unknown significance (VUS). The significance of this primary screening system was to collect subjects with VUSs; further studies must be done to investigate the possibility of a causative relationship between these variants and HL.
SLC26A4 (OMIM* 605646) plays a key role in maintaining the endo-cochlear potential and is the second most common causative gene for ARNSHL [3–5]. According to a previous study, SLC26A4 bi-allelic pathogenic variants account for approximately 11% of Chinese Han probands with severe-to-profound HL and analysis of SLC26A4 in 176 unrelated Chinese patients with ARNSHL demonstrated that 13.6% (24/176) of patients carried at least one mutant allele [15]. In this study, 23.6% patients were found to have genetic defects in SLC26A4, out of which, 20.1% carried bi-allelic pathogenic variants and 3.5% carried one mutant allele. The 5 most prevalent pathogenic variants, c.919-2A > G, c.2168A > G, c.1975G > C, c.1174A > T and c.1707+5G > A, are detected in 77.0% of all mutant SLC26A4 alleles.
Sensorineural hearing loss has been described as a monogenic disease, encompassing bi-allelic pathogenic variants in patients with ARNSHL. Genetic testing has previously been completed and a final molecular diagnosis has been made upon the identification of bi-allelic pathogenic variants within one gene. Inherited as a classic Mendelian trait, typical ARNSHL is caused by bi-allelic pathogenic variants in one gene, giving rise to a 25% risk of ARNSHL in the siblings of an HL subject. However, in the current study, 2 independent genes were involved in HL, indicating an increased risk prediction for siblings and further offspring of patients. In this study 3 patients carried bi-allelic GJB2 variants and heterozygous SLC26A4 variants and 2 patients carried bi-allelic SLC26A4 variants and heterozygous GJB2 variants. Simultaneous analysis of common HL genes yields more accurate genetic findings related to HL and has vital implications for improving genetic counseling and risk prediction. Furthermore, it’s important to recognize that it cannot be assumed that having pathogenic variants in more than 1 gene is equivalent to di-genic inheritance. Without functional evidence, the conclusion for di-genic inheritance cannot be made.
MT-RNR1
The mitochondrial gene, MT-RNR1 encodes 12S ribosomal RNA and is considered to be a chief cause for non-syndromic HL and aminoglycoside-induced HL [16]. Homoplasmic m.1555A > G and m.1494C > T, located at the highly conserved decoding site of the 12S ribosomal RNA gene, have been identified as a cause of maternally inherited non-syndromic HL. HL inherited through the mitochondria accounts for less than 1% of all HL, though the frequency of m.1555A > G in Chinese non-syndromic HL patients can vary from 1.67%-15.5% [17–20]. In this study, 5 cases (0.72%,5/695) were found to carry homoplasmic m.1555A > G, out of which, 1 had a history of aminoglycoside antibiotic usage and another had a recognizable audio-profile of aminoglycoside-induced HL. Thus, MT-RNR1 m.1555A > G is one possible reason for hearing loss in these 2 cases. However, it is important to recognize that even though MT-RNR1 m.1555A > G is not necessarily causative for HL, mitochondrial genes are transmitted maternally and therefore, identification of MT-RNR1 m.1555A > G and m.1494C > T may have substantial implications for genetic counselling and offer an early warning for maternal members in the family pedigree.
SNPscan technique
Single nucleotide polymorphisms (SNPs), the third generation of DNA genetic markers after Microsatellites, have been widely used in molecular genetics. In the current study, multiplex genotyping of 115 variants in 695 patients was successfully performed using an SNPscan technique developed by our team. It is a rapid and efficient molecular diagnostic strategy for identifying common genes related to deafness in Chinese populations and could be used clinically as a primary screening system, leading to advances in terms of power and cost efficiency.
This screening approach has been confirmed using Next Generation Sequencing and unpublished data from our lab showed that 18 pathogenic GJB2 variants and 77 pathogenic SLC26A4 variants in our panel detected 98% of all known GJB2 and 96% of all known SLC26A4 mutant alleles in our cohort. SNPscan analysis demonstrated that the cause of hearing loss in 61.3%(425/695) of subjects was unidentified; further analysis of all known genes related to deafness will be carried out using targeted genomic enrichment (TGE) and massively parallel sequencing (MPS).
Acknowledgments
We sincerely thank all the patients and everyone for their participation and support in this study. These investigations were supported by Key Clinical Department of Ministry of Health Foundation (62450–10) to Furong Ma.
Data Availability
All relevant data are within the paper.
Funding Statement
These investigations were supported by Key Clinical Department of Ministry of Health Foundation (62450-10) to Furong Ma.
References
- 1.Rennels M, Pickering LK: Sensorineural hearing loss in children. The Lancet, 365:2085–2086. [DOI] [PubMed] [Google Scholar]
- 2.Kenneson A, Van Naarden Braun K, Boyle C: GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med 2002, 4:258–274. 10.1097/00125817-200207000-00004 [DOI] [PubMed] [Google Scholar]
- 3.Hilgert N, Smith RJ, Van Camp G: Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res 2009, 681:189–196. 10.1016/j.mrrev.2008.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, et al. : Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat Genet 1997, 17:411–422. 10.1038/ng1297-411 [DOI] [PubMed] [Google Scholar]
- 5.Runge-Samuelson C, Olivier M: The rocky road toward clinical genetic testing: insights into the physio-genetic basis of hearing loss. Physiol Genomics 2009, 39:83–84. 10.1152/physiolgenomics.00125.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kupka S, Toth T, Wrobel M, Zeissler U, Szyfter W, Szyfter K, et al. : Mutation A1555G in the 12S rRNA gene and its epidemiological importance in German, Hungarian, and Polish patients. Hum Mutat 2002, 19:308–309. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Li Z, Zhu Y, Yang A, Li R, Zheng J, et al. : Mitochondrial 12S rRNA variants in 1642 Han Chinese pediatric subjects with aminoglycoside-induced and nonsyndromic hearing loss. Mitochondrion 2010, 10:380–390. 10.1016/j.mito.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du W, Cheng J, Ding H, Jiang Z, Guo Y, Yuan H: A rapid method for simultaneous multi-gene mutation screening in children with nonsyndromic hearing loss. Genomics 2014, 104:264–270. 10.1016/j.ygeno.2014.07.009 [DOI] [PubMed] [Google Scholar]
- 9.Dai P, Yu F, Han B, Liu X, Wang G, Li Q, et al. : GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J Transl Med 2009, 7:26 10.1186/1479-5876-7-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtsuka A, Yuge I, Kimura S, Namba A, Abe S, Van Laer L, Van Camp G, Usami S: GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum Genet 2003, 112:329–333. 10.1007/s00439-002-0889-x [DOI] [PubMed] [Google Scholar]
- 11.Roux AF, Pallares-Ruiz N, Vielle A, Faugere V, Templin C, Leprevost D, et al. : Molecular epidemiology of DFNB1 deafness in France. BMC Med Genet 2004, 5:5 10.1186/1471-2350-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabriel H, Kupsch P, Sudendey J, Winterhager E, Jahnke K, Lautermann J: Mutations in the connexin26/GJB2 gene are the most common event in non-syndromic hearing loss among the German population. Hum Mutat 2001, 17:521–522. [DOI] [PubMed] [Google Scholar]
- 13.Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, et al. : Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N Engl J Med 1998, 339:1500–1505. 10.1056/NEJM199811193392103 [DOI] [PubMed] [Google Scholar]
- 14.Abe S, Usami S, Shinkawa H, Kelley PM, Kimberling WJ: Prevalent connexin 26 gene (GJB2) mutations in Japanese. J Med Genet 2000, 37:41–43. 10.1136/jmg.37.1.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Chen J, Shan XJ, Li Y, He JG, Yang BB: Prevalence and range of GJB2 and SLC26A4 mutations in patients with autosomal recessive nonsyndromic hearing loss. Mol Med Rep 2014, 10:379–386. 10.3892/mmr.2014.2148 [DOI] [PubMed] [Google Scholar]
- 16.Kokotas H, Petersen MB, Willems PJ: Mitochondrial deafness. Clin Genet 2007, 71:379–391. 10.1111/j.1399-0004.2007.00800.x [DOI] [PubMed] [Google Scholar]
- 17.Dai P, Liu X, Han D, Qian Y, Huang D, Yuan H et al. : Extremely low penetrance of deafness associated with the mitochondrial 12S rRNA mutation in 16 Chinese families: implication for early detection and prevention of deafness. Biochem Biophys Res Commun 2006, 340:194–199. 10.1016/j.bbrc.2005.11.156 [DOI] [PubMed] [Google Scholar]
- 18.Ji YB, Han DY, Lan L, Wang DY, Zong L, Zhao FF, Liu Q, Benedict-Alderfer C, Zheng QY, Wang QJ: Molecular epidemiological analysis of mitochondrial DNA12SrRNA A1555G, GJB2, and SLC26A4 mutations in sporadic outpatients with nonsyndromic sensorineural hearing loss in China. Acta Otolaryngol 2011, 131:124–129. 10.3109/00016489.2010.483479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, You Y, Huang D, Cui J, Wang Y, Wang Q, Yu F, Kang D, Yuan H, Han D, Dai P: Comprehensive molecular etiology analysis of nonsyndromic hearing impairment from typical areas in China. J Transl Med 2009, 7:79 10.1186/1479-5876-7-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu XZ, Angeli S, Ouyang XM, Liu W, Ke XM, Liu YH, Liu SX, Du LL, Deng XW, Yuan H, Yan D: Audiological and genetic features of the mtDNA mutations. Acta Otolaryngol 2008, 128:732–738. 10.1080/00016480701719011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.