Abstract
Selective immunoglobulin A deficiency (IgAD) is the most common primary immunodeficiency in Europeans. Our GWAS meta-analysis of 1,635 IgAD patients and 4,852 controls identified four new significant (P < 5x10−8) loci and association with a rare IFIH1 variant (Ile923Val). Peak novel variants (PVT1 P = 4.3x10−11, ATG13–AMBRA1 P = 6.7x10−10, AHI1 P = 8.4x10−10 and CLEC16A P = 1.4x10−9) overlapped with autoimmune markers (3/4) and correlated with 21 putative regulatory variants, including eQTLs for AHI1 and DEXI and DNase hypersensitivity in FOXP3+ T regulatory cells. A pathway analysis of the meta-analysis results showed a striking association with the KEGG pathway for IgA production (pathway P < 0.0001): where 22 of 30 annotated pathway genes contained at least one variant with a P-value ≤0.05 in the IgAD meta-analysis. These data suggest that a complex network of genetic effects, including genes known to influence the biology of IgA production, contribute to IgAD.
IgAD is the most common primary immunodeficiency and is defined by serum IgA level <0.07 g/L.1 The prevalence in Europeans is 1:600.1 Secretory IgA (sIgA) is important for mucosal immunity and gut commensalism2,3, and clinical features of IgAD include recurrent mucosal infections. In IgAD, B cells fail to terminally differentiate into IgA+ plasma cells, however IL-21 can restore B cell IgA production in vitro4,5. IgAD is strongly associated with HLA6 and aggregates in families with autoimmunity.7 The prevalence of Celiac disease is 35 times higher in IgAD patients whereas the prevalence of systemic lupus erythematosus (SLE) and type 1 diabetes (T1D) is 10 times higher.8
Ferreira et al.6 previously imputed HLA-B, HLA-DRB1 and HLA-DQB1 in IgAD cases and controls (>2,700 individuals).9 The primary signal mapped to HLA-DQB1*02 (OR = 2.8, P = 7.7x10−57), due to combined independent effects of the HLA-B*08:01-DRB1*03:01-DQB1*02 and HLA-DRB1*07:01-DQB1*02 haplotypes.9 There was a secondary signal at HLA-DRB1*01:02 (OR = 4.28, P = 5.86x10−17) and a protective effect for HLA-DRB1*15:01 (OR = 0.13, P = 2.24x10−35).9 HLA-DQB1*02:01 is protective for IgA nephropathy (OR = 0.71, P = 2.61x10−13). 10 None of the non-MHC IgAD loci overlapped with IgA nephropathy.10
A previous GWAS identified the common allele IFIH1 Thr946Ala (OR = 0.62, control allele frequency = 0.39) as the first and, to date, only non-HLA genome-wide significant IgAD locus.9 Thr946Ala is also protective for T1D11, SLE12, psoriasis13 and vitiligo (see URLs). IFIH1 encodes MDA5, a cytosolic receptor that recognizes dsRNA and initiates interferon pathway activation.
To expand our understanding of IgAD risk, we studied four new IgAD cohorts, and performed a GWAS meta-analysis of ~9.5M SNPs in 1,635 cases and 4,852 controls (Table 1). Genotypes for untyped markers were imputed for each cohort separately (1000 Genomes Project) and genotypes for variants fully typed in the entire cohort. Up to four controls per new case were iteratively selected based on ancestry eigenvectors14 to minimize population substructure (Online Methods). Genomic inflation factor values were minimal (Table 1), indicating that population substructure was adequately addressed. Association analyses were conducted with logistic regression (additive model), accounting for genotype uncertainty and using ancestry eigenvectors as covariates (Online Methods).
Table 1. Case-Control Cohorts for IgAD GWAS.
| Cohortsa | Cases (N) | Controls (N) | Total (N) | Significant Ancestry Eigenvectors (N) | Genomic Inflation Factor (λGC)b | SNPs Genotyped (N) | SNPs after Imputation (N) | Median Info Score |
|---|---|---|---|---|---|---|---|---|
| Meta | 1,635 | 4,852 | 6,487 | 556,344a | 9,464,381 | |||
|
| ||||||||
| 2016 (New) | ||||||||
| Swedish | 483 | 1,932 | 2,415 | 2 | 1.03 | 423,694 | 9,464,381 | 0.988 |
| Spanish | 150 | 230 | 380 | 0 | 1.04 | 339,552 | 8,534,763 | 0.974 |
| Italian | 91 | 364 | 455 | 4 | 1.03 | 218,770 | 8,365,602 | 0.948 |
| Czech | 151 | 602 | 753 | 4 | 1.05 | 112,822 | 6,282,267 | 0.929 |
|
| ||||||||
| 2010 | ||||||||
| Swedish | 421 | 1,080 | 1,501 | 5 | 1.03 | 289,843 | 8,765,152 | 0.979 |
| Finnish | 86 | 344 | 430 | 3 | 1.01 | 314,756 | 8,723,593 | 0.984 |
| Spanish | 253 | 300 | 553 | 3 | 1.00 | 538,800 | 8,794,151 | 0.985 |
Variants genotyped in >1 cohort.
The genomic inflation factor listed is calculated from genotyped variants. However, genomic inflation factors for each cohort were also estimated for imputed SNPs separately, and results did not differ (e.g. maximum genomic inflation factor for imputed variants was 1.06).
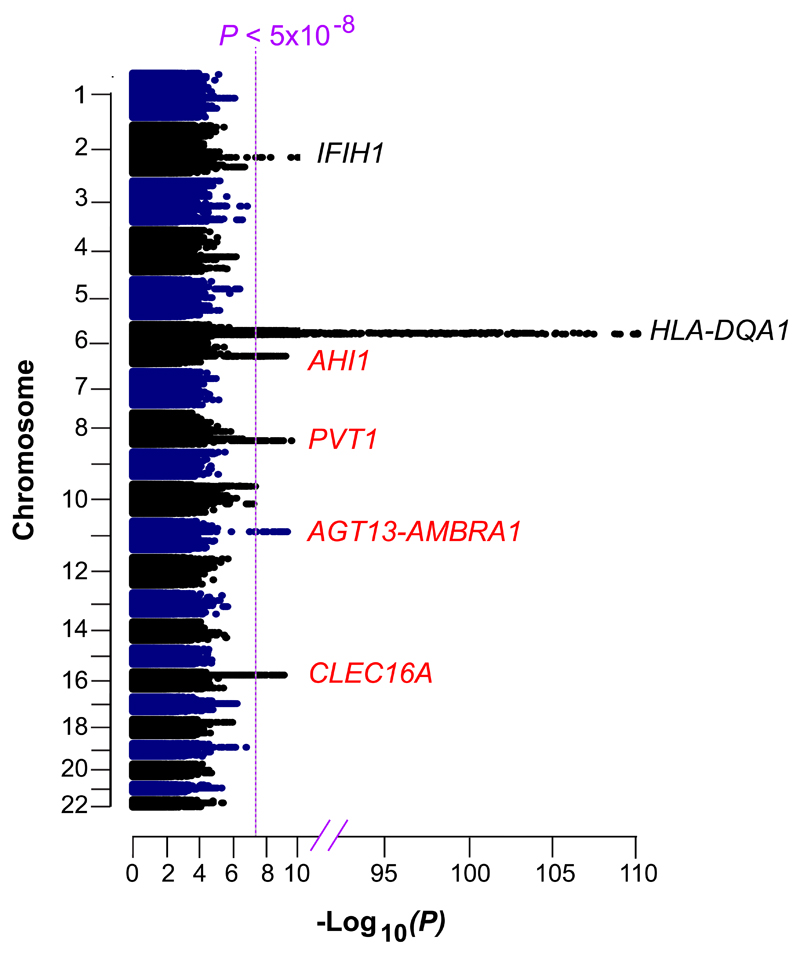
The strongest association was with the MHC region, 2.7kb upstream of HLA-DQA1 (P = 3.3x10−92) (Table 2). Protective association between IgAD and IFIH1 Thr946Ala9 was confirmed (P = 3.7x10−15), and association with the rare loss of function IFIH1 Ile923Val variant (P = 2.6x10−8) was subsequently identified (Supplementary Table 1). Val923 has previously been shown to be protective for T1D11 and psoriasis13 and abrogates interferon signaling.15 A rare gain of function IFIH1 Arg779His variant has also been reported in an IgAD case with SLE and a type I interferon signature.16
Table 2. Genome-wide Significant Results (P < 5x10−8) for IgAD GWAS.
| Closest Gene(s) | Varianta | Position (hg19) | Minor Allele | % Frequency in Swedish Sample | P | FDR q values | OR | Type | Immune Diseases Sharing this Locusb | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||||
| HLA-DQA1 c | rs116041786 | 6:32602396 | C | 14.4 | 38.7 | 3.3e–92 | 1.0e–87 | 0.38 | Intergenic | Celiac Graves’ IBD Sjögren’sd |
| IFIH1 | rs1990760 | 2:163124051 | G | 29.3 | 37.3 | 3.7e–15 | 2.7e–12 | 0.70 | Missense | T1D SLE Psoriasis Vitiligoe |
| PVT1 | rs11299600 | 8:129204573 | 1 bp deletion | 18.9 | 25.3 | 4.3e–11 | 2.4e–8 | 0.73 | Intergenic | RAf |
| ATG13 – AMBRA1 | rs4565870 | 11:46349869 | C | 28.9 | 23.6 | 6.7e–10 | 3.5e–7 | 1.38 | Intergenic | |
| AHI1 | rs7773987 | 6:135707486 | C | 51.3 | 44.6 | 8.4e–10 | 4.3e–7 | 1.30 | Intron | MS |
| CLEC16A | rs34069391 | 16:11161214 | 1 bp insertion | 14.3 | 19.6 | 1.4e–09 | 6.9e–7 | 0.71 | Intron | T1D MS PBCg |
Peak variants were imputed, except for IFIH1 and CLEC16A, which were genotyped in all but the Czech cohort. Three loci had suggestive association in our previous GWAS: PVT1 (P = 4x10−6), DGKZ (P = 2x10−6) and CLEC16A (P = 2x10−7).
Autoimmune diseases reporting a genome-wide significant variant (P < 5x10−8) in Immunobase and the GWAS Catalog (see URLs) with at least a modest effect (OR ≥ 1.1) in the same direction as IgAD results and on the same haplotype as a peak IgAD variant (r2 > 0.45).12 IBD, inflammatory bowel disease; MS, multiple sclerosis; SLE, systemic lupus erythematosus; PBC, primary biliary cirrhosis; RA, rheumatoid arthritis; T1D, type 1 diabetes.
Genetic association of IgAD with HLA class II is well-established, particularly with HLA-DQB1*02:01 and HLA-DRB1*01:02 and the highly protective HLA-DRB1*15:01 allele.6 Few of the cases (2.1%) were carrying two copies of rs116041786*C (versus 15.7% of controls).
MS, RA, SLE, T1D, vitiligo, allergy and asthma also have some associations reported at this locus.
IBD is also associated with this locus.
MS, celiac disease, eczema and allergy also have some associations reported at this locus.
Celiac disease and asthma also have some associations reported at this locus.
Our GWAS identified four new significant (P < 5x10−8) loci: PVT1, ATG13-AMBRA1, AHI1 and CLEC16A (Figure 1, Table 2). Peak novel IgAD variants were (1) a protective one bp deletion 91kb downstream of PVT1 (P = 4.3x10−11); (2) a variant 289kb upstream of ATG13 (P = 6.7x10−10); (3) an intronic AHI1 variant (P = 8.4x10−10); and (4) a protective one bp intronic insertion in CLEC16A (P = 1.4x10−9) (Table 2, Supplementary Table 2, Supplementary Figures 1-6). 28 additional loci contained at least one variant with 5x10−8 < P < 1x10−5, and half (denoted with *) were known autoimmune loci: FAS*, GATA3*, MYO9B, CD86*, BCL6, IKZF2, PTGER4*, IKZF3*, EGR2*, FCRL3*, IL2RA*, PTPN2*, IL2*/IL21*, CDH23*, MAST4, FOXP1, MIR605, LINC01098, DRMT1, LINC00299, C22orf42, FAM171A1, BACH1, ZCCHC24, NCKAP5, LOC100505887, TMEM72 and CD28* (Supplementary Table 3, Supplementary Figure 7).
Figure 1. Genome-Wide Significant Loci in the IgAD Meta-Analysis.
Manhattan plot of P-values from a genome-wide meta-analysis of ~9.5M variants in 1,635 IgAD patients and 4,852 controls. The vertical line indicates genome-wide significance (P < 5x10−8). Loci with genome-wide significant results are labeled; red labels indicate novel genome-wide significant loci.
The heritability of IGAD in the Swedish cohort based on genome-wide imputed variants was 0.49 (standard error [S.E.] = 0.047). This estimate was reduced from 0.49 to 0.39 (S.E. = 0.059) when we conditioned on the five peak non-MHC variants, the peak MHC variant (rs116041786), and the peak MHC variant after conditioning on the peak MHC variant (rs116350876). When we excluded the MHC region altogether from the input, the estimated heritability dropped to 0.14 (S.E. = 0.054).
As one approach to compile a list of potential causative gene(s) and allele(s) in the new loci,17 we cross-referenced the peak variants plus 160 correlated variants (r2 ≥ 0.7) (44 variants in PVT1; 21 variants in ATG13–AMBRA1; 65 variants in AHI1; and 30 variants in CLEC16A; total N = 164), against epigenetic data, eQTLs, DNase I hypersensitive sites, transcription factor binding sites (ChIP-Seq), consensus motifs, GWAS loci, and promoter regions using RegulomeDB18, ENCODE19, Roadmap Epigenomics, GEO, the WashU EpiGenome Browser, ImmunoBase, and the GWAS Catalog (see URLs). The RegulomeDB catalogues lists regulatory evidence for ~60M variants, and about 5% (~3M) are categorized as likely to affect binding (score = 2) or likely to affect binding and linked to expression of a gene target (score = 1).
In our dataset, 13.1% (21/160) of the correlated variants were likely to affect binding (RegulomeDB score < 3), and were located in regions of open chromatin in an immune cell (as determined by DNase hypersensitivity) and/or in transcription factor binding sites (as determined by DNase footprints, ChIP-Seq, and/or binding site motifs) (Supplementary Table 4). Many of these variants (15/21) were also located in histone marks for active transcription (H3k4me3, H3k27ac or H3k4me1) in T regulatory cells (Treg) and all of the novel loci, except for AHI1, encompassed at least one Treg enhancer and active transcription start site. A comparison of the overlap between eQTL data from lymphoblast B cells and IgAD meta P-values using the Bayesian statistical framework Sherlock identified three cis eQTLs in AHI1 and DEXI associated with IgAD (Supplementary Table 4).
The PVT1 locus contains no protein coding genes, and the lncRNA PVT1 appears to be the most likely causative genetic element. PVT1 has been shown to be important for expression and copy number increase of MYC in tumors.20 The peak PVT1 variant was in moderate LD with four potential regulatory variants. The most interesting of these was rs7001706 (r2= 0.70), an intronic variant that lies in a FOXP3 transcription factor binding motif and in an H3K4me1 histone mark in Tregs. When we conditioned the regression analyses on either the peak variant or the potential regulatory variant rs7001706, they accounted for part but not all of the locus effect (data not shown), indicating that PVT1 contains more than one independent signal.
AHI1 is highly expressed in hematopoietic cells and overexpressed in blood cancers.21 AHI1 stabilizes BCR-ABL in leukemia cells by recruiting JAK2, and can regulate phosphorylation of JAK2-STAT5.22 JAK inhibitors are useful in treating rheumatoid arthritis (RA), psoriasis and alopecia areata. The peak variant was in LD with 8 eQTLs, all on the same haplotype. Rs2064430 (r2= 0.85) had the strongest evidence for regulatory binding (RegulomeDB score = 1d) (Supplementary Table 4). A comparison of the overlap between eQTL data from lymphoblast B cells and IgAD Pmeta using the Bayesian statistical framework Sherlock highlighted two cis eQTLs in AHI1 (rs2179781, rs9647635) with strong positive Log Bayes Factor (LBF) scores (6.83, 6.63) (Supplementary Table 4). Of the variants highlighted in Supplementary Table 4, rs2179781 had the smallest Pmeta value. Rs2179781*A was associated with reduced risk for IgAD and reduced AHI1 expression (Supplementary Figure 8).
The peak SNP upstream of ATG13 (autophagy 13) and AMBRA1 (autophagy/beclin-1 regulator 1) was correlated with two likely regulatory variants (Supplementary Table 4). ATG13 and AMBRA1 physically interact 23 and share the same chromatin interaction domain. Autophagy plays a role in autoimmunity24, plasmablast differentiation25, and immunoglobulin production in plasma cells.26 ATG13 and AMBRA1 are the most likely risk genes in this interval, but the causal variant remains to be determined.
CLEC16A contains several eQTLs for DEXI,27 and T1D-associated CLEC16A variants28 are located in an intron that shows chromatin interaction with the DEXI promoter, as determined by 3C27 and Hi-C37,38 mapping. The peak variant was in LD with 7 likely regulatory variants (Supplementary Table 4), including rs35300161, rs34972832 and the DEXI eQTL rs17806299 (r2 = 0.80). Rs35300161 and rs34972832 are present in open chromatin in germinal center and naïve B cells (Supplementary Table 4) and in an enhancer in various T cell types and tissues. A comparison of the overlap between eQTL data from lymphoblast B cells and IgAD Pmeta with Sherlock highlighted one cis eQTL in DEXI (rs741175) with a strong positive Log Bayes Factor (LBF) score (6.16) (Supplementary Table 4).
Clec16a knockdown mice show reduced numbers of B cells,29 and are protected from autoimmunity.30 Further work is required to determine whether CLEC16A or DEXI is the relevant gene in this interval.
We next applied a pathway analysis to the meta-analysis results using genomic randomization (PARIS)31 to estimate empirical significance (N = 10,000 randomizations) and an alternate pathway algorithm based on LD-independent intervals for replication (INRICH).32 PARIS reduces bias from gene size, pathway size, LD blocks and SNP coverage. Preliminary testing with PARIS v1.1 using imputed data has not been completed and thus, only the GWAS chip content was selected for this experiment. Using PARIS, we grouped variants into LD features and single SNPs into linkage equilibrium (LE) features. The LD and LE features were grouped by KEGG pathways and also by our user-defined Treg signature pathway. The features in each pathway were permuted 10,000 times with a randomly selected set of features of similar size. The empirical P is based on the number of features with a P < 0.05 in a pathway, compared to the number of features with a P < 0.05 in the permuted pathway. IgAD was associated (empirical P < 0.0001) with 7/221 (3%) KEGG pathways (total genes queried N = 5,701) (Supplementary Table 5).
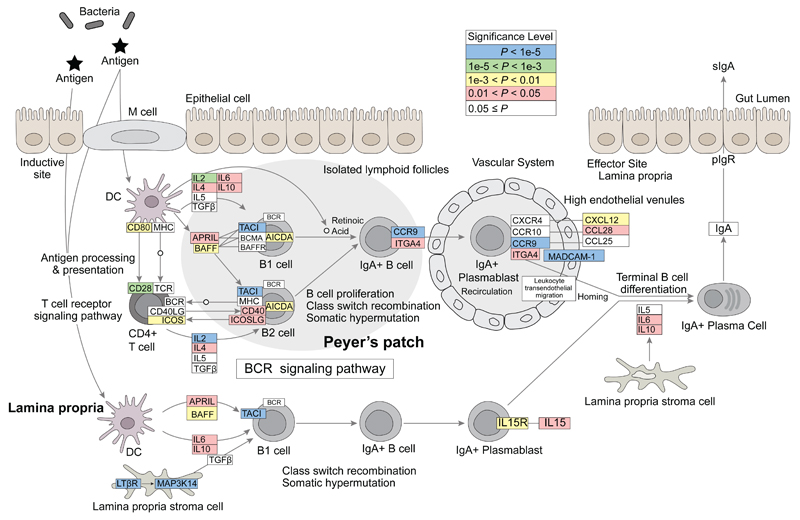
Of particular interest, one of the top pathways identified was the intestinal immune network for IgA production (empirical P < 0.0001), with 22/30 genes in the pathway containing variants with a P value < 0.05 in the IgAD meta-analysis (Figure 2, Supplementary Table 6). These include genes encoding inflammatory and immune regulatory cytokines (IL-2, IL-4, IL-6, IL-10, IL-15, APRIL, BAFF), key cell surface molecules on B and T cells (TACI, CD40, IL-15R, CD28, ICOS, ICOSLG), molecules essential for the homing of lymphocytes to the gut (ITGA4, CCR9, MADCAM-1, CXCL12, CCL28, LTβR, MAP3KI4) and a DNA deaminase required for B cell class switch recombination (AICDA). These data suggest that the genetic contribution to IgA deficiency includes many genes known to regulate IgA production.
Figure 2. KEGG Pathway for IgA Production Associated with IgAD.
A GWAS-based pathway analysis of IgAD meta-analysis P-values was performed of all 221 KEGG database pathways using PARIS and INRICH software (see Methods). A top identified pathway (P < 0.0001 based on 10,000 permutations) was the “Intestinal Immune Network for IgA Production” where 22/30 annotated pathway genes contained variants with a P value < 0.05 in the IgAD meta-analysis. The colors of the boxes around the gene names correspond to the smallest P value observed in the IgAD meta-analysis within that gene: blue, P < 1x10−5; green, 1x10−5< P < 1x10−3; yellow, 1x10−3< P < 0.01; red, 0.01 < P < 0.05, white P ≥ 0.05.
Similar to RA and T1D GWAS SNPs, which are enriched for active histone marks (H3k4me3) in Tregs17 and lymphoid gene enhancers33, respectively, IgAD variants were enriched for active histone marks and enhancers in Tregs. When we applied a set of Treg signature genes (27 autosomal genes that are significantly upregulated in FOXP3+ Tregs versus conventional FOXP3– CD4+ T cells34) as a user-defined pathway in a PARIS pathway analysis, we found that the Treg signature pathway was significantly enriched for association with IgAD (empirical P < 1x10−5) (Supplementary Table 7). FOXP3 is an important transcriptional regulator of Treg differentiation,35 and germline mutations of FOXP3 cause IPEX,36 a disease characterized by impaired Treg function and life-threatening autoimmunity. Tregs in B cell germinal centers of Peyer’s patches have been shown to influence the ability of sIgA to maintain microbial diversity.37
Using GWAS summary statistics for T1D and RA, we tested for shared genetics between these two diseases and IGAD. Applying GPA,38 we found significant shared genetics both between T1D and IGAD, and between RA and IGAD. GPA uses a likelihood ratio test to compare a model of the pairwise summary statistics with no shared genetics to a model with shared genetics. Results indicate significant shared genetics, even when variants in the MHC region were omitted (P < 1.6x10−7).
In conclusion, we identified four new susceptibility loci for IgAD – PVT1, AHI1, AMBRA1–ATG13 and CLEC16A. Interrogation of peak IgAD variants and SNPs in LD revealed 21 putative regulatory variants. Pathway analyses highlighted enrichment for association in the intestinal immune network for IgA production and in Treg signature genes. Further work is needed to validate novel associations via genotyping, to identify causal variants in the new loci, to explore the regulatory role of associated variants through functional studies, and to investigate autoimmune mechanisms that contribute to IgAD pathogenesis.
Online Methods
The GWAS study tested ~9.5M variants in 1,635 IgAD patients (clinical diagnosis defined as serum IgA ≤0.07 g/L with normal IgM and total IgG)39 and 4,852 controls from seven independent case-control cohorts (see Table 1).
Samples
Only IgAD cases without comorbid autoimmune diseases (e.g. Type 1 diabetes, celiac disease) were included in this study. Furthermore, IgAD cases were screened for celiac disease (auto-antibodies to tissue transglutaminase) and excluded from the study if positive.
Genotyping
Illumina, Inc. (San Diego, CA, USA) developed all of the genotyping arrays. We genotyped cases on Omni1-Quad and Omni2.5, Swedish controls (Swedish TwinGene Project)40 on Omni Express, Spanish controls on Omni1-Quad, Italian controls on 1Mb (Milan Hypergenes Study)41 except for 42 Italian controls genotyped on the 550K (New York Cancer Project (NYCP))42, and Czech controls on HumanHap1M, Omni1-Quad, and Omni 2.5 (National Cancer Institute Division of Cancer Epidemiology and Genetics) Imputation Reference Dataset). 64 Czech controls were genotyped on HumanHap500 (NCI CGEMS (Cancer Genetic Markers of Susceptibility) Project). Ferreira et al. genotyped the Swedish cohort43, which included 34 Icelandic cases, on Hap300 (except for 175 cases genotyped on Human610), Finnish cases on 610-Quad, Finnish controls on CNV370 arrays, and the Spanish cohort on 610-Quad v1. See the “SNPs Genotyped (N)” column in Table 1 for the number of SNPs that overlapped between the arrays for each of the 7 cohorts. All SNPs were mapped to build hg19 coordinates using liftOver.
QC Post-Genotyping and Pre-Imputation
Prior to imputation, we removed variants with a genotyping rate <98%, ambiguity (A/T SNPs and C/G SNPs), evidence of deviation from Hardy-Weinberg equilibrium in controls (P < 1x10−4), and MAF < 1x10−6. Subsequently, we removed individuals who were missing >1% of genotypes and who had heterozygosity ±6 standard deviations (s.d.) from the mean. To estimate identity-by-descent (IBD) and conduct principal components analysis (PCA) (EIGENSOFT v3.044), we randomly selected 100,000 LD-pruned variants (LD pruning was based on r2 > 0.2; excluding insertion/deletions, variants in regions of long range LD45 or in the MHC (chr6:24–36Mb) or in a large inversion on chromosome 8,46 and variants with MAF < 0.05) and included 878 ancestry informative markers.47 We estimated and plotted IBD for all pairs of samples and removed duplicate samples and cryptic related individuals (P(IBD=0)<0.8). For the PCA analysis, we used 5 outlier removal iterations and 10 principal components (PCs) along which to remove outliers during each outlier removal iteration. Any sample exceeding 6 s.d. along one of the PCs was considered a population outlier and was removed. We then reran PCA without the ancestry outliers.
For the four new cohorts, we iteratively selected up to four controls similar to a previously described approach (developed by M.F.S.) to minimize population substructure differences between cases and controls.48 Each control could only be selected once. For the matching we sequentially selected a control that passed QC and best matched the case, considering the EIGENVALUES results from PCs contributing at least 1% of variance determined from PCs with significant P–values (P < 0.05) in the Tracy-Wisdom test44. We estimated percent variance explained by subtracting the eigenvector value of the first non-significant PC from each significant PC, summing this value, and dividing each eigenvector value by the sum. EIGENVALUES for each PC were adjusted for the EIGENVECTOR scale (i.e. adjusted for the relative contribution of each EIGENVECTOR in describing the data). The lowest cumulative sum of the absolute differences (Manhattan distance for each PC after adjustment based on relative EIGENVALUES) between all cases and all controls was scored as the best match.
Subsequently, we reran PCA a third time for each cohort using the final set of case-control samples. PCs contributing at least 1% of variance determined from PCs with significant P-values (P < 0.05) in the Tracy-Wisdom test44 were used as covariates in the association analyses. Significant PCs were also plotted with cases and controls color coded differently for visual inspection. We tested for differential genotyping rates between cases and controls but did not observe any.
Statistical Analysis Before Imputation
We conducted χ2 tests of association on genotypes for each cohort separately, using only variants that overlapped between the arrays used to genotype in the cohort, and estimated a genomic inflation factor (λGC) 49 based on the median χ2 for non-MHC genotyped variants adjusted for ancestry (PLINK v1.0749). Prior to meta-analysis and imputation, we plotted P-values in a quantile-quantile (Q-Q) plot (excluding the MHC) and in a Manhattan plot in R.
Imputation
We imputed genotypes for each cohort separately using the 1000 Genomes Project Phased I integrated variant set release as a reference panel (SHAPEIT v2.r72750, IMPUTE2 v2.3.051). We imputed cases and controls from the same cohort together, using only variants that overlapped between the arrays used to genotype the cohort. Genotypes were imputed in 5 MB chunks, excluding centromeres. We used an effective size of the population of 20,000; 80 haplotypes as templates when phasing observed genotypes; and performed 30 MCMC iterations, where the first 10 MCMC iterations were discarded as burn-in.
Statistical Analysis After Imputation
We excluded variants with an imputation info score <0.6 or a MAF<0.01 in the controls or evidence of deviation from HWE (P < 1x10−6), and tested association for each case-control cohort separately with logistic regression (additive model), adjusted for ancestry (SNPTEST v2.5-beta452). The median info score for each cohort was: Swedish 0.988; Spanish 0.974; Italian 0.948; Czech 0.929; Swedish 2010 0.979; Finnish 2010 0.984; and Spanish 2010 0.985. We used a missing data likelihood score test to address genotype uncertainty. We estimated a genomic inflation factor (λGC)49 based on the distribution of P-values for each cohort using the GenABEL estlambda package in R (see URLs). We plotted P-values in a quantile-quantile (Q-Q) plot (excluding the MHC) and in a Manhattan plot in R.
Meta-analysis
We pooled results in a meta-analysis (PLINK v1.0749), and report P values and ORs for a random effects model. We plotted genome-wide results with R v3.0.2 (see URLs) and regional results in LocusZoom v1.3, using LD estimated in PLINK with the 1000 Genomes EUR data (release 20130502 v.5).53 Meta-analysis P values were adjusted for multiple testing with the false discovery rate (FDR) in R.54
Pathway Analysis of IgAD GWAS Results
We conducted a GWAS based pathway analysis on IgAD GWAS meta-analysis P-values after imputation for a subset of variants (~540K variants from the Illumina 550K array) using PARIS (v 1.1.0b)55. PARIS groups SNPs into LD features and single SNPs into linkage equilibrium features. The LD features can extend up to 50KB past gene boundaries. It then groups these features by pathway, and permutes the genomic structure of interrogated pathways to determine the significance of the pathway while accounting for differences in LD, gene size, pathway size and SNP coverage between pathways. The total number of features with a significant P-value (defined as P < 0.05), is compared with the number of significant features in the permuted pathway. The software was tested and validated using genotype data from the Illumina 550K. We only used a subset of variants because PARIS (v 1.1.0b) has been demonstrated to be robust when used on this SNP array (Yaspan, pers. commun.).
Variants with significant P-values (P < 0.05 from the GWAS meta-analysis data) in Kyoto Encyclopedia of Genes and Genomes (KEGG)56 pathways (v. 3/2011) were identified, and the genome was permuted (10K permutations) to estimate pathway significance (P < 0.0001). Out of 10,000 randomizations, none of the randomized pathways had more variants with P < 0.05 than the identified pathways. A follow up pathway-based analysis was conducted with Inrich (v.1.0) using only pathways that were significant in the PARIS analysis (P < 0.0001).57 We identified clumps (LD independent intervals) using P < 0.0001 as the significance threshold for index SNPs and P < 0.05 as the secondary significance threshold for clumped SNPs and r2 > 0.5 as the LD threshold for clumping (PLINK). There were 72 non-overlapping LD independent genomic intervals of enriched association, based on the subset of meta-analysis P-values used in the PARIS analysis. Pathways that contained at least two non-MHC genes and had both a PARIS P < 0.0001 and an Inrich P < 0.05 were considered statistically significant.
Regulatory Variation
For each of the five non-MHC IgAD loci reaching genome-wide significance (P < 5x10−8), SNPs that were in LD (r2 > 0.5, EUR) with the peak SNP were identified. The peak SNP was defined as the SNP with the lowest P-value. LD was estimated using whole genome sequence (WGS) data (mean 30x coverage) available for 583 European Americans (PLINK v1.90b20). Significant peak variants, and variants correlated with the peak variants, were queried for known and predicted regulatory DNA elements (RegulomeDB v1.1, dbSNP141).
Overlap between expression QTLs and GWAS
The overlap between IgAD meta-analysis P-values and expression QTL data from lymphoblast B cells 58,59 was assessed using the Bayesian statistical framework Sherlock (see URLs).60
Heritability Estimates
The program GCTA-GREML (see URLs) was used to estimate the variance between the cases and controls by the entire genome, and the variance explained by the IgAD genome-wide significant variants. The heritability of IgAD for the entire genome was estimated with GCTA in the Swedish IgAD cohort (n = 1,214,325 imputed genotypes).61 The other IgAD cohorts were underpowered for heritability estimates (gctaPower). We estimated the heritability of IgAD for the 7 peak IgAD variants by including the 7 peak IgAD variants as covariates. This analysis was then repeated without the MHC region (chr6:24–36 Mb).
Shared Genetics with T1D and RA
The program Genetic analysis incorporating Pleiotropy and Annotation (GPA)62 was used to estimate the extent of the genetic overlap between IgAD and half a million T1D variants63 and between IgAD and 6 million RA variants64. Summary statistics for T1D were publicly available from dbGaP (accession number: phs000180.v1.p1). Summary statistics for RA were from the European subset of a recent meta-analysis of RA64 (see URLs). There were 5,966,608 and 501,001 variants in the IgAD-RA and IgAD-T1D pairwise analysis, respectively. Using these genome-wide summary statistics, we estimated the proportion of shared variants (variants associated to both IgAD and RA, or to both IgAD and T1D). We tested for significance of shared genetics between pairs of diseases using a likelihood ratio test62. GPA estimates the proportion of variants associated to both diseases and uses a likelihood ratio test to assess statistical significance. This analysis was conducted genome-wide, and then repeated with the exclusion of the MHC region (chr6: 24–36 Mb) due to the extensive LD and highly significant variants in this region. After removing variants in the MHC, 5,944,048 and 498,320 variants remained in the IgAD-RA and IgAD-T1D analyses, respectively.
Supplementary Material
Note: Supplementary information is available on the Nature Genetics website.
Acknowledgements
We thank the cases and controls that participated in this study. We thank Brian Yaspan, Jeong Kim, Jonathan Sitrin and Tiffany Hung for insightful discussion, Jennifer Tom and Oleg Mayba for helpful feedback on a manuscript draft, and Allison Bruce for sharing her graphical expertise. Genentech, Inc. funded the current GWAS. Annette Lee and Peter Gregersen performed the sample genotyping at the Laboratory of Genomics and Human Genetics at the Feinstein Institute for Medical Research. These studies were supported by the US National Institutes of Health (U19 AI067152 and AR043274), the Swedish Research Council, the European Research Council (242551-ImmunoSwitch), EURO-PADnet grant 201549, CETOCOEN PLUS and the Fondazione C. Golgi, Brescia, Italy. Financial support was also provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet. The population allele and genotype frequencies were obtained from a data source funded by the Nordic Center of Excellence in Disease Genetics based on regional samples from Finland and Sweden. The Helsinki 4 study was supported by the Yale Center for Human Genetics and Genomics and the Yale Program on Neurogenetics and US National Institutes of Health grants R01NS057756 and U24 NS051869.
Footnotes
URLs
Hi-C Mapping, promoter.bx.psu.edu/hi-c/index.html
LocusZoom, genome.sph.umich.edu/wiki/LocusZoom_Standalone
RegulomeDb, regulome.stanford.edu
Immunobase, immunobase.org
NHGRI-EBI GWAS Catalog, ebi.ac.uk/gwas
GenABEL, genabel.org/GenABEL/estlambda.html
Gene Expression Omnibus (GEO), ncbi.nlm.nih.gov/geo
GCTA-REML, cnsgenomics.com/software/gcta/reml.html
Encyclopedia of DNA Elements (ENCODE), genome.gov/encode
WashU EpiGenome Browser, epigenomegateway.wustl.edu/browser
Roadmap Epigenomics, roadmapepigenomics.org
Sherlock, sherlock.ucsf.edu
STRING, string-db.org
RA GWAS summary statistics, plaza.umin.ac.jp/~yokada/datasource/software.htm
Plink, pngu.mgh.harvard.edu/purcell/plink/
Eigensoft, hsph.harvard.edu/alkes-price/software
Kegg, genome.jp/kegg/pathway.html
PARIS, ritchielab.psu.edu/software/paris-download
Inrich, atgu.mgh.harvard.edu/inrich
SHAPEIT, mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html
IMPUTE2, mathgen.stats.ox.ac.uk/impute/impute_v2.html
SNPTEST, mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
liftOver, genome.ucsc.edu/cgi-bin/hgLiftOver
gctaPower, cnsgenomics.com/shiny/gctaPower
Accession Codes
GWAS data generated in this study are accessible through Genentech, Inc. at http://research-pub.gene.com/bronson_et_al_2016.
Author Contributions
P.G.B. and D.C. carried out the analyses for this study. P.G.B., L.H., T.W.B., R.R.G and T.B. conceived and directed this study. L.H., Q.P.-H, A.P., V.L., T.F., J.L., E.A., L.F.P., V.F. and V.T. performed subject diagnosis, coordinated the enrollment of subjects and provided access to genotyping datasets. M.F.S. and J.M. provided access to genotypes for healthy controls. M.F.S. provided guidance for addressing population structure due to ancestry. M.F.S., Q.P.-H., R.C.F., T.B., R.R.G., and W.O. contributed to data access and analysis. P.G.B., T.W.B., L.H., and R.R.G. wrote the manuscript with collaboration from coauthors. All authors discussed the results and commented on the manuscript.
Competing Financial Interests
The authors do not declare competing financial interests.
References
- 1.Pan-Hammarstrom Q, Hammarstrom L. Antibody deficiency diseases. Eur J Immunol. 2008;38:327–33. doi: 10.1002/eji.200737927. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–6. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–61. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borte S, et al. Interleukin-21 restores immunoglobulin production ex vivo in patients with common variable immunodeficiency and selective IgA deficiency. Blood. 2009;114:4089–98. doi: 10.1182/blood-2009-02-207423. [DOI] [PubMed] [Google Scholar]
- 5.Cao AT, et al. Interleukin (IL)-21 promotes intestinal IgA response to microbiota. Mucosal Immunol. 2015 doi: 10.1038/mi.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira RC, et al. High-density SNP mapping of the HLA region identifies multiple independent susceptibility loci associated with selective IgA deficiency. PLoS Genet. 2012;8:e1002476. doi: 10.1371/journal.pgen.1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oen K, Petty RE, Schroeder ML. Immunoglobulin A deficiency: genetic studies. Tissue Antigens. 1982;19:174–82. doi: 10.1111/j.1399-0039.1982.tb01437.x. [DOI] [PubMed] [Google Scholar]
- 8.Ludvigsson JF, Neovius M, Hammarstrom L. Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol. 2014;34:444–51. doi: 10.1007/s10875-014-0009-4. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira RC, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet. 2010;42:777–80. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 10.Kiryluk K, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nature Genetics. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunninghame Graham DS, et al. Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with systemic lupus erythematosus. PLoS Genet. 2011;7:e1002341. doi: 10.1371/journal.pgen.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Carriers of rare missense variants in IFIH1 are protected from psoriasis. J Invest Dermatol. 2010;130:2768–72. doi: 10.1038/jid.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregersen PK, et al. Risk for myasthenia gravis maps to a (151) Pro-->Ala change in TNIP1 and to human leukocyte antigen-B*08. Ann Neurol. 2012;72:927–35. doi: 10.1002/ana.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shigemoto T, et al. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284:13348–54. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Eyck L, et al. Brief Report: IFIH1 Mutation Causes Systemic Lupus Erythematosus With Selective IgA Deficiency. Arthritis Rheumatol. 2015;67:1592–7. doi: 10.1002/art.39110. [DOI] [PubMed] [Google Scholar]
- 17.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle AP, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng YY, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–6. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringrose A, et al. Evidence for an oncogenic role of AHI-1 in Sezary syndrome, a leukemic variant of human cutaneous T-cell lymphomas. Leukemia. 2006;20:1593–601. doi: 10.1038/sj.leu.2404321. [DOI] [PubMed] [Google Scholar]
- 22.Zhou LL et al. AHI-1 interacts with BCR-ABL and modulates BCR-ABL transforming activity and imatinib response of CML stem/progenitor cells. J Exp Med. 2008;205:2657–71. doi: 10.1084/jem.20072316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–16. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 24.Pierdominici M, et al. Role of autophagy in immunity and autoimmunity, with a special focus on systemic lupus erythematosus. FASEB J. 2012;26:1400–12. doi: 10.1096/fj.11-194175. [DOI] [PubMed] [Google Scholar]
- 25.Clarke AJ, et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Annals of the Rheumatic Diseases. 2015;74:912–920. doi: 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pengo N, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 27.Davison LJ, et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum Mol Genet. 2012;21:322–33. doi: 10.1093/hmg/ddr468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, et al. Association of CLEC16A with human common variable immunodeficiency disorder and role in murine B cells. Nat Commun. 2015;6:6804. doi: 10.1038/ncomms7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuster C, et al. The Autoimmunity-Associated Gene CLEC16A Modulates Thymic Epithelial Cell Autophagy and Alters T Cell Selection. Immunity. 2015;42:942–52. doi: 10.1016/j.immuni.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaspan BL, et al. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011;129:563–71. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee PH, O'Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–6. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidl C, et al. The enhancer and promoter landscape of human regulatory and conventional T-cell subpopulations. Blood. 2014;123:e68–78. doi: 10.1182/blood-2013-02-486944. [DOI] [PubMed] [Google Scholar]
- 35.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–41. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–1. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 37.Kawamoto S, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–65. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 38.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. doi: 10.1371/journal.pgen.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.International Union of Immunological Societies Expert Committee on Primary, I et al. Primary immunodeficiencies: 2009 update. J Allergy Clin Immunol. 2009;124:1161–78. doi: 10.1016/j.jaci.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magnusson PK, et al. The Swedish Twin Registry: establishment of a biobank and other recent developments. Twin Res Hum Genet. 2013;16:317–29. doi: 10.1017/thg.2012.104. [DOI] [PubMed] [Google Scholar]
- 41.Salvi E, et al. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension. 2012;59:248–55. doi: 10.1161/HYPERTENSIONAHA.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell MK, et al. The New York Cancer Project: rationale, organization, design, and baseline characteristics. J Urban Health. 2004;81:301–10. doi: 10.1093/jurban/jth116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira RC, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nat Genet. 2010;42:777–80. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 44.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 45.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83:132–5. doi: 10.1016/j.ajhg.2008.06.005. author reply 135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kosoy R, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregersen PK, et al. Risk for myasthenia gravis maps to a (151) Pro-->Ala change in TNIP1 and to human leukocyte antigen-B*08. Ann Neurol. 2012;72:927–35. doi: 10.1002/ana.23691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 51.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 53.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 55.Yaspan BL, et al. Genetic analysis of biological pathway data through genomic randomization. Hum Genet. 2011;129:563–71. doi: 10.1007/s00439-011-0956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee PH, O'Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–9. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 59.Duan S, et al. Genetic Architecture of Transcript-Level Variation in Humans. The American Journal of Human Genetics. 82:1101–1113. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He X, et al. Sherlock: Detecting Gene-Disease Associations by Matching Patterns of Expression QTL and GWAS. American Journal of Human Genetics. 2013;92:667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. doi: 10.1371/journal.pgen.1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–81. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.