Abstract
Stem rust is one of the most potentially harmful wheat diseases, but has been effectively controlled in China since 1970s. However, the interest in breeding wheat with durable resistance to stem rust has been renewed with the emergence of Ug99 (TTKSK) virulent to the widely used resistance gene Sr31, and by which the wheat stem rust was controlled for 40 years in wheat production area worldwide. Yunnan Province, located on the Southwest border of China, is one of the main wheat growing regions, playing a pivotal role in the wheat stem rust epidemic in China. This study investigated the levels of resistance in key wheat cultivars (lines) of Yunnan Province. In addition, the existence of Sr25, Sr26, Sr28, Sr31, Sr32, and Sr38 genes in 119 wheat cultivars was assessed using specific DNA markers. The results indicated that 77 (64.7%) tested wheat varieties showed different levels of resistance to all the tested races of Puccinia graminis f. sp. tritici. Using molecular markers, we identified the resistance gene Sr31 in 43 samples; Sr38 in 10 samples; Sr28 in 12 samples, and one sample which was resistant against Ug99 (avirulent to Sr32). No Sr25 or Sr26 (effective against Ug99) was identified in any cultivars tested. Furthermore, 5 out of 119 cultivars tested carried both Sr31 and Sr38 and eight contained both Sr31 and Sr28. The results enable the development of appropriate strategies to breed varieties resistant to stem rust.
Introduction
Stem rust (caused by Puccinia graminis Pers. f. sp. tritici Eriks. & E. Henn.) is one of the most serious diseases of wheat, worldwide [1, 2]. In China, it has been effectively controlled through the development of resistant cultivars and deployment of effective resistance genes, especially 1B/1R translocation gene Sr31 in different epidemiological regions since 1970s [3, 4]. However, in 1998, a new race of wheat stem rust pathogen designated as Ug99 (TTKSK), expressing virulence to Sr31, was first identified in Uganda [5, 6]. It has spread throughout the major wheat growing regions of Africa such as Ethiopia, Zimbabwe, Mozambique, Kenya, Sudan, Yemen, Egypt, and Tanzania [7, 8]. The variants exhibited stronger virulence and could rapidly spread worldwide. For example, variants with virulence against common stem rust-resistance genes Sr24, Sr38, and Sr36 have also been detected [9]. According to the Food and Agriculture Organization (FAO) forecasts, this disease may spread eastward from Iran into countries of Central Asia [10]. TTKSK has been detected in Iran [11] and may soon threaten wheat production in the Indian sub-continent [2, 12]. The spread of Ug99 and its variants to India is a threat to the security of wheat production in China.
Yunnan Province, located on the Southwest border of China, is close to India, increasing the risk of Ug99 contamination [3]. The annual urediospores life cycle of Pgt is completed in the region, playing a key role in the large-scale epidemic of wheat stem rust [13]. Therefore, the resistance level of the cultivars in Yunnan province has a direct impact on epidemiology. In addition, because of the rarity of the wheat stem rust since the 1970s, the resistance of wheat cultivars has not been taken seriously. Therefore, due to the imminent risk in Chinese wheat production posed by Ug99, analysis of resistance against stem rust and delineation of the resistance genes in the cultivars (lines) locally are of great significance in evaluation of the risk, because 60 percent of wheat varieties contain Sr31 in China. It also raises the possibility of crop rotation as well as development of new rust-resistant sources. Wheat protection and breeding of resistant cultivars using conventional methods are time-consuming, intricate, and slow, and are influenced by the environment eventually. Currently, plant breeding is updated using molecular markers. Development of molecular markers has led to efficient methods of plant breeding [14]. Various studies have been conducted to confirm the presence of Sr genes in wheat cultivars. A collection of 54 wheat cultivars and 11 breeding lines from South Africa was screened to identify Sr2, Sr24 and Sr31 genes using DNA markers [15]. PCR-based DNA markers were used to check rust resistance genes among the 20 wheat genotypes and 22 markers were amplified [16]. Kokhmetova and Atishova [17] studied the presence of Sr genes (Sr2, Sr22, Sr24, Sr36, and Sr46), which are effective against Ug99 in 88 cultivars of spring soft wheat in Kazakhstan. Haile et al. [18] screened out 30 Sr genes using SSR and STS markers in 58 tetraploid wheat accessions of Ethiopia. In China, molecular markers closely linked with Sr22, Sr25, and Sr33 are used for the detection of wheat cultivars in Heilongjiang Province, and four cultivars may contain Sr22 and three probably carry Sr33 [19–21].
In this study, on the basis of resistance levels to Chinese stem rust in wheat cultivars of Yunnan province, the reported molecular markers closely linked to three major resistance genes Sr31, Sr32, Sr38 and another three resistance genes effective against Ug99 races (Sr25, Sr26, and Sr28) were used to assess the prevalence of stem rust resistance in Yunnan wheat cultivars. Breeders may use this information to genetically engineer new and potentially durable combinations of stem rust resistance cultivars.
Materials and Methods
Wheat cultivars and near-isogenic lines
A total of 119 tested wheat cultivars including the primary cultivars and reserve lines in Yunnan province were provided by Dr. Mingju Li at Yunnan Provincial Institute of Agricultural Environment and Resources.
Six Sr genes were tested: Sr25, Sr26, Sr28, Sr31, Sr32, and Sr38. The near-isogenic lines carrying these resistance genes were provided by Dr. Yue Jin from USDA-ARS, Cereal Disease Laboratory, University of Minnesota.
P. graminis f. sp. tritici races
The tested Pgt races include the dominant 21C3CTHTM and 34MKGSM, and 34C3RTGQM. The differentials of races are composed of three parts: four Stakman’s (Little club, Reliance, Einkorn, and Vernal), five Chinese supplemental differentials (Mianzi 52, Huadong 6, Mini 2761, Orofen, and Rulofen), and five sets of 20 single Sr-gene lines (Sr5, Sr21, Sr9e, Sr7b, Sr11, Sr6, Sr8a, Sr9g, Sr36, Sr9b, Sr30, Sr17, Sr9a, Sr9d, Sr10, SrTmp, Sr24, Sr31, Sr38, and McN). At present, the races were identified and designated using three parts of differentials by Wheat Disease Laboratory, Shenyang Agricultural University, namely: part 1 uses four of Stakman’s hosts and is given Arabic numerals, such as 21 or 34 etc.; the second part uses 5 Chinese supplemental lines, and was given a ‘C’ plus Arabic numerals, C1 or C2 or C3 etc.; and the third part uses 20 single Sr-gene lines and given five-letter-code as currently used in the United States and many other countries, for examples: HTTTM or RKGQM. The full names of the races and their virulence/avirulence patterns are shown in Table 1 [3]. They were isolated and identified by Wheat Disease Laboratory, Shenyang Agricultural University, China.
Table 1. Virulence/avirulence patterns of 3 races of Puccinia graminis f. sp. tritici.
| Race | Ineffective Sr genes | Effective Sr genes |
|---|---|---|
| 21C3CTHTM | 6, 7b, 8a, 9a, 9b, 9d, 9f, 9g, 10, 11, 12, 13, 14, 15, 16, 17, 18, 24, 28, 29, 34, 35, Tmp, McN | 5, 9e, 19, 20, 21, 22, 23, 25, 26, 27, 30, 31, 32, 33, 36, 37, 38, 47 |
| 34MKGSM | 5, 6, 7b, 8a, 9a, 9b, 9d, 9f, 9g, 10, 12, 15, 16, 20, 24, 27, 28, 29, McN | 9e, 11, 13, 14, 17, 18, 19, 21, 22, 23, 25, 26, 30, 31, 32, 33, 34, 35, 36, 37, 38, 47, Tmp |
| 34C3RKGQM | 5, 6, 7b, 8a, 9a, 9b, 9d, 9f, 9g, 12, 16, 19, 21, 23, 24, 27, 28, 29, McN | 9e, 10, 11, 13, 14, 15, 17, 18, 20, 22, 25, 26, 30, 31, 32, 33, 34, 35, 36, 37, 38, 47, Tmp |
DNA extraction and fragment analysis
DNA was extracted from young leaves of seedlings according to the method described by Lagudah et al. [22]. The DNA quality was determined using 1.2% (w/v) agarose gel. DNA quantification was performed using NanoDrop-1000 version 3.3.1 spectrophotometer. PCR primers were synthesized by Sangon Biotech (http://www.sangon.com/, China). PCR assays were performed according to the published protocols (Table 2). PCR amplifications were carried out in 25 μL volume, including 2.5 μL 10×buffer (Mg2+), 0.5 μL 10 mmol·L-1 dNTPs, 1 μL 10 μmol·L-1 of each primer, 0.2 μL 5 U·μL-1 Taq-polymerase, and 2 μL 30 ng·μL-1 DNA. De-ionized water was used to obtain 25 μL. 1.2% (w/v) agarose gel was used to detect the fragments of target gene. The agarose gels were analyzed by Uvitec cambridge. The incubation time and voltage used for analysis of fragments were 200V and 30 min. All above reagents were provided by Sangon Biotech (http://www.sangon.com/, China). Further, the ddH2O and reaction system lacking the template DNA were used as controls. The specific reaction conditions were based on published studies.
Table 2. The primers linked to resistance genes Sr25, Sr26, Sr28, Sr31, Sr32 and Sr38.
| Tagged Sr genes | Marker | Size of markers (bp) | Primer sequence | References |
|---|---|---|---|---|
| Sr25 | Xwmc221 | 190 |
F-5′ACGATAATGCAGCGGGGAAT
R-5′ GCTGGGATCAAGGGATCAAT |
21 |
| Sr26 | Sr26#43 | 207 |
F-5′AATCGTCCACATTGGCTTCT
R-5′ CGCAACAAAATCATGCACTA |
24 |
| Sr28 | Wpt-7004 | 194 |
F-5′CTCCCACCAAAACAGCCTAC
R-5′ AGATGCGAATGGGCAGTTAG |
25 |
| Sr31 | SCSS30.2576 | 576 |
F-5′GTCCGACAATACGAACGATT
R-5′ CCGACAATACGAACGCCTTG |
26 |
| Sr32 | csSr32#1 | 184 |
F-5’GGTTTGGTGGCAACTCAGGT
R-5’ CATAAGCCAAAGAGGCACCA |
27 |
| Sr38 | Xcmwg682 | 259 |
F-5’AGGGGCTACTGACCAAGGCT
R-5’TGCAGCTACAGCAGTATGTACACAAAA |
29 |
Resistance determination
The whole cultivars (lines) were planted in 12 cm diameter porcelain pots. The Little Club (LC) was used as the control to ascertain the viability of spores or successful inoculation of races to cultivars. Leaves of 7-days-old seedlings were moistened by water with 0.1% Tween 20 using an atomizer, then sprayed 1 g of fresh urediospores and dried talc in a ratio of 1:20 (V:V). The inoculated seedlings were kept in a dew chamber for 16~20 h dark at 18~22°C and RH of 95% before transferring to a greenhouse adjusted at 18~22±1°C. Infection types (ITs) were assessed 14 days after inoculation using 0–4 IT scale described by Stakman et al. [23]. ITs were then grouped into two categories. ITs ‘0’, ‘;’, ‘1’, ‘1+’, ‘2’, ‘2+’ and X were considered as low infection types (resistance) while ITs ‘3-’, ‘3’, ‘3+’ and ‘4’ as high infection types (susceptible) [23].
Results
Wheat seedling resistance
The reaction of 119 main wheat cultivars in Yunnan to the Pgt races are shown in Table 3. Seventy-seven (64.7%) of the tested wheat varieties showed varying levels of resistance to races 21C3CTHTM, 34MKGSM and 34C3RKGQM. The remaining 42 wheat cultivars (35.3%) showed varying levels of susceptibility.
Table 3. Resistant proportion of 119 wheat cultivars to wheat stem rust.
| Race | Susceptible | Resistance | ||
|---|---|---|---|---|
| Number of cultivars | Percentage/% | Number of cultivars | Percentage/% | |
| 34MKGSM | 24 | 20.2 | 95 | 79.8 |
| 21C3CTHTM | 22 | 18.5 | 97 | 81.5 |
| 34C3RKGQM | 36 | 30.3 | 83 | 69.7 |
| All tested races | 77 | 64.7 | 42 | 35.3 |
A total of 97 cultivars showed different levels of resistance to race 21C3CTHTM, accounting for 81.5% of all the tested cultivars. Ninety-five cultivars were resistant to race 34MKGSM, and 24 showed susceptibility. A total of 83 cultivars showed resistance to 34C3RKGQM, accounting for 69.7% of all the tested cultivars, which was relatively low compared to resistance to 34MKGSM and 21C3CTHTM.
Sr25 screening
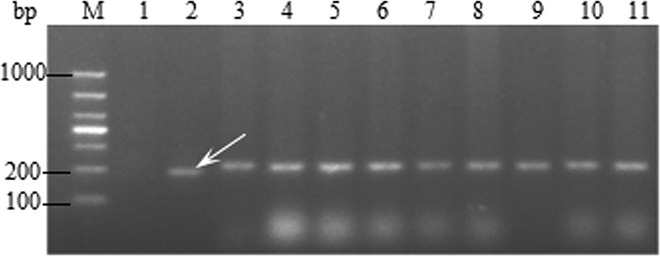
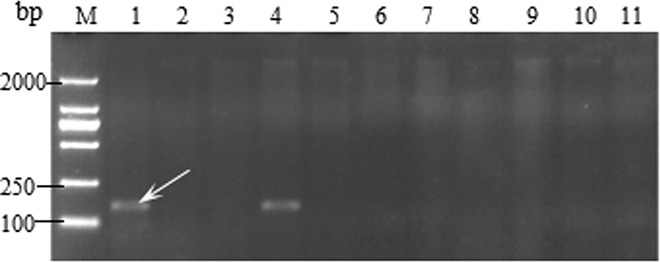
LC was used as the negative control and the monogenic Sr25 served as the positive control. A 190 bp specific band was amplified in the positive control using the primer Xwmc221. This result showed that no amplification of the 190 bp band in the 119 wheat cultivars of Yunnan Province (Fig 1).
Fig 1. Amplification result for parts of wheat varieties with Xwmc221.
Lane 1–11, Water control, Monogenic Sr25, Little Club, Kunmai 4, Kunmai 5, Jingmai 8, Jingmai 9, Jingmai 10, Jingmai 11, Jingmai 12, and Demai 3. ‘M’ indicates 1000 bp DNA ladder and white arrow indicates the position of the specific band.
Sr26 screening
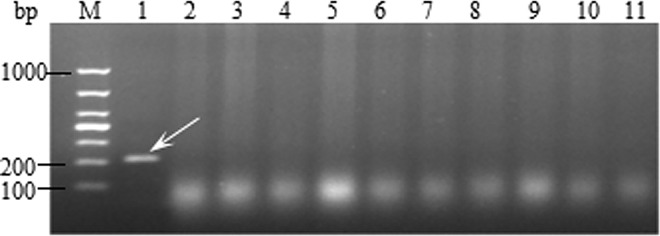
Mago et al. [24] developed a pair of RFLP markers for detection of wheat stem rust resistance gene Sr26, and the effectiveness of the specific marker Sr26#43 was evaluated. The results showed that the primer Sr26#43 amplified a 207 bp fragment, and was used to detect Sr26 in the cultivars (Fig 2). LC was used as a negative control and monogenic Sr26 was used as the positive control. Primer Sr26#43 amplified a 207 bp band in the positive control Sr26, while no bands were amplified in the remaining materials, indicating that the tested cultivars do not contain the resistance gene Sr26.
Fig 2. Amplification result for parts of wheat varieties with Sr26#43.
Lane 1–11, Monogenic Sr26, Little Club, Water control, Jingmai 8, Jingmai 9, Jingmai 10, Jingmai 11, Jingmai 12, Jingmai 14, Jing 0202, and Jing 04–6. ‘M’ indicates 1000 bp DNA ladder and white arrow indicates the position of the specific band.
Sr28 screening
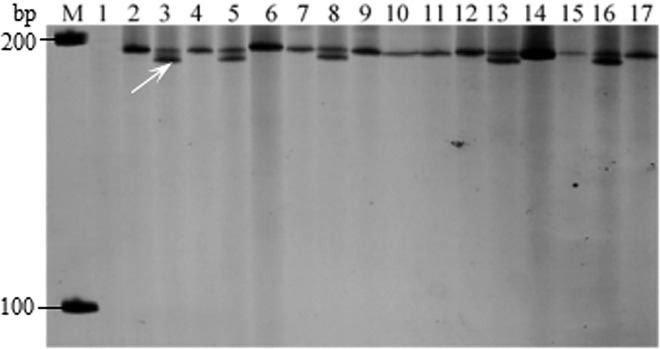
Rouse et al. [25] showed that the marker wPt-7004 amplified 194 bp and 166 bp fragments in the cultivars containing Sr28 resistance genes. Further analysis confirmed that the amplification of the 194 bp fragment represented the specific band for Sr28. Polyacrylamide gel electrophoresis was used for detection of Sr28. As shown in Fig 3, the primer wPt-7004 amplified a band measuring 194 bp to 200 bp in the negative control, but an extra 194 bp band was amplified in the positive control Sr28. In this study, a similar 194 bp band was detected in Nanyuan 1, Feng 0103, Jing 07–2, Jingmai 10, Jing 05–1, 088–16, E33, Linmai 15, Jing 0202, Jingmai 10, Yunmai 54, Yunmai 47, and Feng 615, indicating that the 12 tested cultivars carried Sr28 (Table 4).
Fig 3. Amplification result for parts of wheat varieties with Wpt-7004.
Lane 1–11, Water control, Little Club, Monogenic Sr28, Jing 04–6, Jing 0202, Jingmai 14, Jingmai 11, Jingmai 10, Jingmai 8, Yunza 7, Yunza 6, Yunxuan 2, Yunmai 54, Yunmai 53, Yunmai 52, Yunmai 47, and Yunmai 39. ‘M’ indicates 1000 bp DNA ladder and white arrow indicates the position of the specific band.
Table 4. Results for wheat seedling resistance and molecular detection.
| Cultivars/lines | 21C3CTHTM | 34MKGSM | 34C3RKGQM | Sr25 | Sr26 | Sr28 | Sr31 | Sr32 | Sr38 |
|---|---|---|---|---|---|---|---|---|---|
| Yunmai 39 | 1* | ; | 1+ | - | - | - | + | - | - |
| Yunmai 42 | 4 | 3 | 4 | - | - | - | - | - | - |
| Yunmai 43 | 3 | 4 | 4 | - | - | - | - | - | - |
| Yunmai 47 | 0 | 1 | ; | - | - | + | + | - | - |
| Yunmai 48 | 1+ | 4 | 3- | - | - | - | - | - | - |
| Yunmai 51 | 3+ | 3 | 3+ | - | - | - | - | - | - |
| Yunmai 52 | 4 | 3 | 4 | - | - | - | - | - | - |
| Yunmai 53 | 3+ | ; | 3 | - | - | - | - | - | - |
| Yunmai 54 | ; | 0 | 1+ | - | - | + | + | - | - |
| Yunmai 56 | 3 | 1+ | 1 | - | - | - | - | - | - |
| Yunxuan 2 | 1 | 0 | 0 | - | - | - | - | - | - |
| Yunxuan 3 | 0 | 0 | 0 | - | - | - | - | - | - |
| Yunxuan 11–12 | 0 | 1 | 1+ | - | - | - | + | - | - |
| Yunza 5 | 4 | 3 | 3+ | - | - | - | - | - | - |
| Yunza 6 | 3 | 3 | 4 | - | - | - | - | - | - |
| Yunza 7 | 3- | 3- | 4 | - | - | - | - | - | - |
| Kun 022-222-1 | 0 | ;1- | 1 | - | - | - | + | - | + |
| Kunmai 4 | 0 | ; | 0 | - | - | - | - | - | - |
| Kunmai 5 | 0 | 0 | 0 | - | - | - | + | - | - |
| Jingmai 8 | 0 | 0 | ; | - | - | - | + | - | - |
| Jingmai 9 | 0 | 0 | 1 | - | - | - | + | - | - |
| Jingmai 10 | 0 | 0 | 0 | - | - | + | + | - | - |
| Jingmai 11 | 0 | 1 | ; | - | - | - | + | - | - |
| Jingmai 12 | 0 | 0 | 0 | - | - | - | + | - | - |
| Jingmai 14 | 0 | 0 | 1 | - | - | - | + | - | + |
| Jing 0202 | ; | 1 | 0 | - | - | + | + | - | - |
| Jing 04–6 | 0 | 1 | 0 | - | - | - | + | - | - |
| Demai 3 | 2 | 0 | 3 | - | - | - | - | - | - |
| Demai 7 | 0 | 0 | 0 | - | - | - | + | - | - |
| De 05–81 | 0 | 0 | 1 | - | - | - | - | - | + |
| Linmai 6 | 1 | 3- | 3 | - | - | - | - | - | - |
| Linmai 15 | 0 | 0 | 0 | - | - | + | + | - | - |
| Fengyin 03–2 | 0 | 1 | 3 | - | - | - | - | - | - |
| Feng 05–394 | ;1- | 0 | ; | - | - | - | + | - | - |
| Yunmai 101 | 0 | ;1- | 0 | - | - | - | + | - | - |
| Feng 1124 | ; | 0 | 0 | - | - | - | - | - | + |
| Yumai 1 | 0 | 0 | 0 | - | - | - | + | - | - |
| Yumai 2 | ;1 | ; | 1 | - | - | - | + | - | - |
| Yumai 3 | 0 | 0 | 0 | - | - | - | + | - | - |
| Fengmai 31 | 1 | ; | 3- | - | - | - | - | - | - |
| Fengmai 32 | ; | 0 | 1 | - | - | - | + | - | - |
| Fengmai 33 | ;1- | ;1 | ; | - | - | - | + | - | - |
| Wenmai 12 | ; | 0 | 1+ | - | - | - | + | - | - |
| Chumai 12 | ;1 | 1 | ; | - | - | - | + | - | - |
| Liangmai 4 | 0 | 0 | 0 | - | - | - | + | - | + |
| Mian 1971–98 | 3 | 1 | 3 | - | - | - | - | - | - |
| De 0716 | 0 | 0 | 0 | - | - | - | + | - | - |
| Dianmai 34 | 0 | 0 | 3 | - | - | - | - | - | - |
| Chu 2008 jian-4 | 0 | 0 | ; | - | - | - | + | - | - |
| 91E001 | 0 | 1+ | 2 | - | - | - | - | + | - |
| R101 | ; | 0 | 0 | - | - | - | - | - | - |
| R57 | 0 | 1 | 1 | - | - | - | + | - | + |
| E33 | 0 | 0 | 0 | - | - | + | + | - | - |
| 06D6-6 | 1 | 0 | 1+ | - | - | - | + | - | - |
| Yu 09–5 | 0 | 0 | 2 | - | - | - | - | - | - |
| Feng 615 | ; | ;1- | 0 | - | - | + | - | - | - |
| Yimai 1 | 3 | 4 | 4 | - | - | - | - | - | - |
| Yixuan A03-2 | ; | 1+ | 0 | - | - | - | - | - | + |
| Yixi 96–6 | 0 | 4 | 4 | - | - | - | - | - | - |
| Yixi 2003–64 | 0 | 3 | 4 | - | - | - | - | - | - |
| Jing 05–1 | 0 | 0 | 0 | - | - | + | + | - | - |
| Jing 07–2 | 1 | 0 | 1 | - | - | + | - | - | - |
| Linmai 17 | 0 | 0 | 0 | - | - | - | + | - | - |
| Chu 06–9 | 1 | ; | 1 | - | - | - | - | - | - |
| Feng 1128 | 0 | 1 | 0 | - | - | - | - | - | + |
| SH 710 | 2 | 0 | 3 | - | - | - | - | - | - |
| De 07–19 | 0 | 0 | 0 | - | - | - | + | - | - |
| De 07–20 | 0 | 0 | 0 | - | - | - | + | - | - |
| De 07–21 | 0 | 0 | 0 | - | - | - | + | - | - |
| De 08–3 | ; | 0 | 1 | - | - | - | + | - | + |
| De 08–4 | 0 | 0 | 0 | - | - | - | - | - | - |
| De 08–11 | 0 | 0 | 0 | - | - | - | - | - | - |
| Wen 05–1 | 0 | 0 | 1+ | - | - | - | - | - | - |
| Wen 06–3 | 0 | 0 | 0 | - | - | - | - | - | - |
| 084–12 | 3- | 2 | 3+ | - | - | - | - | - | - |
| Feng 0483 | 0 | 0 | 0 | - | - | - | - | - | - |
| Feng 0103 | 1+ | 3 | 4 | - | - | + | - | - | - |
| Feng 0230 | 0 | ; | 0 | - | - | - | - | - | - |
| Mosha | 0 | 0 | 0 | - | - | - | - | - | - |
| Jingxuan 9 | 0 | 0 | 0 | - | - | - | - | - | - |
| Guoji 13 | ; | 0 | ; | - | - | - | - | - | - |
| Yunmai 29 | 0 | 1 | 0 | - | - | - | - | - | - |
| Mianyang 19 | 0 | 4 | 3 | - | - | - | - | - | - |
| I1 | 3+ | 4 | 4 | - | - | - | - | - | - |
| I18 | 0 | 0 | 1+ | - | - | - | - | - | - |
| Fengmai 13 | 3 | ;1 | 4 | - | - | - | - | - | - |
| Fengmai 24 | 1 | 1+ | 0 | - | - | - | - | - | - |
| Jingmai 7 | 0 | 4 | 0 | - | - | - | - | - | - |
| Demai 4 | 0 | 0 | 0 | - | - | - | + | - | - |
| Nanyuan 1 | 1 | 4 | 3 | - | - | + | - | - | - |
| Mianyang 20 | 3 | 4 | 4 | - | - | - | - | - | - |
| Kunming chunmai | 0 | 1+ | 0 | - | - | - | - | - | - |
| Nanda 2419 | 3 | 4 | 4 | - | - | - | - | - | - |
| Fengmai 34 | 0 | 0 | 0 | - | - | - | - | - | - |
| Fengmai 35 | 4 | 0 | 4 | - | - | - | - | - | - |
| Fengmai 36 | 0 | 0 | 1 | - | - | - | - | - | - |
| Fengmai 37 | 0 | 0 | 3 | - | - | - | - | - | - |
| Fengmai 38 | 0 | 0 | 0 | - | - | - | - | - | - |
| Fengmai 39 | 0 | 0 | 0 | - | - | - | - | - | - |
| Yimai line 2003–27 | 4 | 4 | 4 | - | - | - | - | - | - |
| Yimai 10 | 0 | 4 | 4 | - | - | - | - | - | - |
| K07-295 | 0 | ; | 1 | - | - | - | + | - | + |
| K042-39 | 1+ | 0 | 1 | - | - | - | + | - | - |
| 4–12 | 3+ | 1 | 2 | - | - | - | - | - | - |
| 4–8 | 0 | 0 | 0 | - | - | - | - | - | - |
| 05–1 | ;1- | 0 | 3 | - | - | - | - | - | |
| 088–16 | 0 | 0 | ; | - | - | + | + | - | - |
| 09D4-1 | 0 | 1 | 0 | - | - | - | + | - | - |
| 09D4-6 | 4 | 4 | 1+ | - | - | - | - | - | - |
| 08 yu F-5 | 0 | 3- | 0 | - | - | - | - | - | - |
| 066–3 | 0 | 0 | 1+ | - | - | - | + | - | - |
| 017–10 | 0 | 1 | 2 | - | - | - | - | - | - |
| HX-06-1 | 0 | 0 | 4 | - | - | - | - | - | - |
| 098–2 | 3 | 0 | 0 | - | - | - | - | - | - |
| 098–4 | 0 | 0 | 0 | - | - | - | - | - | - |
| 098–6 | 1 | 0 | 2 | - | - | - | - | - | - |
| 098–10 | 3 | 3 | 4 | - | - | - | - | - | - |
| 098–12 | 1 | ; | 3 | - | - | - | - | - | - |
| 098–13 | 0 | 1 | 3 | - | - | - | - | - | - |
*Infection types (ITs) are based on a 0-to-4 scale, ITs of 0,;, ; 1, 1, and 2 are indicative of a resistant (low) response and ITs of 3 or 4 are a susceptible (high) response.
Sr31 screening
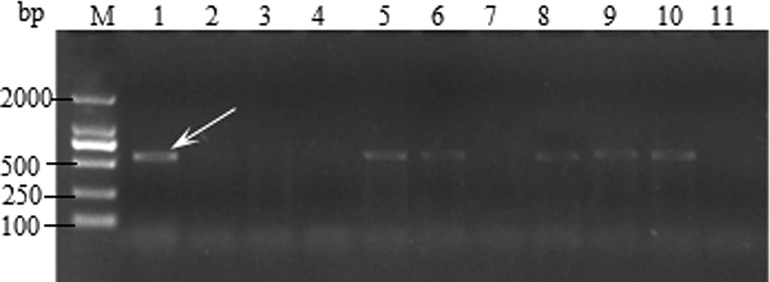
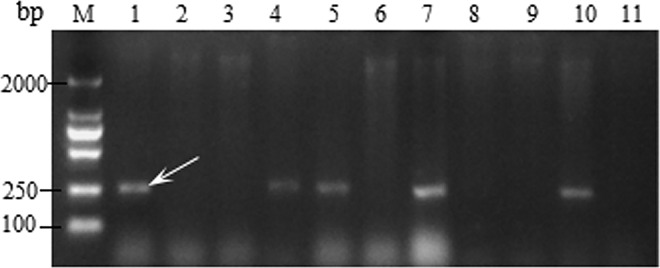
The SCSS30.2576 marker is linked to stem rust resistance gene Sr31. The Sr31 genes transferred from rye into wheat (Triticum aestivum L.) contributes to resistance in all the virulent pathotypes of stem rust (P. graminis f. sp. tritici) found in China. The SCSS30.2576 marker amplified an expected PCR product of approximately 576 bp [26] in the positive control Sr31 (Fig 4). It acted as a specific marker with no product generated in lines that do not carry Sr31. Among the tested cultivars, 43 cultivars tested positive for the Sr31 marker (Table 4).
Fig 4. Amplification result for parts of wheat varieties with SCSS30.2576.
Lane 1–11, Monogenic Sr31, Little Club, Water control, Fenyin 03–2, Feng 05–394, Yunmai 101, Feng 1124, Yumai 1, Yumai 2, Yumai 3, and Fengmai 31. ‘M’ indicates 2000 bp DNA ladder and white arrow indicates the position of the specific band.
Sr32 screening
Sr32 is located on the short arm of chromosome in Sears’ wheat Aegilops speltoides 2D-2S#1 translocation line C82.2 [27]. A dominant marker, csSr32#1, derived from an AFLP fragment [27] amplified 184bp PCR product in wheat lines carrying Sr32. In this study, the marker csSr32#1 was used to determine the presence of Sr32 in 119 wheat materials. The band was amplified only in the positive control Sr32 and one wheat cultivar 91E001, but none was detected in the remaining cultivars (Fig 5).
Fig 5. Amplification result for parts of wheat varieties with csSr32#1.
Lane 1–11, Monogenic Sr32, Little Club, Water control, 91E001, R101, R57, E33, 06D6-6, Yu 09–5, Feng 615, and Yimai 1. ‘M’ indicates 2000 bp DNA ladder and white arrow indicates the position of the specific band.
Sr38 screening
Rust resistance genes Lr37, Sr38, and Yr17 are located within a segment of T. ventricosum (Tausch) Cess. chromosome 2NS translocated to the short arm of bread wheat chromosome 2AS [28]. Helguera et al. [29] developed polymerase chain reaction (PCR) assays based on RFLP marker cMWG682 to facilitate the transfer of this cluster of rust resistance genes into commercial wheat (T. aestivum L.) cultivars. The marker played a major role in molecular assisted breeding (MAS) [30]. Li et al. [31] validated the marker cMWG682 using 126 wheat lines and indicated that cMWG682 amplified a 259 bp fragment in the positive control Yr17/6*Avocet S and 12 cultivars. In this study, monogenic Sr38 was used as a positive control and LC as a negative control. A 259 bp fragment was amplified in the positive control Sr38 and 10 of the tested wheat cultivars, using primer cMWG682 (Fig 6), suggesting that the 10 cultivars (lines) carried Lr37-Sr38-Yr17 (Table 4).
Fig 6. Amplification result for parts of wheat varieties with Xcmwg682.
Lane 1–11, Monogenic Sr38, Little Club, Water control, De 05–81, Feng 1124, Yumai 1, Liangmai 4, De 0716, Dianmai 34, Feng 1128, and De 07–19. ‘M’ indicates 2000 bp DNA ladder and white arrow indicates the position of the specific band.
Discussion
Gene Sr25 transfer to wheat Thinpyrum ponticum, occurs on the long arm of chromosome 7DL. It is usually closely linked to leaf rust resistance gene Lr19. Another Th. ponticum-derived gene resulted in undesirable yellow flour [32], and improved the yield [21]. The Sr25 resistance is affected by growth periods and temperature. The resistance in seedling stage is higher than in adult stage and is more easily susceptible under high temperature conditions [33]. Due to its resistance to Ug99, the specific primer Xwmc221 was used for detection of 150 cultivars in China, but no Sr25 was identified [21]. In this study, the same primer pair was used for detection of Sr25 in119 wheat cultivars (lines) from Yunnan Province. The results also indicated that none of the tested wheat cultivars contained the gene. Therefore, the gene was targeted in the future breeding programs to improve the resistance level of wheat cultivars to Ug99 in China.
The wheat stem rust resistance gene Sr26 (derived from Agropyron elongatum) is derived from foreign chromosome translocated to wheat chromosomes [24]. This gene located on chromosome 6AL, confers resistance to all races of wheat stem rust worldwide [9] and is widely used in Australia [24]. It is reported that the Chinese wheat varieties may contain Sr26. Two out of 448 wheat cultivars contain Sr26 that was identified by researchers [34–36]. In this study, the Yunnan varieties were tested for presence of Sr26. However, the gene was not found in any of the materials tested, which indicated that Yunnan wheat varieties lack the Sr26.
Sr28 originates in T. aestivum L. Kota. Kota has been used as a differential host by Stakman since 1962. However, due to ineffectiveness to most races of Pgt, its application was limited in resistance breeding [25]. With the emergence of Ug99, researchers focused on the discovery of genes or gene combinations resistant to Ug99, to strengthen the resistant cultivars in the international wheat stem rust nursery of Kenya. SD1691 resistant to Ug99 (TTKSK) was studied by Rouse et al. [37], and found to harbor the Sr28 resistance gene [25]. In addition, Sr28 was ineffective to several popular American races of Pgt. However, it was combined with other resistance genes to yield effective resistance gene combinations, and has been used in breeding disease resistance [25]. Therefore, the diversity arrays technology (DArT) [38] marker closely linked to Sr28 was developed for more convenient screening and identification of the effective gene. Similarly, Sr28 was also ineffective to many Pgt races in China [39, 40]. Chen et al. [35] reported that Chinese wheat cultivars carry Sr28. In this study, the DArTmarker wPt-7004 screened by Rouse was used to detect the gene in Yunnan wheat cultivars (lines). The results showed that 12 wheat varieties (lines) carried this gene. These were Nanyuan 1, Feng 0103, Jing07-2, Jing 05–1,088–16, E33, Linmai 15, Jing 0202, Jingmai 10, Yunmai 54, Yunmai 47, and Feng 615.
The gene Sr31, which confers a high degree of effectiveness to stem rust was introgressed into bread wheat from ‘Petkus’ rye as a 1BL/1RS translocation [26]. Sr31 has been widely used in Chinese and global wheat stem rust breeding programs [26, 41, 42]. However, wheat stem rust has re-emerged as a major threat to global wheat production following the emergence of the new race Ug99 virulent to Sr31 in 1999 [5]. To date, no virulent race against Sr31 has been reported in China [41]. In our study, 43 cultivars (lines) probably carried Sr31. Because Sr31 has been widely used in Chinese wheat breeding since 1970s, and identified that many cultivars carry this gene [41, 42]. Our results were consistent with previous reports. For example, Wang et al. [43] and Zhang et al. [44] identified a 1BL/1RS translocation in 211 and 75 wheat cultivars (lines) using the co-dominant PCR marker. Their results showed that 81 and 25 wheat varieties carried this gene, respectively. Conversely, pedigree tracking indicated that resistant materials carrying Sr31 such as ‘Kavkaz’ and ‘Luofu’ lines were widely used in wheat breeding in Yunnan Province [45], suggesting the origin of resistance genes in these wheat varieties. In addition, a specific band was amplified in Yunmai 51 (91B-831/92B-84) and Fengmai 31 (918M40-1) using SCSS30.2576 marker. However, the two cultivars were highly susceptible to Pgt in China and no race virulent to Sr31 was found, suggesting that the two cultivars do not contain the gene Sr31. This finding may be attributed to false-positive results, which are common challenges associated with MAS. For example, Yu et al. reported that the variety Thatcher does not carry Sr2 but tested positive [46].
Sr32 present on the short arm of chromosome 2S#1 was transferred to wheat from Aegilops speltoides Tausch [47]. Sr32 exhibited strong resistance against the predominant race group 21C3 in China [13, 39]. In this study, we found that 91E001 carried Sr32. The wheat cultivar 91E001 used in breeding programs was introduced from Mexico to China, with strong resistance against wheat stripe rust [48]. Additionally, Sr32 was shown to be effective against seven race variants of Ug99 lineage [9], and was used in future resistance breeding. The durability of resistance genes is enhanced by deploying pyramids in cultivars.
The translocation line VPM1 of Triticum aestivum L. containing 2NS chromosome segments (25~38cM) from Ae. ventricosa was cultivated by Maia in 1967 [32]. The fragment carried three effective genes against rust: Lr37 against leaf rust, Sr38 against stem rust and Yr17 against stripe rust. These three genes have been widely used by breeders worldwide [32, 41]. Sr38 is no longer resistant to new races related to Ug99. No virulent Pgt race to Sr38 has been found in China. PCR analysis was used for effective screening of resistance genes Lr37-Yr17-Sr38 established by Helguera [29], which played an important role in MAS [30], and was used in this study for identification of genes in wheat cultivars of Yunnan Province. The results showed 10 wheat cultivars harboring the gene. The resistance of these cultivars against the Chinese races 34MKGSM and 21C3CTHTM may be attributed to the Lr37-Yr17-Sr38 genes.
In this study, the molecular genetic markers of Sr25, Sr26, Sr28, Sr31, Sr32 and Sr38 in wheat cultivars against Pgt races were used to identify the 119 wheat cultivars (lines) in Yunnan province. Forty three and 10 wheat cultivars (lines) are likely to carry Sr31 and Sr38, respectively. Sr31 and Sr38 have been widely used in Chinese stem rust resistance breeding [41]. Although Sr31 and Sr38 are no longer resistant to new races related to Ug99, they are still useful when combined or used as pyramids with other genes effective against Ug99. Overall, wheat cultivars in Yunnan province lack the genes Sr25 and Sr26, which are effective against Ug99. Sr28 alone was detected in 12 wheat cultivars including Nanyuan 1, Feng 0103, Jing 07–2, Jing 05–1, 088–16, E33, Linmai 15, Jing 0202, Jingmai 10, Yunmai 54, Yunmai 47, and Feng 615. Sr32 was detected in one cultivar 91E001. Spread of Ug99 into China will most likely lead to colonization of the southwest area such as Yunnan and Sichuan provinces, where Pgt is already prevalent [41]. Therefore, the widespread cultivation of these wheat cultivars susceptible to Ug99 in Yunnan greatly increases the risk of its epidemic. Fortunately, the two genes Sr28 and Sr32 might play a role in the prevention of colonization and invasion of Ug99. The 12 cultivars containing Sr28 and the 91E001 carrying Sr32 identified in this study will be useful for resistance breeding and propagation of cultivars.
Acknowledgments
We appreciate very much to Prof. Yuanhu Xuan from College of Plant Protection, Shenyang Agricultural University for critical reading of our manuscript, and Dr. Mingju Li from Yunnan Provincial Institute of Agricultural Environment and Resources for providing whole wheat cultivars.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported by the National Key Basic Research Program of China (2013CB127701), the Special Fund for Agro-scientific Research in the Public Interest (201303016), and the National Natural Science Found of China (31171829).
References
- 1.Pardey PG, Beddow JM, Kriticos DJ, Hurley TM, Park RF, Duveiller E, et al. Right-sizing stem-rust research. Science. 2013; 340: 147–148. 10.1126/science.122970 [DOI] [PubMed] [Google Scholar]
- 2.Singh RP, Hodson DP, Jin Y, Huerta-Espino J, Kinyua MG, Wanyera R, et al. Current status, likely migrations and strategies to mitigate the threat to wheat production from raceUg99 (TTKS) of stem rust pathogen. CAB Rev Perspectives Agric Veterinary Sci Nutr Nat Resour. 2006; 1:13. [Google Scholar]
- 3.Cao YY, Chen WQ. Stepwise shift of differential hosts and racial designation of Puccinia graminis f. sp. tritici. Journal of Triticeae Crops. 2010; 30: 167–162 (in Chinese). [Google Scholar]
- 4.Wu XX, Li TY, Chen S, Wang GQ, Cao YY, Ma SL, et al. Stem rust resistance evaluation and Ug99-resistance gene detection of 139 wheat cultivars. Scientia Agricultura Sinica. 2014; 47: 4618–4626 (in Chinese). [Google Scholar]
- 5.Pretorius ZA, Singh RP, Wagoire WW, Payne TS. Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 2000; 84: 203. [DOI] [PubMed] [Google Scholar]
- 6.Botma V, Liezel H, Robert FP, Haydar K, Cornelia MB, Zacharias AP. Characterization of two new Puccinia graminis f. sp. tritici races within the Ug99 lineage in South Africa. Euphytica. 2011; 179: 119–127. [Google Scholar]
- 7.Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Peter N, Ruth W, et al. Will stem rust destroy the world’s wheat crop? Advances in Agronomy. 2008; 98: 271–309. [Google Scholar]
- 8.Patpour M, Hovmøller MS, Justesen AF, Newcomb M, Olivera P, Jin Y, et al. Emergence of virulence to SrTmp in the Ug99 race group of wheat stem rust, Puccinia graminis f. sp. tritici, in Africa. Plant Dis. 2016; 2: 552. [Google Scholar]
- 9.Jin Y, Szabo LJ, Pretorius ZA, Singh RP, Ward RW, Fetch TJ. Detection of virulence to resistance gene Sr24 within race TTKS of Puccinia graminis f. sp. tritici. Plant Dis. 2008; 92: 923–926. [DOI] [PubMed] [Google Scholar]
- 10.FAO. Available: http://www.fao.org/agriculture/crops/rust/stem/stem-pathotypetracker/Stem-effectivesrgenes/en/. Accessed 7 Feb 2013.
- 11.Nazari K, Mafi M, Yahyaoui A, Singh RP, Park RF. Detection of wheat stem rust race (Puccinia graminis f. sp. tritici) TTKSK (Ug99) in Iran. Plant Dis. 2009; 93: 317. [DOI] [PubMed] [Google Scholar]
- 12.Stokstad E. Deadly wheat fungus threatens world’s breadbaskets. Science. 2007; 315: 1786–1787. 10.1126/science.315.5820.1786 [DOI] [PubMed] [Google Scholar]
- 13.Li TY. Genetic control approaches to wheat (oat) stem rusts and Ug99 in China. M.Sc. Thesis, Shenyang Agricultural University. 2014. Available: http://cdmd.cnki.com.cn/Article/CDMD-10157-1014295098.htm. (in Chinese).
- 14.Umesh G, Sarvjeet K, Rakesh Y, Neha S, Kajal T, Aakash KG. Recent trends and perspectives of molecular markers against fungal diseases in wheat. Front Mic. 2015. August 25 10.3389/fmicb.2015.00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pretorius ZA, Jin Y, Bender CM, Herselman L, Prins R. Seedling resistance to stem rust race Ug99 and marker analysis for Sr2, Sr24 and Sr31 in South African wheat cultivars and lines. Euphytica. 2012; 186: 15–23. [Google Scholar]
- 16.Asma H, Tayyaba S, Tahsin G, Mahmood R, Fatima J, Summera S, et al. Study of rust resistance genes in wheat germplasm with DNA markers. Bioinformation. 2014; 10: 371–3770. 10.6026/97320630010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokhmetova AM, Atishova MN, et al. Identification of sources of resistance to wheat stem using molecular markers. Russ J Genet Appl Res. 2012; 2: 486–493. [Google Scholar]
- 18.Haile JK, Hammer K, Badebo A, Nachit MN, Roder MS. Genetic diversity assessment of Ethiopian tetraploid wheat landraces and improved durum wheat varieties using microsatellites and markers linked with stem rust resistance. Genetic Resources and Crop Evolution. 2013; 60: 513–527. [Google Scholar]
- 19.Ma Y. Preliminary molecular detection for the stem rust resistant gene Sr22 in part of the wheat germplasms. Journal of Triticeae Crops. 2013; 1: 7–10 (in Chinese). [Google Scholar]
- 20.Ma Y, Shao LG, Wang Y, Li CH, Che JY, Gao FM, et al. Molecular detection of the stem rust resistant gene Sr33 in spring wheat cultivars. Journal of Triticeae Crops. 2013; 33: 34–38 (in Chinese). [Google Scholar]
- 21.Han JD. Resistant gene control and related mechanism to the invasion of race Ug99 of Puccinia graminis f. sp. tritici. M.Sc. Thesis, Shenyang Agricultural University. 2009. Available: http://cdmd.cnki.com.cn/Article/CDMD-10157-2009190472.htm (in Chinese).
- 22.Lagudah ES, Appels R, Brown AH, McNeil D. The molecular genetic analysis of Triticum tauschii-the D genome donor to hexaploid wheat. Genome. 1991; 34: 375–386. [Google Scholar]
- 23.Stakman EC, Stewart DM, Loegering WQ. Identification of physiologic races of Puccinia graminis var. tritici. US Department of Agric. 1962; ARS E-617, p 53.
- 24.Mago R, Bariana HS, Dundas IS, Spielmeyer W, Lawrence GJ, Pryor AJ, et al. Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor Appl Genet. 2005; 111: 496–504. 10.1007/s00122-005-2039-z [DOI] [PubMed] [Google Scholar]
- 25.Rouse MN, Nava IC, Chao S, Anderson JA, Jin Y. Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor Appl Genet. 2012; 125: 877–885. 10.1007/s00122-012-1879-6 [DOI] [PubMed] [Google Scholar]
- 26.Das BK, Saini A, Bhagwat SG, Jawali N. Development of SCAR markers for identification of stem rust resistance gene Sr31 in the homozygous or heterozygous condition in bread wheat. Plant Breeding. 2006; 125: 544–549. [Google Scholar]
- 27.Mago R, Verlin D, Zhang P, Bansal U, Bariana H, Jin Y, et al. Development of wheat–Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theor Appl Genet. 2013; 126: 2943–2955. [DOI] [PubMed]
- 28.Seah S, Bariana H, Jahier J, Sivasithamparam K, Lagudah ES. The introgressed segment carrying rust resistance genes Yr17, Lr37 and Sr38 in wheat can be assayed by a cloned disease resistance gene-like sequence. Theor Appl Genet. 2012; 102: 600–605. [Google Scholar]
- 29.Helguera M, Khan IA, Kolmer J, Lijavetzky D, Zhong-qi L, Dubcovsky J. PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Published in Crop Sci. 2003; 43: 1839–1847. [Google Scholar]
- 30.Kuchel H, Ye G, Fox R, Jefferies S. Genetic and economic analysis of a targeted marker-assisted wheat breeding strategy. Molecular Breeding. 2005; 16: 67–78. [Google Scholar]
- 31.Li FQ, Han JD, Wei GR, Zeng QD, Kang ZS. Identification of Lr37-Yr17-Sr38 in wheat cultivars of Huanghuai wheat region using molecular markers. Journal of Northwest A&F University. 2009; 37: 151–158 (in Chinese). [Google Scholar]
- 32.McIntosh RA, Wellings CR, Park F. Wheat Rusts, an Atlas of Resistance Genes 1st ed. Australia: CSIRO Press; 1995. [Google Scholar]
- 33.Dyck PL. Transfer of gene for stem rust resistance from Triticum araticum to hexaploid wheat. Genome. 1992; 35: 788–792. [Google Scholar]
- 34.Zhang SK, Qiu YC, Yao P. Postulation of resistant genes to stem rust in 94 cultivars of wheat important resistant resources. Journal of Shenyang agricultural university. 1998; 29: 117–122 (in Chinese). [Google Scholar]
- 35.Chen WQ, Wang JX. Preliminary analysis for the resistance genes of 76 wheat germplasms to stem rust and leaf rust. Acta Agronomica sinica. 1997; 23: 655–663 (in Chinese). [Google Scholar]
- 36.Li MJ, Chen WQ, Duan XY, Xu SC, Yu YX, Bi YL, et al. Analysis of wheat cultivars adult resistance to wheat strip rust and powdery mildew in Yunnan Province. Yunnan Agricultural Science and Technology. 2012; 2: 4–7 (in Chinese). [Google Scholar]
- 37.Rouse MN, Wanyera R, Njau P, Jin Y. Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis. 2011; 95: 762–766. [DOI] [PubMed] [Google Scholar]
- 38.Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang SY, et al. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Gene. 2006; 113: 1409–1420. [DOI] [PubMed] [Google Scholar]
- 39.Han JD, Cao YY, Sun ZG. 2007–2008 Race dynamics of Puccinia graminis f. sp. tritici in China and the virulence of CIMMYT wheat germplasm resistant to Ug99. Journal of Triticeae Crops. 2010; 30: 163–166 (in Chinese). [Google Scholar]
- 40.Wu YS, Huang ZT. Twenty years’ racial identification and fluctuation analysis of Puccinia graminis Var. tritici in China. Journal of Shenyang Agricultural University. 1987; 18: 105–138 (in Chinese). [Google Scholar]
- 41.Cao YY, Han JD, Zhu GQ, Zhang L. Ug99, a new virulent race of Puccinia graminis f. sp. tritici, and its effect on China. Plant Protection. 2007; 33: 86–89 (in Chinese). [Google Scholar]
- 42.He ZH, Xia XC, Chen WQ. Breeding for resistance to new race Ug99 of stem rust pathogen. Journal of Triticeae Crops. 2008; 28: 170–173 (in Chinese). [Google Scholar]
- 43.Wang XJ, Feng GH, Liu DH, Wang J, Zhang HY, Zhao JH. Molecular detection of 1BL/1RS translocation in partial wheat cultivars (Lines) from Huang-Huai wheat region of China. Journal of Triticeae Crops. 2008; 28: 381–386 (in Chinese). [Google Scholar]
- 44.Zhang YW, Liu B, Liu TG, Gao L, Chen WQ. Molecular detection of Yr10 genes and 1BL/1RS translation in wheat cultivars. Plant protection. 2014; 40: 54–59 (in Chinese). [Google Scholar]
- 45.Wu YS, Feng YB, Yang YH. Excellent cultivars and their genetic relationship in Yunnan Province. Yunnan Agricultural Science and Technology. 1998; 2: 9–12 (in Chinese). [Google Scholar]
- 46.Yu LX, Liu S, Anderson JA, Singh RP, Jin Y, Dubcovsky J, et al. Haplotype diversity of stem rust resistance loci in uncharacterized wheat lines. Mol Breeding. 2010; 26: 667–680. [Google Scholar]
- 47.Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS. Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica. 1996; 91: 59–87. [Google Scholar]
- 48.Li WH. Establishment of new surveillance stem for Chinese races and Ug99 of Puccinia graminis f. sp. tritici, resistant genes defection in commercial wheat varieties. M.Sc. Thesis, Shenyang Agricultural University. 2012. Available: http://cdmd.cnki.com.cn/Article/CDMD-10157-1012442449.htm (in Chinese).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.