Abstract
During development, neurons switch between growth states, such as initial axon outgrowth, axon pruning, and regrowth. By studying the stereotypic remodeling of the Drosophila mushroom body (MB), we found that the heme-binding nuclear receptor E75 is dispensable for initial axon outgrowth of MB γ neurons, but required for their developmental regrowth. Genetic experiments and pharmacological manipulations on ex-vivo cultured brains indicate that neuronally-generated nitric oxide (NO) promotes pruning but inhibits regrowth. We found that high NO levels inhibit the physical interaction between the E75 and UNF nuclear receptors, likely accounting for its repression of regrowth. Additionally, NO Synthase (NOS) activity is downregulated at the onset of regrowth, at least partially, by short inhibitory NOS isoforms encoded within the NOS locus, indicating how NO production could be developmentally regulated. Taken together, these results suggest that NO signaling provides a switching mechanism between the degenerative and regenerative states of neuronal remodeling.
Introduction
Neuronal remodeling is an evolutionarily conserved strategy used to refine neural circuits (Luo and O'Leary, 2005). Remodeling can include degenerative events, such as neurite pruning, as well as regrowth of axons and dendrites to form of new connections. Classical examples include the formation of ocular dominance columns in the mammalian visual cortex, refinement of visual projections at the superior colliculus and large scale axon elimination of layer 5 corticospinal neurons (Luo and O'Leary, 2005; Schuldiner and Yaron, 2015). Defective remodeling has been suggested to play a role in both schizophrenia and autism (Cocchi et al., 2015; Thomas et al., 2015). How neurons switch between developmental growth states, such as initial axon outgrowth, pruning and regrowth is a fundamental question that is mostly unknown.
The Drosophila mushroom body (MB) provides a unique platform to study the cellular and molecular aspects of remodeling due to its temporal and spatial stereotypy as well as the wide spectrum of genetic tools available. During metamorphosis, bifurcated axons of larval MB γ neurons prune up to the branching point and dendrites are completely eliminated, both of which later regrow to adult specific areas (Figure 1A). While our understanding of the molecular mechanisms underlying pruning has dramatically increased in the last decade, it is far from complete (Yu and Schuldiner, 2014). Moreover, developmental regrowth has only recently been identified as a unique, genetically regulated growth process that is distinct from initial axon outgrowth (Yaniv et al., 2012). MB γ neuron remodeling occurs within a very defined and short time window, suggesting the existence of a tightly regulated switch that occurs at the transition between pruning and regrowth. However, whether and how pruning and regrowth are co-regulated is currently unknown.
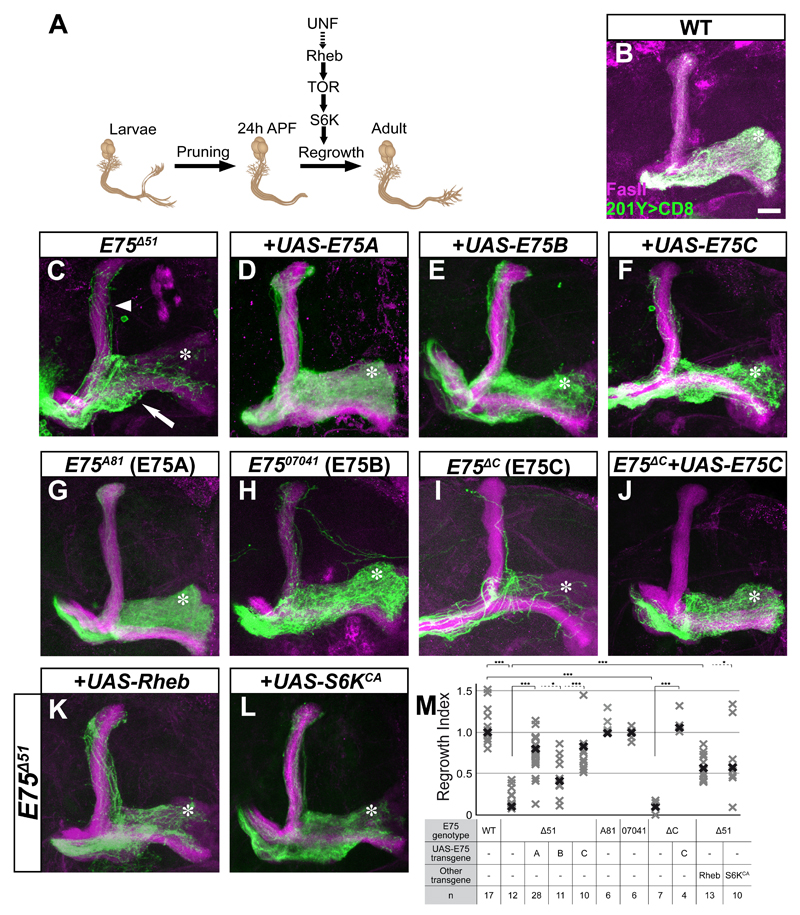
Figure 1. E75C is required for developmental axon regrowth.
(A) Scheme of MB neuronal remodeling in which UNF promotes developmental regrowth in a TOR pathway dependent process. Adapted from Yaniv et al, 2012.
(B-L) Confocal Z-projections of adult MARCM γ neuron MB neuroblast clones of the indicated genotypes labeled by 201Y-Gal driving the expression of membrane bound GFP (CD8-GFP). While WT (B) adult MB clones neurons fully innervate the medial lobe (asterisk) most E75Δ51 (C) axons stall at the branch point (arrow). The regrowth defect is rescued by expression of E75A (D), E75B (E) or E75C (F) within the mutant clone. An E75A specific deletion (E75A81, G) and an E75B specific insertion (E7507041, H) display a WT phenotype. An E75C specific deletion (E75ΔC, I) displays a strong regrowth defect that is rescued by UAS-E75C (J, see Figure S1A for allele information). E75Δ51 regrowth defect is partially suppressed by Rheb, a regulator of the TOR pathway (K) as does expression of S6KCA (L).
(M) Quantification of the developmental regrowth defects of the genotypes shown in B-L. Bold shapes represent the median in each group. One-way ANOVA was performed with Dunnett’s T3 post-hoc test *** p<0.001; *p<0.05
Magenta represents anti-FasII staining. Green is 201Y-Gal4 driven mCD8:GFP. Asterisks demarcate the distal tip of the medial adult γ lobe. The scale bar represents 20 μm.
See also Figure S1.
We have previously demonstrated that the nuclear receptor UNF (also known as Hr51 and Nr2e3) is required for developmental axon regrowth in a process that is mediated, at least in part, by the TOR pathway (Yaniv et al., 2012). Here we report that another nuclear receptor, E75 (Ecdysone induced protein 75B, Eip75B), is also required for developmental regrowth of MB γ axons but not for their initial outgrowth. The fact that E75 is attached to a heme moiety that can bind monovalent gases led us to investigate the role of nitric oxide (NO) during remodeling. We found that while NO synthase (NOS) promotes pruning of MB γ axons, NO levels must be attenuated to allow for UNF/E75 mediated axon regrowth to occur. Taken together, our study has identified NO as a switching mechanism between axon degeneration and regrowth during remodeling.
Results
The nuclear receptor E75 is required for developmental regrowth
We have previously shown that the orphan nuclear receptor UNF is required for the developmental regrowth of MB γ-axons following pruning but not for their initial outgrowth (Yaniv et al., 2012). The mammalian ortholog of UNF, photoreceptor specific nuclear receptor (PNR), has been shown to dimerize and function in vivo with another nuclear receptor, Rev-erb-α (Nr1d1; Cheng et al., 2004; Mollema et al., 2011). Therefore, we tested whether the closest Drosophila homolog of Rev-erb-α, E75, is also involved in developmental regrowth of MB γ neurons. We generated MARCM (Lee and Luo, 1999) clones that are positively labeled and homozygous mutant for E75 on a heterozygous background. We used the E75Δ51 allele, in which four out of the five major E75 protein isoforms are deleted (Figure S1A). Indeed, E75Δ51 mutant MARCM clones did not fully innervate the adult γ lobe (compare axons reaching to asterisk in Figure 1C to 1B, 1M). Interestingly, we also observed a weak pruning defect in some of the E75Δ51 MARCM clones (Figure 1C, arrowhead; see more later).
To determine which E75 isoform is required for full innervation of the adult γ lobe, we performed two complimentary experiments focusing on the three classical E75 proteins (E75A, E75B and E75C): 1) isoform specific rescue experiments within E75Δ51 MARCM clones; 2) isoform specific MARCM analysis. Overexpression of E75A, -B or -C (Figure 1D-F, S1B) transgenes within E75Δ51 MB clones all significantly rescued the regrowth defect (Figure 1M), suggesting functional redundancy between the different isoforms. Isoform specific mutation analysis revealed that MARCM clones homozygous for an E75A specific deletion (Figure 1G, S1A; E75A81) or E75B specific P-element insertion (Figure 1H, S1A; E7507041) display a WT phenotype (Figure 1M). In contrast, MARCM MB clones homozygous for an E75C specific deletion did not fully innervate the adult γ lobe, similar to E75Δ51 mutants (E75ΔC; Figure 1I, S1A, 1M). Interestingly, E75ΔC clones also contained fewer axons, a phenotype that seems unrelated to MB remodeling and is normalized in our quantifications. Importantly, overexpression of E75C within E75Δc MARCM clones completely rescued both regrowth and proliferation mutant phenotypes (Figure 1J, 1M).
Time course analyses of both E75Δ51 and E75ΔC mutants illustrated that γ axons initially extend normally (Figure S1C-E), but fail to undergo axon regrowth following pruning (Figure S1F-H). Taken together, these results suggest that E75C, like UNF, is required for developmental regrowth but not initial outgrowth of MB γ axons.
We next checked whether E75 promotes regrowth via the TOR pathway. Indeed, overexpression of Rheb, the upstream activator of TOR, significantly suppressed the regrowth defect of E75Δ51 mutants (Figure 1K, 1M) as did overexpression of the constitutively active form of the TOR downstream effector, S6 kinase (S6KCA, Figure 1L, 1M). This suggests that E75 and UNF both promote regrowth, at least partially, via the TOR pathway.
This finding suggests that, like in mice, these two nuclear receptors may act as a functional heterodimer (Cheng et al., 2004; Mollema et al., 2011). Genetic epistatic experiments are indeed consistent with this hypothesis: MB MARCM clones doubly mutant of unf and E75Δ51 did not display increased severity of regrowth defect as compared to each single mutant (Figure S1K). Likewise, the regrowth defect in unf mutant clones was not rescued by overexpression of E75 isoforms nor did overexpression of UNF within E75Δ51 mutant clones (Figure S1K). These results suggest that UNF and E75 function within the same pathway at the same level. Furthermore, we have previously shown that overexpression of UNF within MB neurons results in a severe pruning defect by an unknown mechanism (Figure S1I, Yaniv et al., 2012). This gain of function defect is suppressed by co-expression of E75C (Figure S1J) in addition to UNF, suggesting that the increased levels of E75C might sequester the high UNF protein levels, consistent with a model in which the two nuclear receptors physically interact. However, despite numerous attempts, we were unable to show that E75 and UNF form a complex in co-immunoprecipitation (co-IP) experiments from Drosophila whole cell lysates or nuclear extracts under basal conditions (see more below).
Taken together, our results suggest that E75C is required for developmental axon regrowth but not initial outgrowth, likely functioning together with UNF via the TOR pathway.
Reducing NO levels promotes developmental axon regrowth ex vivo
Heme has been proposed as an endogenous ligand of E75 (Reinking et al., 2005) and its mammalian homologs, Rev-erb-α and –β (Raghuram et al., 2007) as well as for UNF (De Rosny et al., 2008). Studies suggest that the heme bound to UNF (De Rosny et al., 2008) or E75 (Reinking et al., 2005) can bind NO and carbon monoxide (CO) in vitro. Furthermore, NO levels were found to modulate the activity of E75 in vivo by changing its affinity to binding partners (Caceres et al., 2011; Johnston et al., 2011), thus changing its’ transcriptional activity. We therefore wanted to determine whether NO is also important for the function of E75 and UNF in regulating axon regrowth.
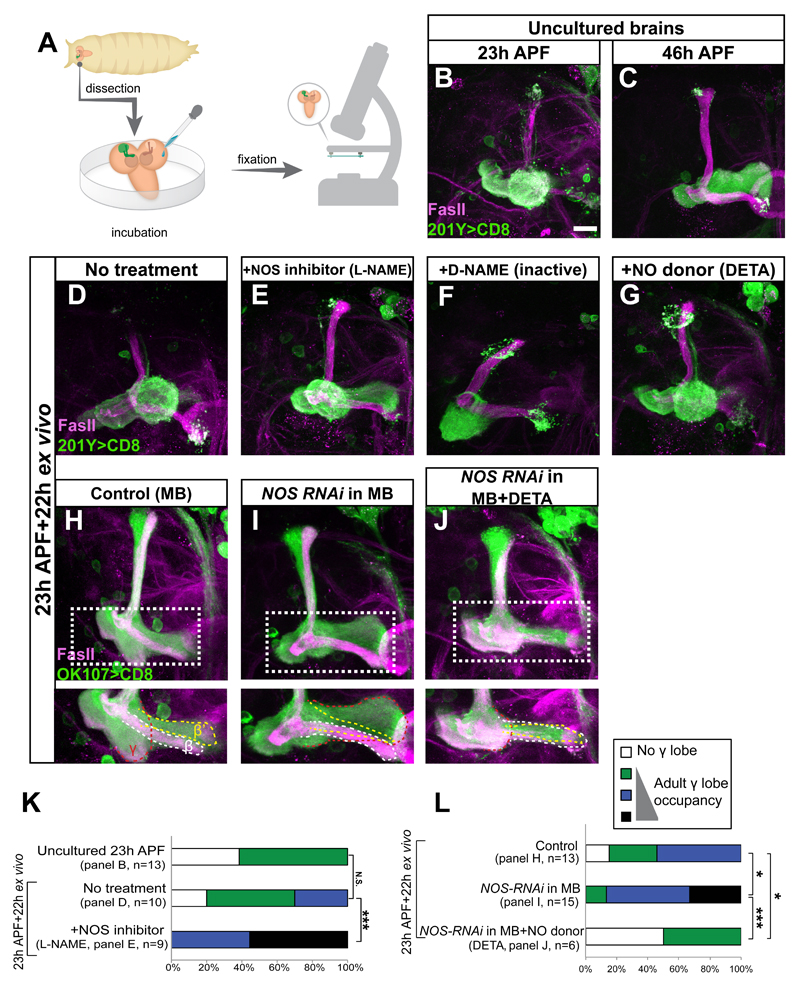
In order to determine whether NO levels affect developmental axon regrowth, we set up an ex vivo long-term culture system for whole pupal Drosophila brains that allows pharmacological manipulation of the media conditions (Figure 2A). In this system axon pruning proceeds normally and we have been able to image pruning of MB neurons by time-lapse microscopy (Rabinovich et al., 2015). During normal development, MB γ neurons begin to regrow axons by 23-24h after puparium formation (APF; Figure 2B) and complete their growth by 46h APF (Figure 2C). However, while γ axons prune normally in culture, we observed little spontaneous regrowth ex vivo (Figure 2D), suggesting that this system either lacks an inducing signal for regrowth or that an inhibitory signal is not cleared properly.
Figure 2. Decrease in NO levels promotes developmental axon regrowth ex vivo.
(A) Scheme of the ex vivo culturing procedure. Brains were removed at 23h APF and cultured for an additional 22h before being fixed and stained. Modified from Rabinovich et al 2015.
(B-C) Confocal Z-projections of 23h APF (B) or 46h APF (C) pupal brains expressing CD8-GFP driven by 201Y-Gal4.
(D-G) Confocal Z-projections of brains dissected at 23h APF and cultured ex vivo for 22h in the indicated conditions. Culturing brains that were dissected at the onset of developmental regrowth results in minimal regrowth ex-vivo (D). While supplementing the media with 10μM L-NAME (NOS inhibitor) promotes regrowth (E) treatment with 100μM DETA (NO donor, G) or 10μM of D-NAME (inactive isomer, F) does not.
(H-J) Confocal Z-projections of brains of the indicated genotype and treatment expressing CD8-GFP driven by the pan-MB driver OK107, dissected at 23h APF and cultured for 22h ex vivo. Knocking down NOS levels in MB neurons (I, compare to H) promotes regrowth but adding back the NO donor DETA blocks this regrowth (J). Note that the OK107-Gal4 driver, used in panels H-J, labels all MB neurons types. Lower panels show a high magnification of Z-projections of selected slices from the medial lobe. Dashed red line demarcates the regrown γ lobe, dashed white line demarcates the β and dashed yellow line demarcates the β’ MB lobes.
Magenta represents FasII. Green is 201Y (B-G) or OK107 (H-J) – Gal4 driven mCD8:GFP. The scale bars represent 20 μm
(K-L) Quantification of developmental regrowth of the genotypes shown in B, D, and E (K) or H-J (L). *** p<0.001; *p<0.05; NS, not significant. One-way ANOVA was performed with a Dunnett’s post-hoc test.
See also Figure S2.
We set out to determine whether NO may be such a signal by modulating NO levels pharmacologically. Indeed, decreasing NO levels by adding the NOS inhibitor N-nitro-L-Arginine Methyl Ester (L-NAME), but not its nonactive isomer D-NAME, significantly and robustly induced regrowth of the adult γ lobe (Figure 2E-F, 2K). Increasing NO levels by adding the NO donor diethylenetriamine (DETA) did not promote regrowth ex vivo (Figure 2G). Taken together, our results demonstrate that attenuating NO production promotes axon regrowth of MB γ neurons in cultured brains.
Inhibiting NOS in MB neurons promotes axon regrowth ex vivo
Since NO levels dramatically affected regrowth in culture, we next wanted to determine whether they were produced by endogenous NOS and in which cells. We performed RNAi experiments in which the single Drosophila NOS (dNOS) gene was knocked down in various cell populations using two different and non-overlapping RNAi lines and regrowth was assayed ex vivo (RNAi lines: TRiP.HMC03076, see Figure 2 and S2; IR-X, Caceres et al., 2011, see Figure S2). We assayed the efficiency of the RNAi TRiP line by expressing it in the prothoracic gland (PG) and staining with the fluorescent NO sensor DAF-2 and with an antibody against dNOS (Lacin et al., 2014), both of which were significantly reduced (Figure S2A-B). Knocking down NOS in glia (by the pan-glial driver Repo-Gal4) did not promote regrowth (Figures S2C, S2E, S2H) while knocking down NOS in all neurons (by the pan-neuronal driver c155-Gal4, Figures S2D, S2F, S2H) or only in MB neurons (using the OK107-Gal4 driver; compare Figure 2I to 2H, 2L; Figure S2G-H) significantly promoted regrowth. Remarkably, elevating NO levels pharmacologically by adding the NO donor DETA repressed the growth promoting effect of knocking down NOS in the MB (Figure 2J compare to 2I, 2L), demonstrating that high NO levels inhibit axon regrowth. Taken together, our data pinpoint that NOS activity within the MB itself is the source of the axon regrowth inhibitory signal.
Neuronal NO represses axon regrowth in vivo
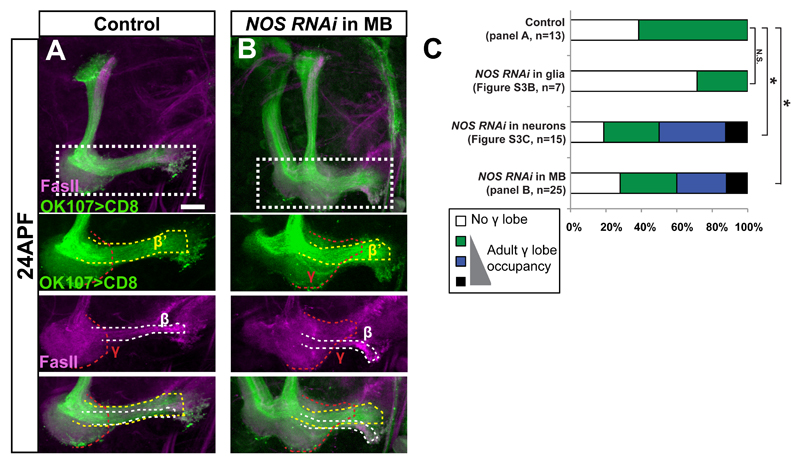
The data presented so far suggests that inhibiting NOS enhances regrowth in culture. We next wanted to determine whether NOS is physiologically important in vivo. We speculated that if NO is important to inhibit, and perhaps even time, developmental regrowth, then downregulating NOS should result in precocious axon regrowth. We focused our attention to 24h APF, a developmental time point in which WT γ neurons have only just begun to regrow (Figure 3A). Reducing NOS levels in glia had no effect on γ axons regrowth (Figure 3C, S3B, S3D-F). In contrast, knocking down NOS levels in neurons (Figure S3C) and even specifically within MB neurons (Figure 3B) resulted in significant precocious regrowth of the adult γ lobe in vivo (Figure 3C) suggesting that neuronally derived NO is required for normal regrowth during development and functions in a MB autonomous manner.
Figure 3. NOS represses developmental regrowth in vivo.
(A-B) Confocal Z-projections of brains dissected at 24h APF, expressing UAS-NOS-RNAi with no driver (A), or driven by OK107-Gal4 (MB driver) (B). Lower panels show a high magnification of Z-projections of selected slices from the medial lobe. The dashed red, white and yellow lines demarcate γ, β and β’ neurons, respectively. While expressing NOS-RNAi in glia (Figure S3B) does not affect developmental regrowth when compared to a control (A), knocking down NOS expression in all neurons (Figure S3C) or just in MB neurons (B) results in precocious regrowth.
Magenta represents FasII. Green is OK107–Gal4 driven mCD8:GFP. The scale bars represent 20 μm
(C) Quantification of the developmental regrowth phenotypes in A-B and Figure S3A-C. One-way ANOVA was performed with a Dunnett’s post-hoc test. NS - not significant *p<0.05
See also Figure S3.
The effect of NO on regrowth is mediated by E75 and UNF
We next investigated the mechanism by which NO inhibits regrowth. NO canonical signaling involves the activation of the soluble guanylate cyclase (sGC) pathway (Figure S4G). If NO inhibits regrowth by activating sGC, then adding the sGC inhibitor 1H-[1,2,4]Oxadiazolo[4,3-a]quinoxalin-1-one (ODQ) should recapitulate the L-NAME phenotype and result in increased regrowth of γ axons in culture. Conversely, adding an analog of the sGC product, cyclic GMP (8-bromo-cGMP) should cancel the regrowth promoting effect of L-NAME. Neither of these treatments altered axon regrowth (compare Figure S4B to S4A and S4D to S4C). To investigate whether the sGC pathway plays a role in axon regrowth in vivo, we looked at whole animal mutants for the α subunit of the Drosophila sGC, dgcα1 (Riedl et al., 2005). We observed no precocious regrowth in these dgcα1207 mutants (Figure S4F). Together, these results indicate that, NO does not inhibit regrowth via the sGC pathway.
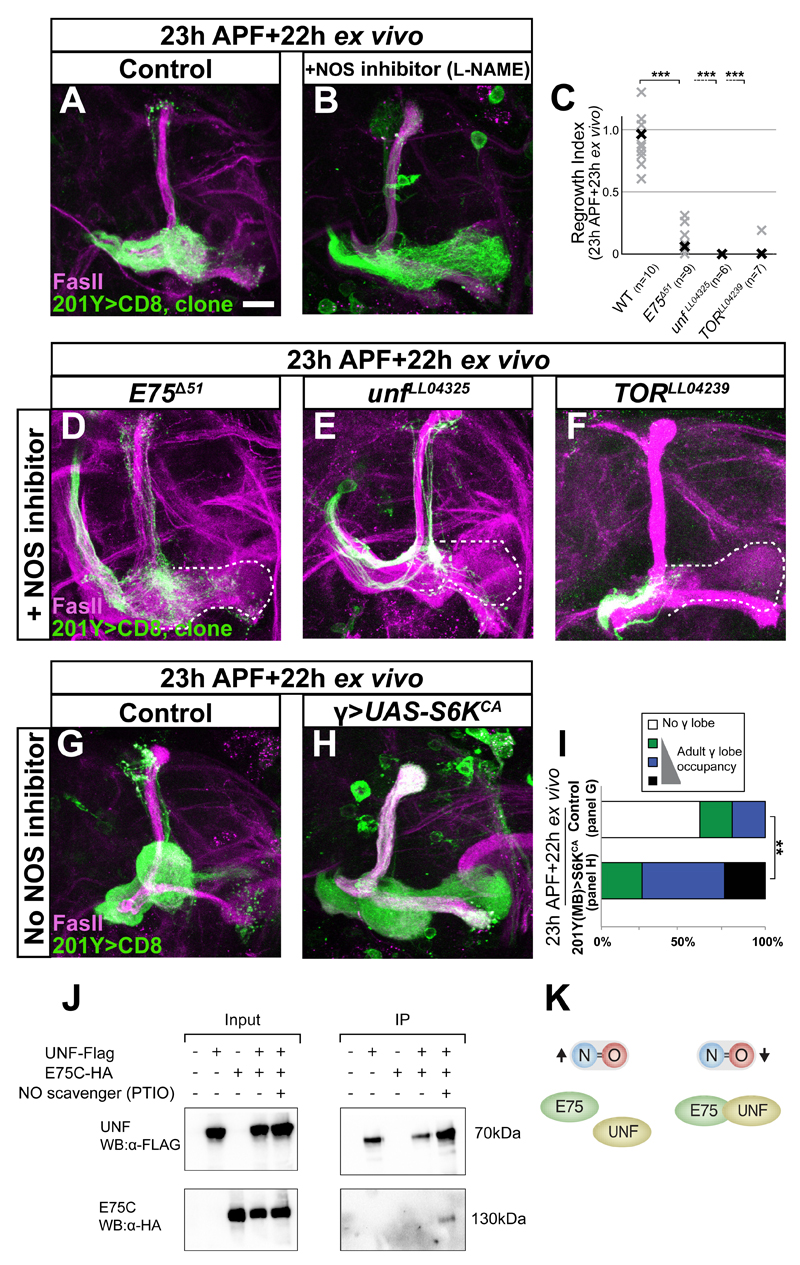
We next examined whether the effect of NO is mediated by the heme binding nuclear receptors UNF and E75 and the TOR pathway. We generated WT and mutant MARCM clones and cultured the brains in media containing L-NAME (Figure 4A, B). Indeed, E75, unf or TOR MARCM cloned failed to regrow in culture even with L-NAME while the non-clonal (control) neurons within the same brain regrew as expected (compare Figure 4B to D-F, non-clonal γ neurons visualized by FasII staining and outlined in white, quantified in 4C).
Figure 4. NO inhibits developmental regrowth via E75 and UNF likely by interfering with the creation of a stable heterodimer.
(A-B, D-F) Confocal Z-projections of brains of the indicated genotypes labeled by CD8-GFP driven by 201Y-Gal4, dissected at 23h APF and cultured for 22h either untreated (A) or incubated with 10μM L-NAME (B, D-F).
While WT axons treated with L-NAME regrew normally (B compare to A), E75Δ51 (D), unfLL04325 (E) or TORLL04239 (F) mutant axons (labeled by GFP) did not regrow (quantified in C). Note that non-clonal axons within these mutants did regrow (as shown by FasII staining).
(C) Quantification of regrowth defects in A-B, D-F. One way ANOVA with Dunnett’s T3 post hoc test; ***p<0.001
(G-H) Confocal Z-projections of brains expressing CD8-GFP (G) or additionally UAS-S6KCA (H) dissected at 23h APF and cultured for 22h. Overexpression of constitutively active S6K promotes regrowth even when cultured without L-NAME (H, compare to G).
Magenta represents FasII. Green is 201Y-Gal4 driven mCD8:GFP. The scale bars represent 20 μm.
(I) Quantification of the developmental regrowth phenotypes in G-H. **p<0.01
(J) UNF and E75C co-IP only when NO levels are low. BG3 Drosophila neuronal cells were transfected with the indicated transgenes. UNF-Flag was immunoprecipitated using anti-Flag beads and E75C-HA was detected using anti-HA antibody. UNF and E75C do not form a complex under basal conditions (lane 4) but co-IP when NO levels were reduced by the NO scavenger PTIO (lane 5).
(K) Scheme demonstrating our finding in J that UNF and E75C are present within the same complex only when NO levels are low.
See also Figure S4.
We next asked whether activation of the TOR pathway is sufficient to promote regrowth in culture even without NOS inhibition. Overexpression of S6KCA induced regrowth in culture without L-NAME (Figure 4H, compare to 4G, 4I) or even when cultured with DETA, and thus exposed to high NO levels (Figure S4H-I). Taken together, our results indicate that the E75/UNF/TOR pathway is required to mediate the effect of NO on regrowth and that activating the TOR pathway is sufficient to promote regrowth in culture even without lowering NO levels.
The recognition that NO functions via E75 and UNF, led us to revisit the issue of the physical interaction between them. We repeated the co-IP experiments while pharmacologically manipulating NO levels. Indeed, while E75C and UNF failed to co-IP under basal conditions when transfected into Drosophila BG3 cells (Figure 4J, lane 4) or when NO levels were elevated (data not shown), they did co-IP when we added the NO scavenger 2-Phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl 3-oxide (PTIO; Figure 4J, lane 5). These data strongly suggest that UNF and E75C physically interact only in low NO levels (Figure 4K). Taken together, the simplest and most likely interpretation for these data is that high NO levels inhibit developmental regrowth by perturbing the E75-UNF heterodimer, which is required to promote regrowth via the TOR pathway.
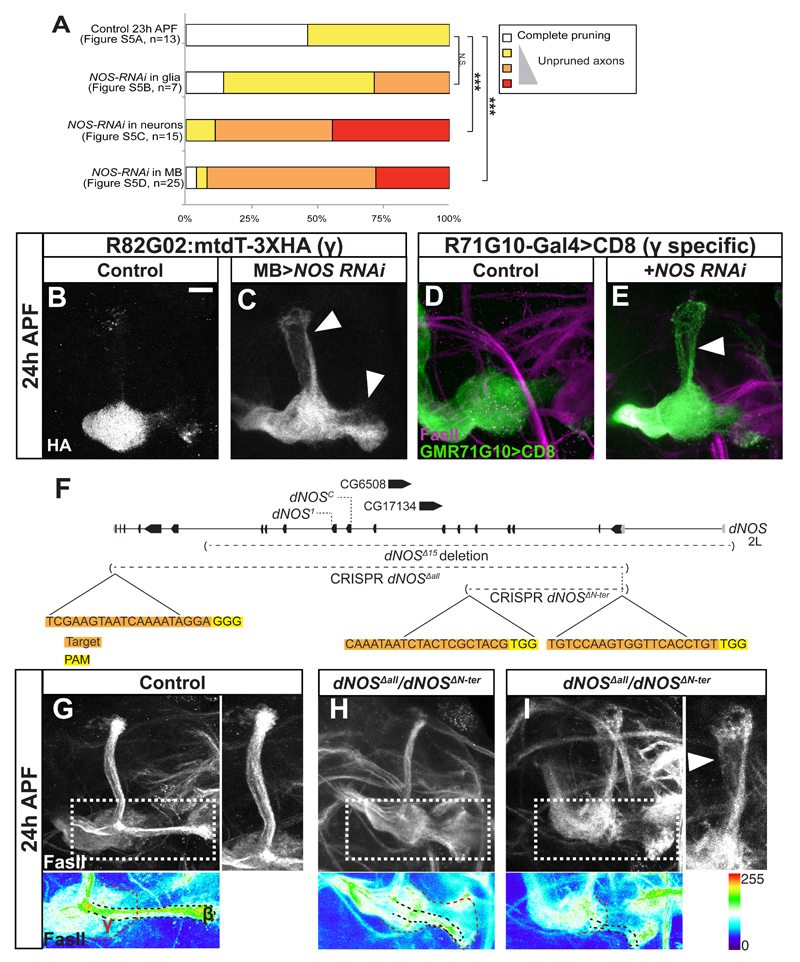
NO is required for the efficient axon pruning of MB γ neurons
While analyzing images from animals expressing NOS RNAi in MB neurons, we noticed that in addition to precocious regrowth, some brains also displayed a pruning defect in which the larval γ neurons were not fully pruned at 24h APF (Figures S5A-D, Figure 5A). To visualize these unpruned axons directly we developed a new Gal4-independent MB marker, R82G02:mtdT:3XHA by exploiting the FlyLight Gal4 collection (Jenett et al., 2012; Figure S5). Using this tool we demonstrated that knocking down NOS in all neurons (Figure S5F) or specifically in MB neurons (Figure 5C) but not in glia (Figure S5E) resulted in a pruning defect when compared to WT (Figure 5B).
Figure 5. NOS is required for developmental axon pruning.
(A) Quantification of the pruning defect in brains expressing NOS RNAi in different cell populations (the raw data is in Figure S5A-D). Pruning efficacy was assayed using FasII staining. One-way ANOVA was performed with a Dunnett’s post-hoc test. ***p<0.001
(B-C) Confocal Z-projections of brains dissected at 24h APF expressing R82G02:mtdT-3XHA to visualize γ neurons independent of Gal4, and OK107-Gal4 alone (B) or additionally expressing NOS RNAi (C). Arrowheads in C mark unpruned γ axons.
(D-E) Confocal Z-projections of brains dissected at 24h APF expressing CD8-GFP driven by the γ specific R71G10-Gal4 (D) or additionally expressing NOS RNAi (E).
Knocking down NOS is MB neurons (C), or specifically in γ neurons (E) inhibits axon pruning.
(F) Model of the NOS locus demarcating several mutant alleles used in this study, as well as the gRNAs used to generate two NOS null alleles using CRISPR technology. Boxes depict exons while lines depict introns. Black are coding while gray are non-coding exons. CG6508 and CG17134 are predicted to encode for two unstudied peptidases that are encoded within a NOS intron.
(G-I) Confocal Z-projections of brains dissected at 24h APF of control (G) or of dNOSΔall/dNOSΔN-ter transheterozygous brains (H-I). Lower panels show the FasII staining using the ‘thermal’ look up table (LUT) in a Z-projection of selected slices from the medial lobe. Red dashed line demarcates the regrown γ lobe and black dashed line demarcates the β lobe.
Gray in B-C represents HA staining of 82G02:HA:mtdt. Magenta in D-E and gray or thermal LUT in G-I represent FasII staining. Green is R71G10-Gal4 driven mCD8:GFP in D-E. The scale bars represent 20 μm.
See also Figures S5 and S6, and Table S1.
We next asked whether NOS was required within the γ neurons themselves. Indeed, knocking down NOS only in γ neurons using a γ specific driver (R71G10-Gal4; Jenett et al., 2012; Figure S5M-O) resulted in a significant pruning defect (Figure 5E compare to 5D), suggesting that the function of NOS to promote pruning is autonomous to γ neurons.
If NOS is necessary for efficient pruning and for timing regrowth then a dNOS fly should exhibit both incomplete pruning and precocious regrowth. Indeed, analysis of a homozygous mutant allele (dNOSΔ15, Figure 5F, S6A-B) (Yakubovich et al., 2010) as well as various transheterozygous allele combinations of dNOSΔ15, dNOS1 (Lacin et al., 2014) and dNOSC (Regulski et al., 2004) displayed both phenotypes associated with NOS knockdown (Figure 5F, S6A-J, Table S1). Within the Drosophila NOS field there has been a long-standing controversy in regards to whether NOS is an essential gene or not. While our data supports the notion that NOS is a non-essential gene (also see our RT-PCR on dNOSΔ15 in Figure S6L), to conclusively solve this issue we generated two new dNOS null alleles using CRISPR technology (Figure 5F, S6K-L, S7H). In dNOSΔall we deleted the entire gene including two intronic genes encoding proteases with unknown functions. To distinguish between the role of NOS and these unknown proteases, we also generated dNOSΔN-ter in which we deleted the first 5 coding exons of NOS (encoding for the oxygenase domain and glutamine rich sequence, that contributes to protein-protein interactions, Figure 5F). We found that these two CRISPR mutants are homozygous viable and display both a pruning defect and precocious regrowth at 24h APF (Figure 5H-I compare with 5G, Table S1). Using these new null alleles, we wanted to check if NOS function is cell-autonomous and not only MB autonomous. MARCM clones of dNOSΔall and of dNOSΔN-ter appeared WT at 24h APF (Figure S6M-O, 7K). This suggests that NO does not act cell autonomously, possibly due to the fact that NO is diffusible. Interestingly, while most of the brains expressing NOS RNAi or mutant for NOS exhibited defects in either pruning or regrowth, only a small percentage of brains exhibited both phenotypes (Table S1). While these two processes are mutually exclusive at the single neuron level, since an axon that isn’t pruned cannot regrow, this data suggests that these processes are largely mutually exclusive at the population level as well. Taken together, our results suggest that NOS generates a “switch” signal, which promotes axon pruning whilst inhibiting developmental axon regrowth in vivo.
High NO levels promote axon pruning
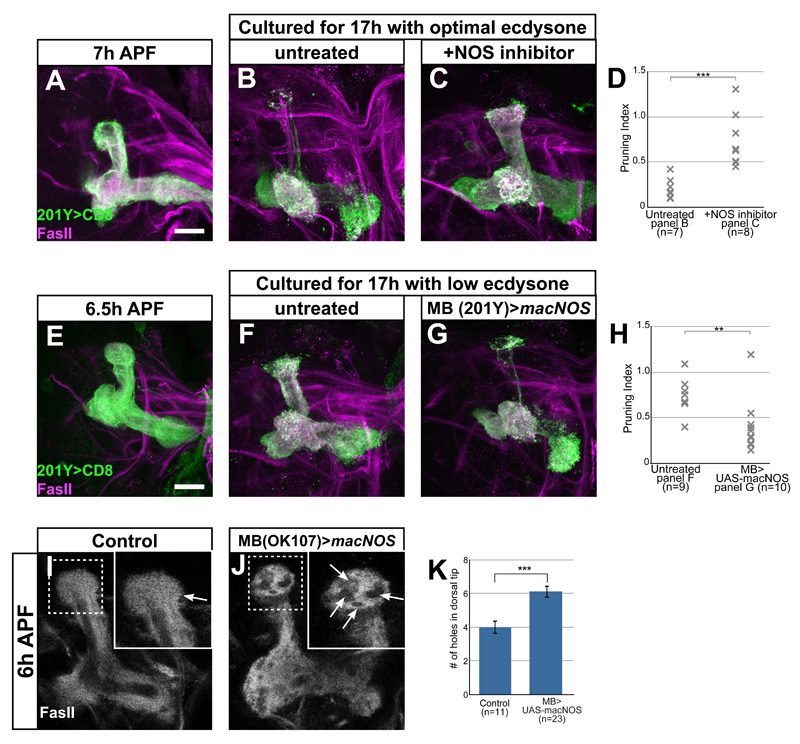
To directly test whether NO promotes axon pruning, we manipulated NO levels in our ex vivo culturing system in two complementary experiments. First, we decreased NO levels by adding L-NAME and found that it significantly blocked pruning (Figure 6A-D; Rabinovich et al., 2015), suggesting that high NO levels are required for pruning progression. Second, we wanted to determine if high NOS activity was sufficient to drive pruning in culture conditions in which pruning does not normally occur. Indeed, overexpression of a constitutively active NOS transgene (macNOS) within MB neurons promotes pruning in low ecdysone conditions (Figure 6E-H). Taken together, our results demonstrate that high NO levels promote axon pruning in cultured brains.
Figure 6. NO promotes pruning ex vivo and in vivo.
(A-C, E-G) Confocal Z-projections of brains expressing CD8-GFP driven by 201Y-Gal4 dissected at ~6.5 h APF and cultured for 17h with the indicated treatments. While WT brains dissected at ~6.5h APF (A, E) and cultured for 17h in the optimal 10µM ecdysone undergo pruning (B), adding the NOS inhibitor 10µM L-NAME blocks pruning (C). Culturing brains in 5µM ecdysone levels does not induce pruning (F), but expressing a constitutively active NOS (macNOS) in MB neurons in these culture conditions promoted pruning (G).
(D, H) Quantification of pruning in A-C (D) or E-G (H). Pruning index was calculated as the intensity of GFP at the dorsal tip/intensity of GFP at the peduncle. An index of 1 demonstrates no pruning and 0 demonstrates complete pruning. *p<0.05 ***p<0.001 using one-way ANOVA with Tukey post-hoc test
(I-J) Confocal Z-projections of brains dissected at 6h APF labeled by FasII in brains expressing OK107-Gal4 alone (I) or additionally expressing UAS-macNOS (J). Overexpression of NOS by OK107-Gal4 causes an increase in areas within the dorsal lobe lacking FasII staining (arrows in high-mag insets).
(K) Quantification of the number of ‘holes’ in FasII staining in the dorsal lobe in I and J. ***p<0.005 using a two-tailed independent sample T-test.
Magenta and grey represent FasII staining. Green is 201Y-Gal4 (A-C, E-G) driven mCD8:GFP. The scale bars represent 20 μm.
Since knocking down NOS inhibits pruning, overexpressing NOS should induce early in vivo. One of the early morphological signs of MB pruning is the appearance of “holes” in FasII staining due to the infiltration of astrocytes into the degenerating lobe (Awasaki et al., 2006; Hakim et al., 2014). Overexpression of macNOS in all neurons (data not shown) or specifically in MB neurons (compare Figure 6J to 6I) promoted axon pruning at 6h APF as highlighted by increased number of ‘holes’ in FasII expression (Figure 6K).
We next explored if NO promotes pruning via the canonical sGC pathway. We co-cultured early pupal brains with L-NAME and 8-bromo-cGMP with the expectation that if NO functions by activating the sGC, then adding back an analog of its product cGMP will bypass the pruning inhibition. In fact, these brains displayed strong inhibition of axon pruning similar to L-NAME alone (data not shown). Finally, pruning also occurred normally in dgcα1 mutants (Figure S4I), suggesting that NO does not promote pruning via the sGC pathway. Interestingly, E75Δ51 MARCM clones display a modest pruning defect in addition to the regrowth defect discussed above (see Figure 1, dorsally projecting axons in C-F), consistent with an additional possible role for E75 in mediating the NO effect on pruning. Further studies on how NO promotes pruning and whether this involves E75 are therefore necessary.
NOS activity is tightly regulated during neuronal remodeling
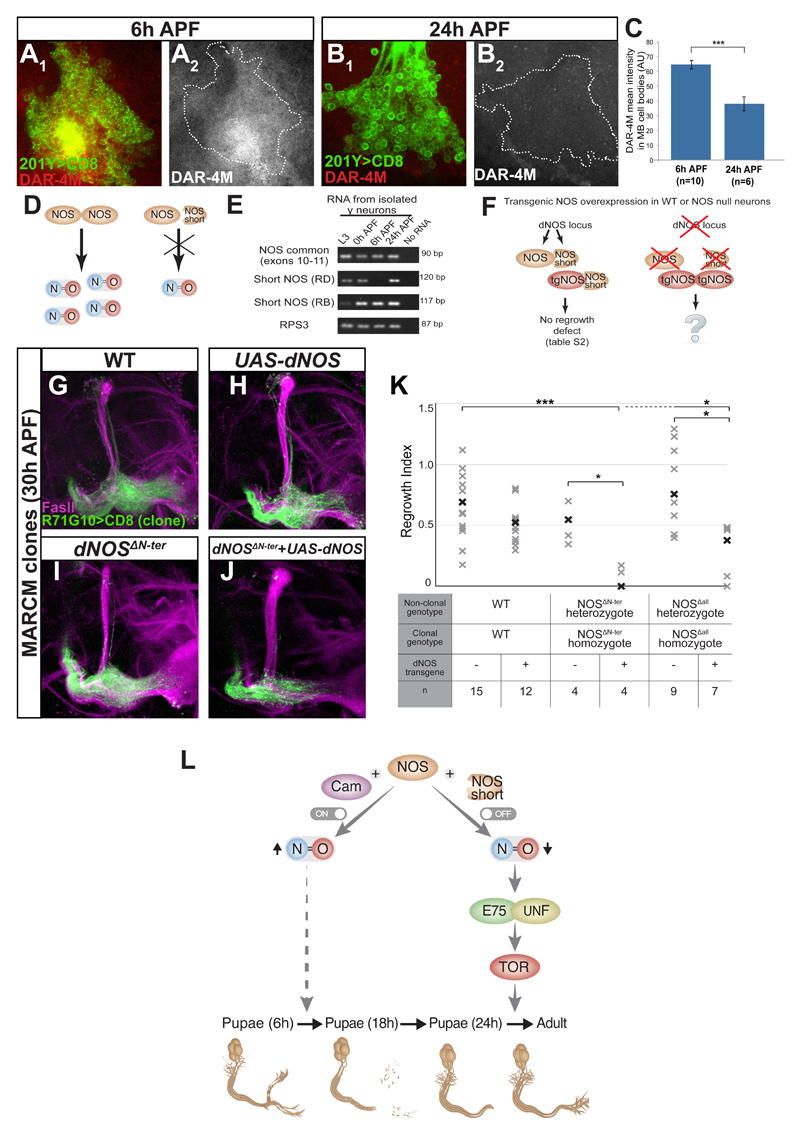
The data thus far suggests that endogenous MB NO levels are dynamic and must be high at the onset pruning and subsequently low to enable axon regrowth via UNF and E75. To explore potential developmental changes in NOS mRNA levels, we isolated MB γ neuron cell bodies at different developmental time points using FACS and extracted RNA for analysis. NOS mRNA levels appear unchanged throughout development (top lane in Figure 7E). We next wanted to determine protein levels using dNOS antibody staining (Lacin et al., 2014). Although not allowing detailed high-resolution analysis in the CNS, antibody staining showed no gross changes in protein expression between 6h APF (the onset of pruning) and 24h APF (the onset of regrowth; data not shown). Finally, we turned to check NO levels directly using the fluorescent NO detectors DAR-4M and DAF-2DA. As predicted, NO levels within MB cell bodies are high at 6h APF and low at 24h APF (Figure 7A-C and data not shown). Therefore, our data indicate that while NOS mRNA and protein levels remain relatively unchanged, NOS activity is tightly regulated during development.
Figure 7. NO levels are developmentally regulated via dNOS inhibition by short inhibitory isoforms.
(A-B) Confocal single slices of MB neuronal cell bodies expressing CD8-GFP driven by 201Y-Gal4 at 6h APF (A) or 24h APF (B) and stained using the NO indicator DAR-4M. NO levels are low at 24h APF, at the onset of axon regrowth (compare B to A)
(C) Quantification of DAR-4M intensity in A-B using a two-tailed independent sample T-test. ***p<0.001
(D) Proposed model of NOS regulation by truncated NOS proteins. When full-length dNOS forms an active homodimer, the complex produces NO. If full-length dNOS forms a heterodimer with a short dNOS isoform, the complex is inactive.
(E) RT-PCR of isolated MB γ neurons at L3, 0h, 6h and 24h APF. All samples expressed similar levels of both the positive control RPS3 (4th row) as well as RNA that is common for all NOS isoforms (exons 10-11, 1st row). In addition, they all expressed, at different levels, RNA species specific for isoform RD (2nd row) and for isoform RB (3rd row). See Figure S7 for NOS isoforms annotation and primer location.
(F) Model of transgenic NOS (tgNOS) overexpression in WT and NOS mutant brains. In WT brains the dNOS locus encodes for the full legth dNOS protein as well as for short inhibitory isoforms (NOS short). Overexpression of tgNOS in WT neurons did not inhibit regrowth (left part of model; see Table S2). To determine the consequence of expressing tgNOS in NOS null animals (right part of the model), we performed experiments described in G-J and S7I.
(G-J) Confocal Z-projections of brains expressing CD8-GFP driven by 201Y-Gal4 MARCM clones of the following genotypes: WT (G), WT also expressing UAS-dNOS (H), dNOSΔN-ter(I) and dNOSΔN-ter also expressing UAS-dNOS (J) dissected at 30h APF.
(K) Quantification of regrowth defect in dNOSΔN-ter and dNOSΔAll mutant clones expressing a dNOS transgene. Analysis was done using one way ANOVA with Dunnett’s T3 post hoc test; *p<0.05 ***p<0.001
(L) A working model of the pivotal role of NO on MB neuronal remodeling. At the onset of pruning NOS is active and, together with Cam, leads to increased NO levels that stimulate axon pruning via a yet unknown mechanism. In order for the MB γ axons to switch from pruning to regrowth, NOS is switched off, at least in part via heterodimerization with short inhibitory dNOS isoforms, leading to reduced NO levels which enable the formation of a stable E75/UNF complex that subsequently promotes axon regrowth in a TOR dependent manner.
Interestingly, in contrast to its ability to promote pruning in vivo, overexpressing NOS from three independent transgenes, did not significantly inhibit or delay axon regrowth (Table S2). We generated an additional optimized dNOS transgene (UAS-dNOSOS) and while its expression in MB neurons did appear to mildly delay regrowth, this was not statistically significant (Table S2), suggesting additional mechanisms regulating NOS activity beyond protein expression. It has been shown both in mammals (Bredt and Snyder, 1990) and in flies (Ray et al., 2007) that the calcium sensor Calmodulin (Cam) binds to NOS and increases its activity by facilitating electron transfer. We next tested the involvement of Cam in our system. Indeed, expressing RNAi against Cam in the PG, which normally expresses high levels of dNOS, resulted in dramatically reduced NO levels (Figure S7A-B). Moreover, Cam knockdown in MB neurons phenocopied the loss of NOS: pruning defect and precocious regrowth at 24h APF (Figure S7C-G, Table S1). These results suggest that Cam indeed facilitates dNOS activity during MB remodeling. If Cam is a major factor governing NOS activity within the MB, then perhaps in order to inhibit developmental axon regrowth both NOS and Cam must be overexpressed. However, overexpression of Cam together with dNOS or macNOS was also unable to significantly inhibit or delay axon regrowth (Table S2) suggesting that while Cam is important for dNOS function, it is not sufficient to promote its activity in pupal MB γ neurons. Taken together, this suggests that the activity of dNOS at the onset of regrowth is down regulated by yet another mechanism.
Previous analysis of the dNOS locus has uncovered that it encodes up to 10 different transcripts giving rise to 7 different polypeptides (Stasiv et al., 2001) out of which only one, the full length dNOS1, encodes for the enzymatically active protein. Because NOS functions as a dimer, it has been suggested that these truncated dNOS proteins function as dominant negatives (Stasiv et al., 2004) and therefore may provide an additional layer of dNOS activity regulation (Figure 7D). We first wanted to determine whether these short dNOS mRNA transcripts are expressed within MB γ neurons during development. By isolating RNA from dissociated and FACS sorted MB γ neurons, we were able to amplify using RT-PCR two short dNOS transcripts: dNOS-RD and RB (Figure 7E, see S7H for gene structure). Due to low expression levels and because we purified only 1000 cells for the RT-PCR experiment, these experiments were difficult to quantify. Nonetheless, while the levels of dNOS-RB seemed unaltered during development, the levels of RD might be developmentally regulated: While we were never able to amplify RD from early pupal cDNA (6h APF; no product in 6/6 trials), we could readily detect it in samples prepared from 24 APF pupae (in 4 out of 6 trials). These results are consistent with short dNOS isoforms being expressed and perhaps even upregulated at the onset of axon regrowth, therefore providing a possible mechanism by which the cell can rapidly shut down dNOS activity (model in Figure 7D).
Our inability to inhibit regrowth by overexpressing transgenic NOS may thus be due to its’ dimerization with a short dNOS protein that would render the complex inactive (Figure 7F). To explore this hypothesis, we wanted to express transgenic NOS in mutants lacking the entire dNOS locus and thus unable to generate short inhibitory dNOS isoforms (Figure 7F). While dNOS homozygous mutants are viable and expression of NOS transgenes in the MB does not affect viability, expression of NOS transgenes in MB neurons within a homozygous dNOS mutant animal resulted in early pupal lethality (Figure S7I). To overcome the lethality problem, we expressed transgenic NOS within dNOS MARCM clones. Indeed, while both dNOSΔN-ter and dNOSΔall clones regrew similar to WT at 30h APF (Figure 7I, compare to 7G, 7K), clones additionally expressing dNOS displayed a dramatic delay in regrowth (Figure 7J, compare to 7H and 7I, 7K). This indicates that the dNOS locus also includes elements that negatively regulate NOS activity and removing the entire locus releases this inhibition on transgenic NOS (see right part of model in Figure 7F). Therefore, the existence of short dNOS isoforms may be an important mechanism by which dNOS activity is down regulated following pruning leading to the rapid decrease in NO levels to enable axon regrowth.
Taken together, these results demonstrate that elevating NOS activity, in a mechanism likely facilitated by Cam, promotes pruning and inhibits axon regrowth. The rapid shut down of NOS activity at the onset of axon regrowth seems to be mediated, at least in part, by short inhibitory dNOS isoforms. Thus, we propose that NO levels act as a robust switching mechanism between two contradictory developmental modules, degeneration and regrowth, during neuronal remodeling (Figure 7L).
Discussion
Neuronal remodeling involves a cellular switch between different growth and degeneration states. These different cellular states must be tightly controlled not only to ensure that decisions are taken in an accurate and timely manner but also to circumvent the potential damaging effects of opposing forces. How neurons switch between these different growth states during development was unknown. Here we show that NO provides a switching mechanism between axon pruning and regrowth during neuronal remodeling of Drosophila MB neurons. dNOS activity promotes axon pruning while at the same time inhibits the onset of regrowth. NO levels are high just before axon pruning and low at the onset of regrowth. Our data strongly suggests that the regulation of dNOS activity is mediated, at least in part, by transcription of short inhibitory dNOS isoforms. Importantly, we found that NO inhibits developmental axon regrowth by interfering with the generation of a stable E75/UNF heterodimer. We thus propose that NO acts as a molecular switch to actively transition neurons between the degenerative state of axon pruning to the regrowth state and ensures that both states do not occur simultaneously (Figure 7L).
Modulation of nuclear receptor activity by NO
Up to this point, neuronal involvement of NO, such as in promoting neuroplasticity, has been mostly attributed to changes in sGC activity (Gallo and Iadecola, 2011). In contrast, we show here that the mechanism by which NO regulates neuronal remodeling is not via the canonical sGC pathway.
Previous work has shown that UNF, as well as E75, like its mammalian homologs Rev-erb-α and -β, can bind heme (Raghuram et al., 2007; Reinking et al., 2005). Heme is known to bind to monovalent gases, such as NO, thus providing a tempting mechanism for direct regulation of E75 and UNF by NO. Indeed, E75 has been shown to block the transcriptional activity of another Drosophila nuclear receptor, Hr3, in an NO dependent manner both in vitro (Reinking et al., 2005) and in vivo (Caceres et al., 2011). In our study, we found that UNF and E75 are both required to promote developmental axon regrowth and are present within the same complex only in low NO levels, suggesting that NO inhibits regrowth by interfering with the creation of a stable E75/UNF heterodimer.
Interestingly, a recent report has suggested that UNF and E75 function together to regulate the various aspects of the development and function of the s-LNvs clock neurons (Jaumouille et al., 2015). While Jaumouillé and colleagues have nicely shown that both UNF and E75 perturb circadian rhythms in similar ways and that both NRs bind to several promoters of interest, they have not demonstrated physical interactions between E75 and UNF.
We believe that our finding the NO regulates the formation of a stable nuclear receptor complex opens up new avenues of research to study sGC independent roles for NO signaling. The large number and versatility of heme binding proteins highlights the potential targets for regulation by NO signaling.
NOS activity is tightly regulated during neuronal remodeling
Our observation that NO levels in MB γ neuron cell bodies are high at the onset of pruning and low at the beginning of regrowth is consistent with our model that high NO levels promote pruning but inhibit regrowth. Our data suggests that the regulation of NOS activity appears to be multi-faceted. Similar to the mechanisms regulating NOS in mammals, we show here that Cam is required for efficient NO production and that knocking down Cam expression phenocopies NOS mutant defects. Cam likely regulates NOS activity by facilitating electron transfer as previously shown (Bredt and Snyder, 1990; Ray et al., 2007).
In mammals, the ability of CaM to bind nNOS is regulated by Ca2+ levels (Zhou and Zhu, 2009). Interestingly, Ca2+ transients precede and likely trigger dendrite pruning of Drosophila dendritic arborization (da) neurons (Kanamori et al., 2013). It is therefore plausible that Ca2+ may also play a role in activating dNOS in MB neurons.
However, overexpressing NOS does not result in inhibition of axon regrowth, even when expressed together with Cam, leading to the conclusion that NOS activity is down regulated before regrowth in an alternative mechanism. We therefore tested a hypothesis raised by Stasiv and colleagues (2004; 2001) that the dNOS locus encodes for short inhibitory isoforms. Indeed, NOS overexpression in neurons that are null mutant for the entire NOS locus (including the short isoforms) results in delayed regrowth. This, together with our findings that at least two of these short dNOS mRNAs are expressed in the MB during development, supports the notion that short dNOS isoforms play a crucial role in dNOS activity regulation during MB remodeling. However, in these experiment we observed a delay but not a complete block in axon regrowth, suggesting there are yet alternative mechanisms for down-regulating dNOS activity. Interestingly, the human nNOS (NOS1) genomic locus also encode for multiple isoforms (Wang et al., 1999), suggesting that this mechanism of dNOS regulation may be evolutionarily conserved.
NO as a neurodevelopmental switch
Although developmental neuronal remodeling has been observed in a myriad of animals and systems, a comprehensive view is still lacking (Riccomagno and Kolodkin, 2015; Schuldiner and Yaron, 2015). While several mechanisms have been shown to regulate neuronal remodeling such as neuronal competition, semaphorin signaling, caspase activation, endocytosis, ecdysone and TGF-β signaling as well as JNK signaling, how they function together, and which pathways are general or cell specific is not currently known (reviewed in Riccomagno and Kolodkin, 2015; Schuldiner and Yaron, 2015). It will be interesting to test how NO signaling might interact with these known and newly discovered mechanisms to promote developmental pruning and potentially also degeneration following injury.
While Ramon y Cajal identified that the dendritic arbor of Purkinje and granule cells (Garcia-Lopez et al., 2010) undergo what he called ‘process resorption’ followed by regrowth to form the mature dendrites, regrowth has not been extensively studied. Only recently we have demonstrated that the regrowth phase is genetically regulated and distinct from initial outgrowth (Yaniv et al., 2012).
When neurites undergo pruning followed by regrowth in a very structured fashion, they have to switch between degeneration and regrowth. While this switch can either result from competition between two forces (pruning and regrowth) or an active switch of developmental programs, our data is more consistent with the latter. First of all, if the switch would consist of competition, then any mutation that inhibits pruning should result in accelerated or precocious regrowth but this is something we have not seen in any of the pruning defect mutants. Second, the identification of several nuclear receptors including Ecdysone Receptor B1 (EcR-B1) and UNF, which regulate different aspects of remodeling (Boulanger and Dura, 2015), suggests an active switch between different transcriptional programs.
NO has been implicated in various forms of neuronal remodeling and plasticity. For example, NO is necessary for both activity mediated synapse formation (Nikonenko et al., 2013) and for the expression of neuroplasticity associated protein expression (Gallo and Iadecola, 2011) but can also cause synapse disassembly in motoneurons (Moreno-López et al., 2010). NO signaling is also important for neurite growth and synaptic remodeling after nerve crush in the pond snail L. stagnalis (Cooke et al., 2013). In nNOS knockout mice there is a regenerative delay, possibly due to decreased Wallerian degeneration, that is also associated with an increase in post-injury sprouts (Keilhoff et al., 2002). This increased sprouting seen in nNOS knockout mice could be due to decreased pruning of unnecessary neurites, which may also explain the decrease in functional recovery following injury. High levels of NO and of protein S-nitrosylation have already been associated with Alzheimer’s disease and Parkinson’s disease and neuronal NOS (nNOS) inhibitors have been suggested as therapies for neurodegenerative disease (Trippier et al., 2013). Thus, our proposed function of NOS in MB neuronal remodeling is consistent with high levels of NO promoting pruning and neurodegeneration while low levels promote growth. Taken the fact that E75, UNF and NOS are all conserved, raises the possibility that some of the pathways and principles that we have described here could be conserved in developmental as well as injury mediated axon regeneration in other organisms. Because NO is a diffusible gas, it does not necessarily have to be cell-autonomously generated but could be provided by neighboring cells. Furthermore, other monovalent gases might also regulate heme-binding proteins such as E75 and UNF.
Delineating the molecular mechanisms that underlie the ability of neurons to switch between degenerative and regenerative developmental programs should, in the long run, provide a platform to better understand neurodegenerative diseases and the mechanisms that normally limit regeneration following injury.
Experimental procedures
Drosophila strains
E75Δ51, E75A81, E7507041, UAS-Rheb, UAS-S6k.STDE, TRiP.HMC03076 (UAS-NOS RNAi), Amnesiac-Gal4, R71G10-GAL4, TRiP.HMS01318 (UAS-Cam RNAi), and UAS-Cam.W were obtained from the Bloomington Drosophila stock center (Indiana University, USA). UAS-NOS-RNAi (IR-X) and 2 alleles of UAS-macNOS (macrophage NOS) were kindly provided by H. Krause (Caceres et al., 2011). dNOS C and UAS-dNOS (UAS-dNOSGE) were kindly provided by M. Regulski. dNOSΔ15 was kindly provided by P. O’Farrell. dNOS1 was kindly provided by J. Skeath. unfLL04325, UAS-unf-FLAG and TORLL04329 were previously described (Yaniv et al., 2012).
Generation and imaging of MARCM Clones
MB MARCM neuroblast clones were generated at NHL and examined later, as described previously (Lee and Luo, 1999). Brains were mounted on Slowfade (Invitrogen) and imaged on Zeiss LSM710 confocal microscopes.
Antibody Staining Conditions
Rat monoclonal anti-mouse CD8 α subunit, 1:100 (Invitrogen); mouse monoclonal anti-FasII (1D4), 1:25 (Developmental Studies Hybridoma Bank); rat anti-HA, 1:250 (Roche); guinea pig anti-NOS (1:100 kindly provided by J. Skeath). Alexa 405, Alexa 488 or Alexa 647 conjugated secondary antibodies were used at 1:300 (Invitrogen).
Ex vivo brain culturing system
The full details of this culturing system are provided in Rabinovich et al (2015). In brief, pupae were collected at puparium onset (white pupae) and aged until the required developmental stage in 25ºC. The brains were quickly (<2 min/brain) and carefully dissected in a filtered basic culture medium (Rabinovich et al., 2015). All the brains for a single experiment were dissected simultaneously in the basic media, and were randomly divided to wells containing modified medium according to the conditions and controls tested. For the pharmacological manipulations 10µM L-NAME, 100µM DETA, 10µM D-NAME, 10µM ODQ, or 100nM 8-Bromo-cGMP (all from Sigma-Aldrich) were added. The cultured brains were kept in a humidified incubator at 25°C until fixation and processing for immunohistochemistry according to standard protocols.
DAF2-DA and DAR-4M staining
Brains were dissected in ringer solution and incubated with 10μM 4,5-diaminofluorescein diacetate in PBS (DAF2-DA, Sigma-Aldrich) for 1h at RT or with 10μM of Diaminorhodamine-4M AM (DAR-4M, Sigma-Aldrich) in Schenider’s Drosophila media for 2h at RT followed by fixation for 20 min in PBS containing 4% paraformaldehyde and 0.3% Triton X-100 for later analysis.
Supplementary Material
Acknowledgements
We thank H. Krause, P. O’Farrell, M. Regulski, J. Skeath, C. Thummel, Exelixis at Harvard Medical School and the Bloomington Stock Center for reagents; M. Amosi, O. Fuchs and O. Mayseless for technical assistance; R. Rotkopf for statistical analyses; A. Halme, K. King-Jones and H. Krause for discussions and sharing of unpublished observations; M. Schuldiner, A. Yaron and the members of the Schuldiner lab for discussions and critical readings of the manuscript; the FasII (1D4) antibody monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa. This work was supported by the Israeli Science Foundation (ISF), grant #686/11 and the European Research Council (erc), consolidator grant “AxonGrowth”. The Zeiss LSM710 microscope was purchased with the help of the Adelis foundation. O.S. is an incumbent of the Rothstein Career Development Chair of Genetic Diseases.
References
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K. Essential Role of the Apoptotic Cell Engulfment Genes draper and ced-6 in Programmed Axon Pruning during Drosophila Metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Boulanger A, Dura JM. Nuclear receptors and Drosophila neuronal remodeling. Biochimica et biophysica acta. 2015;1849:187–195. doi: 10.1016/j.bbagrm.2014.05.024. [DOI] [PubMed] [Google Scholar]
- Bredt DS, Snyder SH. Isolation of nitric oxide synthetase, a calmodulin-requiring enzyme. Proc Natl Acad Sci U S A. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L, Necakov AS, Schwartz C, Kimber S, Roberts IJH, Krause HM. Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev. 2011:1–11. doi: 10.1101/gad.2064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh ECT, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Cocchi E, Drago A, Serretti A. Hippocampal Pruning as a New Theory of Schizophrenia Etiopathogenesis. Mol Neurobiol. 2015 doi: 10.1007/s12035-015-9174-6. [DOI] [PubMed] [Google Scholar]
- Cooke RM, Mistry R, Challiss RAJ, Straub VA. Nitric Oxide Synthesis and cGMP Production Is Important for Neurite Growth and Synapse Remodeling after Axotomy. J Neurosci. 2013;33:5626–5637. doi: 10.1523/JNEUROSCI.3659-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rosny E, De Groot A, Jullian-Binard C, Borel F, Suarez C, Le Pape L, Fontecilla-Camps JC, Jouve HlnM. DHR51, the Drosophila melanogasterHomologue of the Human Photoreceptor Cell-Specific Nuclear Receptor, Is a Thiolate Heme-Binding Protein. Biochemistry. 2008;47:13252–13260. doi: 10.1021/bi801691b. [DOI] [PubMed] [Google Scholar]
- Gallo EF, Iadecola C. Neuronal Nitric Oxide Contributes to Neuroplasticity-Associated Protein Expression through cGMP, Protein Kinase G, and Extracellular Signal-Regulated Kinase. J Neurosci. 2011;31:6947–6955. doi: 10.1523/JNEUROSCI.0374-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lopez P, Garcia-Marin V, Freire M. Dendritic spines and development: towards a unifying model of spinogenesis--a present day review of Cajal's histological slides and drawings. Neural plasticity. 2010;2010:769207. doi: 10.1155/2010/769207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim Y, Yaniv SP, Schuldiner O. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLoS One. 2014;9:e86178. doi: 10.1371/journal.pone.0086178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumouille E, Machado Almeida P, Stahli P, Koch R, Nagoshi E. Transcriptional regulation via nuclear receptor crosstalk required for the Drosophila circadian clock. Curr Biol. 2015;25:1502–1508. doi: 10.1016/j.cub.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-Driver Line Resource for Drosophila Neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A. Ecdysone- and NO-Mediated Gene Regulation by Competing EcR/Usp and E75A Nuclear Receptors during Drosophila Development. Mol Cell. 2011;44:51–61. doi: 10.1016/j.molcel.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T, Kanai MI, Dairyo Y, Yasunaga Ki, Morikawa RK, Emoto K. Compartmentalized Calcium Transients Trigger Dendrite Pruning in Drosophila Sensory Neurons. Science. 2013;340:1475–1478. doi: 10.1126/science.1234879. [DOI] [PubMed] [Google Scholar]
- Keilhoff G, Fansa H, Wolf G. Differences in peripheral nerve degeneration/regeneration between wild-type and neuronal nitric oxide synthase knockout mice. J Neurosci Res. 2002;68:432–441. doi: 10.1002/jnr.10229. [DOI] [PubMed] [Google Scholar]
- Lacin H, Rusch J, Yeh RT, Fujioka M, Wilson BA, Zhu Y, Robie AA, Mistry H, Wang T, Jaynes JB, et al. Genome-wide identification of Drosophila Hb9 targets reveals a pivotal role in directing the transcriptome within eight neuronal lineages, including activation of nitric oxide synthase and Fd59a/Fox-D. Dev Biol. 2014;388:117–133. doi: 10.1016/j.ydbio.2014.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Luo L, O'Leary DD. Axon retraction and degeneration in development and disease. Ann Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- Mollema NJ, Yuan Y, Jelcick AS, Sachs AJ, Von Alpen D, Schorderet D, Escher P, Haider NB. Nuclear Receptor Rev-erb Alpha (Nr1d1) Functions in Concert with Nr2e3 to Regulate Transcriptional Networks in the Retina. PLoS ONE. 2011;6:e17494. doi: 10.1371/journal.pone.0017494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-López B, Sunico CR, González-Forero D. NO Orchestrates the Loss of Synaptic Boutons from Adult “Sick” Motoneurons: Modeling a Molecular Mechanism. Mol Neurobiol. 2010;43:41–66. doi: 10.1007/s12035-010-8159-8. [DOI] [PubMed] [Google Scholar]
- Nikonenko I, Nikonenko A, Mendez P, Michurina TV, Enikolopov G, Muller D. Nitric oxide mediates local activity-dependent excitatory synapse development. Proc Natl Acad Sci U S A. 2013;110:E4142–4151. doi: 10.1073/pnas.1311927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich D, Mayseless O, Schuldiner O. Long term ex-vivo culturing of Drosophila brain as a method to live image pupal brains: insights into the cellular mechanisms of neuronal remodeling. Front Cell Neurosci. 2015;9 doi: 10.3389/fncel.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, Khorasanizadeh S, Burris TP, Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray SS, Sengupta R, Tiso M, Haque MM, Sahoo R, Konas DW, Aulak K, Regulski M, Tully T, Stuehr DJ, et al. Reductase domain of Drosophila melanogaster nitric-oxide synthase: redox transformations, regulation, and similarity to mammalian homologues. Biochemistry. 2007;46:11865–11873. doi: 10.1021/bi700805x. [DOI] [PubMed] [Google Scholar]
- Regulski M, Stasiv Y, Tully T, Enikolopov G. Essential function of nitric oxide synthase in Drosophila. Curr Biol. 2004;14:R881–882. doi: 10.1016/j.cub.2004.09.068. [DOI] [PubMed] [Google Scholar]
- Reinking J, Lam MMS, Pardee K, Sampson HM, Liu S, Yang P, Williams S, White W, Lajoie G, Edwards A, et al. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Riccomagno MM, Kolodkin AL. Sculpting Neural Circuits by Axon and Dendrite Pruning. Annu Rev Cell Dev Biol. 2015 doi: 10.1146/annurev-cellbio-100913-013038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl CAL, Neal SJ, Robichon A, Westwood JT, Sokolowski MB. Drosophila soluble guanylyl cyclase mutants exhibit increased foraging locomotion: behavioral and genomic investigations. Behav Genet. 2005;35:231–244. doi: 10.1007/s10519-005-3216-1. [DOI] [PubMed] [Google Scholar]
- Schuldiner O, Yaron A. Mechanisms of developmental neurite pruning. Cell Mol Life Sci. 2015;72:101–119. doi: 10.1007/s00018-014-1729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiv Y, Kuzin B, Regulski M, Tully T, Enikolopov G. Regulation of multimers via truncated isoforms: a novel mechanism to control nitric-oxide signaling. Genes Dev. 2004;18:1812–1823. doi: 10.1101/gad.298004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiv Y, Regulski M, Kuzin B, Tully T, Enikolopov G. The Drosophila nitric-oxide synthase gene (dNOS) encodes a family of proteins that can modulate NOS activity by acting as dominant negative regulators. J Biol Chem. 2001;276:42241–42251. doi: 10.1074/jbc.M105066200. [DOI] [PubMed] [Google Scholar]
- Thomas MS, Davis R, Karmiloff-Smith A, Knowland VC, Charman T. The over-pruning hypothesis of autism. Dev Sci. 2015 doi: 10.1111/desc.12303. [DOI] [PubMed] [Google Scholar]
- Trippier PC, Jansen Labby K, Hawker DD, Mataka JJ, Silverman RB. Target- and Mechanism-Based Therapeutics for Neurodegenerative Diseases: Strength in Numbers. J Med Chem. 2013;56:3121–3147. doi: 10.1021/jm3015926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Newton DC, Robb GB, Kau C-L, Miller TL, Cheung AH, Hall AV, VanDamme S, Wilcox JN, Marsden PA. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc Natl Acad Sci U S A. 1999;96:12150–12155. doi: 10.1073/pnas.96.21.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakubovich N, Silva EA, O'Farrell PH. Nitric oxide synthase is not essential for Drosophila development. Curr Biol. 2010;20:R141–142. doi: 10.1016/j.cub.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv SP, Issman-Zecharya N, Oren-Suissa M, Podbilewicz B, Schuldiner O. Axon Regrowth during Development and Regeneration Following Injury Share Molecular Mechanisms. Curr Biol. 2012;22:1774–1782. doi: 10.1016/j.cub.2012.07.044. [DOI] [PubMed] [Google Scholar]
- Yu F, Schuldiner O. Axon and dendrite pruning in Drosophila. Curr Opin Neurobiol. 2014;27:192–198. doi: 10.1016/j.conb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhu D-Y. Neuronal nitric oxide synthase: Structure, subcellular localization, regulation, and clinical implications. Nitric Oxide. 2009;20:223–230. doi: 10.1016/j.niox.2009.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.