Abstract
Rationale
There is a critical need to develop robust, mechanistic strategies to identify patients at increased risk of cancer therapeutics-related cardiac dysfunction (CTRCD).
Objective
We aimed to discover new biomarkers associated with doxorubicin and trastuzumab-induced CTRCD using high-throughput proteomic profiling.
Methods and Results
Plasma, echocardiograms, and clinical outcomes were collected at standardized intervals in breast cancer patients undergoing doxorubicin and trastuzumab cancer therapy. Thirty-one longitudinal plasma samples from three cases with CTRCD and four age- and cancer-matched controls without CTRCD were processed and analyzed using label-free liquid chromatography- mass spectrometry (LC-MS). From these analyses, 862 proteins were identified from case/control pairs 1 and 2, and 1,360 proteins from case/control pair 3. Proteins with a greater than 1.5-fold change in cases compared to controls with a p<0.05 either at the time of CTRCD diagnosis or across all timepoints were considered candidate diagnostic or predictive biomarkers, respectively. The protein that demonstrated the largest differences between cases and controls was immunoglobulin E (IgE), with higher levels detected at baseline and across all timepoints in controls without CTRCD as compared to matched CTRCD cases (p<0.05). Similarly, in a validation study of 35 participants treated with doxorubicin and trastuzumab, high baseline IgE levels were associated with a significantly lower risk of CTRCD (p=0.018).
Conclusions
In patients receiving doxorubicin and trastuzumab, high baseline IgE levels are associated with a lower risk of CTRCD. These novel findings suggest a new paradigm in cardio-oncology, implicating the immune system as a potential mediator of doxorubicin and trastuzumab-induced cardiac dysfunction.
Keywords: CTRCD, immune mediators, proteomics, plasma biomarkers, label-free quantitation, cardiomyopathy
INTRODUCTION
Doxorubicin and trastuzumab (Herceptin®) are used widely in the treatment of breast cancer, are highly effective, and have led to important survival gains.1 However, these agents carry a substantially increased risk of cardiovascular morbidity and mortality. Doxorubicin-induced cardiac dysfunction occurs in 9% of treated patients at dosages of 250 mg/m2.2 Trastuzumab, a highly effective humanized monoclonal antibody used in the treatment of Her2 (ErbB2)-positive breast cancer, also causes significant declines in left ventricular ejection fraction (EF), resulting in potential delays or cessation of necessary therapy.3, 4 When anthracyclines and trastuzumab are used in combination, up to 18% of patients develop cancer therapeutics-related cardiac dysfunction (CTRCD) and 2-4% develop severe, symptomatic heart failure.5
There is an important need to identify patients at increased risk of developing CTRCD, particularly prior to the development of overt disease. Cardiovascular biomarkers have been widely studied as potential tools to risk stratify patients; however, many of these markers are neither specific nor sensitive for diagnosing CTRCD or predicting which patients are at increased cardiovascular risk. Moreover, our lack of mechanistic understanding of the underlying pathophysiology of doxorubicin and trastuzumab cardiac dysfunction has hindered the development of newer markers. As such, discovery proteomics represents a unique opportunity to identify novel pathways and biomarkers.
Traditionally, there have been a number of hurdles that have hindered advances in proteomics discovery. These include the great complexity of the plasma or serum proteome; the presence of a few very high-abundant proteins; the wide dynamic range of protein concentrations; and the molecular heterogeneity of many proteins. However, considerable advances have been made in liquid chromatography-mass spectrometry (LC-MS) based biomarker discovery and validation methods over the past several years that have substantially reduced these barriers. Now, rigorous and powerful proteomic-based technologies can be used to successfully discover and validate new biomarkers.
The overall purpose of this study was to apply current proteomics techniques to discover new circulating biomarkers that can be used in diagnosis and risk prediction in CTRCD. In breast cancer patients undergoing therapy with doxorubicin and trastuzumab, we hypothesized that patterns of change over time in protein markers would differ between participants who experience CTRCD and those who do not, and specifically sought to discover markers that demonstrated significant changes at the time of CTRCD diagnosis (‘diagnostic markers’) and prior to the onset of CTRCD (‘predictive markers’). To determine the answer to this question, we used a cohort of well-phenotyped breast cancer patients undergoing doxorubicin and trastuzumab therapy with longitudinal blood sampling coupled to cardiac dysfunction outcomes, derived from clinical and quantitative echocardiography data. We first performed a matched case-control discovery study in longitudinal samples and then validated our most promising biomarker in a cohort analysis of baseline samples using independent assays.
METHODS
Study population
Cases and controls were selected from the Cardiotoxicity of Cancer Therapy (CCT) study, an ongoing, National Heart Lung and Blood Institute (NHLBI)-funded prospective longitudinal cohort study of women with breast cancer recruited from the Rena Rowan Breast Cancer Center of the Abramson Cancer Center at the University of Pennsylvania (Philadelphia, PA). The primary inclusion criteria were women at least 18 years of age diagnosed with breast cancer and prescribed doxorubicin and/or trastuzumab therapy. The only exclusion criterion was pregnancy. Cases and controls all received doxorubicin (240 mg/m2) and cyclophosphamide followed by paclitaxel and trastuzumab, the latter as per standard prescribing algorithms.
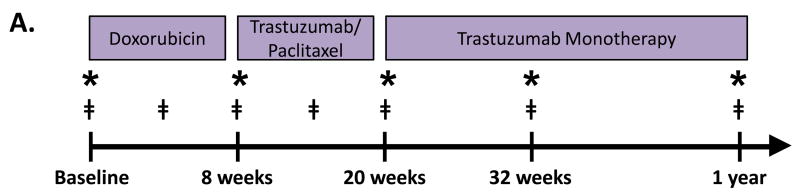
At baseline, prior to initiation of chemotherapy, and at each follow-up visit, each participant provided detailed clinical data via standardized questionnaires. Clinical data were verified via review of medical records. Blood samples were obtained at baseline, during doxorubicin, after doxorubicin completion, and every 6 weeks during trastuzumab. Transthoracic echocardiograms were performed at standardized intervals, and participants underwent an echocardiogram at baseline, after doxorubicin completion, and every 3 months during trastuzumab therapy (Figure 1A). This study was approved by the University of Pennsylvania Institutional Review Board, and all participants provided written informed consent.
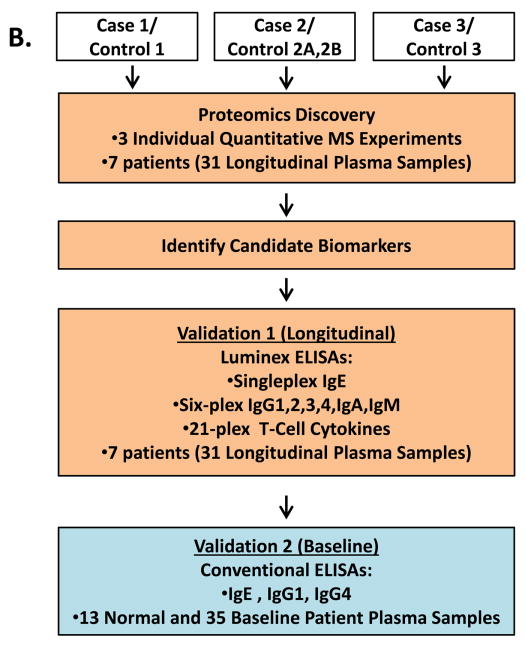
Figure 1. Study Cohort and Experimental Approach.
A. Blood draw and echocardiography protocol for patients who were treated with doxorubicin and trastuzumab therapy. * denotes when transthoracic echocardiograms were performed. ‡ denotes when blood samples were collected. B. Strategy for proteomic discovery and validation of candidate biomarkers. Steps highlighted in orange were performed using longitudinal plasma samples (including baseline). Step highlighted in blue was performed using only baseline plasma samples.
Transthoracic echocardiography
Transthoracic echocardiograms were acquired by a dedicated sonographer team at an Intersocietal Accreditation Commission laboratory according to a specific protocol at baseline and standardized time intervals. Two-dimensional images were acquired using Vivid 7 or E9 machines (GE Healthcare, Milwaukee, WI).
Echocardiograms were quantitated at the University of Pennsylvania Center for Quantitative Echocardiography (Philadelphia, PA). Quantitation was performed using the Tomtec Cardiac Performance Analysis (Unterschleissheim, Germany). Apical 4-chamber LV end-diastolic volume (EDV) and end-systolic volume (ESV) were calculated using the Simpson’s method of discs as recommended by the American Society of Echocardiography (ASE), and used to derive left ventricular ejection fraction.6
Identification of cancer therapy related cardiac dysfunction cases and controls
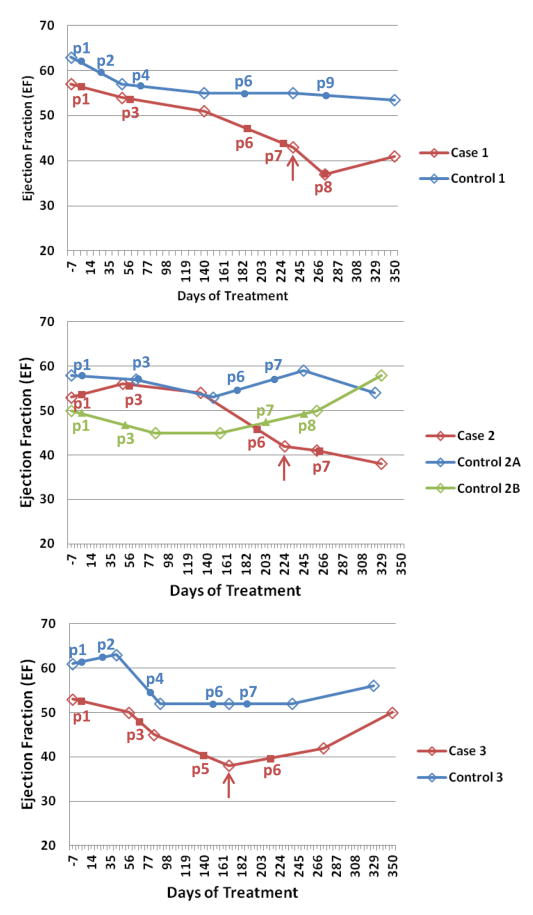
Of the participants who experienced CTRCD, as defined as cardiac dysfunction with a reduction in EF by ≥10% from baseline to an absolute value of <50% at any subsequent visit, three were selected for proteomics analyses. Cases also had to experience symptoms of heart failure as adjudicated by a cardiologist, and all were started on cardiac medications.
Controls were selected based upon the lack of significant EF change during the entire duration of follow-up (<10% absolute change in EF and EF>50%) were matched to the cases based upon specific selection criteria. These criteria for matching included: age (+/- 10 years), hormonal status, cancer stage, and race. In one instance, a patient meeting all of the criteria was not available. For this reason, two controls were selected for Case 2 (Table 1).
Table 1.
Clinical Characteristics of Case/Control Pairs used in Discovery Proteomics
| Patient | Age (yrs) | Race | Nodal Status | Hormone Status | Breast Cancer Side | Baseline EF (%) | Nadir EF (%) | CTRC D Timing (days) | Cardiac Medication |
|---|---|---|---|---|---|---|---|---|---|
| Case 1 | 48 | Cauc | N1 | ER+ | Right | 57 | 37 | 238 | Enalapril, Carvedilol |
| Ctrl 1 | 40 | Cauc | N1 | ER+ | Right | 63 | 55 | None | |
| Case 2 | 46 | Cauc | N3C | ER- | Right | 53 | 42 | 226 | Enalapril, Carvedilol |
| Ctrl 2A | 43 | Asian | N2 | ER- | Left | 58 | 53 | None | |
| Ctrl 2B | 53 | Cauc | N2 | ER+ | Left | 50 | 45 | None | |
| Case 3 | 43 | Asian | N0 | ER- | Right | 53 | 38 | 167 | Lisinopril, Carvedilol |
| Ctrl 3 | 36 | Cauc | N0 | ER- | Left | 61 | 52 | None |
Plasma sample collection, timepoints, and processing
For all participants, blood was collected from venipuncture in the presence of EDTA, and this plasma was processed at 3353 RPM for 20 minutes, aliquoted and stored at -80°C. Longitudinal plasma samples for each case and control were selected for the proteomics study to evaluate changes over time in the proteome between cases and controls. In total, 31 samples from three cases and four controls in the cohort of participants receiving doxorubicin followed by trastuzumab were selected for discovery analyses. Samples used in the discovery analysis were derived from the following timepoints: prior to any chemotherapy, during chemotherapy, at the time of CTRCD diagnosis, and after the CTRCD diagnosis (Figure 1A). Timepoints for the matched controls were selected to match the case specimens. Samples used in subsequent validation analysis were derived from baseline only.
Proteomics discovery and data analysis
The 31 longitudinal case and control plasma samples used for discovery were processed using a 3-dimensional plasma proteome analysis strategy previously developed by the Speicher laboratory7 (Supplemental Methods, Supplemental Figure I). Briefly, 80 μl aliquots of plasma samples were depleted of 20 abundant plasma proteins using a ProteoPrep20 Immunodepletion Column (Sigma-Aldrich, St. Louis, MO) and run for a short distance (2 cm) on 1-D SDS gels. Three lanes representing the immunodepleted fraction from 10 μl of original plasma were run for each sample, and each lane was sliced into twenty 1-mm slices. Corresponding slices for the triplicate lanes of each sample that represented a total of 30 μl original plasma were combined and digested with trypsin.
Each set of case/control samples was analyzed in a separate label-free LC-MS/MS experiment. The first two case/control longitudinal sample sets were analyzed on an LTQ Orbitrap XL (Thermo Scientific, Waltham, MA) mass spectrometer and the third case/control sample set was analyzed on a Q Exactive Plus mass spectrometer (Thermo Scientific). Both instruments were equipped with Nano-Acquity (Waters, Milford, MA) pumps and a column heater maintained at 40°C. Tryptic digests were injected onto a UPLC Symmetry trap column (180 μm i.d. × 2 cm packed with 5 μm C18 resin; Waters), and peptides were separated by RP-HPLC on a BEH C18 nanocapillary analytical column (75μm i,d, × 25 cm, 1.7 μm particle size, Waters) at a flow rate of 200 nL/min. Solvent A was Milli-Q (Millipore, Billerica, MA) water containing 0.1% formic acid, and Solvent B was acetonitrile containing 0.1% formic acid. The 85 minute gradient used for samples analyzed on the Oribitrap XL was as previously described7, while a slightly extended 95 minute gradient consisting of 5-30% B over 75 min, 30-80% B over 10 min, 80%B for 10 min, before returning to 5% B over 1 min was used for analysis of samples on the Q Exactive Plus. The HPLC, peak retention times, and mass spectrometer were carefully monitored throughout each experiment to ensure that performance was within tight tolerances in order to facilitate comparisons of LC-MS signals.
Raw mass spectrometric data were processed by MaxQuant software (Ver. 1.4.1.2) as previously described8, 9(Supplemental Methods). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository10 with the dataset identifier PXD004058.
Validation of findings
Luminex Assay
To verify our most promising proteomics discovery findings, singleplex IgE and multiplex IgG1, IgG2, IgG3, IgG4, IgA and IgM Isotyping assays were performed using Luminex kits (Millipore) according to the manufacturer’s protocol (Figure 1B). The 31 longitudinal plasma samples from the 3 case/control pairs were diluted 1:50 or 1:16,000 for IgE and Isotyping kits, respectively, and added in duplicate on 96 well plates. Multiplex assays (Millipore) for Th1 and Th2 associated cytokines (IFNγ, IL-4, IL-5, IL-6, IL-10, IL-13, GM-CSF, TNF-α, IL-2, IL-12, IL-1β, IL-7, IL-8, IL-17A, IL-21, IL-23, MIP-1α, MIP-1β, MIP-3α, Fractalkine, ITAC) were also performed to further evaluate changes in IgE associated immune markers. Beads were read using a MAGPIX instrument (Millipore) and data were analyzed with Milliplex Analyst software (version 5.1) (Supplemental Methods).
Plasma samples from healthy participants
In order to define the immunoglobulin levels amongst healthy participants, thirteen plasma EDTA samples were obtained from healthy female donors at the University of Pennsylvania (n=5) and the Wistar Institute (n=8). Participants were without evidence of comorbid conditions, including cardiovascular or oncologic disease. Plasma samples were processed and stored in an identical manner to the cancer cohort, and tested on the platforms as detailed above
Standard colorimetric ELISA assay on baseline samples from the Doxorubicin and Trastuzumab cohort and normal healthy participants
Sandwich ELISA assays (Affymetrix eBioscience, San Diego, CA) were used to measure human IgE, IgG1 and IgG4 plasma levels at baseline (prior to treatment) for the entire cohort of 35 participants who received doxorubicin and trastuzumab, plus 13 normal female plasma donors (Figure 1B and Supplemental Methods). ELISA plates were coated with each respective primary anti-human monoclonal antibodies and assays were performed according to manufacturer’s instructions.
Statistical methods
In two separate discovery analyses, we sought to identify diagnostic and predictive biomarkers. Diagnostic biomarkers were defined as those proteins exhibiting a significant change in protein level at the same time as the onset of CTRCD. Candidate diagnostic biomarkers were selected by initially considering rate of change for individual cases; specifically, the level of a given protein was significantly higher or lower (two-tailed Student’s t-test p-value <0.05 and fold change >1.5) at one or more pre-CTRCD timepoints compared with timepoints after diagnosis of CTRCD.
Predictive biomarkers were defined as those that exhibited overall significant differences between case and control at baseline or at all timepoints; that is, there was a consistent difference between case and control starting at baseline and persisting throughout the study. Significantly changed proteins were defined as having both >1.5 fold change between the average of all timepoints for each case and control pair, and a Student t-test p-value < 0.05.
For Luminex validation studies, differences between case and control samples were calculated at baseline using a two-tailed Student’s t-test. For the ELISA data derived from the 35 baseline samples, the non-parametric Wilcoxon Rank Sum test was used to compare baseline IgE levels from individuals in the doxorubicin/trastuzumab cohort who developed CTRCD with those individuals who did not develop CTRCD. Logistic regression models were then used to determine the associations between baseline levels of biomarkers and odds of CTRCD. Baseline IgE levels and IgE/IgG1 ratios were transformed on the log2 scale for these analyses. Area under the receiver operating characteristic curves (AUC) were calculated to assess the discriminative ability of each biomarker. The biomarker cutpoint at which the optimal sensitivity and specificity could be achieved was also calculated. Statistical significance was set at p<0.05 for all analyses.
RESULTS
Patient characteristics of discovery cohort
Cases and controls selected for the proteomics discovery analyses had the following characteristics, as detailed in Table 1. All participants received a regimen containing doxorubicin (240 mg/m2), cyclophosphamide, followed by paclitaxel and trastuzumab for 4 cycles, followed by 1 year of trastuzumab therapy. All participants also received radiotherapy. No participants had any history of cardiovascular disease or risk factors prior to cancer therapy.
In the cases, CTRCD occurred between 167 to 238 days. Moreover, cases all complained of heart failure symptoms including dyspnea on exertion and fatigue, and were all started on cardiac medications after diagnosis, including angiotensin converting enzyme-inhibitors (ACE-I) and beta blockers. Controls did not have any evidence of significant or sustained declines in EF (i.e., CTRCD) or symptoms of heart failure. Timeline plots of clinical assessment of cardiac function by EF and the relationship to analyzed plasma fractions are shown in Figure 2.
Figure 2. Longitudinal Plots of Ejection Fraction (EF) for Cases and Controls.
Cases are shown in red, and controls in blue (or green for control 2B). Red arrows indicate the point of clinical diagnosis of cardiac dysfunction for each case. The selected subset of longitudinal plasma draws analyzed in this study are numbered (p1, p2…, etc.) and indicated by solid markers. Note that blood draws typically precede echocardiogram measurements (open diamonds on the plots).
Proteomics biomarker discovery
As previously noted, case/control pairs 1 and 2 were analyzed using an Orbitrap XL mass spectrometer, while case/control 3 was analyzed on a Q Exactive Plus instrument when this newer, higher performance instrument became available. Approximately 862 proteins were identified from case/control 1 and 2 plasma proteomes while analysis of case/control 3 resulted in the identification of 1,360 proteins. The increased depth of analysis achieved in case/control 3 was seen as potentially valuable, because most of the additional ~500 proteins identified in this dataset should be very low abundance proteins that were below the detection limit of the Orbitrap XL mass spectrometer.
Diagnostic biomarkers were expected to be those proteins that showed an increase or decrease in protein abundance specifically associated with the timeframe of CTRCD development. Surprisingly, while a number of proteins exhibited large changes in abundance over time, none of these protein changes were significantly associated with onset of CTRCD.
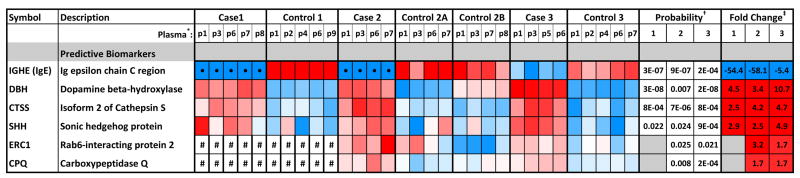
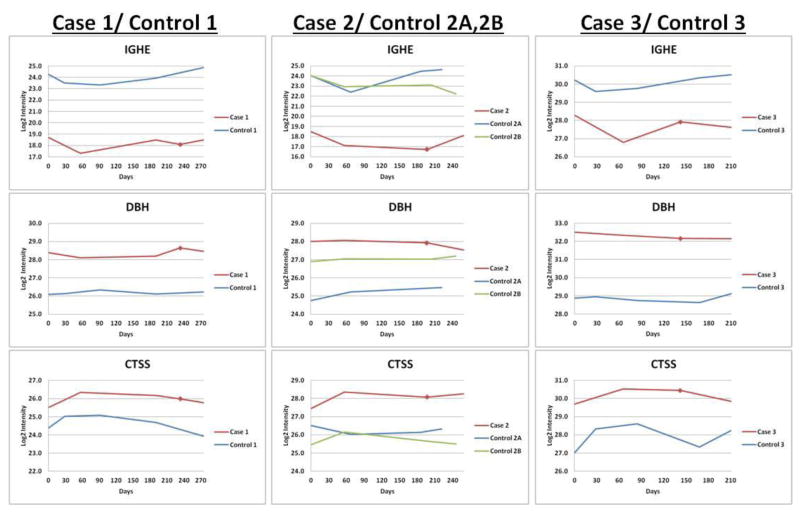
Predictive biomarkers were proteins which exhibited differences in the level between case and control either prior to treatment (baseline, at the time of first plasma collection) or at all timepoints for cases and controls (greater than 1.5-fold change between averages and p<0.05). The six best scoring candidate predictive biomarkers are summarized in the heatmap of Figure 3A. Longitudinal trends were also evaluated for each patient analyzed in the discovery experiments. Interestingly, the three proteins with the largest overall case/control differences (Figure 3B) were either consistently lower in all case timepoints as compared with matched controls, i.e., immunoglubulin E (IgE), or higher in cases as compared with matched controls, i.e., dopamine beta- hydroxylase (DBH) and cathepsin S (CTSS). We focused upon baseline immunoglobulin E (IgE) for validation because it showed the largest differences, from 5- to 58-fold, between cases and controls (Figure 3A).
Figure 3. Top Candidate Predictive Biomarkers.
A. Heat map of relative protein levels, fold change differences and its significance between cases and controls. Color-coded z-scores calculated for protein intensity for the top predictive markers are shown. Color scheme for individual plasma samples: red= high relative protein intensity; blue=low intensity; white= average intensity (no change); • = zero values that were replaced by imputed values for statistical analysis (see Methods); # = not detected in this experiment. * Plasma samples analyzed (see Figure 2). † p-value calculated from two-tailed Student’s t-test. ‡ Fold change between the average of all timepoints for each case and control pair. Red= fold change was >1.5-fold higher in cases; blue= fold change was >1.5-fold lower in cases. B. Longitudinal data from quantitative proteome analysis of three case and control pairs for the top three predictive markers, which have significant differences from the start of treatment and at all timepoints throughout the study. Inflection points indicate the positions of blood draws. Red squares=point of clinical CTRCD diagnosis.
Validation of baseline IgE as a biomarker of cardiac dysfunction
Our most promising biomarker, IgE, was identified in the proteome comparisons based on the epsilon chain C region of immunoglobulin. IgE is the lowest abundance immunoglobulin in plasma, with concentrations in the low ng/ml range.11 Importantly, unlike other immunoglobulins, IgE was not a target of the antibody column used to deplete abundant proteins. Both the heat maps and the longitudinal trend plots show that this protein was consistently low at all timepoints in all three cases in our discovery proteomics analyses (Figures 3A and 3B).
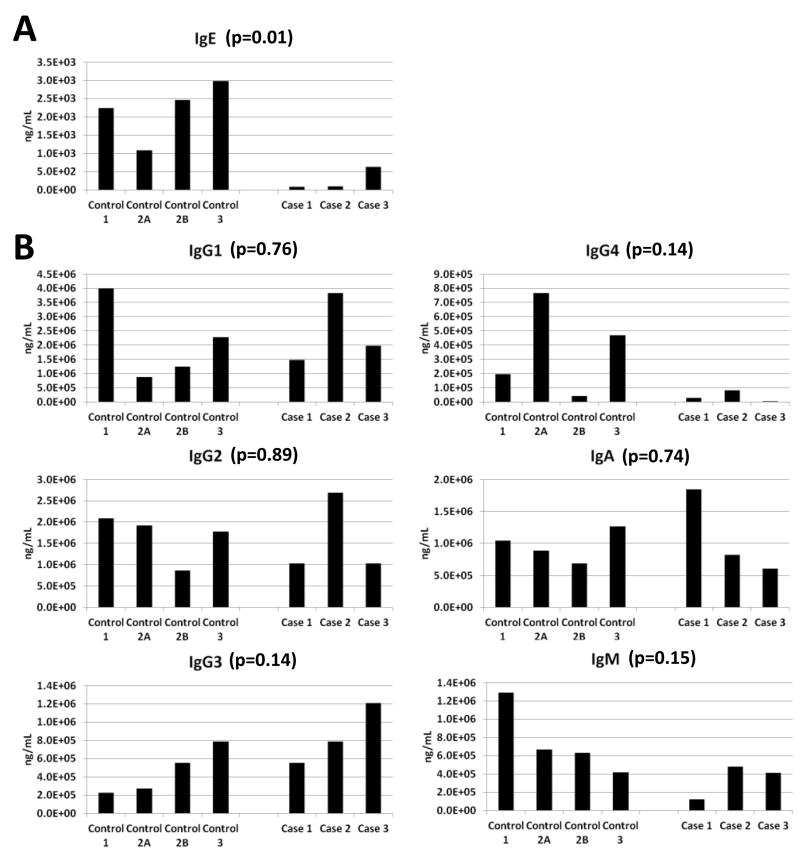
To verify the discovery results for IgE and to determine whether the observed lower IgE levels in cases were indicative of a general suppression of the immune system, we quantitated all immunoglobulin subtype levels using a multiplexed Luminex assay platform in the 3 cases and 4 matched controls. These results are displayed graphically in Figure 4 which details baseline immunoglobulin data only and in Supplemental Figure IIB which includes data from all timepoints. There was no evidence of a general immunosuppression in cases either at baseline (Figure 4B) or throughout therapy in the longitudinal plasma samples (Supplemental Figure IIB). Importantly, the IgE differences at baseline (Figure 4A) and the longitudinal trends observed in the proteome discovery experiments (Supplemental Figure IIA) were verified using this independent Luminex assay. When baseline levels in cases and controls were compared, there was a highly significant difference between groups with controls being higher (p=0.01). An additional interesting observation that arose from these initial validation experiments is that IgG4 levels were also low at baseline for all cases (Figure 4B). To further investigate the role of the immune system in doxorubicin and trastuzumab CTRCD, we also evaluated IgE related Th1 and Th2 cytokine profiles for cases and controls. Interestingly, a number of IgE related cytokines such as IL4, IL5, IL17, and fractalkine were also elevated in controls as compared to cases at baseline12-14 (Supplemental Figure III).
Figure 4. Baseline Levels of Immunoglobulin Subtypes Using Luminex Assays.
A. Baseline measurements of IgE (A.) and other Ig subtypes (B.) measured in the 3 case and 4 control samples from the proteomics discovery. Two-tailed Student’s t-test p-values are reported.
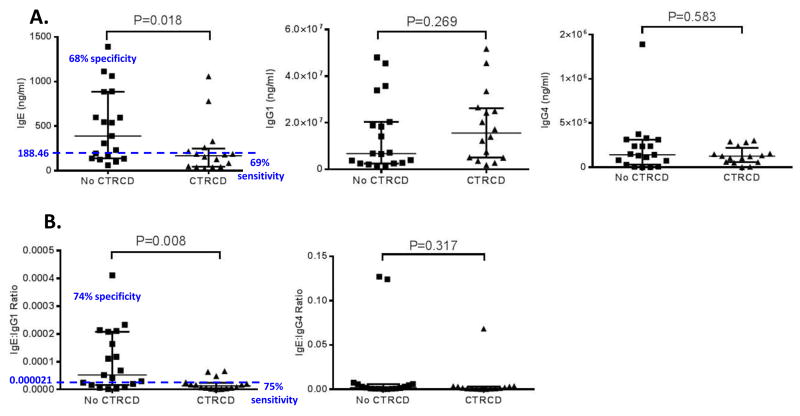
Based on these results, a more comprehensive evaluation of IgE, IgG1 and IgG4 levels was performed by assaying the baseline plasma samples of all thirty-five participants in the doxorubicin/ trastuzumab cohort (Table 2). Conventional colorimetric ELISA assays were used for this second set of validation experiments. The results demonstrated that baseline IgE levels were significantly higher (p=0.018) in participants who did not experience CTRCD [mean 498.8 ng/ml ±401.0; median 389.3 ng/ml with range 60.5-1392.1] compared to those who suffered from CTRCD [mean 234.9 ng/ml ±285.9; median 167 ng/ml with range 23.2-1059.2], suggesting a potential protective role for elevated IgE at baseline in cancer patients undergoing cardiotoxic therapy (Figure 5A). In this cohort, 9 of the thirty-five participants had a history of allergies or asthma, or were taking allergy medications. However, there was no relationship between allergy and baseline IgE levels, and the distribution of participants with a history of allergies or asthma was similar across participants with and without CTRCD.
Table 2.
Clinical Characteristics of Validation Cohort (N=35)
| Clinical Characteristics | N (%) or Median (IQR) |
|---|---|
| Age, years | 45 (39, 57) |
| Race | |
| Caucasian | 21 (60) |
| Black | 10 (29) |
| Other or unknown | 4 (11) |
| Breast cancer side | |
| Left | 16 (46) |
| Right | 14 (40) |
| Bilateral | 5 (14) |
| Breast cancer nodal status | |
| N0 | 15 (43) |
| N1 | 13 (37) |
| N2 | 5 (14) |
| N3 | 2 (6) |
| Breast cancer hormonal status | |
| ER+ | 18 (51) |
| Left-sided Radiotherapy | 16 (46) |
| Baseline EF, % | 55 (54, 59) |
| Cancer Therapeutics Related Cardiac Dysfunction, % | 18 (51) |
Figure 5. ELISA Validation for Baseline IgE, IgG4, IgG1 Levels and Ratios in the Doxorubicin and Trastuzumab Cohort Plasma.
A. Standard sandwich ELISA results for baseline (prior to treatment) plasma for all 35 participants in the doxorubicin/trastuzumab cohort. B. Ratios of the immunoglobulins assayed. No CTRCD: participants treated with doxorubicin and trastuzumab who did not develop cancer therapy- related cardiac dysfunction. CTRCD: participants treated with doxorubicin and trastuzumab who were diagnosed with cancer therapeutics-related cardiac dysfunction. Wilcoxon Rank Sum test was used and corresponding p-values are reported. In each graph, the bars represent the median and interquartile range. The blue dotted line is the threshold observed which reaches the highest specificity and sensitivity for IgE or IgE/IgG1.
We also analyzed samples from a cohort of healthy female volunteers without asthma or allergies and without evidence of any comorbidity including cardiovascular disease or cancer to determine IgE levels in normal individuals [mean 179.6 ng/ml ±231.7; median 97.0 ng/ml (range 36.9-853.8)]. This comparison did not reveal any significant difference between cases who developed CTRCD with normal individuals (p=0.99). In contrast, controls who did not experience CTRCD were significantly higher in comparison to non-cancer, healthy female volunteers (p=0.007). These results suggest that cancer patients who did not experience CTRCD had elevated IgE levels at baseline that may offer a cardioprotective benefit.
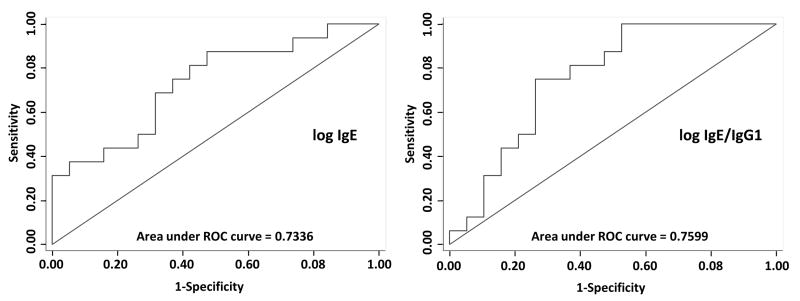
IgG1 and IgG4 levels were not significantly different between individuals with or without CTRCD, nor was the ratio between IgE/IgG4 (p>0.05 for all) (Figure 5B). However, when the ratio of IgE/IgG1 was evaluated, the differences in patients who experienced CTRCD compared to those who did not were most pronounced (p=0.008). Similarly, logistic regression models of IgE and the IgE/IgG1 ratio demonstrated a significant association with high IgE levels and a decreased probability of cardiac dysfunction. Each doubling of IgE was associated with an odds ratio (OR) of 0.52 (95% CI 0.31, 0.90, p=0.018) for the development of CTRCD. For the ratio of IgE/IgG1, each doubling was associated with an OR of 0.63 (95% CI 0.43, 0.92 p=0.017). Moreover, area under the receiver operating characteristics curve (AUC) analyses suggest strong discriminative ability with IgE and IgE/IgG1 with AUCs of 0.73 and 0.76, respectively (Figure 6). We also evaluated the sensitivities and specificities at various cutpoints of IgE and IgE/IgG1. For IgE, a measure of 188.5 ng/ml had a combined sensitivity of 69% and specificity of 68%. The IgE/IgG1 ratio at a cut-point of 2.05×10-5 demonstrated the highest combined sensitivity of 75% and specificity of 74%.
Figure 6. ROC Curves for IgE and IgE/IgG1 Ratio.
Receiver operator characteristic (ROC) curves for logistic regression models with IgE and IgE/IgG1 for the prediction of CTRCD. The area under the ROC curve (AUC) is 0.73 for the model with predictor of log2 IgE at baseline and 0.76 for the model with predictor of log2 (IgE/IgG1) ratio.
Overall, these findings suggest that elevated IgE or IgE/IgG1 levels prior to doxorubicin and trastuzumab therapy may have a protective effect against cardiac dysfunction in patients undergoing this therapy.
DISCUSSION
In this study, we used state-of-the-art proteomic profiling techniques to uncover potential new biomarkers for doxorubicin and trastuzumab CTRCD. Using a carefully phenotyped cohort of breast cancer patients, we discovered differences in baseline circulating IgE levels between patients who suffer from doxorubicin and trastuzumab-induced cardiac dysfunction compared to those who do not. These novel findings present a new paradigm in CTRCD, and implicate a specific, under-studied component of the immune system as a potential mediator of doxorubicin and trastuzumab-induced cardiac dysfunction.
Few studies have quantitated circulating IgE levels in humans, and the results have often been conflicting. Some have noted differences according to demographics, with decreases in IgE with age and increased levels in males,15 and associations between IgE-associated markers and an increased risk of breast cancer, as compared to controls.16 The main established roles of IgE are in the defense against parasitic diseases and pathogenesis of allergic diseases. Upstream, control of IgE expression occurs primarily through two subtypes of CD4+ T-helper cells: Th1 and Th2.17 Th1 and Th2 have counter-regulatory effects, as cytokines secreted by Th1 cells such as interferon-γ inhibit Th2 cytokine production, and vice versa. Th2 cells secrete a number of interleukins, including IL-4, IL-5, IL-6, IL-10, and IL-13. IL-4 and IL-13, which are important for B-cell isotype switching to IgE and IL-4-dependent IgE synthesis. Downstream, mast cells are hematopoietic inflammatory cells that are stimulated by IgE.
We postulate that IgE levels may represent some type of dysregulation between Th1 and Th2. In an exploratory analysis of T-helper cell cytokines and chemokines, several Th2 cytokines also showed trends where baseline levels were higher in controls compared to cases who developed CTRCD (Supplemental Figure III). Patients with high IgE levels at baseline may have a more Th-2 skewed T cell subset composition or a tendency to make a greater Th-2 response.
Of note, a large body of literature supports a role for the immune system in maintaining myocardial homeostasis in heart failure.18, 19 Immune mediators influence myocyte hypertrophy, fetal gene expression, and myocyte loss through apoptosis, and patients with heart failure have been reported to have higher levels of Th1, as opposed to Th2-associated cytokines. However, dysregulation of the inflammatory response has been shown to be both detrimental as well as beneficial to cardiac repair in response to various stressors, emphasizing the dual nature of the inflammatory system.20 In animal models, experimental induction of the Th1 response in mice led to worse left ventricular stiffness, dilation, and fibrosis.21, 22 Moreover, mast cell deficient mice, presumably indicative of an impaired acute inflammatory response, demonstrated improved survival and less myocardial necrosis in the setting of viral myocarditis.23 Conversely, in radiation-induced cardiac injury, emerging data also support a protective role of the immune system in the cardiovascular system.24-26 Here, mast-cell deficient mice demonstrated worse cardiac injury when exposed to radiation as compared to controls, suggesting that the absence of mast cells result in worsened cardiac remodeling in this setting. Although IgE levels in this setting are unknown, it is biologically plausible that mast cell deficient mice also have low IgE levels. Our data strongly suggest the need for further study of IgE and the immune system in the pathogenesis of cancer therapy induced cardiac dysfunction, and in particular, IgE deficient animal models could shed important insight into this area.
Although initially counterintuitive, it is tempting to speculate that high IgE levels in cancer patients, prior to exposure to cardiotoxic therapies, results in a beneficial, protective response, with improvement of cardiac myocyte survival and stabilization of cardiac function with exposure to various stressors, including doxorubicin, trastuzumab, or possibly even chest radiation therapy. It is thus also possible that low circulating IgE levels in the cancer setting result in or reflect a lack of the ability of the cardiovascular system to appropriately compensate in response to cardiac stress and injury induced by these therapies. It is interesting to note that the breast cancer patients who are at increased risk of developing cardiac dysfunction had IgE levels similar to healthy donors, and would suggest that elevation of IgE through unknown mechanism results in cardioprotection and possibly greater cardiac reserve. Perhaps patients with low IgE levels may also have a more susceptible, anergic state. Clearly, additional research is needed to characterize the potential role of IgE in cancer therapy associated cardiac dysfunction in greater detail, including the time course and duration of IgE associated immune system activation and subsequent downstream mediators.27
For the specific application of assessing risk of cardiac dysfunction in response to breast cancer therapies, the current results suggest that IgE and IgG1 levels could be measured prior to initiation of the therapeutic regimen as a method for stratifying patients for cardiac dysfunction risk. Those participants with low IgE and low IgE/IgG1 ratios appear to be at higher risk of developing cardiac dysfunction and might be more closely monitored for development of cardiac dysfunction. Further studies are needed to determine whether baseline or longitudinal monitoring of IgE, IgG1, and possibly other Ig subtypes could confer any benefit in identifying risk or onset of cardiac dysfunction.
We note that using powerful proteomic technology, we identified at least two additional interesting candidate markers, DBH and CTSS. Given the lack of robust high throughput assays for verification of these proteins, we were unable to validate these findings. However, future studies will focus on the development of appropriate assays for further evaluating these candidate biomarkers.
While the identification of baseline IgE and IgE/IgG1 ratios as predictive biomarkers of risk for CTRCD in breast cancer patients receiving doxorubicin and trastuzumab are intriguing, these findings need to be validated in additional patient populations, and mechanisms need to be further elucidated in basic studies. Similarly, the potential predictive value of these biomarkers for other cardiotoxic therapeutic regimens must be evaluated, as it remains unclear if these findings represent a susceptibility in response to exposure to doxorubicin, trastuzumab --- a humanized monoclonal IgG antibody, or possibly even radiation therapy. In summary, although IgE has not previously been directly implicated in cancer therapeutics-related cardiac dysfunction, our findings suggest that it may be indicative of a previously unknown mechanistic connection with cardiac dysfunction in breast cancer patients undergoing therapy with doxorubicin and trastuzumab therapy.
Supplementary Material
Novelty and Significance.
What Is Known?
Anthracyclines, such as doxorubicin, when used in combination with trastuzumab, result in a substantial decline in left ventricular ejection fraction (EF) in ~18% of treated patients, and debilitating heart failure in 2-4%.
There are currently no effective blood biomarkers for detecting early cardiotoxicity from cancer therapy.
Proteomics-based discovery of plasma biomarkers is challenging but feasible with the most recent advances in mass-spectrometry.
What New Information Does This Article Contribute?
Baseline IgE or the IgE/IgG1 ratio may be promising new biomarkers in doxorubicin and trastuzumab-induced cardiac dysfunction.
The immune system is a potential mediator of cardiac dysfunction following such therapy.
Doxorubicin and trastuzumab are two commonly used breast cancer therapies that have led to improvements in cancer survival, but carry a significant risk of cardiac dysfunction. Early identification of subclinical cardiac dysfunction would enable the initiation of cardio-protective strategies, prevent the interruption or discontinuation of necessary cancer therapy, and reduce the development of potential subsequent symptomatic heart failure. Current proteomic-based technologies can be used to successfully discover and validate new biomarkers, and while there are a number of existing biomarkers for cardiac failure in other settings, there is no prior research in the area of blood biomarkers for cardiotoxicity induced by cancer therapy. Our findings show that in patients treated with doxorubicin and trastuzumab, elevated IgE levels prior to treatment are associated with a lower risk of cardiac dysfunction, and implicate the immune system as a potential mediator of cardiac dysfunction in breast cancer patients.
Acknowledgments
The authors would like to thank Peter Hembach for assistance in performing protein depletions and SDS-PAGE, as well as the Wistar Institute Bioinformatics and Proteomics Core Facilities.
SOURCES OF FUNDING
This work was supported by NHLBI R01-HL118018 (Ky), WW Smith Charitable Trust Grants H1205 and H1305 (Speicher), PA Cure Health Grant (Ky), McCabe Fellow Award (Philadelphia, PA, Ky), American Cancer Society Institutional Research Grant -78-002-30 (Atlanta, Georgia, Ky), and NHLBI K23-HL095661 (Ky), and CA10815 (NCI core grant to the Wistar Institute).
Nonstandard Abbreviations and Acronyms
- CI
Confidence interval
- CTRCD
Cancer therapeutics-related cardiac dysfunction
- CTSS
Cathepsin S
- DBH
Dopamine beta-hydroxylase
- ELISA
Enzyme-linked immunosorbent assay
- FDR
False discovery rate
- HR
Hazard ratio
- IgE
Immunoglobulin E
- LC-MS
Liquid chromatography-mass spectrometry
- EF
Ejection fraction
- RP-HPLC
Reverse phase high-pressure liquid chromatography
Footnotes
DISCLOSURES
Dr. Ky is a consultant for Roche Diagnostics.
References
- 1.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against Her2 for metastatic breast cancer that overexpresses Her2. The New England Journal of Medicine. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Swain SM, Whaley FS, Ewer MS. Congestive heart failure in patients treated with doxorubicin: A retrospective analysis of three trials. Cancer. 2003;97:2869–2879. doi: 10.1002/cncr.11407. [DOI] [PubMed] [Google Scholar]
- 3.Suter TM, Procter M, van Veldhuisen DJ, Muscholl M, Bergh J, Carlomagno C, Perren T, Passalacqua R, Bighin C, Klijn JG, Ageev FT, Hitre E, Groetz J, Iwata H, Knap M, Gnant M, Muehlbauer S, Spence A, Gelber RD, Piccart-Gebhart MJ. Trastuzumab-associated cardiac adverse effects in the herceptin adjuvant trial. Journal of Clinical Oncology : official journal of the American Society of Clinical Oncology. 2007;25:3859–3865. doi: 10.1200/JCO.2006.09.1611. [DOI] [PubMed] [Google Scholar]
- 4.Tan-Chiu E, Yothers G, Romond E, Geyer CE, Jr, Ewer M, Keefe D, Shannon RP, Swain SM, Brown A, Fehrenbacher L, Vogel VG, Seay TE, Rastogi P, Mamounas EP, Wolmark N, Bryant J. Assessment of cardiac dysfunction in a randomized trial comparing doxorubicin and cyclophosphamide followed by paclitaxel, with or without trastuzumab as adjuvant therapy in node-positive, human epidermal growth factor receptor 2-overexpressing breast cancer: Nsabp b-31. Journal of Clinical Oncology : official journal of the American Society of Clinical Oncology. 2005;23:7811–7819. doi: 10.1200/JCO.2005.02.4091. [DOI] [PubMed] [Google Scholar]
- 5.Telli ML, Hunt SA, Carlson RW, Guardino AE. Trastuzumab-related cardiotoxicity: Calling into question the concept of reversibility. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 2007;25:3525–3533. doi: 10.1200/JCO.2007.11.0106. [DOI] [PubMed] [Google Scholar]
- 6.Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, Lancellotti P, Muraru D, Picard MH, Rietzschel ER, Rudski L, Spencer KT, Tsang W, Voigt JU. Recommendations for cardiac chamber quantification by echocardiography in adults: An update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr. 2015;28:1–39. e14. doi: 10.1016/j.echo.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Beer LA, Tang HY, Sriswasdi S, Barnhart KT, Speicher DW. Systematic discovery of ectopic pregnancy serum biomarkers using 3-d protein profiling coupled with label-free quantitation. J Proteome Res. 2011;10:1126–1138. doi: 10.1021/pr1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox J, Mann M. Maxquant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 10.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Perez-Riverol Y, Reisinger F, Rios D, Wang R, Hermjakob H. The proteomics identifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prabhu A, Ruzek MC, Kamat DM, Mathur A. Plasma IgE levels, activation marker expression, and cytokine production in non-atopic individuals. Ann Allergy Asthma Immunol. 1997;78:45–53. doi: 10.1016/S1081-1206(10)63371-6. [DOI] [PubMed] [Google Scholar]
- 12.Chong S, Lan H, Zeng K, Zhao X. Serum fractalkine (cx3cl1) concentration correlates with clinical severity in pediatric atopic dermatitis patients. Ann Clin Lab Sci. 2016;46:168–173. [PubMed] [Google Scholar]
- 13.Milovanovic M, Drozdenko G, Weise C, Babina M, Worm M. Interleukin-17a promotes IgE production in human b cells. J Invest Dermatol. 2010;130:2621–2628. doi: 10.1038/jid.2010.175. [DOI] [PubMed] [Google Scholar]
- 14.Reubsaet LL, Meerding J, Scholman R, Arets B, Prakken BJ, van Wijk F, Knol EF. Allergen-specific Th2 responses in young children precede sensitization later in life. Allergy. 2014;69:406–410. doi: 10.1111/all.12366. [DOI] [PubMed] [Google Scholar]
- 15.Gergen PJ, Arbes SJ, Jr, Calatroni A, Mitchell HE, Zeldin DC. Total IgE levels and asthma prevalence in the us population: Results from the national health and nutrition examination survey 2005-2006. The Journal of Allergy and Clinical Immunology. 2009;124:447–453. doi: 10.1016/j.jaci.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petridou ET, Chavelas C, Dikalioti SK, Dessypris N, Terzidis A, Nikoulis DI, Markopoulos C, Papadiamantis Y, Germenis AE. Breast cancer risk in relation to most prevalent IgE specific antibodies: A case control study in Greece. Anticancer Res. 2007;27:1709–1713. [PubMed] [Google Scholar]
- 17.Bacharier LB, Geha RS. Molecular mechanisms of IgE regulation. The Journal of Allergy and Clinical Immunology. 2000;105:S547–558. doi: 10.1016/s0091-6749(00)90059-9. [DOI] [PubMed] [Google Scholar]
- 18.Torre-Amione G. Immune activation in chronic heart failure. The American Journal of Cardiology. 2005;95:3C–8C. doi: 10.1016/j.amjcard.2005.03.006. discussion 38C-40C. [DOI] [PubMed] [Google Scholar]
- 19.Mann DL. Inflammatory mediators and the failing heart: past, present, and the foreseeable future. Circ Res. 2002;91:988–998. doi: 10.1161/01.res.0000043825.01705.1b. [DOI] [PubMed] [Google Scholar]
- 20.Coggins M, Rosenzweig A. The fire within: cardiac inflammatory signaling in health and disease. Circ Res. 2012;110:116–125. doi: 10.1161/CIRCRESAHA.111.243196. [DOI] [PubMed] [Google Scholar]
- 21.Cheng X, Ding Y, Xia C, Tang T, Yu X, Xie J, Liao M, Yao R, Chen Y, Wang M, Liao YH. Atorvastatin modulates Th1/Th2 response in patients with chronic heart failure. Journal of Cardiac Failure. 2009;15:158–162. doi: 10.1016/j.cardfail.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Yamaoka-Tojo M, Tojo T, Inomata T, Machida Y, Osada K, Izumi T. Circulating levels of interleukin 18 reflect etiologies of heart failure: Th1/Th2 cytokine imbalance exaggerates the pathophysiology of advanced heart failure. Journal of Cardiac Failure. 2002;8:21–27. doi: 10.1054/jcaf.2002.31628. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi H, Hara M, Yamamoto K, Miyamoto T, Kinoshita M, Yamada T, Uchiyama K, Matsumori A. Mast cells play a critical role in the pathogenesis of viral myocarditis. Circulation. 2008;118:363–372. doi: 10.1161/CIRCULATIONAHA.107.741595. [DOI] [PubMed] [Google Scholar]
- 24.Boerma M. Experimental radiation-induced heart disease: past, present, and future. Radiat Res. 2012;178:1–6. doi: 10.1667/rr2933.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boerma M, Wang J, Wondergem J, Joseph J, Qiu X, Kennedy RH, Hauer-Jensen M. Influence of mast cells on structural and functional manifestations of radiation-induced heart disease. Cancer Res. 2005;65:3100–3107. doi: 10.1158/0008-5472.CAN-04-4333. [DOI] [PubMed] [Google Scholar]
- 26.Boerma M, Zurcher C, Esveldt I, Schutte-Bart CI, Wondergem J. Histopathology of ventricles, coronary arteries and mast cell accumulation in transverse and longitudinal sections of the rat heart after irradiation. Oncol Rep. 2004;12:213–219. doi: 10.3892/or.12.2.213. [DOI] [PubMed] [Google Scholar]
- 27.Fildes JE, Shaw SM, Yonan N, Williams SG. The immune system and chronic heart failure: Is the heart in control? J Am Coll Cardiol. 2009;53:1013–1020. doi: 10.1016/j.jacc.2008.11.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.