Abstract
Androgen receptor (AR) splice variants (AR-Vs) are constitutively active transcription factors that function in the absence of ligand. AR-Vs represent one of several AR re-activation mechanisms utilized by prostate cancer to circumvent first-line androgen deprivation therapy. Second line therapies such as enzalutamide and abiraterone are treatments that re-target components of the androgen/AR axis. However, these second line therapies do not benefit all patients, and patients that do receive initial benefit can develop resistance rapidly. Alterations in components of the androgen/AR axis, including expression of AR-Vs, appear to be linked to primary as well as secondary resistance to second line therapies. However, some key conclusions appear to differ depending on the tissue compartment and measurement platform utilized for analysis. In this review, alterations in AR and the broader AR pathway will be examined in the context of primary prostate cancer tissue, metastatic castration-resistant prostate cancer tissue, circulating tumor cells, and circulating cell-free tumor DNA. Questions regarding the utility of AR-V measurements to provide prognostic information or predict patient responses to AR-targeted therapies will be addressed.
Keywords: androgen receptor splice variant, prostate cancer, castration-resistant prostate cancer, circulating tumor cell
1. Introduction
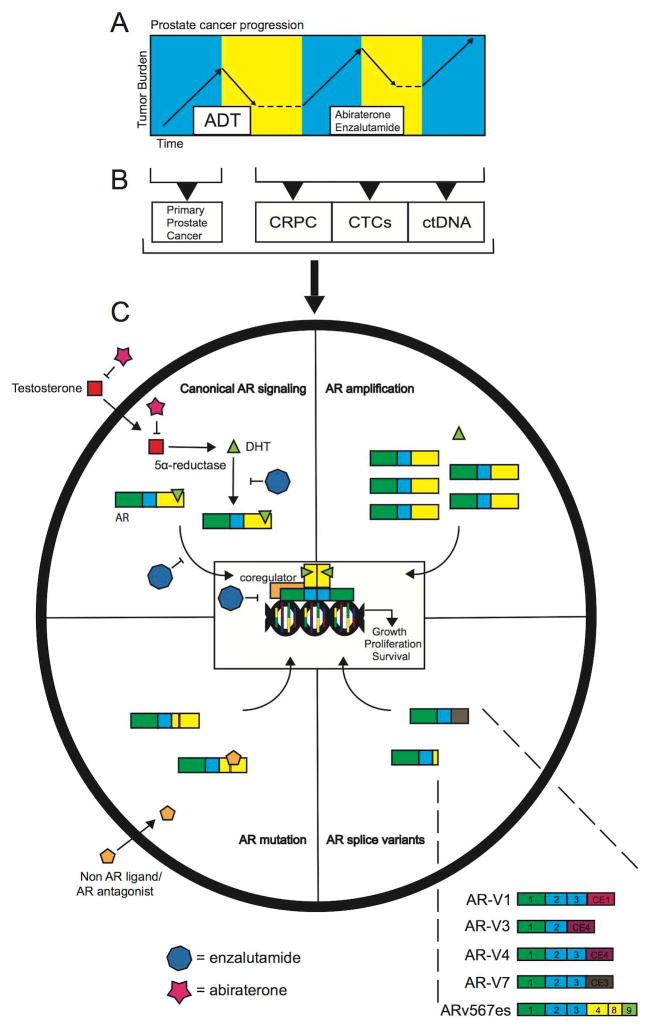
Prostate cancer accounts for a quarter of all new cancer diagnoses, and one in seven men in the United States will develop the disease. Of those men, roughly 27,000 will die each year, making prostate cancer the second leading cause of cancer related death in the United States. [1]. Prostate cancer is referred to as an androgen-dependent disease, which is based on the requirement of androgen-mediated activation of the androgen receptor (AR) for growth and survival of prostate cancer cells. As a result of androgen-dependence, the androgen/AR axis has been a target of treatment since the early 1940s when Huggins et al. established the beneficial effects of castration in men with metastatic prostate cancer [2]. Contemporary therapies for prostate cancer continue to rely on reducing the levels of circulating androgens and inhibiting AR function via orchiectomy or luteinizing hormone releasing hormone (LHRH) agonists. Antiandrogens such as bicalutamide are also used to competitively block the ability of androgens to bind and activate the AR. Patients that fail this first-generation of androgen depletion therapies (ADT) and progress to a disease stage referred to as castration-resistant prostate cancer (CRPC) are treated with second-generation AR-targeting therapies such as abiraterone acetate and enzalutamide (Fig. 1A). Abiraterone acetate is an inhibitor of the cytochrome P450 c17 (CYP17) testosterone synthesis enzyme [3], which can achieve a more robust inhibition of androgen production than castration. Enzalutamide is an antagonist that binds the AR with a higher affinity than bicalutamide, and can partially inhibit AR translocation to the nucleus, reduce binding of AR to DNA, and prevent recruitment of coactivators required for active transcription [4]. Despite these advances in treatment, CPRC remains a uniformly fatal disease. Importantly, persistent AR signaling can be maintained during and after treatment with these androgen/AR-targeted therapies. The focus of this review will be observations made in clinical tissues collected at various disease stages (Fig. 1B) that have illuminated key mechanisms of AR re-activation that can occur in CRPC (Fig. 1C), with an emphasis on AR splice variants (AR-Vs).
Fig. 1. Canonical/resistance AR signaling and AR-V structure.
A. Hypothetical timeline representing progression of primary prostate cancer to castration resistant prostate cancer. B. Tissues and other clinical material (CTCs-circulating tumor cells; ctDNA-cell free, circulating tumor DNA) available for interrogation of alterations in the androgen/AR pathway. C. Illustration of normal and aberrant modes of androgen/AR signaling. The upper left quadrant summarizes canonical AR signaling that occurs in normal development and prostate cancer. In this setting, DHT binds AR, which promotes AR translocation to the nucleus and transcriptional activation. Components of this axis targeted by abiraterone and enzalutamide are indicated. The top right quadrant illustrates how amplification of the AR locus results in AR protein overexpression, which is highly sensitive to castrate levels of androgen. The bottom right quadrant illustrates the domain structure of AR-Vs. The expanded view displays exon composition of 5 separate mRNAs that encode discrete AR-Vs that are detailed within the text. The bottom left quadrant shows how mutations in the AR ligand binding domain can convert non AR ligands and AR antagonists to agonists, leading to transcription of AR target genes.
1.1 Brief Introduction to AR Structure/Function
The AR is a member of the class I nuclear steroid receptor family. Within this family of steroid receptors are transcription factors regulated by ligands including mineralocorticoids (MR), estrogens (ER), progestins (PR) and glucocorticoids (GR). All of these steroid receptors are activated upon binding ligand, after which they engage their cognate hormone response element DNA sequences located in promoter and enhancer regions throughout the genome. The AR gene is located on the X chromosome at cytogenetic position Xq11-12. This gene contains eight exons and encodes a modular 110kDa protein [5]. Exon 1 of AR encodes the NH2-terminal domain (NTD), which is intrinsically disordered and harbors the transcriptional activation function-1 (AF-1) domain essential for the bulk of AR transcriptional activity. Exons 2 and 3 encode the DNA-binding domain. The 5′ section of exon 4 encodes the hinge region, which is a flexible domain containing a nuclear localization signal (NLS) that becomes accessible upon ligand binding. The 3′ section of exon 4 along with exons 5–8 encode the AR ligand-binding domain (LBD) and an AF-2 domain that is transcriptionally active but with weaker activity than AF-1 [5]. Upon binding to androgen, AR undergoes a conformational change, which exposes the NLS, and results in translocation to the nucleus. Once in the nucleus, AR binds to androgen response elements (AREs) and recruits a wide variety of coregulatory proteins with various scaffolding, enzymatic, and chromatin-modifying functions, ultimately leading to a finely-tuned level of transcriptional output for target genes. These features of canonical AR signaling are illustrated in Figure 1C.
1.2 Brief Introduction to AR Splice Variants (AR-Vs)
Over the past 8 years, AR-Vs have emerged as an important component of resistance to therapies targeting the androgen/AR axis. Approximately 20 discrete AR-Vs have been identified in cell- and animal-based models of CRPC progression, as well as tissues from patients with prostate cancer [6, 7, 8, 9]. AR-Vs share the same NTD/DBD modular domain structure as the full-length AR, but commonly lack the COOH-terminal LBD/AF-2 module. Figure 1C illustrates the structure of select AR-Vs that will be detailed in this review. Truncation of the AR LBD results in constitutive transcriptional activity of the AR NTD/DBD core in the absence of ligand. Thus, most AR-Vs have been reported to function as ligand-independent transcription factors, although discrepancies have been noted [10]. Originally, AR-Vs were reported to arise from proteolytic cleavage of full-length AR by calpains, which are calcium-dependent proteinases [11]. These early studies found that the AR hinge region harbored a calpain cleavage site, which could be targeted to eliminate the AR LBD. However, subsequent studies in the 22Rv1 cell line, which was the same model used to develop the proteolysis mechanism, revealed that truncated AR protein species were differentially sensitive to small interference RNAs (siRNAs) compared with full-length AR. This finding of differential siRNA sensitivity formed the basis for the concept that AR-Vs were encoded by mRNAs that were separate from the mRNA encoding full-length AR [12]. Subsequent studies demonstrated that different AR-V proteins were encoded by mRNAs containing exons 1–3 or exons 1–4 of the AR gene, but variable 3′ terminal exons harboring in-frame translation termination signals [13]. Subsequent studies defined an additional class of AR-V proteins that were encoded by skipping of exons encoding the AR LBD, but terminating with an out-of-frame 3′ terminal AR exon 8 [13].
One mechanism thought to underlie AR-V expression in prostate cancer is alternative splicing. RNA co-immunoprecipitation (RIP) assays using enzalutamide-treated VCaP cells revealed that splicing regulatory factors ASF/SF2, and U2AF65 were recruited to pre-mRNA sequences aligning with a splice acceptor site in AR exon CE3, which is the 3′ terminal exon in the mRNA encoding AR-V7 [14]. In this scenario, it was concluded that increased recruitment, but not expression, of splicing factors contributed to AR-V7 in these cells. In line with this, the AR-V7-negative LNCaP cell line did not display the same pattern of engagement with splicing factors seen in VCaP cells. Further, siRNA-mediated knockdown of splicing factors in this model system reduced the expression of AR-V7 [14]. However, one caveat with comparing splicing factor recruitment to AR pre-mRNAs in VCaP vs. LNCaP cells is that VCaP cells harbor approximately 20 more copies of the AR gene than the hypotetraploid LNCaP cell line due to massive amplification of the AR gene [15]. In this case, VCaP cells express much higher levels of AR mRNA and protein, and it would be assumed that there would be higher sensitivity for detection of splicing regulatory factors bound to AR pre-mRNA in VCaP cells.
An additional mechanism that has been shown to underlie AR-V expression in prostate cancer is genomic rearrangements within the AR gene. For example, a 35kb tandem duplication containing AR exon 3 as well as several cryptic exons including CE3 is linked to the creation of truncated AR variants in the androgen-independent 22Rv1 cell line. This tandem duplication extends the genomic span between exons 3 and 4, which may increase the probability of exon 3 splicing to exon CE3 or other cryptic exons, thereby increasing expression of AR-V7 as well as other AR-Vs relative to full-length AR [16, 17]. Moreover, patient derived xenograft models LuCaP 86.2 and LuCaP 136 express high levels of the ARv567es exon skipping AR splice variant resulting from splicing of AR exon 4 directly to AR exon 8. These patient derived xenografts harbor a deletion or inversion, respectively, of a genomic segment containing AR exons 5–7. These specific rearrangements were demonstrated to cause ARv567es expression following targeted manipulation of the AR gene in prostate cancer cell lines using genome engineering technology [17].
2. Alterations in the AR and AR-V pathways in primary prostate cancer tissue
2.1 Alterations in AR in primary prostate cancer tissue
Alterations in the AR gene are very rare in primary prostate cancer tissues, which is consistent with the lack of use of ADT for patients with localized disease. In a targeted sequencing study of archival, formalin-fixed specimens, only one AR point mutation was detected in primary prostate cancer tissue [18]. Other studies have reported a slightly higher mutational burden in primary prostate cancer specimens, but rates still remain relatively low [19]. The most comprehensive study of primary prostate cancer tissues to date, the cancer genome atlas (TCGA) project, reported zero somatic AR point mutations across a series of 333 primary prostate cancer tissues [19]. On the other hand, this TCGA study did report rare instances of AR gene amplification, but at less than 1% frequency. Overall, these studies provide evidence of minimal direct AR alterations in primary prostate cancer.
2.2 AR-Vs in normal and primary prostate cancer tissue
Although mutation and amplification of the AR gene are rarely observed in primary prostate cancer, expression of AR-Vs has been observed in non-malignant prostate tissue as well as primary prostate cancer. Targeted qRT-PCR analysis of AR-V7 mRNA revealed that AR-V7 was expressed in 5 of 17 benign prostate tissues [8]. Analysis of RNA-seq data from peritumoral prostate tissues in the TCGA project also revealed frequent expression of AR-V7 as well as other AR-Vs in normal tissue [19]. The significance of AR-V mRNA expression in benign tissue is not clear, but is consistent with another study where mRNA expression of several AR-Vs was detected in normal prostate tissue from men who had been castrated with an LHRH receptor agonist or treated with DHT gel in a study testing male contraception [9]. Analysis of RNA-seq data from cancer tissues in the TCGA cohort indicated that AR-V7 was the most frequently expressed AR-V mRNA in primary prostate cancer tissue [19]. Additionally, a range of other AR-V mRNAs arising from cryptic exon inclusion as well as exon skipping events were also detected frequently in primary prostate cancer tissues from the TCGA study [19]. An unexpected finding from analysis of TCGA data was an overall lack of expression of the ARv567es exon skipping AR splice variant [19]. This finding was unexpected because a previous targeted RT-PCR study found that ARv567es expression mirrored that of AR-V7 and AR-V1 in normal prostate tissue as well as primary prostate cancer samples, with levels detectable but low relative to full length AR [9, 20]. In contrast, targeted qRT-PCR analysis of AR-V7 mRNA was largely in-line with findings from the TCGA study, revealing that 34 of 82 hormone naïve prostatectomy specimens were positive for AR-V7 mRNA expression [8]. Interestingly, AR-V7 mRNA expression greater than the median across this cohort was associated with lower probability of PSA-progression free survival following surgical treatment [8]. Conversely, expression of AR-V1, composed of contiguously-spliced AR exons 1, 2, 3, and CE1, was not found to be associated with PSA-progression free survival. However, a subsequent study that evaluated AR-V1 and AR-V7 mRNA expression using a branched chain assay failed to identify an association between either of these AR-V species and prostate cancer progression [21]. As a result of these discordant findings, it remains unclear whether AR-V7 mRNA levels in primary prostate cancer tissue provide prognostic information.
The unique COOH-terminal extensions encoded by the unique 3′ terminal exons in AR-Vs such as AR-V7 have provided opportunities for development of AR-V-specific antibodies [7, 8]. For example, staining of tissue microarrays with an antibody specific for AR-V7 revealed that AR-V7 protein was expressed in both the nucleus and cytoplasm of hormone naïve primary prostate cancer tissues [7]. Tissue microarrays also exhibited cytoplasmic AR-V7 staining in benign tissue but no nuclear signal [7]. AR-V7 cytoplasmic staining in human prostate tissue microarrays was found to be associated with shorter time to PSA recurrence [7]. The finding of cytoplasmic AR-V7 expression is unexpected, as biochemical studies have demonstrated this species to be a constitutively nuclear protein [22]. This raises concern about the performance of AR-V7 antibody reagents in tissue-based studies, particularly in benign prostate and hormone naïve prostate cancer, where AR-V7 expression is extremely low.
2.3 Alterations in the broader AR pathway in primary prostate cancer tissue
Primary prostate cancer has recently been classified based on seven distinct molecular subtypes. Four of these subtypes are defined by genomic fusions of ETS family genes (ERG, ETV1, ETV4, or FLI1), or mutations in SPOP, FOXA1, or IDH1. In the analysis of 333 primary tumors from the TCGA study, 74% of samples fell into one of these subclasses [19]. Interestingly, the majority of these molecular subclasses are mechanistically linked to the broader androgen/AR pathway in prostate cancer. For example, the most frequent ERG fusion is TMPRSS2:ERG, which results in androgen/AR-mediated overexpression of ERG mRNA and protein via the androgen/AR-regulated TMPRSS2 enhancer and promoter [19, 23]. TMPRSS2:ETV1 is the most frequent ETV fusion, which has a similar regulatory outcome of androgen/AR-mediated ETV1 mRNA and protein overexpression via the TMPRSS2 enhancer and promoter. Additional fusions to diverse promoters and enhancers have been characterized for ERG, ETV1, ETV4, and FLI1, the majority of which are highly-active in prostate cells by virtue of androgen/AR regulation. Impaired expression of these genetic drivers following inhibition of the androgen/AR axis provides a molecular basis for the robust response of prostate cancer cells to ADT. However, evaluation of whether TMPRSS2:ERG or other ETS fusions relay any prognostic information has not yielded clear results [24, 25].
In addition to gene fusion subclasses, the FOXA1 and SPOP mutant subclasses can also be considered as molecular alterations affecting the broader AR pathway. For instance, the FOXA1 gene encodes a pioneer transcription factor that facilitates the relaxation of compacted chromatin structures for AR and other transcription factors [19, 26, 27, 28]. Mutations in FOXA1 include missense, inframe deletions, and truncating mutations, which are concentrated in the exons encoding the FOXA1 forkhead DNA binding domain [19]. Similarly, the SPOP gene, which encodes the substrate-binding subunit of a Cullin-based E3 ubiquitin ligase [29, 30], is also frequently mutated in primary prostate cancer [19, 28]. SPOP mutations are concentrated in exons encoding the MATH domain, which is responsible for substrate recruitment. Two substrates identified for SPOP are AR and the AR co-activator SRC-3, with mutant versions of SPOP displaying impaired binding to these substrates [31, 32, 33]. Therefore, AR and SRC-3 proteins are stabilized in cells harboring mutant SPOP. Interestingly, the SPOP MATH domain interacts with the AR LBD, and therefore AR-Vs appear to be resistant to SPOP-mediated degradation [31]. However, a separate study indicated that AR-Vs may remain susceptible to SPOP-dependent ubiquitination and degradation by virtue of heterodimerization with full-length AR [34]. Assignment of an AR activity score to primary prostate cancer tissues, which was based on expression levels of a set of 20 AR regulated genes, revealed that FOXA1 and SPOP sub-classes had the highest levels of AR transcriptional output. This reinforces how intimately FOXA1 and SPOP are tied to the normal function of AR and also the disease-associated function of AR in primary prostate cancer.
3. Alterations in the AR and AR-V pathways in CRPC tissue
Most prostate tumors have an initial response to ADT because of the androgen-dependent nature of the disease. However, virtually all patients will eventually develop resistance. In this CRPC stage of the disease, most tumors remain dependent on activity of the androgen/AR axis [35]. Mechanisms underlying persistent activity of the androgen/AR axis in CRPC have been reviewed in detail [36], and include AR mutations, AR amplification, expression of AR splice variants, and alterations in AR regulators (Fig. 1C). As discussed below, a critical role in CRPC is supported by the finding that these alterations occur with higher frequency in CRPC than in primary prostate cancer, although there are some notable discrepancies. Additionally, the proportion of the genome that is altered through copy number alteration and/or mutation is greater in patients with CRPC, indicating that androgen/AR-independent mechanisms also contribute to resistance [19]. Overall, CRPC is a complex disease state with a myriad of genomic alterations that enable continued growth and disease spread in the face of therapeutic pressure.
3.1 Alterations in AR in CRPC tissue
Point mutations in AR are more frequent in CRPC than primary prostate cancer. In a whole exome sequencing study of 25 tissues from patients with CPRC, AR point mutations were found to occur in 20% of samples [18]. Other studies with larger sample sizes detected AR point mutations at a frequency of roughly 10% [37, 38]. Among the mutations found in these studies were T878A, W742C, and L702H, which are located in the AR LBD and have been shown to convert a variety of AR antagonists into agonists [18, 38]. Although single point mutations are the most frequent occurrence, there have been reports where multiple point mutations have been detected in a single tissue sample, such as T878A and Q903H in a recent whole exome sequencing study of CRPC biopsies [38]. This study did not establish if these mutations were concurrent on the same AR allele, or whether this reflected intra-tumor heterogeneity. However, the latter scenario is likely given that the Q903H mutation was present at a lower allelic fraction than T878A.
The most common AR alteration in CRPC is AR gene amplification. In a biopsy-based whole exome sequencing study of 150 metastatic CRPC bone and soft tissue tumor samples, AR amplification occurred in more than 50% of samples [38]. In another study of CRPC samples, AR copy number gain was detected in 25 of 50 cases [37]. Mechanistically, AR amplification leads to overexpression of AR mRNA and protein, which is thought to facilitate AR activation by a mass action effect under low androgen conditions [39, 40]. Given that AR amplification in primary prostate cancer is exceedingly rare, this highlights the clinical importance of increased AR protein expression in CRPC [19, 41, 42, 43]. AR gene amplification appears to occur in a mutually exclusive pattern with AR point mutations, although rare exceptions have been noted that may reflect intra-tumor heterogeneity [38].
3.2 AR-Vs in CRPC tissue
Because AR-Vs were discovered in cell line model systems of CRPC and were shown in these models to function as ligand-independent transcription factors, most studies interrogating AR-Vs in clinical material have focused on CPRC tissues. Although expression of numerous AR-Vs has been detected in CRPC tissues [7, 8, 9, 20, 38], quantitative assessments have often revealed that AR-Vs are expressed at a much lower level in clinical material than in cell lines where they were initially discovered and characterized functionally [7, 8, 9, 12]. This has complicated efforts to clearly define the importance of AR-Vs in mediating resistance to ADT and second-generation AR-targeted therapies such as abiraterone and enzalutamide. Despite these challenges, there is compelling evidence in support of a strong link between AR-V expression and CRPC progression. For example, Hu and colleagues evaluated 124 clinical samples representing normal prostate, hormone-naïve primary prostate cancer, and CRPC for mRNA expression of AR-V7 (1/2/3/CE3 isoform) using an RT-PCR assay. They observed that 84% of CPRC specimens displayed AR-V7 mRNA expression vs. 41% of hormone-naïve primary prostate cancer specimens [8]. This mRNA-based finding was supported by a subsequent immunohistochemistry analysis of a human prostate tissue microarray consisting of hormone-naïve primary prostate cancer and CRPC specimens [7]. Staining with an antibody specific for AR-V7 demonstrated a significant increase in AR-V7 nuclear expression in CRPC tumors relative to hormone-naïve primary prostate tumors. Surprisingly, 86% of hormone-naïve primary prostate cancer tissues had AR-V7 protein expression in the cytoplasm compared to 100% in CRPC samples [7]. However, as discussed earlier, cytoplasmic AR-V7 staining may reflect non-specific reactivity in tissues expressing AR-V7 protein at a level below the threshold of accurate detection. Overall, these data demonstrate that AR-V7 mRNA and protein is expressed in treatment-naïve primary prostate cancer samples, but the transcriptional effects of nuclear AR-V7 may not materialize until the disease has progressed to the CRPC stage.
More recent RT-PCR-based studies have interrogated expression of multiple AR-Vs simultaneously in CRPC tissues. For example, Sun and colleagues reported that 31 of 69 metastatic CRPC specimens were positive for either AR-V7 or ARv567es mRNA expression [9]. Of note, ARv567es was detected at twice the frequency as AR-V7 in this study, which is in opposition to more recent RNA-seq based studies that have concluded AR-V7 is the most frequently-expressed AR splice variant in CRPC tissue [19, 38]. As expanded below, these discrepancies may reflect the use of targeted RT-PCR vs. RNA-seq based assessments. In this same study by Sun and colleagues, AR-V7 and ARv567es were co-expressed in 6 of 69 metastatic samples [9]. Hörnberg and colleagues performed a similar RT-PCR-based analysis of AR-V1 and AR-V7 transcript expression in bone metastases from hormone-naïve and CRPC patients [20]. In this study, AR-V1 expression was consistently detected in CRPC bone metastases. AR-V1 positivity was also consistently observed in hormone-naïve bone metastases and primary prostate tumors, although, AR-V1 expression in CRPC bone metastases samples was higher relative to the other samples. Moreover, AR-V7 mRNA displayed a similar expression pattern in this study, but was detected less frequently than AR-V1 in both primary prostate tumor and hormone-naïve bone metastases [20]. Importantly, patients with bone metastases displaying the highest quartile of AR-V7 mRNA expression had reduced cancer-specific survival compared with patients having lower AR-V7 expression in bone metastases [20]. Collectively, these studies employing RT-PCR detection have provided strong support for an important disease role of AR-Vs in CRPC.
Interestingly, RNA-seq assessments of AR-Vs in clinical CRPC have not been confirmatory of the RT-PCR-based conclusions that AR-Vs play an important role in CRPC progression. The common strategy for assessment of AR-V expression in RNA-seq datasets has been to quantify the number of individual RNA-seq reads spanning exon/exon boundaries that are common to all AR mRNA species as well as the number of individual RNA-seq reads spanning exon/exon boundaries that are unique to AR and AR-Vs, and developing ratios to reflect the fractional amount of overall AR expression that is attributable to any particular AR species. This quantification strategy was used to analyze RNA-seq data from 125 pre-abiraterone and -enzalutamide CRPC biopsy samples as well as 333 primary prostate cancer specimens [19, 38]. These data demonstrated that AR-V7 (defined by the exon 3/CE3 splice junction) and AR-V3 (defined by the exon 2/CE4 splice junction) were the most frequently expressed AR-V mRNAs in CRPC tissues, and displayed the highest fractional expression of all AR-V mRNAs as a function of overall AR expression. Remarkably, AR-V7 and AR-V3 were also the most frequently detected AR-Vs in primary prostate cancer specimens, and their fractional expression levels as a function of overall AR expression appeared to be largely unchanged compared to the measurements in CRPC tissues [38]. Additionally, ARv567es, defined by the exon 4/8 splice junction, was only detected in 4/125 CRPC specimens, despite having being found in previous RT-PCR studies of CRPC metastases to display a trend towards association with reduced cancer-specific survival [20]. Understanding the basis for this discordance in conclusions following RT-PCR measurements vs. RNA-seq measurements of AR-Vs in prostate cancer tissues will be important for developing a clearer picture of their role in CRPC progression. One possibility is that RT-PCR measurements typically measure the expression of a specific AR-V relative to an internal housekeeping gene whereas RNA-seq data has been evaluated based on fractional expression of an AR-V relative to overall AR expression. It is possible this RNA-seq data analysis strategy is not highlighting those tissues where overall AR expression is increased, leading to higher expression of AR-Vs as well as full-length AR. Additionally, data comparing the change in expression of AR-Vs after introduction of second line AR inhibitors would provide great insight into how AR-Vs respond, and thereby contribute to CRPC progression in different treatment contexts.
3.2 Alterations in the broader AR pathway in CRPC tissue
As is the case in primary prostate cancer tissue, alterations in AR-related pathway components are also observed in CRPC tissue. For example, FOXA1 has been found to be mutated in CRPC samples, at a slightly higher frequency than in primary prostate cancer [28, 37, 38]. Interestingly, there also appears to be a shift in the distribution of FOXA1 mutations in CRPC compared with primary prostate cancer. For example, in primary prostate cancer, FOXA1 mutations were concentrated in the Forkhead domain [19] whereas a study of CRPC tissue found that four of seven FOXA1 mutations were in the tail-anchored (TA) domain and only one (G87R) mutation was detected in the Forkhead domain [37]. Similar non-Forkhead domain mutations, most of which localized to a region between the Forkhead domain and tail-anchored domain, were also frequently observed in a biopsy-based study of 150 CRPC specimens [38]. SPOP mutations, like FOXA1 aberrations, are also present at similar frequency in CRPC as primary prostate cancer, but with no apparent differences in distribution [28, 38]. Beyond FOXA1 and SPOP, several other AR activators and repressors within the AR pathway are altered in CRPC. For instance, nuclear receptor coactivators 1 and 2 (NCOA1/2), which are known to facilitate AR transcriptional activity, are frequently overexpressed at the mRNA and protein levels in metastatic CRPC [41], while nuclear receptor corepressors 1 and 2 (NCOR1/2), which are AR repressors, are commonly inactivated by mutation (nonsense, frameshift, missense), gene fusion or decreased mRNA expression [38, 41]. The importance of these specific AR pathway alterations is underscored by the finding that they are more frequent in metastatic CRPC samples compared to primary prostate cancer [19, 28]. Taking into account the cumulative total of all the alterations identified in the AR and the broader AR pathway has indicated that AR signaling is altered in 100% of metastatic CRPC cases [41]. These data underscore the critical role of AR in prostate cancer progression and therapeutic resistance.
4. Detection of Alterations in the AR and AR-V Pathways in Blood
Circulating tumor cells (CTCs) and cell-free circulating tumor DNA (ctDNA) provide great potential for determining cancer progression in real time with methods that are non-invasive compared to biopsies, where sample yield is typically low. Observing proteomic, transcriptomic, miRNA expression, and genotypic changes in CTCs or genomic changes in ctDNA in response to various therapeutics enables analysis and understanding of tumor evolution. As this technology matures, it may also lead to more nuanced care in the developing age of personalized medicine. In the context of CRPC, CTC and ctDNA are thought to represent cells or cellular material shed from metastases. This section of the review will discuss how alterations in the AR and broader AR pathway have been evaluated in CTCs and ctDNA, with emphasis on how major findings have been in concordance or discordance with the tissue-based findings discussed in previous sections.
4.1 Alterations in the AR in CTCs
An early examination by Miyamoto et al into AR alterations in CTCs focused on transcriptional function of androgen-activated AR [44]. Given that PSA is a gene target transcriptionally activated by the AR, and PSMA is a gene target transcriptionally repressed by AR, immunophenotyping for expression of these AR gene targets was used to classify individual CTCs as “AR active” (PSA+/PSMA−), “AR inactive” (PSA−/PSMA+), or “mixed AR signaling” (PSA+/PSMA+). Within this study, initial ADT resulted in a robust switch from activated AR signaling to inactive AR signaling, whereas second line treatment with abiraterone induced a more mixed signal [44]. These trends in AR activity within CTCs suggest an evolution toward a more complex and heterogeneous disease state. Understanding this evolution with respect to alterations in the AR gene and expression profiles may reveal a mechanistic basis for this heterogeneity.
Shaffer et al. performed one of the earliest analyses of AR alterations in CTCs from patients with CRPC. Within this study, Shaffer and colleagues used fluorescence in situ hybridization (FISH) to examine chromosomal aberrations in nine patients. Among the nine patients with CRPC, five of them exhibited amplification of AR [45]. Even within this small set, AR amplification frequency in CTCs mirrored the frequencies seen in larger scale tissue studies [38]. Related to this finding was a study that examined the AR gene in CTCs from patients treated with abiraterone. In this study, AR amplification was heterogeneous after second line therapy [46]. More recently, other AR alterations such as missense mutations, deletions, and insertions were examined in CTCs. One study found an unusually high percentage of missense mutations (15 of 35 CRPC patients) [47]. This frequency of AR mutation was much higher than observed in tissue-based analyses of CRPC tissue. One possible explanation is that tissue-based studies have routinely sampled a single metastatic site, whereas the cells that are enriched during CTC collection procedures are thought to originate from multiple metastatic sites. Given that sampling of several CRPC tissue sites from individual patients has revealed multiple independent AR amplification and mutation events [48], it is possible that CTC-based analysis reflects the totality of AR mutational burden within a patient, leading to a higher estimate of AR mutation frequency.
In a separate study that performed single-cell RNA-seq of CTCs, the T877A point mutation associated with ligand promiscuity and therapy resistance was only detected in CTCs from 1 of 13 patients with CRPC. An E709E mutation was also detected in 1/13 CRPC patients in this study [49]. This AR mutational frequency (2/13 patients) is concordant with the AR mutational frequency observed in larger-scale studies with CRPC tissues. However, there was insufficient depth of AR mRNA sequencing coverage for many of the individual CTCs in this study to rule out the possibility of a higher AR mutation frequency. Overall, these studies have demonstrated proof-of-principle for detection of specific AR gene alterations using various analysis platforms in CTCs, but the results have been highly variable. Improving the sensitivity and specificity of these platforms for AR analyses may provide a more accurate picture going forward.
4.2 Alterations in the AR in ctDNA
In line with the non-invasive principles of CTC analysis, interrogation of ctDNA is another strategy for evaluating tumor evolution and tracking responses to therapy in patients with CRPC. Romanel et al. reported targeted next-generation sequencing of ctDNA from plasma samples collected longitudinally from 97 CRPC patients before and during therapy with abiraterone [50]. Of 274 plasma samples analyzed, 41 displayed AR point mutations known to associate with ADT resistance [50], which is in concordance with the frequency observed in studies of CRPC tissue [37, 38]. Only 217 plasma samples from this cohort had a sufficient cancer cell fraction to enable analysis of AR gene copy number. Of these 217 samples, 80 demonstrated an AR copy number gain [50]. Importantly, detection of AR point mutations or copy number gain in plasma ctDNA from patients pre-therapy was associated with less favorable clinical outcomes including reduced PSA response rates, shorter progression-free survival, and shorter overall survival compared to patients where no AR alterations were detected [50].
4.3 AR-Vs in CTCs
Single-cell RNA-seq of CTCs from patients with CRPC has also enabled examination of AR splicing patterns and detection of AR-V expression in this context. For instance, in a study of individual CTCs isolated from 13 patients, 33/73 CTCs analyzed displayed expression of at least one type of AR-V [49]. Consistent with RNA-seq studies of CRPC tissue [38], AR-V7 was the most frequently detected AR-V (26 of 73 CTCs; 8 of 11 patients) [49]. Expression of ARv567es was also detected with high frequency (18 of 73 CTCs; 8 of 11 patients) [49], which is in disagreement with the overall lack of ARv567es mRNA expression observed in RNA-seq studies of CRPC tissue. Expression of AR-V1, AR-V3, and AR-V4 (contiguously-splice AR exons 1/2/3/CE4) has also been detected by single-cell RNA-seq of CTCs [49]. Interestingly, 13 of 73 individual CTCs expressed two or more different AR-Vs, which demonstrates that multiple AR splicing patterns can exist in a single cell. However, development of approaches for detection of AR-V protein expression in individual CTCs will be necessary to understand whether these mRNA species are translated to generate functional AR-V protein. Finally, partitioning the patients in this study into those that had been treated with enzalutamide vs. those that had not been treated with enzalutamide did not reveal enrichment for AR-V expression in the enzalutamide-treated cohort [49].
While there is evidence of heterogeneous AR-V expression in CTCs before and after treatment with enzalutamide, there is strong evidence that simple detection by RT-PCR of AR-V7 mRNA in CTCs may serve as a predictive biomarker for patients treated with second-generation AR-targeted therapies. Specifically, in a study of 62 CRPC patients initiating therapy with abiraterone (31 patients) or enzalutamide (31 patients), Antonarakis and colleagues reported that the presence of CTCs positive for AR-V7 mRNA expression was associated with lower PSA response rates, diminished progression-free survival, and reduced overall survival in both treatment groups [51]. These data imply a clear association of AR-V7 mRNA with resistance to second line AR-targeted therapies. Results in this study are in agreement with a study examining AR-V7 protein expression in bone marrow aspirates from CRPC patients [52] as well as an independent study evaluating CTCs from patients treated with enzalutamide [53]. Interestingly, these studies have also reported that increased AR-V7 mRNA expression in bone marrow aspirates and CTCs directly correlated with the number of prior therapies patients had received [51, 52, 53]. Indeed, patients with an AR-V7 mRNA negative CTC profile pre-therapy have been shown to convert to an AR-V7 mRNA positive CTC profile during treatment with enzalutamide or abiraterone [51, 53, 54, 55].
While AR-V7 mRNA positivity in CTCs may be predictive of lack of response to abiraterone or enzalutamide therapy, AR-V7 positivity in CTCs does not appear to be associated with lack of response to taxane chemotherapy. Specifically, patients with an AR-V7 mRNA positive CTC profile pre-therapy displayed superior clinical outcomes when treated with taxane chemotherapy compared with abiraterone or enzalutamide. Conversely, clinical outcomes were indistinguishable for all three treatments in patients with an AR-V7 mRNA negative CTC profile per-therapy [55]. This result may be unexpected in light of mechanistic cell-based studies reporting that microtubule-dependent translocation of AR to the nucleus can be impaired by taxane chemotherapy, whereas AR-V7 translocation to the nucleus is taxane insensitive [56, 57]. However, subsequent studies demonstrated this AR translocation effect may be an artifact of treating prostate cancer cells with concentrations of taxanes higher than can be achieved clinically [58]. Interestingly, it has also been reported that patients with an AR-V7 mRNA positive CTC profile pre-therapy can undergo reversion to AR-V7 mRNA negative CTC profile during taxane chemotherapy [54,55]. This plasticity in AR-V7 expression further highlights the complexity of AR-V expression in prostate cancer, and shows how various selection pressures can modulate resistance mechanisms.
5. Functional Changes in AR and AR-V signaling during PCa progression
Since AR is a transcription factor, understanding functional changes in regard to gene expression repertoire (ie the set of transcriptional targets regulated by AR, or the AR “transcriptome”) and chromatin binding patterns (ie the collection of cis-elements bound by AR, or the AR “cistrome”) during cancer progression is important. Understanding how AR gene and splicing alterations, disease context, co-regulator expression, and treatment history can influence AR chromatin binding sites and transcriptional targets is expected to provide further insight to how aberrations in AR and the broader AR signaling pathway drive cancer progression. Fine-tuned and expanded usage of techniques such as ChIP-seq, and related ChIP-seq variations, has allowed for more advanced and context specific mapping of AR binding. Furthermore, the complex nature of AR binding is inherent in binding sites where AREs can be found at sites distant from target genes. In this section, AR and AR-V cistromics and transcriptional programs will be discussed in further detail.
5.1 AR cistrome and transcriptome in normal tissue, primary prostate cancer, and CRPC
ChIP-seq and transcriptional profiling in prostate cancer cell lines has been a popular avenue for researchers to understand functional changes in AR action [59]. Moreover, these studies have highlighted mechanistic aspects of the AR cistrome and transcriptional regulation by FOXA1 and ERG [59, 60, 61]. In order to fully understand functional changes in AR signaling, it is important to analyze the transcriptome and cistrome of AR in prostate cancer tumor tissue. This allows for the verification of gene expression and binding data themes found in model systems, which, as a result, relay a more robust understanding of AR functional behavior in a variety of disease settings. For instance, comparison of AR ChIP-seq datasets derived from prostate cancer cell lines and clinical tissues demonstrated that AR chromatin binding events in prostate cancer cell lines more closely reflect the AR chromatin binding events that occur in CRPC tissues than the AR chromatin binding events that occur in primary prostate cancer. This is expected, given that the prostate cancer cell lines used in this study were derived from patients with CRPC. However, over 50% of AR chromatin binding sites observed in CRPC tissues were not detected in prostate cancer cell lines, highlighting the importance of studying tissues directly [62]. Recently, Pomerantz and colleagues used ChIP-seq to compare AR chromatin binding sites in primary prostate cancer and adjacent normal tissue [63]. Hierarchical clustering of AR chromatin binding events revealed distinct groups of AR cistromes between primary tumor samples and matched normal tissues. Inclusion of the AR positive prostate cancer cell lines LNCaP and VCaP in hierarchical clustering resulted in classification of a third distinct group, which most closely resembled primary tumors [63]. Overall, this study identified 9,179 AR chromatin binding sites that were enriched in tumor tissue and 2,690 AR chromatin binding sites that were enriched in normal tissue. Importantly, this study identified FOXA1 and HOXB13 as key AR-associated transcription factors that ultimately determined tumor-enriched AR chromatin binding events. Consistent with this finding, transfection of a normal prostate epithelial cell line with lentivirus encoding FOXA1 and HOXB13 resulted in a pattern of AR chromatin binding that resembled primary prostate cancer tissue [63]. Building on these same themes, comparison of AR chromatin binding events in primary prostate cancer vs. CRPC specimens revealed differences in AR chromatin binding associated with disease progression [64]. These findings have indicated that AR chromatin binding undergoes extensive reprogramming at various stages of disease progression. Understanding the mechanisms by which alterations in the AR and broader AR pathway intersect with these chromatin binding alterations will provide important insights to the genome-wide impacts of AR alterations in prostate cancer.
5.2 AR versus AR-V cistrome and transcriptome
There has also been great interest in understanding the similarities and differences in cistromes and transcriptomes regulated by full-length AR and AR-Vs. One early observation was that AR-Vs uniquely regulated a set of G2/M-phase regulatory genes including UBE2C [65]. However, more recent analysis of this set of G2/M-phase cell cycle regulator genes revealed they were biphasic targets that could be induced by both androgen/AR as well as AR-Vs at low levels of AR transcriptional output, but were repressed when AR transcriptional output was increased [66]. A study by Lu and colleagues compared the cistromes and transcriptomes regulated by AR and AR-Vs in the 22Rv1 prostate cancer cell line manipulated with siRNA to achieve cell preparations that were either full length AR-/AR-V+ or full length AR-/AR-V− [67]. In this study, AR-Vs were observed to regulate a unique gene set separate from full length AR in the absence of androgen. Additionally, binding of AR-Vs didn’t appear to be affected by loss of full length AR or androgens [67]. However, it should be noted that full-length AR in 22Rv1 cells harbors a tandem duplication of a zinc finger module in the AR DNA binding domain [16]. Therefore it is not clear whether the differences in AR vs. AR-V chromatin binding observed in this study was due to differential DNA recognition by a larger 3-zinc finger AR DBD module vs. a normal 2-zinc finger AR DBD module. Nevertheless, this study also found that AR-V chromatin binding sites had 75% overlap with full-length AR chromatin binding sites [67]. In a separate ChIP-seq study with cell lines engineered to express either full-length AR or ARv567es from the same endogenous AR promoter, ARv567es was found to engage with the same genomic sites engaged by full-length AR, albeit with lower affinity [66]. This study further revealed that AR-Vs required dimerization to engage with androgen response elements [66], which has since been confirmed by a separate study [68]. A more complete picture of how AR-Vs engage chromatin will require analysis of additional cell line models as well as clinical tissues expressing various levels of AR-Vs relative to full-length AR.
6. Conclusion
Interrogating multiple tissue compartments in patients at various stages of prostate cancer progression has provided key insights to the alterations that occur in the AR and broader AR signaling pathway. While mutation and amplification of the AR gene do not appear to play a major role in prostate tumorigenesis, there is ample evidence that other common genomic alterations including ETS gene fusions, SPOP mutations, and FOXA1 mutations can alter transcriptional regulation of the AR signaling axis. These alterations may be intimately linked to differences in transcriptional output and/or chromatin binding that have been noted in the AR in studies with primary prostate cancer and adjacent normal tissue. Alternatively, mutation and amplification of the AR gene are more frequent in CRPC tissue, and appear to be responses to therapeutic stress as opposed to alterations that underlie metastasis. Altered splicing of the AR gene, manifesting in expression of AR-Vs appears to occur at all stages of disease progression as well as normal prostate tissue, raising questions about the specific context in which these species may play a role in disease progression. Nevertheless, targeted interrogation approaches have revealed a trend towards increased expression of specific AR-Vs, such as AR-V7, in CRPC vs. primary prostate cancer tissue. This is underscored by the potential predictive utility of detecting AR-V7 mRNA in CTCs from patients with CRPC. Reconciling the discrepancies noted in AR-V7 expression patterns in tissue profiling (ie RNA-seq) vs. targeted (ie RT-PCR) studies with tissues or CTCs will be critical for future studies. Additionally, understanding the mechanistic role of AR-Vs in disease progression will require their evaluation in the context of other AR and AR pathway alterations known to occur during disease progression, as well as alterations in AR independent pathways. Moreover, development of inhibitors that can selectively target the transcriptional activity of AR-Vs will provide important clinical information on their contributions at various disease stages.
Highlights.
Reactivation of androgen receptor is critical in prostate cancer disease progression.
Androgen receptor splice variants contain the same N-terminal domain and DNA binding domain as full-length AR, but lack the COOH-terminal ligand binding domain rendering them constitutively active in the absence of ligand.
Androgen receptor splice variant transcripts are detectable in benign prostate tissue, primary prostate cancer, castration-resistant prostate cancer, and circulating tumor cells.
Androgen receptor splice variant-7 may serve as a predictive biomarker for patients treated with second generation androgen receptor targeted therapies.
Acknowledgments
SMD is currently funded by a Movember/Prostate Cancer Foundation Challenge Award, American Cancer Society Research Scholar Grant RSG-12-031-01-TBE, NIH grant R01CA174777, US Department of Defense Prostate Cancer Research Program grants W81XWH-12-2-0093, W81XWH-13-1-0518, W81XWH-15-1-0633, and W81XWH-15-1-0501, and a grant from the Minnesota Partnership for Biotechnology and Medical Genomics.
Abbreviations
- LHRH
luteinizing hormone releasing hormone
- ADT
androgen depletion therapy
- CRPC
castration-resistant prostate cancer
- CYP17
cytochrome P450 c17
- MR
mineralocorticoid receptor
- ER
estrogen receptor
- PR
progestins receptor
- GR
glucocorticoid receptor
- NTD
NH2-terminal domain
- AF-1
activation function 1
- LBD
ligand bind domain
- ARE
androgen response element
- RIP
RNA co-immunoprecipitation
- NCOA
nuclear receptor coactivator
- NCOR
nuclear receptor corepressor
- CTC
circulating tumor cell
- ctDNA
circulating tumor DNA
- FISH
fluorescence in situ hybridization
- PSA
prostate-specific antigen
- PMSA
prostate-specific membrane antigen
Footnotes
Conflict of Interest
SMD has served as a paid consultant/advisor for Astellas/Medivation
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. 22:232–40. doi: 10.3322/canjclin.22.4.232. http://www.ncbi.nlm.nih.gov/pubmed/4625049. [DOI] [PubMed] [Google Scholar]
- 3.Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368:138–48. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20:3001–15. doi: 10.1200/JCO.2002.10.018. http://www.ncbi.nlm.nih.gov/pubmed/12089231. [DOI] [PubMed] [Google Scholar]
- 6.Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759–65. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305–13. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun S, Sprenger CCT, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–30. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656–67. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libertini SJ, Tepper CG, Rodriguez V, Asmuth DM, Kung HJ, Mudryj M. Evidence for calpain-mediated androgen receptor cleavage as a mechanism for androgen independence. Cancer Res. 2007;67:9001–5. doi: 10.1158/0008-5472.CAN-07-1072. [DOI] [PubMed] [Google Scholar]
- 12.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469–77. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–96. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33:3140–50. doi: 10.1038/onc.2013.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Hwang TH, Oseth LA, Hauge A, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–67. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Alsagabi M, Fan D, Bova GS, Tewfik AH, Dehm SM. Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res. 2011;71:2108–17. doi: 10.1158/0008-5472.CAN-10-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KAT, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–7. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–6. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hörnberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H, Coram MA, Nolley R, Reese SW, Young SR, Peehl DM. Transcript levels of androgen receptor variant AR-V1 or AR-V7 do not predict recurrence in patients with prostate cancer at indeterminate risk for progression. J Urol. 2012;188:2158–64. doi: 10.1016/j.juro.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015;43:5880–97. doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 24.Barwick BG, Abramovitz M, Kodani M, Moreno CS, Nam R, Tang W, et al. Prostate cancer genes associated with TMPRSS2-ERG gene fusion and prognostic of biochemical recurrence in multiple cohorts. Br J Cancer. 2010;102:570–6. doi: 10.1038/sj.bjc.6605519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minner S, Enodien M, Sirma H, Luebke AM, Krohn A, Mayer PS, et al. ERG status is unrelated to PSA recurrence in radically operated prostate cancer in the absence of antihormonal therapy. Clin Cancer Res. 2011;17:5878–88. doi: 10.1158/1078-0432.CCR-11-1251. [DOI] [PubMed] [Google Scholar]
- 26.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Grasso CS, Cani AK, Hovelson DH, Quist MJ, Douville NJ, Yadati V, et al. Integrative molecular profiling of routine clinical prostate cancer specimens. Ann Oncol. 2015;26:1110–8. doi: 10.1093/annonc/mdv134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagai Y, Kojima T, Muro Y, Hachiya T, Nishizawa Y, Wakabayashi T, et al. Identification of a novel nuclear speckle-type protein, SPOP. FEBS Lett. 1997;418:23–6. doi: 10.1016/s0014-5793(97)01340-9. http://www.ncbi.nlm.nih.gov/pubmed/9414087. [DOI] [PubMed] [Google Scholar]
- 30.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An J, Wang C, Deng Y, Yu L, Huang H. Destruction of full-length androgen receptor by wild-type SPOP, but not prostate-cancer-associated mutants. Cell Rep. 2014;6:657–69. doi: 10.1016/j.celrep.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geng C, Rajapakshe K, Shah SS, Shou J, Eedunuri VK, Foley C, et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74:5631–43. doi: 10.1158/0008-5472.CAN-14-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci U S A. 2013;110:6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsiades N, Sung CC, Schultz N, Danila DC, He B, Eedunuri VK, et al. Distinct patterns of dysregulated expression of enzymes involved in androgen synthesis and metabolism in metastatic prostate cancer tumors. Cancer Res. 2012;72:6142–52. doi: 10.1158/0008-5472.CAN-12-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonkhoff H, Berges R. From pathogenesis to prevention of castration resistant prostate cancer. Prostate. 2010;70:100–112. doi: 10.1002/pros.21042. [DOI] [PubMed] [Google Scholar]
- 36.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–11. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linja MJ, Savinainen KJ, Saramäki OR, Tammela TL, Vessella RL, Visakorpi T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001;61:3550–5. http://www.ncbi.nlm.nih.gov/pubmed/11325816. [PubMed] [Google Scholar]
- 40.Ford OH, Gregory CW, Kim D, Smitherman AB, Mohler JL. Androgen receptor gene amplification and protein expression in recurrent prostate cancer. J Urol. 2003;170:1817–21. doi: 10.1097/01.ju.0000091873.09677.f4. [DOI] [PubMed] [Google Scholar]
- 41.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bubendorf L, Kononen J, Koivisto P, Schraml P, Moch H, Gasser TC, et al. Survey of gene amplifications during prostate cancer progression by high-throughout fluorescence in situ hybridization on tissue microarrays. Cancer Res. 1999;59:803–6. http://www.ncbi.nlm.nih.gov/pubmed/10029066. [PubMed] [Google Scholar]
- 43.Koivisto P, Kononen J, Palmberg C, Tammela T, Hyytinen E, Isola J, et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Res. 1997;57:314–9. http://www.ncbi.nlm.nih.gov/pubmed/9000575. [PubMed] [Google Scholar]
- 44.Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, et al. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:2023–9. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 46.Attard G, Swennenhuis JF, Olmos D, Reid AHM, Vickers E, A’Hern R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Y, Palma JF, Agus DB, Wang Y, Gross ME. Detection of androgen receptor mutations in circulating tumor cells in castration-resistant prostate cancer. Clin Chem. 2010;56:1492–5. doi: 10.1373/clinchem.2010.143297. [DOI] [PubMed] [Google Scholar]
- 48.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature. 2015;520:353–357. doi: 10.1038/nature14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romanel A, Tandefelt DG, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015;7:312re10–312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Efstathiou E, Titus M, Wen S, Hoang A, Karlou M, Ashe R, et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol. 2015;67:53–60. doi: 10.1016/j.eururo.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinestel J, Luedeke M, Arndt A, Schnoeller TJ, Lennerz JK, Wurm C, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget. 2015 doi: 10.18632/oncotarget.3925. http://www.ncbi.nlm.nih.gov/pubmed/25970787. [DOI] [PMC free article] [PubMed]
- 54.Nakazawa M, Lu C, Chen Y, Paller CJ, Carducci MA, Eisenberger MA, et al. Serial blood-based analysis of AR-V7 in men with advanced prostate cancer. Ann Oncol. 2015;26:1859–65. doi: 10.1093/annonc/mdv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Nakazawa M, et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015;1:582–91. doi: 10.1001/jamaoncol.2015.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK, et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer Res. 2011;71:6019–29. doi: 10.1158/0008-5472.CAN-11-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thadani-Mulero M, Portella L, Sun S, Sung M, Matov A, Vessella RL, et al. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res. 2014;74:2270–82. doi: 10.1158/0008-5472.CAN-13-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Leeuw R, Berman-Booty LD, Schiewer MJ, Ciment SJ, Den RB, Dicker AP, et al. Novel actions of next-generation taxanes benefit advanced stages of prostate cancer. Clin Cancer Res. 2015;21:795–807. doi: 10.1158/1078-0432.CCR-14-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19:1023–9. doi: 10.1038/nm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma NL, Massie CE, Ramos-Montoya A, Zecchini V, Scott HE, Lamb AD, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 63.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47:1346–1351. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stelloo S, Nevedomskaya E, van der Poel HG, de Jong J, van Leenders GJ, Jenster G, et al. Androgen receptor profiling predicts prostate cancer outcome. EMBO Mol Med. 2015;7:1450–64. doi: 10.15252/emmm.201505424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457–62. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736–49. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu J, Lonergan PE, Nacusi LP, Wang L, Schmidt LJ, Sun Z, et al. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. J Urol. 2015;193:690–8. doi: 10.1016/j.juro.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu D, Zhan Y, Qi Y, Cao B, Bai S, Xu W, et al. Androgen Receptor Splice Variants Dimerize to Transactivate Target Genes. Cancer Res. 2015;75:3663–71. doi: 10.1158/0008-5472.CAN-15-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]