Abstract
Purpose:
Genome and exome sequencing can identify variants unrelated to the primary goal of sequencing. Detecting pathogenic variants associated with an increased risk of a medical disorder enables clinical interventions to improve future health outcomes in patients and their at-risk relatives. The Clinical Genome Resource, or ClinGen, aims to assess clinical actionability of genes and associated disorders as part of a larger effort to build a central resource of information regarding the clinical relevance of genomic variation for use in precision medicine and research.
Methods:
We developed a practical, standardized protocol to identify available evidence and generate qualitative summary reports of actionability for disorders and associated genes. We applied a semiquantitative metric to score actionability.
Results:
We generated summary reports and actionability scores for the 56 genes and associated disorders recommended by the American College of Medical Genetics and Genomics for return as secondary findings from clinical genome-scale sequencing. We also describe the challenges that arose during the development of the protocol that highlight important issues in characterizing actionability across a range of disorders.
Conclusion:
The ClinGen framework for actionability assessment will assist research and clinical communities in making clear, efficient, and consistent determinations of actionability based on transparent criteria to guide analysis and reporting of findings from clinical genome-scale sequencing.
Genet Med 18 12, 1258–1268.
Keywords: clinical actionability, exome sequencing, genome sequencing, incidental findings, secondary findings
Introduction
Over the past decade, the cost of genome-scale sequencing (i.e., genome and exome sequencing) has dropped dramatically, which has led to its increased application in clinical and research settings.1,2,3 The generation of genome-wide sequencing data enables identification of findings in addition to the variants related to the primary reason for sequencing, such as variants in genes that are unanticipated (incidental findings) or purposefully sought (secondary findings).4,5 Identification of a pathogenic variant associated with an increased risk of a significant, but preventable, medical outcome affords the opportunity to implement interventions that prevent or mitigate future clinical manifestations in patients and their at-risk family members.6,7 Although some genes and associated disorders have a considerable evidence base for clinical actionability, the evidence supporting clinical actionability for most genetic disorders varies widely. As such, there is a need to develop and implement standardized, evidence-based methods to characterize the clinical actionability of genomic findings.8 These actionability profiles of genetic disorders could be used to facilitate decision making regarding which findings should be considered for return of results to improve future health outcomes.
The Clinical Genome Resource (ClinGen) is a National Institutes of Health–funded consortium of researchers and clinicians who are building an open-access and centralized resource to define the clinical relevance and actionability of genomic variants (http://clinicalgenome.org/).9 The ClinGen Actionability Working Group (AWG) is developing practical methods to identify genetic disorders with the greatest clinical utility when detected in previously undiagnosed adults. To this end, the ClinGen AWG developed a standardized protocol to generate evidence-based profiles of clinical actionability of genes and associated disorders in the form of summary reports and semiquantitative metric (SQM) scores. Although the methods to identify the evidence base are transparent and standardized, it is noncomprehensive in order to allow for practical and efficient consideration of numerous genes and disorders. Thus, the summary reports generated by this protocol are focused and are intended to provide the initial guidance regarding the return of findings to patients based on clinical actionability and are not intended to provide guidance on clinical care that would occur downstream.
We applied the protocol generated by the ClinGen AWG to the list of 56 genes and associated disorders recommended by the American College of Medical Genetics and Genomics (ACMG) to be returned as secondary findings to patients when a pathogenic or likely pathogenic variant is detected during clinical genome-scale sequencing.10,11 We describe the issues that arose during the process, many of which have led to improvements in the protocol, scoring metric, and ability to determine clinical actionability. These issues illustrate the challenges of the task and provide insight for future endeavors involving the characterization of clinical actionability. The ClinGen AWG framework provides a structure to enable research and clinical communities to make clear, streamlined, and consistent determinations of clinical actionability based on transparent criteria to guide analysis and reporting of genomic variation.
Materials and Methods
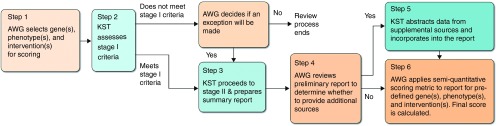
The ClinGen AWG developed a protocol for the standardized synthesis of available evidence and the generation of consensus scores of clinical actionability of genes and associated disorders using reproducible and transparent methods (Figure 1). We adapted methods to identify relevant evidence and abstract information on clinical actionability into summary reports from the Evaluation in Genomic Applications in Practice and Prevention working group's protocol to evaluate the evidence base for clinical actionability of incidental or secondary findings.12 The SQM scoring methodology was based on a framework generated by the North Carolina Clinical Genomic Evaluation by Next-generation Exome Sequencing project (NCGENES).13
Figure 1.
ClinGen Actionability Working Group workflow. AWG, Actionability Working Group; KST, Knowledge Synthesis Team.
Clinical scenario
The clinical scenario under consideration for this protocol was an adult patient who had undergone genome-scale sequencing as part of clinical care. This patient had an incidental or secondary finding in the form of a known or likely pathogenic variant causally associated with a genetic disorder unrelated to the original indication for sequencing and had not been previously diagnosed with this genetic disorder but may have been symptomatic with associated clinical manifestations. For example, a pathogenic variant in the HFE gene may be discovered as a secondary finding in a patient known to have joint pain and who might have undergone treatment for it but had not been previously diagnosed with familial hemochromatosis.
Clinical actionability summary reports
For the purposes of protocol development, clinical actionability was defined as clinically prescribed interventions specific to the genetic disorder under consideration that are effective for prevention or delay of clinical disease, lowered clinical burden, or improved clinical outcomes in a previously undiagnosed adult. Interventions included patient management (e.g., risk-reducing surgery), surveillance (e.g., colonoscopy), or specific circumstances the patient should avoid (e.g., certain types of anesthesia). Actionability included interventions to improve outcomes for at-risk family members (e.g., increased surveillance). Genetic testing recommendations for at-risk family members alone, however, were not considered sufficient to meet the criteria for actionability. In addition, for the purposes of this protocol, actionability did not include reproductive decision making.
We assessed the clinical actionability for each topic (i.e., genes and associated disorders) in two stages. Stage I was a rapid rule-out step to eliminate topics from further consideration that did not meet the three criteria for minimal clinical actionability: (i) the genetic disorder is clinically actionable in an undiagnosed adult; (ii) if penetrance or relative risk is known, then there is at least one pathogenic variant with moderate penetrance (≥40%) or relative risk (≥2) in any population; and (iii) the resulting genetic disorder is a significant health condition (stage I dashboard of the summary report template is available in the Supplementary Materials online). A single reference was sufficient to satisfy any of the stage I criteria, so an exhaustive literature search was not necessary at this stage. The topic automatically proceeded to stage II if all three stage I criteria were met. If a topic did not meet stage I criteria, then the AWG could decide by consensus to make an exception and move the topic to stage II. No further action was taken if a topic did not meet stage I criteria and was not recommended for exception by the AWG.
The purpose of stage II was to document and synthesize key evidence related to the clinical actionability of a pathogenic genomic finding (Table 1). The AWG Knowledge Synthesis Team identified the most current literature on clinical actionability for a given topic by using the standardized search methods outlined by Goddard et al.12 Briefly, online resources (such as Online Mendelian Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/omim), GeneReviews (http://www.ncbi.nlm.nih.gov/books/NBK1116/), OrphaNet (http://www.orpha.net), PubMed (http://www.ncbi.nlm.nih.gov/pubmed), and the National Guideline Clearinghouse (http://www.guideline.gov/)) were targeted and filtered to identify clinical guidelines and secondary literature, such as systematic reviews and meta-analyses. Primary literature and narrative reviews were not considered. This search method was transparent, reproducible, and appropriate for the current application, but it was limited in scope for efficiency and practicality and thus was not comprehensive. Each reference identified during the search was examined for relevance to actionability in the context of the clinical scenario. Any references deemed irrelevant were excluded.
Table 1. Knowledge Synthesis Team's stage II methods for generating summary reports.

Once we had identified all relevant references, each reference was assigned a quality tier based on predefined criteria (Table 1). Information from the highest-tiered source was abstracted and condensed into a summary report with standardized information documented for each topic (the summary report template is available in the Supplementary Materials online). The information abstracted addressed five of the most important aspects of clinical actionability: (i) the nature of the threat to health for an individual carrying a pathogenic allele of the given gene or genes; (ii) the effectiveness of available interventions for preventing or mitigating harm; (iii) the likelihood that the threat will materialize (i.e., the penetrance of that trait attributable to the variant); (iv) the nature of the intervention in terms of the burdens or risks placed on the individual with the genomic finding; and (v) the chance that the underlying risk or disorder could escape detection prior to harm in the setting of standard care. Information for this last domain was abstracted only for the summary reports and was not incorporated into the scores for clinical actionability.
AWG review of summary reports
Although the methods for identifying evidence on clinical actionability facilitate the transparency and feasibility of addressing a large number of topics, these reports do not reflect a comprehensive overview of all primary literature, and they thus limit our ability to inform all aspects of clinical actionability. As such, discrepancies may emerge between the summary reports and primary study publications or widely accepted expert clinical practice regarding clinical actionability of a particular topic. To address this issue, AWG members reviewed preliminary summary reports and nominated additional sources for consideration, such as primary literature, when applicable (tier 5, Table 1). This important contribution made the process more comprehensive and enabled the scoring of all domains of clinical actionability.
Semiquantitative scoring metric application
Selection of outcome–intervention sets for scoring. Some genetic conditions are associated with a variety of distinct clinical outcomes that vary in terms of severity, actionability, or likelihood. Likewise, interventions vary with regard to effectiveness, risk, or burden. For example, hereditary breast and ovarian cancer syndrome (HBOC) is associated with breast and ovarian cancers, which are distinct in likelihood and actionability, and the interventions for these outcomes vary in effectiveness, risk, and burden (e.g., surveillance vs. risk-reducing surgery). Thus, assessing all possible outcomes and interventions simultaneously to generate a single clinical actionability score would be misleading. The AWG addressed this issue by considering specific outcome–intervention pairs within the defined clinical scenario and scoring each pair separately for clinical actionability. The outcomes selected for scoring referred to significant and likely actionable clinical manifestations of the genetic disorder, which were then paired with relevant interventions. In the example of HBOC, three outcome and intervention pairs were identified for scoring: (i) breast cancer and surveillance; (ii) breast cancer and risk-reducing surgery; and (iii) ovarian cancer and risk-reducing surgery.
Scoring method. Assuming a pathogenic variant in the gene or genes under consideration, the AWG members applied the SQM to the information in the corresponding summary report to score each topic for four domains of clinical actionability: (i) severity of the outcome; (ii) likelihood of the disease; (iii) effectiveness of the intervention; and (iv) nature of the intervention in terms of burden and risk to the patient (Table 2). Each domain was scored from 0 to 3, resulting in a final “actionability score” from 0 (least clinically actionable) to 12 (most clinically actionable). In addition, scorers rated the knowledge base regarding the likelihood of the disease and the effectiveness of the intervention; this scale ranged from substantial evidence (“A”) to poor evidence (“D”) as well as nonsystematically identified sources (“E”).
Table 2. Semiquantitative metric to score clinical actionability.
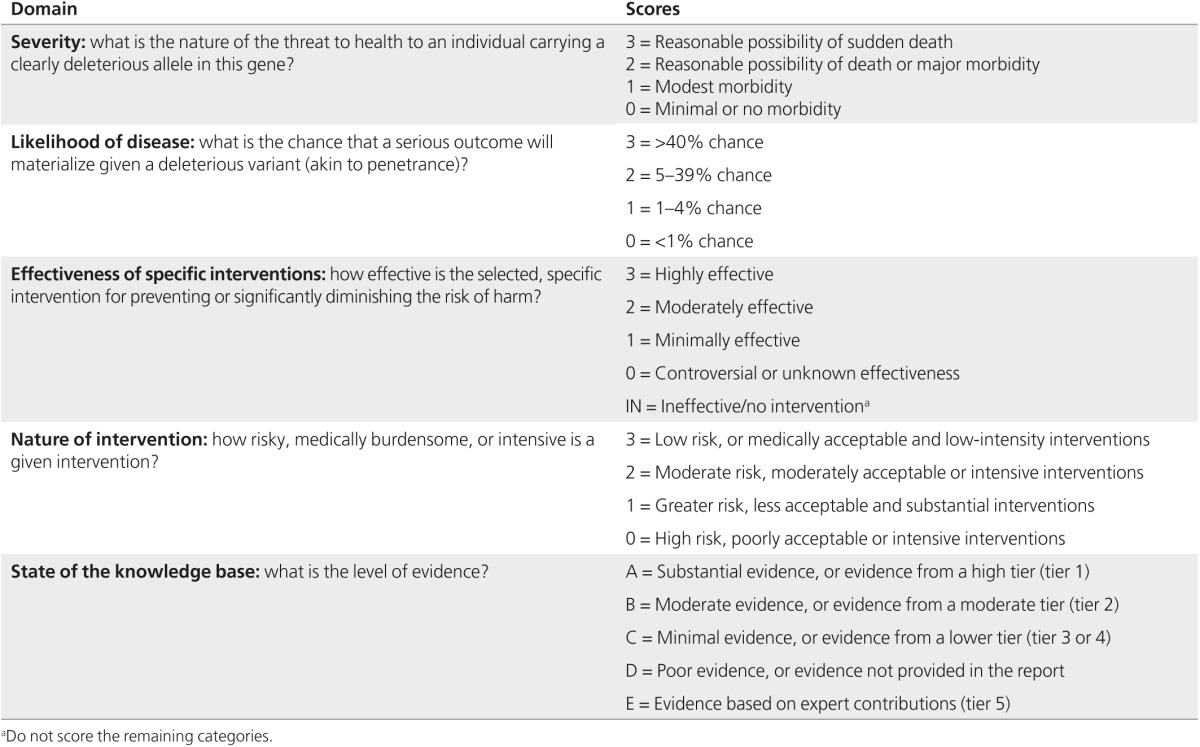
Each topic was independently scored by multiple AWG members, and then the scores were discussed among the larger AWG. We used consensus for assigning a single actionability score to each outcome–intervention pair.
Results
Clinical actionability summary reports and scores
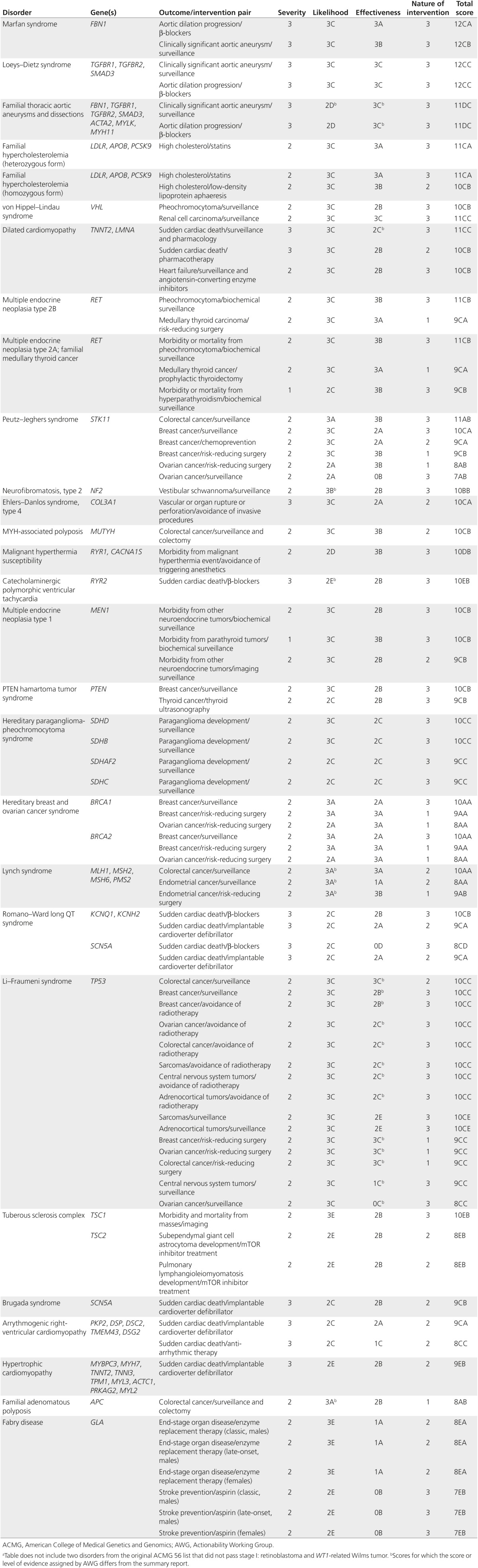
The AWG applied the current protocol to the list of 56 genes recommended for return as secondary findings by ACMG.10,11 Three topics did not pass stage I of the assessment for clinical actionability based on evidence identified in the preliminary literature search. Two of the three that did not pass stage I, retinoblastoma and WT1-related Wilms tumor, were not determined to be actionable in an undiagnosed adult, and the AWG did not make an exception for these topics to proceed to stage II. The third disorder that did not pass stage I based on evidence in the identified literature, Brugada syndrome, was determined to be actionable in an undiagnosed adult and a significant medical problem, but it did not meet the criteria for moderate penetrance or relative risk, because only 20–30% of people with this syndrome experience syncope and 8–12% have at least one cardiac arrest that could cause sudden death.14 However, the AWG made an exception for Brugada syndrome to proceed to stage II due to the severity of the clinical outcomes. Thus, Brugada syndrome and the remaining topics from the 56 genes assessed by the AWG (ACMG 56) that passed stage I have completed the AWG workflow and have been assigned consensus scores for clinical actionability (Table 3). These scores indicate mid- to high levels of actionability and a variety of available evidence to inform clinical actionability with regard to clinical interventions and the likelihood of clinical manifestations. The summary reports generated by the AWG are available on the ClinGen website (www.clinicalgenome.org).
Table 3. Clinical actionability scores for the ACMG 56a.
AWG challenges with scoring clinical actionability
The design and application of the protocol to characterize clinical actionability have presented thought-provoking challenges. For example, defining the outcomes and interventions for scoring clinical actionability was straightforward for some topics (e.g., familial adenomatous polyposis, which is associated with colorectal cancer as its major life-threatening manifestation that can largely be prevented with colectomy15) but difficult for others (e.g., Li–Fraumeni syndrome, a disorder associated with a vast array of neoplasms, including breast cancer, colorectal cancer, brain tumors, and osteosarcomas16). In the latter case, it was difficult to narrow down all potential outcomes and interventions for scoring. Some strategies the AWG considered included scoring only outcome–intervention sets for the most common outcomes or the most severe outcomes. In addition, interventions selected for scoring and their associated risk or burdens were often highly contextual and thus difficult to score. For example, for a woman with a pathogenic BRCA1 variant, the risk and burden of a recommended oophorectomy may be greater during childbearing years than during postmenopausal years. One potential option would be to generate scores across different contexts. In the example of BRCA1 and oophorectomy, this approach could entail generating scores by age (e.g., 25, 40, and 55 years); both the penetrance and burden of the intervention could vary significantly.
Assessment of the effectiveness of an intervention has been discussed extensively among the AWG. Some interventions overtly prevent an outcome, whereas others aim to minimize morbidity through early detection. For example, in women with BRCA1 and BRCA2 variants associated with HBOC, risk-reducing surgeries (i.e., prophylactic mastectomy and oophorectomy) are effective for reducing breast and ovarian cancer incidence, whereas breast cancer surveillance, such as mammography and magnetic resonance imaging (MRI), are designed for early disease detection to reduce morbidity.17 Because the AWG pursues both types of interventions, they are scored separately for actionability and the surveillance option is judged by its effectiveness to ultimately prevent a poor clinical outcome rather than its effectiveness in detecting cancer. Similarly, patients with neurofibromatosis type 2, who are at high risk for bilateral vestibular schwannomas, are advised to undergo regular MRI surveillance.18,19 MRI surveillance, of course, does not directly prevent vestibular schwannomas, but it is effective in triggering and directing distal and definitive clinical treatment (e.g., tumor excision). Therefore, when the AWG assessed clinical actionability, the effectiveness of both the proximal and distal effects of the intervention were considered. Thus, the concept of effectiveness with regard to our protocol could be fluid across outcomes and interventions and include both reduction in mortality and decreased or delayed morbidity.
The AWG also considered circumstances to avoid while evaluating the clinical actionability of a gene–disease pair. For example, patients with the vascular form of Ehlers–Danlos syndrome should avoid elective surgery and other invasive procedures (e.g., arteriograms) because of the increased risk of vascular rupture or organ perforation.20 The difficulty in scoring the effectiveness of such an intervention lies in the fact that avoiding the circumstance does not improve the health of the patient per se but not avoiding the circumstance can worsen it.
The protocol developed by the AWG relied heavily on published practice guidelines, with priority given to guidance based on evidence review. Paradoxically, many published guidelines do not provide evidence summaries for their recommendations. Thus, the actual effectiveness of a recommended intervention is sometimes unclear or unknown. For example, a recommendation for patients with Fabry disease is to take aspirin or other platelet-inhibiting agents to reduce their risk of stroke, although the effectiveness of this intervention was not described in the original source.21 AWG members were not comfortable scoring the effectiveness of this recommendation without evidence of stroke reduction for these patients, and the absence of supporting evidence limited the ability to measure effectiveness. This issue may eventually be addressed by new guideline criteria submitted to the National Guidelines Clearinghouse (http://www.guideline.gov/about/inclusion-criteria.aspx) as well as the Institute of Medicine's recommendations for developing trustworthy guidelines.22 These criteria would require guidelines to be transparent regarding the methods used, to be based on a systematic review of evidence (where it exists), and to provide an assessment of the benefits and harms of recommended care and alternative care options.
For rarer disorders, guidelines often base recommendations on evidence extrapolated from more common and related disorders. For example, evidence of the effectiveness of breast surveillance and risk-reducing surgery among women with BRCA1 and BRCA2 mutations has been used to recommend these interventions for women with rarer disorders that also increase the risk of breast cancer (e.g., Peutz–Jegher and Li–Fraumeni syndromes).17 To account for this extrapolation in such cases, the AWG adjusted the rating for the knowledge base when scoring actionability, such as reducing the score of the knowledge base even if the guideline even if the guideline meets criteria for a tier 1 or tier 2 reference (Table 2). Similarly, AWG members often gave the knowledge base a higher score than what the guideline tier warranted if their clinical opinion or experience indicated a discrepancy. For example, a tier 3 source for the penetrance of colorectal cancer on familial adenomatous polyposis was identified, but the knowledge base for likelihood was scored as an “A” based on discussion by the AWG.
Discussion
The AWG applied a standardized protocol to assess clinical actionability to the list of 56 genes recommended by the ACMG to return as secondary findings.10,11 As we expected, these disorders had scores at moderate to high levels of actionability. However, the actionability scores for different outcome–intervention pairs within genetic disorders varied. For HBOC, for example, breast cancer and surveillance had a higher actionability score than did ovarian cancer and oophorectomy (10 and 8, respectively). These scores highlight the nuance of generating consensus scores on actionability that account for both the effectiveness and nature of the intervention. In the HBOC example, the intervention deemed more effective for eliminating the harm (i.e., risk-reducing surgery) received a lower actionability score than the intervention deemed less effective for eliminating the harm (i.e., breast cancer surveillance) that was also less burdensome and invasive. In addition, although some disorders have substantial evidence for clinical actionability (e.g., HBOC), others have limited evidence (e.g., hereditary paraganglioma–pheochromocytoma syndrome).
The SQM, which enabled us to balance qualitative and quantitative scoring, was useful because it provided a predefined range of scores rather than the presence or absence of potential clinical actionability. This approach also allowed for scorers' subjectivity, a necessary criterion for any metric that must deal with issues that, like actionability, are not entirely quantifiable and incorporate subjective perceptions. The fact that the AWG scoring framework and the Multi-Criteria Decision Analysis developed by EVIDEM23 have many parallels and were developed independently reinforces the applicability of the scoring approach used by ClinGen.
The actionability scores generated by the AWG protocol should inform ongoing efforts by the ACMG and other groups to maintain and update lists to guide return of secondary or incidental findings. In addition, external stakeholders can apply the scores to prioritize genes for reporting incidental or secondary findings or for other purposes for which actionability is important. For example, the range of scores generated across a variety of genes and associated disorders could be used to identify a threshold indicative of a certain level of actionability to warrant the return of findings to patients. In addition, the end user could decide a priori whether a single outcome and the associated actionable intervention for a disorder and associated gene or genes are sufficient for the return of the finding. Actionability scoring methods may also inform other contexts in which actionability is important, such as primary screening.
The limitations of the AWG protocol are worth noting. For example, the recommendations added to the summary reports were deliberately not comprehensive because focusing on evidence-based and expert-based clinical resources and secondary literature made it possible to generate brief reports in a practical amount of time and in a consistent format. Thus, the protocol is not designed to identify all available evidence regarding actionability for a particular topic. However, this process identified gaps in synthesized knowledge and the need for future research and clinical translation to improve the knowledge base for clinical actionability of genetic disorders. It also highlights the need for evidence-based guidelines, as recommended by the National Academy of Medicine (formerly the Institute of Medicine).22 This would provide access to evidence summaries supporting clinical recommendations to better describe the effectiveness of clinical interventions, an important aspect of clinical actionability.
The current protocol was limited in scope and did not address certain patient-specific factors that are important to clinical care, such as personal utility, patient perspectives on the burden of interventions, or how the risks of outcomes and burdens of interventions vary with certain patient characteristics (e.g., age). In addition, the current protocol is limited to genetic disorders with clinical actionability among adults and does not address only childhood-onset disorders. Although the limited scope of the protocol was based on feasibility, it can be expanded in future iterations.
The SQM developed for this protocol enabled us to assign a range of scores rather than the presence or absence of clinical actionability. It should be recognized that the scoring assignments for various aspects of actionability necessarily have an inherently subjective component (Table 2). For example, a disorder with the potential for sudden death (e.g., Brugada syndrome) was assigned a higher score for severity than a disorder that may lead to a more delayed mortality (e.g., Lynch syndrome). The rationale for this was that medical events that could lead to sudden death (e.g., aortic aneurysm) may have little or no clinical warning or window of time after onset to prevent death. Thus, for these conditions, a higher score was assigned to account for the arguably greater need to identify patients early in order to implement preventative interventions. In addition, a more clinically invasive intervention (e.g., prophylactic mastectomy) was assigned a lower score for the nature of the intervention than a less invasive intervention (e.g., chemoprevention). Although some may argue that the invasive nature of the intervention may not make a condition more or less actionable, we predetermined that our assessment of clinical actionability should account for the risk and burden to the patient. Overall, it is important to note that assessments of issues such as actionability are meant to provide a starting point for discussion.
Generation of the summary reports for clinical actionability of additional genetic disorders is ongoing. Advances in health care will provide evidence regarding clinical actionability, such as new or more effective interventions. In addition, our understanding of penetrance, phenotypes, and mutational prevalence of many genes and associated disorders is likely to change. Our knowledge is currently derived, for the most part, from populations accrued through biased ascertainment due to clinical manifestations or prior diagnosis. Study designs that can reduce this ascertainment bias will improve estimates of the prevalence and penetrance of genetic disease. Efforts to expand this protocol will probably include plans to ensure that summary reports stay up to date and remain a reliable resource.
In summary, the AWG protocol provides a structured, transparent framework for summary reviews of evidence regarding the clinical actionability of genomic variation that could be used in a variety of contexts. The ClinGen AWG framework provides needed support to the research and clinical communities in general for making clear, streamlined, and consistent determinations of clinical actionability based on transparent criteria to guide the analysis and reporting of variations detected in genome-scale sequencing.
Disclosure
The authors declare no conflict of interest.
Acknowledgments
This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. The ClinGen consortium is also funded by the National Human Genome Research Institute and the Eunice Kennedy Shriver National Institute of Child Health and Human Development through contracts 1U41HG006834-01A1, 1U01HG007437-01, 1U01HG007436-01, and 1UO1HG006487-01. The efforts of L.G.B. were supported by the Intramural Research Program of the National Human Genome Research Institute.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
We thank Kevin Lutz and Elizabeth Hess for their editorial support.
Supplementary Material
References
- Hayden EC. Technology: the $1,000 genome. Nature 2014;507:294–295. [DOI] [PubMed] [Google Scholar]
- Green ED, Guyer MS; National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature 2011;470:204–213. [DOI] [PubMed] [Google Scholar]
- Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med 2014;370:2418–2425. [DOI] [PubMed] [Google Scholar]
- Kohane IS, Masys DR, Altman RB. The incidentalome: a threat to genomic medicine. JAMA 2006;296:212–215. [DOI] [PubMed] [Google Scholar]
- Presidential Commission for the Study of Bioethical Issues. Anticipate and Communicate: Ethical Management of Incidental and Secondary Findings in the Clinical, Research, and Direct-to-Consumer Contexts. Presidential Commission for the Study of Bioethical Issues: Washington, DC, 2013. [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Amendola LM, Turner EH, et al.; National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. Actionable, pathogenic incidental findings in 1,000 participants' exomes. Am J Hum Genet 2013;93:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Rubinstein WS, Facio FM, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet 2012;91:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Meslin EM, Marteau TM, Caulfield T. Genomics. Deflating the genomic bubble. Science 2011;331:861–862. [DOI] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, et al.; ClinGen. ClinGen–the Clinical Genome Resource. N Engl J Med 2015;372:2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Berg JS, Grody WW, et al.; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American College of Medical Genetics and Genomics. Incidental findings in clinical genomics: a clarification. Genet Med 2013;15:664–666. [DOI] [PubMed] [Google Scholar]
- Goddard KA, Whitlock EP, Berg JS, et al. Description and pilot results from a novel method for evaluating return of incidental findings from next-generation sequencing technologies. Genet Med 2013;15:721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Foreman AK, O'Daniel JM, et al. A semiquantitative metric for evaluating clinical actionability of incidental or secondary findings from genome-scale sequencing. Genet Med 2015; e-pub ahead of print 13 August 2015. [DOI] [PMC free article] [PubMed]
- Napolitano C, Prior S. Brugada Syndrome. OrphaNet Encyclopedia, 2009. http://www.orpha.net/. Accessed 22 September 2015.
- Burt RW, Cannon JA, David DS, et al.; National comprehensive cancer network. Colorectal cancer screening. J Natl Compr Canc Netw 2013;11:1538–1575. [DOI] [PubMed] [Google Scholar]
- Schneider K, Zelley K, Nichols KE, et al. Li-Fraumeni syndrome. 19 January 1999 (updated 11 April 2013). In: Pagon RA, Adam MP, Ardinger HH, et al. (eds). GeneReviews® [Internet]. University of Washington: Seattle, WA, 1993–2015. http://www.ncbi.nlm.nih.gov/books/NBK1311/. Accessed 22 September 2015. [Google Scholar]
- National Collaborating Centre for Cancer. Familial Breast Cancer. Classification and Care of People at Risk of Familial Breast Cancer and Management of Breast Cancer and Related Risks in People with a Family History of Breast Cancer. National Institute for Health and Care Excellence (NICE): London, 2013:57 (Clinical guideline; no. 164). https://www.nice.org.uk/guidance/cg164. [PubMed]
- Gutmann DH, Aylsworth A, Carey JC, et al. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA 1997;278:51–57. [PubMed] [Google Scholar]
- Evans DG, Baser ME, O'Reilly B, et al. Management of the patient and family with neurofibromatosis 2: a consensus conference statement. Br J Neurosurg 2005;19:5–12. [DOI] [PubMed] [Google Scholar]
- Burcharth J, Rosenberg J. Gastrointestinal surgery and related complications in patients with Ehlers-Danlos syndrome: a systematic review. Dig Surg 2012;29:349–357. [DOI] [PubMed] [Google Scholar]
- Eng CM, Germain DP, Banikazemi M, et al. Fabry disease: guidelines for the evaluation and management of multi-organ system involvement. Genet Med 2006;8:539–548. [DOI] [PubMed] [Google Scholar]
- Graham R, Mancher M, Miller Wolman D, Greenfield S, Steinberg E, et al. Clinical Practice Guidelines We Can Trust. National Academies Press: Washington, DC, 2011. [PubMed] [Google Scholar]
- Goetghebeur MM, Wagner M, Khoury H, Levitt RJ, Erickson LJ, Rindress D. Evidence and value: impact on DEcisionMaking–the EVIDEM framework and potential applications. BMC Health Serv Res 2008;8:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.