Abstract
Using synchronized cells, one can directly measure delay in mitosis brought about by the G2 DNA damage checkpoint in response to exposure to exogenous DNA damaging agents. Scoring mitosis in the fission yeast Schizosaccharomyces pombe is relatively simple. Many techniques exist for synchronizing cells for such assays. We present a detailed explanation of the setup and use of centrifugal elutriation to synchronize cells in G2, exposure of cells to DNA damage, and measurement of mitotic progression and delay.
Keywords: G2 DNA damage checkpoint, mitosis, septation index, elutriation, bleomycin, ionizing radiation.
1. Introduction
The G2 DNA damage checkpoint responds to DNA damage during G2, after replication is completed. This checkpoint prevents the G2 to M phase transition in the presence of DNA damage allowing cells sufficient time to repair DNA damage before undertaking nuclear and cellular division (1). Much of what is known about this checkpoint has been discovered in the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe (2, 3). This chapter presents a protocol for studying the response of fission yeast to G2 DNA damage.
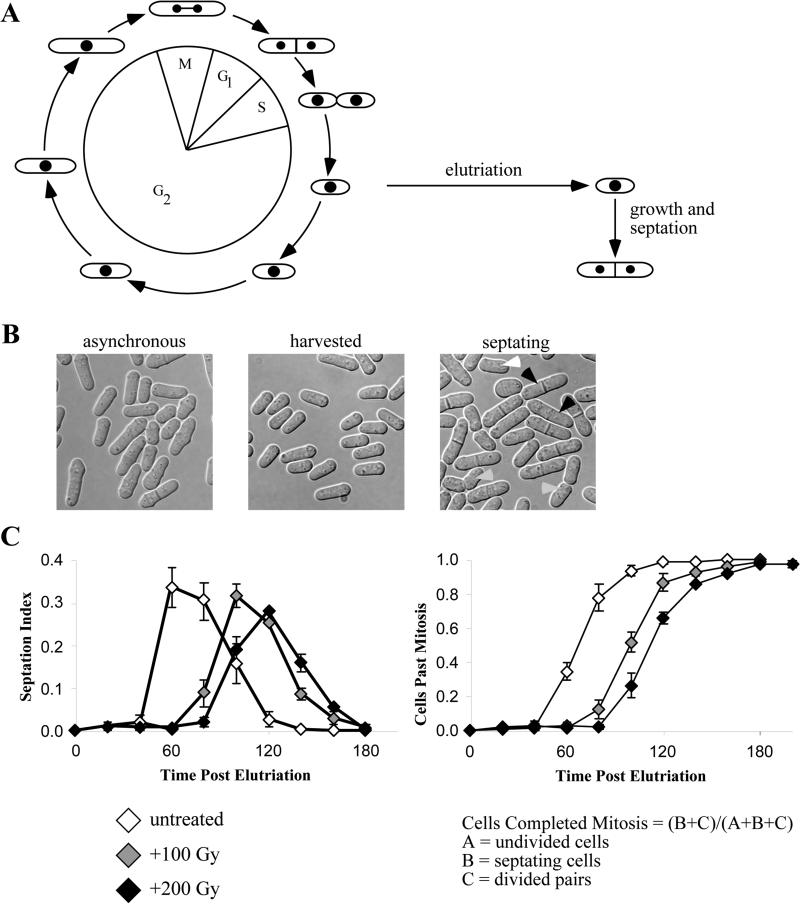
Fission yeast is easily manipulated and offers a short cell cycle allowing for easy determination of the G2 DNA damage cell cycle response. In particular, it is straightforward to follow the passage from G2 though mitosis by microscopy. Fission yeast is a rod shaped organism that divides by medial fission. After mitosis, cytokinesis is accomplished by the construction of a septum through the middle of the cell. Septum formation may be scored in live yeast since the septum is easily recognized under phase microscopy (See Figure 2B). Timing of mitosis and delay due to exposure to DNA damage may be measured by scoring synchronous cells as the population progresses through M phase and septates.
Figure 2.2. G2 DNA Damage Response.
(A) Cell cycle position is tied to cell morphology in fission yeast. Completion of mitosis and replication occur simultaneously so the smallest cells in an asynchronous culture will be cells that have just entered G2 phase. Continued cell growth but no further septation is an indication of G2 DNA damage checkpoint mediated arrest. (B) DIC images of an asynchronous culture, freshly elutriated population and actively engaged in septation. Black arrowheads indicate septating cells, gray arrowheads indicate divided cell pairs and the white arrowhead indicates half of a divided pair, a cell too short to be a undivided cell, but one that has detached from its division partner. (C) Examples of septation and mitotic progression plots and delays in mitosis brought about by exposure to increasing dose of ionizing radiation. Plots represent three independent experiments. Error bars represent the standard error of the mean.
Several techniques allow for the synchronization of yeast in G2. They can be divided into two broad categories: block-and-release and selection synchronization. Block-and-release approaches include nutrient starvation, drug-induced arrest and use of cell division cycle (cdc) mutants, all of which block cells at a particular point in the cell cycle and allow for the release of a synchronous culture. Selection synchronization techniques rely on separation of a synchronous subset of a culture. Centrifugal elutriation, lactose gradient centrifugation and ficoll step gradient centrifugation all represent techniques allowing the selection of small and synchronized cells from an asynchronous culture (4-6). The selection techniques have two important advantages over block-and-release synchronization. First, they do not severely disrupt the cell cycle, making them more sensitive and less artifact prone. Second, they may be employed to separate healthy small cells from an otherwise sick population containing dead and permanently arrested cells. This advantage allows the analysis of strains, particularly repair defective strains, that display a degree of growth heterogeneity that would otherwise confound cell cycle analysis.
Centrifugal elutriation allows cell synchronization without significant known artifacts and with minimal cell cycle delay (7). In particular, elutriation allows recovery of cells with a minimum of cell stress and without the use of cdc alleles or drug exposure. Although elutriation suffers from limitations, including that only single cultures may be synchronized at a time and that a limited number of cells can be recovered, we find it an efficient and robust approach to synchronizing cells.
We present here how to assemble and operate a centrifugal elutriation system, methods permitting sterile harvesting of cells and basic protocols to expose synchronized cells to increasing doses of ionizing and ultra-violet radiation. Additionally, we describe in detail how to score and plot septation and mitotic indices for elutriated cultures.
2. Materials
2.1 Elutriator and Pump Setup
Tygon R-3603, 1/8” ID, 1/4” ED, 1/16” wall, VWR cat# 63009-170.
Kontes 3-way Stopcock, VWR cat# KT420163-4503.
WPA biowave Cell Density Meter, cat# CO8000, www.biochrom.co.uk/product/20/co8000-cell-density-meter.html.
Masterflex L/S Economy Digital Drive and Easy-Load II Pump Head, Cole-Parmer Instrument Company, cat# C-07524-40, and C-77200-60.
Beckman J-20 with a JE-5 series rotor and 4 ml elutriation chamber, Beckman Instruments.
Flow cell cat# 73.4/SOG, www.starna.com.
Silicone pump tubing.
2.2 G2 Elutriation, DNA Damage and Timecourse
-
8.
Graduated flasks for cell loading, recirculation, and collection.
-
9.
YES rich media (yeast extract with supplements): 5 g/l yeast extract, 30 g/l glucose, 75 mg/l leucine, 75 mg/l uracil, 75 mg/l adenine, 75 mg/l histidine, autoclaved.
-
10.
Bleomycin, Sigma cat # B-2434.
-
11.
Stratalinker UV source, or equivalent.
-
12.
Faxitron RX-650 X-ray source, www.faxitron.com, or equivalent.
-
13.
Levy hemacytometer counting chamber, VWR cat# 15170-208.
-
14.
Pall Life Sciences Supor Disk Filter, 25 mm2 0.2 μn, VWR cat # 28147-956.
-
15.
Pall Life Sciences Supor Disk Filter, 47 mm2 0.2 μn, VWR cat # 28148-551.
-
16.
Pall Life Sciences Vacuum manifold compatible with 25 or 47 mm2 filters.
2.3 Materials - Sterile Elutriation
-
17.
Two, 2 liter flasks containing 1.0 liter water, autoclaved.
-
18.
One 500 ml bottle containing 150 ml water, autoclaved.
-
19.
50 ml Bleach.
-
20.
500 ml 70% ethanol.
3. Methods
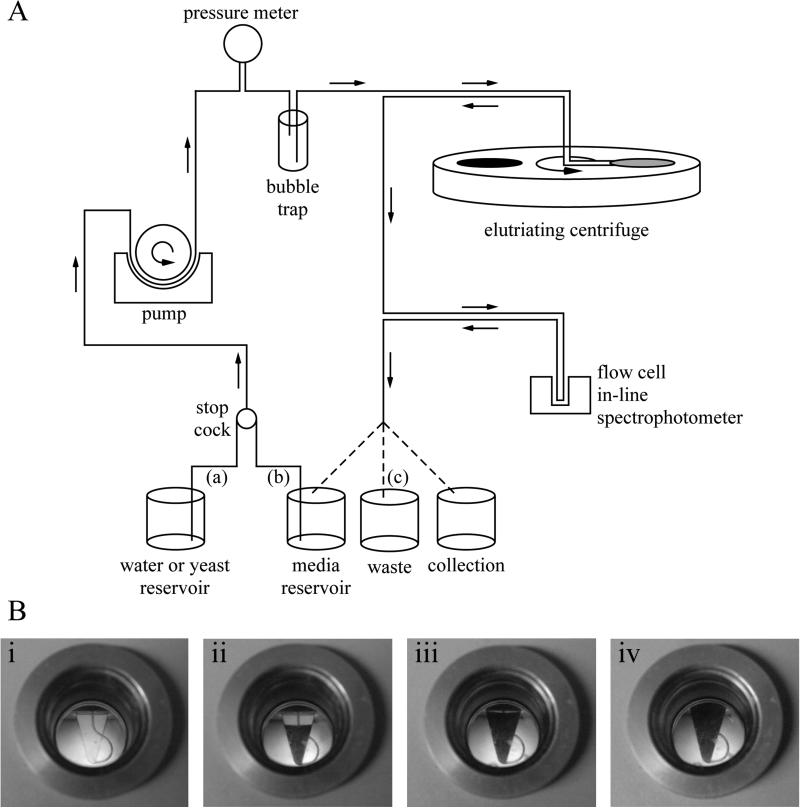
Elutriation selects cells based on size. It procedure depends on a special centrifuge rotor that allows liquid to be pumped through a spinning centrifuge chamber (Figure 2.1). The centrifugal force experienced by cells as they are pumped through the chamber forces them down to the bottom of the chamber. This force is countered by the flow of the medium, which forces the cells to the top of the chamber. The hydrodynamics in the chamber sorts the cells by size, with smaller cells raising to the top. A synchronized strobe light and viewing window allows one to observe the cells in the chamber. By adjusting the opposing forces, it is possible to trap cells in the chamber and move them up to the point where only the smallest cell are washed out of the chamber. These small cells can be collected as a synchronous culture. Due to a particularity of the fission yeast cell cycle, the smallest cells in the culture are those that have just entered G2. These cells have just completed septation and replication and thus are early in the G2 phase of the cell cycle (See Figure 2.2A).
Figure 2.1. The General Setup for Elutriation.
(A) Two inlet tubes (a, b) are setup allowing quick switching between media, yeast or water. Media from either reservoir is flowed through the system using a pump. The pressure meter allows determination of whether the system is blocked. The bubble trap smooths pressure pulses created by the pump and prevents air bubbles from entering and blocking the system with the centrifuge running. An in-line spectrophotometer is useful for monitoring cells escaping the centrifuge chamber when cells are recirculated and harvested. The outlet tubing (c) allows the user to recirculate media, remove waste or collect cells. (B) The elutriator chamber, viewed during a run. i) An empty chamber. ii) A partially full chamber during cell loading. iii) An almost full chamber; only a small gap remains between the cell front and the top of the chamber. At this point, no more cells need to be loaded, and the rotor speed can be reduced to allow cells to escape. Alternatively, the pump speed can be reduced, which will cause the cell front to retreat and make room for more cells. iv) A full chamber with cells escaping.
3.1 Initial Elutriator Setup
Assemble the 4 ml centrifugation chamber according to manufacturer's instructions (See Figure 1) (See Note 1).
Place a single inlet tube (a) in the water reservoir, the second inlet tube (b) in fresh media, the flow cell in the in-line spectrophotometer, and the outlet tube (c) in the waste (See Note 2).
Start the pump, set to 100 ml/min (See Note 3).
Fill the bubble trap and flood the entire system with water (See Note 4).
Start the centrifuge at 4100 rpm and set the centrifuge temperature to 30°C (See Notes 5, 6 and 7).
Switch inlet source to (b) and flow fresh media into the system. Once the water is flushed out, transfer the outlet tubing (c) in the same reservoir of media permitting recirculation (See Note 8).
Adjust the strobe lamp to visualize the centrifugation chamber through the centrifuge port hole and reference the in line spectrophotometer against the recirculating media.
3.2 Growth, Loading and Elutriation of Cells
-
8.
500 ml cells should be grown to mid-log phase (OD600 ~ 1.0) (See Note 9).
-
9.
Transfer a inlet tube (a) to a reservoir containing the strain to be loaded.
-
10.
Switch inlet source to (a) to load cells into the small 4 ml chamber at 30 ml/min flow rate at 4100 rpm until the cell front nears the top of chamber (See Note 10).
-
11.
When chamber is full switch stopcock to recirculate from the media reservoir.
-
12.
Decrease centrifuge speed in 50 rpm increments to eventually allow the smallest cells to escape the elutriation chamber. Monitor the escaping cell density with the inline spectrophotometer. (See Note 11).
-
13.
When the escaping cell density is between OD600 0.1-0.2, switch outlet tube (c) to an appropriate container to collect cells (See Note 12).
-
14.
Check harvested cells under 40x power using hemacytometer to observe cell size (See Note 13).
-
15.
Place the synchronous culture in an appropriate flask and incubate for the timecourse at 30°C, 200 rpm.
3.3 Cell Counting and Septation Index Determination
-
16.
For each timepoint, pipette 10 μl cells onto a hemacytometer and count cells (See Note 14).
-
17.
Count cells every 20 minutes for 3-5 hours (See Note 15).
3.4 Flushing out the Tubing, Centrifugation Chamber and Housing Assembly
-
18.
Stop the centrifuge and increase pump speed to 90 ml/min.
-
19.
Flush the system with water and then purge with air.
-
20.
Disconnect chamber and wash separately with mild detergent.
-
21.
Connect the chamber inlet and outlet tubes to one another and flood the tubing and bubble trap with 70% ethanol followed by an excess of water (See Note 16).
-
22.
Purge out all liquid from tubing.
3.5 Sterile Elutriation Setup (See Note 17)
-
23.
Flush the system with nonsterile water with the pump set to 100 ml/min.
-
24.
Setup and start the centrifuge set to 2000 rpm.
-
25.
Add 50 ml bleach to one flask containing 1 liter of autoclaved water.
-
26.
Flush out the water with the dilute bleach solution and allow recirculation for at least 20 minutes.
-
27.
Flush out bleach with 500 ml 70% ethanol and allow recirculation for 20 minutes (See Note 18).
-
28.
Flush out the alcohol using the remaining 1 liter of sterile water (See Note 19).
-
29.
Flood the system with sterile media to flush out the water.
-
30.
Load samples as above.
-
31.
Collect cells and sample timepoints using sterile technique (See Note 20).
3.6 Treatment with Ionizing Radiation
-
32.
Prepare 5 ml samples of G2 elutriated yeast for each dose to be tested.
-
33.
Expose cells to appropriate doses of ionizing radiation (See Note 21).
-
34.
Transfer irradiated cultures to culture tubes and incubate shaking at 30°C for the timecourse duration.
-
35.
Count undivided cells, septating cells and divided pairs every 20 minutes for 3-5 hours (See Notes 14 and 15).
3.7 Treatment with Ultraviolet Radiation
-
36.
Setup a vacuum apparatus and prewet 25 mm2 filters with yeast media (See Note 22).
-
37.
Transfer up to 10 OD G2 elutriated culture and concentrate onto filters by vacuum.
-
38.
Place filters yeast side up on 3M blotting paper and expose in a Stratalinker (See Note 23).
-
39.
Fold filters slightly and drop into culture tubes.
-
40.
Add 5 ml media and vortex to release yeast into the medium.
-
41.
Shake tubes to a 30°C for the timecourse.
-
42.
Count undivided cells, septating cells and divided pairs every 20 minutes for 3-5 hours (See Notes 14 and 15).
4. Notes
4.1 Initial Elutriator Setup
Two elutriation chambers are available with 4 and 40 ml capacities. The following procedure described utilizes the smaller 4 ml chamber. For fission yeast, the 4 ml chamber holds approximately 400 ODs allowing 40 ODs to be synchronously harvested (see Note 9). The 40 ml chamber capable of holding 4000 ODs may be used to harvest up to 400 ODs if more cells are required.
The stop-cock is used to switch input between the two inlet sources (a, b) allowing seamless switching between various reservoirs of cells or media. Media recirculation (outlet (c) and inlet tube (b) are in the same vessel) allows the user increased preparation or lag time if needed.
Never shut off the pump while the centrifuge is running. If collecting cells in the centrifugation chamber, loss of pump pressure will result in cells being pushed “back” into the loading channel and clogging the system. If a clog of this nature occurs, first stop the centrifuge, then restart the pump and set the flow rate to 100 ml/min to attempt to force the cells out of the loading channel. If this method fails, try firmly tapping the side of the loading chamber to dislodge the clogging cells. If the clog cannot be cleared, the chamber must be dissembled and cells removed by hand.
Fill the bubble trap to about ¾ full; the air pocket in the bubble trap acts as a pressure buffer to dampen the flow pulses from the pump, which reduces turbulence in the elutriation chamber.
As the centrifuge speeds up, the remaining air bubbles with be forced from the rotor.
Monitor back pressure at the pressure meter. It should be a few psi when the chamber is empty of cells. When using the large, 40 ml chamber the psi may approach 10 psi when the chamber is almost full of cells. A back pressure of greater than 10 psi indicates a block in the system, possibly due to bubbles trapped in the rotor. If the pressure remains above 10 psi, stop the centrifuge to allow bubbles to escape. Stopping the centrifuge will cause any loaded cells to be washed out of the chamber, but they can be collected and reloaded. If the pressure remains significantly above 10 psi, a tube joint will fail.
For temperature or cold sensitive strains, set the temperature of the elutriating centrifuge chamber to the appropriate temperature. The centrifuge is refrigerated but not heated, so to reach temperatures above ambient, one must allow the centrifuge to run, to warm the chamber using heat generated from the centrifuge motor itself. Achieving 30°C from room temperature takes approximately 20 minutes centrifuge run time when set to 4100 rpm.
Always use fresh media and remove as much water as possible. Reuse of used media from the main culture or excessive recirculation with a chamber full of cells may cause cells to run out of sugar and begin respiration, producing CO2 bubbles, which will reduce harvested cell homogeneity.
4.2 Growth, Loading and Elutriation of Cells
-
9.
Grow 500 ml culture in YES at 30°C, 200 rpm to an optical density at 600 nm (OD600) of about 1, starting from a mid-log 50 ml starter culture. OD600 is used to follow yeast cell proliferation. Optical density is also used approximate cell number; 1 OD unit (abbreviated OD) is the number of cells required to give 1 ml of culture an optical density of 1. Therefore, a 100 ml culture at OD600 of 0.1, a 10 ml culture at OD600 of 1 and a 1 ml culture at OD600 of 10 all contain 10 ODs of cells. 1 OD is approximately 2×107 cells. The 4 ml and 40 ml chambers hold approximately 400 and 4000 OD yeast cells, respectively. For the large chamber, several liters of cells may be grown to an optical density of 2.0. Sick strains may require more cells to fill the chamber, in excess of 800 or 8000 ODs of cells for the small and large chambers respectively. For cultures that are extremely sick or flocculate at low OD, sonicate the culture using twenty 50 msec pulses at maximum power. Sonication also helps to break up divided pairs, thus increasing enrichment of small cells for any strain.
-
10.
Begin loading the 40 ml chamber at 100 ml/min flow rate, 4100 rpm. To save time loading, after cells begin to accumulate in the chamber increase the pump speed to 130 ml/min until the chamber is close to full. When using the small chamber, it may be loaded at 100 ml/min for several minutes before the pump is reduced to 30 ml/min.
-
11.
The pump and rotor speeds need to be balanced to bring the cell boundary to the top of the chamber and allow the smallest cells to escape. For course adjustment, to get the cell boundary to the top of the chamber, it is easiest to adjust the pump speed. Once cells begin to escape, adjusting the rotor speed gives one much finer control. Decreasing centrifuge speed subtly reduces forces on cells allowing small changes in cell elutriation while increasing pump speeds even by 1 ml/min causes dramatic changes and may force a heterogeneous population of cells out of the chamber. The flow cell and in line spectrophotometer are used to follow the optical density of the media exiting the centrifuge. Remember to tare the spec with the flow cell inserted and only after media is recirculating. Depending on how many cells are loaded in the chamber, they may escape between 3700-2900 rpm. The pump speed should be kept below 40 ml/min when harvesting cells. High pump flow causes turbulence within the chamber, reducing harvested cell synchrony. Once the collection of cells in begun, it is necessary to occasionally decrease the rotor speed to maintain the desired rate of cell elutriation, as the number of cells escaping will gradually decrease over time.
-
12.
The optical density of the escaping cells should not exceed an of 0.4; to harvest the most synchronous population, aim to collect between at an OD600 of 0.1 and 0.2. Practically, it is best to limit recovery of cells is 10% of the loaded, asynchronous culture.
-
13.
Strive for less than 1% of the collected cells to be septated on in divided pairs, more than 1% indicates a poor synchronization.
4.3 Cell Counting, Septation and Mitotic Index Determination
-
14.
Count 100 cells. Typically, observed cells fall into three categories; undivided cells, septating cells, and divided pairs. Undivided cells appear small at the beginning of the timecourse and will grow and increase in cell length over time. Septating cells display a septum across the cell midsection and are typically longer. Divided pairs are those cells which have completed septation and show dimples around the septum (Figure 2.1B). The septation index is simply the percentage of cells septating observed at any single timepoint. In a well synchronized population, septation will peak with a value between 30 and 60%. For cultures incubated at 30°C, septation will peak around 80 minutes post elutriation, while cultures incubated at 25°C will peak around 120 minutes. Progression through mitosis may also be represented by plotting the total number of cells which have completed septation, the percentage of cells that are septating or have completed septation (both sepatated and divided pair populations). Just after the septation index peaks, this index should approach 100%. This index may be calculated using the following equation (Figure 2B).
Cells Completed Mitosis = (B+C)/(A+B+C)
A = undivided single cells
B = septating cells
C = divided pairs
Undivided cells, septating cells and divided pairs are each counted as a single cell. Divided pairs are counted as a single event because the pair of cells arose from a single mother cell. At later timepoints, undivided cells will be long and some divided pairs will have separated into single cells, thus count pairs of these small cells as single events (See Figure 2B, right hand panel). For counting multiple rounds of mitosis, after most cells have completed septation and are divided pairs, sonicate the culture to separate divided pairs and continue counting assuming single cells are now undivided cells.
-
15.
Septation takes approximately 20 minutes; thus, if counted every 20 minutes, each septation should be counted once. Therefore, the cumulative septation count across every time point should be around 100%; a greater or lesser value suggests over or under counting of septa.
4.4 Flushing out the Elutriator Tubing, Chamber and Housing Assembly
-
16.
To eliminate cell debris buildup inside the bubble trap and tubing, occasionally recirculate dilute bleach for 30 minutes following by flushing the system with two liters of water.
4.5 Sterile Elutriation Setup
-
17.
The elutriator is a nonsterile system harboring bacteria and fungi. If plating cells after synchronization to compare viability between treated and untreated samples, sterilize the system prior to loading cells. Be sure when sterilizing to fill the bubble trap completely to sterilize the entire inner surface. Wear gloves at all times when handling the inlet and outlet tubes. Use sterile glassware for sample incubation, preparation and collection.
-
18.
Ethanol will produce bubbles. If the system gets blocked and over-pressurizes, stop the centrifuge and shake tubing and chamber to dislodge those bubbles. Restart the centrifuge and continue ethanol recirculation. Two phases may form in the air trap, be sure to invert the air trap to ensure the phases mix.
-
19.
Vegetative yeast are extremely sensitive to both ethanol and bleach, be sure to allow the entire liter of water to flush out the system.
-
20.
If plating cells, use media containing ampicillin or carbenicilin to prevent bacteria from growing and out competing the yeast.
4.6 Treatment with Ionizing Radiation
-
21.
Samples may be exposed using a Faxitron X-ray cabinet or other ionizing radiation source. For exposure in a Faxitron cabinet, it is convenient to transfer 5ml samples to 35mm Petri dishes and irradiate cells while suspended in liquid culture. Regardless of the source used, begin by exposing the sample to be irradiated to the greatest degree, then place samples to be exposed at lower doses in at later time points during the irradiation. This strategy will stagger all of the irradiated samples to finished simultaneously. 0, 50, 100 and 200 Gy doses are good starting points for healthy strains. As a control, cells may be exposed to 3 mU Bleomycin, which acts as an IR mimetic and will arrest checkpoint proficient strains.
4.7 Treatment with Ultraviolet Radiation
-
22.
Filters should be prelabeled using pencil. Filter capacity is approximately 2 OD/cm2. Thus, up to 10 OD and 40 OD yeast may be deposited on 25 mm and 47 mm filters respectively.
-
23.
A Stratalinker UV Crosslinker is a convenient way to deliver a defined UV dose. For repair proficient strains, doses up to 50 J/m2 are useful.
References
- 1.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 2.Stewart E, Enoch T. S-phase and DNA-damage checkpoints: a tale of two yeasts. Curr Opin Cell Biol. 1996;8:781–787. doi: 10.1016/s0955-0674(96)80078-0. [DOI] [PubMed] [Google Scholar]
- 3.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards RJ, Carr AM. Analysis of radiation-sensitive mutants of fission yeast. Methods Enzymol. 1997;283:471–494. doi: 10.1016/s0076-6879(97)83038-8. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker GM. Synchronization of yeast cell populations. Methods Cell Sci. 1999;21:87–93. doi: 10.1023/a:1009824520278. [DOI] [PubMed] [Google Scholar]