Abstract
Developmental neuronal remodeling is a crucial step in sculpting the final and mature brain connectivity in both vertebrates and invertebrates. Remodeling includes degenerative events, such as neurite pruning, that may be followed by regeneration to form novel connections during normal development. Drosophila provides an excellent model to study both steps of remodeling since its nervous system undergoes massive and stereotypic remodeling during metamorphosis. Although pruning has been widely studied, our knowledge of the molecular and cellular mechanisms is far from complete. Our understanding of the processes underlying regrowth is even more fragmentary. In this review, we discuss recent progress by focusing on three groups of neurons that undergo stereotypic pruning and regrowth during metamorphosis, the mushroom body γ neurons, the dendritic arborization neurons and the crustacean cardioactive peptide peptidergic neurons. By comparing and contrasting the mechanisms involved in remodeling of these three neuronal types, we highlight the common themes and differences as well as raise key questions for future investigation in the field. WIREs Dev Biol 2016, 5:618–635. doi: 10.1002/wdev.241
For further resources related to this article, please visit the WIREs website
INTRODUCTION
Neuronal remodeling is an essential step in the formation of the adult nervous system. This conserved process is crucial in order to form the precise connectivity required for the organism to properly function and survive. For the sake of this review, we will define developmental neuronal remodeling as the phenomenon in which exuberant connections that were formed during early developmental stages are eliminated at later stages and often further refined by regrowth to adult specific targets. The initial discovery that normal development involves regressive events that do not include cell death occurred only about 40 years ago,1 identified in insects,2 and in mammals3, 4 at around the same time. Since then its relevance has been appreciated in many systems ranging from invertebrates such as flies and worms to mammalian model organisms and even humans.5, 6, 7 There are several mechanisms by which the nervous system can ‘tweak’ its connectivity throughout development, usually in tightly regulated and perfectly timed processes. These regressive events occur on different scales, from single synapses and up to the elimination of entire dendritic trees or long stretches of axons in which the cell body remains intact.6, 8 The picture emerging from studying various vertebrate and invertebrate models is that small scale pruning occurs via retraction while large scale pruning occurs via localized degeneration of axons and dendrites.9
In retrospect, developmental neuronal remodeling was already identified by Ramon y Cajal at the turn of the 20th century. He, among his many discoveries relating to nervous system development, found that spinal motor neurons, as well as Purkinje and granule cells, initially form a large dendritic tree that is pruned in what he called ‘process resorption.’10 Remarkably, he also noticed that subsequent to the pruning process, the dendrites regrow to form the mature connections.
Defects in remodeling are expected to result in excessive and improper neuronal connections. Indeed, dysregulated pruning has been suggested to underlie several neuropsychiatric diseases such as schizophrenia and autism,11, 12, 13, 14, 15 at least in part due to the disruption of the balance between excitatory and inhibitory pathways, but the molecular and physiological mechanisms are not well understood. One fascinating phenomena that is thought to arise from improper pruning is synesthesia, in which the activation of one sense causes the involuntarily stimulation of another sense. Grapheme‐color synesthetes, for example, see certain letters in specific colors while in chromesthesia people associate sounds with colors. One of the most prevalent hypotheses for explaining synesthesia, which affects up to 5% of the population, is that we are all born with cross connectivity between cortical areas that is pruned during postnatal remodeling, but this does not occur properly in synesthetes.16, 17 Finally, because neuronal remodeling is essentially developmentally regulated neurite degeneration followed by developmentally regulated regeneration, understanding the mechanisms that regulate neuronal remodeling could provide a broader insight into the mechanisms of axon degeneration during development, disease and following injury and increase our knowledge on the mechanisms that limit regeneration following injury.
Although there has been much progress in delineating the molecular mechanisms and chronological progression of axon and dendrite pruning in several systems, our knowledge is far from being complete. For example, it is still unclear which genes and pathways are globally required for remodeling and which are cell or perhaps even compartment specific. We also don't understand how the temporal and spatial specificity of remodeling is achieved. Finally, the interactions between neurons and other cells, such as glia, are just beginning to be uncovered. While pruning has been studied in several systems, the subsequent regrowth to mature targets has remained a ‘black box’, most likely because it was assumed to utilize the same molecular mechanisms as initial neurite outgrowth. Until recently, it was not clear whether developmental regrowth following pruning is genetically encoded or whether it occurs naturally after the pruning pathway is arrested. Studies in the fly now highlight developmental regrowth as a unique and central process that occurs during neuronal remodeling.
Drosophila melanogaster has emerged as a leading model system to understand the molecular and cellular mechanisms of axon and dendrite remodeling in both the CNS and the PNS.7 Among the many reasons that make Drosophila a unique model system is the fact that it undergoes stereotypic remodeling during metamorphosis. Additionally, the cutting edge genetic tools that are routinely available in Drosophila allow the visualization and manipulation of small and identifiable populations of neurons in vivo. In this focus article, we will outline the current state of the neuronal remodeling field in the fly, focusing on three neuronal systems, and use the insights gained in these systems to raise open questions.
THREE MAJOR DROSOPHILA SYSTEMS UNDERGO STEREOTYPIC NEURONAL REMODELING DURING METAMORPHOSIS
The Drosophila mushroom body (MB) is a central nervous system (CNS) structure involved in learning and memory of olfactory cues in both larvae and adults.18 It provides an attractive model to study remodeling as it undergoes stereotypical remodeling during metamorphosis that involves pruning of axons and dendrites followed by developmental regrowth to form adult specific connections. The MB is comprised of three types of neurons, γ, α′/β′ and α/β, that are born sequentially from four identical neuroblasts per hemisphere, out of which only the γ neurons undergo remodeling.19 Initially, the γ neurons extend bifurcated axons to form a dorsal and a medial lobe (Figure 1). During the early stages of pupation, the dendrites prune completely and the axons prune up to a stereotypic branch point. Axon fragmentation is usually complete by 18 h after puparium formation (APF). By 24 h APF axon regrowth is evident and at 48 h APF, the γ neurons have re‐extended their axons to form a new, adult specific, medial lobe and a new dendritic tree is apparent.
Figure 1.
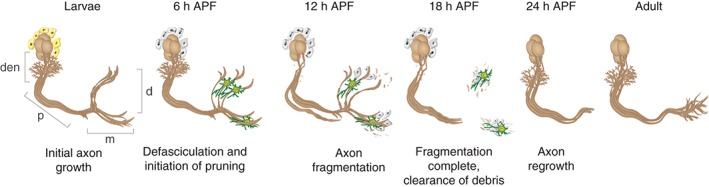
Mushroom body (MB) γ neurons undergo stereotypic remodeling: at larval stage, MB γ neurons project a single axon that branches to form dendrites (den), a tightly fasciculated axon peduncle (p) that bifurcates to form the dorsal (d) and medial (m) lobes. Cortex glia (yellow) instruct MB axon pruning by secreting the TGF‐β ligand Myoglianin. At 6 h after puparium formation (APF), MB γ dendrites are mostly eliminated and axons begin to undergo defasciculation, allowing the astrocytes (green) to infiltrate the dorsal and medial lobes. The role of the remaining unidentified glia or cortex glia during this and later time points is not known (gray). At 12 h APF, axon fragmentation and blebbing are apparent. At 18 h APF, fragmentation is complete and axonal fragments are being engulfed mainly by astrocytes. At 24 h APF, the MB γ neurons begin to regrow toward the adult targets, forming the adult γ lobe. [Reprinted with permission from Ref 7. Copyright 2014]
Dendritic arborization (da) neurons are another popular model for studying neuronal remodeling in the peripheral nervous system (PNS).20 These neurons extend stereotypic and highly branched dendrites underneath the epidermis that forms the larval body wall. By precise tiling between individual neurons of the same type, they form a network that innervates the entire body wall. The da neurons are classified into four classes, I–IV, according to their increasing complexity of dendritic morphology. Both class I (ddaD/ddaE) and class IV (ddaC) neurons completely prune their larval dendrites, but not axons, by localized degeneration during metamorphosis (class IV depicted in Figure 2). In contrast, class II and III da neurons undergo developmentally regulated apoptosis.21, 22 Dendrite pruning of da neurons occurs in a similar time window as that of MB neurons, with dendrites being completely pruned by 20 h APF. These neurons begin to regrow their dendritic arbor at about 36 h APF for class I21 and 24 h APF for class IV.21, 23
Figure 2.
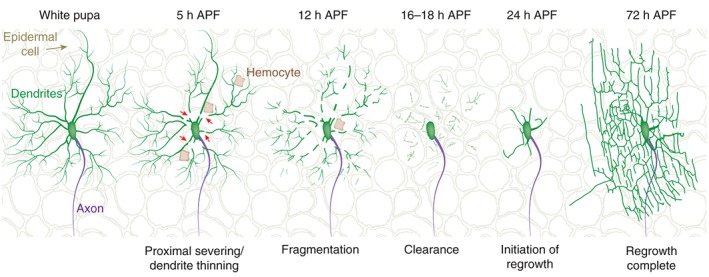
Dendrite remodeling of da class IV during metamorphosis: at the white prepupal stage (0 h APF), da class IV neurons extend a complex dendritic arbor (green) as well as a single axon projecting ventrally to the ventral nerve cord (purple). Dendrites undergo blebbing, thinning, and proximal severing at about 5 h APF, followed by fragmentation (12 h APF). By 16–18 h APF, all dendrites have been fragmented and debris are engulfed and degraded largely by epidermal cells (background open brown silhouettes) but also by phagocytic hemocytes (brown). At 24 h APF, the dendrite begins to regrow by extending primary branches. By 72 h APF, the entire elaborate dendritic tree has regrown. Arrows point to areas of proximal severing of the dorsal dendrite branches or of dendrite thinning.
Crustacean cardioactive peptide (CCAP) expressing neurons (called CCAP or Bursicon neurons because they also express the Bursicon peptide) are mostly located in the ventral nerve cord and also undergo massive neuronal remodeling during metamorphosis. CCAP is a peptide conserved in crustaceans and insects where it was found to affect the function of cardiac and reproductive systems as well as promote ecdysis.24 CCAP neurons in Drosophila are important for pupal development and adult maturation, and their proper function and connectivity was found to affect wing expansion and cuticle tanning.24, 25 There are two major types of CCAP neurons in the ventral nerve chord: interneurons and efferent neurons, both of which undergo remodeling during metamorphosis (Figure 3). Pruning of both axons and dendrites, almost up to the cell body, is first apparent at 3 h APF, with the peak of pruning occurring at 9–12 h APF.25, 26 The first signs of neurite regrowth appear at 18 h APF with adult connections being fully apparent by 60 h APF.26 In addition, developmental changes are also seen in cell size and position. The somata of the CCAP neurons located in the abdominal segments more than double in size and the cells migrate to or are pushed down to the tip of the ventral nerve cord when the abdomen reorganizes itself during metamorphosis.26
Figure 3.
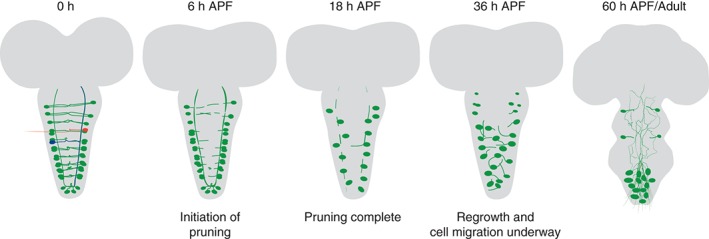
Remodeling of CCAP/Bursicon neurons during metamorphosis: in the larval and prepupal stage, CCAP expressing neurons line the entire ventral nerve cord (VNC) and form an intricate network. There are two major types of neurons, efferent neurons, and interneurons. Efferent neurons (one example highlighted in red) send projections from the cell bodies, that are located on both sides of the VNC, cross to the midline where they then exit the VNC through the dorsal part of the VNC. The neurites of the interneurons (one example highlighted in blue) cross the midline and then bifurcate to send projections to both the anterior and posterior areas of contralateral side of the VNC. At 6 h APF pruning begins with the neurites in the center of the VNC disappearing first followed by the longitudinal tracts. At the onset of regrowth (36 APF), cell somas are also beginning to migrate to the abdominal area of the VNC. At the adult stage, cell somas are up to twice the size of larval somas and they send projections through the adult VNC. By 60 h APF, the CCAP neurons have acquired their final adult connectivity with the unipolar neurons sending their dendrites up along the VNC while their axons enter the VNC at the midline and exit from the ventral side of the VNC to innervate their targets. Labels are major time occurances of processes. For more detailed time analysis, see Figure 4 (based on Refs 25 and 26).
Drosophila motoneurons (MNs) also undergo extensive remodeling which takes place at different stages of development. Unlike the mammalian MNs, where neuronal competition drives pervasive branch removal to give rise to monosynaptically innervated muscles,3, 27 remodeling of the fly NMJ appears to be developmentally regulated and no evidence for competition has yet been identified.28, 29 In the Drosophila MNs, remodeling occurs via distinctive mechanisms at different developmental stages. In the late embryo/early larva, ectopic connections undergo selective elimination in an activity dependent manner, mediated by repulsive factors.30 During metamorphosis further remodeling of the MN occurs: the elaborate dendritic tree of MN 1–4 is refined via the pruning and regrowth of the dendrites.31 In addition, the connections between the MN and the muscle are remodeled when some MNs eliminate their axons presumably by retraction,32 concomitant with dismantling of synapses.33 Later, MNs form new connections via regrowth.34 In essence, the remodeling of different MNs at different stages of development likely involves distinct mechanisms and therefore warrants a dedicated review due to the complexity of the system. Here, we will not focus on MN remodeling but will relate to this process for the sake of comparison and contrasting with other remodeling systems.
The emerging theme from the three neuronal systems that we have described is that they all involve pruning followed by regrowth and that both of these processes globally occur at similar time frames throughout the animal (Figure 4).
Figure 4.
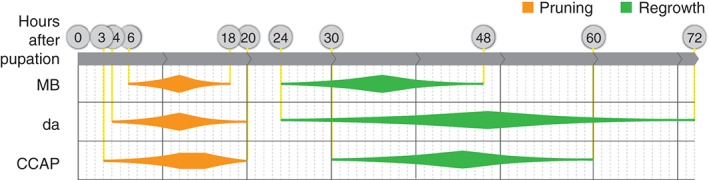
Remodeling of MB γ neurons, da dendrites and CCAP neurons occur on similar time scales. Pruning (orange) of MB γ axons, da dendrites, and CCAP neurites all begin close to pupation onset and ends by 20 h APF. Regrowth (green) begins around 24 h APF and lasts between 24 and 48 h.
However, the three systems that we have decided to focus on in this article are not a comprehensive list. In fact, a recent study looking at the active zone protein Bruchpilot suggested an entire erasure of the larval synaptic connectivity during metamorphosis,35 consistent with previous assumptions.36 For example, embryonic born projection neurons (PNs), which relay olfactory information from the antennal lobe to the MB and the lateral horn, undergo stereotypic remodeling during metamorphosis in a similar time frame as the other neurons described here.37 However, it is also important to mention that not all axons are remodeled during metamorphosis, even within the same neural structure—e.g., MB α′/β′ neurons extend axons toward the end of larval life and retain their axonal connections throughout metamorphosis. Furthermore, da neurons retain their axon while the dendrites are remodeled. Therefore, it appears that we are just looking at the tip of the iceberg and future studies will likely uncover new neurons that undergo remodeling and also highlight neurons that retain their processes. A comparison between such neurons should provide additional insights into what commits a neuron to remodeling.
Neuronal remodeling had also been shown in a variety of vertebrates ranging from mice to humans. For example, large‐scale pruning of murine layer V corticospinal axons is responsible for segregating the connections of motor and visual cortical neurons.6 Defects in pruning have been associated with several neurological abnormalities, such as autism, in which it has been hypothesized that there is over‐pruning.11
NEURONAL REMODELING INVOLVES DISTINCT MORPHOLOGICAL CHANGES
As mentioned, all of the aforementioned systems undergo neurite pruning and regrowth within a similar developmental time frame (Figure 4), and also share several morphological attributes during remodeling.
One of the first morphological signs of pruning in all three systems is neurite blebbing followed by degeneration. In both the MB and in da neurons, the degeneration is preceded by a loss of microtubules (MTs). In the MB, dendrite pruning precedes axon pruning that begins with the disappearance of MTs, likely leading to the subsequent axon blebbing and fragmentation.38 Similarly, proximal severing of the dendritic tree in da neurons also occurs following MT disappearance.39 However, how MT elimination is regulated and whether this elimination is required or sufficient to induce pruning is less understood, especially in MB neurons.
The careful morphological characterization of da neuronal remodeling has been easier to study than MB remodeling, due to the ease in which live imaging can be conducted. da neurons are easy to visualize because they innervate the body wall and can be imaged through the epidermis using time‐lapse imaging even during the early stages of metamorphosis. Furthermore, da neurons do not grow in clusters and rather tile the entire body wall so that each neuron occupies its own domain and it is therefore easy to achieve single cell resolution. Using such time‐lapse imaging, Lee and colleagues (2009) have demonstrated that the pruning of da dendrites begins with several cuts close to the cell body by which the proximal branches are severed all at once.39 This is followed by degeneration of the entire dendritic arbor, although it remains unclear whether the degeneration propagates from the proximal arbor out or rather the rest of the arbor disintegrates at once. The loss of MTs precedes axon severing and begins close to the future cut site in the proximal arbor and afterward propagates outward toward more distal branches.39 This destabilization of MTs in proximal dendrites is mediated, at least in part, by Katanin p60‐like 1, which belongs to the Katanin family of MT severing proteins.39 So, in theory, the spatial control of da neuron dendrite pruning might be achieved by this initial severing event that divides the cell to arbors that will and will not undergo pruning. In the case of MB axon pruning, the picture of how the spatial specificity is achieved is much less understood. While the entire dendritic tree of MB γ neurons prune, the axons prune only up to their original branching point, while the pedunclar axon remains intact (Figure 1). By following the distribution of tagged tubulin molecules during developmental axon pruning, Watts and colleagues (2003) have shown that tubulin disappears mainly from the axonal areas that will subsequently undergo pruning but remains localized to the axon peduncle.38 These data highlight MT disruption as an early morphological distinguisher between domains that will and will not be eliminated and suggest that MT disassembly might function as a mechanism to differentiate between the two axonal compartments. Since the disappearance of MTs precedes pruning in both the MB and da neurons, this implies a conserved mechanism among different cell types. The identity of MT severing or destabilizing proteins has not yet been shown in the MB or in the CCAP neurons and should be further investigated. Furthermore, MT disassembly has not been shown to be required for axon pruning of MB neurons and has not been shown to be sufficient to induce pruning in any of the systems discussed here. Interestingly, MT disassembly is also evident in pruning of cultured mammalian neurons in response to trophic deprivation40 and the stabilization of MT protects axon fragmentation.41 In an elegant study, Maor‐Nof and colleagues found that the MT destabilizing protein kinesin 2A (KIF2A) is at least partially responsible for MT destabilization followed by pruning of cultured neurons due to trophic deprivation and also in vivo for normal sensory axon patterning.41
Unlike in the da neurons, following the progression of pruning in the MB has not been explored by live imaging because the pupa is encased in a dark cuticle and more importantly the brain itself is surrounded by opaque fat bodies. Therefore, the cellular mechanisms of MB neuronal remodeling have not been explored in great depth. In order to address this limitation, we have recently developed an ex vivo culturing system in which pupal brains can be dissected and cultured. Remarkably, in the right culture conditions pupal brains continue to develop and undergo axon pruning in a similar time scale as in vivo. Live imaging of MB γ neuronal pruning has revealed that axon blebbing precedes the formation of 1–2 cuts in each of the dorsal and medial parts of the axon.42 Interestingly, the location of the axonal nicks does not appear to correlate with the future tip of axon fragmentation. Similarly, these 1–2 nicks occur at different times in different axons arguing against an extracellular signal that initiates the program with a precise location and time. This ex vivo culture system should be powerful in further understanding the cellular mechanism of MB remodeling by examining, the dynamics of tagged proteins and organelles such as MTs and mitochondria.
In both the MB and the da neurons, the final adult morphology following regrowth is dramatically different than the initial larval morphology. The regrown MB γ axons form only an adult medial lobe that is in a distinct location from the larval medial lobe (Figure 1). The reformed MB dendritic calyx is organized in a different manner than the larval one. While in the larva the dendrites of cells originating from different neuroblasts are intermingled, following remodeling these dendrites are segregated with each group of γ neurons related by lineage occupying only one area of the calyx.43 In addition, the adult dendrites are more complex, usually extending three or more primary dendrites and a spread out dendritic tree, while the larval neurons usually only extend one primary dendrite which forms a small dendritic tree.43 These massive changes in morphology and connectivity between the larva and the adult make sense when we think about the different olfactory roles in these stages. The regrowth of da dendrites depends upon the class of neurons. While the total branch length in class IV neurons is the same in larva and in the adult, the shape of the dendritic tree changes dramatically from a radial projection to a lattice‐like structure (Figure 2).44 However, the dendritic arbors in class I are actually smaller in the adult than in the larva but are more complex, having more than tripled the number of terminals.44
CCAP neurons are the least studied of this group, although they present a much more intricate process of remodeling that includes not just pruning and regrowth but also the migration and enlargement of the soma.26 Live imaging of this process has been hindered by the same issues as live imaging of the MB. Modifying the brain culturing protocol that we have developed42 to include the CCAP neurons in the ventral nerve cord will allow better time resolution of the remodeling process and allow for the investigation of basic questions as to the morphological changes during remodeling.
ECDYSONE SIGNALING IS A MAJOR GATEKEEPER OF PRUNING
The first key regulator of pruning was discovered in a large mosaic forward genetic screen. Focusing on the X chromosome, Lee and colleagues (2000) screened for genes that affect axon pruning of MB neurons. The screen involved generating MARCM (Mosaic Analysis with a Repressible Cell Marker) clones mutant for randomly generated EMS mutations and searching for clones in which pruning was defective. Two lethal alleles mapped to the same gene—Ultraspiracle (usp), the ortholog of retinoic acid receptor.45 USP is part of a heterodimer of nuclear receptors that bind the steroid molting hormone 20‐hydroxyecdysone (ecdysone). Ecdysone is a master regulator of various biological changes necessary for proper development such as morphogenesis, apoptosis and reproduction.46 An ecdysone pulse induces the transition between the three larval instar stages and another is required for pupation. The nuclear receptor complex that binds ecdysone consists of USP and one of the Ecdysone receptor isoforms—EcR‐A, −B1, and ‐B2. Indeed, inhibiting EcR function within MB neurons by expressing its dominant negative version (EcRDN) inhibits neurite pruning, suggesting that the USP/EcR complex is required for axon and dendrite pruning of MB neurons in a cell autonomous manner (Figure 5). Furthermore, by performing detailed immunostaining combined with isoform specific rescue experiments, the authors found that EcR‐B1 expression within the MB is restricted to neurons at the onset of pruning, suggesting that it provides a key regulatory aspect.45
Figure 5.
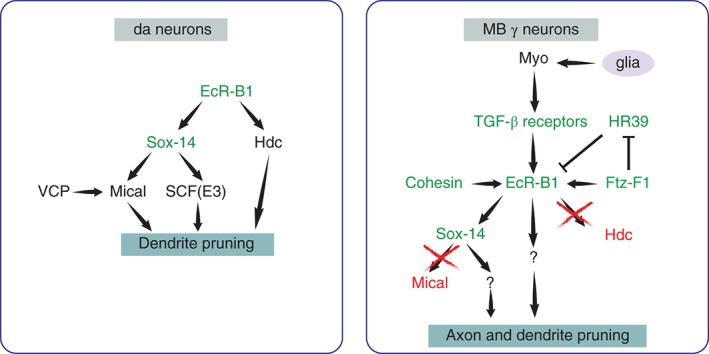
Mechanisms of ecdysone receptor B1 (EcR‐B1) signaling to trigger pruning in MB γ and da neurons. In da neurons, EcR‐B1 triggers dendrite pruning via Sox‐14 and Hdc. Sox‐14 promotes pruning via Mical and Cullin1‐based SCF E3 ligase (see text for more information). In MB neurons cortex, and perhaps also astrocyte‐like, glia secrete Myoglianin (Myo) a TGF‐β ligand that binds to neuronal TGF‐β receptors that induce the expression of EcR‐B1 in MB γ neurons. Cell autonomous functions of Cohesin and Ftz‐F1 include the activation of EcR‐B1. EcR‐B1 activates Sox‐14, which functions in a non‐Mical dependent pathway, and probably other pathways, to induce pruning. Green indicates common factors in both systems and red are targets that were tested and found not to be involved.
Following the discovery that ecdysone signaling was required for MB axon pruning, it was discovered that remodeling of da dendrites also requires ecdysone and the activation of EcR‐B122, 47 in a cell autonomous manner. In the case of remodeling of CCAP neurons, our knowledge is very sparse. In fact, the signal to induce pruning has yet to be identified. Since CCAP neurons are themselves crucial for ecdysis,24 it is technically difficult to ascertain whether ecdysone exerts a feedback mechanism to induce pruning. A possible way to separate these two processes is to knock down the expression of the ecdysone receptor in CCAP neurons only after pupation, assuming that this will not interfere with normal pupal development. Alternatively, testing putative EcR‐B1 targets (see more below) can also determine whether the EcR‐induced pruning program is relevant for CCAP remodeling too. The involvement of ecdysone in developmental pruning has also been identified in an additional Drosophila system that undergoes dendrite pruning during metamorphosis, the thoracic ventral (Tv) neurosecretory cells36, 48 as well as in MNs.32 In the TVs, EcR has also been shown to be cell autonomously required for pruning,49 further supporting the key role of ecdysone as a major regulator of pruning in Drosophila, regardless of the system studied.
The paramount role of EcR in regulating pruning is also evident by the fact that its transcription is tightly regulated by at least three independent pathways in the context of MB axon pruning (Figure 5). EcR‐B1 expression was found to be regulated by the Cohesin complex,50 a complex usually involved in maintaining sister chromatid cohesion during meiosis. The mechanism by which the Cohesin complex regulates expression of EcR‐B1 and how the Cohesin complex itself is regulated during development is not yet understood. EcR expression was also shown to be regulated by the orphan nuclear receptor Ftz‐F1,51 initially thought to function downstream of EcR, suggesting that a complicated regulatory loop might still need to be uncovered. Ftz‐F1 is thought to induce the expression of EcR‐B1 both directly and indirectly by inhibiting the expression of another nuclear receptor, HR39, that inhibits EcR‐B1 expression.51 Finally, the expression of EcR‐B1 is also tightly regulated by extrinsic signals that culminate in the activation of the TGF‐β pathway that was found to be cell autonomously required for EcR‐B1 expression and pruning in the MB.52 In an elegant paper Awasaki and colleagues (2011) have demonstrated that the TGF‐β ligand, Myoglianin (Myo), is secreted by cortex glial cells53 and induces TGF‐β signaling by binding to the TGF‐β receptor type I Baboon (Babo) and either one of the type II receptors Punt (Put) or Wishful thinking (Wit).52, 53 We have recently shown that, in addition to classical TGF‐β receptors, MB γ neurons also express an immunoglobulin (Ig) superfamily transmembrane receptor, Plum, that facilitates TGF‐β signaling, EcR‐B1 expression, and the induction of pruning.54 Interestingly, the participation of TGF‐β in pruning appears to be conserved. Drosophila MNs undergo various types of remodeling, including the elimination of ectopic synapses during early larva and remodeling during metamorphosis. Although initially thought to be unrelated to pruning of longer stretches of axons, we found that the removal of ectopic synapses is also regulated by the TGF‐β pathway, mediated by Plum and acting upstream of the EcR.54 Similarly, NMJ dismantling during metamorphosis appears to depend upon the expression of EcR‐B1 in both the presynaptic MN and the postsynaptic muscle, with TGF‐β inducing neuronal EcR‐B1 expression in a mechanism likely involving the Ftz‐F1 nuclear receptor.32 Likewise, TGF‐β released from mammalian astrocytes has been identified as a key regulator in synaptic pruning in retinal ganglion cells although in this case TGF‐β signaling appears to regulate the expression of C1q.55
Although the function of EcR‐B1 is essential to induce pruning in all system tested so far,22, 37, 45, 47 its expression is not sufficient to induce pruning, at least in the MB. Overexpressing any of the Ecdysone receptors in the MB has no effect on MB morphology, such as early or more extensive axon fragmentation45 but whether this is sufficient to induce intracellular signaling is not known.
Many targets of EcR‐B1 transcriptional activity have been proposed in the fly, but in the context of remodeling we only know a few (Figure 5). In da neurons EcR‐B1 was found to activate the transcription factor Sox‐14 that, in turn, induces the expression of Mical, a cytoskeletal regulator which is important for pruning of da dendrites.56 Recently, it was shown that splicing of mical by Valosin‐containing protein (VCP) is required for its expression and for dendrite pruning of da neurons.57 Sox‐14, but not Mical, also appears to participate in MB pruning.56 What are the downstream targets of Sox14 that regulate pruning in the MB and whether it represents the major EcR target driving MB pruning is currently unknown. Headcase (Hdc) is an additional downstream target of EcR‐B1 that regulates da dendrite severing but not MB pruning.58 Interestingly, Hdc functions in a Sox‐14 independent pathway,58 thus illustrating that there are at least two separate pathways activated by EcR‐B1 in da neurons that induce pruning. The full extent of the EcR targets that are required for pruning of MB or da neurons is not yet fully appreciated. Furthermore, it is not known whether EcR‐B1 might also play a role in regrowth following pruning. While the levels of EcR‐B1 in the CNS are very low at the onset of regrowth,59 perhaps such a rapid decrease in the MB is instrumental in signaling to the cells to begin regrowth. One way in which this could happen is by freeing up USP and allowing it to dimerize and function with a different NR, at least two of which, hormone receptor 38 (HR38) and Seven‐up (Svp), have been identified.60, 61
UPREGULATION OF DEGRADATION PATHWAYS IS NECESSARY FOR EFFICIENT PRUNING
One common aspect to all systems undergoing pruning is that they involve local degeneration of neurites. Since this includes the active destruction of cellular machinery in a tightly regulated temporal and spatial manner, a significant question is how this occurs mechanistically.
A major mechanism by which the cell regulates protein half‐life and protein turnover is via the ubiquitin‐proteasome system (UPS).62 Proteins that are to be degraded become ‘tagged’ with ubiquitin through the actions of three enzymes, the ubiquitin‐activating E1, the ubiquitin conjugating E2 and the ubiquitin ligating E3. By over‐expressing the yeast ubiquitin carboxyl‐terminal hydrolase 2 (UBP2) in MB neurons, essentially functioning to reverse the function of the UPS in these neurons, Watts and colleagues (2003) showed that the UPS system was required for axon pruning (Figure 6). By performing mosaic analyses for mutants in the sole Drosophila E1, uba1, or in different subunits of the proteasome, they also showed that the UPS was cell autonomously required for pruning.38 Likewise, inhibiting UPS function by overexpressing Ubp2 or by mutating uba1 inhibited dendrite pruning in da neurons.47 By extracting RNA from laser‐captured cells at different times during development, Hoopfer and colleagues (2008) found that the levels of various UPS components are upregulated within the MB neurons during metamorphosis,63 suggesting an activation of broad program for destruction of cellular components.
Figure 6.
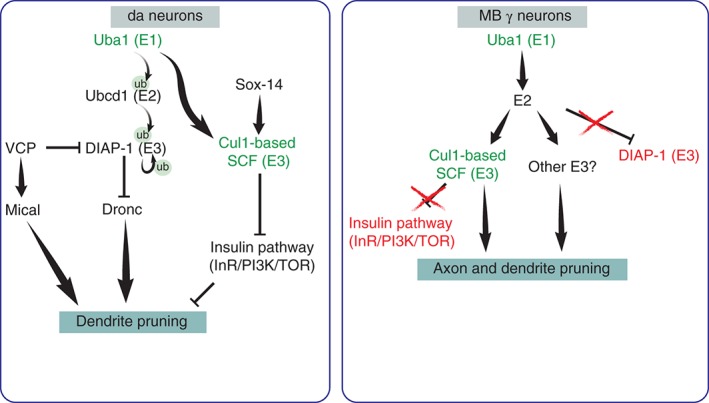
Degradation pathways inducing da dendrite and MB γ neurons pruning: in da neurons, Uba1, the sole Drosophila E1, activates the E2 UbcD1, which then activates the E3 ligase DIAP‐1 and causes it to undergo self‐promoting degradation. The degradation of DIAP‐1 disinhibits the expression of the caspase Dronc and allows the subsequent elevation of caspase activity, which in turn promotes dendrite pruning by unknown mechanisms. At the same time a Cullin1 (Cul1)‐based E3 complex is activated by an unknown E2 and promotes pruning by inhibiting the insulin pathway. In addition, Valosin‐containing protein, a ubiquitin sensitive chaperone, promotes the degradation of DIAP‐1 and in parallel allows for the correct splicing of mical mRNA. In the MB, Uba1 is required to for axon pruning but the relevant E2 complexes are largely unknown. The Cul1‐based SCF E3 ligase complex promotes axon pruning in the MB by an unknown mechanism. Green indicates common factors in both systems and red are targets that were tested and found not to be involved.
How does the activation of the UPS system promote pruning? In da neurons, there are at least two UPS‐dependent processes that were shown to promote dendrite pruning (Figure 6): By conducting a screen of E2 and E3 molecules, Kuo and colleagues (2006) found that the UbcD1 E2 is required for the ubiquitin mediated degradation of Drosophila Inhibitor of Apoptosis‐1 (DIAP1), a caspase antagonizing E3 ligase. Degradation of DIAP results in the accumulation and local activation of caspases, which subsequently promote dendrite pruning.64, 65 It was also suggested that VCP, a ubiquitin‐dependent ATPase, is also required for da pruning. Two mechanisms have been suggested for VCP actions: the degradation of DIAP166 and the correct splicing of the pruning inducer Mical,57 but how VCP is activated and how this relates to the core UPS machinery is not yet clear. Additionally, an RNAi screen has also highlighted the role of another E3 ubiquitin ligase, the SCF complex that comprises Cullin1, Roc1a, SkpA, and Slimb.67 This complex was shown to negatively regulate the insulin/Target of Rapamycin (InR/PI3K/TOR) pathway, whose activity was found to inhibit dendrite pruning.67 Whether these are the only UPS dependent processes that promote pruning and what is the relationship between them is not currently understood.
In the MB, however, the UPS targets for degradation have not yet been identified. Current data does not support a role for caspases in the induction of pruning since overexpression of either of caspase inhibitors p35 or DIAP‐1 does not inhibit pruning.38, 68 Furthermore, no active caspases have been detected in the MB γ neurons during pruning.68 However, it is not possible to rule out the possibility that caspases do function during MB axon pruning but their effect is masked by redundant pathways. The insulin pathway does not appear to participate in γ neuron pruning since overexpressing TOR does not affect pruning.69 The UPS system has also been shown to be required for Wallerian degeneration in Drosophila and in mammals.40, 70 In the Drosophila system, it was suggested that the E3 ligase Highwire/phr1 is required to downregulate the levels of NMNAT, an inhibitor of axon degradation following axotomy, in both mammals and Drosophila 71, 72 and additionally downregulate the levels of Wallenda (Wnd, the fly ortholog of DLK). However, current data does not support a role for Highwire, NMNAT or Wnd in axon pruning of MB neurons.38, 63, 70 Therefore, the identity of the E2/E3 and targets for degradation in the context of MB axon pruning remains unknown.
Endocytosis represents an additional mechanism by which membrane‐bound proteins are removed and then recycled or targeted for degradation. A forward genetic screen has led to the discovery that the fly UV radiation resistance‐associated gene (UVRAG) is required for MB axon pruning.73 UVRAG was previously shown to participate in a variety of complexes, among them the phosphatidylinositol 3‐kinase class III (PI3K‐cIII) complex, which promotes both autophagy and endocytosis. The PI3K‐cIII complex was shown to phosphorylate phosphatidylinositol lipids on endocytic membranes to give rise to PI(3)P which subsequently recruits a wide variety of proteins containing an FYVE domain. Genetic analyses showed that MB pruning is an autophagy‐independent process, suggesting a role for endocytosis.73 Indeed, the FYVE containing ESCRT‐0 (Endosomal Sorting Complex Required for Transport) complex was also required for pruning, suggesting that endosome maturation resulting in the attenuation of a membranal signal is a crucial step in pruning promotion. By performing a suppressor screen, Issman‐Zecharya and colleagues (2014) have shown that UVRAG functions, at least in part, by mediating the endolysosomal degradation of Patched (Ptc), that appears to inhibit pruning in a Smoothened (Smo) and Hedghog (Hh)‐independent manner.73 How Ptc inhibits pruning and whether it is the only inhibitory membranal protein that is targeted via the endolysosomal pathway remains to be further explored.
Interestingly, the endocytic machinery has also been implicated in dendrite pruning of da neurons. An RNAi screen in these neurons has revealed that Rab5‐mediated endocytosis is required for dendrite pruning by downregulating the L1‐cell adhesion molecule (CAM) ortholog Neuroglian (Nrg).74 Indeed, overexpression of Nrg was sufficient to inhibit dendrite pruning, suggesting that its removal from the membrane is essential for dendrite pruning. While these two studies have shown that ESCRT complexes are required for pruning because of their role in the maturation of endosomes to multivasicular bodies,73, 74 it has also been suggested that the ESCRT‐3 complex might play an independent role as a membrane remodeling complex within the dendrites themselves.75 Finally, localized endocytosis has also been suggested to play an additional role in directly regulating dendrite thinning.76 In essence, dendrite thinning allows the compartmentalization of the dendrites, allowing for localized calcium signaling, which has been shown to play a crucial role in dendrite pruning by activation of calpain.77 Interestingly, elevated calcium and calpains have also been implicated in axon degeneration mediated by trophic deprivation.78 How dendrite thinning helps to activate calcium signaling is not understood. Likewise, the relationship between the endocytosis mediated dendrite thinning and dendrite severing, which is thought to be initiated by IK239 is not yet resolved. Finally, whether calcium signaling plays a role in MB or CCAP neurite pruning has yet to be determined.
Taken together, studies have shown that degradation of cytoplasmic targets by the UPS system as well as the attenuation of membranal derived signals by endocytosis play important roles in promoting pruning. The cross talk between the UPS and endocytic machinery is an important aspect of the regulation of pruning, which merits further exploration.
DESTABILIZING ADHESION AS A MECHANISM TO INDUCE PRUNING
The regulation of adhesion molecules has recently been implicated in remodeling of both da and MB neurons. As discussed above, endocytosis mediated downregulation of the adhesion molecule Nrg has been shown to be required for dendrite pruning of da neurons.74 Remarkably, while overexpression of Nrg, or disruption of endocytosis, prevents proper pruning, mutating Nrg results in precocious pruning.74 Although the exact function of Nrg has not yet been shown, it stands to reason that it mediates an interaction between the da neurons and the extracellular matrix or neighboring cells and that this interaction, or adhesion, must be disrupted for pruning to occur. Finding the binding partner of Nrg as well as its source will provide important insights into how cell‐cell interactions regulate da pruning. Likewise, a structure function analysis of the required domains in Nrg should provide an insight into the intracellular signaling pathways that inhibit pruning.
By performing a large‐scale mosaic forward genetic screen, it was recently discovered that Drosophila c‐Jun N‐terminal Kinase (dJNK, also known as Basket, Bsk) mediated destabilization of Fasciclin II (FasII), the ortholog of neural CAM (NCAM) is required for MB pruning.79 In the MB, FasII mediates homophilic interactions between adjacent γ axons, causing them to fasciculate and form an axonal bundle. Interestingly, overexpression of FasII was sufficient to inhibit pruning.79 Furthermore, expression of FasII lacking its intracellular domain or even overexpression of other cell adhesion proteins was sufficient to inhibit axon pruning, suggesting that pruning depends on the destabilization of cell adhesion and not necessarily FasII signaling.79 Remarkably, mutations in dJNK, or overexpression of FasII affected only axon but not dendrite pruning. This is the first example of a gene differentially affecting axon and dendrite pruning within the same neuron, suggesting the existence of distinct mechanisms to regulate the pruning of different neurites. Additionally, the fact that reduction of adhesion is required during the pruning of both da dendrites and MB axons raises a speculative hypothesis that different parts of the neurons might depend on different adhesion molecules. Thus, differential localization of distinct CAMs to different compartments, localized destabilization of cell adhesion, or both, could provide a spatial regulation of neurite pruning. It will thus be interesting to uncover the ‘adhesion code’ for different parts of the neuron. Finally, how dJNK regulates the membrane stability of FasII is only partially understood. Whether dJNK regulates the stability of other adhesion molecules remains to be determined.
NEURONS INTERACT WITH SURROUNDING CELLS THROUGHOUT THE PRUNING PROCESS
Neuronal remodeling occurs in a complex tissue that comprises many cell types. Therefore, neuron‐neuron as well as glia‐neuron and epithelial cell‐neuron interactions could be instrumental at different stages of remodeling.
Glial cells play at least two important roles in MB pruning. First, they are the major source of TGF‐β that activates the pruning program within MB neurons.53 Second, they act to clear axonal debris following pruning. Glial cells recognize and engulf cellular debris68, 80 in a mechanism that depends on the engulfment receptor Draper (CED‐1 homolog).70, 81 Debris is then internalized and degraded by the lysosomal pathway.68, 80 Although many types of glial cells surround the MB, astrocytes are specifically recruited to the degenerating lobes and play a major role in debris clearance.35, 82 Interestingly, maturation of astrocytes so that they infiltrate the MB γ lobe is also dependent upon ecdysone signaling.82 Whether neuron–glia interactions play additional roles in axon pruning such as providing spatial information as to where pruning should stop, remain to be determined.
In the context of da neurons, epidermal cells, and not glia, have been identified to act as the primary phagocytes to clear neuronal debris.83 However, glia in the da system may still play an important role. A study in Drosophila embryos has identified an interaction between da neurons and glia both expressing Nrg that controls da sprouting and branching.84 Since Nrg must be downregulated in order for da pruning to occur,74 perhaps this homophilic physical interaction between the dendrites and glia has to be disrupted to allow for pruning. Furthermore, glial cells appear to enwrap the cell body and axon of da neurons. It was previously suggested that these glial cells might provide a signal that is required for dendrite severing85 but the possibility that glia actually protect the cell body and axon has not been fully explored. Interestingly, both microglia and astrocytes, have been shown to actively participate in mammalian synaptic pruning through phagocytosis suggesting that the role of glia as phagocytic cells is conserved.86, 87
REGROWTH FOLLOWING PRUNING
In all three systems described here, neuronal remodeling involves pruning of dendrites or axons followed by developmental regrowth (Figure 7). How is this regrowth following pruning regulated? Theoretically, the cessation of a pruning program might be sufficient to allow regrowth in some sort of tug‐of‐war between opposing forces of growth and degeneration. However, the first clue that developmental regrowth is regulated by a distinct genetic program came from our finding that the orphan nuclear receptor UNF is required for developmental axon regrowth of MB neurons.69 While UNF was previously implicated in axon growth of MB neurons,88, 89 detailed MARCM analysis, which distinguishes between cell autonomous and non‐cell autonomous roles of UNF, has revealed that UNF is cell‐autonomously specifically required for the regrowth of MB neurons following pruning but not for the initial growth of any MB neuron type.69 Since nuclear receptors function as ligand activated transcription factors, a transcriptional program likely underlies axon regrowth following pruning, and since UNF is only required for regrowth, that this program is distinct from that underlying initial axon growth. By exploring known targets of the mammalian UNF ortholog, photoreceptor specific nuclear receptor (PNR, also known as Nr2e3), we found that UNF positively regulates the Target of Rapamycin (TOR) pathway, although the precise mechanism remains to be uncovered. Mutating the TOR inhibitor Tuberous Sclerosis 1 (Tsc1) suppressed the UNF mediated regrowth defect as did expressing a constitutively active version of the downstream TOR target S6 kinase (S6KCA).69 Surprisingly, TOR is also specifically required for developmental axon regrowth but not for the initial axon outgrowth of any MB neuron type. This was especially unexpected since TOR had been implicated in a wide variety of growth paradigms. Because mammalian TOR (mTOR) was shown to be required in axon regeneration following injury in mice,90, 91 these findings suggest that regrowth following pruning is not only distinct from initial axon outgrowth but might also share molecular mechanisms with regeneration following injury. By combining mosaic genetics and primary cell culture to explore neurite sprouting of isolated MB neurons we have further demonstrated that initial axon outgrowth, developmental regrowth and sprouting after injury share some mechanisms but each process also utilizes distinct mechanisms.92
Figure 7.
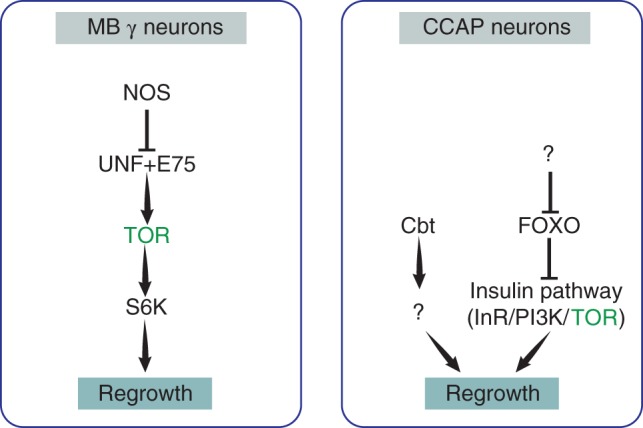
Regrowth in MB γ and CCAP neurons requires TOR signaling: in the MB, nitric oxide synthase (NOS) inhibits the formation of a stable E75/UNF complex. Under low nitric oxide conditions, the nuclear receptors E75 and UNF function together to induces regrowth via the TOR and S6K pathway. In CCAP neurons, unknown signals inhibit the transcription factor FOXO and activate the transcription factors Cabut (Cbt). The downregulation of FOXO allows derepression of the insulin/TOR pathway, which, in turn, induces regrowth. Cbt functions via an unknown mechanism. Green indicates common factors in both systems.
By further delving into the mechanism by which UNF regulates MB γ axon regrowth we found an additional nuclear receptor, Ecdysone inducible protein at 75B (Eip75, hereafter referred to as E75), that also cell autonomously regulates regrowth following pruning without affecting initial axon regrowth. Furthermore, E75, like UNF, exerts its’ function via the TOR pathway.93 This, together with genetic epistasis experiments and co‐immunoprecipitation studies, have led us to propose that UNF and E75 function as nuclear receptor dimers in order to promote axon regrowth.93 The identity of the full spectrum of UNF downstream targets as well as the pathways functioning downstream of the TOR pathway that induce regrowth have yet to be discovered. However, recent work has identified IGF‐II mRNA‐binding protein (Imp) as a regulator of profilin mRNA transport and localization during regrowth.94 Profilin is know to play a major role in actin polymerization,95 suggesting that altering actin dynamics may promote regrowth.
While very little is known about the pruning phase of CCAP neurons, more information is available regarding neurite regrowth. Zhao et al (2008) conducted an extensive gain‐of‐function screen, using defective wing expansion as an output for improper CCAP wiring. Among the genes identified in this gain of function screen were the transcription factor Cabut (Cbt) and the kinesin binding protein Klarsicht (Klar).26 In another, loss of function screen, Chen and colleagues found that downregulation of the RNA binding protein Alan shepard (Shep) resulted in specific defects in metamorphic growth, without affecting larval growth or neurite pruning.96 How these three proteins affect metamorphic growth or whether they are important for axon regrowth following pruning of MB neurons is currently not known.
Defects in CCAP neurite regrowth and soma size increase during neuronal remodeling have also been observed following perturbations in the insulin signaling pathway.97 An increase in insulin signaling promotes neuronal growth as well as axon branching following pruning. This effect is mediated by the TOR pathway and, at least partially, by the FOXO transcription factor, a negative effector of the insulin pathway.97 This result is especially interesting since it was previously shown that axon regrowth in the MB is also mediated by the TOR pathway.69 Furthermore, one of the most well‐defined phenotypes in adult flies due to disruption of CCAP neurons is a defect in wing expansion. A similar phenotype has been observed for homozygous mutants of UNF98 but the neuronal defect for this was not explored. It is therefore plausible to that UNF may also regulate the regrowth of CCAP neurons, thus explaining the lack of wing expansion. Together, these results suggest that remodeling of CCAP neurons and of MB neurons may share molecular mechanisms and collectively highlights the TOR pathway as required for developmental regrowth and not initial outgrowth.
The only identified regulator of da dendrite regrowth and guidance following pruning to the best of our knowledge is ecdysone‐induced cysteine‐proteinase 1 (Cp1).23 Cp1 is responsible for the cleavage and activation of the transcription factor Cut and promotes the formation of higher order dendrites during the regrowth phase. da neurons mutant for Cp1 undergo pruning normally but during regrowth only extend a primary branch that does not elaborate into high‐order branches or adhere to the pupal body wall.23 It is unclear whether failure to adhere to the epidermal cells results in lack of dendrite growth or vice versa and therefore this needs to be further examined.
Since several transcription factors have been implicated in the regrowth of MB, da and CCAP neurons, it would be interesting to compare a list of putative targets of UNF, Cut, and Cbt, and find commonalities that may pinpoint proteins or pathways important for regrowth and for increasing the intrinsic growth ability of neurons.
SWITCHING BETWEEN PRUNING AND REGROWTH MUST BE TIGHTLY REGULATED
In all of the systems we describe here, the switch between pruning and regrowth occurs quite rapidly (Figure 4). Is this switch achieved by a changing balance between two competing and contradicting processes? Or is it an active switch between two distinct growth programs? If the net outcome would result from a balance between growth and fragmentation then one would expect to see increased growth in all mutants in which pruning is defective and likewise excessive pruning in all mutants in which regrowth is defective. This has not been described in any of the studies focusing on remodeling of MB neurons. Strengthening this notion is the fact that both steps in remodeling—pruning and regrowth—are regulated by nuclear receptors that in essence function as ligand dependent transcription factors. Thus, it will be interesting to uncover the entire repertoire of genetic targets for the nuclear receptors EcR‐B1 and UNF in the MB, and for other identified transcription factors in the other systems. The recent advances in RNA‐seq suggest that advances in this direction are just a matter of time.
Nitric oxide (NO) levels were recently identified as a possible switching mechanism between pruning and regrowth of MB neurons. As previously mentioned, the NRs UNF and E75 both promoted regrowth via the TOR pathway.69, 93 E75 is the closest fly ortholog to the mammalian Rev‐erb‐α and has been shown to bind a heme moiety that may function as a sensor for monovalent gases, such as NO.99, 100 Indeed, previous work has shown that E75 functions differentially in distinct NO environments by altering its’ affinity to binding partners.101, 102 This, together with the fact that UNF has also been shown to bind heme,103 suggests a role of NO in axon regrowth of MB γ axons. Indeed, a rapid reduction in the levels of NO, generated by neuronal NO synthase (NOS), is required to promote axon regrowth. Lowering NO levels facilitates the formation of a stable UNF/E75 complex which, presumably, results in the activation of a set of genes that promote regrowth.93 However, high NO levels, generated by neuronal NOS together with calmodulin, are required for normal progression of axon pruning. The decrease in NO levels from pruning to regrowth is developmentally regulated and occurs rapidly in a highly timed and regulated way.93 However, the precise regulation of the activity of NOS and how high levels of NO promote pruning are open questions that remain to be further investigated.
Since all three neuronal systems discussed here undergo remodeling during similar time frames, it is likely that similar switching mechanisms might also be involved. Although it would be extremely interesting to test the role of NO in switching between pruning and regrowth in these other systems, other mechanism might also play a role.
CONCLUDING NOTES
It is quite apparent that the field of developmental axon pruning and regrowth is still in its infancy. Drosophila presents an excellent model to study the various stages of neuronal remodeling since the nervous system undergoes sterotypic remodeling during metamorphosis. Most of what we know about remodeling in Drosophila comes from two neuronal systems—MB axon and da dendrite pruning. Studies in the last decade have revealed common players such as ecdysone signaling, protein degradation via the ubiquitin proteasome system, and destabilization of cell adhesion. However, several important differences have also been highlighted. Most notably, while caspases have been implicated in da dendrite remodeling, there is still no evidence for their role in MB axon pruning. While it is plausible that caspases do not play a role in MB remodeling, it is also possible that parallel and redundant pathways exist thereby masking the importance of the apoptotic machinery. Similarly, while ecdysone signaling has been shown to be required in both MB and da neurons, the full spectrum of the downstream targets of EcR and their precise role is not yet fully appreciated. Likewise, our knowledge about developmental regrowth emerged from studies in MB and CCAP neurons, in both of which TOR signaling appears to play a central role, although how this pathway is regulated might differ. Therefore, we believe that it is important to study more neuronal systems in Drosophila to allow a more comprehensive comparison that will allow us to highlight genes and pathways that are globally required for neuronal remodeling and those that are specific to the CNS or PNS, to axon versus dendrite or to specific cell types. Obviously, extensive knowledge in Drosophila will allow us to make comparisons with what is known about neuronal remodeling in vertebrates. Not in all cases these comparisons are straightforward—e.g., ecdysone signaling is specific to invertebrates but the role of nuclear hormone receptors might actually be conserved.104
Our knowledge of both stages of neuronal remodeling, pruning and regrowth, is still fragmentary. While we know quite a bit about the regulation of these processes, the downstream machinery of how axons dismantle themselves or regrow is not well understood. The further exploration of MT and actin stability and dynamics should provide us with important clues to these questions. Likewise, the interaction between neurons and their surrounding cells is likely to yield more interesting insights into the spatial and temporal regulation of remodeling. Finally, the recent discovery that cell adhesion plays a role in axon pruning offers an opportunity to link extracellular signals to changes in cytoskeleton dynamics.
Drosophila genetics has been the driving force of recent progress in the field. We envision that genomic strategies will likely soon be added to the discovery toolbox, due to recent advances in cell isolation and RNA‐seq from small quantities of RNA. Likewise, live imaging of intact pupae or ex‐vivo cultured brains will allow the field to focus on dynamic aspects of neuronal remodeling.
Research into the mechanisms underlying neuronal remodeling will not only allow us to further understand the development and refinement of the nervous system but may also allow us to delineate mechanisms underlying neuron degeneration during disease and following injury.
ACKNOWLEDGMENTS
We thank R. Hewes for insightful comments and suggestions regarding Figure 3 and T. Mosca for assistance with NMJ remodeling. Work in our laboratory is mainly supported by grants from the EU (ERC consolidator grant), the Israel Science Foundation (ISF), and the Minerva Foundation.
Conflict of interest: The authors have declared no conflicts of interest for this article.
REFERENCES
- 1. Cowan WM, Fawcett JW, O'Leary DD, Stanfield BB. Regressive events in neurogenesis. Science 1984, 225:1258–1265. [DOI] [PubMed] [Google Scholar]
- 2. Truman JW, Reiss SE. Dendritic reorganization of an identified motoneuron during metamorphosis of the tobacco hornworm moth. Science 1976, 192:477–479. doi:10.1126/science.1257782. [DOI] [PubMed] [Google Scholar]
- 3. Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci 1999, 22:389–442. doi:10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 4. Innocenti GM. Growth and reshaping of axons in the establishment of visual callosal connections. Science 1981, 212:824–827. [DOI] [PubMed] [Google Scholar]
- 5. Innocenti GM, Price DJ. Exuberance in the development of cortical networks. Nat Rev Neurosci 2005, 6:955–965. doi:10.1038/nrn1790. [DOI] [PubMed] [Google Scholar]
- 6. Luo L, O'Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci 2005, 28:127–156. doi:10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- 7. Yu F, Schuldiner O. Axon and dendrite pruning in Drosophila. Curr Opin Neurobiol 2014, 27:192–198. doi:10.1016/j.conb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cheng H‐J. Axon pruning: an essential step underlying the developmental plasticity of neuronal connections. Philos Trans R Soc Lond B Biol Sci 2006, 361:1531–1544. doi:10.1098/rstb.2006.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schuldiner O, Yaron A. Mechanisms of developmental neurite pruning. Cell Mol Life Sci 2015, 72:101–119. doi:10.1007/s00018-014-1729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García‐López P, García‐Marín V, Freire M. Dendritic spines and development: towards a unifying model of spinogenesis—a present day review of Cajal's histological slides and drawings. Neural Plast 2010, 2010:769207–769229. doi:10.1155/2010/769207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas MSC, Davis R, Karmiloff‐Smith A, Knowland VCP, Charman T. The over‐pruning hypothesis of autism. Dev Sci 2016, 19:284–305. doi:10.1111/desc.12303. [DOI] [PubMed] [Google Scholar]
- 12. Sekar A, Bialas AR, de Rivera H, et al.** Schizophrenia risk from complex variation of complement component 4. Nature 2016, 530:177–183. doi:10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang G, Gudsnuk K, Kuo S‐H, et al. Loss of mTOR‐dependent macroautophagy causes autistic‐like synaptic pruning deficits. Neuron 2014, 83:1131–1143. doi:10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cocchi E, Drago A, Serretti A. Hippocampal pruning as a new theory of schizophrenia etiopathogenesis. Mol Neurobiol 2016, 53:2065–2081. doi:10.1007/s12035-015-9174-6. [DOI] [PubMed] [Google Scholar]
- 15. Feinberg I. Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res 1982, 17:319–334. [DOI] [PubMed] [Google Scholar]
- 16. Rouw R, Scholte HS. Increased structural connectivity in grapheme‐color synesthesia. Nat Neurosci 2007, 10:792–797. doi:10.1038/nn1906. [DOI] [PubMed] [Google Scholar]
- 17. Bargary G, Mitchell KJ. Synaesthesia and cortical connectivity. Trends Neurosci 2008, 31:335–342. doi:10.1016/j.tins.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 18. Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci 2003, 4:266–275. doi:10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 19. Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 1999, 126:4065–4076. [DOI] [PubMed] [Google Scholar]
- 20. Kanamori T, Togashi K, Koizumi H, Emoto K. Dendritic remodeling: lessons from invertebrate model systems. Int Rev Cell Mol Biol 2015, 318:1–25. doi:10.1016/bs.ircmb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 21. Williams DW, Truman JW. Mechanisms of dendritic elaboration of sensory neurons in Drosophila: insights from in vivo time lapse. J Neurosci 2004, 24:1541–1550. doi:10.1523/JNEUROSCI.4521-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Williams DW, Truman JW. Cellular mechanisms of dendrite pruning in Drosophila: insights from in vivo time‐lapse of remodeling dendritic arborizing sensory neurons. Development 2005, 132:3631–3642. doi:10.1242/dev.01928. [DOI] [PubMed] [Google Scholar]
- 23. Lyons GR, Andersen RO, Abdi K, Song W‐S, Kuo CT. Cysteine proteinase‐1 and cut protein isoform control dendritic innervation of two distinct sensory fields by a single neuron. Cell Rep 2014, 6:783–791. doi:10.1016/j.celrep.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park JH, Schroeder AJ, Helfrich‐Förster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide‐containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development 2003, 130:2645–2656. [DOI] [PubMed] [Google Scholar]
- 25. Luan H, Lemon WC, Peabody NC, Pohl JB, Zelensky PK, Wang D, Nitabach MN, Holmes TC, White BH. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci 2006, 26:573–584. doi:10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao T, Gu T, Rice HC, McAdams KL, Roark KM, Lawson K, Gauthier SA, Reagan KL, Hewes RS. A Drosophila gain‐of‐function screen for candidate genes involved in steroid‐dependent neuroendocrine cell remodeling. Genetics 2008, 178:883–901. doi:10.1534/genetics.107.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tapia JC, Wylie JD, Kasthuri N, Hayworth KJ, Schalek R, Berger DR, Guatimosim C, Seung HS, Lichtman JW. Pervasive synaptic branch removal in the mammalian neuromuscular system at birth. Neuron 2012, 74:816–829. doi:10.1016/j.neuron.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 28. Jarecki J, Keshishian H. Role of neural activity during synaptogenesis in Drosophila. J Neurosci 1995, 15:8177–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Featherstone DE, Broadie K. Surprises from Drosophila: genetic mechanisms of synaptic development and plasticity. Brain Res Bull 2000, 53:501–511. doi:10.1016/S0361-9230(00)00383-X. [DOI] [PubMed] [Google Scholar]
- 30. Carrillo RA, Olsen DP, Yoon KS, Keshishian H. Presynaptic activity and CaMKII modulate retrograde semaphorin signaling and synaptic refinement. Neuron 2010, 68:32–44. doi:10.1016/j.neuron.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Consoulas C, Restifo LL, Levine RB. Dendritic remodeling and growth of motoneurons during metamorphosis of Drosophila melanogaster. J Neurosci 2002, 22:4906–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boulanger A, Farge M, Ramanoudjame C, Wharton K, Dura J‐M. Drosophila motor neuron retraction during metamorphosis is mediated by inputs from TGF‐β/BMP signaling and orphan nuclear receptors. PLoS One 2012, 7:e40255. doi:10.1371/journal.pone.0040255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Z, Chen Y, Wang D, Wang S, Zhang YQ. Distinct presynaptic and postsynaptic dismantling processes of Drosophila neuromuscular junctions during metamorphosis. J Neurosci 2010, 30:11624–11634. doi:10.1523/JNEUROSCI.0410-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hebbar S, Fernandes JJ. Pruning of motor neuron branches establishes the DLM innervation pattern in Drosophila. J Neurobiol 2004, 60:499–516. doi:10.1002/neu.20031. [DOI] [PubMed] [Google Scholar]
- 35. Tasdemir‐Yilmaz OE, Freeman MR. Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev 2014, 28:20–33. doi:10.1101/gad.229518.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Truman JW. Metamorphosis of the central nervous system of Drosophila. J Neurobiol 1990, 21:1072–1084. doi:10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- 37. Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development 2005, 132:725–737. doi:10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- 38. Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin‐proteasome system. Neuron 2003, 38:871–885. [DOI] [PubMed] [Google Scholar]
- 39. Lee H‐H, Jan LY, Jan Y‐N. Drosophila IKK‐related kinase Ik2 and Katanin p60‐like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci USA 2009, 106:6363–6368. doi:10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhai Q, Wang J, Kim A, Liu Q, Watts R, Hoopfer E, Mitchison T, Luo L, He Z. Involvement of the ubiquitin‐proteasome system in the early stages of Wallerian degeneration. Neuron 2003, 39:217–225. doi:10.1016/S0896-6273(03)00429-X. [DOI] [PubMed] [Google Scholar]
- 41. Maor‐Nof M, Homma N, Raanan C, Nof A, Hirokawa N, Yaron A. Axonal pruning is actively regulated by the microtubule‐destabilizing protein kinesin superfamily protein 2A. Cell Rep 2013, 3:971–977. doi:10.1016/j.celrep.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 42. Rabinovich D, Mayseless O, Schuldiner O. Long term ex vivo culturing of Drosophila brain as a method to live image pupal brains: insights into the cellular mechanisms of neuronal remodeling. Front Cell Neurosci 2015, 9:6010. doi:10.3389/fncel.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu S, Chiang A‐S, Lee T. Development of the Drosophila mushroom bodies: elaboration, remodeling and spatial organization of dendrites in the calyx. Development 2003, 130:2603–2610. doi:10.1242/dev.00466. [DOI] [PubMed] [Google Scholar]
- 44. Shimono K, Fujimoto A, Tsuyama T, et al. Multidendritic sensory neurons in the adult Drosophila abdomen: origins, dendritic morphology, and segment‐ and age‐dependent programmed cell death. Neural Dev 2009, 4:37. doi:10.1186/1749-8104-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee T, Marticke S, Sung CS, Robinow S, Luo L. Cell‐autonomous requirement of the USP/EcR‐B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron 2000, 28:807–818. [DOI] [PubMed] [Google Scholar]
- 46. King‐Jones K, Thummel CS. Nuclear receptors—a perspective from Drosophila. Nat Rev Genet 2005, 6:311–323. doi:10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- 47. Kuo CT, Jan LY, Jan Y‐N. Dendrite‐specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin‐proteasome, and ecdysone signaling. Proc Natl Acad Sci USA 2005, 102:15230–15235. doi:10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schubiger M, Wade AA, Carney GE, Truman JW, Bender M. Drosophila EcR‐B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 1998, 125:2053–2062. [DOI] [PubMed] [Google Scholar]
- 49. Schubiger M, Tomita S, Sung CS, Robinow S, Truman JW. Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech Dev 2003, 120:909–918. doi:10.1016/S0925-4773(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 50. Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac‐based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell 2008, 14:227–238. doi:10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boulanger A, Clouet‐Redt C, Farge M, Flandre A, Guignard T, Fernando C, Juge F, Dura J‐M. ftz‐f1 and Hr39 opposing roles on EcR expression during Drosophila mushroom body neuron remodeling. Nat Neurosci 2011, 14:37–44. doi:10.1038/nn.2700. [DOI] [PubMed] [Google Scholar]
- 52. Schubiger M, Wade AA, Carney GE, Truman JW, Bender M. Drosophila EcR‐B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 1998, 125:2053–2062. [DOI] [PubMed] [Google Scholar]
- 53. Awasaki T, Huang Y, O'Connor MB, Lee T. Glia instruct developmental neuronal remodeling through TGF‐β signaling. Nat Neurosci 2011, 14:821–823. doi:10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu XM, Gutman I, Mosca TJ, Iram T, Özkan E, Garcia KC, Luo L, Schuldiner O. Plum, an immunoglobulin superfamily protein, regulates axon pruning by facilitating TGF‐β signaling. Neuron 2013, 78:456–468. doi:10.1016/j.neuron.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bialas AR, Stevens B. TGF‐β signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci 2013, 16:1773–1782. doi:10.1038/nn.3560. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, Low BC, Kolodkin AL, Wang H, Yu F. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci 2009, 12:1497–1505. doi:10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rumpf S, Bagley JA, Thompson‐Peer KL, Zhu S, Gorczyca D, Beckstead RB, Jan LY, Jan Y‐N. Drosophila valosin‐containing protein is required for dendrite pruning through a regulatory role in mRNA metabolism. Proc Natl Acad Sci USA 2014, 111:7331–7336. doi:10.1073/pnas.1406898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Loncle N, Williams DW. An interaction screen identifies headcase as a regulator of large‐scale pruning. J Neurosci 2012, 32:17086–17096. doi:10.1523/JNEUROSCI.1391-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Truman JW, Talbot WS, Fahrbach SE, Hogness DS. Ecdysone receptor expression in the CNS correlates with stage‐specific responses to ecdysteroids during Drosophila and Manduca development. Development 1994, 120:219–234. doi:10.1101/SQB.1974.038.01.070. [DOI] [PubMed] [Google Scholar]
- 60. Sutherland JD, Kozlova T, Tzertzinis G, Kafatos FC. Drosophila hormone receptor 38: a second partner for Drosophila USP suggests an unexpected role for nuclear receptors of the nerve growth factor‐induced protein B type. Proc Natl Acad Sci USA 1995, 92:7966–7970. doi:10.1073/pnas.92.17.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zelhof AC, Yao TP, Chen JD, Evans RM, McKeown M. Seven‐up inhibits ultraspiracle‐based signaling pathways in vitro and in vivo. Mol Cell Biol 1995, 15:6736–6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin‐proteasome system and onto human diseases and drug targeting. Bioorg Med Chem 2013, 21:3400–3410. doi:10.1016/j.bmc.2013.01.056. [DOI] [PubMed] [Google Scholar]
- 63. Hoopfer ED, Penton A, Watts RJ, Luo L. Genomic analysis of Drosophila neuronal remodeling: a role for the RNA‐binding protein Boule as a negative regulator of axon pruning. J Neurosci 2008, 28:6092–6103. doi:10.1523/JNEUROSCI.0677-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kuo CT, Zhu S, Younger S, Jan LY, Jan Y‐N. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 2006, 51:283–290. doi:10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 65. Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci 2006, 9:1234–1236. doi:10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 66. Rumpf S, Lee SB, Jan LY, Jan Y‐N. Neuronal remodeling and apoptosis require VCP‐dependent degradation of the apoptosis inhibitor DIAP1. Development 2011, 138:1153–1160. doi:10.1242/dev.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wong JJL, Li S, Lim EKH, et al. A cullin1‐based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol 2013, 11:e1001657. doi:10.1371/journal.pbio.1001657.s023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol 2004, 14:668–677. doi:10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 69. Yaniv SP, Issman‐Zecharya N, Oren‐Suissa M, Podbilewicz B, Schuldiner O. Axon regrowth during development and regeneration following injury share molecular mechanisms. Curr Biol 2012, 22:1774–1782. doi:10.1016/j.cub.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 70. Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DD, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron 2006, 50:883–895. doi:10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 71. Xiong X, Hao Y, Sun K, et al. The highwire ubiquitin ligase promotes axonal degeneration by tuning levels of nmnat protein. PLoS Biol 2012, 10:e1001440. doi:10.1371/journal.pbio.1001440.g009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Babetto E, Beirowski B, Russler EV, Milbrandt J, Diantonio A. The Phr1 ubiquitin ligase promotes injury‐induced axon self‐destruction. Cell Rep 2013, 3:1422–1429. doi:10.1016/j.celrep.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Issman‐Zecharya N, Schuldiner O. The PI3K class III complex promotes axon pruning by downregulating a Ptc‐derived signal via endosome‐lysosomal degradation. Dev Cell 2014, 31:461–473. doi:10.1016/j.devcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 74. Zhang H, Wang Y, Wong JJL, Lim K‐L, Liou Y‐C, Wang H, Yu F. Endocytic pathways downregulatethe L1‐type cell adhesion molecule neuroglian to promote dendrite pruning in Drosophila. Dev Cell 2014, 30:463–478. doi:10.1016/j.devcel.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 75. Loncle N, Agromayor M, Martin‐Serrano J, Williams DW. An ESCRT module is required for neuron pruning. Sci Rep 2015, 5:8461. doi:10.1038/srep08461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kanamori T, Yoshino J, Yasunaga K‐I, Dairyo Y, Emoto K. Local endocytosis triggers dendritic thinning and pruning in Drosophila sensory neurons. Nat Commun 2015, 6:6515. doi:10.1038/ncomms7515. [DOI] [PubMed] [Google Scholar]
- 77. Kanamori T, Kanai MI, Dairyo Y, Yasunaga KI, Morikawa RK, Emoto K. Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science 2013, 340:1475–1478. doi:10.1126/science.1234879. [DOI] [PubMed] [Google Scholar]
- 78. Yang J, Weimer RM, Kallop D, Olsen O, Wu Z, Renier N, Uryu K, Tessier‐Lavigne M. Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron 2013, 80:1175–1189. doi:10.1016/j.neuron.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 79. Bornstein B, Zahavi EE, Gelley S, Zoosman M, Yaniv SP, Fuchs O, Porat Z, Perlson E, Schuldiner O. Developmental axon pruning requires destabilization of cell adhesion by JNK signaling. Neuron 2015, 88:926–940. doi:10.1016/j.neuron.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 80. Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol 2004, 14:678–684. doi:10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 81. Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced‐6 in programmed axon pruning during Drosophila metamorphosis. Neuron 2006, 50:855–867. doi:10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 82. Hakim Y, Yaniv SP, Schuldiner O. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLoS One 2014, 9:e86178. doi:10.1371/journal.pone.0086178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Han C, Song Y, Xiao H, Wang D, Franc NC, Jan LY, Jan Y‐N. Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron 2014, 81:544–560. doi:10.1016/j.neuron.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yamamoto M, Ueda R, Takahashi K, Saigo K, Uemura T. Control of axonal sprouting and dendrite branching by the Nrg‐Ank complex at the neuron‐glia interface. Curr Biol 2006, 16:1678–1683. doi:10.1016/j.cub.2006.06.061. [DOI] [PubMed] [Google Scholar]
- 85. Han C, Jan LY, Jan Y‐N. Enhancer‐driven membrane markers for analysis of nonautonomous mechanisms reveal neuron‐glia interactions in Drosophila. Proc Natl Acad Sci USA 2011, 108:9673–9678. doi:10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Schafer DP, Stevens B. Phagocytic glial cells: sculpting synaptic circuits in the developing nervous system. Curr Opin Neurobiol 2013, 23:1034–1040. doi:10.1016/j.conb.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chung W‐S, Clarke LE, Wang GX, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature November 2013, 504:394–400. doi:10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bates KE, Sung CS, Robinow S. The unfulfilled gene is required for the development of mushroom body neuropil in Drosophila. Neural Dev 2010, 5:4. doi:10.1186/1749-8104-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lin S, Huang Y, Lee T. Nuclear receptor unfulfilled regulates axonal guidance and cell identity of Drosophila mushroom body neurons. PLoS One 2009, 4:e8392. doi:10.1371/journal.pone.0008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu K, Tedeschi A, Park KK, He Z. Neuronal intrinsic mechanisms of axon regeneration. Annu Rev Neurosci 2011, 34:131–152. doi:10.1146/annurev-neuro-061010-113723. [DOI] [PubMed] [Google Scholar]
- 91. Park KK, Liu K, Hu Y, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 2008, 322:963–966. doi:10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Marmor‐Kollet N, Schuldiner O. Contrasting developmental axon regrowth and neurite sprouting of Drosophila mushroom body neurons reveals shared and unique molecular mechanisms. Dev Neurobiol 2016, 76:262–276. doi:10.1002/dneu.22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rabinovich D, Yaniv SP, Alyagor I, Schuldiner O. Nitric oxide as a switching mechanism between axon degeneration and regrowth during developmental remodeling. Cell 2016, 164:170–182. doi:10.1016/j.cell.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Medioni C, Ramialison M, Ephrussi A, Besse F. Imp promotes axonal remodeling by regulating profilin mRNA during brain development. Curr Biol 2014, 24:793–800. doi:10.1016/j.cub.2014.02.038. [DOI] [PubMed] [Google Scholar]
- 95. Prokop A, Beaven R, Qu Y, Sánchez‐Soriano N. Using fly genetics to dissect the cytoskeletal machinery of neurons during axonal growth and maintenance. J Cell Sci 2013, 126(Pt 11):2331–2341. doi:10.1242/jcs.126912. [DOI] [PubMed] [Google Scholar]
- 96. Chen D, Qu C, Bjorum SM, Beckingham KM, Hewes RS. Neuronal remodeling during metamorphosis is regulated by the alan shepard (shep) gene in Drosophila melanogaster. Genetics 2014, 197:1267–1283. doi:10.1534/genetics.114.166181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gu T, Zhao T, Hewes RS. Insulin signaling regulates neurite growth during metamorphic neuronal remodeling. Biol Open 2014, 3:81–93. doi:10.1242/bio.20136437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sung CS, Wong LE, Chang Sen LQ, Nguyen E, Lazaga N, Ganzer G, Mcnabb SL, Robinow S. The unfulfilled/DHR51 gene of Drosophila melanogaster modulates wing expansion and fertility. Dev Dyn 2009, 238:171–182. doi:10.1002/dvdy.21817. [DOI] [PubMed] [Google Scholar]
- 99. Reinking J, Lam MMS, Pardee K, et al. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell 2005, 122:195–207. doi:10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 100. Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV‐ERBalpha and REV‐ERBbeta. Nat Struct Mol Biol 2007, 14:1207–1213. doi:10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Caceres L, Necakov AS, Schwartz C, Kimber S, Roberts IJH, Krause HM. Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev 2011, 25:1476–1485. doi:10.1101/gad.2064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A. Ecdysone‐ and NO‐mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol Cell 2011, 44:51–61. doi:10.1016/j.molcel.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. De Rosny E, De Groot A, Jullian‐Binard C, Borel F, Suarez C, Le Pape L, Fontecilla‐Camps JC, Jouve HM. DHR51, the Drosophila melanogaster homologue of the human photoreceptor cell‐specific nuclear receptor, is a thiolate heme‐binding protein. Biochemistry 2008, 47:13252–13260. doi:10.1021/bi801691b. [DOI] [PubMed] [Google Scholar]
- 104. Liu Y, Rutlin M, Huang S, Barrick CA, Wang F, Jones KR, Tessarollo L, Ginty DD. Sexually dimorphic BDNF signaling directs sensory innervation of the mammary gland. Science 2012, 338:1357–1360. doi:10.1126/science.1228258. [DOI] [PMC free article] [PubMed] [Google Scholar]


