Abstract
Rhodopsin is a prototypical G-protein coupled receptor (GPCR) that initiates phototransduction in the retina. The receptor consists of the apoprotein opsin covalently linked to the inverse agonist 11-cis retinal. Rhodopsin and opsin have been shown to form oligomers within the outer segment disc membranes of rod photoreceptor cells. However, the physiological relevance of the observed oligomers has been questioned since observations were made on samples prepared from the retina at low temperatures. To investigate the oligomeric status of opsin in live cells at body temperatures, we utilized a novel approach called FRET spectrometry, which previously has allowed the determination of the stoichiometry and geometry (i.e., quaternary structure) of various GPCRs. In the current study, we have extended the method to additionally determine whether or not a mixture of oligomeric forms of opsin exists and in what proportion. Application of this improved method revealed that opsin expressed in live Chinese hamster ovary (CHO) cells at 37 °C exists as oligomers of various sizes. At lower concentrations, opsin existed in an equilibrium of dimers and tetramers. The tetramers were in the shape of a near-rhombus. At higher concentrations of the receptor, higher order oligomers began to form. Thus, a mixture of different oligomeric forms of opsin is present in the membrane of live CHO cells and oligomerization occurs in a concentration-dependent manner. The general principles underlying the concentration-dependent oligomerization of opsin may be universal and apply to other GPCRs as well.
Keywords: Förster resonance energy transfer, G protein-coupled receptor, membrane protein, oligomerization, quaternary structure
INTRODUCTION
Our molecular concept of a G protein-coupled receptor (GPCR) has expanded in recent years to now include oligomeric forms [1-3]. Oligomerization of GPCRs has the potential to impact all aspects of the signaling cycle including receptor biogenesis, activation, and desensitization [4]. The physiological relevance of oligomers has been demonstrated in vivo in mice or in native tissue and cells [5-8]. Rhodopsin, the light receptor in rod photoreceptor cells of the retina, is in many regards a prototypical GPCR. In contrast to other GPCRs, however, rhodopsin consists of the apo-protein opsin covalently linked to the inverse agonist 11-cis retinal. In regards to receptor oligomerization, rhodopsin or opsin appear to behave like other members in the GPCR family.
Whether rhodopsin/opsin exists as monomers or forms oligomers has been a longstanding question and the topic has proven to be controversial, especially after the publication of atomic force microscopy (AFM) images of native murine rod outer segment (ROS) disc membranes. These high-resolution images revealed an oligomeric arrangement of rhodopsin and opsin in the form of rows of dimeric receptor [9, 10]. These AFM studies overcame the ambiguity inherent in classical biochemical studies of detergent-extracted rhodopsin that assessed quaternary structure [1, 11]. However, their physiological relevance was questioned, due to possible lipid phase separation effects incurred during sample preparation and their apparent disagreement to classical biophysical studies monitoring the diffusion of rhodopsin in native membranes [12-17]. Recent studies have shown that lipid phase separation does not appear to induce the formation of rhodopsin oligomers in the membrane [18]. Additionally, classical biophysical studies do not rule out the existence of rhodopsin/opsin oligomers since, in some studies, an appreciable population of rhodopsin molecules have been shown to be immobile, and estimates of the lateral diffusion rate of rhodopsin may have been overestimated [16, 19, 20]. Furthermore, the sizes of rhodopsin oligomers observed in native membranes may not be inconsistent with updated estimates of the diffusion constant for rhodopsin [21].
Several additional lines of evidence from in vitro studies support the notion of rhodopsin oligomers existing in native tissue. The G protein transducin binds rhodopsin in the ROS in a cooperative manner with a Hill coefficient greater than 1, which indicates that at least some rhodopsin in the ROS is in an oligomeric arrangement [22, 23]. Cryo-electron tomography studies of cryo-sectioned native ROS reveal similar types of oligomeric complexes of rhodopsin as observed by AFM [24]. Rhodopsin-transducin and rhodopsin-arrestin complexes from native bovine retina reveal that two rhodopsin molecules can bind either a single transducin or arrestin molecule and that the rhodopsin molecules in the complexes are functionally asymmetric [25-28]. Oligomerization does not appear to be a prerequisite for binding and activating signaling molecules, however, since in vitro studies show that monomeric rhodopsin is sufficient to bind and activate transducin [29-32], couple to arrestin [33-36], and be phosphorylated by rhodopsin kinase [35]. Rhodopsin oligomerization may be necessary to provide a platform to make single photon detection in photoreceptor cells possible and asymmetry in rhodopsin oligomers may contribute to signaling efficiency and play a protective role in photoreceptor cells under intense lighting conditions [25-28, 37, 38].
The experimental evidence for rhodopsin oligomers as the physiological structure of the light receptor comes largely from in vitro studies. Thus, the nature of rhodopsin oligomers formed in live cells is unclear. Moreover, we do not yet understand the mechanisms underlying the association of rhodopsin monomers to form oligomeric complexes. Experimental approaches are limited for quantitatively characterizing the organization of opsin in the membrane of living cells. A tractable system is therefore required to investigate the oligomeric potential of opsin in living cells in the absence of artifacts. To answer fundamental questions about the oligomerization of rhodopsin in live cells, we have tagged the apoprotein opsin with fluorescent proteins and applied a novel spectrometric method – ‘Förster resonance energy transfer (FRET) spectrometry’ [39]. Oligomers of rhodopsin or opsin have previously been detected in transfected cells and lipid vesicles using standard FRET methods [29, 40-47]. Bulk approaches, however, are unable to distinguish between different arrangements and sizes of oligomers formed by the receptors, unless structural information is already known from separate experiments, which is then inserted into suitable theoretical models for FRET in oligomeric complexes. A fluorescence cross-correlation spectroscopy study of opsin in live cells has indicated that the receptor is exclusively dimeric and in equilibrium with monomers [48]; however, this arrangement is inconsistent with observations in native ROS disc membrane. Moreover, this method relies critically on the mobility of the receptor within the membrane and cannot probe oligomeric structures with low mobility because the fluorescent tags would become photobleached during the measurements.
In the present study, the FRET spectrometry method [39, 49, 50] and the analysis of data with theoretical models allowed for the determination of the size and shape of all opsin oligomers present in live cells. This structural information allowed for further analysis using an ensemble FRET approach [51, 52] to quantitatively assess the proportion of the different oligomeric forms of opsin embedded in the membrane of live cells. Our novel approach provides a more complete structural view of rhodopsin oligomerization over previous studies and reveals insights into the mechanism by which rhodopsin and other GPCRs form oligomers in the membrane.
MATERIALS AND METHODS
DNA Constructs
SYFP2 and mTurquoise (mTq) are variants of the widely used yellow fluorescent protein (YFP) and cyan fluorescent protein [53, 54]. The vectors pmRho-SYFP2-1D4 and pmRho-mTq-1D4 were generated as described previously [46, 47]. The full CMV promoter in these vectors was replaced by a truncated CMV promoter to decrease the expression level of rhodopsin. The truncated CMV promoter, CMVd3, was generated by PCR using the primers 5′ - ACGATGATTAATATGGGCGGTAGGCGTGTACG and 5′ – GGTAGCGCTAGCGGATCTGA, and included an AseI restriction site at the 5′ end and an NheI restriction site at the 3′ end. The amplified CMVd3 replaced the endogeneous full CMV promoter in pmRho-SYFP2-1D4 and pmRho-mTQ-1D4 at the AseI and NheI restriction sites to generate the vectors pCMVd3-mRho-SYFP2-1D4 and pCMVd3-mRho-mTQ-1D4.
Fluorescent Protein Purification
Fluorescent proteins were purified using similar procedures described previously and as briefly described herein [55]. The vectors pRSET-SYFP2 and pRSET-mTq were generated as described previously [53, 54]. BL21 (DE3) competent cells (NEB, Ipswich, MA) were transformed with pRSET-SYFP2 or pRSET-mTq and plated on LB agar with 50 αg/mL ampicillin and incubated overnight at 37°C. 5 tubes of 6 mL LB broth with 50 αg/mL ampicillin were inoculated with bacteria picked from individual colonies and incubated overnight at 37°C with orbital shaking. Fluorescent protein expression was induced by adding 40 αL of IPTG (100 mM stock) and incubating further for 4 h at 37 °C with orbital shaking. The cultures were spun down at 4,500 × g for 10 min and placed at – 80 °C for 1 h. Cells were lysed using the CelLytic B Plus kit (Sigma-Aldrich, St. Louis, MO). Cell pellets were pooled from the 5 tubes, resuspended in 2 mL of CelLytic B Plus working solution, and incubated at room temperature for 10 min with shaking. The cell lysate was centrifuged at 1,900 × g for 10 min. The supernatant containing the fluorescent protein was collected and loaded by gravity flow on a 1 mL HisPur Ni-NTA spin column (ThermoScientific, Waltham, MA), which was equilibrated with 2 ml 10 mM imidazole in phosphate-buffered saline (PBS), and incubated for 30 min at room temperature. The column was centrifuged at 700 × g for 2 min. The column was washed 3 times by addition of 2 mL 25 mM imidizole in PBS and centrifugation at 700 × g for 2 min. The bound fluorescent protein was eluted with 1 mL 250 mM imidizole in PBS and centrifugation at 700 × g for 2 min. Imidizole was removed using a 2 mL ZebaSpin desalting spin column (ThermoScientific, Waltham, MA). 700 αL of the eluant was loaded on the desalting column equilibrated with PBS. Purified fluorescent protein was collected by centrifugation at 1,000 × g for 2 min. The concentration of purified fluorescent protein was determined by UV/Vis absorbance spectroscopy on a Nanodrop2000c (ThermoScientific, Waltham, MA). The λmax and extinction coefficient for SYFP2 was 515 nm and 101 × 103 M−1 cm−1, respectively, and that for mTq was 434 nm and 30 × 103 M−1 cm−1, respectively.
Cell Sample Preparation
Chinese hamster ovary (CHO) cells were cultured and maintained in T-25 flasks in 5 ml of Dulbecco’s Modified Eagle Medium (DMEM)–high glucose (product number 11965-092, Thermo Fisher Scientific, Waltham, MA) containing 10% fetal bovine serum and 1% non-essential amino acids. Cell lines were maintained at 37 °C in a humidified incubator with 5% CO2. Cells were seeded at approximately 40% confluency into 35 mm Delta T heated culture dishes (Bioptechs Inc., Butler, PA) with 0.17 mm thick bottom-bonded coverslips, which were previously coated with poly-D-lysine. The cells were incubated for 48 hours, and then washed with Dulbecco’s Phosphate-Buffered Saline. 2 ml of Opti-MEM® (Life Technologies, Carlsbad, CA) was added to the cells in each dish. Cells were transfected with the vectors pCMVd3-mRho-SYFP2-1D4 and pCMVd3-mRho-mTQ-1D4 in the presence of Lipofectamine® (Invitrogen, Carlsbad, CA). When cells were transfected with both vectors, the total amount of DNA added was 4 μg and the ratio of vectors was varied (i.e., 3:1, 1:1 and 1:3). When cells were transfected with only a single vector, 2 μg of DNA was added. Cells were incubated for 24 hours and then imaged.
Fluorescence-Based Imaging
Cells were transported from the incubator to the imaging system in a portable incubator (Darwin Chambers Company, St. Louis, MO). The microscope was equipped with a thermostated microscope stage, Delta-T temperature-control system (Bioptechs Inc., Butler, PA), to maintain the temperature of cells at 37 °C ± 2 °C throughout the image acquisition process. Samples were excited by a tunable femtosecond laser (Mai Tai™, Spectra Physics, Santa Clara, CA), and imaged using a two-photon optical micro-spectroscope OptiMiS™ TruLine (Aurora Spectral Technologies, Milwaukee, WI) coupled to an inverted microscope (Nikon Eclipse Ti™, Nikon Instruments Inc., Melville, NY) equipped with a 100X oil-immersion objective. Each sample was line-scanned [56], applying an average power of 350 mW of laser light per line (0.23 mW per excitation voxel), successively at two wavelengths: 815 nm and 960 nm. The image integration time was set to 30 milliseconds per line and a spectral resolution of ~ 1 nm was used to collect the fluorescence emission.
Protein Standards Measurements
For fluorescent protein solutions measurements, the same type of dishes as described above were coated with 1% Bovine Serum Albumin (BSA) in PBS (1 h incubation at 37 °C) to prevent binding of the protein molecules to the dish bottom. Fluorescent protein standard solutions of mTurquoise and SYFP2 (stored at −70°C), prepared as mentioned above, were diluted with deionized water, to bring the final concentrations to 5, 10, 20 or 40 μM. These samples were then imaged under the same imaging conditions as the opsin samples were imaged. The average fluorescence intensity was plotted vs. protein concentration for each of the two fluorescent proteins. The points in each graph were best fit with a straight line whose slope corresponded to the number of fluorescence counts per μM of solution. The slopes for the two samples were used to estimate the concentration of the opsin molecules attached with either type of fluorescent markers in the CHO cell membranes.
Spectral Unmixing and Determination of Apparent FRET Efficiencies (Eapp)
The composite emission spectrum of cells co-expressing donor- and acceptor-tagged opsin (i.e., mTurquoise-opsin and SYFP2-opsin, respectively) was unmixed using donor and acceptor spectra obtained by imaging of cells expressing only one type of fluorescent protein. The unmixing was performed using a MATLAB routine written in house [56], which provided two sets of unmixed images for each acquired image (i.e., donor emission in presence of acceptor (kDA) and acceptor emission in presence of donor (kAD)). These images were then used to compute the apparent FRET efficiency per pixel (Eapp), via the following equation [49]:
| (1) |
where QA and QD are quantum yields of donor and acceptor, respectively, which were obtained from the literature [53, 54]. WD and WA are the areas under the individual donor and acceptor spectra, which were computed from the measured mTurquoise and SYFP2 spectra.
Receptor Concentration Determination
The total number of donor and acceptors per pixel were determined in each cell by comparing the average donor and acceptor fluorescence intensities to those of solutions of known concentrations of donor or acceptor fluorescent proteins (see Protein Standards Measurements above). The donor and acceptor concentrations were then expressed as molecules/μm2 by dividing the number of molecules per pixel by the area of the membrane, which fits along an appropriate cross section of the excitation voxel corresponding to every pixel. The voxel size, which depends on the laser excitation wavelength, was estimated from this volume using previously derived equations [57]. Briefly, the donor-only emission intensity following excitation with the wavelength λ1 = 815 nm and acceptor-only emission intensity following excitation at λ2 = 960 nm were computed from the unmixed donor and acceptor intensities for the samples coexpressing donors and acceptors, after correcting for acceptor direct excitation by measuring the ratios of the rates of the de-excitations for the donor and acceptor at the two excitation wavelengths and applying the set of equations and methods as described elsewhere [51].
Theoretical Modeling of FRET Efficiency
(a) The Meta-Histogram Approach
Regions of interest (ROI) approximately coincided with the outlines of individual cells. Eapp were selected and the values from the pixels comprising the ROI were binned in intervals of 0.01 of FRET efficiency to obtain Eapp histograms (or FRET spectrograms). The positions of the main or dominant peak in each histogram were then collected and binned in intervals of 0.02 to generate a meta-histogram of FRET efficiencies [39, 58]. The experimental Eapp meta-histograms were then fitted to theoretical models of FRET efficiency, developed for oligomers of different stoichiometry and shapes [39, 49]. The prediction of the average value of FRET efficiency per donor for each geometric configuration, q, of an oligomeric complex consisting of k donors is given by:
| (2) |
where rij represents the distance between the ith donor and the jth acceptor and the summation indices i and j are over all donors and acceptors in a given configuration, respectively. The geometric configurations pertaining to a particular oligomeric model are related through a common parameter, Ep, which is the FRET efficiency of a donor acceptor pair separated by a distance r1, as is seen in equation (2), or pairwise FRET efficiency. For a given oligomer complex, there exist a number of degenerate geometric configurations which have the same value of Ek,q; this degeneracy leads to a number of well-defined peaks in a histogram representation of Ek,q values. The number and positions of the histogram peaks are constrained by the (i) distance and (ii) orientation of each donor with respect to the acceptors in the complex (see Supplementary Figure 3 and Supplementary Tables 1-4).
The experimentally obtained meta-histogram of Eapp values was simulated with a model which represented the oligomer complex in the form of a parallelogram-shaped tetramer; this particular model contained seven unique Ek,q values each of which was represented by a Gaussian distribution in the fitting function, as follows:
| (3) |
where Aq represents the amplitude of the qth Gaussian distribution and σq its variance. The positions of each Gaussian were dependent on the same three fitting parameters: Ep, the ratio of the lengths of the two adjacent sides of the parallelogram r1/r2, and the angle α between the sides (see Supplementary Figure 3).
(b) The statistical ensemble approach
The average FRET efficiency, Eave, for each ROI was computed using equation (1) and the average kDA and kAD values within the ROI. The acceptor mole fraction, XA, corresponding to each ROI was also computed by using the following equation:
| (4) |
where nD and nA represent the average number of donor and acceptor molecules, respectively, residing in the pixels of the given ROI (see Receptor Concentration Determination). A plot of the average Eapp vs. XA was constructed and simulated with a theoretical model consisting of a mixture of oligomeric species of different sizes, as given by:
| (5) |
where n is the number of monomers in an oligomer (e.g. for dimers, n=2), k the number of donors in said oligomer, ND,tot the total number of donors in the mixture, and Ek,q defined in Equation 2. Note that the summation over n allows for oligomers of different sizes to be incorporated into the model with μn representing the total number of complexes of size n within the mixture.
The three parameters needed to compute each of the Ek,q values (Ep, r1/r2, and α) were extracted from fitting the meta-histogram of Eapp values. The fitting of Equation 5 to the experimental data was achieved by adjusting the concentrations, μn, of the various oligomer populations to minimize the fitting residual given by Equation 4.
For both the spectrometric method and the statistical ensemble method, the theoretical model was fitted to the experimental curves by adjusting the fitting parameters in order to minimize the fitting residual given by the following formula [50, 59]:
| (6) |
where “Experimental” stands for the experimentally measured Eapp values and “Simulated” for the Eapp values simulated using the theoretical models and the model adjustable (or fitting) parameters, and f is the number of degrees of freedom.
Molecular Modeling
The structure of a rhodopsin tetramer was generated from AFM data of native ROS disc membranes (PDB: 1N3M) [9, 10]. The atomic coordinates of YFP were taken from the crystal structure (PDB: 2YFP) [60]. Both proteins were fused so that YFP was bound to the C-terminus of rhodopsin. The model of the fused protein was constructed and optimized using the program YASARA version 11.11 (YASARA Biosciences, Vienna, Austria) [61], employing Yasara2 forcefield. For the relaxation procedure, the backbone of the new protein was constrained except for the C-terminus of rhodopsin, which served as a linker between rhodopsin and YFP. During short molecular dynamics simulation in continual annealing mode (gradual diminishing and rising of temperature between 300 K and 0 K) the linker attained the optimal conformation. Only the last part of the C-terminus (residues 344-348) changed positions compared to the initial structure of untagged rhodopsin.
RESULTS
Initial assessment of opsin oligomerization by FRET efficiency histogram analysis
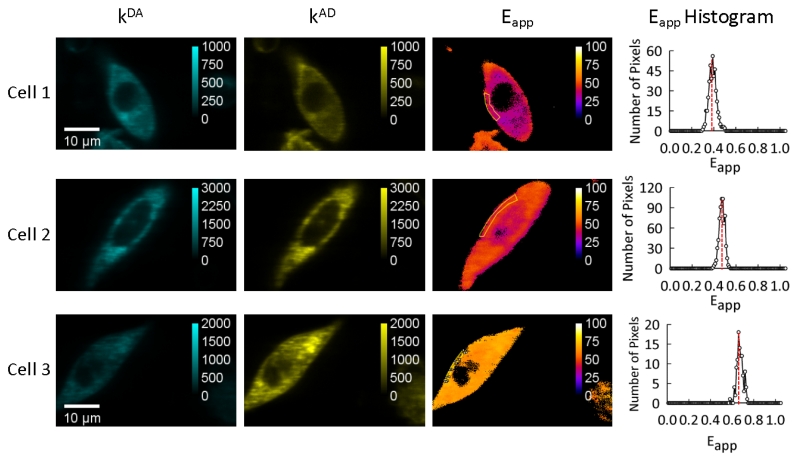
Opsin was tagged with the fluorescent proteins mTurquoise and SYFP2, which are suitable donor and acceptor pairs for FRET studies [54]. Opsin-mTurquoise (energy donor, D) and opsin-SYFP2 (energy acceptor, A) were coexpressed in CHO cells at 37 °C. Individual cells were imaged using a two-photon optical microspectroscope (Figure 1). Composite emission spectra were collected for each pixel of the image corresponding to the plasma membrane region, which were spectrally unmixed to compute pixel-level FRET efficiencies (i.e., Eapp), as described in the Materials and Methods. In contrast to typical FRET assays that only provide an average of FRET behavior for an ensemble of protein complexes [62], the method used here provides FRET efficiencies for each pixel in images of a single cell. Figure 1 shows the results of spectral unmixing and the apparent FRET efficiency maps (Eapp) computed from those images for a random selection of cells.
Figure 1. Typical results obtained from imaging individual cells co-expressing opsin-mTurquoise and opsin-SYFP2.
Spectral unmixing (see Materials and Methods) provided separate maps of the fluorescence signals of donors in the presence of acceptors (kDA) and acceptors in the presence of donors (kAD). Apparent FRET efficiency (Eapp) values were determined for each pixel in images of individual cells from the pixel-level values of kDA and kAD, as described in the Materials and Methods section. Contours of regions of interest (ROI) corresponding to the plasma membrane were drawn based on the kAD images and then transferred to the Eapp maps (shown as orange lines). Histograms of Eapp were generated for each ROI using a bin size of 0.01. The positions of the maxima of these histograms (indicated by vertical red lines) were used to generate the meta-histograms shown in Figure 2.
The distribution of Eapp values in histograms generated from individual cells can provide an initial assessment about whether or not opsin forms oligomers and the nature of those oligomers. Histograms of Eapp (or FRET spectrograms) generated for each cell revealed narrow distributions, essentially characterized by a single peak (Figure 1). The position of this peak along the horizontal axis varied from cell to cell because of variations in the ratio of donor- and acceptor-tagged opsins expressed in individual cells. In fact, this variation was intentionally enhanced by using three different ratios of transfected DNA coding for the donor- or acceptor-tagged receptor (3:1, 1:1, and 1:3), which provided the desired effect of maximizing coverage along the horizontal axis necessary for analyses presented in the following sections. Stochastic FRET occurring from non-interacting opsin molecules are predicted to result in a majority of FRET efficiency values close to zero in Eapp histograms with only a minor number of values with higher values [62]. Since the peak position in histograms are much greater than Eapp values of zero, the FRET detected in experiments indicate that opsin-mTurquoise and opsin-SYFP2 oligomerize in the membrane of live cells.
The shape of the distribution in Eapp histograms provides an initial assessment about the size of opsin oligomers present in CHO cells. Histograms exhibited narrow distributions of Eapp values (Figure 1). These are indicative of the presence of either (a) a single dominant type of small oligomer at every image pixel (e.g., dimers or tetramers), if the expression level is at most one oligomer per pixel, or (b) a mixture of different oligomeric forms (e.g., monomers, dimers, and/or higher order oligomers), if a high concentration of receptor is present in each image pixel [58]. The expression of opsin in the current study is relatively high resulting in an average concentration of 1,350 molecules/μm2. Thus, the narrow distributions in Eapp histograms observed in Figure 1 point to a mixture of quaternary forms in individual CHO cells. We can also rule out the presence of exclusively large oligomers, which would have generated relatively broad histograms presenting several peaks (e.g., five for rhombus tetramers [49, 59]) at low expression levels of opsin.
The Eapp histograms can be represented in two ways to gain more detailed information on quaternary structure:
Meta-histograms of histogram peak positions [58]; these are created by first determining the Eapp value corresponding to the center of the peak position in each Eapp histogram of an individual cell (see Figure 1) and then generating a histogram of those peak positions (Figure 2).
Averages of the histograms plotted vs. either [D]/[A] [52] or the molar fraction of the acceptors, XA [51, 63] (Figure 3).
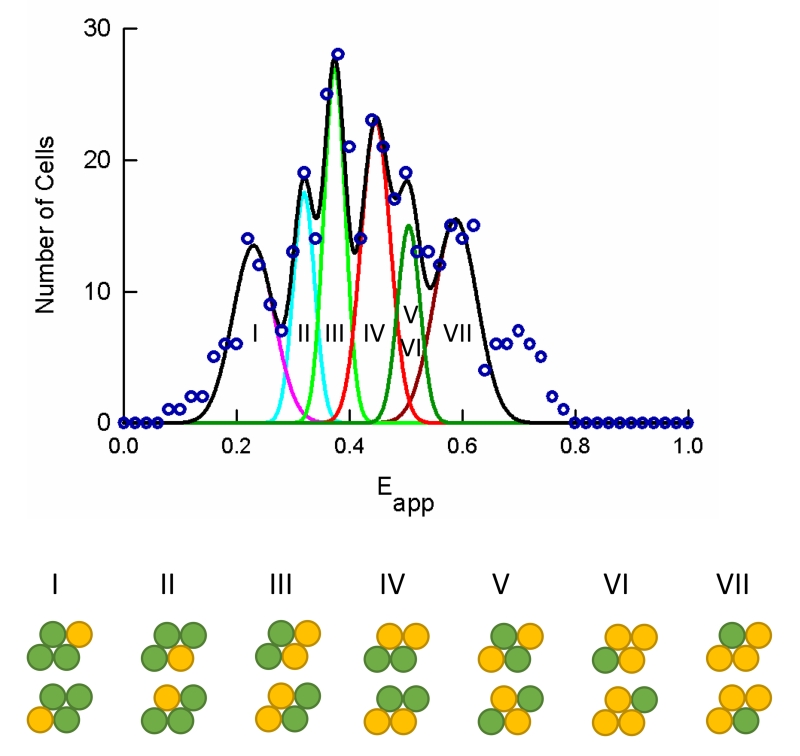
Figure 2. Meta-histogram analysis.
Meta-histograms were generated by collecting the peak positions from individual cell Eapp histograms (e.g., Figure 1) at a bin size of 0.02 (open circles). The data were fit with a theoretical model for a parallelogram-shaped tetramer. The fitted curve is shown as correlated Gaussian functions shown individually (pink, cyan, green, red, dark-green, and dark-red thin lines) and as a sum (thick black line). The configurations of donors (green) and acceptors (yellow) within a parallelogram-shaped tetramer corresponding to each Gaussian is shown. The best-fit parameters obtained by fitting the theoretical model to the experimental data were: Ep = 0.380, α = 62.2°, and r1/r2 = 0.93.
Figure 3. Average apparent FRET efficiency (Eapp) versus acceptor mole fraction (XA).
The average Eapp and XA were computed from pixel-level FRET efficiencies for plasma membranes of individual CHO cells. Data were fitted with models assuming the presence of only dimers (solid black line) or a mixture of dimers and parallelogram tetramers (solid red line), as described in the theoretical section. The best-fit value of the FRET efficiency for a model consisting of only dimers was 0.550 and the corresponding fitting residual was 2.212. When incorporating tetramers to the fitting model, the values of Ep, the main parallelogram angle (α), and the ratio between the lengths of the sides were fixed to the values obtained from the meta-histogram analysis displayed in Figure 2. The best-fit value for the tetramer to dimer ratio (ρ) was 1.47 and the corresponding fitting residual was 2.127.
The first method allows determination of the quaternary structure [58], while the latter allows for the determination of the proportion of oligomeric forms present if the quaternary structure is known [64]. Both methods are used herein to extract complete information about the oligomeric states of opsin in living cells, as described below.
Characterization of the quaternary structure of opsin by meta-histogram analysis
In order to determine the most probable quaternary structure for opsin oligomers, we analyzed the distribution of FRET efficiencies for the entire population of cells examined. Meta-histograms were generated by plotting a histogram of the dominant peak position in each cell-level Eapp histogram (e.g., Figure 1) [50, 58]. The resulting meta-histogram from all cells imaged in the current study (at all expression levels) is shown in Figure 2. Data in the meta-histogram was fit with models for different quaternary structures. The data were inconsistent with an oligomer that was exclusively a dimer, trimer, or square tetramer, which would result in only 3 peaks or less in the meta-histogram [39]. A parallelogram-shaped tetramer model provided a visually good fit to the data in the metahistogram (Figure 2) (except for a peak appearing at Eapp greater than 60%, which will be discussed momentarily), and a fitting residual better than those obtained for all the other models tested, including dimers and trimers. Although the angle and the ratios of the sides of the parallelogram were such that the modeled parallelogram only slightly deviated from a rhombus, it nevertheless led to a significantly improved fit compared to a rhombic tetramer model, as revealed by simple visual inspection (see Supplemental Figure 1) and a lower fitting residual (0.0076 for parallelogram versus 0.0282 for rhombus). Thus, a major oligomeric species of tagged opsin in the membrane of CHO cells is a tetramer in the shape of a parallelogram.
While consistent with the majority of the data, the parallelogram tetramer model could not account for meta-histogram features occurring at Eapp values greater than 0.6 (Figure 2). The simplest explanation for this discrepancy is the occurrence of oligomers larger than tetramers, such as arrays of dimers with similar geometry (in terms of inter-protomeric distances and angles) to those of the tetramers discussed above, at comparatively high expression levels of the receptor. This hypothesis was tested by splitting the data in the meta-histogram into separate meta-histograms corresponding to four different expression levels of opsin. Consistent with our hypothesis, the last peak in the meta-histogram was absent for concentrations lower than 150 molecules/pixel (804 molecules/μm2), started to appear at average concentrations of 220 molecules/pixel (1147 molecules/μm2) and was more prominent at concentrations of 675 molecules/pixel (3,400 molecules/μm2) (see Supplemental Figure 2). Thus, significant levels of higher order oligomers, must form at comparatively high expression levels of the receptor.
To more explicitly assess the size of higher order oligomers, we first tried one-dimensional arrays of dimers, or oligomers that form by successive addition of dimers to one end of the array. For instance, addition of a dimer to a tetramer leads to a hexamer, or a linear array of three dimers (see Supplemental Figure 3). This arrangement is predicted to result in a large number of peaks in meta-histograms of Eapp (see Supplemental Tables 2 and 3). Numerically, however, the positions of the Eapp peaks predicted by this model fall within the range defined by those predicted for the parallelogram tetramer model (see supplemental Table 1). Thus, a hexamer model cannot explain the right-most peak in the meta-histogram presented in Figure 2. In order to account for that peak, a two-dimensional expansion of the parallelogram, to form a structure of at least 3 × 4 receptors (i.e., 12 receptors or a 12-mer) was needed (see Supplemental Figure 4 and Supplemental Table 4). Of the several possible combinations of donors and acceptors within a 12-mer, only those corresponding to one or two donors surrounded by acceptors (see Supplemental Table 4) were evaluated numerically since they were the ones that predicted Eapp values high enough to match those generating the right-most peak in the meta-histogram shown in Figure 2.
Mixture of oligomeric forms of opsin characterized by statistical ensemble FRET analysis
While the meta-histogram analysis points to tetramers as the major oligomeric species of opsin in the membrane of CHO cells at lower expression levels, with larger oligomers appearing at higher expression levels, we cannot not rule out the coexistence of tetramers and higher order oligomers with dimers. If tetramers have finite lifetimes and dissociate into dimers, then dimers can contribute to the amplitude of the peak in meta-histograms corresponding to the pairwise FRET efficiency, Ep, without affecting peak positions derived from tetramers [59]. Thus, our analysis of the peak positions in the meta-histogram in Figure 2 is oblivious to the presence of dimers even if they are present. In order to assess whether dimers are also present in our cells, we computed the cellular average of Eapp detected at the plasma membrane of CHO cells and plotted them against the acceptor molar fraction (XA) (Figure 3). This analysis represents the statistical ensemble approach to FRET analysis.
The Eapp vs. XA plot for all the cells investigated was first fitted with a dimer model (described in the theoretical section) to confirm results from the meta-histogram analysis that indicated that opsin did not exclusively form dimers. By fitting the dimer model to the Eapp vs. XA plots, we obtained an Ep value of 0.550, which is significantly lower than the highest values of Eapp observed in the histograms of Eapp (Figure 1). For a system of dimers, the highest FRET efficiency in a histogram (i.e., the right-most peak in the histogram) corresponds to DA dimers (Ed), while all the other peaks resulting from different combinations of DA and DD dimers appear to the left of that peak (i.e., lower Eapp values) [62]. Since the main peaks of several of the experimental histograms were located beyond the Ed value determined from the dimer model fit of the data in Figure 3, this indicates that the dimer model is inconsistent with our data. Similar to the meta-histogram analysis, the statistical ensemble approach also indicates that opsin does not form exclusively dimers in live CHO cells.
The data were then fitted with a model that assumes opsin forms a mixture of dimers and tetramers (described in the methods section) by fixing the three geometrical parameters to the values given in the caption to Figure 2 and allowing the ratio of tetramers to dimers, ρ =[t]/[d], to vary. The fitting was not only significantly better, as revealed by a lower fitting residual value, but it was also consistent with the meta-histogram analysis. Thus, the statistical ensemble FRET analysis indicates that dimers and tetramers of opsin coexist in live CHO cells and that the average ratio of tetramers to dimers in all cells investigated here is 1.47 (Figure 3).
Quantification of the equilibrium between dimers and tetramers of opsin
The analysis of the Eapp vs. XA plots was refined to investigate the dependence of the oligomer size on the expression level of the receptor. Data were split into four subsets according to the concentration of receptors in the cell membrane (Figure 4). The data were fit with models for a dimer (blue), tetramer (green), or a mixture of dimers and tetramers (red). In each case, the mixture model fit the data best, which indicated that opsin exists as a mixture of oligomeric forms at all concentrations of the receptor tested. The ratio between the concentrations of tetramers and dimers, ρ =[t]/[d], was determined from the fit of each subset of data to the mixture model. The tetramer to dimer ratio increased with increasing concentrations of the receptor. In cells expressing opsin at the lowest concentrations tested (227 molecules/μm2), opsin is predicted to be 71% dimeric and 29% tetrameric. At concentrations of 591 molecules/μm2, opsin is predicted to form dimers and tetramers in roughly equal proportions. At higher concentrations, opsin is predicted to form more tetramers than dimers. At concentrations of 1147 molecules/μm2, opsin is predicted to be 36% dimeric and 64% tetrameric. At the highest concentrations tested (3400 molecules/μm2), the level of dimeric opsin is predicted to be negligible. Consistent with observations in meta-histogram analysis for higher order oligomers, the formation of tetramers also appears to occur in a concentration-dependent manner.
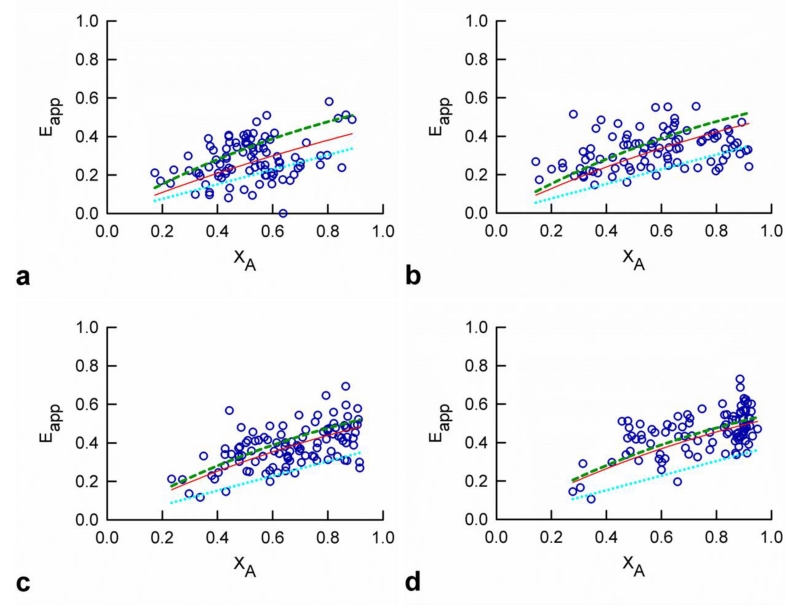
Figure 4. Average apparent FRET efficiency (Eapp) versus acceptor molar fraction (XA) determined from cells expressing different concentrations of opsin.
Data in Figure 3 were split into four subsets according to the concentration of opsin in the plasma membrane of the cell. The range of concentrations of opsin represented in each subset are as follows: (a) less than 402 molecules/μm2 (average, 227 molecules/μm2), (b) 402 - 804 molecules/μm2 (average, 591 molecules/μm2), (c) 804 - 1607 molecules/μm2 (average, 1147 molecules/μm2), and (d) greater than 1607 molecules/μm2 (average, 3400 molecules/μm2). The data were fit with a model assuming a mixture of parallelogram-shaped tetramers and dimers (red solid line). The values of Ep, the main parallelogram angle (α), and the ratio between the lengths of the sides were fixed to the values obtained from the meta-histogram analysis displayed in Figure 2. The best-fit values for the tetramer to dimer ratios (ρ) were: (a) 0.401, (b) 1.08, (c) 1.79 and (d) 484. The corresponding fitting residuals were: (a) 1.165, (b) 1.049, (c) 0.937 and (d) 0.903. The individual lines corresponding to the dimer component (dotted blue line) and the tetramer component (dashed green line) are also indicated for each concentration range.
To quantify the equilibrium between dimeric and tetrameric receptors, we plotted ρ as a function of the total expression level of the receptor (Figure 5). The data were fitted to the following quadratic equation, which follows from the Law of Mass Action:
| (7) |
where the dissociation constant, Kt→d is defined as,
| (8) |
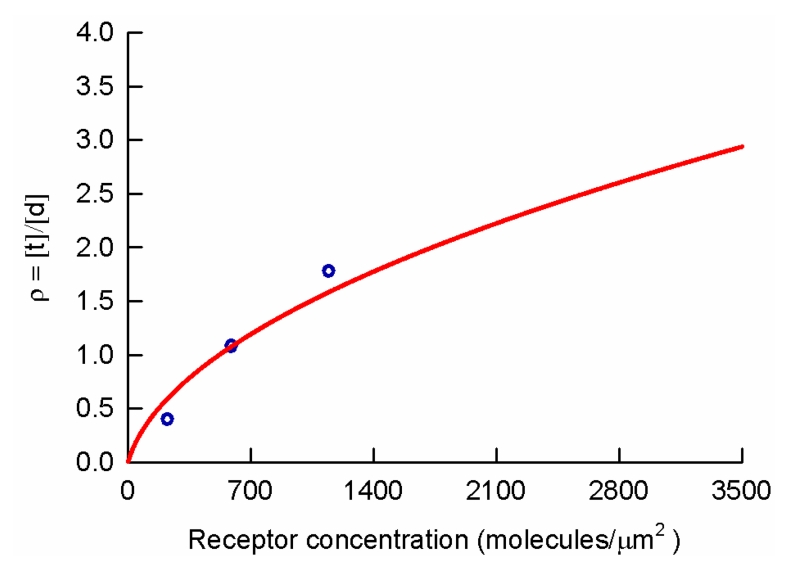
The theoretical fit of equation (7) to the first three values of ρ determined above provided a best-fit value of 87 molecules/μm2 for Kt→d. The fourth data point (corresponding to ρ =484) obtained from the theoretical fit of the tetramer and dimer mixture model to the subset of data with the highest expression level, was much higher than that predicted by extrapolating the curve predicted by equation (7) to the average concentration of receptors (3,400 molecules/μm2) of this data group; the predicted value was 2.89. This again indicated that a sizeable amount of higher order oligomers must be present in the cells for the highest concentration range, in agreement with the results of the meta-histogram analysis (Supplemental Figure 2). Taken together, our results show that opsin expressed in CHO cells is present in multiple oligomeric forms. The formation of the oligomers occurs in a concentration-dependent manner, with larger oligomers forming at higher concentrations of receptor.
Figure 5. Tetramer to dimer ratio (ρ) versus average opsin concentration.
The open circles represent the experimental data from the three subsets with the lowest concentration of receptor in Figure 4. The best-fit value for ρ is plotted against the average concentration of opsin in cells represented in each subset. The data was fit with equation (7). The best-fit value for Kt→d was 87 molecules/μm2.
DISCUSSION
Concentration-dependent oligomerization of opsin in live cells
The investigation of fluorescently tagged opsin in live CHO cells by spectral FRET has provided a quantitative assessment of the quaternary structure and complement of oligomeric forms of opsin present in the membrane of live cells. The analysis over a range of opsin concentrations in CHO cells revealed that oligomerization is stochastic and concentration dependent. Our data analyzed by multiple methods were incompatible with opsin being present exclusively in a dimeric form at any concentration tested. In addition, the existence of trimers and higher order oligomers with an odd number of protomers can also be excluded because the smallest unit seems to be the dimer. At concentrations below 804 molecules/μm2, opsin is predominantly in equilibrium between dimeric and tetrameric forms, favoring the latter with increasing concentrations of the receptor. Tetramers represented 29 % and 52% of the oligomeric complexes at average receptor concentrations of 227 and 591 molecules/μm2, respectively. At concentrations above 804 molecules/μm2, higher order oligomers began to manifest. Thus, the size of oligomers formed by opsin is concentration dependent and increases with increasing concentration of the receptor.
The concentration-dependent oligomerization of opsin observed in the current study suggests an equilibrium scheme presented in Figure 6. Within such a scheme, the oligomeric status of the receptor is determined by the individual equilibrium constants and the concentration of the receptor in the membrane. Our approach cannot directly detect monomers and therefore we cannot assess the equilibrium between monomers and dimers of opsin explicitly. A previous study using fluorescence cross-correlation spectroscopy to infer the oligomeric status of tagged opsins in COS-7 cells derived an equilibrium dissociation constant of 1,010 molecules/μm2 for the monomer-dimer equilibrium [48]. This value is much greater than the equilibrium dissociation constant of 87 molecules/μm2 we have derived for the dimer-tetramer equilibrium. Moreover, only dimers were observed in the fluorescence cross-correlation spectroscopy study, which is at odds with our study where there were no conditions where opsin was exclusively dimeric. In contrast to our study, estimates of receptor concentration in the fluorescence cross-correlation spectroscopy study were only indirectly inferred, which may have led to incorrect estimates of receptor concentration and determination of equilibrium constant. Those studies may have been conducted at concentrations favoring dimers over tetramers or higher order oligomers or a difference in the cell system studied may also contribute to these differences.
Figure 6. Equilibria of different oligomeric forms of the receptor.
The scheme shows the equilibria between different sized oligomers of the receptor (R). The superscripts represent the number of receptor molecules in the oligomer. The dissociation constants defining each equilibrium will dictate the complement of oligomeric forms present for a given concentration of the receptor.
A monomer-dimer equilibrium dissociation constant of 3.6 molecules/μm2 has been derived previously for the GPCR N-formyl peptide receptor [65]. This value is over two orders of magnitude smaller than the equilibrium constant derived for opsin by fluorescence cross-correlation spectroscopy [48], but is consistent for a case where a dimer-tetramer equilibrium occurs with an equilibrium constant of 87 molecules/μm2. Moreover, a monomer-dimer equilibrium dissociation constant of 3.6 molecules/μm2 would also be roughly the same magnitude in affinity for β1 and β2 adrenergic receptor monomers to form dimers and weaker in affinity than that for GABAB receptor monomers to form dimers [66]. In the current study, tagged opsins were expressed in the membrane of CHO cells at concentrations of 50 – 16,000 molecules/μm2 with an average of 1,350 molecules/μm2. Thus, under the conditions of our study, if dimerization occurs with a similar affinity as in the dimerization of the N-formyl peptide receptor, we would expect no monomers to be present in the membrane. Although limitations in our procedure precluded an estimate of the equilibrium of tetramers and higher order oligomers, we can provide an estimate of the lower limit. Higher order oligomers only begin to be detected at an average concentration of 1,147 molecules/μm2. Thus, the tetramer-higher order oligomer equilibrium dissociation constant will be greater than 1,147 molecules/μm2.
Comparison of opsin oligomers in CHO cells versus photoreceptor cells
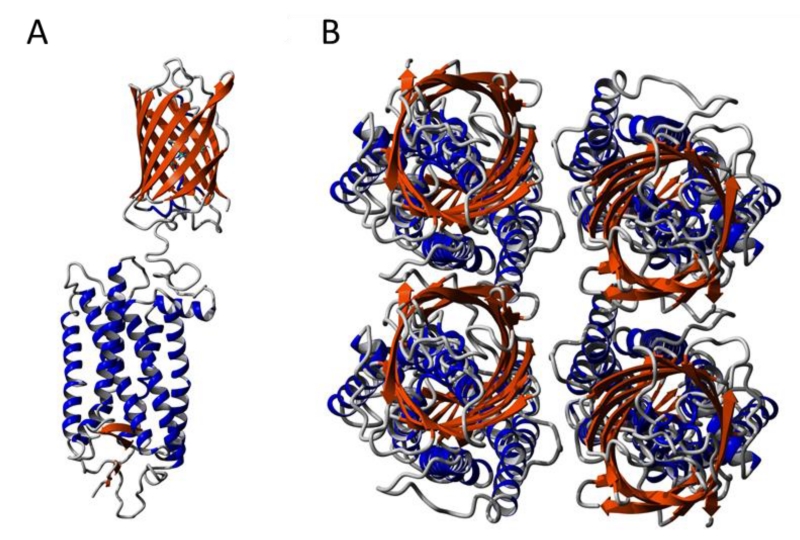
The major oligomeric species observed under the conditions of our experiments in CHO cells is a tetramer with the fluorescent proteins in a near-rhombic arrangement. A theoretical model of a rhodopsin tetramer based on AFM data from native ROS disc membranes (PDB: 1N3M) was examined to determine if the current FRET data are consistent with data derived from AFM on native photoreceptor cell membranes. Cryoelectron microscopy studies of cryosectioned ROS also display a similar oligomeric arrangement of rhodopsin in ROS disc membranes [24]. The tetramer model consists of two symmetrical dimers with the intradimeric interface involving helices in transmembrane domains 4 and 5. A YFP molecule was added to the C-terminus of rhodopsin (Figure 7 A) to determine the relative position of YFP in the model of a rhodopsin tetramer (Figure 7 B). The positions of the YFP molecules are in a near-rhombic arrangement as observed in our spectral FRET studies. Thus, the tetramers of tagged opsin in CHO cells are predicted to be similar to the tetrameric unit of rhodopsin oligomers in native ROS disc membranes in photoreceptor cells.
Figure 7. Model of a rhodopsin tetramer.
YFP (orange, PDB: 2YFP) was added to the C-terminus of rhodopsin (blue) in a tetramer model based on AFM data (PDB: 1N3M). The structure of the fusion proteins was constructed and optimized using Yasara version 11.11 (YASARA Biosciences). (A) Side view of a single rhodopsin-YFP fusion protein. (B) Birds eye view of the cytoplasmic surface of a tetramer of rhodopsin-YFP fusion protein.
Limitations in the method precluded an explicit description of the higher order oligomers detected in CHO cells expressing high opsin concentrations in the membrane. The precise size and complement of the higher order oligomers could not be determined with accuracy because of ambiguities in the data obtained at higher expression levels and FRET pairs used. The meta-histogram analysis of data from cells expressing high concentrations of opsin points to an oligomer that is at least a dodecamer in a 3 × 4 array. This general configuration for higher order oligomers is consistent with the arrays of dimers observed in AFM images of native ROS disc membranes [10], albeit smaller in size.
Both CHO cells and photoreceptor cells exhibit a concentration dependence on the size of oligomers formed by the light receptor. AFM of single native ROS disc membranes shows that higher concentrations of rhodopsin in the membrane result in larger nanodomains made up of oligomeric receptor [21, 67]. Despite consistency in the structures of oligomeric forms of opsin in the membrane of CHO cells and in native ROS disc membranes, the difference in the concentration of the receptor in the membrane in the two systems results in different sizes of oligomers. The concentration range of 50 – 16,000 molecules/μm2 with an average of 1,350 molecules/μm2, investigated in CHO cells is well below the concentration of rhodopsin found in native ROS disc membranes. In native membranes, the concentration of rhodopsin can range from 8,000 – 35,000 molecules/μm2 with an average of 20,000 molecules/μm2 [67]. Based on the equilibria defined in the current study, monomer, dimers, and tetramers are predicted to be absent in ROS disc membranes at concentrations of rhodopsin found natively. Also predicted by the scheme presented above is that a distribution of different oligomeric sizes would be present in ROS disc membranes. The size of nanodomains of rhodopsin in ROS disc membranes, which reflects oligomeric size, exhibit a Log Gaussian distribution with the most frequent size being 335 molecules/μm2 [21, 67] (Supplemental Fig. 5), which corresponds to an oligomer with 24 rhodopsin molecules. Thus, the high concentrations of rhodopsin present in the membrane of photoreceptor cells, results in a predominant oligomeric species being a 24-mer whereas the comparatively modest concentration of opsin in CHO cells results in tetramers as the predominant oligomeric species.
Studies here using FRET spectrometry demonstrate that opsin in CHO cells and rhodopsin/opsin in native ROS disc membranes form similar oligomeric structures and have an intrinsic propensity to form oligomers according to the scheme presented in Figure 6. Despite these similarities on the basic mechanism of opsin oligomerization, differences may arise because the individual equilibrium constants may be influenced by external factors. Although both opsin and rhodopsin form similar types of oligomeric complexes in native ROS disc membranes [10], it is unclear how binding the inverse agonist 11-cis retinal affects the individual equilibrium constants. Light activation of rhodopsin is not predicted to disrupt oligomerization [44], however, we cannot rule out the possibility that it modulates the complement of oligomeric forms present in the membrane. Moreover, the ROS disc membranes contain a unique complement of lipids compared to plasma membranes [68, 69]. Further studies are required to determine the impact external factors such as ligands and lipid composition play on the equilibria that dictate the oligomeric status of opsin.
A common mechanism for the oligomerization of GPCRs
The large oligomers formed by rhodopsin in native ROS disc membranes may represent a unique case among GPCRs due to the unparalleled high expression levels of the light receptor in photoreceptor cells. The underlying principles for oligomerization, however, are likely conserved among GPCRs. The concentration-dependent oligomeric properties of opsin observed in the current study is consistent with observations made for other GPCR systems that were quantitatively assessed. A monomer-dimer equilibrium has been observed for the M1 muscarinic receptor and N-formyl peptide receptor [65, 70], a dimer-tetramer equilibrium has been observed for the M3 muscarinic receptor [59], a monomer-dimer-tetramer equilibrium has been observed for the β1 and β2 adrenergic receptors and 5-HT2C receptor [66, 71], and a dimer-tetramer-octamer equilibrium has been observed for the GABAB receptor [72]. As observed in the current study, oligomerization occurs in a concentration-dependent manner. The oligomerization of all GPCRs may therefore be described by a common phenomenon according to the scheme presented in Figure 6.
The intrinsic propensity to form different sizes of oligomeric complexes is likely different for different GPCRs and determined by the individual equilibrium constants. For instance, β1 adrenergic receptors are predominantly monomeric at concentrations of 0.15 – 0.3 molecules/μm2 whereas β2 adrenergic receptors are predominantly dimeric and GABAB receptors are almost exclusively dimeric at similar concentrations of receptor [66]. Thus, the equilibrium constants defining the oligomerization of different receptors and concentrations at which the receptors are expressed in the cell will dictate the oligomeric status of the receptor. In addition to the intrinsic propensity to form oligomers, binding ligand may modulate the equilibrium constants and change the oligomeric status of the receptor (e.g., [73]).
The question of whether rhodopsin and other GPCRs form oligomers continues to be a topic of debate [74, 75]. The unifying scheme presented may provide an explanation for why varying observations are made about the oligomeric status of GPCRs, even for the same receptor. Since the concentration of the receptor is rarely controlled and we do not yet fully understand all the variables that can modulate oligomerization equilibria, differences in receptor expression and yet to be determined modulatory factors may lead to apparently disparate observations on the oligomeric status of GPCRs. Moreover, while all GPCRs may have the intrinsic ability to form oligomers, even large oligomers, they may never do so at concentrations typically observed in cells membranes in nature. Thus, special attention must be given for studies carried out in heterologous expression systems where the receptor is often overexpressed because of the transfection procedure and non-native promoters used.
Functional implications of the observed concentration-dependent oligomerization of opsin
Investigating the oligomerization of opsin in CHO cells demonstrates that opsin can exist as smaller oligomers such as dimers and tetramers similarly to other GPCRs. The higher order oligomers of the light receptor observed in native photoreceptor cell membranes appear to be a result of the high concentrations of the receptor present in the ROS disc membrane. This concentration-dependent oligomerization predicts that reducing the expression of rhodopsin would alter the complement of oligomeric forms of rhodopsin found in native ROS disc membranes of photoreceptor cells. Surprisingly, this does not turn out to be the case. Studies on heterozygous rhodopsin knockout mice reveal that photoreceptor cells adjust the size of the ROS to maintain a constant concentration of rhodopsin in the ROS disc membrane, thereby maintaining a constant oligomeric size of rhodopsin when rhodopsin expression is diminished [67]. This suggests that the higher order oligomers are important for the role rhodopsin plays in photoreceptor cells and therefore the cell adapts to maintain an optimal concentration of the receptor in the membrane.
Communication between protomers resulting in asymmetric properties among receptor molecules within small oligomers such as dimers and tetramers has been documented for both rhodopsin and other GPCRs [25, 28, 76, 77]. For some GPCRs, this communication within a dimer or tetramer appears to play a central role in receptor signaling and underlies the efficacy of the system [78, 79]. Rhodopsin appears to have additional requirements compared to other GPCRs because of its function as a dim light receptor that must facilitate the exquisite sensitivity of rod photoreceptor cells, which cannot be accomplished by smaller oligomers. The higher order oligomers of rhodopsin can provide a platform to achieve the signaling efficiency necessary for the single-photon detection capabilities of rod photoreceptor cells [24, 37, 80, 81]. Thus, the high concentrations and supramolecular structure of rhodopsin appear to work in tandem to accomplish its role as a dim light receptor.
Concluding remarks
Overall, our studies in a heterologous expression system highlight the intrinsic property of opsin to form oligomers in live cells, the size of which increases in a concentration-dependent manner. The equilibrium scheme that describes the oligomerization of opsin appears to be generalizable to other GPCRs as well. All GPCRs appear to have the intrinsic propensity to oligomerize, however, whether they appear in nature as monomers, dimers, or larger oligomers is determined by the concentration of the receptor in the membrane and the equilibrium constants. Rhodopsin/opsin is no different than other GPCRs except in the high concentrations at which it is present in nature. Concentrations of 20,000 molecules/μm2 for rhodopsin is orders of magnitude higher than the concentrations of other typical GPCRs present in nature, which can range from 5 to 30 molecules/μm2 [8, 82].
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Joachim Goedhart (University of Amsterdam, Amsterdam, Netherlands) for providing the vectors pRSET-SYFP2 and pRSET-mTq.
FUNDING INFORMATION
The optical microspectroscopy imaging facility used for this research was developed with support of the National Science Foundation, Major Research Instrumentation Program (Grant No. PHY-1126386 awarded to V.R.). Cell culture preparation, imaging experiments, and FRET data analysis and interpretation were funded by the National Science Foundation, Physics of Living Systems Program (Grant No. PHY-1058470 awarded to V.R.). The generation of vectors for expression of tagged opsin in CHO cells and the expression and purification of fluorescent proteins were funded by grants from the National Institutes of Health (R01EY021731, P30EY011373, and T32EY024236) and Research to Prevent Blindness (Unrestricted Grant).
Abbreviations
- AFM
atomic force microscopy
- CHO
Chinese hamster ovary
- FRET
Förster resonance energy transfer
- GPCR
G protein-coupled receptor
- mTq
mTurquoise
- PBS
phosphate-buffered saline
- ROI
regions of interest
- ROS
rod outer segment
Footnotes
DECLARATIONS OF CONFLICT OF INTEREST
Dr. Raicu is a co-funder of Aurora Spectral Technologies LLC (AST), which commercializes OptiMiS technology used in this work. Dr. Stoneman was employed by AST in the past and occasionally consults for them. All the other authors declare no competing financial interests.
REFERENCES
- 1.Park PS-H, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferre S. The GPCR heterotetramer: challenging classical pharmacology. Trends Pharmacol Sci. 2015;36:145–152. doi: 10.1016/j.tips.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milligan G. The prevalence, maintenance, and relevance of G protein-coupled receptor oligomerization. Mol Pharmacol. 2013;84:158–169. doi: 10.1124/mol.113.084780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, Brabet I, Roux T, Bazin H, Bourrier E, Lamarque L, Breton C, Rives ML, Newman A, Javitch J, Trinquet E, Manning M, Pin JP, Mouillac B, Durroux T. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat Chem Biol. 2010;6:587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivero-Muller A, Chou YY, Ji I, Lajic S, Hanyaloglu AC, Jonas K, Rahman N, Ji TH, Huhtaniemi I. Rescue of defective G protein-coupled receptor function in vivo by intermolecular cooperation. Proc Natl Acad Sci U S A. 107:2319–2324. doi: 10.1073/pnas.0906695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldhoer M, Fong J, Jones RM, Lunzer MM, Sharma SK, Kostenis E, Portoghese PS, Whistler JL. A heterodimer-selective agonist shows in vivo relevance of G protein-coupled receptor dimers. Proc Natl Acad Sci U S A. 2005;102:9050–9055. doi: 10.1073/pnas.0501112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrick-Davis K, Grinde E, Lindsley T, Teitler M, Mancia F, Cowan A, Mazurkiewicz JE. Native serotonin 5-HT2C receptors are expressed as homodimers on the apical surface of choroid plexus epithelial cells. Mol Pharmacol. 2015;87:660–673. doi: 10.1124/mol.114.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 10.Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chabre M, le Maire M. Monomeric G-protein-coupled receptor as a functional unit. Biochemistry. 2005;44:9395–9403. doi: 10.1021/bi050720o. [DOI] [PubMed] [Google Scholar]
- 12.Chabre M, Cone R, Saibil H. Biophysics: is rhodopsin dimeric in native retinal rods? Nature. 2003;426:30–31. doi: 10.1038/426030b. [DOI] [PubMed] [Google Scholar]
- 13.Liebman PA, Entine G. Lateral diffusion of visual pigment in photorecptor disk membranes. Science. 1974;185:457–459. doi: 10.1126/science.185.4149.457. [DOI] [PubMed] [Google Scholar]
- 14.Cone RA. Rotational diffusion of rhodopsin in the visual receptor membrane. Nat.New Biol. 1972;236:39–43. doi: 10.1038/newbio236039a0. [DOI] [PubMed] [Google Scholar]
- 15.Poo M, Cone RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247:438–441. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- 16.Wey CL, Cone RA, Edidin MA. Lateral diffusion of rhodopsin in photoreceptor cells measured by fluorescence photobleaching and recovery. Biophys.J. 1981;33:225–232. doi: 10.1016/S0006-3495(81)84883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downer NW, Cone RA. Transient dichroism in photoreceptor membranes indicates that stable oligomers of rhodopsin do not form during excitation. Biophys J. 1985;47:277–284. doi: 10.1016/S0006-3495(85)83917-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakshit T, Senapati S, Sinha S, Whited AM, Park PS-H. Rhodopsin forms nanodomains in rod outer segment disc membranes of the cold-blooded Xenopus laevis. PLoS ONE. 2015;10:e0141114. doi: 10.1371/journal.pone.0141114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Govardovskii VI, Korenyak DA, Shukolyukov SA, Zueva LV. Lateral diffusion of rhodopsin in photoreceptor membrane: a reappraisal. Mol Vis. 2009;15:1717–1729. [PMC free article] [PubMed] [Google Scholar]
- 20.Najafi M, Haeri M, Knox BE, Schiesser WE, Calvert PD. Impact of signaling microcompartment geometry on GPCR dynamics in live retinal photoreceptors. J Gen Physiol. 2012;140:249–266. doi: 10.1085/jgp.201210818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whited AM, Park PS. Nanodomain organization of rhodopsin in native human and murine rod outer segment disc membranes. Biochim Biophys Acta. 2015;1848:26–34. doi: 10.1016/j.bbamem.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willardson BM, Pou B, Yoshida T, Bitensky MW. Cooperative binding of the retinal rod G-protein, transducin, to light-activated rhodopsin. J Biol Chem. 1993;268:6371–6382. [PubMed] [Google Scholar]
- 23.Wessling-Resnick M, Johnson GL. Transducin interactions with rhodopsin. Evidence for positive cooperative behavior. J Biol Chem. 1987;262:12444–12447. [PubMed] [Google Scholar]
- 24.Gunkel M, Schoneberg J, Alkhaldi W, Irsen S, Noe F, Kaupp UB, Al-Amoudi A. Higher-order architecture of rhodopsin in intact photoreceptors and its implication for phototransduction kinetics. Structure. 2015;23:628–638. doi: 10.1016/j.str.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013;27:1572–1584. doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jastrzebska B, Ringler P, Palczewski K, Engel A. The rhodopsin-transducin complex houses two distinct rhodopsin molecules. J Struct Biol. 2013;182:164–172. doi: 10.1016/j.jsb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sommer ME, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286:7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst OP, Gramse V, Kolbe M, Hofmann KP, Heck M. Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit. Proc Natl Acad Sci U S A. 2007;104:10859–10864. doi: 10.1073/pnas.0701967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee S, Huber T, Sakmar TP. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 31.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 32.Whorton MR, Jastrzebska B, Park PS, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc Natl Acad Sci U S A. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang Y, Zhou XE, Gao X, He Y, Liu W, Ishchenko A, Barty A, White TA, Yefanov O, Han GW, Xu Q, de Waal PW, Ke J, Tan MH, Zhang C, Moeller A, West GM, Pascal BD, Van Eps N, Caro LN, Vishnivetskiy SA, Lee RJ, Suino-Powell KM, Gu X, Pal K, Ma J, Zhi X, Boutet S, Williams GJ, Messerschmidt M, Gati C, Zatsepin NA, Wang D, James D, Basu S, Roy-Chowdhury S, Conrad CE, Coe J, Liu H, Lisova S, Kupitz C, Grotjohann I, Fromme R, Jiang Y, Tan M, Yang H, Li J, Wang M, Zheng Z, Li D, Howe N, Zhao Y, Standfuss J, Diederichs K, Dong Y, Potter CS, Carragher B, Caffrey M, Jiang H, Chapman HN, Spence JC, Fromme P, Weierstall U, Ernst OP, Katritch V, Gurevich VV, Griffin PR, Hubbell WL, Stevens RC, Cherezov V, Melcher K, Xu HE. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cangiano L, Dell’Orco D. Detecting single photons: a supramolecular matter? FEBS Lett. 2013;587:1–4. doi: 10.1016/j.febslet.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Dell’Orco D. A physiological role for the supramolecular organization of rhodopsin and transducin in rod photoreceptors. FEBS Lett. 2013;587:2060–2066. doi: 10.1016/j.febslet.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Raicu V, Singh DR. FRET spectrometry: a new tool for the determination of protein quaternary structure in living cells. Biophys J. 2013;105:1937–1945. doi: 10.1016/j.bpj.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borochov-Neori H, Fortes PA, Montal M. Rhodopsin in reconstituted phospholipid vesicles. 2. Rhodopsin-rhodopsin interactions detected by resonance energy transfer. Biochemistry. 1983;22:206–213. doi: 10.1021/bi00270a030. [DOI] [PubMed] [Google Scholar]
- 41.Borochov-Neori H, Montal M. Rhodopsin in reconstituted phospholipid vesicles. 1. Structural parameters and light-induced conformational changes detected by resonance energy transfer and fluorescence quenching. Biochemistry. 1983;22:197–205. doi: 10.1021/bi00270a029. [DOI] [PubMed] [Google Scholar]
- 42.Botelho AV, Huber T, Sakmar TP, Brown MF. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kota P, Reeves PJ, Rajbhandary UL, Khorana HG. Opsin is present as dimers in COS1 cells: Identification of amino acids at the dimeric interface. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0510982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansoor SE, Palczewski K, Farrens DL. Rhodopsin self-associates in asolectin liposomes. Proc Natl Acad Sci U S A. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajan RS, Kopito RR. Suppression of wild-type rhodopsin maturation by mutants linked to autosomal dominant retinitis pigmentosa. J Biol Chem. 2005;280:1284–1291. doi: 10.1074/jbc.M406448200. [DOI] [PubMed] [Google Scholar]
- 46.Miller LM, Gragg M, Kim TG, Park PS. Misfolded opsin mutants display elevated beta-sheet structure. FEBS Lett. 2015;589:3119–3125. doi: 10.1016/j.febslet.2015.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gragg M, Kim TG, Howell S, Park PS. Wild-type opsin does not aggregate with a misfolded opsin mutant. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbamem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comar WD, Schubert SM, Jastrzebska B, Palczewski K, Smith AW. Time-resolved fluorescence spectroscopy measures clustering and mobility of a G protein-coupled receptor opsin in live cell membranes. J Am Chem Soc. 2014;136:8342–8349. doi: 10.1021/ja501948w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raicu V, Stoneman MR, Fung R, Melnichuk M, Jansma DB, Pisterzi LF, Rath S, Fox M, Wells JW, Saldin DK. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nat Photon. 2009;3:107–113. [Google Scholar]
- 50.Mishra AK, Mavlyutov T, Singh DR, Biener G, Yang J, Oliver JA, Ruoho A, Raicu V. The sigma-1 receptors are present in monomeric and oligomeric forms in living cells in the presence and absence of ligands. Biochem J. 2015;466:263–271. doi: 10.1042/BJ20141321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.King C, Stoneman M, Raicu V, Hristova K. Fully quantified spectral imaging reveals in vivo membrane protein interactions. Integr Biol (Camb) 2016;8:216–229. doi: 10.1039/c5ib00202h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raicu V, Jansma DB, Miller RJ, Friesen JD. Protein interaction quantified in vivo by spectrally resolved fluorescence resonance energy transfer. Biochem J. 2005;385:265–277. doi: 10.1042/BJ20040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kremers GJ, Goedhart J, van Munster EB, Gadella TW., Jr. Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Forster radius. Biochemistry. 2006;45:6570–6580. doi: 10.1021/bi0516273. [DOI] [PubMed] [Google Scholar]
- 54.Goedhart J, van Weeren L, Hink MA, Vischer NO, Jalink K, Gadella TW., Jr. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat Methods. 2010;7:137–139. doi: 10.1038/nmeth.1415. [DOI] [PubMed] [Google Scholar]
- 55.Bindels DS, Goedhart J, Hink MA, van Weeren L, Joosen L, Gadella TW., Jr. Optimization of fluorescent proteins. Methods Mol Biol. 2014;1076:371–417. doi: 10.1007/978-1-62703-649-8_16. [DOI] [PubMed] [Google Scholar]
- 56.Biener G, Stoneman MR, Acbas G, Holz JD, Orlova M, Komarova L, Kuchin S, Raicu V. Development and experimental testing of an optical micro-spectroscopic technique incorporating true line-scan excitation. Int J Mol Sci. 2014;15:261–276. doi: 10.3390/ijms15010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zipfel WR, Williams RM, Christie R, Nikitin AY, Hyman BT, Webb WW. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A. 2003;100:7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh DR, Mohammad MM, Patowary S, Stoneman MR, Oliver JA, Movileanu L, Raicu V. Determination of the quaternary structure of a bacterial ATP-binding cassette (ABC) transporter in living cells. Integr Biol (Camb) 2013;5:312–323. doi: 10.1039/c2ib20218b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patowary S, Alvarez-Curto E, Xu TR, Holz JD, Oliver JA, Milligan G, Raicu V. The muscarinic M3 acetylcholine receptor exists as two differently sized complexes at the plasma membrane. Biochem J. 2013;452:303–312. doi: 10.1042/BJ20121902. [DOI] [PubMed] [Google Scholar]
- 60.Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 61.Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- 62.Singh DR, Raicu V. Comparison between whole distribution- and average-based approaches to the determination of fluorescence resonance energy transfer efficiency in ensembles of proteins in living cells. Biophys J. 2010;98:2127–2135. doi: 10.1016/j.bpj.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Del Piccolo N, Placone J, Hristova K. Effect of thanatophoric dysplasia type I mutations on FGFR3 dimerization. Biophys J. 2015;108:272–278. doi: 10.1016/j.bpj.2014.11.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raicu V. Efficiency of resonance energy transfer in homo-oligomeric complexes of proteins. J Biol Phys. 2007;33:109–127. doi: 10.1007/s10867-007-9046-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kasai RS, Suzuki KG, Prossnitz ER, Koyama-Honda I, Nakada C, Fujiwara TK, Kusumi A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calebiro D, Rieken F, Wagner J, Sungkaworn T, Zabel U, Borzi A, Cocucci E, Zurn A, Lohse MJ. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci U S A. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rakshit T, Park PS. Impact of reduced rhodopsin expression on the structure of rod outer segment disc membranes. Biochemistry. 2015;54:2885–2894. doi: 10.1021/acs.biochem.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boesze-Battaglia K, Schimmel R. Cell membrane lipid composition and distribution: implications for cell function and lessons learned from photoreceptors and platelets. J Exp Biol. 1997;200:2927–2936. doi: 10.1242/jeb.200.23.2927. [DOI] [PubMed] [Google Scholar]
- 69.Fliesler SJ, Anderson RE. Chemistry and metabolism of lipids in the vertebrate retina. Prog Lipid Res. 1983;22:79–131. doi: 10.1016/0163-7827(83)90004-8. [DOI] [PubMed] [Google Scholar]
- 70.Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JE, Lazareno S, Molloy JE, Birdsall NJ. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc Natl Acad Sci U S A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward RJ, Pediani JD, Godin AG, Milligan G. Regulation of Oligomeric Organization of the Serotonin 5-Hydroxytryptamine 2C (5-HT2C) Receptor Observed by Spatial Intensity Distribution Analysis. J Biol Chem. 2015;290:12844–12857. doi: 10.1074/jbc.M115.644724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Calebiro D, Nikolaev VO, Gagliani MC, de Filippis T, Dees C, Tacchetti C, Persani L, Lohse MJ. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol. 2009;7:e1000172. doi: 10.1371/journal.pbio.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pediani JD, Ward RJ, Godin AG, Marsango S, Milligan G. Dynamic Regulation of Quaternary Organization of the M1 Muscarinic Receptor by Subtype-Selective Antagonist Drugs. J Biol Chem. 2016 doi: 10.1074/jbc.M115.712562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lambert NA, Javitch JA. CrossTalk opposing view: Weighing the evidence for class A GPCR dimers, the jury is still out. J Physiol. 2014;592:2443–2445. doi: 10.1113/jphysiol.2014.272997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bouvier M, Hebert TE. CrossTalk proposal: Weighing the evidence for Class A GPCR dimers, the evidence favours dimers. J Physiol. 2014;592:2439–2441. doi: 10.1113/jphysiol.2014.272252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shivnaraine RV, Kelly B, Sankar KS, Redka DS, Han YR, Huang F, Elmslie G, Pinto D, Li Y, Rocheleau JV, Gradinaru CC, Ellis J, Wells JW. Allosteric modulation in monomers and oligomers of a G protein-coupled receptor. Elife. 2016;5 doi: 10.7554/eLife.11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han Y, Moreira IS, Urizar E, Weinstein H, Javitch JA. Allosteric communication between protomers of dopamine class A GPCR dimers modulates activation. Nat Chem Biol. 2009;5:688–695. doi: 10.1038/nchembio.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Redka DS, Heerklotz H, Wells JW. Efficacy as an intrinsic property of the M(2) muscarinic receptor in its tetrameric state. Biochemistry. 2013;52:7405–7427. doi: 10.1021/bi4003869. [DOI] [PubMed] [Google Scholar]
- 79.Redka DS, Morizumi T, Elmslie G, Paranthaman P, Shivnaraine RV, Ellis J, Ernst OP, Wells JW. Coupling of g proteins to reconstituted monomers and tetramers of the M2 muscarinic receptor. J Biol Chem. 2014;289:24347–24365. doi: 10.1074/jbc.M114.559294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dell’Orco D, Koch KW. A dynamic scaffolding mechanism for rhodopsin and transducin interaction in vertebrate vision. Biochem J. 2011;440:263–271. doi: 10.1042/BJ20110871. [DOI] [PubMed] [Google Scholar]
- 81.Schoneberg J, Heck M, Hofmann KP, Noe F. Explicit spatiotemporal simulation of receptor-g protein coupling in rod cell disk membranes. Biophys J. 2014;107:1042–1053. doi: 10.1016/j.bpj.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hegener O, Prenner L, Runkel F, Baader SL, Kappler J, Haberlein H. Dynamics of beta2-adrenergic receptor-ligand complexes on living cells. Biochemistry. 2004;43:6190–6199. doi: 10.1021/bi035928t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.