Abstract
This systematic review is written to investigate the outcome of cervical cancer. A comprehensive search of PubMed and EMBASE was performed to identify eligible studies. Nineteen studies from thirteen articles with a total of 1,310 participants were included in this meta-analysis. Overall survival (OS), disease-free survival (DFS), and recurrence-free survival (RFS) as a prognosis for cervical cancer were extracted and calculated, if available. Pooled hazard ratios (HRs) and 95% confidence intervals (CIs) were calculated using STATA (version 12.0), resulting in the pooled HRs 0.70 (95% CI: 0.51–0.97) for OS, 1.02 (95% CI: 0.53–1.98) for DFS, and 0.56 (95% CI: 0.40–0.77) for RFS. The results indicated that cervical cancer patients with decreased microRNA expression were associated with shorter OS and RFS. It suggested that microRNAs might be promising markers for predicting the survival rate of cervical cancer.
Keywords: microRNA, prognosis, cervical carcinoma, meta-analysis
INTRODUCTION
Cervical carcinoma is the third most common female cancer that is closely related to human papilloma virus (HPV) infection. It was estimated that there were half a million new cases and 200, 000 deaths from cervical cancer every year [1]. Additionally, the five-year survival rate is less than 40%, especially for patients with advanced cancer [2]. Poor prognosis in cervical cancer is a chief public health problem and leads to vast hospitalization costs [3]. Although chemotherapy, radiotherapy, and related surgery have already been used as conventional treatment for cervical cancer patients, the clinical outcomes vary obviously among different patients and they are hard to be predicted. Therefore, new biomarkers which can estimate the prognosis of patients with cervical cancer are urgently required.
MicroRNAs are small noncoding RNAs, about 22–24 nucleotides long, that promote or inhibit tumor growth, progression, and metastasis [4]. They also have special expression within the tissue and are stable in the blood [5]. The expressions of various microRNAs were recently discovered to be associated with cervical cancer, indicating that they may be valuable prognostic biomarkers [6–18]. Therefore, we performed this meta-analysis to evaluate the correlation between microRNAs expression and survival rate in patients with cervical cancer.
RESULTS
Study characteristics
Nineteen studies from thirteen articles with a total of 1,310 cervical cancer patients from China, Iran, and Korea were included in this meta-analysis [6–18]. These studies were all retrospective cohort studies published during 2012–2015, and they reported the prognostic value of nineteen different microRNAs in cervical cancer patients. Of the nineteen studies, eighteen reported microRNAs as a prognostic factor of OS, four discussed the association between microRNA expression and DFS, and two assessed the association of microRNA with RFS. A flow diagram of the study selection process was summarized in Figure 1. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect microRNAs in all studies, although the cutoff values varied. The patients in the included studies were clinically staged I–IV according to the International Federation of Gynecology and Obstetrics (FIGO) staging criteria. Most patients were obtained for the studies after treatment. The low and high microRNA expression groups of each study had both lymph node metastasis and lymph node negative status. However, there was no significant difference among the included studies. The main features and extracted data of all the studies were summarized in Tables 1 and 2.
Figure 1. Flow diagram of the study selection process.
Table 1. The main features of enrolled studies.
| Author | Year | Population | Sample size | Method | Cut-off | miRNA | Survival analysis | Source of HR | Follow-up (month) |
|---|---|---|---|---|---|---|---|---|---|
| Luo | 2015 | China | 88 | qRT-PCR | Median | miR-26b | OS, RFS | Reported | mean 74 (5.12-98.5) |
| Safari | 2015 | Iran | 40 | qRT-PCR | Median | miR-20a | OS | DE | 100 |
| miR-10a | OS | DE | |||||||
| Wang C | 2015 | China | 138 | qRT-PCR | Median | miR-335 | OS, RFS | Reported, DE | Mean 71(23-117) |
| Wang Q | 2015 | China | 114 | qRT-PCR | Median | miR-145 | OS | Reported | median 47(11-69) |
| He | 2014 | China | 73 | qRT-PCR | ROC curve | miR-107 | OS, DFS | DE | median 68.4 |
| miR-130a | OS, DFS | DE | |||||||
| Liang | 2014 | China | 335 | qRT-PCR | X-tile algorithm | miR-215 | DFS | Reported | 60 |
| Ma | 2014 | China | 60 | qRT-PCR | ROC curve | miR-205 | OS | Reported | 60 |
| Park | 2014 | Korea | 45 | qRT-PCR | 2.5-fold | miR-363-3p | OS | Reported | 60 |
| Wang | 2014 | China | 54 | qRT-PCR | Median | miR-31 | OS | Reported | 60 |
| Yang | 2014 | China | 133 | qRT-PCR | Median | miR-126 | OS | Reported | 60 |
| Luo | 2013 | China | 60 | qRT-PCR | Mean | miR-497 | OS, DFS | Reported, DE | 60 |
| Shen | 2013 | China | 126 | qRT-PCR | Median | miR-224 | OS | DE | median 51.9 |
| Huang | 2012 | China | 44 | qRT-PCR | Mean | miR-125b | OS | Reported | mean 23.6 (2-70) |
| miR-100 | OS | Reported | |||||||
| miR-143 | OS | DE | |||||||
| miR-145 | OS | DE | |||||||
| miR-199a-5p | OS | DE |
HR: hazard ratio; qRT-PCR: quantitative real-time polymerase chain reaction; OS: overall survival; RFS: recurrence-free survival; DFS: disease-free survival.
Table 2. HRs for microRNAs.
| Study | miRNA | Sample size | OS | DFS | RFS | Expression associates with poor prognosis | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| High level | Low level | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |||
| Luo 2015 | miR-26b | 32 | 56 | 0.388 (0.355-0.727) | 0.007 | - | - | 0.475 (0.311-0.573) | 0.013 | Low |
| Safari 2015 | miR-20a | 24 | 16 | 2.47 (1.31-4.66) | 0.005 | - | - | - | - | High |
| miR-10a | 24 | 16 | 2.35 (1.23-4.50) | 0.01 | - | - | - | - | High | |
| Wang C 2015 | miR-335 | 59 | 79 | 0.251 (0.095-0.663) | 0.005 | - | - | 0.66 (0.47-0.92) | 0.015 | Low |
| Wang Q 2015 | miR-145 | 51 | 63 | 0.63 (0.54-0.83) | 0.008 | - | - | - | - | Low |
| He 2014 | miR-107 | 31 | 42 | 1.48 (0.93-2.35) | 0.1005 | 1.89 (1.19-3.00) | 0.0073 | - | - | High |
| miR-130a | 33 | 40 | 1.38 (0.87-2.19) | 0.1723 | 1.74 (1.10-2.77) | 0.018 | - | - | High | |
| Liang 2014 | miR-215 | 199 | 136 | - | - | 0.49 (0.28-0.86) | 0.013 | - | - | Low |
| Ma 2014 | miR-205 | 30 | 30 | 0.33 (0.14-0.76) | 0.009 | - | - | - | - | Low |
| Park 2014 | miR-363-3p | 27 | 18 | 0.1 (0.0-0.4) | 0.006 | - | - | - | - | Low |
| Wang 2014 | miR-31 | 27 | 27 | 1.482 (1.081-2.037) | 0.036 | - | - | - | - | High |
| Yang 2014 | miR-126 | 71 | 62 | 0.252 (0.049-0.498) | 0.003 | - | - | - | - | Low |
| Luo 2013 | miR-497 | 26 | 34 | 0.498 (0.332-0.743) | 0.0167 | 0.64 (0.38-1.06) | 0.085 | - | - | Low |
| Shen 2013 | miR-224 | 66 | 60 | 1.59 (1.12-2.26) | 0.009 | - | - | - | - | High |
| Huang 2012 | miR-125b | 4 | 40 | 0.352 (0.102-1.014) | 0.057 | - | - | - | - | Low |
| miR-100 | 10 | 34 | 0.161 (0.036-0.814) | 0.044 | - | - | - | - | Low | |
| miR-143 | 30 | 14 | 0.55(0.29-1.04) | 0.064 | - | - | - | - | Low | |
| miR-145 | 26 | 18 | 0.58(0.32-1.05) | 0.072 | - | - | - | - | Low | |
| miR-199a-5p | 24 | 20 | 0.56(0.31-1.01) | 0.056 | - | - | - | - | Low | |
HR: hazard ratio; 95% CI: 95% confidence interval; OS: overall survival; RFS: recurrence-free survival; DFS: disease-free survival; -: no reported.
Correlation between microRNA expression and prognosis
Heterogeneity was found in this meta-analysis (OS: I2 = 85.6%, P < 0.001; DFS: I2 = 85.9%, P < 0.001; RFS: I2 = 50.4%, P = 0.156), so the random–effects model was used to calculate the pooled HR and 95% CI values (Figure 2). According to the results displayed in Figure 2A (HR = 0.70, 95% CI: 0.51–0.97, P = 0.034) and Figure 2C (HR = 0.56, 95% CI: 0.40–0.77, P < 0.001), it was concluded that low microRNA expression indicated a poor prognosis in cervical cancer patients. However, the result displayed in Figure 2B (HR = 1.02, 95% CI: 0.53–1.98, P = 0.950) indicated that there was no obvious statistical significance.
Figure 2. A. Forest plot of the correlation between microRNA and OS in cervical cancer patient.
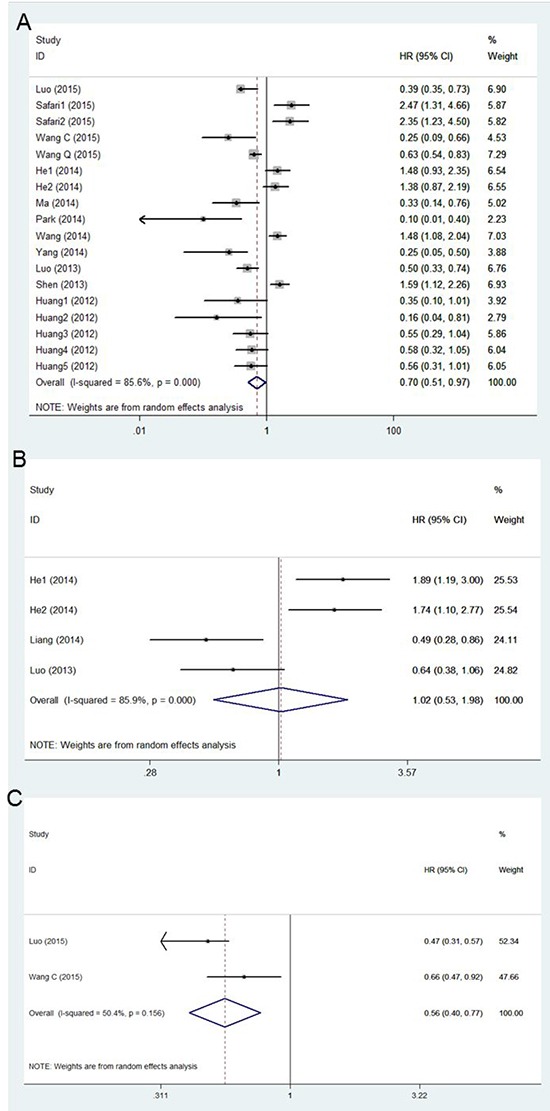
B. Forest plot of the correlation between microRNA and DFS in cervical cancer patient. C. Forest plot of the correlation between microRNA and RFS in cervical cancer patient.
Meta-regression analysis
To explore the source of heterogeneity, a meta-regression analysis was performed based on five variables as shown in Table 3. For the eighteen studies on OS, sample size (coefficient = −0.0194861, P = 0.018) and cutoff value (coefficient = −0.8039943, P = 0.010) were significantly related to heterogeneity. Due to the small size of the studies on DFS and RFS, we did not perform the meta-regression analysis for these studies.
Table 3. Results of meta-regression on OS.
| Variables | Coefficient | Standard error | t | P value | 95%CI |
|---|---|---|---|---|---|
| Year | −0.3586375 | 0.2829042 | −1.27 | 0.229 | −0.9750327, 0.2577577 |
| Country | −0.3432079 | 0.4892228 | −0.70 | 0.496 | −1.409133, 0.7227171 |
| Sample size | −0.0194861 | 0.0070848 | −2.75 | 0.018 | −0.0349225, −0.0040497 |
| Cut-off | −0.8039943 | 0.2634475 | −3.05 | 0.010 | −1.377997, −0.22999915 |
| Sample type | −1.118296 | 0.7370428 | −1.52 | 0.155 | −2.724175, 0.4875821 |
Publication bias and sensitivity analysis
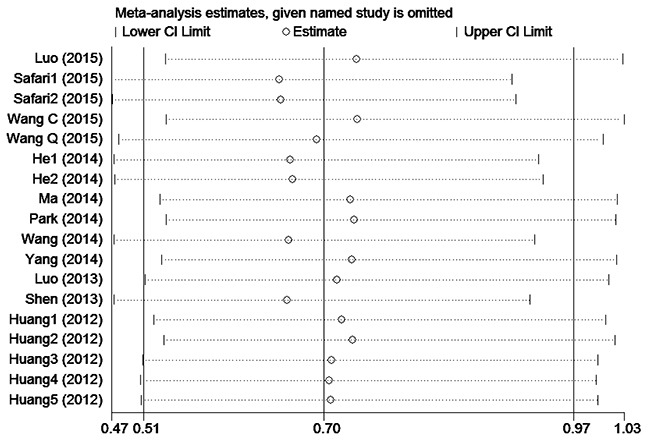
A funnel plot and Egger's test were performed for assessing publication bias (Figure 3). The funnel plot was almost symmetrical, and the p–value of the Egger's test was 0.360 (> 0.05), indicating no obvious publication bias in this meta-analysis of OS. In order to assess whether the results were credible and stable with obvious heterogeneity, sensitivity analysis was carried out by means of omitting each study by turns (Figure 4). The result indicated that there was no obvious influence of one individual study on the pooled HR. Due to the small size of the studies on DFS and RFS, publication bias and sensitivity analysis were not performed for these studies.
Figure 3. Funnel plot of eighteen studies included in this meta-analysis for OS.
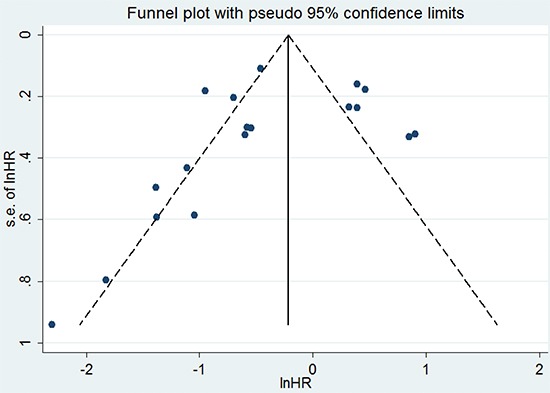
Figure 4. Sensitivity analysis of eighteen studies included in this meta-analysis for OS.
DISCUSSION
This meta-analysis is the first systematic assessment of the correlation between microRNA expression and the prognosis of patients with cervical cancer. As this meta-analysis showed, the pooled HR of OS was 0.70 (95% CI: 0.51-0.97; P = 0.034), which suggested that decreased microRNAs expression were associated with shorter OS in patients with cervical cancer. The forest plot revealed heterogeneity in this meta-analysis (I2 = 85.6%; P < 0.001), so we performed meta-regression analysis to explore the source. The results of the meta-regression analysis indicated that the sample size and cut-off value contributed to heterogeneity across the eighteen studies on OS. Cut-off values, country of origin, sample type, and other factors may have been sources of the heterogeneity in many similar meta-analyses [19]. We also evaluated the relationship between microRNAs expression and RFS and DFS. The pooled HR of RFS from the included studies was 0.56 (95% CI: 0.40–0.77, P < 0.001), which supported a similar conclusion. However, pooled HR of DFS from the included studies was 1.02 (95% CI: 0.53–1.98, P = 0.950). The interval of HR overlapped 1, which suggested no obvious significance.
In order to explore the source of heterogeneity in this meta-analysis, we also considered the type of cervical cancer. The main pathological classifications of cervical cancer are squamous cell carcinoma, adenocarcinoma, and adenosquamous carcinoma. In the thirteen included articles, most of them had no clear classifications, and a paper by Huang et al. revealed that the patients obtained had neuroendocrine small cell cervical carcinoma (SCCC), which is a less common type of squamous cell cervical carcinoma. After excluding this article, the pooled HR of OS was 0.80 (95% CI: 0.54–1.19, P = 0.275; I2 = 88.8%, P < 0.001), which suggested a higher degree of heterogeneity. The results of DFS and RFS did not change. Subgroup analysis on pathology types was not made for their undefined classifications and insufficient related information.
It is well known that a signaling pathway is a kind of enzymatic reaction pathway that can introduce extracellular signal molecules into cells through the cytomembrane. Signaling pathways cooperating with related target genes can activate or inhibit the process of cell growth, development, metabolism, apoptosis, invasion, and proliferation. For this reason, we also systematically investigated microRNAs and their potential targets and pathways included in this meta-analysis (Table 4). They can provide a reference for studying the mechanism of cervical cancer and targeted therapy.
Table 4. Summary of miRs with altered expression, their potential targets and pathways entered in this study.
| microRNA (Ref.) | Expression | Potential target | Pathway |
|---|---|---|---|
| miR-26b (11) | Low | USP9X, TAK1, TAB3, CDK8, PTGS2, SLC7A11 | Cell growth, apoptosis, EMT and NF-κB signaling pathways |
| miR-20a (12) | High | E2F2, E2F3 | Cell proliferation and modulate translation |
| miR-10a (12) | High | E2F2, E2F3 | Cell invasion and metastasis |
| miR-335 (13) | Low | MERTK, Rb1, SP1, BRCA1, RUNX2, PTPRN2, TRIM29 | EMT, PTEN/AKT/mTOR signaling pathways |
| miR-145 (14) | Low | p53 | Cell invasion and transcription |
| miR-107 (15) | High | CCR5 | Cell proliferation and invasion |
| miR-130a (15) | High | Tap63 | Cell migration, invasion and metastasis |
| miR-215 (16) | Low | BRAF, KRAS, TP53, RUNX1 | Cell migration, invasion and malignant progression |
| miR-205 (17) | Low | CYR61, CTGF | Cell proliferation and migration |
| miR-363-3p (18) | Low | CREB1, NOTCH1 | Cell proliferation, migration and apoptosis |
| miR-31 (19) | High | ARID1A | cell proliferation, apoptosis, migration and invasion |
| miR-126 (20) | Low | EGFL7, ADAM9b, VEGF-A, CRK | VEGF/PI3K-AKT signaling pathways |
| miR-497 (21) | Low | IGF-1R | Cell growth, proliferation, migration and invasion |
| miR-224 (22) | High | RKIP | Cell metastasis, growth and proliferation |
| miR-125b (23) | Low | BAK1, ErbB2 | Cell motility, invasion, glucose metabolism and chemosensitivity |
| miR-100 (23) | Low | RPSP3, PLK1, mTOR | Cell growth and migration |
| miR-143 (23) | Low | DNMT3A, KRAS, BCL-2 | Cell proliferation, apoptosis and metastasis |
| miR-145 (23) | Low | BNIP3, IRS, C-MYC, YES, STAT1, MMP-11, ADAM-17 | Cell proliferation, apoptosis and metastasis |
| miR-199a-5p (23) | Low | DDR1, SWI, SNF, PAK4 | Cell invasion and migration |
EMT: epithelial-to-mesenchymal transition; NF-κB: Nuclear factor-κB; VEGF: vascular endothelial growth factor; PI3K: phosphoinositol 3-kinase; AKT: serine/threonine kinase; PTEN: phosphatase and tensin homologue; mTOR: mammalian target of rapamycin.
This meta-analysis had several limitations that should be considered. First, significant heterogeneity existed among the studies included. Although we found that sample size and cutoff value contributed to heterogeneity, there were other potential sources. Due to the various microRNAs enrolled in this meta-analysis, the cutoff values of different microRNAs were divergent. We lacked a standard microRNA cutoff value in spite of the fact that median and mean values were often the primary cutoff value. Moreover, the normalization of the condition of qRT-PCR was also inconsistent. Second, the sample types were heterogeneous. Almost all types tested the microRNA expression in the tumor tissue, but only one was the detection of serum. As suggested in a relevant report, the synchronous detection of microRNA in the serum may conveniently provide additional information about host response and prognosis [20]. Third, part of the data derived from the relevant data extrapolation might be less credible compared with the data obtained from articles directly. Fourth, there was no obvious significance in the pooled HR of DFS that was different from the results for OS and RFS. The limited number of studies could be the possible reason. Fifth, as most of the studies enrolled were derived from Asian, potential publication bias might exist in this meta-analysis. Finally, due to the lack of related research focusing on the same microRNA, we had to calculate the pooled effect of different microRNAs for clinical evaluation. This solution was also used in other meta-analyses that lacked enough studies focusing on the same marker [19, 21]. In addition, because of certain articles without necessary data, we could not perform subgroup analysis based on microRNA category, related therapy, and other clinical characteristics, which may contribute to a portion of the heterogeneity. However, earlier related research indicated that aberrant microRNA expression was correlated with clinical characteristics such as lymph node metastasis, histological grade, and tumor diameter [22, 23].
In conclusion, we found that decreased microRNA expression was an indicator of a poor prognosis in cervical cancer patients, even if the limitations mentioned above existed. For further study, large prospective studies are needed to validate the prognostic role of microRNAs.
MATERIALS AND METHODS
Search strategy
Original articles investigating the prognostic role of microRNAs in cervical cancer were searched in the PubMed and EMBASE databases without time restriction. All of the articles were published before October 8, 2015. Terms such as “cervical cancer or cervical carcinoma or cervical intraepithelial neoplasia or uterine cervix cancer” and “microRNA or miRNA or miR” were jointly searched (Table 5).
Table 5. Search details.
| Electronic databases | PubMed, Embase |
|---|---|
| Search terms # 1 | cervical cancer OR cervical carcinoma OR cervical intraepithelial neoplasia OR uterine cervix cancer |
| Search terms # 2 | microRNA OR miRNA OR miR |
| Search terms # 3 | Search terms # 1 AND Search terms # 2 |
Selection criteria
A study was considered eligible if it met the following criteria: (1) study was written in English; (2) study investigated the prognostic value of cervical cancer patients with survival outcomes; and (3) study detected microRNA expression in tissue, serum, or plasma. An article was excluded in the following circumstances: (1) it was a review, comment, letter, or basic research article; (2) it lacked key information, such as hazard ratio (HR), 95% confidence interval (95% CI), or other useful data for extrapolating; and (3) it was a repeated study including the same samples from the same patients as a study already published. Once overlapping data were used in more than one article, the most complete study was exclusively included in this meta-analysis. Two reviewers independently evaluated the articles identified by the above criteria.
Quality assessment
We systematically evaluated the methodological quality of all the included studies according to a critical review checklist from the Dutch Cochrane Centre proposed by the Meta-analysis of Observational Studies in Epidemiology [24]. The key points included: study population, country of the study, study design, outcome, cut-off value, microRNA detection method, and follow-up period. Each study was required to meet all of these points.
Data extraction
The data of all eligible studies were independently extracted by two investigators. Disagreements were resolved by discussion. The following primary information was collected: name of the first author, year of publication, country of the study, sample size, disease stage, test method, cutoff value, follow-up time, and HRs of microRNA for OS, DFS, and RFS (as well as the 95% CIs and P values). If the HRs and 95% CIs were not available, we calculated them using the relevant data provided in the articles or asked for related information by emailing the authors. If only the Kaplan-Meier curves were available, we extracted the relevant data from the graphed survival plots and calculated the HRs and 95% CIs. All calculations used the methods developed by Parmar, Williamson, and Tierney [25–27].
Statistical analysis
The pooled HRs of microRNA expression for OS, DFS, and RFS were calculated in this meta-analysis. A pooled HR < 1 means a poor prognosis for patients with low microRNA expression. In contrast, a pooled HR > 1 and a lower limit of the 95% CI of a pooled HR > 1 indicate a poor prognosis for patients with high microRNA expression [28]. Heterogeneity was assessed using a Q test and I2 statistic (P < 0.05 and/or I2 > 50% were considered statistically heterogeneous). If there was no obvious heterogeneity, a fixed-effects model was used; otherwise, the random-effects model was used. Publication bias was evaluated using the funnel plot and Egger's test (values of P > 0.05 indicated lack of publication bias) [29]. All of the analyses were performed using STATA (version 12.0). For all the results, values of P < 0.05 were considered statistically significant.
Footnotes
CONFLICTS OF INTEREST
No conflicts of interest exist for any authors.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Tarver T. American Cancer Society (ACS) Atlanta, GA: American Cancer Society; 2012. Cancer Facts & Figures 2012; p. 66. p., pdf. Available from. Journal of Consumer Health on the Internet. 2012; 16:366-367. [Google Scholar]
- 3.Huang EY, Hsu HC, Sun LM, Chanchien CC, Lin H, Chen HC, Tseng CW, Ou YC, Chang HY, Fang FM, Huang YJ, Wang CY, Lu HM, Tsai CC, Ma YY, Fu HC, et al. Prognostic value of pretreatment carcinoembryonic antigen after definitive radiotherapy with or without concurrent chemotherapy for squamous cell carcinoma of the uterine cervix. International journal of radiation oncology, biology, physics. 2011;81:1105–1113. doi: 10.1016/j.ijrobp.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 5.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic acids research. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo M, Shen D, Wang W, Xian J. Aberrant expression of microRNA-26b and its prognostic potential in human cervical cancer. International journal of clinical and experimental pathology. 2015;8:5542–5548. [PMC free article] [PubMed] [Google Scholar]
- 7.Safari A, Seifoleslami M, Yahaghi E, Sedaghati F, Khameneie MK. Upregulation of miR-20a and miR-10a expression levels act as potential biomarkers of aggressive progression and poor prognosis in cervical cancer. Tumour biology. 2015 doi: 10.1007/s13277-015-4064-0. [DOI] [PubMed] [Google Scholar]
- 8.Wang C, Jiang T. MicroRNA-335 represents an independent prognostic marker in cervical cancer. Tumour biology. 2015;36:5825–5830. doi: 10.1007/s13277-015-3252-2. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Qin J, Chen A, Zhou J, Liu J, Cheng J, Qiu J, Zhang J. Downregulation of microRNA-145 is associated with aggressive progression and poor prognosis in human cervical cancer. Tumour biology. 2015;36:3703–3708. doi: 10.1007/s13277-014-3009-3. [DOI] [PubMed] [Google Scholar]
- 10.He L, Wang HY, Zhang L, Huang L, Li JD, Xiong Y, Zhang MY, Jia WH, Yun JP, Luo RZ, Zheng M. Prognostic significance of low DICER expression regulated by miR-130a in cervical cancer. Cell death & disease. 2014;5:e1205. doi: 10.1038/cddis.2014.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang H, Li Y, Luo RY, Shen FJ. MicroRNA-215 is a potential prognostic marker for cervical cancer. Journal of Huazhong University of Science and Technology Medical sciences. 2014;34:207–212. doi: 10.1007/s11596-014-1260-x. [DOI] [PubMed] [Google Scholar]
- 12.Ma Q, Wan G, Wang S, Yang W, Zhang J, Yao X. Serum microRNA-205 as a novel biomarker for cervical cancer patients. Cancer cell international. 2014;14:81. doi: 10.1186/s12935-014-0081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H, Lee MJ, Jeong JY, Choi MC, Jung SG, Joo WD, Lee C, An HJ. Dysregulated microRNA expression in adenocarcinoma of the uterine cervix: clinical impact of miR-363-3p. Gynecologic oncology. 2014;135:565–572. doi: 10.1016/j.ygyno.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Wang N, Zhou Y, Zheng L, Li H. MiR-31 is an independent prognostic factor and functions as an oncomir in cervical cancer via targeting ARID1A. Gynecologic oncology. 2014;134:129–137. doi: 10.1016/j.ygyno.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Song KL, Chang H, Chen L. Decreased expression of microRNA-126 is associated with poor prognosis in patients with cervical cancer. Diagnostic pathology. 2014;9:220. doi: 10.1186/s13000-014-0220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M, Shen D, Zhou X, Chen X, Wang W. MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery. 2013;153:836–847. doi: 10.1016/j.surg.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 17.Shen SN, Wang LF, Jia YF, Hao YQ, Zhang L, Wang H. Upregulation of microRNA-224 is associated with aggressive progression and poor prognosis in human cervical cancer. Diagnostic pathology. 2013;8:69. doi: 10.1186/1746-1596-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L, Lin JX, Yu YH, Zhang MY, Wang HY, Zheng M. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PloS one. 2012;7:e33762. doi: 10.1371/journal.pone.0033762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang ZY, Dai ZL, Yin XW, Li SH, Li SP, Ge HY. Meta-analysis shows that circulating tumor cells including circulating microRNAs are useful to predict the survival of patients with gastric cancer. BMC cancer. 2014;14:773. doi: 10.1186/1471-2407-14-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer science. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao W, Guo Q, Zhang T, Zhong D, Yu Q. Prognostic value of microRNAs in acute myocardial infarction: A systematic review and meta-analysis. International journal of cardiology. 2015;189:79–84. doi: 10.1016/j.ijcard.2015.04.055. [DOI] [PubMed] [Google Scholar]
- 22.Zhao S, Yao DS, Chen JY, Ding N. Aberrant expression of miR-20a and miR-203 in cervical cancer. Asian Pacific journal of cancer prevention. 2013;14:2289–2293. doi: 10.7314/apjcp.2013.14.4.2289. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Yao D, Li Y, Chen H, He C, Ding N, Lu Y, Ou T, Zhao S, Li L, Long F. Serum microRNA expression levels can predict lymph node metastasis in patients with early-stage cervical squamous cell carcinoma. International journal of molecular medicine. 2013;32:557–567. doi: 10.3892/ijmm.2013.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA, Thacker SB. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. Jama. 2000;283:2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 25.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Statistics in medicine. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Williamson PR, Smith CT, Hutton JL, Marson AG. Aggregate data meta-analysis with time-to-event outcomes. Statistics in medicine. 2002;21:3337–3351. doi: 10.1002/sim.1303. [DOI] [PubMed] [Google Scholar]
- 27.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spruance SL, Reid JE, Grace M, Samore M. Hazard ratio in clinical trials. Antimicrobial agents and chemotherapy. 2004;48:2787–2792. doi: 10.1128/AAC.48.8.2787-2792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]


