Abstract
The tertiary structure of the GTPase Center (GAC) of 23S rRNA as seen in cocrystals is extremely compact. It is stabilized by long-range hydrogen bonds and nucleobase stacking, and a triloop that forms within its 3-way junction. Its folding pathway from secondary structure to tertiary structure has not been previously observed, but it was shown in equilibrium experiments to require Mg2+ ions. The fluorescent nucleotide 2-aminopurine was substituted at selected sites within the 60 nucleotide GAC. Fluorescence intensity changes upon addition of MgCl2 were monitored over a time-course from 1 ms to 100 s as the RNA folds. The folding pathway is revealed here to be hierarchical through several intermediates. Observation of the nucleobases during folding provides a new perspective on the process and the pathway, revealing the dynamics of nucleobase conformational exchange during the folding transitions.
Keywords: GTPase center RNA, 2-aminopurine fluorescence, RNA folding kinetics, stopped-flow fluorescence
Graphical Abstract
RNA tertiary structures can often be visualized as interactions between discrete structural elements, but de novo prediction of those interactions is often impossible. Uncertainty and ambiguity arises from the number of potential interactions between known elements (such as a tetraloop/tetraloop receptor) and between elements that have unrecognized contributions (such as loops). Tertiary interactions, such as hydrogen bonding, stacking, ion chelation, can be identified in RNA crystal structures and then modeled into novel RNA sequences. More examples of interactions are necessary to develop a tool box for modeling, but equally necessary is knowledge of how different interactions are temporally ordered during formation of a functional RNA tertiary structure.
The 60 nucleotide GTPase center (GAC) of 23S/28S rRNA is seen in cocrystals and X-ray and cryoEM structures of the ribosome to adopt an intricate tertiary fold. GAC secondary structure is highly conserved among kingdoms[1]. Its tertiary structure is known from two GAC cocrystals with L11 protein[2,3] and several ribosome structures[4],5[6]. Cocrystals show multiple base triples that anchor distal secondary structures, U-turns, a triloop, long range nucleobase stacking, long-range hydrogen bonding, and chelated ions. The GAC facilitates ribosome function through its interactions with EF-G and the L11 protein, which occur when the GAC is folded. The folding process, from secondary structure to tertiary structure, is unknown.
RNA tertiary folding requires charge neutralization of the phosphodiester backbone to allow close approach of phosphates in a packed RNA. Many RNAs use divalent ions to efficiently shield proximal phosphates, and in vivo, Mg2+ ions are the most abundant. Mg2+ ions appear in virtually all x-ray crystal structures of RNAs, though while in solution most Mg2+ ions appear diffusely associated with RNA molecules. In physiological solutions with monovalent ions, Mg2+ will more efficiently neutralize phosphate charges, and so be preferentially associated with an RNA strand. It is difficult to assign specific contributions of Mg2+ ions to the process of RNA tertiary folding.
Structure/function studies of E. coli GAC identified a single nucleotide substitution that conferred a requirement for Mg2+ ions to adopt its tertiary fold[7]. Twelve of the 58 GAC nucleobases are completely conserved throughout all kingdoms (Figure 1), but those sites that vary (and co-vary) allow substitutions to be introduced and tested for structure/function. In particular, position 1061 is 58% uridine, 36% guanine, and 6% adenine[8]. Curiously, the distribution largely follows phylogenetic classifications: uridine is found in eubacteria, guanine in eukaryotes, and some archaebacteria have adenine. Substituting the natural E. coli U1061 with A1061 (Figure 1) produced a GAC that was active in vivo[9], but which in vitro would not adopt a tertiary structure in the absence of Mg2+ ions. Thermodynamic characterization of this E. coli U1061A variant[2,10,11] led to the conclusion that a single Mg2+ ion could make a large contribution to its folding free energy[12]. Combining thermodynamic data with cocrystal structures of the GAC with L11, Draper and colleagues identified a specific chelated Mg2+ ion as responsible for stabilization of the tertiary structure[12]. In the cocrystal structure (pdb 1hc8[2]), the phosphate of nucleotide A1073 appears to chelate both a K+ ion and this specific Mg2+ ion, holding them both in an electronegative pocket. This pocket is formed by tertiary interactions that stabilize the juxtaposition of the two hairpins. Does the Mg2+ ion trap a conformation of the GAC, stabilizing it to anchor the tertiary fold (conformational selection)? Does the Mg2+ ion alter the structure of the GAC to create its own binding pocket (induced fit[13])? Equilibrium methods and crystal structures cannot distinguish between these alternatives, but a kinetic study of GAC folding might provide some insight.
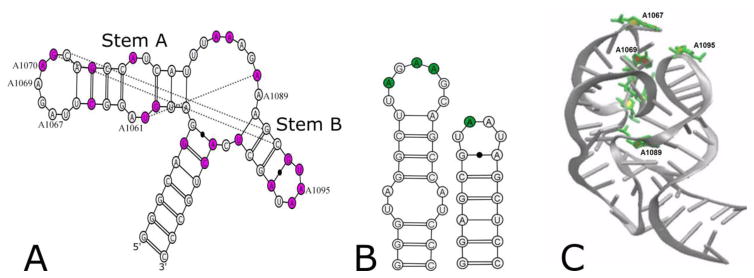
Figure 1.
A) Secondary structure of the E. coli GAC U1061A, with phylogenetically conserved bases in magenta and tertiary contacts observed in the cocrystals indicated by lines. Numbered nucleotides were substituted with 2-aminopurine. B) Sequences of hairpin constructs with sites of 2AP substitution in green. C) Cocrystal structure (1HC8) of E. coli GAC with 2AP substitution sites in green.
RNA tertiary folding has been characterized for several RNA molecules. RNA folding can be initiated by introduction of specific ions (typically Mg2+), then the temporal progress of chemical probing of the phosphodiester backbone is monitored in quenched-flow experiments. Timescales and intermediates during tertiary structure formation have been identified for the 414 nucleotide Tetrahymena Group I intron[14–19],[20], and the 215 nucleotide Azoarcus Group I intron[21–24]. Folding of the Tetrahymena Group I intron has been mapped by time-resolved hydroxyl radical footprinting, revealing a hierarchical folding pathway[25,26]. The timescales of steps in its folding range from milliseconds to tens of seconds; several intermediates have been identified; and misfolding has been consistently observed[17,27–29]. Folding pathways of the Azoarcus Group I intron have also been probed by time-resolved hydroxyl radical footprinting and 2-aminopurine fluorescence[21]. These have been powerful methods for mapping the folding process of Group I introns and their sub-structures.
For the GAC, we wanted to observe the nucleobases directly, to monitor how and when they formed their interactions that created and stabilized the tertiary fold. Here, we took advantage of known properties of the GAC. The E. coli U1061A:B. stearothermophilus L11 cocrystal (pdb 1hc8) allowed us to identify those nucleobases that stacked with other nucleobases but did not make hydrogen bonds. With this information, we substituted six nucleotides with the fluorescent nucleobase 2-aminopurine (2AP), which we predicted should not alter the tertiary structure of the GAC. Mg2+-dependent tertiary folding of these six RNAs was studied by stopped-flow fluorescence, allowing observation of fluorescence intensity on timescales from ms-sec. We find multiple states along a folding pathway that includes several Mg2+-dependent intermediates. Most striking, we observe dynamic conformational exchanges of nucleobases from stacked →unstacked →stacked as the GAC folds. From our time-resolved and steady-state fluorescence data, we constructed a model of how the GAC folds that incorporates its Mg2+-dependence and its conformational changes.
Results
The sequence of the E. coli GAC with the U1061A substitution is given in Figure 1. Six GAC RNAs constructs were chemically synthesized[30] by Agilent Labs, each with a single 2AP substitution. Three substitutions were placed in the 5′ hairpin loop (A1067AP, A1069AP, and A1070AP) which, as shown in the cocrystal[2], arranges the first five bases (U1065 to A1069) in a U-turn conformation and extrudes the 3′ three bases (A1070/G1071/C1072) to create what we refer to here as a T+loop[31],[32]. The A1089AP substitution was designed to monitor the 3-way junction. The A1061AP substitution probes the internal bulge in the 5′ hairpin stem. The A1095AP substitution monitors the 3′ hairpin loop (U1092/A1079), which is a canonical U-turn[33].
2-aminopurine substitution does not alter GAC tertiary structure formation
The six 2AP-GAC constructs were compared to in vitro transcribed GAC to measure thermal stability of their secondary structure, tertiary structure, and response to Mg2+. Thermal denaturation, monitoring absorbance at 260 nm and 280 nm, was previously shown to report the stability of GAC secondary and tertiary structure[7,12]. Transition temperatures of the AP-GAC were nearly identical to the un-modified GAC (Figure 2). Based on those criteria, we conclude that each 2AP-GAC construct adopted the predicted secondary structure and required Mg2+ to form a tertiary structure.
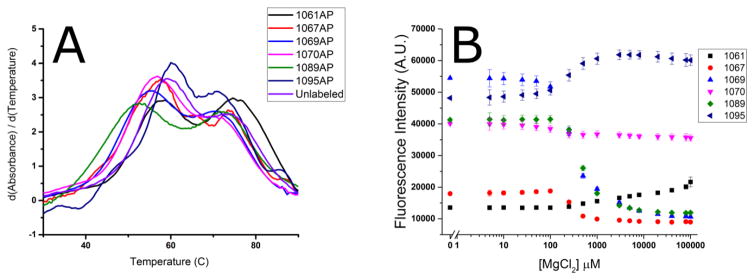
Figure 2.
Mg2+ dependence of 2AP-GAC stability. A. First derivative of absorbance (A260) vs temperature (dA/dT) which reveals the conformational transitions of the RNAs. Melting was measured in 100 mM KCl, 3 mM MgCl2, 10 mM sodium cacodylate, pH 6.5. The lower temperature transition is assigned to tertiary structure melting; the second transition is secondary structure melting. B. MgCl2 titration of 2AP fluorescence at 20°C. [GAC] = 2 μM in 100 mM KCl, 10 mM sodium cacodylate, pH 6.5.
As another measure of structure formation, we used time resolved fluorescence anisotropy[34] to observe the global rotational correlation time of 2AP-GAC molecules. Depolarization of 2AP in the RNAs is dependent on the global rotation of the molecule and on local motions. We predicted that these values in the secondary and tertiary structure would be different.
Measured fluorescence anisotropy decay curves for each 2AP site were fit by one or two exponential components. We globally fit the longest rotational correlation time of the data from every 2AP construct by φ1 = 6.5 ± 0.08 ns in the absence of Mg2+ and by φ1 = 7.5 ± 0.05 ns in the presence of 8 mM Mg2+. The goodness of fit of all sites to these values suggests that each site is reporting on a common global structure. While we anticipated that a compact tertiary fold should result in such agreement, it is rather surprising that the secondary structure values are so consistent. We can account for the result if the GAC arms have relative motion in the secondary structure, which would result in a shorter depolarization time.
2-aminopurine sites report on the GAC tertiary fold
Each 2AP site exhibits a unique fluorescence response to the addition of Mg2+ that reflects its environment. We interpret fluorescence intensity in terms of base stacking: low intensity is consistent with quenching by nucleobase stacking; high intensity corresponds to a more unstacked 2AP[35]. Time-resolved fluorescence lifetimes report on conformational states accessible to 2AP, and time-resolved anisotropy reports on global and local motions. We observe multi-exponential fluorescence lifetimes for 2AP in all sites in the GAC, and most sites also have an anisotropy (depolarization) component consistent with local motion (Supplemental Table 1). These data show that no 2AP nucleobase is rigidly fixed; 2AP in all six sites undergoes ps-ns local conformational sampling.
The Mg2+ concentration-dependence of each 2AP-GAC was measured using steady-state fluorescence. GAC secondary structure was formed prior to MgCl2 titration (Methods). The relative change in fluorescence intensity, for 1 μM GAC, was site specific, as shown in Figure 2. Each construct showed an approximate midpoint of transition from 0.3 – 0.5 mM MgCl2 and reaches a plateau by 3 mM MgCl2. We conclude that 3 mM Mg2+ is saturating.
Steady state 2AP fluorescence intensity varies by site. In Mg2+ titrations, A1061AP fluorescence intensity is consistently low, indicating that it is mostly stacked with or without Mg2+. A1095AP, at the apex of a U-turn, shows an increase in fluorescence intensity with added Mg2+. A1089AP in the 3-way junction loses about a third of its fluorescence intensity in the presence of Mg2+. T+loop A1067AP fluorescence intensity is uniformly low, suggesting it is mostly stacked. In contrast, A1070AP fluorescence is high regardless of Mg2+ concentration, consistent with unstable stacking. A1069 fluorescence has a substantial intensity difference before and after Mg2+ addition. Together, these data suggest that the T+loop and the junction undergo a conformational change.
The local thermal stability at each 2AP site was compared by fluorescence to the thermal stability of the entire tertiary structure, which was measured by UV absorbance. In the presence of Mg2+ the melting transition for the tertiary and secondary structures occur at 56–60 °C and 75 °C, respectively, whereas in the absence of Mg2+ a single broad transition occurs at 43–60 °C[36]. Fluorescence intensity of each 2AP-GAC was measured during thermal denaturation in the presence and absence of 3 mM MgCl2 (Supplemental Figure 1). Secondary structure denaturation in the absence of Mg2+ was only apparent in the 1061AP trace, presumably reporting on melting of the hairpin stem at the internal bulge. In contrast, tertiary structure melting in the presence of Mg2+ was observed for all sites with the exception of GAC-A1095AP, which is at the apex of the U-turn in stemloop B. At higher temperatures, progressive loss of fluorescence is attributed to collisional quenching[34] with solvent or adjacent nucleotides as the RNA structure is disrupted.
Stopped flow fluorescence data reveal multiple intermediate folded states
Steady state experiments showed that each 2AP site was sensitive to environmental changes upon tertiary structure formation. Now, we ask how fluorescence intensity changes during the folding process. For these experiments we use stopped-flow spectroscopy to rapidly mix the GAC with increasing amounts of MgCl2 to observe the tertiary folding process in real time. All stopped flow time course traces (progress curves) are fit to a sum of exponentials to determine the relaxation times that define the kinetics of GAC folding (EQ1).
GAC absorbance at 260 nm decreases upon tertiary structure formation. In stopped-flow absorbance experiments, we observed a trace that was fit by two exponentials[36]. The dominant transition at 20° C in 20 mM MgCl2 had a time constant of ~ 660 ms, which we assign to tertiary folding.
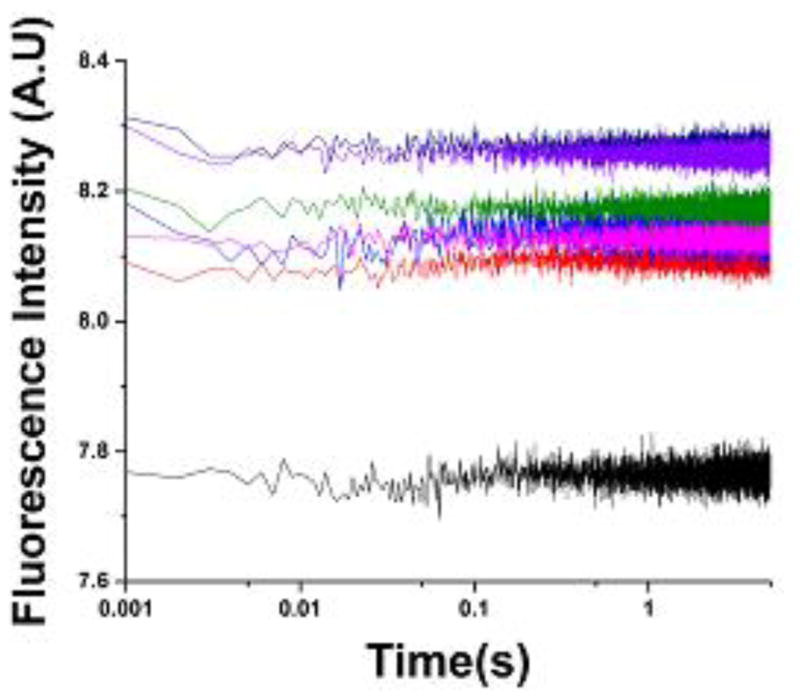
In contrast to the traces from stopped-flow absorbance, stopped-flow fluorescence traces show transitions on four separate timescales. Each 2AP site on the GAC had a unique progress curve, with transitions ranging from less than a millisecond to the order of 10’s of seconds. Each trace was fit by three exponentials (EQ1), giving three time constants (τ1, τ2, τ3) that varied with the amount of Mg2+ added until the system was saturated (Figure 4). Global analysis of five constructs at a given MgCl2 concentration simultaneously fit the progress curves to EQ 1, using the same values of the relaxation time parameters (τ1, τ2, τ3) for all curves (Figure 4).
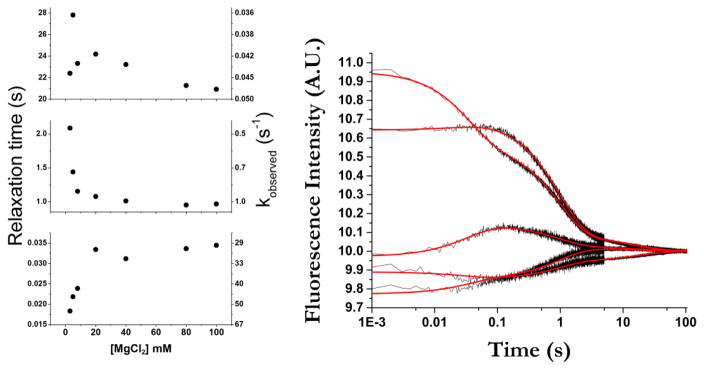
Figure 4.
Relaxation times and corresponding observed rates (1/τ) from global fits. Errors are contained within the data points. The scatter in the first points in the top plot is due to sparse sampling. An example of global fitting of stopped-flow traces for all 2AP-GAC molecules is shown for addition of 20 mM MgCl2. Black traces are data; red lines are fit. All progress curves and fits are shown in Supplemental Figures 2 and 3.
| (EQ1) |
where y is the fluorescence signal, yo is an offset, An are the amplitude of the fluorescence signal change transition, and τn is the relaxation time of the fluorescence change.
The progress curves of five 2AP-GAC molecules were fit by identical relaxation times, suggesting that each RNA followed the same folding pathway (Table 1) (A1089AP is an outlier). To generalize the results, we interpret our data using the framework of classical chemical kinetics to infer the lower limit of states in the folding pathway as one plus the number of observed transitions[37]. By this criterion, we have at least four transition states in the pathway.
Table 1.
Relaxation times for 2AP-GAC folding transitions.
| τ1 (sec) | τ2 (sec) | τ3 (sec) | ||||
|---|---|---|---|---|---|---|
| Construct | Global | 1089AP | Global | 1089AP | Global | 1089AP |
| 3 mM Mg2+ | 0.018 | -- | 2.1 | 3.8 | 22 | 26 |
| 5 mM Mg2+ | 0.022 | -- | 1.4 | 2.7 | 28 | 28 |
| 8 mM Mg2+ | 0.024 | 0.007 | 1.2 | 2.0 | 23 | 25 |
| 20 mM Mg2+ | 0.033 | 0.011 | 1.1 | 1.7 | 24 | 22 |
| 40 mM Mg2+ | 0.031 | 0.014 | 1.0 | 1.4 | 23 | 20 |
| 80 mM Mg2+ | 0.034 | 0.024 | 0.95 | 1.4 | 21 | 21 |
| 100 mM Mg2+ | 0.034 | 0.027 | 0.97 | 1.3 | 21 | 21 |
Relaxation times calculated from global fitting and independent A1089AP curve fitting. The error generated by Origin software from the curve fit is uniformly <5% of the parameter value. 20° C in 100 mM KCl, 10 mM sodium cacodylate, pH 6.5.
The progress curves are fit by three quantifiable transitions that occur from 10’s of ms (τ1) to seconds (τ2, τ3). The shortest relaxation time increases at higher Mg2+ concentrations (in other words, the associated conformational transition is apparently slowed by addition of Mg2+). Because this transition occurs after the initial RNA/Mg2+ interaction (< 1.5 ms; see below), it suggests that the GAC must undergo a conformational change[38] to a new Mg2+ binding competent state. These data support the hypothesis of Draper[12] that the GAC has two distinct Mg2+ interactions. In contrast, the subsequent relaxation times, on the order of single and tens of seconds, become shorter with increasing Mg2+ concentration until they reach a plateau (Figure 4), perhaps reflecting a general shielding of phosphate charges.
A1089AP-GAC was the outlier in the global fits, so its kinetic data were fit independently to EQ 1. Interestingly, the only prominent difference among the RNAs was the value of τ2 of A1089AP-GAC (Table 1). On average, it is 55% longer than the analogous relaxation time of the other molecules. We speculate that it represents an additional intermediate state of the hinge region, possibly formation of the triloop.
Taken together, we propose that the folding pathway of the GAC RNA consists of at least six resolvable species.
Isolating the source of rapid signal change
In addition to the measured transitions states, a first transition occurs during the dead time of our instrument (~1.5 ms), and is not represented in EQ1. The amplitude of this early transition is Mg2+-dependent, and varies with Mg2+ concentration; fluorescence intensity can either increase or decrease with respect to the starting value (Figure 3). We conclude that the GAC undergoes a Mg2+-dependent conformational transition during the rapid mixing process and before fluorescence is measured. This transition is visible in time courses from all molecules, and achieves an equilibrium before recording of the fluorescence signal.
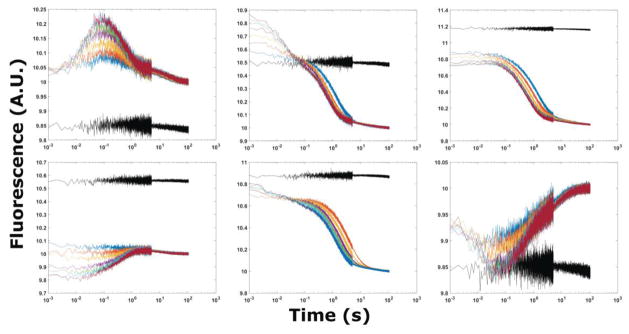
Figure 3.
Stopped flow fluorescence traces of 2AP-GAC at 20° C. Top row from left to right A1061AP, A1067AP, A1069AP. Bottom row from left to right A1070AP, A1089AP, A1095AP. MgCl2 additions from 3 (blue), 5, 8, 20, 40, 80, to 100 (red) mM in 100 mM KCl, 10 mM sodium cacodylate. The black trace is buffer-only mixing control.
The source and nature of rapid conformational changes (< 1 ms) from the association of divalent cations with RNA have been previously observed and attributed to electrostatic relaxation or counterion collapse[20,39–42]. Often this phenomenon is associated with a rapid decrease in the radius of gyration of an RNA molecule, indicating a physical collapse. Indeed, previous SAXS experiments with the GAC have shown that it does undergo a physical compaction, from RG = 25 Å to RG = 15.9 Å in the presence of Mg2+ [10]. The rapid signal change that occurs during our instrument’s dead time could report on a global collapse of the GAC or, alternatively, could be caused by local conformational changes at each probed site in the GAC.
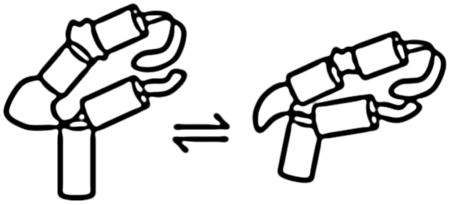
To differentiate the global versus local origins of this fast relaxation process we compared the GAC properties to those of its two hairpins alone. Each hairpin was synthesized with a single 2AP to correspond to a position in the full GAC (Figure 1B). The fluorescence intensity of A1067AP and A1070AP in the hairpin alone are consistent with unstacked nucleobases (Supplemental Figure 4), whereas in the GAC these sites are stacked. With this conformational difference in mind, the hairpin RNAs were also examined with stopped flow fluorescence.
All four hairpin constructs exhibit a rapid change of fluorescence intensity within the dead time of the instrument, but undergo no further transitions. Stopped-flow fluorescence of the 3′ hairpin with A1095AP illustrates these data (Figure 5). Conformational changes within RNA loops have been measured by several methods, and have been shown to occur on the order of microseconds[43,44]. The relative increase or decrease of the hairpin fluorescence intensity corresponds to the analogous GAC construct, and we assign those signals to a local loop conformational change. However, the relative amplitudes of the signal change in hairpins and GAC are different, suggesting additional contributions from other events or interactions in the GAC.
Figure 5.
Stopped-flow fluorescence traces of the A1095AP U-turn hairpin construct. Black trace is control addition of buffer only. Adding MgCl2 to 3, 5, 8, 20, 80,100 mM results in an immediate increase in fluorescence intensity but no subsequent transitions. Experiments with other hairpins showed similar rapid intensity changes. [RNA] = 100 nM in 100 mM KCl, 10 mM sodium cacodylate pH 6.5. 20° C.
Discussion
Direct observation of nucleobases in the GAC reveals their conformational excursions during the folding process, with conformational transitions on timescales from microseconds to tens of seconds. Some of these transitions are local, such as those in the loops, but others report on global rearrangements of the RNA as it forms tertiary structure. This is the first comprehensive study of the nucleobases themselves during RNA tertiary folding.
Defining the structures of the starting and ending states
In their review of 3-way junctions, Lescoute and Westhof[45] included the GAC. The goal of their analysis was to formulate rules that would allow prediction of RNA tertiary folds. Their conclusion, based on a compendium of 3-way junctions, was that the diversity of interactions essentially precludes prediction. However, joint consideration of RNA sequence and structural homology offered some suggestions of possible topologies. For example, if the secondary structure of an RNA includes a loop sequence that is known to form a GNRA tetraloop, then it can be modeled with that structure. Since GNRA tetraloops dock with a receptor, the RNA can then be examined to identify receptors. More generally, predicting the tertiary fold of an RNA requires knowledge of the starting structure in order to identify discrete elements that could interact.
The secondary structure of the U1061A GAC[1] defines our starting state in the folding pathway. Hydroxyl radical footprinting of the GAC secondary structure showed that the hairpin loops and the 3-way junction are unstructured in the absence of Mg2+ [12]. The four nucleotide U-turn that caps the 3′ hairpin was previously investigated by solution NMR[46],[47]. We have extended those studies to compare the loop properties as a function of temperature and Mg2+ (SF 5), and we conclude that the U-turn structure is present in the unfolded RNA and is stabilized by Mg2+. In contrast, nucleotides in the T+loop appear to be unstructured based on previous NMR data[48] and hydroxyl radical probing. In summary, we define the starting structure of the U1061A GAC as two hairpins, one with an unstructured loop and one with a U-turn conformation, joined to a terminal stem by a disordered 3-way junction.
We posit that the ending state is similar to the tertiary structure found in the cocrystals. Structure probing of the GAC under folded conditions (20 mM K-MOPS, 5 mM MgCl2) in the presence and absence of the L11 protein gave nearly identical patterns, supporting this assumption[12]. Our NMR experiments with selectively 15N-labeled GAC are in agreement, as we observed formation of four long-range hydrogen bonds from specific G and U imino protons (15N-1H) only when Mg2+ is present[36], as predicted from the cocrystal structures.
Temporal events in the folding pathway
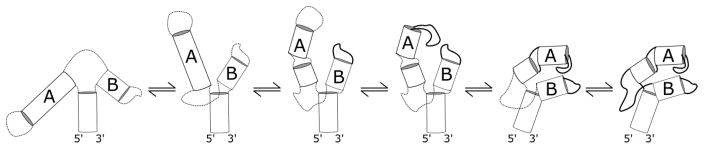
Between these starting and ending states, we propose that the folding pathway contains four resolvable intermediate conformational states of the GAC. Folding is hierarchical, but the populations of each state are not uniquely present as illustrated. There is a dynamic equilibrium of conformational states, particularly in the final tertiary structure (3°) which contains I3 and I4.
2° is the starting conformational state. I1 corresponds to the initial loop relaxation and I2 corresponds to the putative partial global collapse. I3 represents the first state after a second Mg2+ interaction, I4 represents the formation of the tertiary contacts, and 3° represents the last populated state which exists in a dynamic equilibrium.
The first transitions contain information on both local conformations and global folding events which is represented by I1 and I2 in our pathway scheme. The intermediate I1 can be explained as conformational change due to the relaxation of the single-stranded regions. We propose that the second intermediate I2 is the result of a partial electrostatic collapse of the GAC. Previous Mg2+ induced RNA folding experiments by Das et al[20] have shown that partial RNA compaction is identifiable by time-resolved SAXS within milliseconds of interacting with Mg2+.
The first observable transition (τ1 = ~30ms) occurs in every labeled element of the GAC. The relaxation time increases as Mg2+ concentration is increased until approximately 20 mM MgCl2, where it reaches a plateau. This phenomenon, an increase in relaxation time as a function of increasing ligand concentration, is only observed if there is a rate limiting kinetic step preceding a binding event[38]. By coupling this observation with the identification of I1 and I2, we conclude that I2 accepts a Mg2+ ion. Therefore, we assign the third intermediate, I3, to the conformation of the GAC when Mg2+ is chelated.
The presence of two longer relaxation times (τ2, τ3) is evidence that there are three states[37] that are populated after the Mg2+ chelation event. We choose to represent the final three states in our proposed pathway as sequential isomerizations. Each relaxation time does not solely represent single isomerization steps but rather represents multiple kinetic steps. As such, the three latter states are probably all present after chelation and exist in a dynamic equilibrium.
The physical process of GAC folding
The progress curves provide some clues about the structural rearrangements of the GAC as it folds. We separate the traces into three time regimes: 1 to 100 ms, 100 ms to 1 s, and 1 to 100 s. In our folding scheme, I1 and I2 are formed prior to the first time interval, so the progress curves are reporting on formation of I3, I4, and the final tertiary fold.
All sites experience conformational changes within the first 100 ms of folding. A1061AP in the internal bulge of the hairpin moves out of a stacked conformation. Its fluorescence intensity reaches a maximum at 100 ms at 20° C, with an amplitude that is Mg2+-dependent. If we assume that A1061AP is partially stacked in the internal bulge in its initial conformation, then addition of Mg2+ results in a realignment of this bulge/stem, in what could be a direct ion association or a result of a long-range interaction that distorts the bulge. The structure of this internal bulge with or without Mg2+ is not known, but NMR structures of an A:G mismatch juxtaposed to a U:U pair show distorted geometries of both pairs[49].
In the same time interval, nucleobases in the T+loop exhibit a variety of conformational changes. A1070AP begins to recover fluorescence intensity after its intensity loss during the initial rapid phase, but doesn’t reach a maximum value until about 3 s. A1067AP continues to lose fluorescence intensity after its initial rapid Mg2+-dependent loss. A1069AP fluorescence initially drops, but then is constant until ~90 ms when it begins to decrease. The Mg2+-dependence of each T+loop nucleobase is consistent with a Mg2+-dependent conformational change. We cannot determine from our data if the motions of the nucleobases are correlated or independent, or if the T−loop conformational change is a cooperative transition.
In the cocrystal structures[2,3], A1061 is stacked with A1070, forming a long-range stacking interaction. If it forms in solution, without L11 protein, it is not stable, since the fluorescence intensity of A1070AP-GAC is too high to be consistent with a predominantly stacked state. Also, time-resolved anisotropy shows that A1061AP-GAC undergoes local motion on ns timescale (Supplemental Table 1). However, both sites are sensitive to Mg2+ ions, and undergo a rearrangement of their environments during folding.
A1095AP fluorescence intensity decreases from 1 –100 ms. The progress curves indicate that it undergoes a transient conformational change during GAC folding. This U-turn forms without Mg2+, as do several others [50],[51], so we could be seeing a contribution from the entire GAC to this local environment.
During the interval from 100 ms – 1 sec, all 2AP sites exhibit large changes in fluorescence intensity. We assign these transitions to the global fold of the GAC as it approaches its tertiary structure, consistent with stopped-flow absorbance data. Several sites, A1061AP, A1070AP, and A1095AP, have large Mg2+-dependent intensity changes, suggesting that ions are directly involved in their conformational transitions. Fluorescence intensities of other 2AP sites are much less dependent on the Mg2+ concentration. In our folding scheme, we consider I4 to be the first intermediate to contain most tertiary contacts. Based on the progress curves, we conclude that I4 and 3° are most likely to comprise the conformational ensemble after 1 second.
The time courses from the A1089AP GAC are of particular interest since its major large transition is delayed relative to the other sites. It was designed to report on the conformational transition of the 3-way junction from unstructured to a tri-loop, but several explanations could account for this delayed response. It is possible that the tri-loop can only form after the other tertiary contacts have anchored the hairpins. Alternatively, A1089AP could be more sensitive to formation of the conserved U1060-A1088 long-range hydrogen bond, which is formed late in the folding pathway. It is always possible that the A1089AP substitution could interfere with normal tertiary structure formation, perhaps transient misfolding/destabilization that leads to an increased relaxation time. Assuming that it does report on conformational change of the junction, then we would conclude that tri-loop formation is not stable in the absence of other tertiary contacts, and so it forms last.
We hypothesize that final 1–100 s interval represents conformational sampling by the GAC. The amplitude of this transition is small for all sites, suggesting that this state(s) is less populated and/or that its fluorescence signal is only subtlety different from the preceding conformation. Either alone or together these conclusions suggest that the GAC is sampling iso-energetic conformations, and exists not as a single conformation but in a dynamic equilibrium[52,53].
The T+loop
We consider this nine nucleotide loop to be at the heart of GAC folding. It must adopt a contorted structure to display four of its nucleotides for intramolecular interactions. It displays its phosphodiester backbone to create a pocket for ion chelation. Its extruded nucleobases make long-range base stacking interactions or base triples that anchor the 3-way junction. We refer to it as a T+loop, to distinguish it from typical T−loops[31,32].
tRNA T−loops
The first T−loop was observed in x-ray crystal structures of yeast tRNAPhe [54,55], where the seven nucleotide TψC loop [T54ψ55CGA58U59C60] extruded U59C60 by formation of a five nucleotide loop closed by a T54-A58 reverse Hoogsteen pair. A striking feature of the T54ψ55CGA58 loop structure was a sharp turn formed by ψ55CG, and indeed this portion of the TψC loop was identified as a U-turn structure. [U-turn sequences are typically 5′UNR[33], where N is any nucleobase and R is a purine.] The TψC loop is thus a loop within a loop, which is a structural characteristic of T−loops. Subsequent sequence searches for more T−loops found several[31], but more were identified by structural homology to the TψC loop[56], using existing crystal structures.
T−loops are typically involved in intramolecular or intermolecular RNA-RNA interactions, at times using their extruded bases, but also the nucleotides within their U-turn. Their propensity for making intramolecular interactions makes them a common tertiary structure motif, and they are found in phylogenetically conserved regions of RNAs[56]. In yeast tRNAPhe, nucleotides ψ55C56 make hydrogen bonds to nucleotides in the D-loop, effectively anchoring the characteristic tRNA L-shape. When Chan et al.[56] searched for T−loops in noncoding RNAs, they focused analysis on the U-turn portion of the motif.
The T+ loop
The GAC 1065-1073 loop is notable among T−loops for several reasons, which led to our designation as a T+loop. First, it does not use nt1(U1065) and nt5 (A1069) to form a loop-within-a-loop. Instead, it uses nt1 and nt9 (A1073) to form a Hoogsteen pair that closes the loop. It does form a U-turn-like structure with U1066A1067G1068A1069 (nt2-nt5), but then A1070, G1071, and C1072 are extruded and solvent-exposed. G1071 and C1072 make hydrogen bonded base triples with the stem of hairpin B to anchor the tertiary structure. A1070 is stacked with U1061. This structure is observed in both cocrystals of GAC with L11 proteins, one with E. coli GAC (1hc8)[2] and one with Thermotoga maritima GAC (1mms)[3]. The same GAC structure is found in the crystal structure of E. coli 70S ribosome with its bound L11 protein[6] (2.86 Ǻ resolution).
The T+loop sequence differs in prokaryotes and eukaryotes, but is conserved in yeast and humans: A1979UGGA1983AG1985UC1987 (using human rRNA numbering). Statistics of base conservation among all kingdoms shows significant diversity at very few sites (U:68%/A20%. U93%. A77%/G23%. G96%. A92%. A98%. G100%. C80%/U20%. A80%/C18%)[8]. In crystal structures of S. cerevisiae[58] (pdb 3u5b, 3.0 Ǻ resolution) ribosomes and cryoEM structures of human ribosomes[59] (pdb 3j3f, 5 Ǻ resolution), the T+loop backbone structure is similar to that of prokaryotes (Figure 7), and some base orientations vary.
Figure 7.
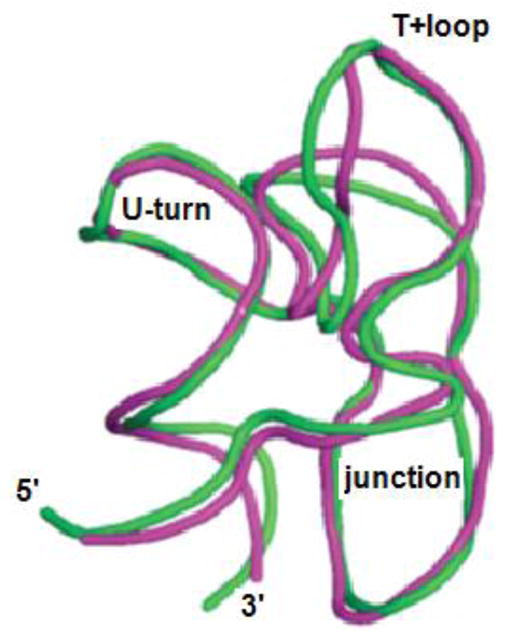
Backbone trace of human[59] (pink) and Thermus[60] (green) GACs from ribosome crystal structures. Most of the variation is in the T+loop and the junction phosphodiester backbone. Thermus ribosome lacked L11, human ribosome includes L12 (equivalent to L11) bound to the GAC.
Significantly, a NMR study of the yeast tRNAPhe TψC loop[61] failed to find any evidence of loop structure in the absence of Mg2+. Those authors described that loop as seriously flexible. If the GAC T+loop is also unstructured in the absence of Mg2+, then when does it adopt its T+ structure? Does addition of Mg2+ cause a conformational change that allows it to dock with stemloop B? Or does it only adopt its T+ structure in the presence of stemloop B and Mg2+ via induced fit? This level of mechanistic detail is necessary to understand how the RNA uses this conserved loop to adopt its tertiary structure.
Properties of a canonical T−loop
The Tetrahymena group 1 intron P5c subdomain includes a 4–5 nucleotide GNRA T−loop[62]. Formation of its canonical U-turn/extruded nucleobase structure is Mg2+-dependent[63], and its conformational change triggers subsequent tertiary interactions of the P5c domain[19]. In kinetics experiments, its folding rate decreases with increasing Mg2+ concentration[64], which we suggest identifies the mechanism of its interaction with its ligand as conformational selection[38]. An NMR investigation of P5c shows that its hairpin loop undergoes a conformational change with a rate of ~323 s−1 (3 ms) in 10 mM sodium phosphate pH 6.4, which becomes faster in 1 mM MgCl2 (964 s−1; 1 ms)[65]. As an isolated hairpin, the native structure is seen as a low abundance state, indicating that it requires tertiary contacts to stabilize its structure.
While T−loops are clearly important tertiary motifs, predicting their structures and identifying their interacting partners will be a challenge. Perhaps their sequence and conformational space are too large to allow such prediction. If loops are disordered in their starting state, then what are the rules that govern their final topology? More examples need to be studied to understand how their nucleotides sample their environments and find their targets.
Methods
RNA samples
Unlabeled GAC RNA was transcribed in vitro with T7 RNA polymerase via run off transcription from plasmid DNA. The 2AP 60mer constructs were chemically synthesized[30] by Agilent laboratories. The hairpin 2AP constructs were purchased from Integrated DNA technologies. All RNAs were dialyzed against 0.5M ethylenediaminetetraacetic acid and then dialyzed into 100 mM KCl, 10 mM sodium cacodylate pH 6.5. We used the Draper laboratory protocol[12] to fold the full GAC RNAs into their proper secondary by heating to 65 °C for 30 minutes in 100 mM KCl, 10 mM sodium cacodylate pH 6.5 and then cooling at room temperature for 15 minutes. We folded the hairpin RNAs by heating to 95 °C for 5 minutes in 100 mM KCl, 10 mM sodium cacodylate pH 6.5 and immediately cooled on ice.
Absorbance Thermal denaturation
Thermal denaturation was monitored in absorbance experiments on a Gilford 260 fitted with a Gilford thermoprogrammer 2527. RNA and MgCl2 concentrations were 2 μM and 3 mM respectively. Temperature was ramped at 0.5 °C per minute from 6 to 90 °C while absorbance was recorded at 260 and 280 nm.
Fluorescence melt/titration
Fluorescence thermal denaturation and titrations experiments were performed on an Photon Technology International spectrofluorometer fitted with Peltier-controlled 4-cuvette turret. Samples were excited at 308 nm and fluorescence emission was measured at 368 nm. Data points are the average of 5 consecutive measurements with an integration time of 1 s. For thermal denaturation experiments the RNA concentration was 2 μM, and MgCl2 concentration was 3 mM in 100 mM KCl, 10 mM sodium cacodylate pH 6.5. Temperature was ramped at 1° C per minute.
For fluorescence titrations RNA concentration was 1 μM and temperature was controlled to 20 °C. The error bars were calculated using the standard deviation of the fluorescence measurements and an assumed 5% uncertainty in pipette volume. Titration data curves were not fit to a thermodynamic binding model as it is impossible to distinguish between specific binding and non-specific Mg2+ interaction.
Fluorescence lifetime/anisotropy
Time correlated single photon counting and time resolved anisotropy measurements were performed on a home built instrument[66],[67]. Lifetime and anisotropy data were fit using Fluofit Pro 4.4 (Picoquant) to a sum of exponentials. For lifetime analysis I(t) is the fluorescence intensity as a function of time t, τi is the lifetime of the ith component and Ai is the pre-exponential factor of the ith component. For anisotropy analysis R0 is the initial steady state anisotropy, βi is the amplitude of the ith component and φi is the depolarization decay time. Anisotropy data for a given set of conditions (i.e. presence or absence of Mg2+) were simultaneously fit where φ1 was constant.
| S. EQ1 |
| S. EQ 2 |
Stopped flow
Stopped-flow experiments were performed on a Applied Photophysics SX-20 stopped-flow spectrometer. Time courses were monitored over 105 seconds, with 5000 points taken uniformly in the first 5 seconds, and 10,000 uniformly spaced points were taken over the next 100 seconds. All measurements were taken at 20 ± 0.1 °C. The time courses are an average of ≥5 independent time courses. Progress curves for A1061AP, A1067AP, A1070AP, and A1095AP, were simultaneously fit to Equation 1 and all relaxation times were globally shared; the A1089AP data were fit separately. There was no significant difference between X2 and residuals generated from the global fits and the individual time courses fit independently. Curve fitting was done using the Origin software package.
Supplementary Material
Figure 6.
Visualization of a possible folding scheme. The first structure is the starting secondary structure. The second is formed in < 1.5 ms and illustrates electrostatic relaxation upon addition of Mg2+ ions. The third structure is the counterion collapsed state. In the fourth structure, the T+loop structure is changed when specific Mg2+ ions associate, leading to a state where the second ion binding occurs when the hairpins interact. Finally, the last state is the tertiary structure.
Highlights.
Mg2+-dependent RNA folding pathways reveal how tertiary structures utilize ions.
GAC tertiary folding pathway has four intermediates; two require Mg2+.
GAC folding kinetics range from milliseconds (local) to seconds (global).
Fluorescent bases reveal transient conformational changes before global folding.
Acknowledgments
We thank Michael Rau for many discussions and NMR experiments. We thank Professor Roberto Galletto for use of his stopped-flow spectrometer. 2AP-GAC RNAs were received from Agilent labs, and we especially thank Dr Laura-Kay Bruhn and Dr Doug Dellinger for making these experiments possible. Research was supported in part from NIH NIGMS to KBH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Robb Welty, Email: robb@wustl.edu.
Kathleen B. Hall, Email: kathleenhal@gmail.com.
References
- 1.Petrov AS, Bernier CR, Gulen B, Waterbury CC, Hershkovits E, Hsiao C, Harvey SC, Hud NV, Fox GE, Wartell RM, Williams LD. Secondary structures of rRNAs from all three domains of life. PLoS One. 2014;9:e88222. doi: 10.1371/journal.pone.0088222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conn GL, Gittis AG, Lattman EE, Misra VK, Draper DE. A compact RNA tertiary structure contains a buried backbone-K+ complex. J Mol Biol. 2002;318:963–73. doi: 10.1016/S0022-2836(02)00147-X. [DOI] [PubMed] [Google Scholar]
- 3.Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 4.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, Korostelev AA. Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure. 2014;22:1210–8. doi: 10.1016/j.str.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu M, Draper DE. Bases defining an ammonium and magnesium ion-dependent tertiary structure within the large subunit ribosomal RNA. J Mol Biol. 1994;244:572–85. doi: 10.1006/jmbi.1994.1753. [DOI] [PubMed] [Google Scholar]
- 8.Bernier CR, Petrov AS, Waterbury CC, Jett J, Li F, Freil LE, Xiong X, Wang L, Migliozzi BLR, Hershkovits E, Xue Y, Hsiao C, Bowman JC, Harvey SC, Grover MA, Wartell ZJ, Williams LD. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 2014;169:195–207. doi: 10.1039/c3fd00126a. [DOI] [PubMed] [Google Scholar]
- 9.Maeder C, Conn GL, Draper DE. Optimization of a ribosomal structural domain by natural selection. Biochemistry. 2006;45:6635–43. doi: 10.1021/bi052544p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grilley D, Misra V, Caliskan G, Draper DE. Importance of partially unfolded conformations for Mg(2+)-induced folding of RNA tertiary structure: structural models and free energies of Mg2+ interactions. Biochemistry. 2007;46:10266–78. doi: 10.1021/bi062284r. [DOI] [PubMed] [Google Scholar]
- 11.Shiman R, Draper DE. Stabilization of RNA tertiary structure by monovalent cations. J Mol Biol. 2000;302:79–91. doi: 10.1006/jmbi.2000.4031. [DOI] [PubMed] [Google Scholar]
- 12.Leipply D, Draper DE. Evidence for a thermodynamically distinct Mg2+ ion associated with formation of an RNA tertiary structure. J Am Chem Soc. 2011;133:13397–405. doi: 10.1021/ja2020923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–7. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 14.Wan Y, Suh H, Russell R, Herschlag D. Multiple unfolding events during native folding of the Tetrahymena group I ribozyme. J Mol Biol. 2010;400:1067–77. doi: 10.1016/j.jmb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–4. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell D, Russell R. Folding pathways of the Tetrahymena ribozyme. J Mol Biol. 2014;426:2300–12. doi: 10.1016/j.jmb.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell D, Jarmoskaite I, Seval N, Seifert S, Russell R. The long-range P3 helix of the Tetrahymena ribozyme is disrupted during folding between the native and misfolded conformations. J Mol Biol. 2013;425:2670–86. doi: 10.1016/j.jmb.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shcherbakova I, Brenowitz M. Perturbation of the hierarchical folding of a large RNA by the destabilization of its Scaffold’s tertiary structure. J Mol Biol. 2005;354:483–96. doi: 10.1016/j.jmb.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 19.Laederach A, Shcherbakova I, Liang MP, Brenowitz M, Altman RB. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J Mol Biol. 2006;358:1179–90. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SGJ, Thiyagarajan P, Doniach S, Pollack L, Herschlag D. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J Mol Biol. 2003;332:311–9. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 21.Chauhan S, Behrouzi R, Rangan P, Woodson SA. Structural rearrangements linked to global folding pathways of the Azoarcus group I ribozyme. J Mol Biol. 2009;386:1167–78. doi: 10.1016/j.jmb.2008.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, Thirumalai D, Woodson SA. Metal ion dependence of cooperative collapse transitions in RNA. J Mol Biol. 2009;393:753–64. doi: 10.1016/j.jmb.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behrouzi R, Roh JH, Kilburn D, Briber RM, Woodson SA. Cooperative tertiary interaction network guides RNA folding. Cell. 2012;149:348–57. doi: 10.1016/j.cell.2012.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleitsman KR, Herschlag DH. A kinetic and thermodynamic framework for the Azoarcus group I ribozyme reaction. RNA. 2014;20:1732–46. doi: 10.1261/rna.044362.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shcherbakova I, Gupta S, Chance MR, Brenowitz M. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J Mol Biol. 2004;342:1431–42. doi: 10.1016/j.jmb.2004.07.092. [DOI] [PubMed] [Google Scholar]
- 26.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–3. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 27.Wan Y, Russell R. Enhanced specificity against misfolding in a thermostable mutant of the Tetrahymena ribozyme. Biochemistry. 2011;50:864–74. doi: 10.1021/bi101467q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinan S, Yuan X, Russell R. The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins. J Biol Chem. 2011;286:37304–12. doi: 10.1074/jbc.M111.287706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–44. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 30.Dellinger DJ, Timár Z, Myerson J, Sierzchala AB, Turner J, Ferreira F, Kupihár Z, Dellinger G, Hill KW, Powell JA, Sampson JR, Caruthers MH. Streamlined process for the chemical synthesis of RNA using 2′-O-thionocarbamate-protected nucleoside phosphoramidites in the solid phase. J Am Chem Soc. 2011;133:11540–56. doi: 10.1021/ja201561z. [DOI] [PubMed] [Google Scholar]
- 31.Nagaswamy U, Fox GE. Frequent occurrence of the T−loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–9. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasilnikov AS, Mondragón A. On the occurrence of the T−loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–3. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gutell RR, Cannone JJ, Konings D, Gautheret D. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J Mol Biol. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- 34.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer; NY: [Google Scholar]
- 35.Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc Natl Acad Sci U S A. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rau MJ, Welty R, Tom Stump W, Hall KB. Formation of Tertiary Interactions during rRNA GTPase Center Folding. J Mol Biol. 2015;427:2799–815. doi: 10.1016/j.jmb.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Starzak M. Mathematical Methods in Chemistry and Physics. 1989. [Google Scholar]
- 38.Vogt AD, Di Cera E. Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry. 2012;51:5894–902. doi: 10.1021/bi3006913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, Chance MR. Principles of RNA compaction: insights from the equilibrium folding pathway of the P4-P6 RNA domain in monovalent cations. J Mol Biol. 2004;343:1195–206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 40.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, Gruner SM, Mochrie SGJ, Pande V, Doniach S, Herschlag D, Pollack L. Rapid compaction during RNA folding. Proc Natl Acad Sci U S A. 2002;99:4266–71. doi: 10.1073/pnas.072589599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pabit SA, Sutton JL, Chen H, Pollack L. Role of ion valence in the submillisecond collapse and folding of a small RNA domain. Biochemistry. 2013;52:1539–46. doi: 10.1021/bi3016636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roh JH, Guo L, Kilburn JD, Briber RM, Irving T, Woodson SA. Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle X-ray scattering. J Am Chem Soc. 2010;132:10148–54. doi: 10.1021/ja103867p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labuda D, Poerschke D. Magnesium ion inner sphere complex in the anticodon loop of tRNAPhe. Biochemistry. 1982;21:49–53. doi: 10.1021/bi00530a009. [DOI] [PubMed] [Google Scholar]
- 44.Ma H, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. Exploring the Energy Landscape of a Small RNA Hairpin. J Am Chem Soc. 2006;128:1523–1530. doi: 10.1021/ja0553856. [DOI] [PubMed] [Google Scholar]
- 45.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fountain MA, Serra MJ, Krugh TR, Turner DH. Structural features of a six-nucleotide RNA hairpin loop found in ribosomal RNA. Biochemistry. 1996;35:6539–48. doi: 10.1021/bi952697k. [DOI] [PubMed] [Google Scholar]
- 47.Huang S, Wang YX, Draper DE. Structure of a hexanucleotide RNA hairpin loop conserved in ribosomal RNAs. J Mol Biol. 1996;258:308–21. doi: 10.1006/jmbi.1996.0252. [DOI] [PubMed] [Google Scholar]
- 48.Wang YX, Huang S, Draper DE. Structure of a U.U pair within a conserved ribosomal RNA hairpin. Nucleic Acids Res. 1996;24:2666–72. doi: 10.1093/nar/24.14.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shankar N, Xia T, Kennedy SD, Krugh TR, Mathews DH, Turner DH. NMR reveals the absence of hydrogen bonding in adjacent UU and AG mismatches in an isolated internal loop from ribosomal RNA. Biochemistry. 2007;46:12665–78. doi: 10.1021/bi700802s. [DOI] [PubMed] [Google Scholar]
- 50.Stallings SC, Moore PB. The structure of an essential splicing element: stem loop IIa from yeast U2 snRNA. Structure. 1997;5:1173–85. doi: 10.1016/s0969-2126(97)00268-2. [DOI] [PubMed] [Google Scholar]
- 51.Gottstein-Schmidtke SR, Duchardt-Ferner E, Groher F, Weigand JE, Gottstein D, Suess B, Wöhnert J. Building a stable RNA U-turn with a protonated cytidine. RNA. 2014;20:1163–72. doi: 10.1261/rna.043083.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DMJ, Ha T. Conformational flexibility of four-way junctions in RNA. J Mol Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 53.McDowell SE, Jun JM, Walter NG. Long-range tertiary interactions in single hammerhead ribozymes bias motional sampling toward catalytically active conformations. RNA. 2010;16:2414–26. doi: 10.1261/rna.1829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quigley GJ, Rich A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 55.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan CW, Chetnani B, Mondragón A. Structure and function of the T−loop structural motif in noncoding RNAs. Wiley Interdiscip Rev RNA. n.d;4:507–22. doi: 10.1002/wrna.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leipply D, Draper DE. Dependence of RNA tertiary structural stability on Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry. 2010;49:1843–53. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science. 2011;334:1524–9. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 59.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–5. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 60.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–8. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao LJ, James TL, Kealey JT, Santi DV, Schmitz U. The dynamic NMR structure of the T psi C-loop: implications for the specificity of tRNA methylation. J Biomol NMR. 1997;9:229–44. doi: 10.1023/a:1018618606857. [DOI] [PubMed] [Google Scholar]
- 62.Wu M, Tinoco I. RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–8. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 64.Koculi E, Cho SS, Desai R, Thirumalai D, Woodson SA. Folding path of P5abc RNA involves direct coupling of secondary and tertiary structures. Nucleic Acids Res. 2012;40:8011–20. doi: 10.1093/nar/gks468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xue Y, Gracia B, Herschlag D, Russell R, Al-Hashimi HM. Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat Commun. 2016;7 doi: 10.1038/ncomms11768. ncomms11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jean JM, Hall KB. 2-Aminopurine electronic structure and fluorescence properties in DNA. Biochemistry. 2002;41:13152–61. doi: 10.1021/bi020308y. [DOI] [PubMed] [Google Scholar]
- 67.Jean JM, Hall KB. Stacking-unstacking dynamics of oligodeoxynucleotide trimers. Biochemistry. 2004;43:10277–84. doi: 10.1021/bi049701p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.