Abstract
To investigate whether ocular albinism type 1 (OA1) is differentially expressed in the skin of mice with different coat colors and to determine its correlation with coat color establishment in mouse, the expression patterns and tissue distribution characterization of OA1 in the skin of mice with different coat colors were qualitatively and quantitatively analyzed by real-time quantitative PCR (qRT-PCR), immunofluorescence staining and Western blot. The qRT-PCR analysis revealed that OA1 mRNA was expressed in all mice skin samples tested, with the highest expression level in brown skin, a moderate expression level in black skin and the lowest expression level in gray skin. Positive OA1 protein bands were also detected in all skin samples by Western blot analysis. The relative expression levels of OA1 protein in both black and brown skin were significantly higher than that in gray skin, but there was no significant difference between black and brown mice. Immunofluorescence assays revealed that OA1 was mainly expressed in the hair follicle matrix, the inner and outer root sheath in the skin tissues with different coat colors. To get further insight into the important role of OA1 in the melanocytes’ pigmentation, we transfected the OA1 into mouse melanocytes and then detected the relative expression levels of pigmentation-related gene. Simultaneously, we tested the melanin content of melanocytes. As a result, the overexpression of OA1 significantly increased the expression levels of microphthalmia-associated transcription factor (MITF), tyrosinase (TYR), tyrosinase-related protein 1 (TRP1) and premelanosome protein (PMEL). However, the tyrosinase-related protein 2 (TRP2) level was attenuated. By contrast, the level of glycoprotein non-metastatic melanoma protein b (GPNMB) was unaffected by OA1 overexpression. Furthermore, we observed a significant increase in melanin content in mouse melanocyte transfected OA1. Therefore, we propose that OA1 may participate in the formation of coat color by regulating the level of MITF and the number, size, motility and maturation of melanosome.
Keywords: ocular albinism type 1 (OA1), microphthalmia-associated transcription factor (MITF), melanosome, coat color
1. Introduction
Melanosomes are the melanocyte-specific organelles within which melanin pigments are synthesized, deposited and transported. Melanosomes derive from endosomal precursors and subsequently go through a series of morphologically- and functionally-defined stages [1]. The first two stages lack pigment, but are characterized by intraluminal matrix fibrils that start to form in the stage I melanosome and are completed in the ellipsoidal stage II melanosome. Deposition of melanin along the matrix fibrils is initiated in the stage III melanosome until all internal structures are obscured in the stage IV melanosome [2]. To date, proteomic analysis of melanosomes has been found about 1500 proteins appearing in all stages of melanosomes [3]. Twelve of these proteins are specifically localized in melanosomes, including tyrosinase (TYR), tyrosinase-related protein 1 (Tyrp1/TRP1), tyrosinase-related protein 2 (Tyrp2/TRP2), ocular albinism type 1 (OA1), melanoma antigen recognized by T-cells (MART-1/MLANA), premelanosome protein (PMEL), vesicle amine transport 1 (VAT-1), oculospanin, syntenin, glycoprotein non-metastatic melanoma protein b (GPNMB), coiled-coil-helix-coiled-coil-helix domain containing 3 (CHCHD3) and flotillin [4]. The function of three melanogenesis-controlling enzymes are well known to us [5], but the roles of other melanosomal proteins are partially understood, such as OA1 [6,7].
Ocular albinism type 1 was initially positionally cloned from the distal short arm of the X chromosome by Bassi [8]. This gene encodes a 404-amino acid internal membrane glycoprotein, named OA1, also identified as G protein-coupled receptor 143 (GPR143) [9]. Previous work displayed that OA1 exhibits structural properties of G protein-coupled receptors (GPCRs) [10,11,12]. Additionally, OA1 shared two fundamental homologies with GPCRs, activating heterotrimeric G proteins and interacting with arrestins [13]. However, differently from a canonical GPCR, OA1 is predominantly localized to the melanosomal membrane rather than the plasma membrane, with its C terminus toward the cytoplasm and with the N terminus toward the lumen of organelle [12]. Based on the localization, a putative ligand should bind OA1 on the luminal side of melanosome compared with receptors at the cell surface. Lopez et al. propose that L-DOPA is an endogenous ligand for OA1 by using a heterologous expression system based on OA1 overexpression in Chinese hamster ovary (CHO) cells [14,15,16,17]. Proteomic analysis of early melanosomes obtained from human MNT1 cells identifies OA1 as a component of these organelles [4]. All of these results suggest that OA1 may play an important role in regulating the signal process related to melanosome function.
Networking analysis of genes by the GeneMANIA algorithm revealed that the TYR, oculocutaneous albinism II (OCA2), OA1, microphthalmia-associated transcription factor (MITF) and cbp/p300 interacting transactivator with glu/asp rich carboxy-terminal domain 1 (CITED1) genes were involved in the biological process of developmental pigmentation, whereas the TYR, OCA2, OA1, dopachrome tautomerase (DCT) and CITED1 genes were implicated in the major biological process of biosynthetic pigment and the pigment metabolic process. The pigmentation process was involved in the interaction of the TYR, OCA2 and OA1 along with the MITF, PMEL and CITED1 genes [18]. The transcriptional regulation of OA1 was controlled by MITF, which belongs to the basic helix-loop-helix–leucine zipper (bHLH-LZ) factor family [19]. To date, MITF has been reported to activate more than 25 pigmentation-related genes, including TYR, tyrosinase-related protein 1 (TRP1), tyrosinase-related protein 2 (TRP2), MART-1, melanocortin 1 receptor (MC1R), endothelin receptor type b (EDNRB), RAB27A, PMEL, GPNMB, solute carrier family 45 member 2 (SLC45A2/MATP), melastatin, and so forth. Therefore, MITF has been regarded as an essential regulator for melanocyte survival, proliferation, development and the expression of melanogenic proteins [20]. In addition, investigations showed that the loss of OA1 function would dramatically reduce the expression of MITF, although the levels are still sufficient to sustain the survival and differentiated status of the pigmented cell [21]. Together, we ask whether the OA1 will participate in the establishment of coat color by regulating the level of MITF.
2. Results
2.1. Expression Profile of Ocular Albinism Type 1 (OA1) mRNA in Mice Skin Samples
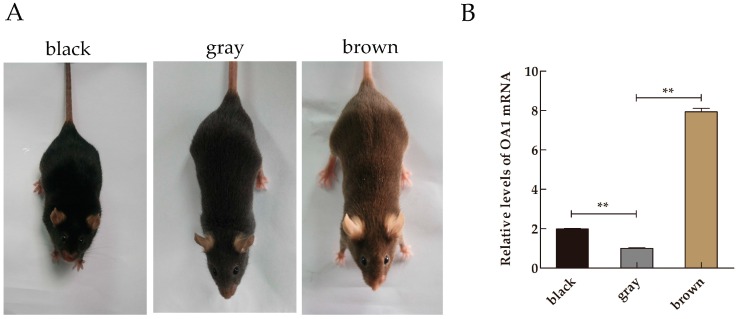
OA1 mRNA expression levels in the skin of mice with different coat colors are shown in Figure 1B. The qRT-PCR results showed that the relative expression levels of OA1 mRNA were 1.9844 ± 0.0554** (p < 0.01) in black skin, 7.9394 ± 0.2998** (p < 0.01) in brown skin and 1.002 ± 0.0742 in the gray skin. These differences correspond to 1.98- and 7.94-fold increases in OA1 mRNA in black skin and in brown skin relative to gray skin, respectively. The increases in OA1 expression in black and brown mice skin compared with gray mice skin were significantly different.
Figure 1.
The ocular albinism type 1 (OA1) mRNA analysis. (A) Photographs of C57BL/6J mice with a variety of coat colors; (B) Relative expression levels of OA1 mRNA in the mice skin samples of different coat colors. The abundance of OA1 was normalized relative to the abundance of β-actin. Data are shown as the mean ± standard errors (n = 3 each), ** p < 0.01.
2.2. Protein Expression of OA1 in Mice Skin Samples
Western blot was performed to further analyze the expression level of OA1 protein in the mice skin tissues with different coat colors. Western blot analysis showed that the total proteins from skin samples of mice with different coat colors were positively immunoreactive to the OA1 anti-rabbit polyclonal antibody, with the target band located at the position of 48 kDa (Figure 2A). Protein band analysis showed that the relative expression levels of OA1 protein were 0.5143 ± 0.0074** (p < 0.01) in black skin, 0.5212 ± 0.0124** (p < 0.01) in brown skin and 0.3300 ± 0.0100 in gray skin. These differences correspond to 1.56- and 1.58-fold higher expression in black and brown skin compared with gray skin, respectively. The protein expression levels of OA1 were significantly affected in different colors (Figure 2B).
Figure 2.
The OA1 protein analysis. (A) Western blot of lysates collected from the skin samples of mice with different coat colors were conducted using OA1 as the primary antibody. β-Actin was used as the loading control; (B) Relative expression levels of OA1 protein in the skin of mice with different coat colors. Data are shown as the mean ± standard errors (n = 3 each), ** p < 0.01; (C) Localization of OA1 protein in hair follicle. The immunofluorescence assay was used to determine the location of OA1 among different coat colors of mice skin. 1: hair follicle matrix; 2: outer root sheath; 3: inner root sheath; (D) Average optical density analysis of OA1 in mice skin with different hair color. Data are shown as the mean ± standard errors (n = 3 each), ** p < 0.01.
2.3. Distribution and Expression of OA1 in Mice Skin Samples
The immunofluorescence assay was performed to determine the location of OA1 in mice of various coat colors. OA1 was found in the inner and outer root sheath, hair follicle matrix and dermal papilla in skin. Different levels and distributions of OA1 were found in various coat colors. In black mice, OA1 staining was strongly positive, especially in the dermal papilla, follicle matrix and outer root sheath melanocytes. However, in brown and gray mice, OA1 was mainly located in the outer root sheath of hair follicle, and it was weakly positive in gray mice (Figure 2C). No positive expression was observed in the negative control (no primary or secondary antibody added). The average optical density (OD) values for OA1 were 0.0418 ± 0.0037** (p < 0.01) in black skin, 0.0525 ± 0.0019* (p < 0.05) in brown skin, respectively, and 0.0257 ± 0.0010 in gray skin. These values indicate that OA1 is expressed at 1.62- and 2.0-fold higher levels in black and brown skin compared with gray skin, respectively (Figure 2D).
2.4. OA1 Controls Not Only Melanosome Structural Protein but Also the Melanin Synthesis in an MITF-Dependent Fashion
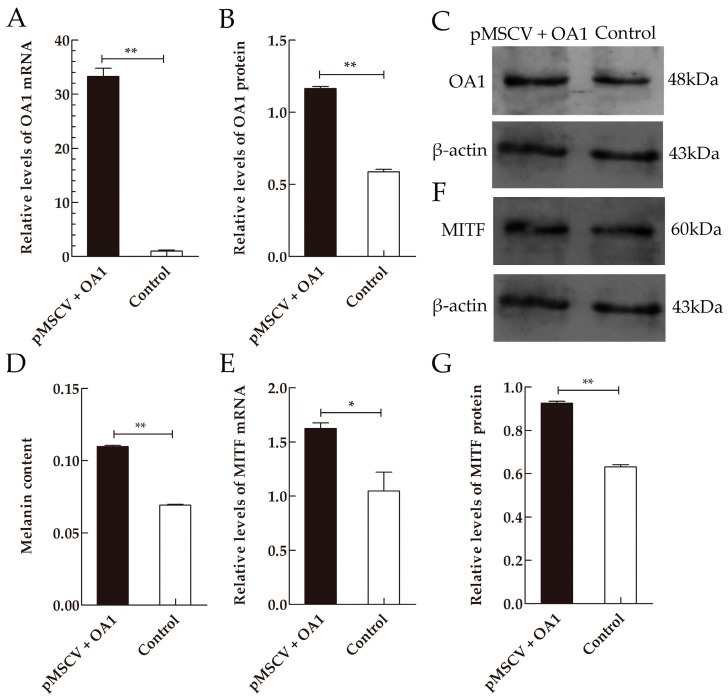
To determine the effect of OA1 on the melanogenesis, we transfected mice melanocytes with the plasmid encoding OA1. There are two groups in this experiment, including the vector-GFP-OA1 group and the mock-vector-GFP group. The qRT-PCR and Western blot analysis showed that the relative expression levels of OA1 mRNA and protein in the OA1-transfected group were 31.90-fold and 1.98-fold higher than that in the control group, respectively (Figure 3A–C). All of these results suggest that OA1 has been transfected into melanocytes efficiently.
Figure 3.
The overexpression of OA1 affects the melanin content in an MITF-dependent fashion. Mice OA1 was PCR amplified and then subcloned into the pMSCV PIG vector with Xho I and EcoR I restriction sites. The pMSCV-OA1-GFP plasmid or empty vector was transfected into the mouse melanocytes. The cells were harvested to extract total RNA and total protein. The expression levels of OA1 were qualitatively and quantitatively analyzed by qRT-PCR and Western blot. (A) Melanin contents in melanocytes transfected with OA1, as well as the control; (B–D) transfection efficiency of OA1 in mouse melanocytes; (E–G) OA1 overexpression influences microphthalmia-associated transcription factor (MITF) mRNA and protein levels in mouse melanocytes. Data are shown as the mean ± standard errors (n = 3 each), * p < 0.05, ** p < 0.01.
To confirm the effect of OA1 on the melanin biosynthesis in melanocytes, the melanin content in cells transfected with pMSCV-GFP-OA1 or pMSCV-GFP was measured. The results demonstrated that the melanin content in the OA1 overexpression melanocytes was 1.59-fold higher than that in the control group (Figure 3D).
Our data indicate that OA1 presents a different expression in mice skin of various coat colors. Therefore, we suspect that OA1 might participate in the formation of coat color, possibly by regulating the expression of some melanosomal protein in an MITF-dependent fashion [22]. To verify this hypothesis, we measured the levels of MITF, TYR, TRP1, TRP2, PMEL and GPNMB. MITF, encoding an integral transcriptional regulator in melanocytes, has been mapped to chromosome 6 in mice. It controls the transcription levels of pigmentation-associated gene and the differentiation, proliferation and survival of melanocytes. TYR is a type I membrane glycoprotein with seven potential N-glycosylation sites [23]. TYR, the first rate-limiting enzyme in melanin synthesis, catalyzes the conversion of l-tyrosine to dopaquinone and the oxidation of 3,4-dihydroxyphenylalanine (DOPA) back to dopaquinone [24]. TRP1 is a 5,6-dihydroxyindole-2-carboxylic acid (DHICA) oxidase, which promotes the oxidation and polymerization of DHICA monomers into melanin [25]. TRP2 possesses dopachrome tautomerase (DCT) activity, which enables a rapid conversion of dopachrome (DC) to DHICA [26,27]. PMEL, also known as PMEL17, is an internal membrane glycoprotein that constitutes the fibrillar matrix of melanosome [28,29,30]. GPNMB is a highly glycosylated type I transmembrane protein with a high level of structural homology to PMEL [31,32].
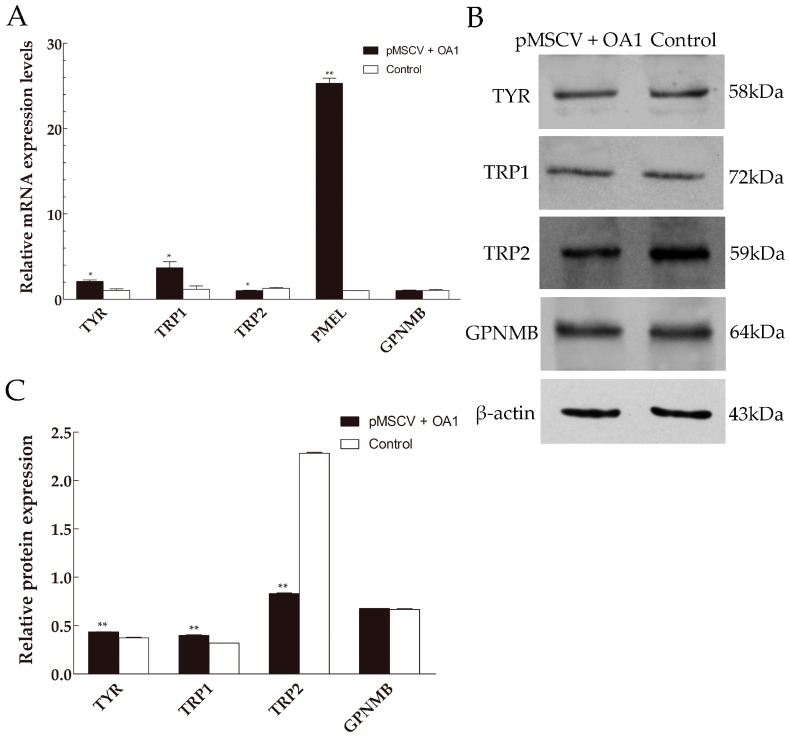
A previous study found that the expression level of MITF reduced correlating with the reduced OA1 expression in OA1−/− or shRNA-depleted melanocytes [22]. Consistently, we observed an increase in MITF mRNA and protein levels when OA1 was overexpressed in the mouse melanocytes (Figure 3E–G). These results revealed that OA1 was a signaling protein controlling the MITF expression levels. To get further insights into the function of OA1 in melanocytes, we measured the expression levels of some pigmentation-related genes. Real-time quantitative PCR analysis demonstrated that the expression levels of TYR, TRP1 and PMEL mRNA were upregulated in the OA1 overexpression melanocytes compared with the control (Figure 4A). Consistent with this, when we analyzed the levels of TYR and TRP1 protein by Western blot, we found that both were modestly increased (Figure 4B,C). However, compared with the control, the expression levels of TRP2 mRNA and protein were attenuated in the OA1 overexpression melanocytes (Figure 4). By contrast, the expression levels of GPNMB mRNA and protein were not affected after OA1 was transfected into the mouse melanocytes. The housekeeping protein β-actin was used as a loading control.
Figure 4.
The overexpression of OA1 affects melanosomal proteins in mouse melanocytes. (A) The pigmentation-related gene mRNA levels were measured by qRT-PCR; (B,C) Western blot of cell lysates probed with primary antibody against the TYR, TRP1, TRP2 and GPNMB. β-actin was used as a protein loading control. Data are shown as the mean ± standard errors (n = 3 each), * p < 0.05, ** p < 0.01.
3. Discussion
The color we see in the skin, hair and eyes of mammals is largely determined by the presence and distribution of melanin. There are two main types of melanin in mammal melanocytes, including brown-black eumelanin and red-yellow pheomelanin [33]. Melanin synthesis exclusively occurs in subcellular organelles termed melanosomes within melanocytes and retinal pigment epithelium (RPE), which is mainly controlled by TYR, TRP1 and TRP2 [34,35,36,37,38]. There are four stages in the development of melanosome: unmelanized immature premelanosome in stage I and II melanosome and melanized stage III and IV melanosome [39].
Melanocytes, which are originally derived from neural crest cells in embryonic skin, are localized in epidermis, as well as in hair follicles to pigment the skin and hair, respectively [40,41,42]. In the hairy skin of mice, melanocytes in the epidermis are found only during the early weeks after birth [43]. Different from the epidermal melanocytes, melanocytes in hair follicles repeatedly proliferate and differentiate for hair pigmentation in every hair cycle. Previous work has shown that there are three morphologically- and functionally-discrete melanocytic populations in hair follicles: melanocyte stem cells, melanocyte progenitor cells and terminally-differentiated melanocytes [44]. The first two localized in the outer root sheath (ORS) of the middle and lower hair follicles express DCT and TRP1, but lack TYR, so they do not produce melanin and belong to the amelanotic melanocytes. The latter residing in the infundibulum and bulb express all enzymes necessary for the melanogenesis, so they belong to the pigmented melanocytes [45,46,47,48]. These ORS melanocytes act as a melanocyte reservoir for the repopulation of epidermal melanocytes and for repigmentation of vitiligo [49,50]. Proteomic analysis of melanosomes found that OA1 was mainly enriched in compartments highly labeled by PMEL17 that correspond to stage I and stage II premelanosomes, although it appeared broadly distributed in all stages of melanosome maturation [51]. In common with this, the immunofluorescence assay revealed that OA1 was primarily expressed in the outer root sheath of hair follicle. In addition, it was also localized in the dermal papilla and hair follicle matrix. These results further support that OA1 plays an important role in the early stages of melanosome biogenesis, as predicted previously.
Here, we demonstrated that different coat colors were associated with different levels of OA1 expression. OA1 mRNA and protein were expressed in all mice skin samples tested, with the highest expression level in brown mice skin, a moderate expression level in black mice skin and the lowest in gray mice skin. These observations revealed that OA1 transcription and translation levels are high in the skin of black and brown mice and low in the skin of gray mice. Therefore, we propose that OA1 may be involved in the establishment of coat color, of which the specific mechanism is as follows. The mutation of OA1 is responsible for the most common form of ocular albinism, in which patients exhibit a reduced number of enlarged melanosomes and visual defects [8,11,52]. OA1 performs two functions in melanogenesis, controlling the rate of melanosome biogenesis at early maturation stages and maintaining the correct melanosomal size at the final stage of the organellogenesis [53]. OA1 regulates not only melanosome biogenesis, but also the motility of melanosomes. Time-lapse video-microscopy and melanosome tracking analyses reveal that the absence of OA1 results in a significant reduction in frequent melanosomal movement along microtubules. However, this observation exclusively occurs in the presence of an intact actin cytoskeleton. Thus, the cytoskeleton represents a downstream effector of OA1 for which the receptor plays a modulatory role in melanosome motility, by switching between MT-based and AF-based systems [54,55]. Burgoyne et al. demonstrated that OA1 expression increased multivesicular endosomes’/bodies’ (MVBs) number. OA1 could inhibit MVB-lysosome fusion through affecting the motility of MVB or lysosome and the luminal pH of MVBs. A delay in lysosomal fusion might allow time for melanin deposition [56]. Taken together, we propose that OA1 may participate in the formation of coat color differences through regulating the number, size, motility and maturation of melanosomes.
OA1 controls the melanosome composition and density at early stages of melanogenesis [51]. Therefore, we thought there was a possible connection between the expression of OA1 and MITF. Our data showed that the overexpression of OA1 in mouse melanocytes caused an increase in the MITF levels. Consistently, the expression level of MITF was attenuated when OA1 expression was absent [22]. These results demonstrate that OA1 could positively control MITF. The role of OA1 in regulating MITF expression participates in the α-MSH-MITF-signaling pathway [22]. The α-melanocyte-stimulating hormone (α-MSH) binding to MC1R activates adenylate cyclase, which results in the activation of cyclic adenosine monophosphate (cAMP) signaling. Additionally, then, cAMP leads to the phosphorylation of cAMP response element binding protein (CREB) that recognizes the MITF promoter [57].
Because OA1 activity positively leads to the regulation of MITF, we asked whether MITF-controlled genes will be affected by its modulation via OA1 [22]. A previous study found that PMEL was an MITF target gene in melanocytes and melanoma, with its mRNA levels being enhanced upon transfer of wt MITF and blocked by a dominant negative form of MITF, which indicated that the expression level of MITF was necessary for the PMEL gene transcription [58]. In addition, the expression level of PMEL showed an increase when MITF was re-expressed in OA1−/− LOa1SN melanocytes [15]. We observed that the relative expression level of the structural protein PMEL mRNA in the OA1 overexpression melanocytes was significantly higher compared with the control. Collectively, these results suggest that OA1 regulates PMEL expression via MITF. By contrast, although GPNMB, encoding a type I transmembrane protein whose transcription is dependent on MITF [59], has been identified as having a high level of structural homology with PMEL, its mRNA and protein expression levels are not changed by OA1 expression. As we all known, loss of function of OA1 causes a decrease in pigmentation and induces the formation of enlarged aberrant melanosome harboring disorganized fibrillar structures. Ultrastructural analysis of the RPE cells reveals that the formation of macromelanosomes may be caused by abnormal growth of single melanosomes rather than the fusion of several [60]. In other words, the decrease of PMEL expression by OA1 absence leads to the slowing down of the rate of new stage II melanosome formation [61]. Collectively, the variation of PMEL expression resulting from OA1 overexpression is likely mediated by MITF. In addition to the structural protein, the melanosomal enzymes are also affected by the MITF level. Our observations showed that the TYR and TRP1 expression was modestly increased in the OA1 overexpression melanocytes compared with the control cells. However, the overexpression of OA1 reduced the TRP2 expression. The transient cotransfection assay displayed that MITF overexpression transactivated the human tyrosinase promoter, as well as the TRP-1 gene promoter, rather than the TRP-2 promoter [62]. Taken together, OA1 not only regulates melanosomal structural protein expression, but also melanin biogenesis in melanocytes in an MITF-independent fashion.
4. Materials and Methods
4.1. Antibody
Rabbit anti-OA1 polyclonal antibody and goat anti-rabbit IgG-FITC were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Rabbit anti-TYR polyclonal antibody, rabbit anti-TRP1 polyclonal antibody, rabbit anti-TRP2 polyclonal antibody and mouse anti-MITF polyclonal antibody were purchased from Abcam (Cambridge, MA, USA). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (IgG) and HRP-conjugated goat anti-mouse IgG were purchased from CWBIO (Beijing, China). Rabbit anti-GPNMB polyclonal antibody were purchased from Proteintech group (Wuhan, China).
4.2. Experimental Animals and Sample Collection
This study was conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals, 27 December 2010, ISBN: 9780309154000).
Three approximately 14-day-old healthy C57BL/6 mice were randomly selected from breeds with black, gray and brown coat colors (Figure 1A). After shaving and then disinfection, three pieces of dorsal skin tissue were collected from each mouse using a skin collector, of which two samples were frozen and stored in liquid nitrogen for total RNA and total protein extraction. The remaining piece of skin was fixed in Bouin’s solution to prepare paraffin sections for immunofluorescence staining.
4.3. Cell Culture and Transfection
Mouse melanocytes (derived from C57BL/6J black mice) were established in the laboratory of alpaca biology, College of Animal Science and Technology, Shanxi Agricultural University (Jinzhong, China) [63]. Mouse melanocytes were cultured in melanocyte medium (MelM) (ScienCell Research Laboratories, Carlsbad, CA, USA) supplemented with 1% melanocyte growth supplement (MelGS) and 1% penicillin-streptomycin and 0.5% fetal bovine serum at 37 °C in a humidified 5% CO2 atmosphere.
The mice OA1 gene was PCR amplified and then was cloned into the pMSCV PIG vector (Addgene, Cambridge, MA, USA) with Xho I and EcoR I restriction sites. Mouse melanocytes were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s guidelines. Then, we harvested the cells and extracted the total RNA and total protein.
4.4. Melanin Content Measurement
Seventy-two hours after transfection, mouse melanocytes were collected and washed in phosphate buffer saline (PBS) (Solarbio, Beijing, China) 3 times. The cells were resuspended in PBS and counted using an automated cell counter (Bio-Rad, Hercules, CA, USA). The cells were centrifuged at 1000 rpm for 10 min at 4 °C, and 1 mL of 1 mol/L NaOH was added followed by mixing and incubation at 80 °C for 30 min. Then, the melanin content was measured at 475 nm. The melanin content was homogenized to the empty vector group.
4.5. RNA Extraction and qRT-PCR Analysis
Total RNA was extracted from mice skin tissues or melanocytes using TRIzol reagent (TAKARA, Dalian, China). The concentration and integrity of total RNA were determined using the NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, NA, USA) and identified by 1% agarose gel electrophoresis, respectively. Complementary (c) DNA synthesis was performed using the PrimeScriptTM RT regent Kit (TAKARA, Dalian, China) following the manufacturer’s instructions. The real-time quantitative PCR was performed using SYBR®Premix Ex TaqTMII (Tli RNaseH Plus) (TAKARA, Dalian, China). All qRT-PCR reactions were performed in triplicate on the StepOne PlusTM Real-Time PCR System (Applied Biosystems, Inc., Foster City, CA, USA). Quantification of OA1 transcript abundance was performed using the comparative threshold cycle (Ct) method. Transcript levels of gene were normalized relative to those of the internal control β-actin. The primer details are shown in Table 1.
Table 1.
Primers used in this study.
| Primer Name | Primer Sequence 5’→3’ | PCR Production (bp) |
|---|---|---|
| Mus-OA1-F-XhoI | CCGCTCGAGGCCACCATGGCCTCCCCGCGCCT | 1218 |
| Mus-OA1-R-EcoRI | CGGAATTCTCAGAGTTCCCCCTGGGCTTG | |
| Mus-OA1-F | ATCAGGGCGTCGATCTGTTG | 193 |
| Mus-OA1-R | AGCAGGCCAAATGTCTGTTG | |
| Mus-MITF-F | AGGACCTTGAAAACCGACAG | 115 |
| Mus-MITF-R | GTGGATGGGATAAGGGAAAG | |
| Mus-TYR-F | ACTTACTCAGCCCAGCATCC | 109 |
| Mus-TYR-R | AGTGGTCCCTCAGGTGTTCC | |
| Mus-TRP1-F | CTTGGAGGTCCGTGTATTTG | 223 |
| Mus-TRP1-R | GACCGCATCAGTGAAAGTGT | |
| Mus-TRP2-F | CCAACGCTGATTAGTCGGA | 213 |
| Mus-TRP2-R | GAAGAAGGGAGGGCTGTCA | |
| Mus-GPNMB-F | GGGCATACATTCCCATCTCG | 215 |
| Mus-GPNMB-R | AGTGTTGTCCCCAAAGTTCCA | |
| Mus-PMEL-F | AGTGTTGTCCCCAAAGTTCCA | 171 |
| Mus-PMEL-R | AGGAGCGGGCTGTTTTCT | |
| Mus-β-actin-F | TTGCTGACAGGATGCAGAAG | 140 |
| Mus-β-actin-R | TTGCTGACAGGATGCAGAAG |
4.6. Western Blotting
Total protein from mice skin tissues or melanocytes was extracted using total protein extraction reagent (Beyotime, Shanghai, China) according to the manufacturer’s instructions, and the concentration was measured using a nucleic acid/protein analyzer. Two hundred micrograms of protein lysate from each sample were size-separated by SDS-PAGE electrophoresis and transferred onto nitrocellulose filter membranes (Boster, Wuhan, China). The membranes were blocked in 5% skimmed milk powder (Boster, Wuhan, China) at room temperature for 1 h and then were probed with the primary antibody diluted in Tris-buffered saline-Tween (TBST) overnight at 4 °C. The next day, the membranes were washed 3 times in TBST for 10 min each and incubated with HRP-conjugated second antibody (1:10,000 (v/v), Boster, Wuhan, China) at 37 °C with horizontal shaking for 1 h. Subsequently, the membranes were washed 6 times in TBST for 5 min each, and the proteins were visualized via a super ECL chemiluminescence solution (Boster, Wuhan, China). The Western blot results were analyzed using Image-ProPlus 6.0 software (Olympus, Hatayaga, Japan) to measure the area and gray value for each target band. The target protein expression level was normalized relative to the corresponding internal reference level in each lane.
4.7. Localization of OA1 by Immunofluorescence Staining
Paraffin-embedded tissues were sectioned, dewaxed for hydration, incubated in 3% H2O2 (Boster, Wuhan, China) at 37 °C for 10 min and then washed 3 times in PBS with shaking for 2 min each. Next, the tissues were blocked with 10% goat serum (Boster, Wuhan, China) for 30 min at room temperate. The excess liquid was then shaken off, and the OA1 anti-rabbit polyclonal antibody (1:100 diluted in PBS) was added dropwise, with no treatment in the negative control. The samples were placed at 4 °C overnight, re-warmed at 37 °C for 30 min the next day and washed 3 times in PBS (3 min per wash). Subsequently, FITC-labeled goat anti-rabbit IgG secondary antibody (1:200 diluted in PBS) was then added dropwise, followed by incubation at 37 °C for 1 h and 6 times in PBS (5 min per wash). The slices were mounted with the anti-fluorescent mounting media and observed under a fluorescence microscope.
4.8. Statistical Analysis
All experiments were carried out in triplicate. Values are presented as the mean ± SD. Statistical analysis was performed by GraphPad Prism 5.0 software (GraphPad Software Inc., La Jolla, CA, USA). Univariate analysis of variance was performed using SPSS 19.0 software (IBM, Armonk, NY, USA), and p-values <0.05 were considered statistically significant and those <0.01 as highly significant.
5. Conclusions
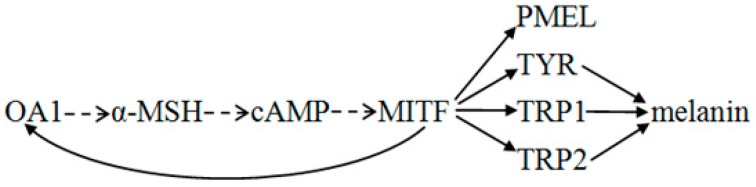
In this study, we found that OA1 was detectably expressed in mice skin of different coat colors, though at significantly different levels. Additionally, OA1 was mainly located in the hair follicle matrix, the inner and outer root sheath in the mice skin tissues with different coat colors. When OA1 was overexpressed in mouse melanocytes, the expression levels of MITF, TYR, TRP1 and PMEL were upregulated, and the expression level of TRP2 was downregulated. By contrast, the levels of GPNMB mRNA and protein were unaffected in melanocyte transfected OA1 compared with the control. Taken together, we propose that OA1 may participate in the formation of coat color by regulating the level of MITF (Figure 5) and the number, size, motility and maturation of melanosomes.
Figure 5.
Potential pathway of OA1 in regulating melanogenesis in mouse melanocytes. Dotted arrows—to be verified; solid arrows—verified.
Acknowledgments
This work was supported by the Aid Program for Innovative Research Team in Shanxi Agricultural University (CXTD201201), the High Technology Research and Development Program of China (863 Program) (2013AA102506), the National Natural Science Foundation of China (31302049), the Specialized Research Fund for the Doctoral Program of Higher Education (20131403120002) and the Special Foundation for Agro-scientific Research in the Public Interest (201303119).
Author Contributions
Tianzhi Chen, Haidong Wang and Changsheng Dong conceived of and designed the experiment together. Tianzhi Chen and Bingling Zhao performed most of the experiments together. Tianzhi Chen and Yu Liu wrote the paper together. Ruiwen Fan and Yuanyuan Zhao supported the technology. Tianzhi Chen, Yu Liu and Pengchao Wang analyzed the data. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Seiji M., Fitzpatrick T.B., Simpson R.T., Birbeck M.S. Chemical composition and terminology of specialized organelles (melanosomes and melanin granules) in mammalian melanocytes. Nature. 1963;197:1082–1084. doi: 10.1038/1971082a0. [DOI] [PubMed] [Google Scholar]
- 2.Hearing V.J. Biogenesis of pigment granules: A sensitive way to regulate melanocyte function. J. Dermatol. Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Chi A., Valencia J.C., Hu Z.Z., Watabe H., Yamaguchi H., Mangini N.J., Huang H., Canfield V.A., Cheng K.C., Yang F., et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J. Proteome Res. 2006;5:3135–3144. doi: 10.1021/pr060363j. [DOI] [PubMed] [Google Scholar]
- 4.Basrur V., Yang F., Kushimoto T., Higashimoto Y., Yasumoto K., Valencia J., Muller J., Vieira W.D., Watabe H., Shabanowitz J., et al. Proteomic analysis of early melanosomes: Identification of novel melanosomal proteins. J. Proteome Res. 2003;2:69–79. doi: 10.1021/pr025562r. [DOI] [PubMed] [Google Scholar]
- 5.Korner A., Pawelek J. Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science. 1982;217:1163–1165. doi: 10.1126/science.6810464. [DOI] [PubMed] [Google Scholar]
- 6.McKay B.S., Schwartz S.G. Pigmentation and macular degeneration: Is there a role for GPR143? J. Ocul. Pharmacol. Ther. 2016;32:3–4. doi: 10.1089/jop.2016.29007.bsm. [DOI] [PubMed] [Google Scholar]
- 7.Giordano F., Simoes S., Raposo G. The ocular albinism type 1 (OA1) GPCR is ubiquitinated and its traffic requires endosomal sorting complex responsible for transport (ESCRT) function. Proc. Natl. Acad. Sci. USA. 2011;108:11906–11911. doi: 10.1073/pnas.1103381108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassi M.T., Schiaffino M.V., Renieri A., de Nigris F., Galli L., Bruttini M., Gebbia M., Bergen A.A., Lewis R.A., Ballabio A. Cloning of the gene for ocular albinism type 1 from the distal short arm of the X chromosome. Nat. Genet. 1995;10:13–19. doi: 10.1038/ng0595-13. [DOI] [PubMed] [Google Scholar]
- 9.Schiaffino M.V., Baschirotto C., Pellegrini G., Montalti S., Tacchetti C., De Luca M., Ballabio A. The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc. Natl. Acad. Sci. USA. 1996;93:9055–9060. doi: 10.1073/pnas.93.17.9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sone M., Orlow S.J. The ocular albinism type 1 gene product, OA1, spans intracellular membranes 7 times. Exp. Eye Res. 2007;85:806–816. doi: 10.1016/j.exer.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Addio M., Pizzigoni A., Bassi M.T., Baschirotto C., Valetti C., Incerti B., Clementi M., de Luca M., Ballabio A., Schiaffino M.V. Defective intracellular transport and processing of OA1 is a major cause of ocular albinism type 1. Hum. Mol. Genet. 2000;9:3011–3018. doi: 10.1093/hmg/9.20.3011. [DOI] [PubMed] [Google Scholar]
- 12.Schiaffino M.V., d’Addio M., Alloni A., Baschirotto C., Valetti C., Cortese K., Puri C., Bassi M.T., Colla C., de Luca M., et al. Ocular albinism: Evidence for a defect in an intracellular signal transduction system. Nat. Genet. 1999;23:108–112. doi: 10.1038/12715. [DOI] [PubMed] [Google Scholar]
- 13.Innamorati G., Piccirillo R., Bagnato P., Palmisano I., Schiaffino M.V. The melanosomal/lysosomal protein OA1 has properties of a G protein-coupled receptor. Pigment Cell Res. 2006;19:125–135. doi: 10.1111/j.1600-0749.2006.00292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez V.M., Decatur C.L., Stamer W.D., Lynch R.M., McKay B.S. L-DOPA is an endogenous ligand for OA1. PLoS Biol. 2008;6:1596. doi: 10.1371/journal.pbio.0060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda N., Naito S., Masukawa D., Kaneda M., Miyamoto H., Abe T., Yamashita Y., Endo I., Nakamura F., Goshima Y. Expression of ocular albinism 1 (OA1), 3, 4-dihydroxy-L-phenylalanine (DOPA) receptor, in both neuronal and non-neuronal organs. Brain Res. 2015;1602:62–74. doi: 10.1016/j.brainres.2015.01.020. [DOI] [PubMed] [Google Scholar]
- 16.Masukawa D., Nakamura F., Koga M., Kamiya M., Chen S., Yamashita N., Arai N., Goshima Y. Localization of ocular albinism-1 gene product GPR143 in the rat central nervous system. Neurosci. Res. 2014;88:49–57. doi: 10.1016/j.neures.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Hiroshima Y., Miyamoto H., Nakamura F., Masukawa D., Yamamoto T., Muraoka H., Kamiya M., Yamashita N., Suzuki T., Matsuzaki S., et al. The protein Ocular Albinism 1 is the orphan GPCR GPR143 and mediates depressor and bradycardic responses to DOPA in the nucleus tractus solitarii. Br. J. Pharmacol. 2014;171:403–414. doi: 10.1111/bph.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamaraj B., Gopalakrishnan C., Purohit R. In silico analysis of mirna-mediated gene regulation in OCA and OA genes. Cell Biochem. Biophys. 2014;70:1923–1932. doi: 10.1007/s12013-014-0152-9. [DOI] [PubMed] [Google Scholar]
- 19.Vetrini F., Auricchio A., Du J., Angeletti B., Fisher D.E., Ballabio A., Marigo V. The microphthalmia transcription factor (MITF) controls expression of the ocular albinism type 1 gene: Link between melanin synthesis and melanosome biogenesis. Mol. Cell. Biol. 2004;24:6550–6559. doi: 10.1128/MCB.24.15.6550-6559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vachtenheim J., Borovansky J. “Transcription physiology” of pigment formation in melanocytes: Central role of MITF. Exp. Dermatol. 2010;19:617–627. doi: 10.1111/j.1600-0625.2009.01053.x. [DOI] [PubMed] [Google Scholar]
- 21.Steingrimsson E., Copeland N.G., Jenkins N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 22.Falletta P., Bagnato P., Bono M., Monticone M., Schiaffino M.V., Bennett D.C., Goding C.R., Tacchetti C., Valetti C. Melanosome-autonomous regulation of size and number: The OA1 receptor sustains PMEL expression. Pigment Cell Melanoma Res. 2014;27:565–579. doi: 10.1111/pcmr.12239. [DOI] [PubMed] [Google Scholar]
- 23.Ujvari A., Aron R., Eisenhaure T., Cheng E., Parag H.A., Smicun Y., Halaban R., Hebert D.N. Translation rate of human tyrosinase determines its N-linked glycosylation level. J. Biol. Chem. 2001;276:5924–5931. doi: 10.1074/jbc.M009203200. [DOI] [PubMed] [Google Scholar]
- 24.Cooksey C.J., Garratt P.J., Land E.J., Pavel S., Ramsden C.A., Riley P.A., Smit N.P. Evidence of the indirect formation of the catecholic intermediate substrate responsible for the autoactivation kinetics of tyrosinase. J. Biol. Chem. 1997;272:26226–26235. doi: 10.1074/jbc.272.42.26226. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi T., Imokawa G., Bennett D.C., Hearing V.J. Tyrosinase stabilization by Tyrp1 (the brown locus protein) J. Biol. Chem. 1998;273:31801–31805. doi: 10.1074/jbc.273.48.31801. [DOI] [PubMed] [Google Scholar]
- 26.Jackson I.J., Chambers D.M., Tsukamoto K., Copeland N.G., Gilbert D.J., Jenkins N.A., Hearing V. A second tyrosinase-related protein, TRP-2, maps to and is mutated at the mouse slaty locus. EMBO J. 1992;11:527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsukamoto K., Jackson I.J., Urabe K., Montague P.M., Hearing V.J. A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 1992;11:519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berson J.F., Theos A.C., Harper D.C., Tenza D., Raposo G., Marks M.S. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 2003;161:521–533. doi: 10.1083/jcb.200302072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi T., Urabe K., Orlow S.J., Higashi K., Imokawa G., Kwon B.S., Potterf B., Hearing V.J. The Pmel 17/silver locus protein. Characterization and investigation of its melanogenic function. J. Biol. Chem. 1994;269:29198–29205. [PubMed] [Google Scholar]
- 30.Theos A.C., Berson J.F., Theos S.C., Herman K.E., Harper D.C., Tenza D., Sviderskaya E.V., Lamoreux M.L., Bennett D.C., Raposo G., et al. Dual loss of ER export and endocytic signals with altered melanosome morphology in the silver mutation of Pmel17. Mol. Biol. Cell. 2006;17:3598–3612. doi: 10.1091/mbc.E06-01-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoashi T., Sato S., Yamaguchi Y., Passeron T., Tamaki K., Hearing V.J. Glycoprotein nonmetastatic melanoma protein b, a melanocytic cell marker, is a melanosome-specific and proteolytically released protein. FASEB J. 2010;24:1616–1629. doi: 10.1096/fj.09-151019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weterman M.A., Ajubi N., van Dinter I.M., Degen W.G., van Muijen G.N., Ruitter D.J., Bloemers H.P. Nmb, a novel gene, is expressed in low-metastatic human melanoma cell lines and xenografts. Int. J. Cancer. 1995;60:73–81. doi: 10.1002/ijc.2910600111. [DOI] [PubMed] [Google Scholar]
- 33.Prota G., Hu D.N., Vincensi M.R., McCormick S.A., Napolitano A. Characterization of melanins in human irides and cultured uveal melanocytes from eyes of different colors. Exp. Eye Res. 1998;67:293–299. doi: 10.1006/exer.1998.0518. [DOI] [PubMed] [Google Scholar]
- 34.Scriver C.R., Stanbury J.B., Wyngaarden J.B., Fredrickson D.S. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York, NY, USA: 1997. pp. 843–844. [Google Scholar]
- 35.Hearing V.J. Unraveling the melanocyte. Am. J. Hum. Genet. 1993;52:1–7. [PMC free article] [PubMed] [Google Scholar]
- 36.Hearing V.J. The melanosome: The perfect model for cellular responses to the environment. Pigment Cell Res. 2000;13:23–34. doi: 10.1034/j.1600-0749.13.s8.7.x. [DOI] [PubMed] [Google Scholar]
- 37.Ito S. The IFPCS presidential lecture: A chemist’s view of melanogenesis. Pigment Cell Res. 2003;16:230–236. doi: 10.1034/j.1600-0749.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 38.Ito S., Wakamatsu K. Human hair melanins: What we have learned and have not learned from mouse coat color pigmentation. Pigment Cell Melanoma Res. 2011;24:63–74. doi: 10.1111/j.1755-148X.2010.00755.x. [DOI] [PubMed] [Google Scholar]
- 39.Fitzpatrick T., Hori Y., Toda K., Seiji M. Melanin 1969: Some definitions and problems. Jpn. J. Dermatol. B. 1969;79:278–282. [Google Scholar]
- 40.Mayer T.C. The migratory pathway of neural crest cells into the skin of mouse embryos. Dev. Biol. 1973;34:39–46. doi: 10.1016/0012-1606(73)90337-0. [DOI] [PubMed] [Google Scholar]
- 41.Nishimura E.K., Yoshida H., Kunisada T., Nishikawa S.I. Regulation of E- and P-cadherin expression correlated with melanocyte migration and diversification. Dev. Biol. 1999;215:155–166. doi: 10.1006/dbio.1999.9478. [DOI] [PubMed] [Google Scholar]
- 42.Rawles M.E. Origin of pigment cells from the neural crest in the mouse embryo. Physiol. Zool. 1947;20:248–266. doi: 10.1086/physzool.20.3.30151958. [DOI] [PubMed] [Google Scholar]
- 43.Hirobe T. Histochemical survey of the distribution of the epidermal melanoblasts and melanocytes in the mouse during fetal and postnatal periods. Anat. Rec. 1984;208:589–594. doi: 10.1002/ar.1092080414. [DOI] [PubMed] [Google Scholar]
- 44.Lin J.Y., Fisher D.E. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 45.Horikawa T., Norris D.A., Johnson T.W., Zekman T., Dunscomb N., Bennion S.D., Jackson R.L., Morelli J.G. DOPA-negative melanocytes in the outer root sheath of human hair follicles express premelanosomal antigens but not a melanosomal antigen or the melanosome-associated glycoproteins tyrosinase, TRP-1, and TRP-2. J. Investig. Dermatol. 1996;106:28–35. doi: 10.1111/1523-1747.ep12326989. [DOI] [PubMed] [Google Scholar]
- 46.Sarin K.Y., Artandi S.E. Aging, graying and loss of melanocyte stem cells. Stem Cell Rev. 2007;3:212–217. doi: 10.1007/s12015-007-0028-0. [DOI] [PubMed] [Google Scholar]
- 47.Slominski A., Wortsman J., Plonka P.M., Schallreuter K.U., Paus R., Tobin D.J. Hair follicle pigmentation. J. Investig. Dermatol. 2005;124:13–21. doi: 10.1111/j.0022-202X.2004.23528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tobin D.J., Bystryn J.C. Different populations of melanocytes are present in hair follicles and epidermis. Pigment Cell Res. 1996;9:304–310. doi: 10.1111/j.1600-0749.1996.tb00122.x. [DOI] [PubMed] [Google Scholar]
- 49.Cui J., Shen L.Y., Wang G.C. Role of hair follicles in the repigmentation of vitiligo. J. Investig. Dermatol. 1991;97:410–416. doi: 10.1111/1523-1747.ep12480997. [DOI] [PubMed] [Google Scholar]
- 50.Ortonne J.P., Pelletier N., Chabanon M., Thivolet J. Vitiligo and cutaneous epitheliomas. Ann. Dermatol. Venereol. 1978;105:1063–1064. [PubMed] [Google Scholar]
- 51.Giordano F., Bonetti C., Surace E.M., Marigo V., Raposo G. The ocular albinism type 1 (OA1) G-protein-coupled receptor functions with MART-1 at early stages of melanogenesis to control melanosome identity and composition. Hum. Mol. Genet. 2009;18:4530–4545. doi: 10.1093/hmg/ddp415. [DOI] [PubMed] [Google Scholar]
- 52.O’Donnell F.E., Hambrick G.W., Green W.R., Iliff W.J., Stone D.L. X-linked ocular albinism: An oculocutaneous macromelanosomal disorder. Arch. Ophthalmol. 1976;94:1883–1892. doi: 10.1001/archopht.1976.03910040593001. [DOI] [PubMed] [Google Scholar]
- 53.Cortese K., Giordano F., Surace E.M., Venturi C., Ballabio A., Tacchetti C., Marigo V. The ocular albinism type 1 (OA1) gene controls melanosome maturation and size. Investig. Ophthalmol. Vis. Sci. 2005;46:4358–4364. doi: 10.1167/iovs.05-0834. [DOI] [PubMed] [Google Scholar]
- 54.Palmisano I., Bagnato P., Palmigiano A., Innamorati G., Rotondo G., Altimare D., Venturi C., Sviderskaya E.V., Piccirillo R., Coppola M., et al. The ocular albinism type 1 protein, an intracellular G protein-coupled receptor, regulates melanosome transport in pigment cells. Hum. Mol. Genet. 2008;17:3487–3501. doi: 10.1093/hmg/ddn241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiaffino M.V. Signaling pathways in melanosome biogenesis and pathology. Int. J. Biochem. Cell Biol. 2010;42:1094–1104. doi: 10.1016/j.biocel.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgoyne T., Jolly R., Martin-Martin B., Seabra M.C., Piccirillo R., Schiaffino M.V., Futter C.E. Expression of OA1 limits the fusion of a subset of MVBs with lysosomes—A mechanism potentially involved in the initial biogenesis of melanosomes. J. Cell Sci. 2013;126:5143–5152. doi: 10.1242/jcs.128561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Busca R., Ballotti R. Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment Cell Res. 2000;13:60–69. doi: 10.1034/j.1600-0749.2000.130203.x. [DOI] [PubMed] [Google Scholar]
- 58.Du J., Miller A.J., Widlund H.R., Horstmann M.A., Ramaswamy S., Fisher D.E. MlANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am. J. Pathol. 2003;163:333–343. doi: 10.1016/S0002-9440(10)63657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loftus S.K., Antonellis A., Matera I., Renaud G., Baxter L.L., Reid D., Wolfsberg T.G., Chen Y., Wang C., Prasad M.K., et al. Gpnmb is a melanoblast-expressed, MITF-dependent gene. Pigment Cell Melanoma Res. 2009;22:99–110. doi: 10.1111/j.1755-148X.2008.00518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Incerti B., Cortese K., Pizzigoni A., Surace E.M., Varani S., Coppola M., Jeffery G., Seeliger M., Jaissle G., Bennett D.C., et al. OA1 knock-out: New insights on the pathogenesis of ocular albinism type 1. Hum. Mol. Genet. 2000;9:2781–2788. doi: 10.1093/hmg/9.19.2781. [DOI] [PubMed] [Google Scholar]
- 61.Burgoyne T., O’Connor M.N., Seabra M.C., Cutler D.F., Futter C.E. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J. Cell Sci. 2015;128:1400–1407. doi: 10.1242/jcs.164400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yasumoto K., Yokoyama K., Takahashi K., Tomita Y., Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J. Biol. Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 63.Shi Z., Ji K., Yang S., Zhang J., Yao J., Dong C., Fan R. Biological characteristics of mouse skin melanocytes. Tissue Cell. 2016;48:114–120. doi: 10.1016/j.tice.2016.01.001. [DOI] [PubMed] [Google Scholar]