Abstract
Cell death is an inherent process that is required for the proper wiring of the nervous system. In the last four decades, it has been found that in a parallel developmental pathway, axons and dendrite are eliminated without the death of the neuron. This developmentally regulated “axonal death” results in neuronal remodeling which is an essential mechanism to sculpt neuronal networks in both vertebrates and invertebrates. Studies across various organisms have demonstrated that a conserved strategy to form adult neuronal circuitry often involves generating too many connections that are later eliminated with high temporal and spatial resolution. Can neuronal remodeling can be perceived as developmentally and spatially regulated neurodegeneration? It has been previously speculated that injury induced degeneration (Wallerian degeneration) share molecular similarities with dying back neurodegenerative diseases. In this opinion piece, we examine the similarities and differences between the mechanisms regulating neuronal remodeling and those being perturbed in dying back neurodegenerative diseases. We focus primarily on ALS and peripheral neuropathies and highlight possible shared pathways and mechanisms. While mechanistic data is just beginning to emerge, and despite the inherent differences between disease oriented and developmental processes, we believe that some of the similarities between these developmental and disease-initiated degeneration warrant closer collaborations and cross talk between these different fields.
Introduction
Developmentally regulated neuronal cell death has been shown to play important roles in development of the vertebrate and invertebrate nervous systems [1–3] and is detected at the final stages of neurodegenerative diseases. In last four decades, accumulating evidence highlight another regressive process during developmental that resembles ‘axonal death’ but does not involve the death of the neuron [4–6]. Axon, dendrite and synapse elimination that result in neuronal remodeling of the neural circuit was found to be a conserved strategy to refine and sculpt neural circuits during the assembly of vertebrate and invertebrate nervous systems. Remodeling includes the elimination of exuberant or improper connections often followed by stabilization or regrowth of new connections to form the adult specific neural circuit (Figure 1A). While pruning of neuronal connections without the death of the neuron was first discovered only about 40 years ago [4–6], reexamining Ramon y Cajal’s images from more than a hundred years ago suggests that he too identified this process in avian Purkinje and mammalian granule cells and termed it process resorption [7, 8]. Following four decades of research and the discovery of a wide variety of neural circuits in different organisms that undergo remodeling, the emerging insight is that a broad set of neural circuits in the central as well as peripheral nervous systems develop by initially forming too many connections that are then refined by various strategies of neuronal remodeling. Defects in remodeling have been attributed to a variety of phsychoneurological conditions including autism, schizophrenia and synesthesia [9–12] although a clear causality has not yet been established and the underlying mechanisms are not well understood.
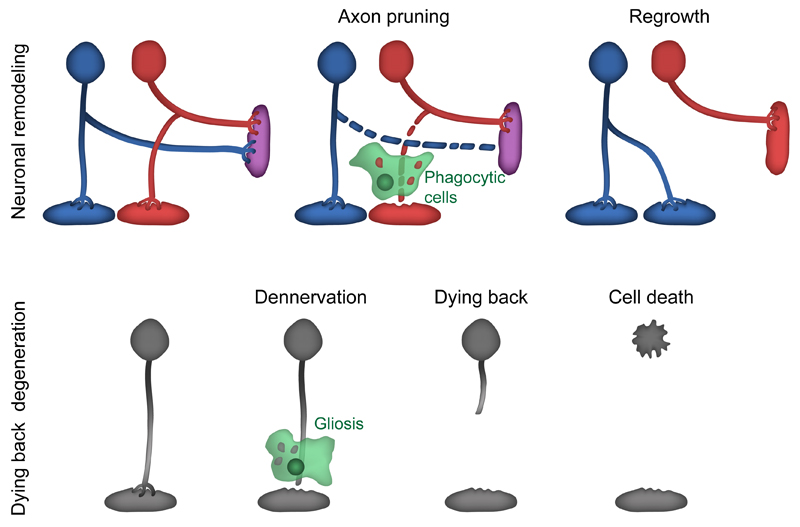
Figure 1. Axonal elimination during development and dying back neurodegeneration.
A) Restricted axonal fragmentation serves as a key mechanism to prune exuberant connections during development. The axonal fragments are engulfed by glia or epidermal cells. Following pruning, axons often regrow to form new connections in a mechanism that resemble axonal regeneration.
B) Genetic lesions or toxic conditions induce axonal loss in a dying back fashion from the target. Dying back is usually accompanied by gliosis and in some cases ends in neuronal cell death.
In this article we chose not to review the current status of neuronal remodeling as several good reviews have been recently published [7, 13–15]. Instead, we decided to explore the idea that understanding the mechanism underlying neuronal remodeling could provide us with insights into the mechanisms of various neurodegenerative diseases. Could neuronal remodeling be perceived as developmentally and spatially regulated neurodegeneration? If this were true, then perhaps some of the machinery that is required to dismantle axons and dendrites in a regulated manner during development is hijacked and exploited due to various insults during neurodegeneration, especially in ‘dying-back’ neurodegenerative diseases in which the breakdown of the axons occurs before and even in the absence of cell death.
Indeed, Raff and colleagues [16] have previously speculated that axon pruning and injury induced axon degeneration (Wallerian degeneration) share molecular similarities with ‘dying-back’ neurodegenerative diseases such as Amyotrophic Lateral Sclerosis (ALS), neuropathies, and spinocerebellar disorders e.g. Friedreich’s ataxia. Up until now, this speculation has mostly been tested by exploring whether expression of Wlds (Wallerian degeneration slow), a fusion protein between Ube4b and Nmnat1 that has been shown to slow Wallerian degeneration, can slow disease onset or progression in different model of neurodegeneration. Results have been mixed – Wlds indeed slows motor neuron loss in some models of motor neuron disease [17] and neuropathy [18] but not in all [19, 20]. Interestingly, Wlds was not able to delay onset or slow disease progression in ALS models [21, 22]. Recent progress in our understanding of Wallerian degeneration and its relevance to disease has been reviewed elsewhere [19, 20, 23–25] and we will therefore not discuss it in depth.
Saxena and Caroni [26] revisit and extend the hypothesis setup by Raff. While they highlight a striking similarity in the sequence of cellular events during developmental remodeling, Wallerian degeneration and ‘dying back’ neurodegenerative diseases, Saxena and Caroni also point out important differences. In essence, they raise a speculative model in which the mechanisms that regulate these three major forms of axon elimination diverge and converge on multiple points. The initiation of axon degeneration, they propose, is unique to each developmental or disease condition but they also propose that the axon dismantling itself involves conserved mechanisms. While this model is highly attractive, hard evidence to support it is still lacking.
In this article we further examine whether insights from the field of developmental neuronal remodeling can complement the progress from the Wallerian degeneration field and enrich our understanding of pathological neurodegeneration. Therefore, we examine here the possible roles of “remodeling genes and pathways” in ‘dying-back’ neurodegenerative diseases, primarily focusing on Amyotrophic Lateral Sclerosis (ALS) and peripheral neuropathy. While mechanistic data is just beginning to emerge, and despite the inherent differences between what happens during disease and development, we believe that the similarities as well as differences warrant closer collaborations and cross talk between these different fields.
Dying back neurodegeneration
It has been long known that the onset of several neurodegenerative diseases begins with axonal atrophy that gradually affects the viability of the neuron (Figure 1B). One of the first investigators to formulate this notion leading to the categorization of several conditions as “dying back” neurodegenerative diseases was Cavanagh [27] who wrote: “…this is in general a slow atrophy affecting individual neurons and their fibers at various moments in time, and the clinical consequences are seen only when the compensatory forces begin to fail in the face of cumulative nerve fiber damage”. He highlighted the “dying back” phenotype in neurodegenerative conditions that result from exposure to neurotoxins, such as Tricresyl phosphate (TCP), and also in diseases including Amyotrophic Lateral Sclerosis (ALS), neuropathies, Friedreich’s ataxia and Werdnig-Hoffmann’s disease, now known as spinal muscular atrophy (SMA) type I.
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig's disease, is a devastating and incurable adult onset neurodegenerative disease. ALS is characterized by rapidly progressive muscular atrophy (‘amyotrophic’) and scarring of tissues in the lateral spinal cord (‘lateral sclerosis’) ultimately resulting in paralysis and death from respiratory failure. The initial symptoms are usually detected in the fifth or sixth decade of life and the average time until death is around 2-3 years. Autopsies from ALS patients as well as ALS mice models suggest that this slow progression of the symptoms lies at least in part due to the fact that neurodegenerative processes begin in distal axons and slowly progress to the cell bodies [28, 29]. The vast majority (~90%) of ALS cases are sporadic (sometimes also called SALS) while the remaining 10% are inherited and thus called familial ALS (FALS) [30–32].
Mutations in the Superoxide Dismutase 1 (SOD1) gene were the first, and until quite recently the only, identified causal mutations driving ALS in human patients and in model animals [33, 34]. In humans, SOD1 mutations, inherited in a dominant fashion, are responsible for ~12% of familial cases but only ~1% of sporadic ALS cases. Despite many decades of intensive research, including the generation of many model animals, the precise mechanism by which mutant SOD1 drives motor neuron disease and even whether it functions cell-autonomous or non-cell-autonomous are not well understood.
Following fifteen years in which mutations in SOD1 was the only genetic lesion linked with ALS, in the recent decade there has been an explosion in our genetic understanding of ALS: mutations in about nine genes have been identified in ~68% of familial and ~11% of sporadic ALS cases [30–32]. The gene that is by far most implicated in ALS is C9ORF72, encoding a protein with an unknown function, in which a hexanucleotide repeat expansion is dominantly inherited [35, 36]. These expansions are likely the cause of ~40% of familial and ~7% of sporadic ALS as well as in ~25% of familial fronto-temporal dementia (FTD), thereby providing a genetic link between ALS and FTD [37].
Taken together, while there has been tremendous progress in our understanding of the genetic basis for ALS, the mechanisms driving motor neuron axonopathy and death remain mostly unclear. The vast majority of the mutations is dominantly inherited and therefore could result in gain of function defects that are often difficult to mechanistically understand.
Peripheral neuropathies
Peripheral nerve degeneration (peripheral neuropathies) is a widespread neuronal disorder that is largely manifested by loss of sensory and/or motor axons of the peripheral nervous system (PNS). This loss leads to ineffective communication of peripheral organs with the central nervous system. Patients with peripheral neuropathies exhibit a variety of symptoms, which include but are not limited to abnormal pain and touch sensations, weakness, and motor deficits as a result of muscle atrophy. Peripheral neuropathies can be divided into heredity neuropathies and acquired neuropathies that are triggered by other systemic diseases or exposure to drugs, chemicals and viral infections [25, 38]. Diabetes is the most common cause of peripheral neuropathy worldwide, and also the disease that is most associated with acquired neuropathies [39]. The most common form of heredity sensory and motor neuropathies is Charcot-Marie-Tooth (CMT) disease that is comprised of a genetically heterogeneous collective assembly of PNS disorders. Moreover, the clinical symptoms of CMT vary in severity and age of onset, even among members of the same family. CMT is classified into several types according to electrophysiological and nerve biopsy measurements, and mode of genetic inheritance. The most frequent types of CMT are CMT1, defined as a demyelinating neuropathy with clear histological evidence of de- and re-myelination, and CMT2, defined as axonal neuropathy [40, 41]. Importantly, almost all types of neuropathies are chronic but exhibit slow progressive deterioration in symptoms as a result of dying back axon degeneration.
Common and Divergent Mechanisms
Role of trophic factors
During development the neurotrophin family of trophic factors, of which Nerve Growth Factor (NGF) is their founding member, regulate various aspects of neuronal development (see Figure 2 for schematic representation of common and divergent pathways throughout the article). In the PNS these trophic factors are produced in limited amounts at peripheral targets and operate through retrograde signaling or locally at the tip of the axons to control multiple aspects of PNS development including cell survival, neuronal specification and target innervation [42]. Studies using in vitro compartmentalized chambers of sensory axons have demonstrated that local axonal deprivation from trophic support elicit restricted axonal degeneration, which morphologically recapitulate axonal pruning [43]. Importantly, several molecular components of axonal degeneration that were discovered in this in vitro setting were also shown to regulate developmental pruning in the CNS [44]. More recently it was demonstrated that male specific developmental masking of the brain derived neurotrophic factor (BDNF) signaling leads to pruning of sensory axons from the mammary gland, establishing sexually dimorphic circuit [45].
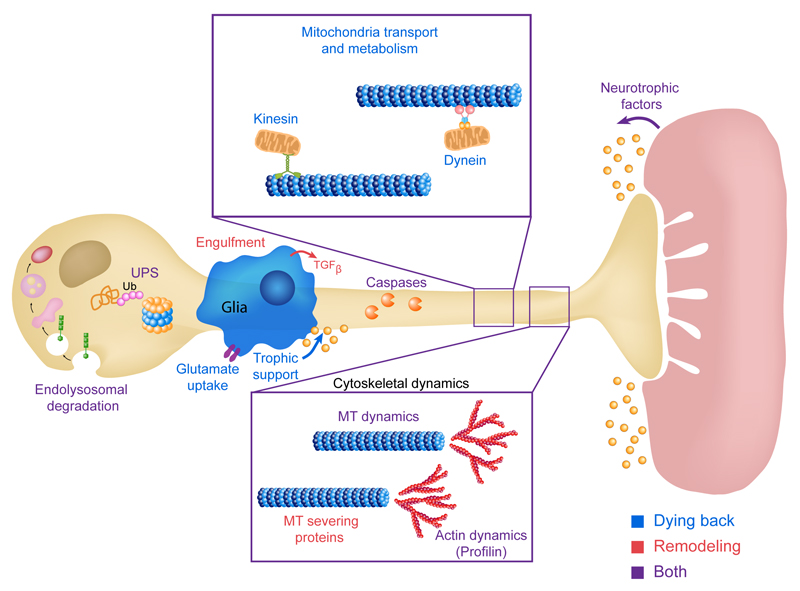
Figure 2. Major pathways that underlie axonal pruning and dying back neurodegeneration.
Illustration of the main pathways that control developmental axon pruning and axon degeneration by dying back, see text for details. Blue, red and purple text – genes and pathways involved in dying back neurodegeneration (blue), developmental neuronal remodeling (red) or both (purple). MT-Microtubules, Ub-Ubiquitin, UPS-Ubiquitin Proteasome System, TGFβ - Transforming growth factor-β, ESCRT -The endosomal sorting complexes required for transport, Caspases - cysteine-aspartic proteases or cysteine-dependent aspartate-directed proteases.
Neurotrophins and other trophic factors are also expressed by adult target tissues and are thought to be required for axonal maintenance and sprouting following injury [46, 47]. Could decreased levels of neurotrophins be an early cause for dying back neurodegenerative diseases? While not much is known mechanistically, it is interesting to mention that several studies have demonstrated a reduction in NGF expression in the skin in diabetic patients, suggesting that reduction in trophic support is at least part of the reason that nociceptive axons, which innervate the skin, are lost during the course of diabetes [48, 49]. While these studies did not determine causality, nevertheless these observations raised high hopes that NGF administration could serve as a potential treatment. Unfortunately, administration of NGF resulted in severe side effects that led to the cessation of the clinical trials [50]. Although reduction in trophic support was not reported in mouse models of ALS, intramuscular application of Ciliary neurotrophic factor (CNTF) delayed but did not prevent axonal loss in the SOD1 mouse model [51]. This study suggests that trophic signaling can counter the degenerative process and that reduction in axonal transport and trophic retrograde signaling may be one force that facilitates axonal loss in the initial phase of the disease.
Protein degradation
Developmental neuronal remodeling involves the activation of an axon or dendrite disintegration and elimination program, which likely involves degradation of proteins, cytoskeleton and membranes. Experiments in flies have shown that the ubiquitin proteasome system (UPS) is required for axon pruning of mushroom body (MB) γ neurons, a CNS structure required for learning and memory, and also for dendrite pruning of the peripheral class IV dendritic arborization (da) sensory neurons that cover the larval body wall. Over-expression of the yeast ubiquitin carboxyl-terminal hydrolase 2 (UBP2), which reverses the addition of ubiquitin molecules, inhibited MB axon pruning [52]. Similarly, mutating genes that encode proteins belonging to the UPS, such as Ubiquitin-Activating Enzyme E1 (UBA1) and proteasome subunits, essentially blocked pruning of MB axons [52] and of da dendrites in a cell autonomous manner [53, 54]. While in da neurons we know of at least two targets for the UPS in the context of pruning, the Drosophila inhibitor of apoptosis (DIAP1) [53] and components of the insulin pathway [55], we do not know which proteins are degraded during developmental pruning of MB axons [13]. Our knowledge of the role of the UPS in neuronal remodeling of mammalian neurons is limited to an in vitro model system of remodeling in which cultured primary neurons (usually from the dorsal root ganglion, DRG) undergo axon elimination following trophic deprivation of the axons but not cell bodies [43]. Indeed, pharmacological inhibition of the UPS inhibited axon elimination in DRG cultures following trophic deprivation [56].
Ubiquitin mediated protein degradation is also involved in ALS. Mutations in at least three UPS related genes, Valosin-containing protein (VCP [57]), Ubiquilin 2 (UBQLN2 [58]) and Sequestosome 1 (SQSTM1 [59]), were found in ALS patients but the mechanisms by which they cause the disease is not well understood. The fact that mutations in the UPS promote ALS pathology and at the same time inhibit developmental pruning seems counterintuitive. However, whether disease-causing mutations are loss of function is still not clear. For example: Ubiquilin2, a protein that is important in linking between ubiquitin ligases and the proteasome is mutated in familial ALS where it is inherited in an X-linked dominant fashion. The prevailing dogma is that the ALS mutations in Ubiquilin2 result in loss of function resulting in the culmination of toxic protein aggregates [32], including that of TDP-43, foci of which are associated in many ALS cases. However a recent study has found that while expression of mutant Ubiquilin 2 in rats resulted in ALS-like phenotypes, the loss of endogenous Ubiquilin 2 did not [60], suggesting that the isolated mutations manifested a gain of function effect. Remarkably, Pun and colleagues [51] have shown that axon degeneration in a mouse SOD1 model requires the UPS. Interestingly, they were able to block SOD1 mediated motor neuron degeneration by applying UPS inhibitors even in rather late stages, when the application of trophic factors (see previous section) was not sufficient to alleviate degeneration. These experiments clearly demonstrate that the axon destruction phase, at least in the SOD1 mouse model, depends on an intact UPS. These data might also hint that ALS associated mutations in UPS genes likely result in gain of function defects but more experiments are necessary to support this speculative idea.
The ALS associated protein VCP, an ATPase from AAA family, is known to interact with ubiquitinated proteins and process them for proteasome degradation, autophagy or endocytosis [61]. However, patient derived fibroblasts containing pathogenic VCP mutations were found to exhibit reduced ATP levels, caused by mitochondrial uncoupling [62]. VCP has been suggested to affect mitochondrial function by sending depolarized mitochondria to degradation [63]. Therefore, the precise mechanism by which mutations in VCP cause ALS and even whether this involves a gain or loss of function, is not well understood. Interestingly, however, mutations in VCP also affect dendrite pruning in Drosophila.
By manipulating the levels of VCP in Drosophila sensory neurons, Rumpf and colleagues [64] have shown that while a strong reduction in VCP levels results in cell death, a moderate reduction results in accumulation of ubiquitinated proteins, including the Drosophila inhibitor of apoptosis (Diap1) whose degradation is required for dendrite pruning of da neurons [53]. Thus, da neurons in which VCP was knocked down do not undergo normal dendrite pruning. In a follow up study the authors found that VCP also affects, in an unknown mechanisms, the splicing of various proteins including the large cytoskeletal protein MICAL that is required for pruning [65, 66]. Thus, VCP might affect pruning of da dendrites by various mechanisms, which are at least partially dose dependent.
Another pathway for protein degradation – the endolysosomal pathway – was also implicated in both dying back neurodegenerative diseases and in neurite remodeling. Three recent studies focusing on neurite pruning in Drosophila have found that the Endosomal Sorting Complexes Required for Transport (ESCRT)-III is cell autonomously required of neurite pruning [67–69]. While Zhang and colleagues [68] found that the endolysosomal pathway was important to downregulate the membranal L1-type cell adhesion molecule Neuroglian (nrg) during dendrite pruning of da neurons, Issman and colleagues [67] found that endolysosomal downregulation of Patched (ptc) was required for MB axon pruning. In contrast, Loncle and colleagues [69] provide evidence for a unique activity of the ESCRT-III complex involving membrane remodeling to promote dendrite pruning in a mechanism that does not involve the endocytic machinery.
Dominantly inherited mutations in the ESCRT-III subunit CHarged Multivesicular body Protein 2B (CHMP2B [70]) have been identified in a small subset of ALS and FTD patients. The mechanism by which these mutations promote ALS is not currently known. Furthermore, dominant mutations in the Phosphatidylinositol 3,5-bisphosphate 5-phosphatase (encoded by FIG4 [71]) were also found in ALS patients. This is especially interesting as we have found that mutations in the Phosphatidylinositol 3 kinase class III inhibit axon pruning of MB neurons [67]. The product of both PI3K-CIII and FIG4 is PI3P, which recruits proteins containing an FYVE domain to endosomal membranes and affect vesicle trafficking.
Two GTPases that regulate endocytosis have been identified as mutated in CMT. Rab7 is a small GTPase, which regulates membrane trafficking during the maturation of the early endosome to the late multivesicular endosome and also the subsequent maturation to the lysosome. CMT2 causing mutations in Rab7 are inherited in a dominant fashion. Interestingly, studies in cultured neurons and cell lines suggest that the mutant forms of Rab7 that cause CMT are fully functional [72, 73]. Moreover, these studies show that these Rab7 mutants are found largely in the GTP bound (active state) and they can drastically reduce the cell surface expression of the neurotrophin receptor TrkA [72, 73]. Lastly, the mutant Rab7 positive vesicles appear more active as they pause less than wild-type vesicles when travelling along the axon [74]. Therefore, hyper activation of the Rab7 regulated endocytosis / trafficking may play a part in promoting CMT. Dynamin 2 (DMN2) is a large GTPase that is involved in clathrin-mediated endocytosis. Mutations in DMN2 have been discovered both in centronuclear myopathies (CME) and CMT; while the CME causing mutations were suggested to be hypermorphic the CMT mutations were found to be hypomorphic [75, 76]. Interestingly, cell autonomous expression of the temperature sensitive form of the Drosophila dynamin ortholog shibire, results in pruning defects [67]. Lastly, mutations in FIG4 have been reported in CMT4J as well, but the deciphering the underlying genetics is complex since patients harbor one null allele, and a second allele that harbors a point mutation [77, 78].
Taken together, it seems that protein degradation via the UPS and endolysosomal pathways seem to be central to both neuronal remodeling and dying back neurodegenerative diseases (Figure 2). Whether this indicates common mechanisms is questionable, especially as both pathways are ‘house keeping’. The mechanisms by which these protein degradation pathways regulate developmental and pathological axon elimination are just beginning to be clarified and therefore more work is necessary to uncover the underlying similarities and differences. These should in turn hint towards the existence of convergent mechanisms underlying neurite degeneration during development and in dying back neurodegeneration.
Caspase activation
Caspases are best known as critical regulators of cell death by apoptosis. As such, caspase activation has been raised as a possible mechanism to trigger neuronal cell death in ALS and in other dying back neurodegenerative diseases. Indeed, caspase activation has been observed in samples from deceased patients [79] and in SOD1 mouse models [80] but surprisingly not in isolated mouse SOD1 motor neuron populations [81]. Manipulating caspase activity by genetic or pharmacological means only slightly improved viability and delayed the onset of neurodegeneration in SOD1 transgenic mice models [80, 82]. A recent study suggested that deleting the proapoptotic protein Bcl-2-associated X protein (Bax) indeed blocked the cell death of MNs expressing mutant SOD1 but only slightly delayed MN denervation [83]. In this study, the authors also identified a delay in events occurring after denervation, such as gliosis, suggesting that Bax has additional non-apoptotic functions. Conditionally deleting both Bax and another Bcl-2 family member, the Bcl-2 homologous antagonist killer (Bak), in the CNS resulted in a more impressive suppression of both neurodegeneration and ALS-like symptoms in SOD1 transgenic mice [84]. Interestingly, up regulation of the anti apoptotic protein, Bcl2a1-a, has been observed at the early stages of axonal loss and before neuronal death in the SOD1 mouse [51]. This up regulation clearly does not protect the axons but may point out that the neurons aim to overcome an aberrant activation of the apoptotic system to circumvent cell death or axonal degeneration.
Thus, the role of the apoptotic machinery in the SOD1 model has yielded confusing results but taken together suggest that in addition to promoting cell death at the final stages of the disease, the apoptotic machinery may play crucial roles in the early stages of axonal atrophy. Surprisingly, to the best of our knowledge, the role of caspases or other apoptotic components has not been examined in more recent models of ALS.
Type I diabetes can be induced in rats by Streptozotocin administration that eliminates the insulin producing pancreatic β-cells. Using this model Cheng and Zochodne [85] have demonstrated that caspase-3 is activated in sensory axons and cell bodies of diabetic rats. Interestingly, despite caspase-3 activation, the neurons did not undergo cell death in this model. In vitro models of DRG neurons treated by the neuropathic agent Cisplatin exhibit reduced axonal outgrowth, which is accompanied with a Bax dependent activation of the apoptotic system [86, 87]. Intriguingly, Bax is also a critical regulator of axonal pruning, where it is required for local caspase activation [88, 89]. Finally, Bcl-w (also known as Bcl-2-like protein 2, Bcl2l2) knock out mice, exhibit peripheral neuropathy as a result of axonopathy without cell death [90]. Interestingly, the authors also found that Bcl-w is the only Bcl2 family member enriched in axons. Taken together, these experiments provide strong evidence that axonal activation of the apoptotic machinery can cause peripheral neuropathy.
While neuronal remodeling does not involve cell death, the roles of numerous components of the apoptotic machinery have been highlighted in various systems [91]. A transgenic reporter for caspase activation has allowed the identification of local caspase activation during dendrite pruning of Drosophila da neurons [54]. Genetic experiments confirmed that the initiator caspase Dronc (caspase-9 ortholog) is cell autonomously required for dendrite pruning [53, 54]. Dronc activation is mediated by the degradation of its inhibitor DIAP1 [53], via the UPS. In mammals, caspases 3, 6 and 9 were shown to be required for trophic-deprivation mediated axon fragmentation of sensory and sympathetic neurons and similarly to flies XIAP1 was suggested to inhibit capses-3 activity. [44, 89, 92, 93]. Caspase-3 and 6 also regulate developmental pruning of retinal ganglion cells (RGCs) in the superior colliculus [44]. In both mammals and flies the mechanism by which caspase activation spares the death of the neuron and how localized activation is achieved are not well understood. Interestingly, sub-threshold non-apoptotic functions of the apoptotic machinery are now becoming evident in various vital cellular processes [94–96]. Importantly, the studies on the role of the apoptotic system in the axonal pruning have pointed out that although cell death and pruning share many elements they are not identical [91, 97].
What could be the underlying cause for the activation of the axonal apoptotic system in dying back neurodegeneration? The prime suspect is mitochondrial damage and dysfunction leading to a cellular energy crisis [98]. Indeed, local induced mitochondrial damage results in local caspase activation leading to dendrite pruning of hippocampal cultured neurons [99]. Furthermore, mitochondria are affected in almost all forms of peripheral neuropathy. Abnormal mitochondria in sensory neurons and axons were detected in murine models of both type I and type II diabetes [100, 101]. Several chemotherapy drugs such as cisplatin and oxaliplatin generate DNA damage and impair mitochondrial function [102]. The taxane family of chemotherapeutics functions by inhibiting tubulin dynamics which leads to impaired axonal mitochondrial trafficking, and subsequently to accumulation of damaged mitochondria [103]. These damaged mitochondria exhibit increased calcium permeability that can result in activation of the apoptotic system. Mitochondria dysfunction is strongly associated with some cases CMT as well. Mutations in the mitochondrial proteins ganglioside-induced differentiation-associated protein 1 (GDAP1) and mitofusin 2 (MFN2), account for different forms of CMT including the most severe cases of CMT2. In both cases the mutations are associated with impairment of mitochondrial activity and calcium homeostasis, and accumulation of axonal defective mitochondria [104, 105].
In ALS, accumulating evidence points to mitochondria playing a larger role in the maintenance of neuronal health, possibly related to trafficking. The aberrant activity and localization of mutant SOD1 can directly affect mitochondrial health and function. Three mechanisms that may play a part in SOD1-related mitochondria toxicity are translocation into the intermembrane space, activity control of SOD1 within the IMS and altered mitochondrial permeability [106, 107].
An additional potential role for mitochondria relates to calcium. Impaired calcium homeostasis, sometimes combined with caspase activation, is known to result in activation of the calcium dependent proteases calpains through the degradation of their intrinsic inhibitor calpastatin. Caplain was found to regulate developmental pruning of retinal ganglion cell axons and axon degeneration in response to trophic deprivation [108]. Furthermore, calcium transients appear to precede dendrite pruning of Drosophila da neurons that likely involves calpain activation [109]. In agreement, calpain inhibition seems to protect against chemotherapy-induced neuropathy [110].
Overall, we think that the accumulating evidence about the non-apoptotic roles of caspases during neuronal remodeling and in other cellular processes warrants reinvestigation of the role of caspase activation during the early phases of dying back neurodegenerative diseases (Figure 2). New tools, such as reporters for caspase activity, as well as more refined and specific genetic perturbations should be useful in clarifying the role of caspases in dying back neurodegenerative diseases.
Neuron-glia interactions
Neuroinflammation is a hallmark of many neurodegenerative diseases including ALS [111, 112]. Neuroinflammation is mostly a consequence of the function of activated microglia and astrocytes, suggesting that they might play an active role in the progression of dying back neurodegenerative diseases. To explore the possible contribution of non-cell-autonomous factors, Clement and colleagues [113] have created chimeric SOD1 mouse models. In these animals, only part of the cells expressed mutant SOD1 while other cells were wild type. Apparently, the existence of non-neuronal wild type cells resulted in extended survival of these ALS mouse models [113]. Furthermore, by ‘floxing’ the mutant SOD1 gene, Boillée and colleagues [114] created a mouse model in which mutant SOD1 expression was exclusively eliminated from microglila. Interestingly, they found that while the onset of neurodegeneration was not affected by excising the mutant SOD1 gene in microglia [114], or in astrocytes [115], disease progression was significantly delayed, suggesting that microglia and astrocytes play a crucial role in disease progression. Finally, mouse astrocytes expressing mutant SOD1 as well as patient derived astrocytes were shown to secrete toxic factors that specifically affect motor neurons [116, 117].
Another link to the potential role of astrocytes in ALS progression is the fact that expression of the excitatory amino acid transporter 2 (EAAT2), which is primarily expressed in astrocytes, is reduced in ALS patients as well as in other neurodegenerative diseases [118]. Reduced activity of glial EAAT2 might lead to increased neuronal glutamatergic excitotoxicity. Indeed, driving the expression of transgenic EAAT2 delays the onset of ALS in SOD1 mice models [119]. Therefore, it seems that mutant glia might play a dual role in disease progression – on one hand, they seem to provide less protection against excitotoxicity but on the other hand, the seem to actively secrete toxic factors [120].
Glia-neuron interaction also plays important roles during neuronal remodeling. In mammals, glia have been shown to be required for small scale synapse remodeling [121]. Microglia were shown to play a key role in synapse elimination in the hippocampus [122] and in the thalamus [123]. Recently, it was also shown that astrocytes are also required for normal synapse remodeling in the thalamus [124] suggesting that the function of microglia and astrocytes complement each other. In Drosophila, glia have been implicated in multiple steps of neuronal remodeling. First, an elegant study has found that cortex glia instruct MB axon pruning by secreting the TGF-β ligand myoglianin [125]. Second, axonal MB fragments are engulfed by glia and processed by the lysosomal degradation pathway [126, 127]. A later study has found that fragment engulfment depends glial expression of the engulfment receptor Draper (the Drosophila Ced-1 ortholog) [126–129]. Recently, it was shown that astrocytes are the main glial type required for fragment clearance despite their low numbers compared to other glial cells [130, 131].
Given the critical role of mammalian Schwann cells for the metabolic homeostasis and maintenance of peripheral axons through trophic support and myelination, it comes as no surprise that Schwann cell dysfunction contributes to peripheral neuropathy. CMT1 mostly results from mutations in myelin-associated genes, such as PMP22, and MPZ (for a more compressive list see [25, 132]). Likewise, other genetic lesions or toxic neuropathic environment that directly damage the nerve might also be deleterious to the Schwann cell, resulting in defects in both cell types. The contribution and crosstalk of the neuronal and glial defects in neuropathies are not well understood [25].
The fact that glia-neuron interactions play key roles in both neurodegenerative diseases and in developmental remodeling (Figure 2) is perhaps not surprising and does not in itself provide evidence for convergent mechanisms. However, the fact that intimate neuron-glia communication affects both development and disease highlights the importance of more mechanistic studies to uncover common mechanisms. For example, recent work has shown that the microglia mediate early synapse loss in Alzheimer’s mouse models by similar molecular mechanisms by which they mediate developmental synapse pruning [133, 134].
Balance between degeneration and regenerative forces
In his seminal review from 1964 Cavanagh wrote: “…the clinical consequences are seen only when the compensatory forces begin to fail in the face of cumulative nerve fiber damage”. What are these compensatory forces and are they involved in a ‘tug-of-war’ with degenerative forces?
This proposed tug-of-was predicts that failure of compensatory pathways should hasten and promote disease progression. Indeed, measurements of axonal regeneration in response to capsaicin-induced denervation of sensory axons were reduced in diabetic patients [135]. Likewise, in vitro studies using tissues from diabetic or healthy mice have suggested that skin-derived axon growth prompting factors, other than NGF, are reduced early after diabetes induction, providing another potential mechanism to the reduced growth potential of these neurons [136]. Finally, ALS causing mutations in Profilin-1 have been identified in human patients [137]. Profilin-1 is a positive regulator of actin nucleation and therefore plays multiple roles during neuronal development including axon growth [138]. Interestingly, axonal transport of the Drosophila profilin mRNA, chic, has been shown to be required for developmental axon regrowth of MB neurons [139].
Axon repulsive cues that restrict axonal growth and induce pruning during development may inhibit the regenerative response as well. An elegant study that began by screening for modifiers of ALS in a zebrafish model but extended the findings to human patients, found that Eph receptor A4 (EphA4) expression is conversely correlated with disease onset and progression [140]. Eph receptors mediate axon repulsion in response to its ligands from the ephrin family and also induce pruning of hippocampal axons during development [141, 142]. Indeed, neurons that express high levels of EphA4 show high susceptibility to ALS causing mutations. Remarkably, knocking down EphA4 in animal models confers relative resistance and delays the onset of disease [140]. Interestingly, another repellent guidance-receptor molecule pair, Plexin-Semaphorin, is known to repel sensory axons and to induce the developmental axon pruning of corticospinal and hippocampal axons [143, 144]. The Semaphorins were also found to be expressed in the adult skin and were shown to inhibit growth of sensory axons in pathological conditions [145]. Therefore, receptors of repellent guidance molecules such as the Semaphorins and Ephrins may play a role in both developmental remodeling and dying back neurodegeneration.
Nitric Oxide (NO) could be an additional player in this tug-of-war. NO is secreted from both microglia and astrocytes [146] and its levels may rise due to mitochondrial dysfunction that is highly prevalence in neuropathy as discussed above. NO was proposed to promote neurodegeneration by multiple possible mechanisms including S-nitrosylation [147, 148]. However, low levels of NO have been shown to support viability and promote growth [147–149]. Interestedly, NO levels have recently been shown to function as a switching mechanism between pruning and regrowth of MB axon in Drosophila [150]. While high NO levels promote pruning by a mechanism that is not well understood, its levels drop rapidly to facilitate developmental axon regrowth via a nuclear receptor complex and the target of Rapamycin (TOR) pathway. Interestingly, the mTOR complex has been shown to positively regulate axon regeneration following injury in mammals as well [151]. Remarkably, mTOR was shown to protect motor neurons expressing mutant SOD1 [152]. Therefore, mechanisms that promote developmental axon regrowth and regeneration following injury might provide neuroprotection to neurons exposed to toxic and pathologic conditions. Whether this ‘tug-of-war’ reflects a genuine convergence between remodeling and disease mediated neurodegeneration remains to be mechanistically investigated.
Concluding remarks
While there has been significant progress in understanding the genetic basis of several dying-back neurodegenerative diseases, such as ALS and peripheral neuropathies, the mechanistic features of these diseases is mostly unknown. Many of the disease mutations are dominantly inherited and therefore might result in a gain of function defect. Likewise, expressing mutant genes in vertebrate and invertebrate models has provided many insights but has not yet been instrumental in providing a mechanistic understanding of these diseases. One reason is that in these models, the expression levels of the mutant genes are often at non-physiological levels which may result in pleiotropic defects, thus making it difficult to find the causal events leading to the disease.
The field of developmental neuronal remodeling has expanded significantly in recent years, many new models have been employed and new concepts and mechanistic understanding have emerged. Nevertheless, we still have much to explore and discover before we can begin to format a comprehensive view of the mechanisms underlying remodeling. Most importantly, a mechanistic understanding of how axons are dismantled during development is still lacking.
In this article, we have explored potential convergence points between what we know about mechanisms regulating developmental remodeling and those associated with dying-back neurodegenerative diseases. Uncovering the mechanisms that drive pathological neurodegeneration during disease is highly changeling. One of the reasons is the fact that disease-causing mutations are complex, involving cell-autonomous and non-cell-autonomous effects. Additionally, the disease-causing mutations must be viable for the organism, or patient, to undergo normal development and therefore often do not reflect a complete loss-of-function but rather hypomorphic mutations or gain-of-function mutations. This means that often mutations are subtle or counterintuitive, especially when involving ‘house keeping genes’, complicating the interpretation of the underlying mechanisms. Furthermore, pathological processes are by nature much less stereotypic than developmental processes. In contrast to disease, developmental remodeling is almost exclusively studied in genetic model organisms under controlled conditions. Furthermore, studying neuronal remodeling involves looking for genes that are required for degeneration, quite the opposite from what is done in research focusing on disease. In other words, those studying disease largely focus on spontaneous mutations that cause neurodegeneration while those studying developmental remodeling largely focus on genetic perturbations that inhibit degeneration. We believe that these differences are at the core of the very little intellectual exchange between disease and developmental axon elimination, but also highlight the potential of more crosstalk between developmental and disease oriented studies. While the similarities discussed in this review are often circumstantial and speculative, we think that there is ample evidence that common mechanisms at certain stages are at least possible. Taken together, we believe that mechanisms that have evolved to dismantle axons and dendrites during normal development have the potential to shed new light about what happens when neuronal structure is comprised during disease and vice versa. Therefore, we believe that more crosstalk between the two fields holds the potential for significant breakthroughs in both.
Acknowledgments
We thank E. Perlson and S. Yaniv for helpful comments on the manuscript and Z. Schoenmann for assistance with the figures. Work in our laboratories is mainly funded by grants from the Israeli Science foundation (ISF) and the Minerva foundation (AY and OS), and the European Research Council (erc) (OS). AY is an incumbent of the Jack & Simon Djanogly Professorial Chair in Biochemistry. O.S. is an incumbent of the Rothstein Career Development Chair of Genetic Diseases.
References
- 1.Yamaguchi Y, Miura M. Programmed Cell Death in Neurodevelopment. Dev Cell. 2015;32:478–490. doi: 10.1016/j.devcel.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Cowan WM, Fawcett JW, O'Leary DD, Stanfield BB. Regressive events in neurogenesis. Science. 1984;225:1258–1265. doi: 10.1126/science.6474175. [DOI] [PubMed] [Google Scholar]
- 3.Truman JW. Metamorphosis of the central nervous system of Drosophila. J Neurobiol. 1990;21:1072–1084. doi: 10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- 4.Truman JW, Reiss SE. Dendritic reorganization of an identified motoneuron during metamorphosis of the tobacco hornworm moth. Science. 1976;192:477–479. doi: 10.1126/science.1257782. [DOI] [PubMed] [Google Scholar]
- 5.Purves D, Lichtman JW. Elimination of synapses in the developing nervous system. Science. 1980;210:153–157. doi: 10.1126/science.7414326. [DOI] [PubMed] [Google Scholar]
- 6.Innocenti GM. Growth and reshaping of axons in the establishment of visual callosal connections. Science. 1981;212:824–827. doi: 10.1126/science.7221566. [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner O, Yaron A. Mechanisms of developmental neurite pruning. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramon y Cajal S. Histology of the Nervous System of Man and Vertebrates. Trans.Oxford Univ. Press; 1995. [Google Scholar]
- 9.Rosenthal R. Of schizophrenia, pruning, and epigenetics: A hypothesis and suggestion. Medical hypotheses. 2011 doi: 10.1016/j.mehy.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 10.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas MSC, Davis R, Karmiloff-Smith A, Knowland VCP, Charman T. The over-pruning hypothesis of autism. Dev Sci. 2016;19:284–305. doi: 10.1111/desc.12303. [DOI] [PubMed] [Google Scholar]
- 12.Tang G, Gudsnuk K, Kuo S-H, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, et al. Loss of mTOR-Dependent Macroautophagy Causes Autistic-like Synaptic Pruning Deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu F, Schuldiner O. Axon and dendrite pruning in Drosophila. Curr Opin Neurobiol. 2014;27:192–198. doi: 10.1016/j.conb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riccomagno MM, Kolodkin AL. Sculpting Neural Circuits by Axon and Dendrite Pruning. Annu Rev Cell Dev Biol. 2015;31:779–805. doi: 10.1146/annurev-cellbio-100913-013038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanamori T, Togashi K, Koizumi H, Emoto K. Dendritic Remodeling: Lessons from Invertebrate Model Systems. Int Rev Cell Mol Biol. 2015;318:1–25. doi: 10.1016/bs.ircmb.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Raff MC, Whitmore AV, Finn JT. Axonal self-destruction and neurodegeneration. Science. 2002;296:868–871. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- 17.Ferri A, Sanes JR, Coleman MP, Cunningham JM, Kato AC. Inhibiting axon degeneration and synapse loss attenuates apoptosis and disease progression in a mouse model of motoneuron disease. Curr Biol. 2003;13:669–673. doi: 10.1016/s0960-9822(03)00206-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang MS, Davis AA, Culver DG, Glass JD. WldS mice are resistant to paclitaxel (taxol) neuropathy. Ann Neurol. 2002;52:442–447. doi: 10.1002/ana.10300. [DOI] [PubMed] [Google Scholar]
- 19.Coleman MP, Freeman MR. Wallerian degeneration, wld(s), and nmnat. Annu Rev Neurosci. 2010;33:245–267. doi: 10.1146/annurev-neuro-060909-153248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman MR. Signaling mechanisms regulating Wallerian degeneration. Curr Opin Neurobiol. 2014;27C:224–231. doi: 10.1016/j.conb.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer LR, Culver DG, Davis AA, Tennant P, Wang M, Coleman M, Asress S, Adalbert R, Alexander GM, Glass JD. The WldS gene modestly prolongs survival in the SOD1G93A fALS mouse. Neurobiol Dis. 2005;19:293–300. doi: 10.1016/j.nbd.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Vande Velde C, Garcia ML, Yin X, Trapp BD, Cleveland DW. The neuroprotective factor Wlds does not attenuate mutant SOD1-mediated motor neuron disease. Neuromolecular Med. 2004;5:193–203. doi: 10.1385/NMM:5:3:193. [DOI] [PubMed] [Google Scholar]
- 23.Gerdts J, Summers DW, Milbrandt J, DiAntonio A. Axon Self-Destruction: New Links among SARM1, MAPKs, and NAD+ Metabolism. Neuron. 2016;89:449–460. doi: 10.1016/j.neuron.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conforti L, Gilley J, Coleman MP. Wallerian degeneration:an emerging axon death pathwaylinking injury and disease. Nat Rev Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 25.Cashman CR, Höke A. Mechanisms of distal axonal degeneration in peripheral neuropathies. Neurosci Lett. 2015;596:33–50. doi: 10.1016/j.neulet.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena S, Caroni P. Mechanisms of axon degeneration: from development to disease. Prog Neurobiol. 2007;83:174–191. doi: 10.1016/j.pneurobio.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Cavanagh JB. The Significance of the “Dying Back” process in experimental and human neurological disease. Int Rev Exp Pathol. 1964:1–49. [PubMed] [Google Scholar]
- 28.Moloney EB, de Winter F, Verhaagen J. ALS as a distal axonopathy: molecular mechanisms affecting neuromuscular junction stability in the presymptomatic stages of the disease. Front Neurosci. 2014;8:252. doi: 10.3389/fnins.2014.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer LR, Culver DG, Tennant P, Davis AA, Wang M, Castellano-Sanchez A, Khan J, Polak MA, Glass JD. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp Neurol. 2004;185:232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 30.Renton AE, Chiò A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17:17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters OM, Ghasemi M, Brown RH. Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 2015;125:1767–1779. doi: 10.1172/JCI71601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- 33.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 34.Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 35.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling S-C, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brewer JR, Morrison G, Dolan ME, Fleming GF. Chemotherapy-induced peripheral neuropathy: Current status and progress. Gynecol Oncol. 2016;140:176–183. doi: 10.1016/j.ygyno.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent AM, Calabek B, Roberts L, Feldman EL. Biology of diabetic neuropathy. Handb Clin Neurol. 2013;115:591–606. doi: 10.1016/B978-0-444-52902-2.00034-5. [DOI] [PubMed] [Google Scholar]
- 40.Tazir M, Bellatache M, Nouioua S, Vallat J-M. Autosomal recessive Charcot-Marie-Tooth disease: from genes to phenotypes. J Peripher Nerv Syst. 2013;18:113–129. doi: 10.1111/jns5.12026. [DOI] [PubMed] [Google Scholar]
- 41.Tazir M, Hamadouche T, Nouioua S, Mathis S, Vallat J-M. Hereditary motor and sensory neuropathies or Charcot-Marie-Tooth diseases: an update. Journal of the Neurological Sciences. 2014;347:14–22. doi: 10.1016/j.jns.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Harrington AW, Ginty DD. Long-distance retrograde neurotrophic factor signalling in neurons. Nat Rev Neurosci. 2013;14:177–187. doi: 10.1038/nrn3253. [DOI] [PubMed] [Google Scholar]
- 43.Campenot RB. Development of sympathetic neurons in compartmentalized cultures. II. Local control of neurite survival by nerve growth factor. Dev Biol. 1982;93:13–21. doi: 10.1016/0012-1606(82)90233-0. [DOI] [PubMed] [Google Scholar]
- 44.Simon DJ, Weimer RM, McLaughlin T, Kallop D, Stanger K, Yang J, O'Leary DDM, Hannoush RN, Tessier-Lavigne M. A caspase cascade regulating developmental axon degeneration. Journal of Neuroscience. 2012;32:17540–17553. doi: 10.1523/JNEUROSCI.3012-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Rutlin M, Huang S, Barrick CA, Wang F, Jones KR, Tessarollo L, Ginty DD. Sexually dimorphic BDNF signaling directs sensory innervation of the mammary gland. Science. 2012;338:1357–1360. doi: 10.1126/science.1228258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson EM, Rich KM, Yip HK. The role of NGF in sensory neurons in vivo. Trends Neurosci. 1986 [Google Scholar]
- 47.Diamond J, Coughlin M, Macintyre L, Holmes M, Visheau B. Evidence that endogenous beta nerve growth factor is responsible for the collateral sprouting, but not the regeneration, of nociceptive axons in adult rats. Proc Natl Acad Sci USA. 1987;84:6596–6600. doi: 10.1073/pnas.84.18.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anand P, Terenghi G, Warner G, Kopelman P, Williams-Chestnut RE, Sinicropi DV. The role of endogenous nerve growth factor in human diabetic neuropathy. Nat Med. 1996;2:703–707. doi: 10.1038/nm0696-703. [DOI] [PubMed] [Google Scholar]
- 49.Hellweg R, Raivich G, Hartung HD, Hock C, Kreutzberg GW. Axonal transport of endogenous nerve growth factor (NGF) and NGF receptor in experimental diabetic neuropathy. Exp Neurol. 1994;130:24–30. doi: 10.1006/exnr.1994.1181. [DOI] [PubMed] [Google Scholar]
- 50.Apfel SC. Nerve growth factor for the treatment of diabetic neuropathy: what went wrong, what went right, and what does the future hold? Int Rev Neurobiol. 2002;50:393–413. doi: 10.1016/s0074-7742(02)50083-0. [DOI] [PubMed] [Google Scholar]
- 51.Pun S, Santos AF, Saxena S, Xu L, Caroni P. Selective vulnerability and pruning of phasic motoneuron axons in motoneuron disease alleviated by CNTF. Nat Neurosci. 2006;9:408–419. doi: 10.1038/nn1653. [DOI] [PubMed] [Google Scholar]
- 52.Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 53.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 Ubiquitinating Enzymes and Caspase Activity Regulating Drosophila Sensory Neuron Dendrite Pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 55.Wong JJL, Li S, Lim EKH, Wang Y, Wang C, Zhang H, Kirilly D, Wu C, Liou Y-C, Wang H, et al. A Cullin1-based SCF E3 ubiquitin ligase targets the InR/PI3K/TOR pathway to regulate neuronal pruning. PLoS Biol. 2013;11:e1001657. doi: 10.1371/journal.pbio.1001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhai Q, Wang J, Kim A, Liu Q, Watts R, Hoopfer E, Mitchison T, Luo L, He Z. Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron. 2003;39:217–225. doi: 10.1016/s0896-6273(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 57.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2012;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deng H-X, Chen W, Hong S-T, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, Zheng JG, Shi Y, Siddique N, Arrat H, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 60.Wu Q, Liu M, Huang C, Liu X, Huang B, Li N, Zhou H, Xia X-G. Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol. 2015;129:417–428. doi: 10.1007/s00401-014-1367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartolome F, Wu H-C, Burchell VS, Preza E, Wray S, Mahoney CJ, Fox NC, Calvo A, Canosa A, Moglia C, et al. Pathogenic VCP Mutations Induce Mitochondrial Uncoupling and Reduced ATP Levels. Neuron. 2013;78:57–64. doi: 10.1016/j.neuron.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim NC, Tresse E, Kolaitis R-M, Molliex A, Thomas RE, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, et al. VCP Is Essential for Mitochondrial Quality Control by PINK1/Parkin and this Function Is Impaired by VCP Mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rumpf S, Lee SB, Jan LY, Jan YN. Neuronal remodeling and apoptosis require VCP-dependent degradation of the apoptosis inhibitor DIAP1. Development (Cambridge, England) 2011;138:1153–1160. doi: 10.1242/dev.062703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirilly D, Gu Y, Huang Y, Wu Z, Bashirullah A, Low B, Kolodkin A, Wang H, Yu F. A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat Neurosci. 2009 doi: 10.1038/nn.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rumpf S, Bagley JA, Thompson-Peer KL, Zhu S, Gorczyca D, Beckstead RB, Jan LY, Jan YN. Drosophila Valosin-Containing Protein is required for dendrite pruning through a regulatory role in mRNA metabolism. Proc Natl Acad Sci USA. 2014;111:7331–7336. doi: 10.1073/pnas.1406898111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Issman-Zecharya N, Schuldiner O. The PI3K Class III Complex Promotes Axon Pruning by Downregulating a Ptc-Derived Signalvia Endosome-Lysosomal Degradation. Dev Cell. 2014;31:461–473. doi: 10.1016/j.devcel.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 68.Zhang H, Wang Y, Wong JJL, Lim K-L, Liou Y-C, Wang H, Yu F. Endocytic Pathways Downregulate the L1-type Cell Adhesion Molecule Neuroglian to Promote Dendrite Pruning in Drosophila. Dev Cell. 2014;30:463–478. doi: 10.1016/j.devcel.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 69.Loncle N, Agromayor M, Martin-Serrano J, Williams DW. An ESCRT module is required for neuron pruning. Sci Rep. 2015;5:8461. doi: 10.1038/srep08461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parkinson N, Ince PG, Smith MO, Highley R, Skibinski G, Andersen PM, Morrison KE, Pall HS, Hardiman O, Collinge J, et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B) Neurology. 2006;67:1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 71.Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, Everett L, Lenk GM, McKenna-Yasek DM, Weisman LS, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang K, Fishel Ben Kenan R, Osakada Y, Xu W, Sinit RS, Chen L, Zhao X, Chen J-Y, Cui B, Wu C. Defective axonal transport of Rab7 GTPase results in dysregulated trophic signaling. J Neurosci. 2013;33:7451–7462. doi: 10.1523/JNEUROSCI.4322-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spinosa MR, Progida C, De Luca A, Colucci AMR, Alifano P, Bucci C. Functional characterization of Rab7 mutant proteins associated with Charcot-Marie-Tooth type 2B disease. J Neurosci. 2008;28:1640–1648. doi: 10.1523/JNEUROSCI.3677-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janssens K, Goethals S, Atkinson D, Ermanoska B, Fransen E, Jordanova A, Auer-Grumbach M, Asselbergh B, Timmerman V. Human Rab7 mutation mimics features of Charcot-Marie-Tooth neuropathy type 2B in Drosophila. Neurobiol Dis. 2014;65:211–219. doi: 10.1016/j.nbd.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 75.Chin Y-H, Lee A, Kan H-W, Laiman J, Chuang M-C, Hsieh S-T, Liu Y-W. Dynamin-2 mutations associated with centronuclear myopathy are hypermorphic and lead to T-tubule fragmentation. Hum Mol Genet. 2015;24:5542–5554. doi: 10.1093/hmg/ddv285. [DOI] [PubMed] [Google Scholar]
- 76.Liu Y-W, Lukiyanchuk V, Schmid SL. Common membrane trafficking defects of disease-associated dynamin 2 mutations. Traffic. 2011;12:1620–1633. doi: 10.1111/j.1600-0854.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicholson G, Lenk GM, Reddel SW, Grant AE, Towne CF, Ferguson CJ, Simpson E, Scheuerle A, Yasick M, Hoffman S, et al. Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P2 phosphatase FIG4. Brain. 2011;134:1959–1971. doi: 10.1093/brain/awr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin LJ. Neuronal death in amyotrophic lateral sclerosis is apoptosis: possible contribution of a programmed cell death mechanism. J Neuropathol Exp Neurol. 1999;58:459–471. doi: 10.1097/00005072-199905000-00005. [DOI] [PubMed] [Google Scholar]
- 80.Li M, Ona VO, Guégan C, Chen M, Jackson-Lewis V, Andrews LJ, Olszewski AJ, Stieg PE, Lee JP, Przedborski S, et al. Functional role of caspase-1 and caspase-3 in an ALS transgenic mouse model. Science. 2000;288:335–339. doi: 10.1126/science.288.5464.335. [DOI] [PubMed] [Google Scholar]
- 81.Perrin FE. No widespread induction of cell death genes occurs in pure motoneurons in an amyotrophic lateral sclerosis mouse model. Hum Mol Genet. 2005;14:3309–3320. doi: 10.1093/hmg/ddi357. [DOI] [PubMed] [Google Scholar]
- 82.Kostic V, Jackson-Lewis V, de Bilbao F, Dubois-Dauphin M, Przedborski S. Bcl-2: prolonging life in a transgenic mouse model of familial amyotrophic lateral sclerosis. Science. 1997;277:559–562. doi: 10.1126/science.277.5325.559. [DOI] [PubMed] [Google Scholar]
- 83.Gould TW, Buss RR, Vinsant S, Prevette D, Sun W, Knudson CM, Milligan CE, Oppenheim RW. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J Neurosci. 2006;26:8774–8786. doi: 10.1523/JNEUROSCI.2315-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reyes NA, Fisher JK, Austgen K, VandenBerg S, Huang EJ, Oakes SA. Blocking the mitochondrial apoptotic pathway preserves motor neuron viability and function in a mouse model of amyotrophic lateral sclerosis. J Clin Invest. 2010;120:3673–3679. doi: 10.1172/JCI42986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng C, Zochodne DW. Sensory neurons with activated caspase-3 survive long-term experimental diabetes. Diabetes. 2003;52:2363–2371. doi: 10.2337/diabetes.52.9.2363. [DOI] [PubMed] [Google Scholar]
- 86.McDonald ES, Windebank AJ. Cisplatin-induced apoptosis of DRG neurons involves bax redistribution and cytochrome c release but not fas receptor signaling. Neurobiol Dis. 2002;9:220–233. doi: 10.1006/nbdi.2001.0468. [DOI] [PubMed] [Google Scholar]
- 87.McDonald ES, Randon KR, Knight A, Windebank AJ. Cisplatin preferentially binds to DNA in dorsal root ganglion neurons in vitro and in vivo: a potential mechanism for neurotoxicity. Neurobiol Dis. 2005;18:305–313. doi: 10.1016/j.nbd.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 88.Nikolaev A, McLaughlin T, O'Leary DDM, Tessier-Lavigne M. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature. 2009;457:981–989. doi: 10.1038/nature07767. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Schoenmann Z, Assa-Kunik E, Tiomny S, Minis A, Haklai-Topper L, Arama E, Yaron A. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J Neurosci. 2010;30:6375–6386. doi: 10.1523/JNEUROSCI.0922-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Courchesne SL, Karch C, Pazyra-Murphy MF, Segal RA. Sensory neuropathy attributable to loss of Bcl-w. J Neurosci. 2011;31:1624–1634. doi: 10.1523/JNEUROSCI.3347-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maor-Nof M, Yaron A. Neurite pruning and neuronal cell death: spatial regulation of shared destruction programs. Curr Opin Neurobiol. 2013;23:990–996. doi: 10.1016/j.conb.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 92.Unsain N, Higgins JM, Parker KN, Johnstone AD, Barker PA. XIAP Regulates Caspase Activity in Degenerating Axons. Cell Rep. 2013;4:751–763. doi: 10.1016/j.celrep.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 93.Cusack CL, Swahari V, Hampton Henley W, Michael Ramsey J, Deshmukh M. Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat Commun. 2013;4:1876. doi: 10.1038/ncomms2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feinstein-Rotkopf Y, Arama E. Can't live without them, can live with them: roles of caspases during vital cellular processes. Apoptosis. 2009;14:980–995. doi: 10.1007/s10495-009-0346-6. [DOI] [PubMed] [Google Scholar]
- 95.Florentin A, Arama E. Caspase levels and execution efficiencies determine the apoptotic potential of the cell. J Cell Biol. 2012;196:513–527. doi: 10.1083/jcb.201107133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Unsain N, Barker PA. New Views on the Misconstrued: Executioner Caspases and Their Diverse Non-apoptotic Roles. Neuron. 2015;88:461–474. doi: 10.1016/j.neuron.2015.08.029. [DOI] [PubMed] [Google Scholar]
- 97.Simon DJ, Pitts J, Hertz NT, Yang J, Yamagishi Y, Olsen O, Tešić Mark M, Molina H, Tessier-Lavigne M. Axon Degeneration Gated by Retrograde Activation of Somatic Pro-apoptotic Signaling. Cell. 2016;164:1031–1045. doi: 10.1016/j.cell.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Erturk A, Wang Y, Sheng M. Local Pruning of Dendrites and Spines by Caspase-3-Dependent and Proteasome-Limited Mechanisms. J Neurosci. 2014;34:1672–1688. doi: 10.1523/JNEUROSCI.3121-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chowdhury SKR, Zherebitskaya E, Smith DR, Akude E, Chattopadhyay S, Jolivalt CG, Calcutt NA, Fernyhough P. Mitochondrial respiratory chain dysfunction in dorsal root ganglia of streptozotocin-induced diabetic rats and its correction by insulin treatment. Diabetes. 2010;59:1082–1091. doi: 10.2337/db09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Edwards JL, Quattrini A, Lentz SI, Figueroa-Romero C, Cerri F, Backus C, Hong Y, Feldman EL. Diabetes regulates mitochondrial biogenesis and fission in mouse neurons. Diabetologia. 2010;53:160–169. doi: 10.1007/s00125-009-1553-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Podratz JL, Knight AM, Ta LE, Staff NP, Gass JM, Genelin K, Schlattau A, Lathroum L, Windebank AJ. Cisplatin induced mitochondrial DNA damage in dorsal root ganglion neurons. Neurobiol Dis. 2011;41:661–668. doi: 10.1016/j.nbd.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Flatters SJL, Bennett GJ. Studies of peripheral sensory nerves in paclitaxel-induced painful peripheral neuropathy: evidence for mitochondrial dysfunction. Pain. 2006;122:245–257. doi: 10.1016/j.pain.2006.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Barneo-Muñoz M, Juárez P, Civera-Tregón A, Yndriago L, Pla-Martin D, Zenker J, Cuevas-Martín C, Estela A, Sánchez-Aragó M, Forteza-Vila J, et al. Lack of GDAP1 induces neuronal calcium and mitochondrial defects in a knockout mouse model of charcot-marie-tooth neuropathy. PLoS Genet. 2015;11:e1005115. doi: 10.1371/journal.pgen.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Strickland AV, Rebelo AP, Zhang F, Price J, Bolon B, Silva JP, Wen R, Züchner S. Characterization of the mitofusin 2 R94W mutation in a knock-in mouse model. J Peripher Nerv Syst. 2014;19:152–164. doi: 10.1111/jns5.12066. [DOI] [PubMed] [Google Scholar]
- 106.Vehviläinen P, Koistinaho J, Gundars G. Mechanisms of mutant SOD1 induced mitochondrial toxicity in amyotrophic lateral sclerosis. Front Cell Neurosci. 2014;8:126. doi: 10.3389/fncel.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu J, Lillo C, Jonsson PA, Vande Velde C, Ward CM, Miller TM, Subramaniam JR, Rothstein JD, Marklund S, Andersen PM, et al. Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria. Neuron. 2004;43:5–17. doi: 10.1016/j.neuron.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 108.Yang J, Weimer RM, Kallop D, Olsen O, Wu Z, Renier N, Uryu K, Tessier-Lavigne M. Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron. 2013;80:1175–1189. doi: 10.1016/j.neuron.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 109.Kanamori T, Kanai MI, Dairyo Y, Yasunaga K-I, Morikawa RK, Emoto K. Compartmentalized calcium transients trigger dendrite pruning in Drosophila sensory neurons. Science. 2013;340:1475–1478. doi: 10.1126/science.1234879. [DOI] [PubMed] [Google Scholar]
- 110.Wang MS, Davis AA, Culver DG, Wang Q, Powers JC, Glass JD. Calpain inhibition protects against Taxol-induced sensory neuropathy. Brain. 2004;127:671–679. doi: 10.1093/brain/awh078. [DOI] [PubMed] [Google Scholar]
- 111.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms Underlying Inflammation in Neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: role of glial activation in motor neuron disease. Lancet Neurol. 2011;10:253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- 113.Clement AM, Nguyen MD, Roberts EA, Garcia ML, Boillée S, Rule M, McMahon AP, Doucette W, Siwek D, Ferrante RJ, et al. Wild-type nonneuronal cells extend survival of SOD1 mutant motor neurons in ALS mice. Science. 2003;302:113–117. doi: 10.1126/science.1086071. [DOI] [PubMed] [Google Scholar]
- 114.Boillée S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, Kollias G, Cleveland DW. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 115.Yamanaka K, Chun SJ, Boillée S, Fujimori-Tonou N, Yamashita H, Gutmann DH, Takahashi R, Misawa H, Cleveland DW. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–253. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, Przedborski S. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–622. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Haidet-Phillips AM, Hester ME, Miranda CJ, Meyer K, Braun L, Frakes A, Song S, Likhite S, Murtha MJ, Foust KD, et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat Biotechnol. 2011;29:824–828. doi: 10.1038/nbt.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.King AE, Woodhouse A, Kirkcaldie MTK, Vickers JC. Excitotoxicity in ALS: Overstimulation, or overreaction? Exp Neurol. 2016;275(Pt 1):162–171. doi: 10.1016/j.expneurol.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 119.Guo H, Lai L, Butchbach MER, Stockinger MP, Shan X, Bishop GA, Lin C-LG. Increased expression of the glial glutamate transporter EAAT2 modulates excitotoxicity and delays the onset but not the outcome of ALS in mice. Hum Mol Genet. 2003;12:2519–2532. doi: 10.1093/hmg/ddg267. [DOI] [PubMed] [Google Scholar]
- 120.Philips T, Rothstein JD. Glial cells in amyotrophic lateral sclerosis. Exp Neurol. 2014;262(Pt B):111–120. doi: 10.1016/j.expneurol.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schafer DP, Stevens B. Phagocytic glial cells: sculpting synaptic circuits in the developing nervous system. Curr Opin Neurobiol. 2013;23:1034–1040. doi: 10.1016/j.conb.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic Pruning by Microglia Is Necessary for Normal Brain Development. Science. 2011 doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 123.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres Ben A, Stevens B. Microglia Sculpt Postnatal Neural Circuits in an Activity and Complement-Dependent Manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chung W-S, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Awasaki T, Huang Y, O'Connor MB, Lee T. Glia instruct developmental neuronal remodeling through TGF-β signaling. Nat Neurosci. 2011;14:821–823. doi: 10.1038/nn.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Awasaki T, Ito K. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol. 2004;14:668–677. doi: 10.1016/j.cub.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 127.Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L. Glia engulf degenerating axons during developmental axon pruning. Curr Biol. 2004;14:678–684. doi: 10.1016/j.cub.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 128.Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 129.Hoopfer ED, McLaughlin T, Watts RJ, Schuldiner O, O'Leary DDM, Luo L. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 130.Hakim Y, Yaniv SP, Schuldiner O. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLoS ONE. 2014;9:e86178. doi: 10.1371/journal.pone.0086178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tasdemir-Yilmaz OE, Freeman MR. Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 2014;28:20–33. doi: 10.1101/gad.229518.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nave K-A, Sereda MW, Ehrenreich H. Mechanisms of disease: inherited demyelinating neuropathies--from basic to clinical research. Nat Clin Pract Neurol. 2007;3:453–464. doi: 10.1038/ncpneuro0583. [DOI] [PubMed] [Google Scholar]
- 133.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016 doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 135.Polydefkis M, Hauer P, Sheth S, Sirdofsky M, Griffin JW, McArthur JC. The time course of epidermal nerve fibre regeneration: studies in normal controls and in people with diabetes, with and without neuropathy. Brain. 2004;127:1606–1615. doi: 10.1093/brain/awh175. [DOI] [PubMed] [Google Scholar]
- 136.Amit S, Yaron A. Novel systems for in vivo monitoring and microenvironmental investigations of diabetic neuropathy in a murine model. J Neural Transm (Vienna) 2012;119:1317–1325. doi: 10.1007/s00702-012-0808-9. [DOI] [PubMed] [Google Scholar]
- 137.Wu C-H, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, Lowe P, Koppers M, McKenna-Yasek D, Baron DM, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. Bioessays. 2008;30:994–1002. doi: 10.1002/bies.20822. [DOI] [PubMed] [Google Scholar]
- 139.Medioni C, Ramialison M, Ephrussi A, Besse F. Imp promotes axonal remodeling by regulating profilin mRNA during brain development. Curr Biol. 2014;24:793–800. doi: 10.1016/j.cub.2014.02.038. [DOI] [PubMed] [Google Scholar]
- 140.Van Hoecke A, Schoonaert L, Lemmens R, Timmers M, Staats KA, Laird AS, Peeters E, Philips T, Goris A, Dubois B, et al. EPHA4 is a disease modifier of amyotrophic lateral sclerosis in animal models and in humans. Nat Med. 2012;18:1418–1422. doi: 10.1038/nm.2901. [DOI] [PubMed] [Google Scholar]
- 141.Xu N-J, Henkemeyer M. Ephrin-B3 reverse signaling through Grb4 and cytoskeletal regulators mediates axon pruning. Nat Neurosci. 2009;12:268–276. doi: 10.1038/nn.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kania A, Klein R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2015.16. [DOI] [PubMed] [Google Scholar]
- 143.Bagri A, Cheng H-J, Yaron A, Pleasure SJ, Tessier-Lavigne M. Stereotyped pruning of long hippocampal axon branches triggered by retraction inducers of the semaphorin family. Cell. 2003;113:285–299. doi: 10.1016/s0092-8674(03)00267-8. [DOI] [PubMed] [Google Scholar]
- 144.Low LK, Liu X-B, Faulkner RL, Coble J, Cheng H-J. Plexin signaling selectively regulates the stereotyped pruning of corticospinal axons from visual cortex. Proc Natl Acad Sci USA. 2008;105:8136–8141. doi: 10.1073/pnas.0803849105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tominaga M, Tengara S, Kamo A, Ogawa H, Takamori K. Psoralen-ultraviolet A therapy alters epidermal Sema3A and NGF levels and modulates epidermal innervation in atopic dermatitis. J Dermatol Sci. 2009;55:40–46. doi: 10.1016/j.jdermsci.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 146.Puentes F, Malaspina A, van Noort JM, Amor S. Non-neuronal cells in ALS: Role of glial, immune cells and blood-CNS barriers. Brain Pathol. 2016 doi: 10.1111/bpa.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nakamura T, Tu S, Akhtar MW, Sunico CR, Okamoto S-I, Lipton SA. Aberrant Protein S-Nitrosylation in Neurodegenerative Diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brown GC. Nitric oxide and neuronal death. Nitric Oxide. 2010;23:153–165. doi: 10.1016/j.niox.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 149.Cárdenas A, Moro MA, Hurtado O, Leza JC, Lizasoain I. Dual role of nitric oxide in adult neurogenesis. Brain research reviews. 2005;50:1–6. doi: 10.1016/j.brainresrev.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 150.Rabinovich D, Yaniv SP, Alyagor I, Schuldiner O. Nitric Oxide as a Switching Mechanism between Axon Degeneration and Regrowth during Developmental Remodeling. Cell. 2016;164:170–182. doi: 10.1016/j.cell.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Saxena S, Roselli F, Singh K, Leptien K, Julien J-P, Gros-Louis F, Caroni P. Neuroprotection through excitability and mTOR required in ALS motoneurons to delay disease and extend survival. Neuron. 2013;80:80–96. doi: 10.1016/j.neuron.2013.07.027. [DOI] [PubMed] [Google Scholar]