Abstract
MicroRNA-21 (miR-21) has been reported as the potential novel diagnostic biomarker for breast cancer in several studies, but their results were inconsistent. Therefore, we conducted a systematic analysis to evaluate the diagnostic value of miR-21 in detecting breast cancer. A comprehensive electronic and manual search was conducted for relevant literatures through several databases up to November 9, 2015. QUADAS-2 was used to assess the quality of the studies included in the study. All statistical analyses were performed using Meta-Disc 1.4 and Stata 12.0. Eleven studies with a total of 918 breast cancer patients and 613 controls were included. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) with their 95% confidence intervals (CIs) were 0.72 (95% CI: 0.69–0.75), 0.80 (95% CI: 0.77–0.83), 3.37 (95% CI: 2.24–5.07), 0.30 (95% CI: 0.19–0.50), and 11.79 (95% CI: 5.23–26.57), respectively. The area under the curve of SROC was 0.8517. In conclusion, our analyses suggested that miR-21 is a promising biomarker in diagnosing breast cancer. For clinical purpose, further large-scale studies are warranted to validate its clinical application.
Keywords: microRNA-21, breast cancer, diagnostic, biomarker, meta-analysis
INTRODUCTION
Breast cancer is the most common cancer among women worldwide. Although breast cancer incidence rates still increases in many Western countries, mortality rates have been decreasing over the past two decades due to early detection and improved treatment [1]. The data from Surveillance, Epidemiology, and End Results Program (SEER) showed that the 5-year relative survival was 98.6% when diagnosed at localized stage as opposed to 23.3% when the disease at distant stage [2]. Thus, early detection and diagnosis has important clinical significances for breast cancer. The previous studies showed that the circulating tumor biomarkers such as carcinoembryonic antigen (CEA) and carbohydrate antigen 153 (CA153) are already applied in clinic, but these biomarkers are not useful to detect early breast cancer due to their low sensitivity and they have long been used as prognostic markers to monitor disease progression or recurrence [3–5].
After the first report of elevated circulating levels of microRNA-21 (miR-21) in patients with diffuse large B-cell lymphoma [6], circulating miRNAs with their stability feature have been postulated as novel biomarkers for cancer processes, such as liver cancer, ovarian cancer, breast cancer [7–9]. Several studies have reported miR-21 as the potential novel diagnostic biomarker for breast cancer, but their results were inconsistent. A recent study suggested that the circulating miR-21 could serve as a potential serum-based biomarker for breast cancer detection in Chinese population, with 80.0% sensitivity and 87.7% specificity [10]. Another study investigated the diagnostic accuracy of single miR-21 and reported a much lower sensitivity with 25.8% [11]. In the Asaga's study, significant up-regulation of miR-21 was detected, but it could not as candidate in the selection criteria at the microarray level [12]. Therefore, we conducted a systematic analysis to evaluate the diagnostic value of miR-21 in detecting breast cancer.
RESULTS
Included studies
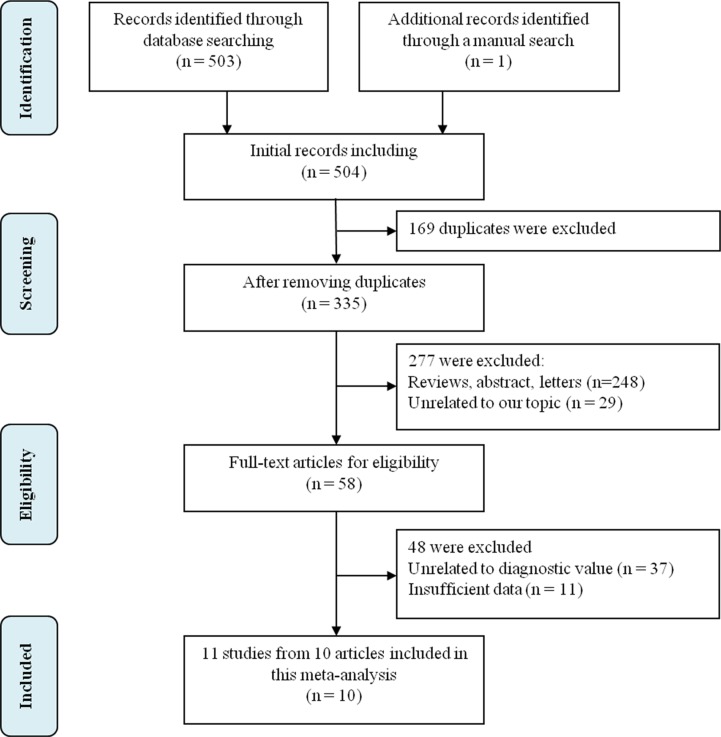
A detailed flowchart of the review process was presented in Figure 1. A total of 504 articles were identified by initial search, with 503 records identified from database searching and 1 record by manual search. Two independent researchers reviewed articles for duplicates, excluding 169 records. After carefully reviewing titles and abstracts of 335 records, as a result, there were 277. Excluded: 248 were reviews, abstract and letters and 29 were not related to our topic, leaving 58 full-text articles for eligibility. Finally, 11 studies from 10 articles were included in this meta-analysis [10–19].
Figure 1. A detailed flowchart of the review process.
Study characteristics and quality assessment
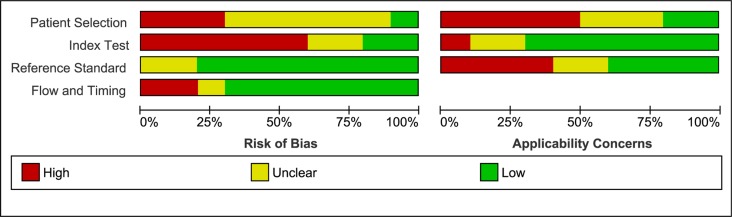
The main characteristics of included studies were summarized in Table 1. Among the 11 studies, 7 studies were conducted in China [10, 11, 13–27], 1 in USA [12], 2 in Mexico [18], and 1 in Egypt [19]. The publication years ranged from 2011 to 2015. A total of 918 breast cancer patients and 613 controls were included. Circulation miR-21 expression levels were measured in serum (n = 8), tumor tissue (n = 2), and plasma (n = 1). In each study, the cutoff values of miR-21 appeared to be different. The quantitative real-time reverse transcription PCR method was used to measure the expression of miR-21. The sufficient data which were used to construct the 2 × 2 table, such as True positive (TP), false positive (FP), false negative (FN), and true negative (TN), were successfully extracted. The quality assessment of the QUADAS-2 tool was shown in Figure 2. Overall, most studies presented they were of high quality relatively.
Table 1. Main characteristics of included studies.
| First author | Year | Country | Ethnicity | Sample size | TP | FP | FN | TN | Cut-off value | Sample types | Reference controls | RNA extraction | Measurements | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||||||||
| Li | 2011 | China | Asian | 33 | 49 | 17 | 3 | 16 | 46 | 18.32 | Serum | miR-16 | TRIzol | SYBR |
| Asaga | 2011 | USA | Caucasian | 102 | 20 | 72 | 3 | 30 | 17 | 3.3-dCq | Serum | miR-16 | TRIzol | SYBR |
| Sun | 2012 | China | Asian | 103 | 55 | 77 | 18 | 26 | 37 | 1.358 2−ΔΔCt | Serum | cel-miR-39 | Filter cartridge | Taqman |
| Wang | 2012 | China | Asian | 50 | 39 | 40 | 5 | 10 | 34 | 4.58 2−ΔΔCt | Serum | miR-16 | TRIzol | SYBR |
| Mar-Aguilar | 2013 | Mexico | Caucasian | 61 | 10 | 58 | 2 | 3 | 8 | 6.48 2−ΔΔCt | Serum | 18S RNA | miRNAeasy kit | Taqman |
| Mar-Aguilar | 2013 | Mexico | Caucasian | 50 | 10 | 38 | 4 | 12 | 6 | 6.48 2−ΔΔCt | Tissue | 18S RNA | miRNAeasy kit | Taqman |
| Gao | 2013 | China | Asian | 89 | 55 | 78 | 7 | 11 | 48 | 13.22 | Serum | CA153, CEA | TRIzol | SYBR |
| Lee | 2013 | China | Asian | 110 | 15 | 99 | 4 | 11 | 6 | 2.5 2−ΔΔCt | Tissue | 18S RNA | TRIzol | SYBR |
| Ng | 2013 | China | Asian | 170 | 100 | 128 | 22 | 42 | 78 | 2.34 2−ΔΔCt | Plasma | miR-145 | TRIzol | Taqman |
| Li | 2013 | China | Asian | 120 | 200 | 31 | 46 | 89 | 154 | NA* | Serum | CA153, CEA | Roche Elecsys | Taqman |
| Toraih | 2015 | Egypt | Caucasian | 30 | 60 | 20 | 8 | 10 | 52 | 7.02 2−ΔΔCt | Serum | RNU6B | Qiagen miRNeasy kit | Taqman |
Data unavailable.
Figure 2. Risk of bias and applicability concerns graph a review of authors' judgments about each domain presented as percentages across included studies.
Diagnostic accuracy and threshold analysis
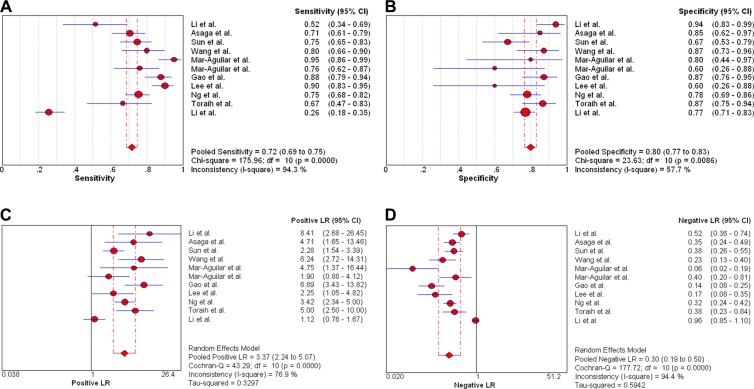
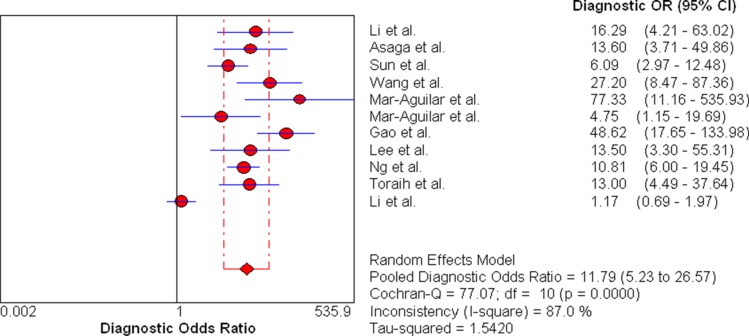
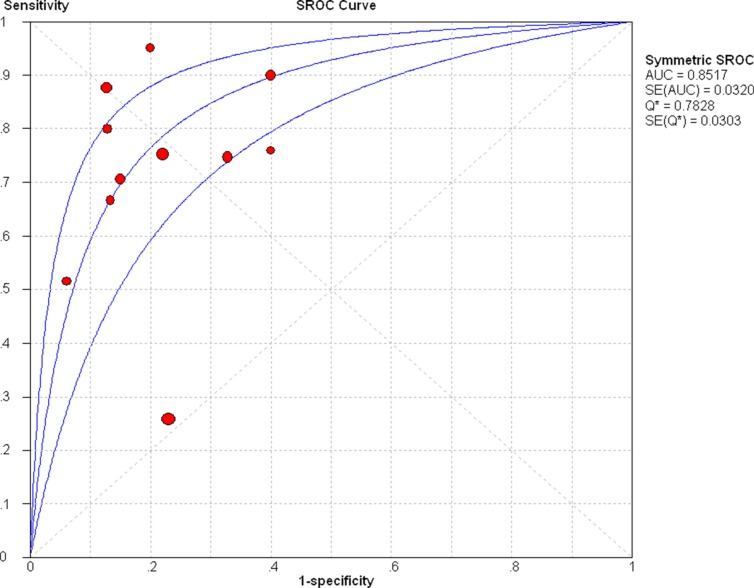
Firstly, we conducted analysis of diagnostic threshold to explore whether the threshold effect was existed in this study, which was an important source of heterogeneity. The results showed that there was no heterogeneity from threshold effect with the spearmen correlation coefficient of sensitivity and 1-specificity of 0.178 (P = 0.601). Then Cochran-Q and inconsistency index (I2) were used to measure whether there was heterogeneity from non-threshold effect in order to choose appropriate calculation model. We used the random effects model to calculate those pooled diagnostic parameters for breast cancer. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) with their 95% confidence intervals (CIs) were 0.72 (95% CI: 0.69–0.75, Figure 3A), 0.80 (95% CI: 0.77–0.83, Figure 3B), 3.37 (95% CI: 2.24–5.07, Figure 3C), 0.30 (95% CI: 0.19–0.50, Figure 3D), and 11.79 (95% CI: 5.23–26.57, Figure 4), respectively. The area under the curve (AUC) of SROC was 0.8517 (Figure 5).
Figure 3.
Forest plots of pooled sensitivity (A), specificity (B), positive likelihood ratio (C), and negative likelihood ratio (D) for miR-21 in the diagnosis of breast cancer.
Figure 4. Forest plots of pooled diagnostic odds ratio for miR-21 in the diagnosis of breast cancer.
Figure 5. Summary receiver operating characteristic (SROC) curve for miR-21 in the diagnosis of breast cancer.
Meta-regression, subgroup analysis and publication bias
We also performed meta-regression analysis to explore source of heterogeneity based on ethnicity, sample size (≥ 100 vs. < 100), sample source, reference controls, RNA extraction, measurements (Table 2). The results showed that none of the above covariates contributed the heterogeneity (all P > 0.05). Then we conducted subgroup analysis based on those covariates. The results of different subgroups were relatively consistent with the major results, which suggested that our results were relatively credible (Table 3). Moreover, Egger' test (P = 0.909) or Begg's test (P = 0.488) was detected and the results showed that there was no significant publication bias in our study.
Table 2. Results of the multivariable meta-regression model for the characteristics with backward regression analysis (Inverse variance weighs).
| Variables | Coefficient | Standard Error | P | RDOR | 95% CI |
|---|---|---|---|---|---|
| Cte | 5.451 | 0.7895 | 0.0023 | −-- | −-- |
| S | 0.205 | 0.1766 | 0.3095 | −-- | −-- |
| Ethnic | 0.424 | 0.4500 | 0.3992 | 1.53 | 0.44–5.33 |
| Sample size | −1.304 | 0.5388 | 0.0519 | 0.27 | 0.08–1.04 |
| Sample types | −0.243 | 0.2440 | 0.3568 | 0.78 | 0.43–1.42 |
| Reference controls | −0.056 | 0.2244 | 0.8178 | 0.95 | 0.46–1.93 |
| RNA extraction | −1.131 | 0.5786 | 0.0863 | 0.32 | 0.08–1.23 |
| Measurements | 2.768 | 1.2297 | 0.0742 | 15.92 | 0.67–375.70 |
Table 3. Results of subgroup analysis in diagnostic accuracy of miR-21 for breast cancer.
| Subgroup | No. of studies (No. of cases) | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | AUC |
|---|---|---|---|---|---|---|---|
| Ethnicity | |||||||
| Asian | 7 (675) | 0.70 (0.66–0.73) | 0.79 (0.76–0.83) | 3.27 (1.92–5.56) | 0.33 (0.17–0.62) | 10.64 (3.66–30.97) | 0.8472 |
| Caucasian | 4 (243) | 0.77 (0.72–0.82) | 0.83 (0.74–0.90) | 3.65 (2.18–6.11) | 0.29 (0.17–0.49) | 13.73 (5.54–34.04) | 0.8625 |
| Sample size | |||||||
| ≥ 100 | 5 (605) | 0.67 (0.63–0.71) | 0.76 (0.71–0.80) | 2.32 (1.41–3.82) | 0.38 (0.19–0.75) | 6.31 (2.09–19.00) | 0.7935 |
| < 100 | 6 (313) | 0.80 (0.75–0.84) | 0.87 (0.82–0.91) | 4.89 (3.13–7.62) | 0.26 (0.15–0.44) | 20.89 (10.14–43.06) | 0.8948 |
| Sample types | |||||||
| Serum | 8 (588) | 0.67 (0.63–0.71) | 0.81 (0.77–0.85) | 3.95 (2.19–7.12) | 0.31 (0.17–0.57) | 13.46 (4.37–41.41) | 0.8865 |
| Tissue | 2 (160) | 0.86 (0.79–0.91) | 0.60 (0.36–0.81) | 2.07 (1.20–3.56) | 0.26 (0.11–0.62) | 8.04 (2.86–22.58) | — |
| Plasma | 1 (170) | — | — | — | — | — | — |
| Reference control | |||||||
| miR-16 | 3 (185) | 0.70 (0.63–0.76) | 0.90 (0.83–0.95) | 6.18 (3.51–10.89) | 0.36 (0.24–0.55) | 18.81 (9.06–39.06) | 0.8954 |
| 18S RNA | 3 (221) | 0.88 (0.83–0.92) | 0.67 (0.47–0.83) | 2.37 (1.44–3.89) | 0.17 (0.06–0.46) | 15.09 (3.49–65.19) | 0.5981 |
| CA153, CEA | 2 (209) | 0.82 (0.74–0.89) | 0.87 (0.79–0.93) | 5.86 (3.59–9.58) | 0.23 (0.09–0.63) | 15.40 (6.97–92.54) | — |
| RNA extraction | |||||||
| TRIzol | 6 (554) | 0.78 (0.75–0.82) | 0.84 (0.79–0.88) | 4.45 (3.02–6.54) | 0.28 (0.19–0.40) | 17.90 (10.63–30.15) | 0.8800 |
| Others | 5 (364) | 0.62 (0.56–0.67) | 0.77 (0.72–0.81) | 2.36 (1.35–4.13) | 0.36 (0.17–0.77) | 6.84 (1.95–23.97) | 0.8058 |
| Measurements | |||||||
| SYBR | 5 (384) | 0.80 (0.75–0.84) | 0.87 (0.81–0.92) | 5.02 (3.09–8.16) | 0.26 (0.16–0.43) | 23.59 (13.66–40.73) | 0.8974 |
| Taqman | 6 (534) | 0.78 (0.73–0.81) | 0.77 (0.71–0.82) | 3.02 (2.17–4.19) | 0.32 (0.22–0.45) | 10.03 (5.50–18.26) | 0.8289 |
DISCUSSION
We performed a systematic review to evaluate the diagnostic value of miR-21 as a potential diagnostic biomarker for breast cancer patients. Our finding suggested that the pooled sensitivity, specificity, PLR, NLR and DOR were 0.72 (95% CI: 0.69–0.75), 0.80 (95% CI: 0.77–0.83), 3.37 (95% CI: 2.24–5.07), 0.30 (95% CI: 0.19–0.50) and 11.79 (95%CI: 5.23–26.57), respectively. The AUC of SROC was 0.8517.
Currently, a number of convenient and novel biomarkers have been established in the routine evaluation of breast cancer. Although estrogen receptor (ER) and human epidermal growth factor receptor-2 (HER2) for predicting the response to endocrine and biological therapies are already available, their performances are far from perfect. For example, there were still some non-responding patients in the assessment of ER and HER2 status [20, 21]. In addition, other molecular biomarkers, such as CEA, cytokeratin fragment (CYFRA 21-1), and neuron specific enolase (NSE), were limited in the clinic with their low sensitivity and specificity [22].
Recently, various studies showed that abnormal expression of miRNAs played an important role in the pathogenesis, metastasis and prognosis for breast cancer [23, 24]. Some studies reported that miR-21 might be as a potential biomarker for breast cancer diagnosis because breast cancer patients had higher serum miR-21 expression than healthy women [25, 26]. In our meta-analysis, the pooled sensitivity and specificity were 0.72 and 0.80, which indicated that the diagnostic accuracy may not be high enough as expected. The results were consistent with the recently published studies by Li et al. and Shen et al. [27, 28]. However, compared with some traditional biomarkers, such as CEA, NSE (with sensitivities of 0.48 and 0.39), miR-21 still had higher diagnostic value in detecting breast cancer. The PLR and NLR were used to estimate the diagnostic accuracy in clinical level. The pooled PLR of 3.37 suggested that breast cancer patients could have about 3.37-fold higher chance of being miR-21 positive compared to healthy controls. The pooled NLR of 0.30 indicated that the possibility of individuals having cancer was 30% if the miR-21 was negative. Moreover, the value of DOR ranged from 0 to infinity, with higher value meaning better test discrimination [29]. The area under curve is another parameter to evaluate the diagnostic value. The ideal SROC curve position is near the upper-left corner which would imply a perfect test [30]. Statistically, if the range of AUC was 0.97 or above which was considered to have excellent accuracy; the range of AUC 0.93–0.96 was considered to be very good; the range of AUC 0.75–0.92 was considered to be good; and a range of AUC less than 0.75 should be cautiously to evaluate the accuracy which might be a random test [31]. Our results of DOR and AUC was 11.79 and 0.8517, respectively, which indicated the overall accuracy in diagnosing breast cancer was high.
Exploring the sources of heterogeneity is important in a meta-analysis. In this study, there was no heterogeneity from threshold effect with the spearmen correlation coefficient of sensitivity and 1-specificity of 0.178. However, substantial heterogeneity was found during the analyzing several parameters. Meta-regression and some subgroup analyses were conducted according to the majors attributes of primary studies. In present study, different measures such as reference controls, RNA extraction and measurement methods, were used to extract miR-21 in different studies. All these variables may influence the heterogeneity. As a result, we failed to find the sources. On the other hand, we also analyzed miR-21 diagnostic efficiency in three different sample types. The subgroup analysis showed that serum-based miR-21 had higher accuracy than miR-21 in tissue for diagnosing breast cancer. Other results of different subgroups were relatively consistent with the major results, which suggested that our results were relatively credible.
There were several potential limitations in our study. Firstly, the results may suffer from publication bias in our study, because studies with null results tend not to be published. Publication languages were limited to English and Chinese. Other potentially eligible studies which met our inclusion criteria may not be included. Secondly, sample sizes of studies included in this meta-analysis were small, which may appear a small-study effect. Thirdly, only Asian and Caucasian populations were considered in subgroup analysis, with no African population involved, which may cause selection bias from population.
In conclusion, our analyses suggested that miR-21 is a promising biomarker in diagnosing breast cancer. For clinical purpose, further large-scale studies are warranted to validate its clinical application.
MATERIALS AND METHODS
Our present study was performed in accordance with the guidelines for the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) [31].
Search strategy
Two researchers independently searched PubMed, Embase, Chinese National Knowledge Infrastructure (CNKI), Wan Fang Data, and VIP database to identify relevant studies which evaluated the diagnostic value of miR-21 for breast cancer patients, up to November 9, 2015. A manual review of relevant publications was also performed to obtain additional studies. The following search terms were used to retrieve articles and abstracts: (microRNA-21 or micro RNA 21 or miRNA-21 or miR-21) and (breast cancer or breast tumor or breast neoplasm or breast carcinoma). Only the most recent or the largest sample size study was included in the final analysis. Publication languages were limited to English and Chinese.
Study selection
Studies included in present meta-analyses should meet the following criteria: (1) diagnostic effect about miR-21 for breast cancer; (2) breast cancer was confirmed by pathological examination; (3) the levels of miR-21 in tissue or serum was determined; (4) sensitivity, specificity, and cut-off values can be found in identified studies or calculated from the provided data. While the exclusion criteria were listed as follow: (1) studies without sufficient data to construct the 2 × 2 table; (2) Meta-analyses, reviews, comments, letters, editorial articles, conference abstracts, meeting, and animal and cell studies; (3) publications were identified as duplicates.
Data extraction
Two researchers reviewed the abstract first independently and then summarized the full selected articles. Any disagreements were resolved by discussion or consulting the third reviewer. The relevant data were extracted as follow: first author, publication years, country of origin, ethnicity, number of patients and controls, true and false positive and negative, cut-off value, sample types, reference control, RNA extraction, measurements.
Quality assessment
Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) [32] was used to assess the quality of the studies included in this meta-analysis independently by the same two researchers. Each of the assessment has seven questions with the answered with “yes”, “no”, or “unclear”. The answer of “yes” means that a study's risk bias can be judged as low, while “no” and “unclear” mean that the risk of bias can be judged as high.
Statistical analysis
Pooled sensitivity, pooled specificity, positive likelihood ratio, negative likelihood ratio, diagnostic odds ratio, and corresponding 95% CIs were calculated to evaluate the diagnostic value of miR-21. Summary receiver operator characteristics which shows the relationship between sensitivity and 1-specificity, was used to evaluate the consistency of results among all studies and the accuracy of the diagnostic test. The Spearman correlation coefficient was used to test the diagnostic threshold effect, which may produce significant heterogeneity (P < 0.05). Additionally, the chi-square, Q value and I2 test were used to assess the heterogeneity from non-threshold effect. A value of P less than 0.1 or an I2 ≥ 50% indicated the existence of significant heterogeneity. Meta-regression and subgroup analyses were conducted to explore sources of heterogeneity. Egger's test [33] and Begg's test [34] were performed to examine the potential publication bias. All statistical analyses were performed using Meta-Disc 1.4 and Stata 12.0 [35].
ACKNOWLEDGMENTS AND FUNDING
This work was supported in part by the Chinese National Key Scientific and Technological Project (Grants No: 2014BAI09B09 and 2015BAI12B15), the Program for Changjiang Scholars and Innovative Research Team in University in China (Grant No: IRT_14R40), and Tianjin Science and Technology Committee Foundation (Grant No: 11ZCGYSY0220).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global Cancer Statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M. SEER Cancer Statistics Review, 1975–2008. National Cancer Institute; Bethesda: 2011. http://seer.cancer.gov/csr/1975_2008/ (last access: 01/09/16) [Google Scholar]
- 3.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25:5287–5312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 4.O'Hanlon DM, Kerin MJ, Kent P, Maher D, Grimes H, Given HF. An evaluation of preoperative CA15-3 measurement in primary breast carcinoma. Br J Cancer. 1995;71:1288–1291. doi: 10.1038/bjc.1995.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uehara M, Kinoshita T, Hojo T, Akashi-Tanaka S, Iwamoto F, Fukutomi T. Long-term prognostic study of carcinoembryonic antigen (CEA) and carbohydrate antigen 15–3 (CA 15–3) in breast cancer. Int J Clin Oncol. 2007;13:447–451. doi: 10.1007/s10147-008-0773-3. [DOI] [PubMed] [Google Scholar]
- 6.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumor-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 8.Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J of Clin Exp Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- 9.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–59. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Zhang QY. The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol. 2012;138:1659–1666. doi: 10.1007/s00432-012-1244-9. [DOI] [PubMed] [Google Scholar]
- 11.Li SX, Guo GL, Chen SX, Jiang YY, Zeng Q. The value of miR-21 combined with CA153, CA125, CA-199 detection in early diagnosis of breast cancer. China Modern Doctor. 2013;51:67–69. [Google Scholar]
- 12.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 13.Li XF, Xu JJ, Zhang QY. Establishment of real-time PCR for detecting serum microRNA-21 and its preliminary application in breast cancer. Chin J Lab Med. 2011;34:920–925. [Google Scholar]
- 14.Gao JJ, Zhang Q, Xu J, Guo L, Li X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin J Cancer Res. 2013;25:743–748. doi: 10.3978/j.issn.1000-9604.2013.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT, Kwong A. Circulating microRNAs as Specific Biomarkers for Breast Cancer Detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee CH, Kuo WH, Lin CC, Oyang YJ, Huang HC, Juan HF. MicroRNA-Regulated Protein-Protein Interaction Networks and Their Functions in Breast Cancer. Int J Mol Sci. 2013;14:11560–11606. doi: 10.3390/ijms140611560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y. National Center for Clinical Laboratory; 2012. Novel serum biomarkers in breast cancer: detection and clinical significance [Master] [Google Scholar]
- 18.Mar-Aguilara F, Mendoza-Ramirez JA, Malagon-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, Rodrigues-Padilla C, Resendez-perez D. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers. 2013;34:163–169. doi: 10.3233/DMA-120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torain EA, Mohammed EA, Farrag S, Ramsis N, Hosny S. Pilot Study of Serum MicroRNA-21 as a Diagnostic and Prognostic Biomarker in Egyptian Breast Cancer Patients. Mol Diagn Ther. 2015;19:179–190. doi: 10.1007/s40291-015-0143-6. [DOI] [PubMed] [Google Scholar]
- 20.Thompson A, Brennan K, Cox A, Gee J, Harcourt D, Harris A, Harvie M, Holen I, Howell A, Nicholson R, Steel M, Streuli C. Evaluation of the current knowledge limitations in breast cancer research: a gap analysis. Breast Cancer Res. 2008;10:R26. doi: 10.1186/bcr1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, et al. Herceptin Adjuvant (HERA) Trial Study Team: Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 22.O'Byrne KJ, Gatzemeier U, Bondarenko I, Barrios C, Eschbach C, Martens UM, Hotko Y, Kortsik C, Paz-Ares L, Pereira JR, von Pawel J, Ramlau R, Roh JK, et al. Molecular biomarkers in non-small-cell lung cancer: a retrospective analysis of data from the phase 3 FLEX study. Lancet Oncol. 2011;12:795–805. doi: 10.1016/S1470-2045(11)70189-9. [DOI] [PubMed] [Google Scholar]
- 23.Gotte M. MicroRNAs in breast cancer pathogenesis. Minerva Ginecol. 2010;62:559–571. [PubMed] [Google Scholar]
- 24.Sreekumar E, Sayan BS, Mirnezami AH, Sayan AE. MicroRNA control of invasion and metastasis pathways. Front Genet. 2011;2:58. doi: 10.3389/fgene.2011.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin cancer Res. 2008;14:360–365. doi: 10.1158/1078-0432.CCR-07-0992. [DOI] [PubMed] [Google Scholar]
- 26.Andorfer CA, Necela BM, Thompson EA, Perez EA. MicroRNA signatures: clinical biomarkers for the diagnosis and treatment of breast cancer. Trends Mol Med. 2011;17:313–319. doi: 10.1016/j.molmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Yang X, Yang J, Zhen J, Zhang D. Serum microRNA-21 as a potential diagnostic biomarker for breast cancer: a systematic review and meta-analysis. Clin Exp Med. 2014 doi: 10.1007/s10238-014-0332-3. [DOI] [PubMed] [Google Scholar]
- 28.Shen L, Wan Z, Ma Y, Wu L, Liu F, Zang H, Xin S. The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumor Biol. 2015;36:1993–2005. doi: 10.1007/s13277-014-2806-z. [DOI] [PubMed] [Google Scholar]
- 29.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56:1129–1135. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 30.Walter SD. Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat Med. 2002;21:1237–1256. doi: 10.1002/sim.1099. [DOI] [PubMed] [Google Scholar]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 33.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 35.Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-Disc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]