Abstract
Leaf color mutants are common in higher plants that can be used as markers in crop breeding or as an important tool in understanding regulatory mechanisms in chlorophyll biosynthesis and chloroplast development. In virescent leaf mutants, young leaves are yellow in color, which gradually return to normal green when the seedlings grow large. In the present study, we conducted phenotypic characterization and genetic mapping of the cucumber virescent leaf mutant 9110Gt conferred by the v-1 locus. Total chlorophyll and carotenoid content in 9110Gt was reduced by 44% and 21%, respectively, as compared with its wild type parental line 9110G. Electron microscopic investigation revealed fewer chloroplasts per cell and thylakoids per chloroplast in 9110Gt than in 9110G. Fine genetic mapping allowed for the assignment of the v-1 locus to a 50.4 kb genomic DNA region in chromosome 6 with two flanking markers that were 0.14 and 0.16 cM away from v-1, respectively. Multiple lines of evidence supported CsaCNGCs as the only candidate gene for the v-1 locus, which encoded a cyclic-nucleotide-gated ion channel protein. A single nucleotide change in the promoter region of v-1 seemed to be associated with the virescent color change in 9110Gt. Real-time PCR revealed significantly lower expression of CsaCNGCs in the true leaves of 9110Gt than in 9110G. This was the first report that connected the CsaCNGCs gene to virescent leaf color change, which provided a useful tool to establish linkages among virescent leaf color change, chloroplast development, chlorophyll biosynthesis, and the functions of the CsaCNGCs gene.
Keywords: cucumber, virescent leaf, chloroplast development, map-based cloning
1. Introduction
In higher plants, there are diverse leaf color mutations such as albino, xanthan, light green, virescent, stripes, zebra, and stay-green [1,2]. Due to the ease of identification, large numbers of leaf-color mutants have been described and characterized in many plant species like Arabidopsis thaliana [3], barley [4], maize [5], rice [6,7,8], soybean [9], sunflower [10], and wheat [11,12]. For example, in rice, more than 200 leaf color mutants have been documented, and 154 responsible genes have been mapped to all 12 chromosomes, of which 53 have been cloned [13]. In soybean, 25 nuclear genes affecting chlorophyll-deficiency have been identified and mapped [9]. Leaf-color mutants have also been reported in tomato [14], pepper [15], and carrots [16,17]. Investigation of these color mutants has contributed significantly to our understanding of chlorophyll metabolism and chloroplast development [18]. On the practical side, these mutations are also useful in plant breeding such as identification of genetic purity in hybrid production [19].
Virescent color is a special color mutation. Plants with this kind of mutation exhibit a delay in greening up to the first true leaf due to a delay in chloroplast development; which is indistinguishable from the wild type (WT) when mature. The first recessive virescent leaf mutant was described in maize in 1912 [20]. Virescent mutants have been characterized in barley (Hordeum vulgare L.) [21], common bean (Phaseolus vulgaris) [22], cotton [23], peanut [24], maize [25], Arabidopsis [20], and rice [26]. The Vir-C mutant of tobacco also displays a virescent phenotype, but it is maternally inherited [27].
In cucumber (Cucumis sativus L.), several leaf color mutants have been reported including chlorophyll deficient (cd), golden leaves (g), golden cotyledon (gc), light green cotyledons-1 (lg-1), light green cotyledons-2 (lg-2), virescent (v), variegated virescent (vvi), yellow cotyledons-1 (yc-1), yellow cotyledons-2 (yc-2), yellow plant (yp), yellow stem (ys) [28], and virescent (v-1) [29], The yp locus has recently been cloned which encodes a magnesium chelatase submit unit I [30]. The cucumber virescent leaf mutant 9110Gt has been phenotypically characterized. Genetically, this mutant was shown to be controlled by a single recessive gene v-1 which was mapped to cucumber chromosome 6 [31]. The objective of the present study was to clone the candidate gene for the v-1 mutant. We carried out high-resolution genetic mapping and identified the most likely candidate gene for the v-1 locus using the cucumber genome and molecular marker resources.
2. Results
2.1. Phenotypic Characterization of the v-1 Mutant
After germination, the virescent mutant 9110Gt showed light yellow cotyledons and first true leaf whereas those on the WT 9110G or 9930 were green (Figure 1a,b). As the true leaves became fully expanded, the light yellow color gradually turned to green. This trait was similar to the virescent leaf color (gene v) described by Pierce and Wehner [28]. The plant of 9110Gt was slightly smaller than that of WT 9110G throughout their development stages. We also found that this color change was temperature dependent. During the summer season, the yellow color in the seedlings was not as obvious as in the winter greenhouse season, and the duration of yellow color in the cotyledons and true leaves was shorter when the temperature was higher.
Figure 1.
Phenotypic characterization of virescent leaf (v-1) mutant 9110Gt, wild type parental line 9930 and their F1. (a) Leaf color of 9110Gt, 9930, and their F1 at the first true leaf stage; (b) Cotyledon color of 9110Gt, 9930, and their F1; (c) Cotyledon colors of 9110Gt × 9930 F2 plants; (d) Structure of mesophyll cells and chloroplasts of 9110G and mutant 9110Gt. g = grana, sg = starch grains, pg = plastoglobulus, scale bar = 1 μm.
We previously showed that the virescent leaf phenotype of 9110Gt was controlled by a single recessive nuclear gene v-1 [31]. In the present study, the F1 plants of both 9110Gt × 9110G and 9110Gt × 9930 had green leaves (Figure 1a,b); the 1800 F2 plants from the 9110Gt × 9930 cross segregated at 446 yellow leaf (v-1v-1) to 1354 green leaf (V-1_) (Figure 1c), which was consistent with the expected 3 green to 1 virescent segregating ratio in the χ2 test (p = 0.83) supporting a single recessive that was responsible for the virescent leaf color in 9110Gt.
We measured pigment (total chlorophyll and carotenoids) contents of 9110G and 9110Gt, and found that, at the virescent stage (cotyledon and first true leaf), the total chlorophyll (Chl) and carotenoids in 9110Gt mutant was only 44% and 21% of the wild-type 9110G, respectively (Table 1). This suggested that the v-1 mutant phenotype is probably due to reduced contents of photosynthetic pigments. The decrease in Chla (46%) was greater than that for Chlb (35%) (Table 1), suggesting that the v-1 mutant exhibits delayed greening during photomorphogenesis because of slower Chl accumulation. When the cotyledons and first true leaves were completely unfolded, mutant plants accumulated substantial quantities of chlorophyll, and the contents of Chl were not significantly different from those in the WT.
Table 1.
Photosynthetic pigment content in virescent leaf mutant 9110Gt and 9110G (WT) at the yellowing stage (Chl = chlorophyll, Car = carotenoid; mg/g FW).
| Materials | Chla (mg/g) | Chlb (mg/g) | Caro (mg/g) | Total Chl (mg/g) | Chla/b |
|---|---|---|---|---|---|
| 9110G | 1.38 ± 0.11 ** | 0.40 ± 0.01 ** | 0.39 ± 0.01 * | 1.78 ± 0.01 ** | 3.45 |
| 9110Gt | 0.74 ± 0.02 | 0.26 ± 0.01 | 0.31 ± 0.06 | 1.00 ± 0.21 | 2.85 |
| % of WT | 46.0 | 35.0 | 21.0 | 44.0 | 17.0 |
*, **: Significant at 5% and 1% level, respectively.
We examined the ultrastructure of chloroplasts in the leaves of 9110Gt and 9110G at the first true leaf stage, which are shown in Figure 1d. As compared with WT, 9110Gt exhibited fewer chloroplasts per cell and few thylakoids per chloroplast (Figure 1d). In addition, there were increased numbers of plastoglobuli, decreased numbers of starch granules, and a lack of granal membranes in the mesophyll chloroplasts of 9110Gt as compared with 9110G. These results indicated that the development of chloroplast was defective in the mutant.
2.2. High-Resolution Mapping of the v-1 Locus
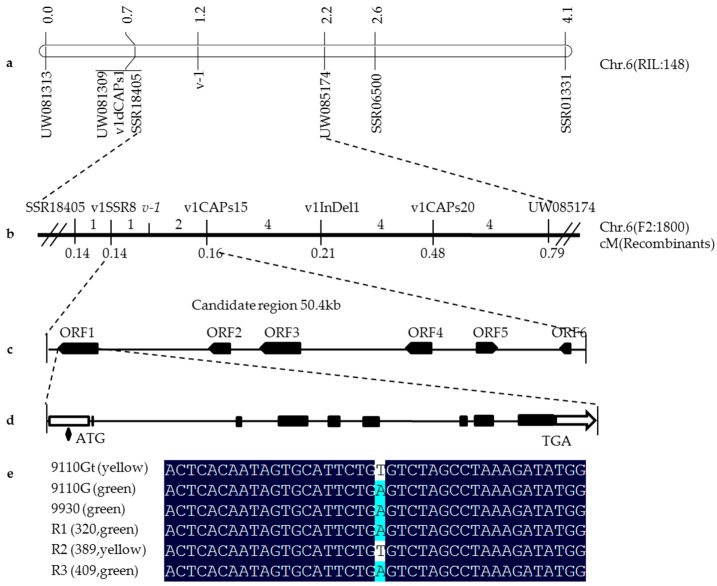
Using 148 RILs of 9110Gt × 9930 cross, we previously mapped the v-1 locus in cucumber chromosome 6 between SSR01331 and SSR18405 with genetic distances 1.6 and 0.3 cM, respectively [31]. Physically, both markers were located in the same scaffold from the 9930 cucumber draft genome assembly scaffold000066 and scaffold03904 in the Gy14 assembly, and the physical distance between the two markers was approximately 493 kb. To identify closer markers, based on the genomic DNA sequences of scaffold3904 and scaffold000066, 148SSR and five dCAPs markers were developed and screened for polymorphisms between 9930 and 9110Gt. Eleven polymorphic markers were identified and mapped with a polymorphism rate of 7.2%. Linkage analysis with 148 RILs suggested that UW085174 and SSR18405 were the two closet flanking markers, which were 1.2 and 0.3 cM away from the v-1 locus, respectively (Figure 2a).
Figure 2.
Fine genetic mapping of v-1 gene. (a) Framework map of v-1 gene with 148 RILs. Numbers above the chromosome are genetic distance in centiMorgan (cM); (b) Fine mapping narrowed the v-1 locus into a 50.4 kb region flanked with markers v1SSR8 and CAPs15 based on 1800 F2 plants. Numbers above the chromosome are recombinants at each interval; (c) Six genes were predicted in the 50.4 kb region; (d) Structure of CsaCNGCs candidate gene. Black boxes are exons; white boxes are UTRs; and black lines indicate introns; and (e) Alleles of a SNP (A/T) in the promoter region of the CsaCNGCs candidate gene in 9110G, 9110Gt, 9930 and 3 recombinant plants.
To fine map the v-1 locus, the 1800 F2 plants were screened with UW081309, UW081313, UW085174, and SSR18405. The v-1 gene was consistently mapped to an interval between UW085174 and SSR18405, and the order of these mapped loci was consistent with their physical positions in the scaffolds. The physical distance between UW085174 and SSR18405 was 171.5 kb in 9930 scaffold000066 and 171.9 kb in Gy14 scaffold03904. Of the 1800 F2 plants, 16 were recombinants between UW085174 and SSR18405. Examination of DNA sequence variations in this 172 kb region identified SNPs or indels that allowed design of 10 SSR, 1 indel, and 25 SNP-derived dCAPA/CAPS, and eventually 4 markers, v1SSR8,v1CAPs15, v1CAPs20, and v1InDel1 were mapped with the 16 F2 recombinant plants (Figure 2b). The names and sequence of all mapped markers on the map are listed in Supplemental Table S1. Now the v-1 gene was delimited by markers v1SSR8 and v1CAPs15 with the genetic distance being 0.14 and 0.16 cM, respectively from v-1. The physical distance was approximately 50.4 kb in 9930 scaffold000066.
2.3. Candidate Gene Annotation and Sequence Analysis
Using the FGENESH program, we annotated the 50.4 kb genomic DNA sequences from 9930 scaffold000066, and six genes (ORF1 to ORF6) were predicted, which are listed in Table 2. None of the six genes have been reported to be involved in leaf color mutations in the literature. Annotation of ORF1 through ORF6 indicated that there were 8, 4, 9, 1, 1, and 2 exons in the coding regions of the six genes, respectively. Alignment of the CDS sequences of the six predicted genes revealed no nucleotide variations in ORF2 to ORF6 between 9110Gt and 9110G, suggesting that these five annotated genes are unlikely to be associated with the leaf color mutation. ORF1 was predicted to encode a cyclic nucleotide-gated channel (CNGC) protein (Table 2) which seemed to be a potential candidate gene for the v-1 locus. Therefore, we designated the gene CsaCNGCs.
Table 2.
Information of six predicted genes in the 50.4 kb region of the v-1 locus.
| Gene | Start | End | Exon | Annotation |
|---|---|---|---|---|
| ORF1 | 263878 | 268887 | 8 | cyclic nucleotide-gated channel |
| ORF2 | 277911 | 280354 | 4 | 3′-5′-exoribonuclease family protein |
| ORF3 | 281291 | 286306 | 9 | receptor-like serine/threonine-protein kinase NCRK |
| ORF4 | 289805 | 293123 | 1 | uncharacterized protein |
| ORF5 | 293407 | 294673 | 1 | uncharacterized protein |
| ORF6 | 299479 | 301054 | 2 | expansin A23 |
BLAST search of the cucumber Gy14 and 9930 draft genomes revealed a single copy of CsaCNGCs in the cucumber genome. We cloned the CsaCNGCs gene sequences from 9110Gt, 9930, and 9110G (Figure S1), which were 5010 bp in length. The coding region of CsaCNGCs gene was predicted to have eight exons and encode a protein with 720-amino acids (Figure 2e). Alignment of the CsaCNGCs candidate gene sequence identified no variations in the coding regions among 9110Gt, 9110G, and 9930 (Figure S1), but there was a single nucleotide mutation (A→T) in the promoter region before the transcription start site (TSS) (Figure 2e). There were three recombinants between the two markers v1SSR8 and v1CAPs15 flanking the v-1 locus (Figure 2b). We genotyped the three recombinants by Sanger sequencing of the promoter region harboring this SNP and the result (Figure 2e) indicated that the SNP alleles were completely consistent with the leaf color phenotype, providing further evidence that CsaCNGCs was indeed the candidate gene for the v-1 locus.
2.4. The Single Nucleotide Mutation in the Promoter Region at the CsaCNGCs Locus Was Unique in Different Cucumber Lines
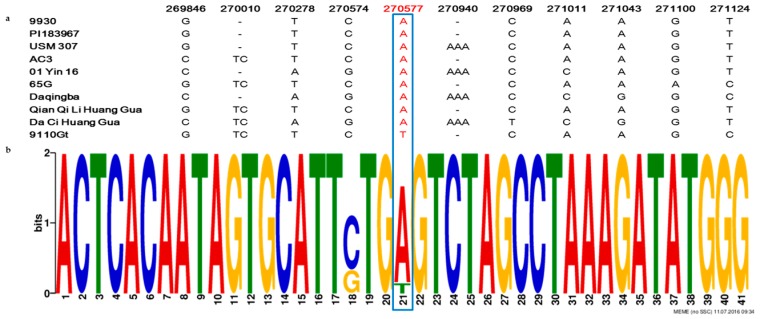
To confirm the identity of the single nucleotide mutation in the promoter region at the CsaCNGCs, we investigated the sequence conservation at CsaCNGCs locus in eight cucumber lines of different ecotypes including a wild cucumber (C. sativus var.hardwickii, PI 193967), a south China type cucumber (USM 307), a North American pickling cucumber (AC3), two European greenhouse cucumbers (01Yin16 and 65G), and three north China type cucumbers (Daqingba, Qian Qi Li Huang Gua, and Da Ci Huang Gua). Motif searches in the 2000 bp sequence upstream of the CsaCNGCs TSS region in the eight cucumber lines revealed, nine SNPs, and two insertion-deletions nucleotide mutations in the promoter region (Figure 3a), butonly 9110Gt had a SNP (A→T) that was consistent with the virescent leaf phenotype among these lines (Figure 3b), supporting CsaCNGCs as the most possible candidate gene for the v-1 locus.
Figure 3.
DNA sequence alignment of CsaCNGCs gene promoter regions among eight cucumber ecotypes. (PI 193967, USM 307, AC3, 01Yin16, 65G, Daqingba, Qian Qi Li Huang Gua, and Da Ci Huang Gua). (a) Among all SNPs or InDels detected in this region, only A→T SNP is consistent with the phenotypes (virescent vs. green leaf). The numbers on top are nucleotide positions in 9930 scaffold000066; (b) The single nucleotide mutation in the promoter region at the CsaCNGCs locus was unique in different cucumber lines. The numbers of sequences which −20 to +20 sequences of the single nucleotide mutation (A/T). Blue boxes = Unique single nucleotide mutation (A/T) in the promoter region of the CsaCNGCs candidate gene in 9110Gt.
2.5. Expression of CsaCNGCs in 9110Gt Mutant and 9110G WT
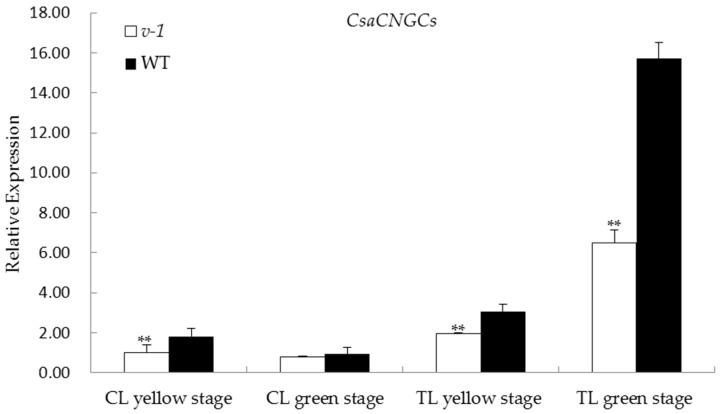
We quantified transcript abundance of CsaCNGCs in the cotyledons and first true leaves of 9110Gt and 9110G with qPCR at both virescent and green leaf stages. The forward and reverse primer sequences (5′-3′) for CsaCNGCs were: GGATTCATGGTTTAATACGAGC and GATTTTGTCCCAAGGAACTGA, respectively. The cucumber EF1α gene was used as the reference (left primer sequence: ACTGTGCTGTCCTCATTATTG; right: AGGGTGAAAGCAAGAAGAGC). The qPCR results are shown in Figure 4. It seemed that CsaCNGCs was constitutively expressed in cotyledons and the first true leaf. No difference in CsaCNGCs expression was found in the green cotyledons between WT and 9110Gt, but its expression was significantly lower in 9110Gt than in WT at both virescent and green first true leaves, as well as the virescent cotyledons. These observations may suggest that the virescent leaf color in 9110Gt is the result of reduced expression of CsaCNGC.
Figure 4.
Real-time PCR expression analysis of CsaCNGCs candidate genes in 9110Gt (v-1) and 9110G (WT) at yellow and green stages. The relative expression level of each candidate gene was measured in the two lines at the cotyledon (CL) and the first true leaf (TL) stages when they were yellow and green in color by qPCR. EF1a was used as the internal control. Data are means ± SD (n = 3). Asterisks indicate statistically significant differences compared with the wild type at ** p < 0.05 by Student’s t test.
3. Discussion
The leaf-color mutation is a good marker for screening the genetic purity of hybrid crops [13]. It has been helpful understanding of chlorophyll anabolism and catabolism, chloroplast development, and photosynthesis [18]. A chloroplast is one of three types of plastids, characterized by its high concentration of chlorophyll. Plants with normal leaf colors have higher chlorophyll content and normal chloroplasts. In the present study, electron microscopic examination revealed delayed formation of thylakoid membranes in the chloroplasts of the 9110Gt virescent mutant (Figure 1d). The levels of chlorophyll a, chlorophyll b, and carotenoids in the cotyledons and first true leaves of 9110Gt at the virescent yellow stage were significantly lower than that in the wild type (9110G and 9930). Chlorophyll a was reduced more than chlorophyll b (Table 1). The malfunctioning chloroplast and reduced chlorophyll biosynthesis in yellow young leaves seem to be a common feature for many temperature-dependent virescent mutants such as those found in Arabidopsis [20], cotton [32]; rice [7,18,26,33,34,35,36,37,38], maize [39], wheat [40], and apple [41]. Biswal et al. [42] showed that the biosynthesis of chlorophyll a is closely linked to chloroplast development. Therefore, chlorophyll deficiency in the cotyledons and first leaves of 9110Gt may be attributed to delayed chloroplast development due to abnormal formation of thylakoid membranes.
In this study, mutant plant 9110Gt carrying the v-1 gene have virescent leaves that exhibit yellowish-green leaves at the seedling stage, and then the color turns to normal green. Through map-based cloning, the v-1 gene was finally delimited in a 50.4 kb region on the short arm of chromosome 6. On the same chromosome, only one gene yp [30] related to the leaf-color mutation has been mapped up to now. C528 mutants carrying the yp gene exhibited golden leaf color throughout their whole growth and development stage. According to the previous reports, this gene does not locate in the fine mapping region of v-1, so we think that these two mutants are controlled by different genes.
The promoter is the non-coding DNA region that occurs upstream of the coding region of a gene and is required for transcription of that specific gene. The promoter plays an important role in the process of plant gene expression and regulation [43]. Analysis of the CsaCNGCs gene sequence identified no variations in the coding regions among mutant and wildtype. We found a single nucleotide mutation from A to T in the promoter region of this candidate gene was the causal SNP for the viresent leaf mutation. In cucumber, the other only leaf color mutation cloned is the “golden leaf” controlled by the yp locus, which exhibited yellow color throughout its development stage [30]. Therefore, the present study represented the first report on map-based locating of a virescent leaf mutation in cucumber. Many genes that are responsible for virescent leaf mutations in different plant species have been cloned. Most of these genes function in chlorophyll biosynthesis and catabolism, or in the developmental regulation of chloroplast, in the metabolism of carotenoids [26,33,34,35,36]. In Arabidopsis [44], rice [36], and maize [39], several virescent mutants were due to nucleotide changes in the chloroplast Clp protease complex which play important roles in plastid development.
In the present study, from multiple lines of evidence, we showed that CsaCNGCs, a gene for the cyclic nucleotide-gated channel, were the only candidate gene for the v-1 locus (Figure 2 and Figure 3). Cyclic-nucleotide-gated ion channels (CNGCs) are nonselective cation channels that were first identified in vertebrate retinal photoreceptors and olfactory sensory neurons [45]. In plants, it was first identified in a screen for calmodulin (CaM) binding partners in barley [46]. Arabidopsis has 20 CNGCs homologs [47], and their functions critically depend on the signaling molecules cAMP and/or cGMP [48]. Both cGMP and cAMP are reported to be involved in a number of physiological processes such as chloroplast development, stomatal functioning, monovalent and divalent cation fluxes, gibberellic acid signaling, pathogen response, and gene transcription [49,50]. The cyclic GMP is able to trigger the production of anthocyanins, and that a combination of cyclic GMP with calcium can regulate the development of chloroplasts and the photosynthetic machinery [51]. CsaCNGCs shared 64% amino acid sequence identity with Arabidopsis AtCNGC15, and the expression level of CsaCNGCs in WT plants was higher than in the mutant (Figure 4).
The mechanism underlying the leaf color change from yellow to green in virescent leaf mutants is an interesting question. In the rice virescent mutant v14, the second and third leaves displayed an albino phenotype, but from the fourth leaf stage, all leaves including the albino ones are recovered to normal green [38]. The cells of the second and third leaves of the v14 mutant lack mature thylakoid membrane systems and starch grain but normal chloroplast structure can be observed in the fourth leaf of the same mutant. Zhang et al. [38] reasoned that there may be different regulation pathways that are involved in chloroplast development in rice. Another possible explanation is that there might be other CsaCNGCs homologs with redundant functional activities in greening stages. The connection between reduced CsaCNGCs expression with chlorophyll deficiency, delayed chloroplast development, and the virescent leaf in the v-1 mutant is not known and merits further investigation.
4. Materials and Methods
4.1. Plant Materials
Two cucumber inbred lines, 9110Gt and 9930, were used for phenotypic characterization of the virescent mutant and development of segregating populations. 9110Gt was a spontaneous virescent leaf mutation found in 9110G, which has a largely European greenhouse cucumber genetic background [29]. 9930 is a typical north China fresh market cucumber, and the draft genome of 9930 is available [52]. For genetic mapping, 148 recombinant inbred lines (RILs) and 1800 F2 individuals from the cross between 9110Gt and 9930 were developed. For phenotyping, all plant materials were grown in the greenhouses of the Institute of Vegetables and Flowers (IVF), Chinese Academy of Agricultural Sciences (Beijing, China) or the Walnut Street Greenhouse of the University of Wisconsin (Madison, WI, USA) in 2012 to 2014 greenhouse seasons. The temperature in the greenhouses was 25–30 °C during the day and 18–25 °C in the night with 30%–85% relative humidity. Leaf-color phenotypes in the RIL and F2 populations were scored before three-true-leaf seedling stage (before as either green (wild type) or virescent yellow (mutant) [31].
4.2. Measurement of Chlorophyll Contents
Fresh leaves (0.2 g) of 9110Gt mutant and 9100G wild type (WT) seedlings were soaked in 80% acetone and the absorption of the extract measured in a Shimadzu UV-1700 UV-visible spectrophotometer at 646, 663, and 470 nm wavelengths for chlorophyll a/b (Chla, Chlb) and carotenoids (Caro), respectively. The concentrations of total chlorophyll and carotenoids were calculated as described by Lichtenthaler [53].
4.3. Transmission Electron Microscopy
Fresh leaf sections were fixed in 3.0% glutaraldehyde solution followed by 1.0% Osmium tetroxide (OsO4). The tissues were dehydrated in a gradient acetone series and embedded in Epon618 epoxy resin prior to thin sectioning. After staining with uranyl acetate and Reynolds’ lead citrate, the sections were observed under a Hitachi H7500 transmission electron microscope (Tokyo, Japan).
4.4. Genomic DNA Extraction, PCR, and Electrophoresis
Genomic DNA was isolated from young leaf tissues with the CTAB method [31]. The DNA was dissolved in ddH2O and kept at −20 °C until use. A UV spectrophotometer (Shanghai, China) was used to measure DNA purity and concentration, and the final concentration of DNA was diluted to 30 ng/μL. The polymerase chain reaction (PCR) system and protocol was the same with Miao et al. [31]. PCR amplified products were separated on 6.0% non-denaturing polyacrylamide gels in 1× TBE buffer at 150 V for 1.5–2 h, and the bands were visualized and photographed after silver staining [54].
4.5. Development of Molecular Markers for Fine Mapping
In our previous study, with 148 RILs, we showed that the virescent leaf of 9110Gt was controlled by a single recessive nuclear gene v-1, which was place in cucumber chromosome 6 flanked with two microsatellite markers SSR01331 and SSR18405 that were 1.6 cM and 0.3 cM away from the v-1 locus, respectively. A scaffold-based chromosome walking strategy was taken to identify more closely linked markers [55]. Draft genome scaffold assemblies from both the 9930 (V2.0) [52] and Gy14 (V1.0) [56] cucumber lines were employed. In the target scaffolds, new markers were selected from the published cucumber genetic maps [57,58] and a collection of 83,689 SSR markers that were developed from the Gy14 draft genome assembly [59].
At the fine mapping stage, SNPs and indels were explored in the next-generation whole genome re-sequencing data. The genomes of 9110Gt, Gy14, and 9930 cucumber inbred lines were re-sequenced with the Illumina Hi-Seq 2000 at >15× coverage each (100 bp paired end). For marker discovery, short sequencing reads were aligned to the 9930 draft genome with the BWA (Burrows-Wheeler Alignment Tool) software package [60]. Indel identification and SNP-calling were performed by SAM tools [61]. For SNP genotyping, dCAPs and CAPs markers were developed with dCAPS Finder 2.0 [62]. A total of 189 new SSR, dCAPs, CAPs, and InDel markers were developed. Primer design was performed with Primer 3.0 (http://bioinfo.ut.ee/primer3-0.4.0) [63]. All primers were synthesized commercially or at the Biotech Center at the University of Wisconsin at Madison. All newly developed markers were first screened for polymorphism with the two parental lines; polymorphic markers were applied to the recombinants in the RIL population for linkage analysis, which were then extended to the larger F2 population.
4.6. Linkage Mapping and Candidate Gene Annotation
Linkage analysis between DNA markers and the v-l locus was performed with JoinMap 4.0 with a LOD threshold of 3.0. The recombination percentages were converted to genetic distances using the Kosambi mapping function [64]. Annotation of DNA sequences in the target genomic DNA region was performed using the program FGENESH (http://linux1.softberry.com/berry.phtml) [65], and the results were checked manually. Gene prediction was conducted with BLASTx at the NCBI (National Center for Biotechnology Information) website (http://blast.ncbi.nlm.nih.gov). Re-sequencing reads of different cucumber lines were aligned with the BWA software using the 9930 draft genome as the reference. To detect potential promoter motifs, sequence regions corresponding to upstream of the TSS positions were scanned by MEME (Motif-based sequence analysis tools) version 4.11.2 (http://meme-suite.org/tools/meme).
4.7. Real-Time Quantitative PCR of Candidate Genes
The cotyledons and first true leaves of 9110Gt and 9110G were flash frozen in liquid nitrogen and used for total RNA extraction using TRIzol® (Thermo Fisher Scientific, Waltham, MA, USA). RNA was pretreated with RNase-free DNase I (Takara, Shiga, Japan), and first-strand cDNA was synthesized from 1.0 μg total RNA using reverse transcriptase SYBR FAST qPCR Kit Master Mix (2×) Universal (KAPA Biosystems, Wilmington, MA, USA).
Quantitative real time PCR (qPCR) was performed in a total volume of 10.0 μL which contained 1.0 μL of first-strand cDNA, 5.0 μL SYBR FAST qPCR Kit Master Mix, 0.4 μL gene-specific primers (10 μM·μL−1), 0.2 μL ROX reference dye, and 3.4 μL ddH2O. Reactions were amplified in a ABI prism 7900 HT Real-Time PCR System with following conditions: pre-incubation at 95 °C for 3 min; 40 cycles of 95 °C for 30 s, 60 °C for 20 s; melting curves 95 °C for 15 s, 60 °C for 15 s, and 95 °C for 15 s. The 2ΔΔCt method [66] was used to calculate relative changes in gene expression. All runs had three technical replicates and three biological replicates.
Acknowledgments
The authors thank Caiqing Xu and Kristin Haider for technical assistance and Graham Collins (Roseworthy Campus of the University of Adelaide in South Australia) for revision of the manuscript. HM’s work in YW’s laboratory was partially funded by the China Scholarship Council. This research was supported by the Natural Science Foundation of China (31101549), China Agriculture Research System (CARS-25), Science and Technology Innovation Program of the Chinese Academy of Agricultural Science (CAAS-ASTIP-IVFCAAS), and the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, China.
Supplementary Materials
Supplementary materials can be found at www.mdpi.com/1422-0067/17/10/1602/s1.
Author Contributions
Han Miao contributed to experimental design, performed the research and data analysis, and wrote the manuscript; Xingfang Gu and Shengping Zhang conceived the study, developed the RIL population, and supervised the experiments; Yiqun Weng contributed to experimental design and data analysis and revised of the manuscript; Min Wang and Ye Wang helped with phenotypic data acquisition. All authors have reviewed and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Khan S., Wani R., Bhat M.U.D., Parveen K. Induced chlorophyll mutations in chickpea (Cicer arietinum L.) Int. J. Agric. Biol. 2005;5:764–767. [Google Scholar]
- 2.Park S.Y., Yu J.W., Park J.S., Li J., Yoo S.C., Lee N.Y., Lee S.K., Jeong S.W., Seo H.S., Koh H.J., et al. The senescence-induced stay green protein regulates chlorophyll degradation. Plant Cell. 2007;19:1649–1664. doi: 10.1105/tpc.106.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagata N., Tanaka R., Satoh S., Tanaka A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell. 2005;17:233–240. doi: 10.1105/tpc.104.027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z.L., Yuan S., Liu W.J., Du J.B., Tian W.J., Luo M.H., Lin H.H. Mutation mechanism of chlorophyll-less barley mutant NYB. Photosynthetica. 2008;46:73–78. doi: 10.1007/s11099-008-0013-0. [DOI] [Google Scholar]
- 5.Asakura Y., Kikuchi S., Nakai M. Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J. 2008;56:1007–1017. doi: 10.1111/j.1365-313X.2008.03659.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F.T., Luo X.D., Hu B.L., Wan Y., Xie J.K. YGL138 (t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice. 2013;6:7. doi: 10.1186/1939-8433-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J.Q., Wang Y.H., Chai J.T., Wang L.H., Wang C.M., Long W., Wang D., Wang Y., Zheng M., Peng C., et al. Green-revertible Chlorina 1 (grc1) is required for the biosynthesis of chlorophyll and the early development of chloroplasts in rice. J. Plant Biol. 2013;56:326–335. doi: 10.1007/s12374-013-0180-9. [DOI] [Google Scholar]
- 8.Huang X.Q., Wang P.R., Zhao H.X., Deng X.J. Genetic analysis and molecular mapping of a novel chlorophyll-deficit mutant gene in rice. Rice Sci. 2008;15:7–12. doi: 10.1016/S1672-6308(08)60013-X. [DOI] [Google Scholar]
- 9.Reed S., Atkinson T., Gorecki C., Espinosa K., Przybylski S., Goggi A.S., Palmer R.G., Sandhu D. Candidate gene identification for a lethal chlorophyll-deficient mutant in soybean. Agronomy. 2014;4:462–469. doi: 10.3390/agronomy4040462. [DOI] [Google Scholar]
- 10.Mashkina E.V., Usatov A.V., Skorina M.V. Comparative analysis of thermotolerance of sunflower chlorophyll mutants. Russ. J. Genet. 2010;46:178–184. doi: 10.1134/S1022795410020079. [DOI] [PubMed] [Google Scholar]
- 11.Hui Z., Tian F.X., Wang G.K., Wang G.P., Wang W. The antioxidative defense system is involved in the delayed senescence in a wheat mutant tasg1. Plant Cell Rep. 2012;31:1073–1084. doi: 10.1007/s00299-012-1226-z. [DOI] [PubMed] [Google Scholar]
- 12.Li N., Jia J., Xia C., Liu X., Kong X. Characterization and mapping of novel chlorophyll deficient mutant genes in durum wheat. Breed. Sci. 2013;63:169–175. doi: 10.1270/jsbbs.63.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X.J., Zhang H.Q., Wang Y., He F., Liu J.L., Xiao X., Wang G.L. Mapped clone and functional analysis of leaf-color gene ygl7 in a rice hybrid (Oryza sativa L. ssp. indica) PLoS ONE. 2014;9:1602. doi: 10.1371/journal.pone.0099564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terry M.J., Kendrick R.E. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green tomato mutants. Plant Physiol. 1999;119:143–152. doi: 10.1104/pp.119.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barry C.S., McQuinn R.P., Chung M.Y., Besuden A., Giovannoni J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008;147:179–187. doi: 10.1104/pp.108.118430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nothnagel T., Straka P. Inheritance and mapping of a yellow leaf mutant of carrot (Daucus carota) Plant Breed. 2003;122:339–342. doi: 10.1046/j.1439-0523.2003.00884.x. [DOI] [Google Scholar]
- 17.Budahn H., Barański R., Grzebelus D., Kiełkowska A., Straka P., Metge K., Nothnagel T. Mapping genes governing flower architecture and pollen development in a double mutant population of carrot. Front. Plant Sci. 2014;5:504. doi: 10.3389/fpls.2014.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z.M., Zhang X., He B., Diao L.P., Sheng S.L., Wang J.L., Guo X.P., Su N., Wang L.F., Jiang L., et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007;145:29–40. doi: 10.1104/pp.107.100321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X.J., Zhang H.Q., Wang Y., Shu Z.F., Wang G.H., Wang G.L. Research advances on rice leaf-color mutant genes. Hybrid Rice. 2012;27:9–14. (In Chinese) [Google Scholar]
- 20.Brusslan J.A., Tobin E.M. Isolation and initial characterization of virescent mutants of Arabidopsis thaliana. Photosynth. Res. 1995;44:75–79. doi: 10.1007/BF00018298. [DOI] [PubMed] [Google Scholar]
- 21.Maclachlan S., Zalik S. Plastid structure, chlorophyll concentration, and free amino acid composition of a chlorophyll mutant of barley. Can. J. Bot. 1963;41:1053–1062. doi: 10.1139/b63-088. [DOI] [Google Scholar]
- 22.Dale J.E., Heyes J.K. A virescent mutant of Phaseolus vulgaris; growth, pigment and plastid characters. New Phytol. 1970;69:733–742. doi: 10.1111/j.1469-8137.1970.tb02458.x. [DOI] [Google Scholar]
- 23.Benedict C.R., McCree K.J., Kohel R.J. High photosynthetic rate of a chlorophyll mutant of cotton. Plant Physiol. 1972;49:968–971. doi: 10.1104/pp.49.6.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benedict C.R., Ketring D.L. Nuclear gene affecting greening in virescent peanut leaves. Plant Physiol. 1972;49:972–976. doi: 10.1104/pp.49.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langdale J.A., Metzler M.C., Nelson T. The argentia mutation delays normal development of photosynthetic cell-types in Zea mays. Dev. Biol. 1987;122:243–255. doi: 10.1016/0012-1606(87)90349-6. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto H., Kusumi K., Noguchi K., Yano M., Yoshimura A., Iba K. The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 2007;52:512–527. doi: 10.1111/j.1365-313X.2007.03251.x. [DOI] [PubMed] [Google Scholar]
- 27.Archer E.K., Bonnett H.T. Characterization of a virescent chloroplast mutant of tobacco. J. Plant Physiol. 1987;83:920–925. doi: 10.1104/pp.83.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pierce L.K., Wehner T.C. Review of genes and linkage groups in cucumber. Hortic. Sci. 1990;25:605–615. [Google Scholar]
- 29.Guo Y.M., Gu X.F., Zhang C.Z., Fang X.J., Zhang S.P., Xu C.Q. Genetic mechanism of the cucumber leaf mutant. Acta Hortic. Sin. 2003;30:409–412. (In Chinese) [Google Scholar]
- 30.Gao M.L., Hu L.L., Li Y.H., Weng Y.Q. The chlorophyll-deficient golden leaf mutation in cucumber is due to a single nucleotide substitution in CsChlI for magnesium chelatase I subunit. Theor. Appl. Genet. 2016 doi: 10.1007/s00122-016-2752-9. [DOI] [PubMed] [Google Scholar]
- 31.Miao H., Zhang S.P., Wang X.W., Zhang Z.H., Li M., Mu S., Cheng Z., Zhang R., Huang S., Xie B., et al. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica. 2011;182:167–176. doi: 10.1007/s10681-011-0410-5. [DOI] [Google Scholar]
- 32.Song M., Yang Z., Fan S., Zhu H., Pang C., Tian M., Yu S. Cytological and genetic analysis of a virescent mutant in upland cotton (Gossypium hirsutum L.) Euphytica. 2012;187:235–245. doi: 10.1007/s10681-012-0666-4. [DOI] [Google Scholar]
- 33.Yoo S.C., Cho S.H., Sugimoto H., Li J., Kusumi K., Koh H.J., Iba K., Paek N.C. Rice Virescent-3 and Stripe-1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol. 2009;150:388–401. doi: 10.1104/pp.109.136648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusumi K., Mizutani A., Nishimura M., Iba K. A virescent gene V1 determines the expression timing of plastid genes for transcription/translation apparatus during early leaf development in rice. Plant J. 1997;12:1241–1250. doi: 10.1046/j.1365-313x.1997.12061241.x. [DOI] [Google Scholar]
- 35.Kusumi K., Sakata C., Nakamura T., Kawasaki S., Yoshimura A., Iba K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 2011;68:1039–1050. doi: 10.1111/j.1365-313X.2011.04755.x. [DOI] [PubMed] [Google Scholar]
- 36.Dong H., Fei G.L., Wu C.Y., Wu F.Q., Sun Y.Y., Chen M.J., Ren Y.L., Zhou K.N., Cheng Z.J., Wang J.L., et al. A rice virescent-yellow leaf mutant reveals new insights into the role and assembly of plastid caseinolytic protease in higher plants. Plant Physiol. 2013;162:1867–1880. doi: 10.1104/pp.113.217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou K.N., Ren Y.L., Lv J., Wang Y.H., Liu F., Zhou F., Zhao S., Chen S., Peng C., Zhang X., et al. Young Leaf Chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta. 2013;237:279–292. doi: 10.1007/s00425-012-1756-1. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q.Y., Xue D.X., Li X.Y., Long Y.M., Zeng X.J., Liu Y.G. Characterization and molecular mapping of a new virescent mutant in rice. J. Genet. Genom. 2014;41:353–356. doi: 10.1016/j.jgg.2014.01.010. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 39.Xing A.Q., Williams M.E., Bourett T.M., Hu W.N., Hou Z.L., Meeley R.B., Jaqueth J., Dam T., Li B.L. A pair of homoeolog ClpP5 genes underlies a virescent yellow-like mutant and its modifier in maize. Plant J. 2014;79:192–205. doi: 10.1111/tpj.12568. [DOI] [PubMed] [Google Scholar]
- 40.Liu X.G., Xu H., Zhang J.Y., Liang G.W., Liu Y.T., Guo A.G. Effect of low temperature on chlorophyll biosynthesis in albinism line of wheat (Triticum aestivum) FA85. Physiol. Plant. 2012;145:384–394. doi: 10.1111/j.1399-3054.2012.01604.x. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-Fernández F., Padmarasu S., Surbanovski N., Evans K.M., Tobutt K.R., Sargent D.J. Characterization of the virescent locus controlling a recessive phenotype in apple rootstocks (Malus pumila Mill.) Mol. Breed. 2014;33:373–383. doi: 10.1007/s11032-013-9956-3. [DOI] [Google Scholar]
- 42.Biswal U.C., Biswal B., Raval M.K. Chloroplast Biogenesis: From Proplastid to Gerontoplast. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2003. pp. 19–77. [Google Scholar]
- 43.Dare A.P., Schaffer R.J., Wang L.K., Allan A.C., Hellens R.P. Identification of a cis-regulatory element by transient analysis of co-ordinately regulated genes. Plant Methods. 2008;4:331–339. doi: 10.1186/1746-4811-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koussevitzky S., Stanne T.M., Peto C.A., Giap T., Sjogren L.L., Zhao Y.D., Clarke A.K., Chory J. An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 2007;63:85–96. doi: 10.1007/s11103-006-9074-2. [DOI] [PubMed] [Google Scholar]
- 45.Chin K., Moeder W., Yoshioka K. Biological roles of cyclic-nucleotide-gated ion channels in plants: What we know and don’t know about this 20 member ion channel family. Botany. 2009;87:668–677. doi: 10.1139/B08-147. [DOI] [Google Scholar]
- 46.Schuurink R.C., Shartzer S.F., Fath A., Jones R.L. Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. USA. 1998;95:1944–1949. doi: 10.1073/pnas.95.4.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mäser P., Thomine S., Schroeder J.I., Ward J.M., Hirschi K., Sze H., Talke I.N., Amtmann A., Maathuis F.J., Sanders D. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nawaz Z., Kakar K.U., Saand M.A., Shu Q.Y. Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genom. 2014;15:1602. doi: 10.1186/1471-2164-15-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talke I.N., Blaudez D., Maathuis F.J., Sanders D. CNGCs: Prime targets of plant cyclic nucleotide signaling. Trends Plant Sci. 2003;8:286–293. doi: 10.1016/S1360-1385(03)00099-2. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Atienza J., van Ingelgem C., Roef L., Maathuis F.J. Plant cyclic nucleotide signaling. Plant Signal. 2007;2:540–543. doi: 10.4161/psb.2.6.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowler C., Neuhaus G., Yamagata H., Chua N.H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 52.Huang S.W., Li R.Q., Zhang Z.H., Li L., Gu X.F., Fan W., Lucas W.J., Wang X., Xie B., Ni P., et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 53.Lichtenthaler H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- 54.Blum H., Beier H., Gross H.J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. doi: 10.1002/elps.1150080203. [DOI] [Google Scholar]
- 55.Li Y.H., Yang L.M., Pathak M., Li D.W., He X.M., Weng Y.Q. Fine genetic mapping of cp: A recessive gene for compact (dwarf) plant architecture in cucumber, Cucumis sativus L. Theor. Appl. Genet. 2011;123:973–983. doi: 10.1007/s00122-011-1640-6. [DOI] [PubMed] [Google Scholar]
- 56.Yang L.M., Koo D.H., Li Y.H., Zhang X.J., Luan F.S., Havey M.J., Jiang J.M., Weng Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012;71:895–906. doi: 10.1111/j.1365-313X.2012.05017.x. [DOI] [PubMed] [Google Scholar]
- 57.Ren Y., Zhang Z.H., Liu J.H., Staub J.E., Han Y.H., Cheng Z.C., Li X.F., Lu J.Y., Miao H., Knag H., et al. Integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE. 2009;4:1602. doi: 10.1371/journal.pone.0005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang L.M., Li D.W., Li Y.H., Gu X.F., Huang S.W., Garcia-Mas J., Weng Y. A 1681-locus consensus genetic map of cultivated cucumber including 67 NB-LRR resistance gene homolog and ten gene loci. BMC Plant Biol. 2013;13:1602. doi: 10.1186/1471-2229-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cavagnaro P.F., Senalik D.A., Yang L.M., Simon P.W., Harkins T.T., Kodira C.D., Huang S.W., Weng Y. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.) BMC Genom. 2010;11:1602. doi: 10.1186/1471-2164-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y.C., Bertil S., Maskell D.L. Cushaw: A cuda compatible short read aligner to large genomes based on the burrows-wheeler transform. Bioinformatics. 2012;28:1830–1837. doi: 10.1093/bioinformatics/bts276. [DOI] [PubMed] [Google Scholar]
- 61.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Neff M.M., Neff J.D., Chory J., Pepper A.E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313X.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 63.Untergrasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosambi D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 65.Salamov A.A., Solovyev V.V. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.