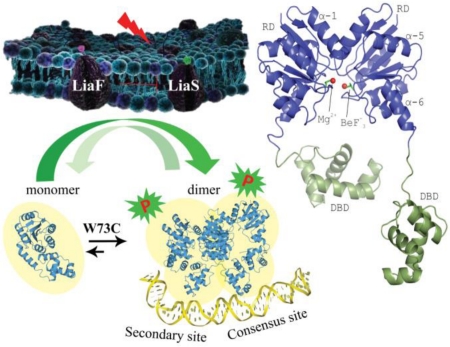
Abstract
The cyclic antimicrobial lipopeptide daptomycin triggers the LiaFSR membrane stress response pathway in enterococci and many other Gram-positive organisms. LiaR is the response regulator that, upon phosphorylation, binds in a sequence specific manner to DNA to regulate transcription in response to membrane stress. In clinical settings, nonsusceptibility to daptomycin by Enterococcus faecium is correlated frequently with a mutation in LiaR of Trp73 to Cys (LiaRW73C). We have determined the structure of the activated Enterococcus faecium LiaR protein at 3.2 Å resolution and, in combination with solution studies, show that activation of LiaR induces formation of a LiaR dimer that increases LiaR affinity at least 40-fold for the extended regulatory regions upstream of the liaFSR and liaXYZ operons. In vitro, LiaRW73C induces phosphorylation-independent dimerization of LiaR and provides a biochemical basis for non-susceptibility to daptomycin by upregulation of the LiaFSR regulon. A comparison of the Enterococcus faecalis LiaR, Enterococcus faecium LiaR and the LiaR homolog from Staphylococcus aureus (VraR) and the mutations associated with daptomycin resistance suggest that physicochemical properties such as oligomerization state and DNA specificity, though tuned to the biology of each organism, share some features that could be targeted for new antimicrobials.
Keywords: LiaR, E. faecium, Response regulator, Daptomycin resistance, X-ray
Graphical abstract
Introduction
The continuing rise and persistence of multi-drug resistant pathogens poses substantial burdens on the clinical community. Vancomycin-resistant enterococci (VRE) are a leading cause of Hospital-Associated Infections (HAI) and are among the most recalcitrant nosocomial pathogens. VRE often develop resistance to a variety of antibiotics making the treatment of infections challenging [1]. In particular, E. faecium is the most multidrug-resistant of the enterococcal species affecting critically ill patients. Indeed, the Infectious Diseases Society of America (IDSA) has included E. faecium as one of the “no-ESKAPE” pathogens [2], against which new drugs are urgently needed. With therapeutic options dwindling, the cyclic lipopeptide daptomycin (DAP) has become one of the last remaining bactericidal options for the treatment of multi-drug resistant enterococcal infections. Unfortunately, the clinical efficacy of DAP is being challenged by the development of resistance which tends to emerge during therapy [3-5].
Daptomycin is a cyclic lipopeptide that targets the cell membrane of Gram-positive bacteria in a calcium-dependent manner becoming a de facto cationic antimicrobial peptide [6]. Recent studies have suggested that the association of the calcium-daptomycin complex to the bacterial cell membrane depends on the presence of phosphatidylglycerol-rich regions in the outer leaflet of the cell membrane with a marked preference for the septa of dividing cells [7]. Upon insertion into the membrane, daptomycin disrupts membrane homeostasis leading to rapid cell death [7-9].
Studies using Bacillus subtilis, enterococci and methicillin-resistant Staphylococcus aureus have suggested that daptomycin disrupts membrane potential via leakage of small ions such as intracellular potassium resulting in alterations in cell physiology and cell division [7, 10-12]. The biochemical mechanism for DAP resistance is still under investigation but it is apparent that there are distinct phenotypic differences associated with the emergence of daptomycin non-susceptibility between E. faecium, E. faecalis and S. aureus [6, 7, 13]. To date, there are at least two distinct biochemical mechanisms for cells to reduce their susceptibility to daptomycin. In S. aureus and B. subtilis changes in the cell envelope appear to increase the overall positive charge of the cell surface, repelling the antibiotic from the membrane target (‘repulsion’) [6, 14, 15]. In contrast, in E. faecalis there is evidence for a ‘diversion’ mechanism in which DAP is diverted away from the division septa [6, 16, 17]. In the case of E. faecium, higher levels of daptomycin resistance appear to be associated more strongly with the repulsion mechanism while strains of E. faecium that are susceptible to lower concentrations seem to be able to divert the antibiotic molecule, suggesting that molecular strategies for resistance between species is more complex than may have been anticipated [16, 17]. One of the most important mediators of daptomycin resistance in enterococci is the three-component LiaFSR signaling pathway [6, 16, 18]. Activation of this system triggers an adaptive response to the antibiotic attack by leading to downstream changes in the transcription of several operons, such as liaFSR and liaXYZ to alter membrane homeostasis resulting in resistance to DAP and antimicrobial peptides [18, 19]. The liaFSR operon encodes LiaS, a membrane bound kinase; LiaF, a protein suggested to inhibit LiaS phosphorylation activity in the absence of membrane stress; and LiaR, a response regulator that is phosphorylated by LiaS to alter transcription from the LiaR regulon. Deletion of liaR in both E. faecalis and E. faecium abolishes the adaptive response and restores susceptibility to daptomycin and antimicrobial peptides, supporting the pivotal role of LiaR in the mechanism of resistance [16]. LiaR is comprised of two functional domains: a receiver domain (residues 1-139) that can be phosphorylated and a DNA binding domain (residues 140-210) with a Helix-Turn-Helix motif that recognizes specific regulatory DNA sequences via formation of a DNA binding domain dimer. Recently, we demonstrated that activation of LiaR in E. faecalis (Efc LiaR) promotes formation of a LiaR tetramer that allows LiaR to recognize complex upstream regulatory regions that extend beyond the predicted consensus sequence. Additionally, we showed that an adaptive mutation isolated in response to DAP exposure (Efc LiaRD191N) “hyperactivates” the liaFSR system leading to resistance [19]. Interestingly, emergence of daptomycin resistance in clinical isolates of E. faecium is highly associated with a Trp73 to Cys substitution in LiaR (Efm LiaRW73C) [17]. Recently published whole genome sequence analysis of 19 E. faecium clinical isolates with DAP MICs between 3 to 32 μg/ml indicate that a Trp73 to Cys substitution in response regulator LiaR was the most frequent change in epidemiologically unrelated clinical isolates of E. faecium with DAP MICs ≥ 3 μg/ml. Efm LiaRW73C is typically found with a second mutation in the kinase LiaST120A suggesting a potential epistatic relationship [17].
A critical difference between the Efc LiaRD191N and Efm LiaRW73C is that the “triggering” mutations associated with daptomycin non-susceptibility are located in different domains of the protein, suggesting that the mechanism of activation associated with LiaR substitutions in DAP resistant clinical strains may differ between E. faecalis and E. faecium.
Using a combination of biophysical and structural approaches we were able to compare and contrast the molecular details of the adaptive mutations that lead to decreased daptomycin susceptibility in E. faecalis and E. faecium. We show that in the absence of phosphorylation, Efm LiaR exists predominately as a monomer and that activation with the chemical phosphomimic BeF3− strongly favors formation of a dimer with high affinity for extended DNA recognition sequences. This finding contrasts with our earlier study of Efc LiaR in which the protein undergoes a dimer to tetramer transition upon phosphorylation [19]. Taken together, our results show that while there are similarities between E. faecium LiaR, E. faecalis LiaR and the LiaR homolog VraR of S. aureus, there are substantial and consequential physicochemical differences. New small molecules that might target LiaR to suppress the membrane stress response of these organisms and thereby extend the efficacy of DAP or other membrane damaging agents should be thoughtfully directed at the common molecular attributes of LiaR and the LiaR homolog VraR to have the broadest applicability.
Results
Beryllofluoride (BeF3−) stabilizes E. faecium LiaR as a dimer
Phosphorylation-dependent oligomerization and consequent activation of response regulators can vary widely between organisms. Based on a comparison of the E. faecium LiaR sequence with that of E. faecalis LiaR and other response regulators from NarL/FixJ subfamily, such as S. aureus VraR, B. subtilis LiaR, E. coli NarL, FixJ, Asp-54 was identified as a potential site for phosphorylation [19, 20].
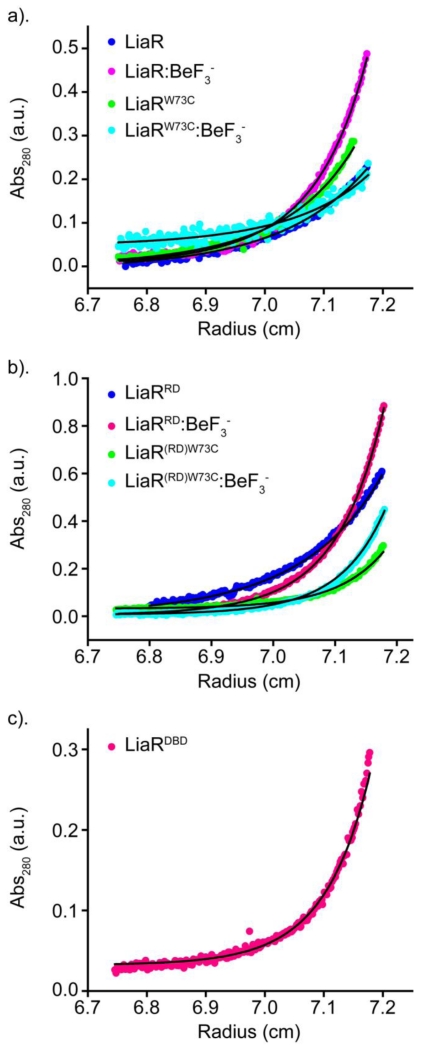
To elucidate the changes in quaternary structure associated with phosphorylation, we used Beryllium trifluoride (BeF3−) binding to LiaR as a functional mimic for phosphorylation [21]. BeF3−, made in situ, is a simple and general method to reproduce a stable phosphorylated-like state [22]. Using sedimentation equilibrium analytical ultracentrifugation (SEQ), we determined the oligomerization state of full length Efm LiaR as well as the isolated receiver domain (residues 1-139 and referred to hereafter as LiaRRD) with and without BeF3− and the DNA binding domain (residues 140-210 and referred to hereafter as LiaRDBD) (Figure 1, S1). Interestingly, the oligomerization of the full-length Efm LiaR differs both qualitatively and quantitatively from what we reported previously for E. faecalis LiaR [19]. Our analytical ultracentrifugation studies suggest that inactivated Efm LiaR exists nearly exclusively as monomer in solution with a dissociation constant of Kd >1600 μM and that addition of BeF3− promotes dimer formation (Kd = 15 μM) (Table 1). We estimated that the propensity towards dimerization is increased at least 2 orders of magnitude by the formation of the BeF3−:LiaR complex. Activation of Efm LiaR by BeF3− produces a monomer↔dimer transition with no observable higher order oligomers like tetramers. To confirm these results we also performed a sedimentation velocity analytical ultracentrifugation experiment with solutions containing 50 μM Efm LiaR or 5 μM Efm LiaR respectively. The sedimentation velocity data was interpreted using a local continuous distribution model searching for species corresponding to 0.5 to 15 Svedbergs. Using this model, we observe two main species with sedimentation coefficients of 1.4 S and 2.0 S respectively. These two species have apparent molecular weights of 23.3 kDa and 40.9 kDa respectively, suggesting the two peaks represent monomeric and dimeric populations of LiaR (Figure S2). By integrating the peaks in the c(S) distribution separately we estimate the fraction of LiaR that is present as a homodimer in the 5 μM and 50 μM samples to be 0% and 11% respectively, providing further evidence that unphosphorylated Efm LiaR has only a very weak propensity to dimerize.
Figure 1. Beryllofluoride (BeF3−) promotes formation of Efm LiaR dimers.
Sedimentation equilibrium analytical ultracentrifugation (AUC) profiles for: (a) full length Efm LiaR (dark blue); Efm LiaR with BeF3− (magenta); adaptive mutant Efm LiaRW73C (green); and adaptive mutant Efm LiaRW73C with BeF3− (cyan). (b) AUC profiles for Efm LiaR(RD) (blue); Efm LiaR(RD) with BeF3− (magenta); Efm LiaR(RD)W73C (green); and Efm LiaR(RD)W73C with BeF3− (cyan): (c) AUC profile for the DNA binding domain of Efm LiaR (LiaR(DBD)) alone (magenta). For each protein, the SEQ profiles were globally fitted to a monomer only or monomer-dimer equilibrium model.
Table 1.
Dimerization affinities of E. faecium LiaR and variants in the absence and presence of BeF3−.
| Protein name | BeF3− | Dimerization Kd (μM) | 95% confidence intervals (μM) |
|---|---|---|---|
| LiaR | − | No dimer observed | Kd> 1600 |
| LiaR | + | 15 | 12 < Kd < 19 |
| LiaRW73C | − | 182 | 119 <Kd < 311 |
| LiaRW73C | + | 236 | 82 < Kd < 1000 |
| LiaRRD | − | 676 | 497 < Kd < 986 |
| LiaRRD | + | 3.4 | 3.2 < Kd < 4.2 |
| LiaRRD(W73C) | − | 94 | 70 < Kd < 128 |
| LiaRRD(W73C) | + | 0.8 | 0.4 < Kd < 1.4 |
| LiaRDBD | − | 0.4 | Kd < 2.0 |
To further characterize the behavior of Efm LiaR in solution, we also analyzed the contribution of isolated LiaR domains to oligomerization in the presence and absence of BeF3−. Sedimentation equilibrium analysis of the domains in the absence of BeF3− showed a weak but detectable propensity for dimerization by the Efm LiaRRD (Kd = 676 μM) and a much stronger dimerization propensity for the Efm LiaRDBD (Kd = 0.4 μM). Addition of BeF3− increased the extent of dimerization formation of Efm LiaRRD by a remarkable 198-fold to a Kd of 3.4 μM. These analyses indicate that while DNA binding domain is capable of homodimerization (Kd = 0.4 μM), the oligomerization of the Efm LiaRRD is strongly dependent on phosphorylation, consistent with a regulatory role in vivo.
LiaRW73C from a daptomycin-resistant clinical isolate of E. faecium promotes phosphorylation-independent dimerization of the LiaR protein
Having established that phosphorylation controls a monomer↔dimer transition for the wild-type Efm LiaR, we sought to investigate whether the substitution of Trp73 to Cys in LiaR (the most common LiaR change associated with daptomycin resistance in E. faecium clinical isolates) [17, 23] has a direct effect on LiaR oligomerization. Using sedimentation equilibrium experiments of the full length Efm LiaRW73C and receiver domain alone (Efm LiaR(RD)W73C) in the presence and absence of BeF3−, we showed that the W73C substitution promotes phosphorylation-independent dimerization of Efm LiaR. Indeed, as mentioned above, in the absence of BeF3− we were unable to measure any significant dimerization of the full length Efm LiaR. In contrast, Efm LiaRW73C showed a readily detectable level of dimerization (Kd=182 μM), suggesting that the Trp73 to Cys substitution strongly stimulates phosphorylation-independent dimerization. This phenomenon was further confirmed by our studies on Efm LiaR(RD)W73C which showed a 7-fold higher propensity for dimerization compared to the wild type Efm LiaRRD. The higher propensity to form a dimer was also observed by sedimentation velocity analytical ultracentrifugation (Figure S3). Using a local continuous c(S) distribution model we could interpret the relative size of the populations of monomer, dimer and larger oligomeric species for the 50 μM and 5 μM samples tested. In agreement with the sedimentation equilibrium results, we observed a greater proportion of the protein in the homodimer state for the W73C mutant of LiaR, relative to the wild type protein. For the 50 μM protein sample 59% of the total protein had a sedimentation coefficient of 1.8 S corresponding to monomeric LiaR (apparent MW of 27 kDa). 34% of the total protein is the homodimer form with a sedimentation coefficient of 2.8 S and apparent MW of 48 kDa. We additionally observed a small fraction of the protein at larger sedimentation coefficients of 4.2 S, 5.5 S and 6.6 S and with apparent molecular weights of 90 kDa, 134 kDa, or 178 kDa respectively that total 8% of the protein population in the 50 μM. These apparent molecular weights correspond to tetramer, hexamer and octamer species of LiaR suggesting the W73C mutation can promote the formation of larger species. As we did not observe these species by size exclusion chromatography it is unclear if these are non specific aggregation effects or a biologically relevant species that might exist in E. faecium. Gel filtration studies further corroborate our finding that Efm LiaRW73C promotes formation of a phosphorylation independent dimer (Supplementary Information, S4).
Interestingly, and unlike the wild type protein, we could not detect any further increase in dimerization of the full length Efm LiaRW73C in the presence of BeF3−. Instead, the best fit for dimerization was with a Kd of 236 μM, which is similar to the dimerization constant observed for Efm LiaRW73C in the absence of BeF3−. This finding is consistent with a model in which the mutation of Trp-73 to cysteine has effectively stimulated the formation of the activated LiaR dimer to the same extent as phosphorylation. It is also possible that the increased propensity for dimerization leads to an increased efficiency of phosphorylation by LiaS. Thermal denaturation studies show that Efm LiaRW73C is 4.7 °C less stable than wild type consistent with a model in which the adaptive mutation has potentially destabilized contacts between the receiver and DNA binding domains (Supplementary Information, S5).
When we tested the effect of BeF3− on the receiver domain alone (Efm LiaR(RD)W73C), we observed a marked increase in dimerization. Indeed, the Kd of dimerization for Efm LiaR(RD)W73C increased from 94 to 0.8 μM. Our results suggest that the DNA binding domain may also have a role in determining the extent of dimerization induced by either phosphorylation or the presence of BeF3−. Thus, in the absence of the DNA binding domain, the extent of dimerization is further enhanced by BeF3− in a manner that is not observed for the intact protein.
The crystal structure of full length Efm LiaR is a dimer poised to make interactions with its target DNA
We determined the X-ray crystal structure of Efm LiaR in the beryllofluoride-activated state to reveal information about the conformational changes induced by phosphorylation. Response regulators have been classified into several common types including the NarL-type that includes a common helix-turn-helix (HTH) motif, OmpR-type that includes a winged HTH motif, Spo0A-type, and Fis-like [24]. The structure of Efm R494 LiaR suggests that this protein belongs to the NarL/FixJ subfamily of proteins [20, 24].
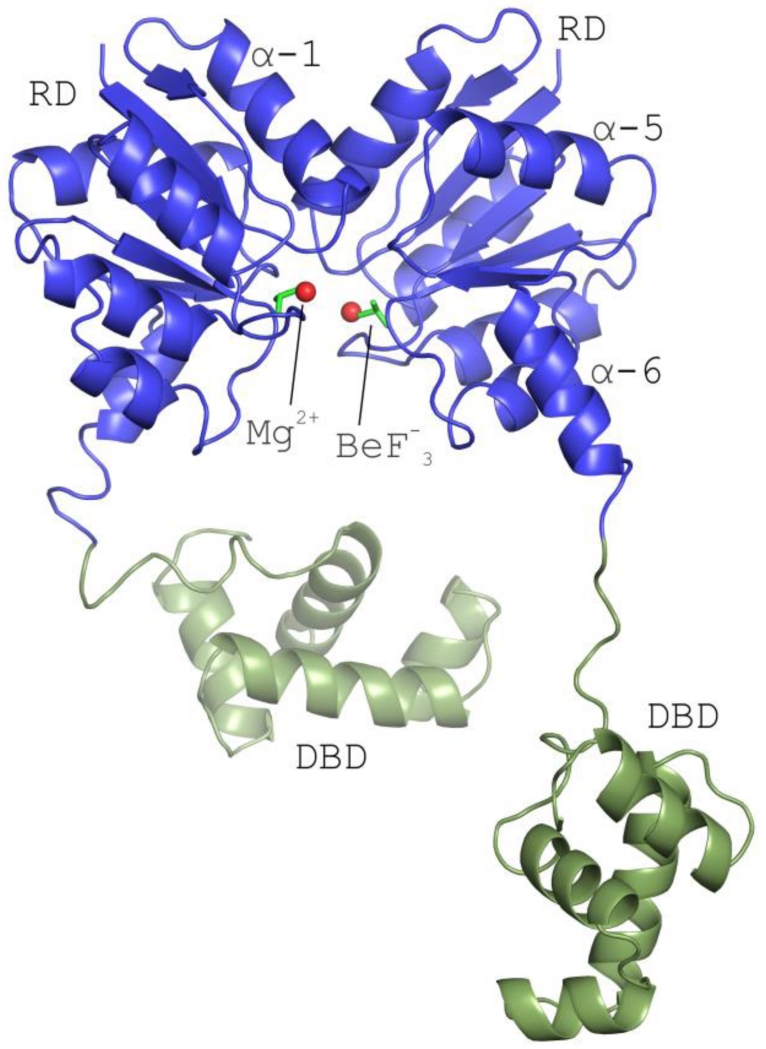
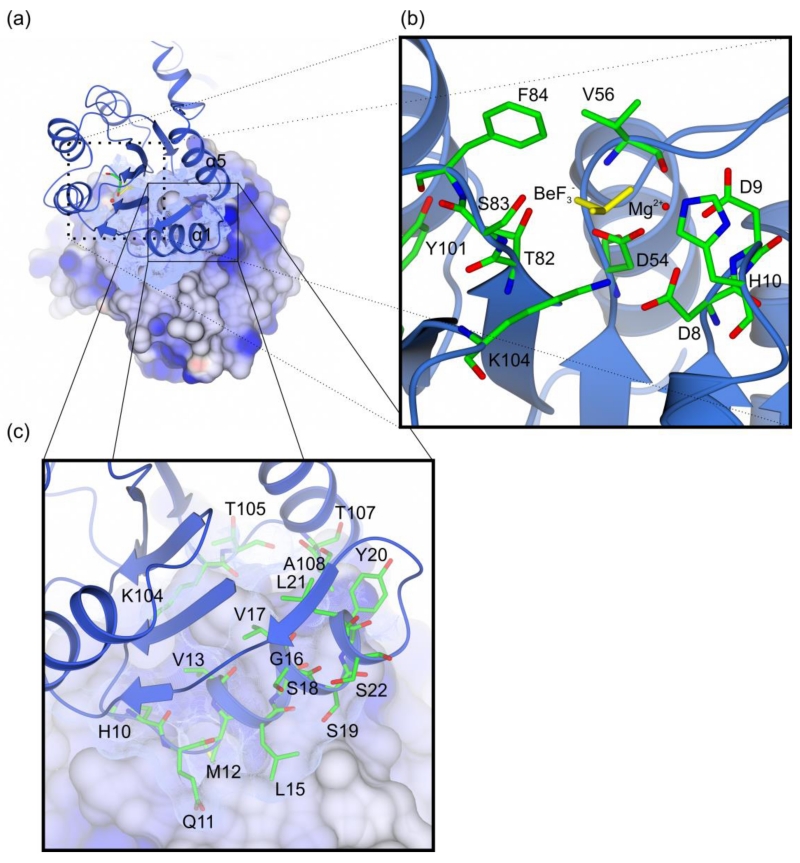
The Efm LiaR:BeF3− structure contains four protomers in the asymmetric unit each with a BeF3− bound to the predicted phosphorylation site at Asp54 (Figure 2, S6). Essentially, a pair of activated dimers are observed in the asymmetric unit. Based on homology to other NarL-type response regulators residue Thr-82 and aromatic residue Tyr-101 would be predicted to play a direct role in the activation mechanism through their hydroxyl groups [25]. In the Efm LiaR:BeF3− structure residues Thr82 and Tyr101 are oriented toward the predicted phosphorylation site, adopting inward positions consistent with BeF3− stimulated formation of the active phosphorylated state (Figure 3). Within the active site, BeF3− is stabilized by a single Mg2+ ion and the backbone amide of Val56. Previous studies have suggested that the residue equivalent to Val56 is one of the structural determinants that influence the auto-dephosphorylation rate of response regulator proteins [26, 27]. BeF3− is also in a position to make hydrogen bonds to Thr82 and Ser83 and a salt bridge to Lys104 (Figure 3a,b). As shown in Figure 2, each protomer has a canonical regulatory domain fold with the (ßα)5 arrangement and Helix-Turn-Helix DNA binding domain seen in other NarL-like proteins [20, 24, 25].
Figure 2. The structure of full-length Efm LiaR in complex with BeF3−/Mg2+ reveals an activated protein poised to bind DNA.
Structural overview of the full length Efm LiaR activated with the BeF3− (PDB ID: 5HEV). For clarity only one of the two pairs of LiaR dimers present in the asymmetric unit are displayed. Receiver domain (blue) showing a molecule of BeF3− bound proximal to the predicted phosphorylation site (Asp54). The LiaR dimerization surface is made up of the α-1 and α-5 helices. Mg2+ and BeF3− are shown as spheres (red) and sticks (dark green), respectively. Helix α-6 makes up part of the flexible linker joining the receiver and DNA-binding domains (light green). The flexible linker allows the DNA binding domains to have a substantial amount of conformational flexibility to engage and bind to DNA.
Figure 3. Overview of the LiaR receiver domain dimer.
(a) The receiver domain forms a dimer through a hydrophobic interface of 692 Å2 that juxtaposes the α1 and α5 helices of two LiaR protomers; (b) LiaR active site in the presence of BeF3− (yellow) and Mg2+ (red). The residues Thr83 and Tyr102, adopt a characteristic active conformation, toward the predicted phosphorylation site at Asp54. BeF3− is also in a position to make hydrogen bonds to Thr82 and Ser83 and a salt bridge to Lys104; (c) Key residues are shown that comprise the dimerization interface in a manner comparable to that observed for VraR. Met12 from one protomer docks into a hydrophobic pocket within the dimerization interface of the second LiaR protomer.
Helix α-6 followed by short loop creates a flexible linker between the receiver and DNA-binding domains. Interestingly, the arrangement of the DNA binding domains is substantially different from the activated form of S. aureus VraR. Presumably this reflects the mobility of the DNA binding domains in both structures when DNA is not present. Our BeF3− activated structure is consistent with full-length Efm LiaR poised in a configuration to interact with DNA. Two of the four DNA binding domains observed in our structure are extended well away from the receiver domain dimer and illustrate the conformational flexibility of the linker regions that join them to the domain (Figure 2).
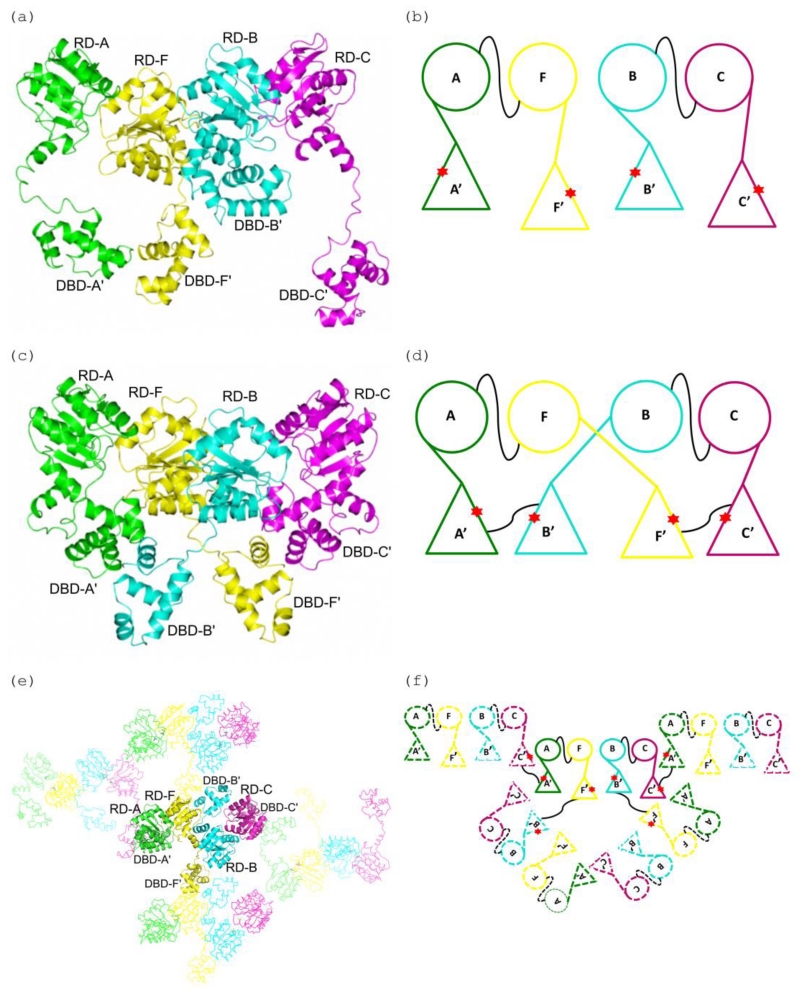
In the Efm LiaR:BeF3− co-structure, two receiver domain dimers (indicated as chains A-F and B-C respectively) are observed in the asymmetric unit (Figure 4a-b). The major interaction surface between neighboring receiver domains buries ~1034 Å2 of solvent accessible surface area through the α4-ß5-α5 interfaces. The protein-protein interactions of the two receiver domains involves 3 potential salt bridges and 3 hydrogen bonds that were also observed in the S. aureus response regulator VraR [19] and are presumed to stabilize the quaternary structure. Like Efm LiaR, S. aureus VraR crystallized as two pairs of receiver domain dimers (Figure 4c-d). In the case of VraR, the DNA binding domains formed additional dimer interfaces but not between the two proteins that comprise the receiver domain dimer but rather as a domain swap (Figure 4d). In the Efm LiaR:BeF3− co-structure, each DNA binding domain also formed additional dimer interfaces (Figure 4e-f) but with DNA binding domains contributed from symmetry related molecules and not to the one that is contributed by the proximal LiaR protomer that comprises the LiaR:LiaR dimer (a domain swap). Thus, the domain swap in the activated VraR tetramer structure and Efm LiaR:BeF3− co-structure are very different and may reflect flexibility of linker regions that join the DNA binding domains to the receiver domain[28]. The arrangement of the Efm LiaR:BeF3− DNA binding domains is likely to be a crystal packing artifact but shows that DNA binding domains can explore a very large volume, a feature that may be important to binding the large extended DNA at the promoter.
Figure 4. A comparison of the DNA domain swapping observed in the E. faecium LiaR and S. aureus VraR structures.
(a), (b) Cartoon depiction of the Efm LiaR structure (PDB ID: 5HEV) showing the receiver domain (RD) and DNA-binding (DBD) domains arrangements in the asymmetric unit. In the presence of BeF3− the crystal contained four protomers of LiaR in the asymmetric unit: A (green), F (yellow), B (cyan) and C (magenta). The star (red) identifies the dimerization interfaces of the DNA-binding domains. Two LiaR protomers (A-F and B-C) form a dimer through dimerization of their receiver domains. The thin black line connecting the monomers identifies the dimerization interfaces of the RDs of two protomers (A-F and B-C).
(c), (d) Cartoon representation of the domain swapped arrangement in the S. aureus VraR activated structure (PDB ID: 4IF4). Two VraR protomers form a dimer through dimerization of their receiver domains (A-F and B-C), while the DNA binding domains form homodimers with a DNA binding domain contributed from a different receiver domain (A’-B’ and F’-C’) resulting in a swapping of domains. The thin black line connecting the monomers identifies the dimerization interfaces.
(e), (f) Cartoon representation of the domain swapping arrangement for the Efm LiaR activated structure (PDB ID: 5HEV). Unlike VraR, the Efm LiaR DNA binding domains make dimer contacts to symmetry related DNA binding domains contributed by other receiver domain pairs. While this arrangement of swapped domains might be an artifact of crystallization it also demonstrates the flexibility of the DNA binding domains in the activated state. The thin black line connecting the monomers identifies the dimerization interfaces.
The phosphorylation-triggered dimerization surface of E. faecium LiaR is similar to that of S. aureus VraR
The receiver domain of Efm LiaR forms a dimer that associates through a hydrophobic interface comprised of α1-α5 that buries 692 Å2 (Figure 3a,c). This dimerization interface, lined by hydrophobic amino acids Val13, Val17, Ser19, Tyr20, Lys104, Ala108 is similar to those found in the structures of B. subtilis DesR [27] and VraR [28] suggesting that these interfacial residues are well conserved among members of NarL subfamily.
Earlier studies have shown that phosphorylation promotes dimerization through different surfaces among different response regulators. While members of the PhoB/OmpR family [29, 30] and FixJ [28, 31] utilize the hydrophobic surface of the α4-β5-α5 interface for phosphorylation triggered dimerization, the NarL transcription factor from E. coli [32], the cell-density-responsive transcription factor LuxR from S. aureus, S. aureus VraR [28] and B. subtilis DesR [27] employ a novel α1-α5 contact surface. An analogous involvement of helix α1 in intermolecular contacts has also been established in the M. tuberculosis NarL [33] and E.coli UhpA [34].
In the VraR structure it has been suggested that movement of α-6 is part of the activation mechanism in which the angle of the helix α-6 changes by 35° in response to phosphorylation [28]. To determine whether the interface for the α-6 helices packing against each other is conserved between VraR and Efm LiaR after activation, we superimposed the receiver domain structures of the two proteins. Based on this analysis, we concluded that the dimerization interaction is similar between S. aureus VraR and E. faecium LiaR with an overall root mean square deviation (r.m.s.d.) of 1.14 Å. [35].
Efm LiaR DNA recognition motifs upstream of the putative promoters for both the Efm liaFSR and the liaXYZ operons are comparable in size and organization to those of E. faecalis
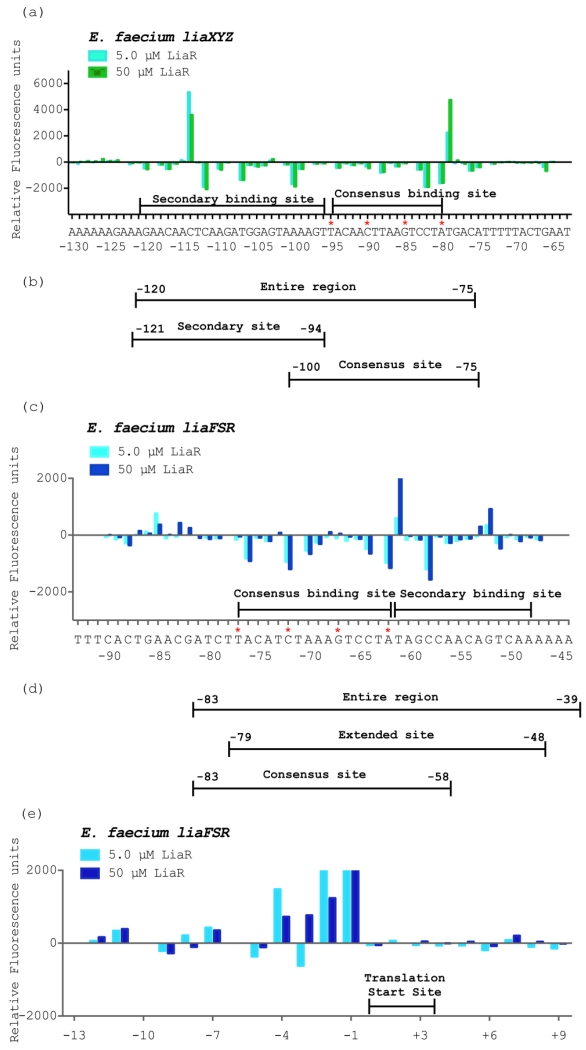
Our previous studies revealed that Efc LiaR recognizes an extended stretch of DNA that extends beyond the proposed canonical consensus sequence T(X)4C(X)4G(X)4A, suggesting a more complex level of regulatory control for target operons [19]. We previously showed that in E. faecalis such consensus sequences are present upstream of the liaFSR and liaXYZ operons [19]. Inspection of the 5’ region of the Efm liaXYZ and liaFSR operons revealed three and four potential consensus sites, respectively. We used DNA Footprint Analysis by Automated Capillary Electrophoresis (DFACE) to identify the DNA sequences protected by activated Efm LiaR. Our initial studies indicated that the unphosphorylated Efm LiaR protein had no measurable affinity for the DNA upstream of either operon. In contrast, when Efm LiaR is activated by formation of the Efm LiaR:BeF3− complex, protection of DNA across extended regions of DNA upstream of the liaXYZ and liaFSR operons could be identified readily. We mapped the LiaR binding region from −292 to +65 for liaXYZ and −281 to +98 for liaFSR. The BeF3− activated protein revealed a concentration dependent region of protection for both operons with no observable protection at 0.5 μM, slight protection at 2.5 μM, (data not shown) and significant protection at 5.0 μM and 50 μM protein (Figure 5a, c). The 50 μM Efm LiaR concentration had a slight increase in protection and cleavage at most bases as compared to 5.0 μM. Interestingly, Efm LiaR protects only one canonical binding motif of T(X)4C(X)4G(X)4A for both operons. As shown in Figure 5, the consensus binding regions starting at positions −80 and −62 respectively for liaXYZ and liaFSR are protected by the Efm LiaR:BeF3− complex. The other potential consensus binding sites did not show protection (data not shown).
Figure 5. Efm LiaR recognizes complex upstream regulatory regions that extend beyond the predicted consensus sequence.
DNase I Footprinting followed by Automated Capillary Electrophoresis (DFACE) was used to establish the Efm LiaR binding footprint within the upstream regions of the liaXYZ (−292 to +65) and liaFSR (−281 to +98) operons of E. faecium R494. Protection studies were performed at 0.5, 5 and 50 μM LiaR activated with the BeF3−. DNaseI digestion patterns are shown as histograms (a, c, e) where negative changes in relative fluorescence units indicate regions of protection and positive changes indicate hypersensitivity. All DNaseI sensitivity data are relative to a no protein negative control.
(a) Efm LiaR binding to the promoter region of the liaXYZ operon. Nucleotide positions refer to the region of LiaR binding (−120 to −75) on the DNA relative to the translation start site of LiaX (cyan, 5 μM LiaR; green, 50 μM LiaR).
(b/d) Oligonucleotides used for DNA binding studies (Table 2).
(c) Efm LiaR binding to the promoter region of the liaFRS operon. Nucleotide positions refer to the region of LiaR binding (−77 to −48) on the DNA relative to the translation start site of LiaF (cyan, 5 μM LiaR; blue, 50 μM LiaR).
(e) Superimposed electropherograms showing a strong and consistent hypersensitive site at positions −4 to −1 (AGCG) relative to the translation start site of LiaF in the presence of Efm LiaR (cyan, 5 μM LiaR; blue, 50 μM LiaR).
While both liaFSR and liaXYZ show binding at a single consensus binding sequence, the overall organization of the protected regions with respect to each other is very different. Based on sequence homology to other response regulators containing a canonical helix-turn-helix DNA binding motif, the active species of Efm LiaR would be expected to dimerize and have site size of ~21 nucleotides. In contrast, the contiguous protection site for the Efm liaXYZ promoter extends from position −120 to −75 (about 45 bp) and includes the predicted consensus region (−95 to −80) and nucleotides from −96 to −120. We refer to this additional region outside the consensus sequence as the “secondary site” and, taken together, make up the extended DNA recognition element recognized by LiaR family members (Figure 5a). In this case the extended protected sequence over a site size of 45 base pairs suggests binding of two dimers or a single tetramer.
For the liaFSR promoter only about 29 (−77 to −48) nucleotides are protected (Figure 5c). The size of the DNase footprint is comparable to that observed for the Efc liaFSR operon. As we observed for the Efc liaFSR operon, in addition to the predicted consensus site (−77 to −62), there is a distinct protection for a secondary site from −63 to −48. Since the protected region for Efm liaFSR is smaller (29 base pairs) than that of Efm liaXYZ (45 base pairs) the evidence for two dimers of LiaR binding to the Efm liaFSR operon is less clear. In the comparable Efc liaFSR operon, the inclusion of the extended region was necessary for high affinity [19]. It is also interesting to note that the organization of the liaFSR and liaXYZ operons is very different. For liaXYZ the secondary site is 5’ of the consensus site while in liaFSR the secondary site is 3’ of the consensus sequence (Figure 5). This organization was also observed in E. faecalis.
While the protection sites of LiaR in E. faecium for both operons are identical in size for the closely related species, E. faecalis [19], the recognition elements for the E. faecium liaFSR and liaXYZ operons are shifted 4 and 16 base pairs respectively closer to the translation start site.
DNase hypersensitive sites suggest long range changes to DNA structure upon LiaR binding
In addition to protection from DNase cleavage, protein binding can also induce changes in DNA structure that increase nuclease sensitivity (hypersensitivity). Hypersensitivity is often correlated with bending or kinks induced in the DNA structure that render the phosphodiester backbone more accessible. As shown in Figure 5, both consensus binding regions share a strong hypersensitive site at the T immediately 3’ of the A in the consensus sequence (T(X)4C(X)4G(X)4A) a feature also observed in the homologous E. faecalis sequences.
Additional evidence for long range DNA structural changes suggestive of DNA bending can be seen as subtle hypersensitive sites at positions −85 and −53 of the liaFSR operon. As in the Efm liaFSR protection pattern, the protected region within liaXYZ operon is flanked by two strong hypersensitive sites at −79 and −114. These additional hypersensitive sites (−85 and −53 within liaFSR operon and −114 within liaXYZ operon) were not observed previously in the E. faecalis species. In E. faecalis, we had previously identified a region of marked hypersensitivity to nuclease digestion located 35 bases downstream of the liaXYZ consensus sequence [19]. Although this specific hypersensitive site was not present in E. faecium there is a strong and consistent hypersensitive site at positions (−4) AGCG (−1) (Figure 5e) of Efm liaFSR. Thus, our findings suggest that LiaR can significantly change the structure of DNA up to 50 bp downstream of the binding site, which could modify transcription rates.
The adaptive mutant Efm LiaRW73C binds more tightly to DNA binding sites within the liaXYZ and liaFSR operons
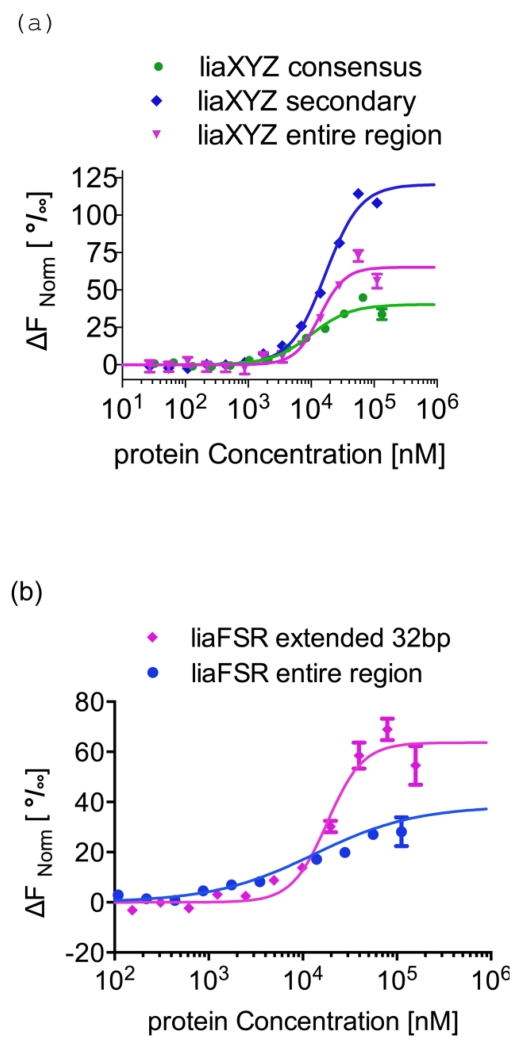
We have reported previously that the adaptive mutant LiaRD191N that is associated with daptomycin resistance in E. faecalis S613, markedly increases LiaR affinity for target DNA sequences and thereby upregulates transcription of LiaR-controlled genes [19]. Here, we tested the hypothesis that Efm LiaRW73C may have a similar effect on binding to specific DNA sequences. Using MicroScale Thermophoresis (MST), we measured the affinity of LiaR for both the consensus and adjoining secondary sites separately as well as for the entire protection region that were recovered from our DNA footprint analysis (Figure 6). We found that Efm LiaRW73C has an increased affinity for DNA target sites compared to wild-type LiaR to both the consensus and secondary sequences of the liaXYZ operon, consistent with our earlier observation of increased affinity for DNA by the adaptive mutant Efc LiaRD191N (Table 2, Figure 6a). However, the affinities of full-length Efm LiaRW73C toward cognate DNA measured in this study are generally weaker than those reported for Efc LiaRD191N. Unlike Efc LiaRD191N, which had higher affinity for the consensus site (0.39 ± 0.03 μM), Efm LiaRW73C binds with comparable affinity to both the consensus and secondary sites (9.9±0.6 μM and 16.7±0.4 μM, respectively).
Figure 6. Adaptive mutant LiaRW73C that confers increased daptomycin resistance in E. faecium binds more tightly to DNA binding sites within the liaXYZ operon.
MicroScale Thermophoresis (MST) was used to measure the strength of Efm LiaR–DNA interactions. To determine the Kd, increasing concentrations of Efm LiaRW73C was added to 40 nM of fluorescently labeled DNAs (Supplementary Information). Fnorm was plotted on the y-axis in per mil [‰] unit (meaning every 1000) against the total concentration of the titrated partner on a log10 scale on the x-axis [45]. The resulting Kd values based on average from four independent MST measurements. Bars indicate the SEM (standard error of the mean)
(a) The binding of LiaRW73C to the consensus (green circles), secondary (blue diamonds), or entire protected region (magenta triangles) sequences within the liaXYZ operon. The DNA sequence for the entire protected region is shown in Figure 5b.
(b) The binding of LiaRW73C to the extended (32 bp, magenta diamonds) or entire (blue circles) protected regions of the liaFSR operon. The DNA sequence for the entire protected region is shown in Figure 5d.
Table 2.
DNA binding affinity of E. faecium LiaR
| Protein |
liaXYZ consensus Kd (μM) |
liaXYZ secondary Kd (μM) |
liaXYZ entire protected Kd (μM) |
liaFSR consensus Kd (μM) |
liaFSR extended Kd (μM) |
liaFSR entire protected Kd (μM) |
Random Kd (μM) |
|---|---|---|---|---|---|---|---|
| LiaRW73C | 9.9±0.6 | 16.7±0.4 | 13.7±1.3 | >100 | 18.2±1.1 | 13.3±1.3 | >200 |
| LiaR | N/Da | N/Da | N/Da | ||||
| LiaRDBD | N/Da | N/Da | N/Da |
no detectable binding my MST up to 450 μM LiaR, 320 μM LiaRDBD
We would have also expected Efm LiaRW73C to bind with higher affinity to the entire contiguous protected region identified by DNA footprinting for the liaXYZ operon. However, the binding affinity was comparable to that for the consensus site alone (liaXYZ: Kd = 13.7±1.3 versus 9.9±0.6 μM) (Table 2). Interestingly, the binding isotherms for the entire protected region indicate a clear positive cooperativity in binding as well. It is not clear whether the apparent cooperativity results from direct dimer-dimer interactions of LiaR dimers at the DNA or from a strong shift of the LiaR monomer-dimer equilibrium in the presence of DNA. We favor the latter as the apparent cooperativity could be observed on the smaller 26 nucleotide DNA substrates that would not accommodate two dimers.
While DNA protection studies revealed that Efm LiaR had regions of protection that extended well beyond the consensus binding sequence, the overall affinity for the liaFSR and liaXYZ consensus sequences was quite different. The affinity of LiaRW73C for a 26 nucleotide DNA sequence extending from −83 to −58 (Figure 5c, d) of liaFSR was at least 10–fold lower than the comparable liaXYZ consensus binding site (Table 2). To test whether the additional sequences protected outside the canonical liaFSR consensus binding site are important for Efm LiaR binding, we extended the DNA sequence an additional 6 nucleotides toward the translation start site and measured the affinity for this extended 32 nucleotides protected region by MST. Efm LiaRW73C binds to the extended sequence at a micromolar concentration Kd=18.2±1.1 μM or about 5-fold higher than the shorter consensus site alone. The affinity for the entire contiguous protected region for Efm liaFSR (46 nucleotides, see Figure 5d) was comparable to that for the extended site Kd=13.3±1.3 μM (Table2). This is consistent with our earlier findings for Efc LiaR binding to the Efc liaFSR operon and confirming our earlier hypothesis that DNA sequences adjoining the canonical consensus sequence of the Efm liaFSR operon are also important for DNA binding.
The DNA binding domain alone is not able to bind regulatory DNA sequences with high affinity
Our analytical ultracentrifugation studies had demonstrated that the Efm LiaRDBD were able to dimerize readily in solution implying that it might have been possible for the DNA binding domains alone to have high affinity for DNA. The binding of Efm LiaRDBD to the liaXYZ and liaFSR operon sequences did not generate enough fluorescence change to obtain reliable Kd suggesting extremely weak protein–DNA interactions. These results support our earlier observation that even though the DNA binding domain itself exists in solution as a dimer, with a Kd of 0.4 μM (Table 1), the receiver domain appears essential for specific DNA binding.
Discussion
Exploration of the membrane stress response pathways of the pathogens E. faecium, E. faecalis and S. aureus reveals many commonalities as well as significant differences that should frame discussions of new therapies and antimicrobials. Examination of LiaR from E. faecium and E. faecalis and its homolog from S. aureus, VraR suggests an interesting evolutionary flexibility in the response regulator quaternary structure. E. faecium LiaR and S. aureus VraR are largely monomeric and become dimeric upon phosphorylation to mediate the cell membrane stress response. In contrast, E. faecalis LiaR exists predominately as a dimer that oligomerizes to an active tetramer upon phosphorylation [19]. DNA footprinting analysis of upstream regulatory regions from all three organisms suggests extended footprints that extend beyond the simple predicted consensus sequence upon phosphorylation or activation by an adaptive substitution. In principle, such extended DNA binding regions could be bound as a pair of dimers or as a single tetramer in the case of Efc LiaR. In the crystal structures of both S. aureus VraR and Efm LiaR there are two pairs of dimers in the asymmetric unit that could reflect how they might bind to extended regions of DNA and produce DNA bending to facilitate recruitment of RNA polymerase. Indeed our DNA footprinting studies for both Efc and Efm LiaR suggest that LiaR binding induces strong DNase hypersensitive sites suggestive of DNA bending. If DNA recognition by this family of response regulators requires four copies of the protein to induce DNA bending and a consequent increase in transcription from target operons, then any mutation that favors oligomerization will effectively “turn on” the system. This has now been observed in the adaptive mutants Efc LiaRD191N and Efm LiaRW73C isolated in response to the antimicrobial peptide daptomycin.
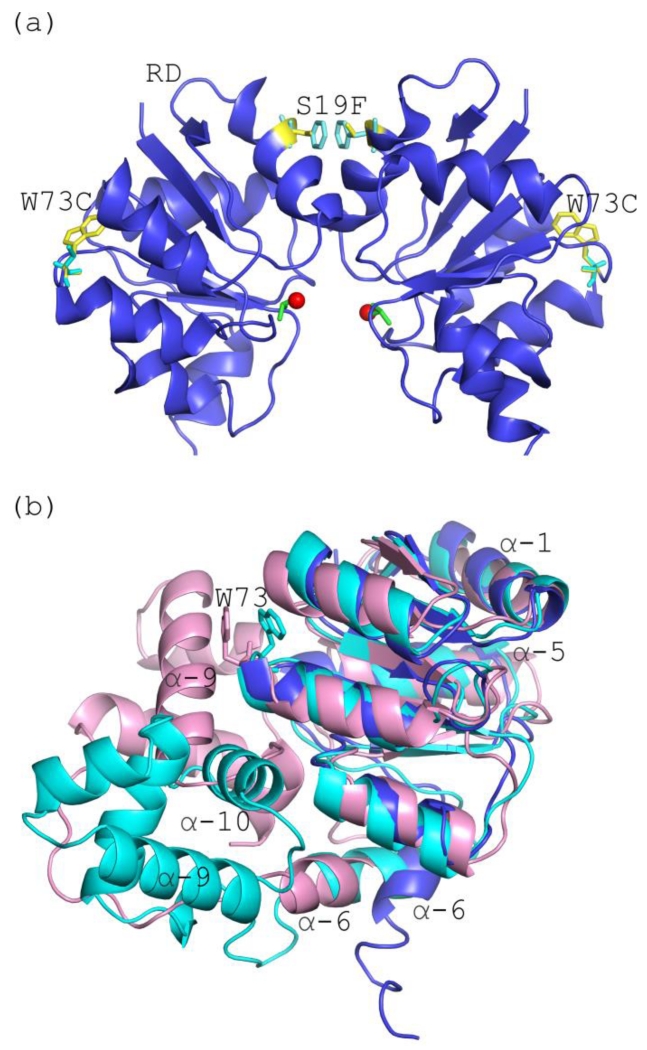
Interestingly, the activating substitution in Efc LiaR occurs at position 73 of the receiver domain and is not at the dimerization surface of the receiver domain dimer. (Figure 7a). The fact that the adaptive mutation W73→C in Efm LiaR is distant from the molecular interfaces comprising a LiaR dimer suggests an indirect role in favoring the dimerization competent monomer (e.g., the phosphorylated state). Based on our SEQ results in the absence of BeF3−, the full length LiaR protein is monomeric. In contrast, the sedimentation equilibrium analysis showed a weak but detectable dimerization propensity (Kd = 676 μM) for the LiaR receiver domain (LiaRRD) and a much stronger propensity for the DNA binding domain (LiaRDBD) to dimerize (Kd = 0.4 μM). Based on these results, we suggest a model where the receiver domain and DNA binding domain interact with each other in the unphosphorylated LiaR protein, stabilizing the monomeric state of the protein and preventing dimerization of either the receiver domains or the DNA binding domain (“closed” state). However, phosphorylation dependent conformational changes or the substitution W73C in the receiver domain may promote the dissociation of the receiver domain from the DNA binding domain, allowing dimerization of both the receiver domain and the DNA binding domains of two LiaR protomers. A closed state has been observed previously in crystal structures of the unphosphorylated VraR and NarL proteins [28, 36]. In these reported structures, the dimerization surface of the DNA binding domain helix α-10 of VraR or helix α-9 of NarL were found to be buried against the receiver domain of the respective protomer to stabilize a closed monomer conformation. Although the exact amino acids involved in this interdomain interface are not conserved across the NarL/FixJ family of response regulators, the general paradigm has been observed in other family members [27].
Figure 7. Mutations associated with daptomycin resistance are located within the receiver domain.
(a) Expanded view of the Efm LiaR receiver domains showing the positions of the mutations W73C and S19F. The S19F mutation (yellow/cyan sticks) is within the dimerization interface, while the adaptive mutation W73C (yellow/cyan sticks) is not located within the molecular surfaces that comprise the LiaR dimer: (b) Structural alignment of the activated Efm LiaR receiver (blue) compared to the inactive state structures of VraR/NarL receiver domains (cyan/light pink). Position 73 in the VraR and NarL structures are depicted as sticks (cyan/light pink, respectively).
In the absence of a structure for the full length unphosphorylated wild type E. faecium LiaR, we mapped the Efm LiaRW73C onto the VraR (PDB ID 4GVP)/NarL (PDB ID 1RNL) structures. As shown in Figure 7b, if our unphosphorylated LiaR structure resembles the inactive NarL but not VraR structure, the Trp73 is in the interfacial contact surface of the receiver to DNA binding domain. Thus the molecular surface comprised in part by Trp73 may be important for stabilizing the closed monomer conformation where the receiver domain interacts with the DNA binding domain of the same protomer chain. The W73C substitution may weaken the interface between the receiver domain and the DNA binding domain, shifting the equilibrium from the closed state to an open state where the receiver and DNA binding domains have a higher propensity to dimerize with another LiaR monomer to form the activated dimer. Alternatively, we cannot rule out the possibility that the molecular dynamics of the receiver domain is altered in a more indirect manner to shift the LiaR ensemble towards a state that more closely resembles the phosphorylated state.
There are at least two biological contexts where increased dimerization can lead to upregulation of the LiaR regulon. The most obvious scenario is one in which LiaRW73C increases phosphorylation independent dimerization to constitutively hyperactivate the LiaFSR stress response pathway. This LiaS independent constitutive hyperactivation mechanism was observed for the Efc LiaRD191N adaptive mutant [19]. It is puzzling however that Efm LiaRW73C does not actually confer the same extent of dimerization as BeF3− stimulated wild type Efm LiaR (wild type Efm LiaR dimerization Kd was 15 μM compared to 236 μM for Efm LiaRW73C). Since in vivo concentrations of LiaR are estimated to be in the low micromolar range [37], the weak in vitro dimerization of Efm LiaRW73C is less than expected for constitutive hyperactivation. Alternatively, the Efm LiaRW73C mutant may interact more efficiently with its cognate kinase LiaS to produce a higher extent of phosphorylation than the comparable wild type LiaR interaction with LiaS. This alternative possibility is made more attractive by the observation that the Efm LiaRW73C mutation has always been identified with a LiaST120A in clinical daptomycin non-susceptible isolates [17]. In the case of Efc LiaRD191N, a concomitant second mutation in the Efc LiaS protein has not been observed. Thus, there may be at least two distinct classes of adaptive mutations through changes in LiaR, a constitutive phosphorylation-independent hyperactivation type (as seen for Efc LiaRD191N) and a second type that promotes more efficient phosphorylation by LiaS that might include Efm LiaRW73C and that may be frequently observed in epistatic combination with sunbstitutions in LiaS.
If adaptive LiaR substitutions that favor oligomerization confer additional resistance to membrane stress, then, it is also likely that any small molecule that destabilizes oligomerization will increase susceptibility. Support for this hypothesis is observed readily in a recently described Efc [16] liaR knockout. When Efc liaR is removed, cells become hypersusceptible to daptomycin suggesting that LiaR could be a good target for new drugs that would complement existing antibiotics to maintain their long term efficacy by blocking the evolution of resistance through liaR [16].
Evolutionarily, there appears to be a spectrum of oligomeric answers to the question of how to bind these specific and extended DNA sequences. We have now observed both a monomer↔dimer and a dimer↔tetramer transition upon phosphorylation depending on the species. In all cases however, the DNA regulatory region is extended. Some organisms like S. aureus and E. faecium may shift the equilibrium towards the monomer while others like E. faecalis start as a dimer (that presumably does not bind DNA with high enough affinity to activate as a dimer alone) and then activate transcription as a tetramer. The facile nature of these equilibrium shifts can be seen readily by the single point mutations that dramatically change the oligomeric state (e.g. LiaRD191N and LiaRW73C in E. faecalis and E. faecium, respectively) (Figure 8)
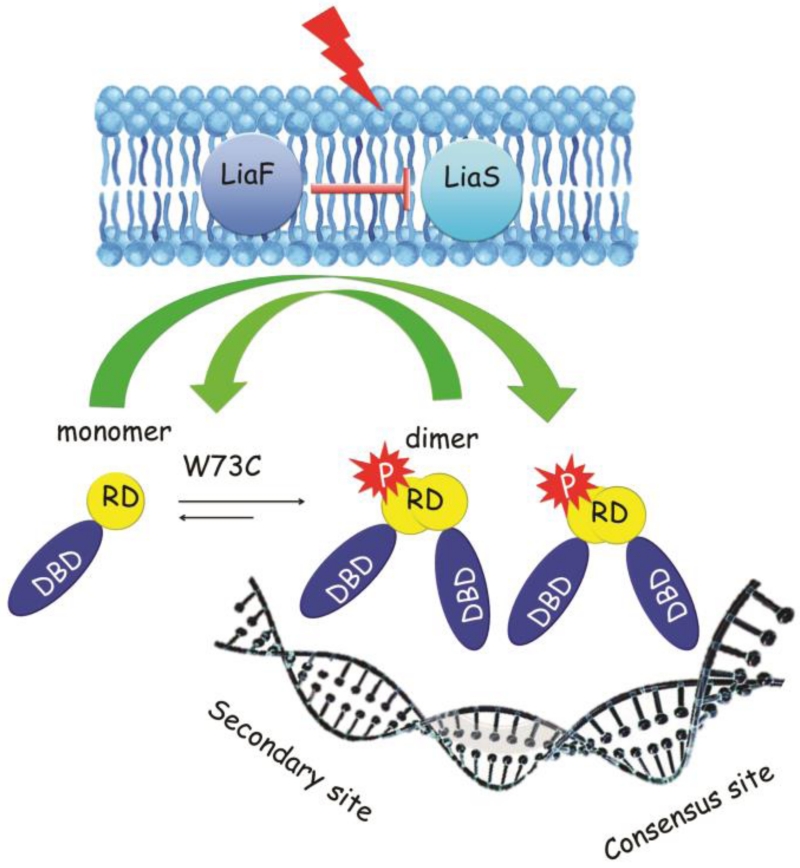
Figure 8. Adaptive mutations in E. faecium LiaR that have been identified during antibiotic selection.
In the absence of the cell membrane acting antibiotics or membrane stress, the three-component regulatory system, LiaFSR, is turned “OFF.” LiaF is thought to down regulate the kinase activity of LiaS. Typically, LiaR is phosphorylated in response to membrane stress to regulate downstream target operons. E. faecium LiaR is present largely as a monomer at physiologically relevant concentrations. Phosphorylation may induce changes in the conformation of the receiver domain leading to release of DNA binding domain and promoting a self-dimerization event to form an active tetramer able to bind extended DNA sequences. The adaptive mutation W73C, even in the absence of phosphorylation, promotes a phosphorylation-like self-dimerization event that increases LiaR affinity for upstream regulatory regions. (RD - receiver domain, DBD - DNA binding domain)
More recently, a new Efm LiaR mutant Efm LiaRS19F has been identified in DAP non-susceptible enterococci isolated from an immunocompromised patient suggesting that mutations within the receiver domain may be more common in E. faecium [38]. Mapping the recently discovered S19F substitution in the response regulator Efm LiaR onto our LiaR three-dimensional crystal structure (Figure 7a) revealed that this mutation is within the receiver domain dimerization interface. Based on our studies, the Efm LiaR S19F substitution may also increase oligomerization of LiaR to form a constitutively activated dimer with high affinity for the extended DNA regulatory sequence even in the absence of phosphorylation, leading to increased daptomycin resistance. If single point mutations can produce such large changes then it is not surprising that evolution has tuned these systems in particular niches. What might be surprising is that E. faecium would be so different from E. faecalis but perhaps this is the bias introduced by nomenclature rather than biology. E. faecium (GenBank accession number: WP_002348688.1) is 60.5 % identical to S. aureus (GenBank accession number: WP_000153535.1) while E. faecalis (GenBank accession number: EFE19096.1) is 59.7 % identical to S. aureus. E. faecalis and E. faecium themselves share 84.3% identity and thus there is substantial genomic variability to allow for these surprising differences (Figure S7). For example, E. faecalis LiaR has a deletion within the N-terminal ß1 strand (Figure S7) that is not present in E. faecium LiaR or S. aureus VraR. The truncation at the N-terminus is unusual within the NarL/FixJ subfamily and further highlights that even among closely related species there may be variability within protein structure with important consequences for protein function.
Interestingly E. faecium in the initial stages (only when the LiaFSR mutation occurs) is able to carry out both diversion and repulsion strategies while S. aureus favors repulsion and E. faecalis favors diversion. These distinctive phenotypic changes suggest E. faecium may have some quite unique responses to membrane stress that contrasts markedly with its closely related peers.
Experimental procedures
Expression and purification of response regulator R494 LiaR and variants from E. faecium
The genes encoding full length LiaRW73C R494, receiver domain (RDW73C, residues 1-139) and DNA binding domain (DBD, residues 140-210) were amplified by PCR from genomic DNA R494, cloned into pETDuet vector and expressed in Escherichia coli BL21 (DE3) cells using LB medium. Site-directed mutagenesis of LiaR was performed by incorporation of mutations into the plasmid by inverse PCR with standard primers designed an overlapping orientation. The proteins were purified with a nickel affinity column (GE Healthcare) followed by the anion-exchange chromatography and gel filtration for the final step as described previously work [19]. N-terminal SUMO peptide (within the 6×His tag) was removed by treatment with His-tagged SUMO protease.
DNA footprint analysis by automated capillary electrophoresis (DFACE)
The 379-bp fluorescent labeled DNA probe for the R494 E. faecium liaFSR operon and 357-bp fluorescent labeled DNA probe for the R494 E. faecium liaXYZ operon were generated with PCR amplification with the primers 5’-(VIC)-aatgattctcctatcgcgcaagca-3’ and 5’-(FAM)-actaaaagcaaaaatccagtattg-3’ for liaFSR operon and 5’-(VIC)-agcgttgatcctctggtttattga-3’ and 5’-(FAM)-aagccttcttcagtcgaaaggatt-3’ liaXYZ operon from Applied Biosystems (Grand Island, NY) as described previously [19, 39, 40]. Briefly, full length of Efm LiaR protein activated with the BeF3− at 0.5, 2.5, 5 μM and 50.0 μM was then incubated with respective fluorescent labeled DNA probe in Binding Buffer (50 mM Tris pH 7.5, 300 mM NaCl, 10 mM MgCl2, and 0.3 mM DTT, 20% (v/v) glycerol, 0.05% (v/v) Tween-20) for 10 min. After this incubation, DNase I (New England Biolabs Inc., Ipswich, MA, 2 U/μl) digestion was performed. The reaction was then stopped by adding EDTA to a final concentration of 5 mM following by incubation at 75°C for 10 min. Control digestions with the probe were performed in the absence of the protein. The DNA fragments were purified with a QIA-quick PCR purification kit (Qiagen, Valencia, CA) and then sent to the Plant-Microbe Genomics Facility for further analysis. The resulting electropherograms from the DFACE assay were analyzed with the software GeneMapper 4.0 set to normalize all samples to the sum of the signal of all samples. In addition, the histograms were further normalized by subtracting the height of the equivalent peak (same Bin) in the negative control from the sample, i.e. each bar is the difference between sample and negative control. Data reported utilized fragments/strand labeled with VIC dye. Nucleotide positions refer to the region of LiaR binding on the DNA relative to the translation start site of LiaX or LiaF, respectively. DNA footprinting was performed at 25°C.
Analytical Ultracentrifugation
Sedimentation equilibrium analytical ultracentrifugation (SEQ) and sedimentation velocity) experiments were performed in an AnTi60 rotor or AnTi50 rotor respectively, at 20°C using a Beckman XL-I instrument. Epon double sector centerpieces and sapphire windows. The protein samples were prepared in either 50 mM Tris-HCl, 300 mM NaCl, 10 mM MgCl2, 2 mM Tris(2-carboxyethyl)phosphine and 10% (v/v) glycerol at pH 7.5 or 50 mM Tris-HCl, 300 mM NaCl, 10 mM MgCl2, 2 mM tris(2-carboxyethyl)phosphine, 10% (v/v) glycerol, 5 mM BeCl2 and 30 mM NaF at pH 7.5 to test the self-association of the protein(s) in the absence and presence of the phosphorylation mimetic Beryllofluoride (BeF3−) respectively. SEQ scans were using absorbance at 280 nm, after 64 h incubation at each rotor speed. The protein partial specific volume, solvent density and solvent viscosity were calculated using Sednterp 1.09 (1). SEQ scans were collected using absorbance at 280 nm, after 64 h incubation at each rotor speed. Sedimentation equilibrium analytical ultracentrifugation analysis for Efm LiaR, LiaR:BeF3−, LiaRW73C, LiaRW73C:BeF3−, LiaRRD, LiaRRD:BeF3−, LiaR(RD)W73C, LiaR(RD)W73C:BeF3−, LiaRDBD recorded at 13000 r.p.m., 20000 r.p.m. and 25000 r.p.m. Data analysis was performed using Sedphat v10.58d (2,3). For each protein, the SEQ profiles were globally fitted to a monomer-dimer equilibrium model with rotor stretch and mass conservation restraints. A F-statistics error mapping approach was used to determine the 95% confidence intervals for the dissociation constant [41-44]. Sedimentation velocity (SV) data, recorded at 50000 r.p.m., was fitted using a local continuous distribution model looking for species between 0.5 and 15 Svedbergs with the frictional ratio, meniscus and bottom position allow to float to find the best fit. A single discrete species was included in the model with a sedimentation coefficient of 0.3 Svedbergs, to account for any buffer component effects. The Tikhonov regularization for the c(S) and c(M) distributions was set to 0.95.
DNA binding affinity determination by Microscale Thermophoresis (MST)
MST measurements were done as described previously [19, 45]. Briefly, a solution of unlabeled LiaR (wild type or mutants) was serially diluted in reaction buffer (50 mM Tris pH 7.5, 300 mM NaCl, 10 mM MgCl2, 0.3 mM DTT, 10% glycerol, 0.05% (v/v) Tween-20) to which an equal volume of Alexa-647 labeled DNA was added to a final concentration of 40 nM. The samples were loaded into standard treated capillaries (Nanotemper). MST experiments were measured on a Monolith NT.115 system (Nanotemper Technologies) using 30% LED and 90% IR-laser power. The resulting Kd values based on average from four independent MST measurements. Temperature of MST experiments was 20°C. Data analyses were performed using Nanotemper Analysis software, v.1.5.41. (Supplementary information).
Structure determination of full length LiaR activated by BeF3−
A beryllofluoride activated LiaR protein crystal suitable for data collection was grown at 10° C in 0.1 M magnesium formate dehydrate, 15 % w/v Polyethylene glycol 3,350 using a ‘microseed matrix screening’ approach [46]. Diffraction data were collected at Argonne National Laboratory’s Advanced Photon Source (ANL APS) beamline 21-ID-D on a MarMosaic 300 CCD detector. X-ray diffraction data were processed using HKL2000 [47]. The crystal belonged to the space group P31 with the unit cell parameters a=31.05, b=76.92, c=76.92, α=β=γ=90°. The 3-D structure was determined by molecular replacement using S. aureus receiver and DNA binding domains as the search probe in Phaser-MR [48].The solution from molecular replacement suggested four molecules in the asymmetric unit and a Matthews coefficient of 3.03 (59.5% solvent content) [49].
The structure was refined to 3.2 Å resolution using the Phenix suite [50] and Coot [51]. The final values for the R=21.86 % and Rfree factors are 27.04 %. A summary of data collection and refinement statistic are reported in Table 3.
Table 3.
Data collection and refinement statistics
| Data collection | |
|
| |
| Wavelength (Å) | 0.97872 |
|
| |
| Resolution (Å)a | 50.0-3.2 (3.26-3.2) |
|
| |
| Space group | P31 |
|
| |
| Molecules per a.u. | 4 |
|
| |
| Cell dimensions a,b,c; (Å); α, β, γ; (°) | 68.03×68.03×276.95; 90×90×120 |
|
| |
| Reflections | 23488 |
| Uniquea | |
|
| |
| Average redundancya | 3.5 (2.9) |
|
| |
| Completeness (%)a | 98.3 (97.9) |
|
| |
| Rmerge (%)a,b | 11.5 (0.83) |
|
| |
| Output <I/sigI>a | 10.1 (1.22) |
|
| |
| Refinement | |
|
| |
| Rwork (%)c | 21.86 |
|
| |
| Rfree (%)d | 27.04 |
|
| |
| r.m.s.d.e from ideality |
|
| Bonds (Å) | 0.003 |
| Angles (°) | 0.569 |
|
| |
| Wilson B-factor (Å2) | 57.6 |
|
| |
| Average B, all atoms (Å2) | 53.0 |
|
| |
| Average B, ligand, BeF3− (4) (Å2) | 42.5 |
|
| |
| Ramachandranf | |
| Favored (%) | 97.10 |
| Allowed (%) | 2.90 |
| Outliers (%) | 0.00 |
|
| |
| PDB ID | 5HEV |
Values for the last shell are in parenthesis
Rmerge = Σ|I − <I> |/ Σ I, where I is measured intensity for reflections with indices of hkl
Rwork = Σ|Fo − Fc |/ Σ| Fo | for all data with Fo > 2 σ (Fo) excluding data to calculate Rfree
Rfree = Σ|Fo − Fc |/ Σ|Fo| for all data with Fo > 2 σ (Fo) excluded from refinement.
Root mean square deviation
Supplementary Material
Research Highlights.
In clinical settings, nonsusceptibility to daptomycin by Enterococcus faecium is correlated frequently with a mutation in LiaR of Trp73 to Cys. To elucidate the physicochemical basis for LiaR mediated changes in daptomycin susceptibility, we used a combination of biophysical and structural approaches. Our study revealed that the adaptive mutation W73C, even in the absence of phosphorylation, activates LiaR to undergo a phosphorylation-like self-dimerization event that allows LiaR to recognize and activate upstream regulatory regions.
Acknowledgements
This work was supported by National Institutes of Health Grant [R01AI080714 to Y.S.]; [R21AI1114961-01 to C.A.]; [R21/331R21AI121519-01 to C.A.]. The Rice University Crystallographic Core Facility is supported by a Kresge Science Initiative endowment grant. We would like to thank the staff at ANL APS for their support during the data collection.
Abbreviations used
- VRE
Vancomycin-resistant enterococci
- HAI
Hospital-Associated Infections
- IDSA
Infectious Diseases Society of America
- SEQ
sedimentation equilibrium analytical ultracentrifugation
- MST
MicroScale Thermophoresis
- DFACE
DNA footprint analysis by automated capillary electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Structure factors and final atomic coordinates have been deposited with the Protein Data Bank (PDB ID: 5HEV)
Conflict of interest
Dr. Cesar Arias is on the speaker’s Bureau for Forest, Theravance, Pfizer, Astra-Zeneca, Cubist, The Medicines Company, Norvartis. He is also consulting for Theravance, Cubist, Bayer and grant investigator for Theravance and Scientific advisor (review panel or advisor committee) for Bayer.
Authors Contributions
MD, CAA and YS designed research, analyzed data, and wrote the paper; MD, AT-Y, KD, PGL, and MRZ performed the experiments. PGL edited the manuscript.
References
- [1].Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol. 2012;10:266–78. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- [3].Munita JM, Murray BE, Arias CA. Daptomycin for the treatment of bacteraemia due to vancomycin-resistant enterococci. Int J Antimicrob Agents. 2014;44:387–95. doi: 10.1016/j.ijantimicag.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fowler VG, Jr., Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med. 2006;355:653–65. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- [5].Humphries RM, Pollett S, Sakoulas G. A current perspective on daptomycin for the clinical microbiologist. Clin Microbiol Rev. 2013;26:759–80. doi: 10.1128/CMR.00030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tran TT, Panesso D, Mishra NN, Mileykovskaya E, Guan Z, Munita JM, et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio. 2013;4:e00281–13. doi: 10.1128/mBio.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194:4494–504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cha R, Rybak MJ. Influence of protein binding under controlled conditions on the bactericidal activity of daptomycin in an in vitro pharmacodynamic model. J Antimicrob Chemother. 2004;54:259–62. doi: 10.1093/jac/dkh259. [DOI] [PubMed] [Google Scholar]
- [9].Fuchs PC, Barry AL, Brown SD. In vitro bactericidal activity of daptomycin against staphylococci. J Antimicrob Chemother. 2002;49:467–70. doi: 10.1093/jac/49.3.467. [DOI] [PubMed] [Google Scholar]
- [10].Cotroneo N, Harris R, Perlmutter N, Beveridge T, Silverman JA. Daptomycin exerts bactericidal activity without lysis of Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:2223–5. doi: 10.1128/AAC.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Silverman JA, Perlmutter NG, Shapiro HM. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:2538–44. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jung D, Rozek A, Okon M, Hancock RE. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol. 2004;11:949–57. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- [13].Tran TT, Munita JM, Arias CA. Mechanisms of drug resistance: daptomycin resistance. Ann N Y Acad Sci. 2015;1354:32–53. doi: 10.1111/nyas.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hachmann AB, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hachmann AB, Sevim E, Gaballa A, Popham DL, Antelmann H, Helmann JD. Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob Agents Chemother. 2011;55:4326–37. doi: 10.1128/AAC.01819-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reyes J, Panesso D, Tran TT, Mishra NN, Cruz MR, Munita JM, et al. A liaR Deletion Restores Susceptibility to Daptomycin and Antimicrobial Peptides in Multidrug-Resistant Enterococcus faecalis. J Infect Dis. 2014 doi: 10.1093/infdis/jiu602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Diaz L, Tran TT, Munita JM, Miller WR, Rincon S, Carvajal LP, et al. Whole-genome analyses of Enterococcus faecium isolates with diverse daptomycin MICs. Antimicrob Agents Chemother. 2014;58:4527–34. doi: 10.1128/AAC.02686-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Miller C, Kong J, Tran TT, Arias CA, Saxer G, Shamoo Y. Adaptation of Enterococcus faecalis to daptomycin reveals an ordered progression to resistance. Antimicrob Agents Chemother. 2013;57:5373–83. doi: 10.1128/AAC.01473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Davlieva M, Shi Y, Leonard PG, Johnson TA, Zianni MR, Arias CA, et al. A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin. Nucleic Acids Res. 2015;43:4758–73. doi: 10.1093/nar/gkv321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Belcheva A, Golemi-Kotra D. A close-up view of the VraSR two-component system. A mediator of Staphylococcus aureus response to cell wall damage. J Biol Chem. 2008;283:12354–64. doi: 10.1074/jbc.M710010200. [DOI] [PubMed] [Google Scholar]
- [21].Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, et al. BeF(3)(−) acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF(3)(−) complex with phosphoserine phosphatase. Proc Natl Acad Sci U S A. 2001;98:8525–30. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Varughese KI, Tsigelny I, Zhao H. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J Bacteriol. 2006;188:4970–7. doi: 10.1128/JB.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Munita JM, Panesso D, Diaz L, Tran TT, Reyes J, Wanger A, et al. Correlation between mutations in liaFSR of Enterococcus faecium and MIC of daptomycin: revisiting daptomycin breakpoints. Antimicrob Agents Chemother. 2012;56:4354–9. doi: 10.1128/AAC.00509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–82. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:626–33. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- [26].Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–65. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Trajtenberg F, Albanesi D, Ruetalo N, Botti H, Mechaly AE, Nieves M, et al. Allosteric activation of bacterial response regulators: the role of the cognate histidine kinase beyond phosphorylation. MBio. 2014;5:e02105. doi: 10.1128/mBio.02105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Leonard PG, Golemi-Kotra D, Stock AM. Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation. Proc Natl Acad Sci U S A. 2013;110:8525–30. doi: 10.1073/pnas.1302819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mack TR, Gao R, Stock AM. Probing the roles of the two different dimers mediated by the receiver domain of the response regulator PhoB. J Mol Biol. 2009;389:349–64. doi: 10.1016/j.jmb.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Barbieri CM, Wu T, Stock AM. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J Mol Biol. 2013;425:1612–26. doi: 10.1016/j.jmb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, et al. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–15. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- [32].Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Cascio D, Gunsalus RP, Dickerson RE. NarL dimerization? Suggestive evidence from a new crystal form. Biochemistry. 1998;37:3665–76. doi: 10.1021/bi972365a. [DOI] [PubMed] [Google Scholar]
- [33].Schnell R, Ågren D, Schneider G. 1.9 Å structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64:1096–100. doi: 10.1107/S1744309108035203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Webber CA, Kadner RJ. Involvement of the amino-terminal phosphorylation module of UhpA in activation of uhpT transcription in Escherichia coli. Mol Microbiol. 1997;24:1039–48. doi: 10.1046/j.1365-2958.1997.4021765.x. [DOI] [PubMed] [Google Scholar]
- [35].Maiti R, Van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;32:W590–4. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baikalov I, Schroder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–61. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- [37].Belcheva A, Verma V, Korenevsky A, Fridman M, Kumar K, Golemi-Kotra D. Roles of DNA sequence and sigma A factor in transcription of the vraSR operon. J Bacteriol. 2012;194:61–71. doi: 10.1128/JB.06143-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sorlozano A, Panesso D, Navarro-Mari JM, Arias CA, Gutierrez-Fernandez J. Characterization of daptomycin non-susceptible Enterococcus faecium producing urinary tract infection in a renal transplant recipient. Rev Esp Quimioter. 2015;28:207–9. [PMC free article] [PubMed] [Google Scholar]
- [39].Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech. 2006;17:103–13. [PMC free article] [PubMed] [Google Scholar]
- [40].Joshi GS, Zianni M, Bobst CE, Tabita FR. Further Unraveling the Regulatory Twist by Elucidating Metabolic Coinducer-Mediated CbbR-cbbI Promoter Interactions in Rhodopseudomonas palustris CGA010. J Bacteriol. 2012:1350–60. doi: 10.1128/JB.06418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. Analytical Ultracentrifugation in Biochemistry and Polymer Science. In: S. E. Harding AJRaJCH, editor. Royal Society of Chemistry; Cambridge: 1992. [Google Scholar]
- [42].Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–24. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- [43].Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–56. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- [44].Johnson ML, Straume M. Modern Analytical Ultracentrifugation. Birkhäuser; Boston: 1994. Comments on the analysis of sedimentation equilibrium experiments. [Google Scholar]
- [45].Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–53. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Obmolova G, Malia TJ, Teplyakov A, Sweet RW, Gilliland GL. Protein crystallization with microseed matrix screening: application to human germline antibody Fabs. Acta Crystallogr F Struct Biol Commun. 2014;70:1107–15. doi: 10.1107/S2053230X14012552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Carter Charles W., Jr., editor. Methods in Enzymology. Academic Press; 1997. pp. 307–26. [DOI] [PubMed] [Google Scholar]
- [48].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–74. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–71. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.