Abstract
Medical imaging has enormous potential for early disease prediction, but is impeded by the difficulty and expense of acquiring datasets prior to symptom onset. UK Biobank aims to address this problem directly by acquiring high quality, consistently acquired imaging data from 100,000 predominantly healthy participants, with health outcomes tracked over coming decades. The brain imaging includes structural, diffusion and functional modalities. Along with body and cardiac imaging, genetics, lifestyle measures, biological phenotyping and health records, this is expected to enable discovery of imaging markers of a broad range of diseases at their earliest stages, as well as provide unique insight into disease mechanisms. We describe UK Biobank brain imaging, and present results derived from the first 5,000 participants’ data release. Although that covers just 5% of the ultimate cohort, it already yields a rich range of associations between brain imaging and other measures collected by UK Biobank.
The primary clinical role of brain imaging to date has been in diagnosis and monitoring of disease progression, rather than providing predictive markers for preventative stratification or early therapeutic intervention. The predominant strategy for finding image-based markers of neurological and psychiatric disease has been to identify patients early in the diagnostic process to maximize statistical power in a small cohort (tens to hundreds of subjects). A key factor motivating the use of small, clinically-defined cohorts is the expense, time and specialized hardware associated with imaging. This approach has been effective in providing markers of disease progression, but identifying imaging markers of early disease requires measurements at the pre-symptomatic stage. Image-based measures of brain structure and function may evolve in a complex way throughout aging and progression of neuropathology. Therefore, markers with utility in monitoring disease progression post-diagnostically may not manifest pre-symptomatically, and conversely the most sensitive early predictors of disease may have plateaued by the time existing diagnoses become accurate.
Nevertheless, when known risk factors have enabled risk-stratified cohorts, imaging has been able to predict disease before symptom presentation. For example, magnetic resonance imaging (MRI) has demonstrated altered brain activity associated with the APOE genotype decades in advance of symptoms associated with Alzheimer’s disease1, and conversion from mild cognitive impairment to Alzheimer’s has been predicted2. These studies suggest that the primary obstacle to identifying early imaging markers is in obtaining data in pre-symptomatic cohorts drawn from the general population.
Alternatively, pre-symptomatic cohorts can be assembled using a prospective approach, in which a large number of healthy participants are intensively phenotyped (including imaging) and subsequently monitored for long-term health outcomes. While this approach is expensive, it is also efficient by capturing early biomarkers and risk factors for a broad range of diseases. It further becomes possible to discover unexpected interactions between risk factors (such as lifestyle and genetics). To date, the largest brain imaging studies have gathered data on a few thousand subjects. While this approach has identified associations between imaging and highly prevalent diseases, existing cohorts are still too small to produce sufficient incidence of many diseases if participants are recruited without identifying risk factors.
UK Biobank is a prospective epidemiological resource gathering extensive questionnaires, physical and cognitive measures and biological samples (including genotyping), in a cohort of 500,000 participants3. Participants consent to access to their full health records from the UK National Health Service, enabling researchers to relate phenotypic measures to long-term health outcomes. This is particularly powerful due to the combination of number of subjects and breadth of linked data. Participants were 40-69 years of age at baseline recruitment; this aims to balance the goals of characterizing subjects prior to disease onset against the delay before health outcomes accumulate. The cohort is particularly appropriate for study of age-associated pathology. All data from UK Biobank are available to researchers world-wide upon application, with no preferential access for scientists leading the study. An imaging extension to the existing UK Biobank study was funded in 2016 to scan 100,000 subjects from the existing cohort, aiming to complete by 2022. Imaging includes MRI of the brain, heart and body, low-dose x-ray bone and joint scans, and ultrasound of the carotid arteries. Identification of disease risk factors should increase over time with emerging clinical outcomes. For example, within the imaged cohort, 1800 participants are expected to develop Alzheimer’s disease by 2022, rising to 6000 by 2027 (diabetes: 8000 rising to 14,000; stroke: 1800 to 4000; Parkinson’s: 1200 to 2800)4.
We present example analytic approaches and studies that will be enabled by UK Biobank. Identification of novel imaging biomarkers of disease risk could support diagnosis, development of therapeutics, and assessment of interventions. The multi-modal, multi-organ imaging enables study of interactions between organ systems, for example, between cardiovascular health and dementia. The breadth of imaging makes this dataset valuable for multi-systemic syndromes such as frailty, accelerated aging characterized by general loss of reserves and poor tolerance to stressors, which indicates increased risk for a range of conditions including dementia5. This kind of resource can also evince hypotheses regarding causal mechanisms of disease that could be tested in follow-up interventional studies. Examples include modifiable risk factors, such as the association of obesity with later life cognitive dysfunction6, and the ability to study complex interactions of risk factors with lifestyle, environment and genetics. Finally, UK Biobank will enable validation and extension of associations identified by smaller-scale studies, including testing of hypotheses that combine results from multiple previous studies.
Results
Design rationale and initial imaging phase
The imaging study was designed to achieve the target of 100,000 subjects, each scanned once, over 5-6 years at three dedicated, identical centers operating 7 days/week, each scanning 18 subjects/day7. This places tight timing constraints, corresponding to one subject imaged every 36 minutes (see Online Methods). The first imaging center was built to establish feasibility and scanned 10,000 subjects over a two-year ramp-up period. Two further identical centers are being commissioned, with the three centers strategically positioned at population hubs: Manchester, Reading and Newcastle.
The goal of capturing imaging phenotypes relevant to the widest possible range of diseases and hypotheses meant that the protocol must deliver data with the broadest predictive power for neuropathology and mental health. We therefore included modalities that drive estimates of anatomical and neuropathological structure (“structural MRI”), brain activity (“functional MRI”, or fMRI), and local tissue microstructure (“diffusion MRI”, dMRI). The resulting imaging protocol (Supplementary Table 1) included: three structural modalities, T1-weighted, T2-weighted and susceptibility-weighted MRI (referred to here as T1, T2 and swMRI); dMRI; and both task and resting-state functional MRI (tfMRI and rfMRI). Recent advances in MRI acquisition technology8 enabled high spatial resolution dMRI and fMRI with high angular and temporal resolution, respectively, despite strict time constraints. For example, the protocol acquires dMRI data with 100 diffusion-encoding directions over two shells in just 7 minutes, enabling advanced model fitting of microstructural parameters that would not have been possible under these time constraints with previous generation technology. Following optimization of acquisition protocols, streamlining of participant preparation and minimization of scanner dead time (see Online Methods), UK Biobank was able to incorporate six neuroimaging modalities in just 36 minutes.
Unlike most of the measurements included in the original UK Biobank resource (e.g., alcohol consumption and cognitive test scores), raw imaging data is not a directly useful source of information. In addition to requiring image processing to remove artefacts and align images across modalities and individuals, most useful image phenotypes are derived through complex calculations combining many voxels and/or images. A fully automated processing pipeline was developed, producing both processed images as well as image-derived phenotypes (IDPs) – currently 2501 distinct individual measures of brain structure and function. Example IDPs include: the volume of specific brain structures; the strength of connectivity between pairs of brain regions; the estimated dispersion of fibers in a given white matter tract. IDPs are intended to be useful for non-imaging experts; however, understanding of the confounds and pitfalls of imaging is required to draw appropriate conclusions.
We present results from the first data release (biobank.ctsu.ox.ac.uk/crystal/label.cgi?id=100), which includes outputs from the processing pipeline for 5285 subjects scanned in 2014-2015. As determined by the processing pipeline, 98% of participants’ datasets resulted in a usable T1, which is crucial for deriving usable information from the other modalities. Of these, data for the other brain imaging modalities were suitable for processing in the following percentages of subjects: T2=97%, swMRI=93%, dMRI=95%, tfMRI=92%, rfMRI=95%. All modalities were acquired and usable in 89% of subjects. Results from this data release are illustrated in Figs 1-4, including a multimodal atlas (separate population-average images for each of the modalities, all aligned to each other), available for download and online browsing (www.fmrib.ox.ac.uk/ukbiobank). Below, we present the following analyses: voxel-by-voxel analyses of age effects; pair-wise correlations of brain IDPs with other subject measures; multivariate analysis identifying dominant population patterns linking brain IDPs to other measured parameters; and an exemplar hypothesis-led study of aging, body mass index and smoking.
Imaging data, atlases and imaging-derived phenotypes
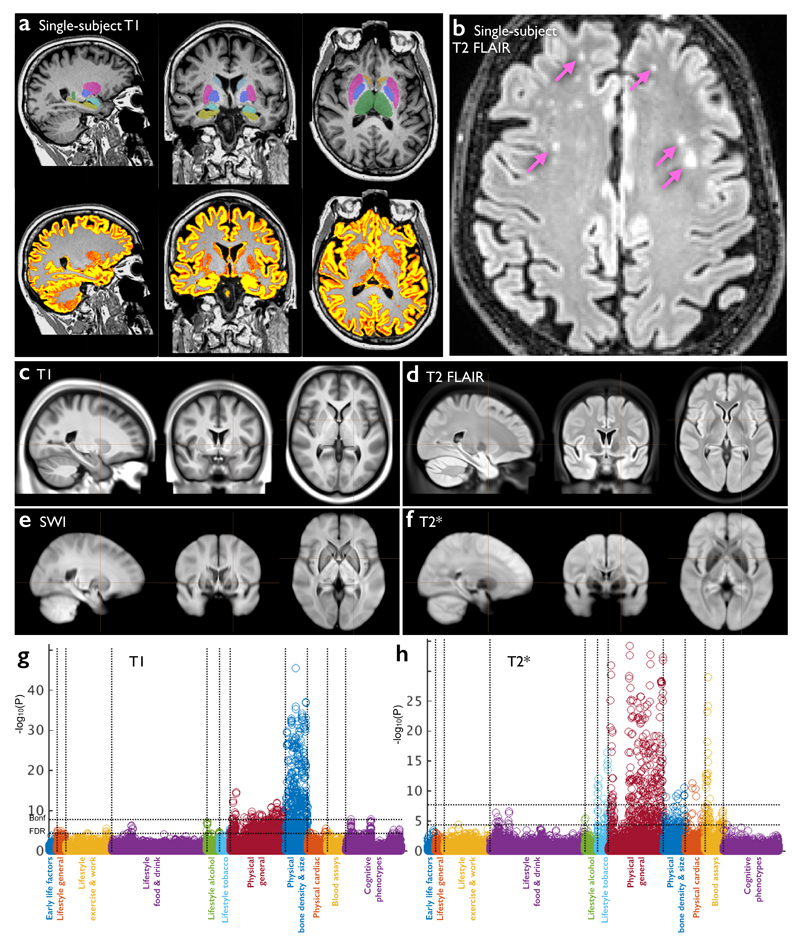
The three structural modalities (Fig 1) provide information about different aspects of the brain’s tissues, structures and neuropathologies. Data quality at the single-subject level is illustrated in Fig 1a,b. The group-averaged images produced for each modality are included in the initial data release as high-quality atlases (Fig 1c-f), depicting strong tissue contrast and excellent fidelity of alignment across subjects. The T1 modality (Fig 1a,c) is the most informative about the basic structure of the brain, including the depiction of the main tissue types (gray and white matter) and gross structure of the brain (main anatomical landmarks). From the T1 data, we derived 25 volumetric IDPs: total tissue volumes (gray, white and ventricular cerebro-spinal fluid), and the volumes of subcortical gray matter structures such as thalamus, caudate, putamen, pallidum, hippocampus and amygdala. The T1 data and T1-derived IDPs provide sensitive markers of atrophy (tissue loss), which can be both global (e.g., thinning of the cortex in aging)9 and local (e.g., reduction of hippocampal volume in Alzheimer’s disease)10. The T2 data (Fig 1b,d) is a FLAIR (fluid-attenuated inversion recovery) acquisition that also depicts basic anatomy, but is valuable primarily for detection of focal “hyperintensities” (i.e., high-signal regions) in white matter. T2 hyperintensities represent white matter lesions that have been associated with a broad range of neuropathological conditions11 (e.g., small vessel ischaemic disease), as well as occurring with increasing incidence in aging populations without (or potentially before) manifestation of neurological symptoms. IDPs relating to the volume of these white matter lesions will be included in future data releases. swMRI is a flexible modality that can be processed in multiple ways, each sensitive to different clinically-relevant properties. The first data release includes T2* signal decay times and enhancement of venous vasculature using susceptibility-weighted image (SWI) filtering12 (Fig 1e,f). swMRI IDPs in the current data release are the median T2* in each of 14 major subcortical gray matter structures, for example reflecting increased iron deposition associated with neurodegeneration13.
Figure 1. Data from the three structural imaging modalities in UK Biobank brain imaging.
(a) Single-subject T1-weighted structural image with minimal pre-processing: removal of intensity inhomogeneity, lower neck areas cropped and the face blanked to protect anonymity. Color overlays show automated modeling of several subcortical structures (above) and segmentation of gray matter (below). (b) Single-subject T2-weighted FLAIR image with the same minimal pre-processing, showing hyperintense lesions in the white matter indicated (arrows). (c) Group-average (n≈4500) T1 atlas; all subjects’ data were aligned together (see Online Methods for processing details) and averaged, achieving higher quality alignment, with clear delineation of deep grey structures and good agreement of major sulcal folding patterns despite wide variation in these features across subjects. (d) Group-average T2 FLAIR atlas. (e) Group-average atlas derived from SWI processing of swMRI phase and magnitude images. (f) Group-average T2* atlas, also derived from the swMRI data. (g) “Manhattan” plot (a layout common in genetic studies) relating all 25 IDPs from the T1 data to 1100 non-brain-imaging variables extracted from the UK Biobank database, with the latter arranged into major variable groups along the x axis (with these groups separated by vertical dotted lines). For each of these 1100 variables, the significance of the cross-subject univariate correlation with each of the IDPs is plotted vertically, in units of –log10(Puncorrected). The dotted horizontal lines indicate thresholds corresponding to multiple comparison correction using false discovery rate (FDR, lower line, corresponding to puncorrected=3.8×10-5) and Bonferroni correction (upper line, puncorrected=1.8×10-8) across the 2.8 million tests involving correlations of all modalities’ IDPs against all 1100 non-imaging measures. Effects such as age, sex and head size are regressed out of all data before computing the correlations. As an indication of the corresponding range of effect sizes, the maximum r2 (fractional variance of either variable explained by the other) is calculated, as well as the minimum r2 across all tests passing the Bonferroni correction. Here the maximum r2 = 0.045 and the minimum r2 = 0.0058. See Online Methods for more details of these analyses. (h) Plot relating all 14 T2* IDPs to 1100 non-imaging variables. Maximum r2 = 0.034, minimum r2 = 0.0063. Marked Bonferroni and FDR multiple comparison threshold levels are the same as in (g).
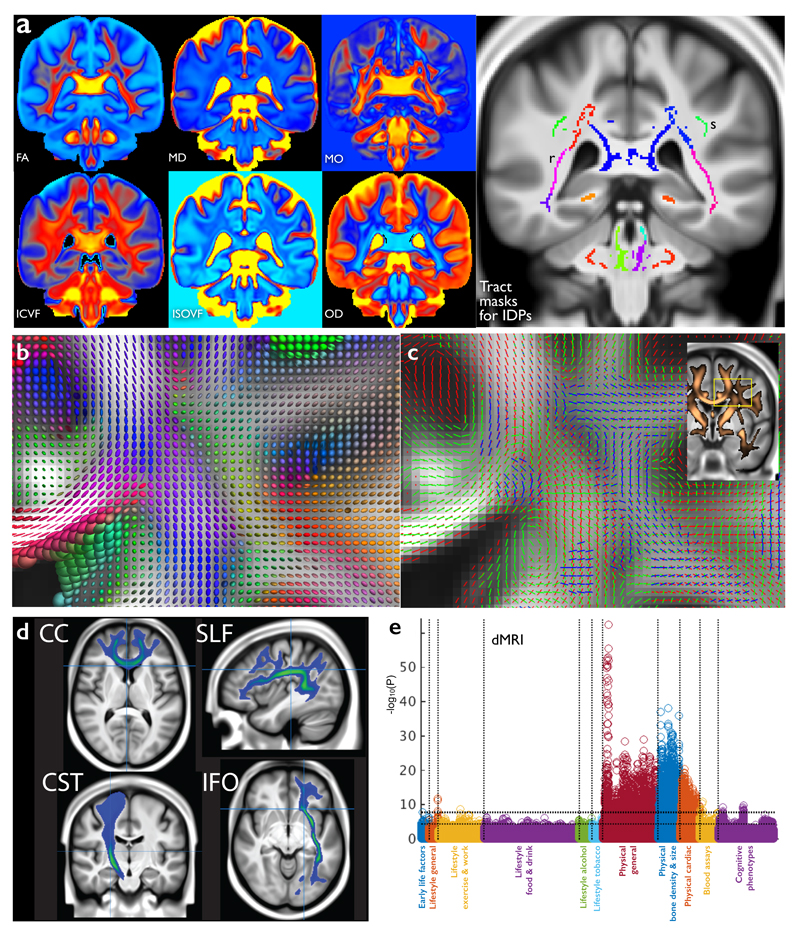
Diffusion MRI (Fig 2) reflects the random diffusion of water molecules, which is affected by the microscopic structure of tissue14, enabling us to infer the local density of cellular compartments in tissue (e.g. neurites). Additionally, axon bundles in white matter create an orientation dependence of water movement due to hindrance of diffusion perpendicular to the long axis of white matter tracts, an effect that can be tracked from voxel to voxel (“tractography”) to derive long-range white matter pathways. Three complementary diffusion models were fit to the signal in each voxel: (i) the diffusion tensor model15, describing the signal phenomenologically as resulting from a 3D ellipsoid profile of water displacement; (ii) the neurite orientation dispersion and density imaging (NODDI) model16, estimating microstructural properties (e.g., neurites vs. extracellular space); and (iii) the ball & sticks model17, estimating the orientation of multiple fiber populations within a voxel for tractography. 675 IDPs were extracted, by averaging parameters estimated by the first two models over 75 different white-matter tract regions based both on subject-specific tractography18 and from population-average white matter masks19.
Figure 2. The diffusion MRI data in UK Biobank.
(a) Group-average (n≈4500) atlases from six distinct dMRI modeling outputs, each sensitive to different aspects of the white matter microarchitecture. See Online Methods for processing details. The atlases shown are: FA (fractional anisotropy), MD (mean diffusivity) and MO (tensor mode); ICVF (intra-cellular volume fraction), ISOVF (isotropic or free water volume fraction) and OD (orientation dispersion index), from the NODDI microstructural modeling. Also shown are several group-average white matter masks used to generate IDPs (e.g., pink (r) are retrolenticular tracts in the internal capsules; upper-green (s) are the superior longitudinal fasciculi). (b) Tensor ellipsoids depicting the group-averaged tensor fit at each voxel for the region shown inset in (c). The shapes of the ellipsoids indicate the strength of water diffusion along three principal directions; long thin tensors indicate single dominant fiber bundles, whereas more spherical tensors (within white matter) generally imply regions of crossing fibers (seen more explicitly modeled in corresponding parts of (c)). (c) Group-averaged multiple fiber orientation atlases, showing up to 3 fiber bundles per voxel. Red shows the strongest fiber direction, green the second, and blue the third. Each fiber bundle is only shown where the modeling estimates that population to have greater than 5% voxel occupancy. Inset shows the thresholded mean FA image (copper) overlaid on the T1, with the region shown in detail in (b) and c) highlighted. (d) Four example group-average white matter tract atlases estimated by probabilistic tractography fed from the within-voxel fiber modeling: corpus callosum (genu), superior longitudinal fasciculus, corticospinal tract and inferior fronto-occipital fasciculus. (e) Plot relating all 675 dMRI IDPs (nine distinct dMRI modeling outputs from tensor and NODDI models × 75 tract masks) to 1100 non-imaging variables (see Fig 1g for details). Maximum r2 = 0.057, minimum r2 (passing Bonferroni) = 0.0065. Dotted horizontal lines (multiple comparison thresholds) are the same as in Fig 1g.
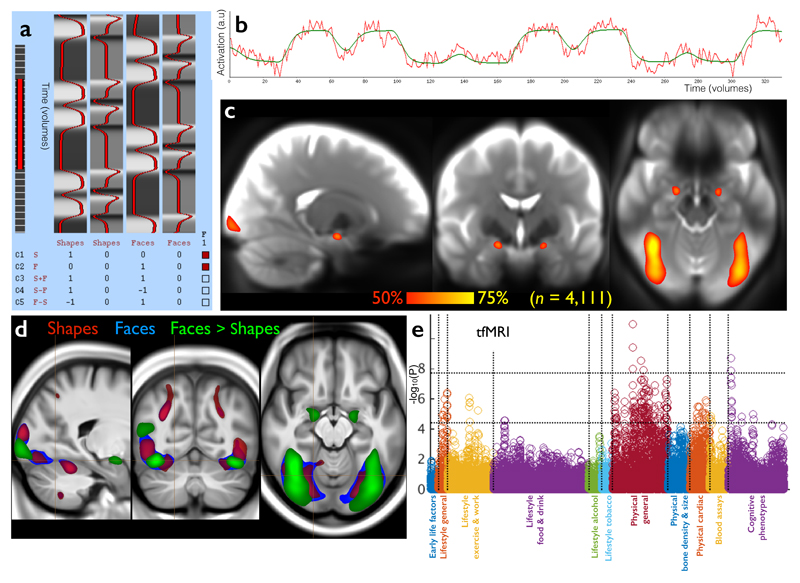
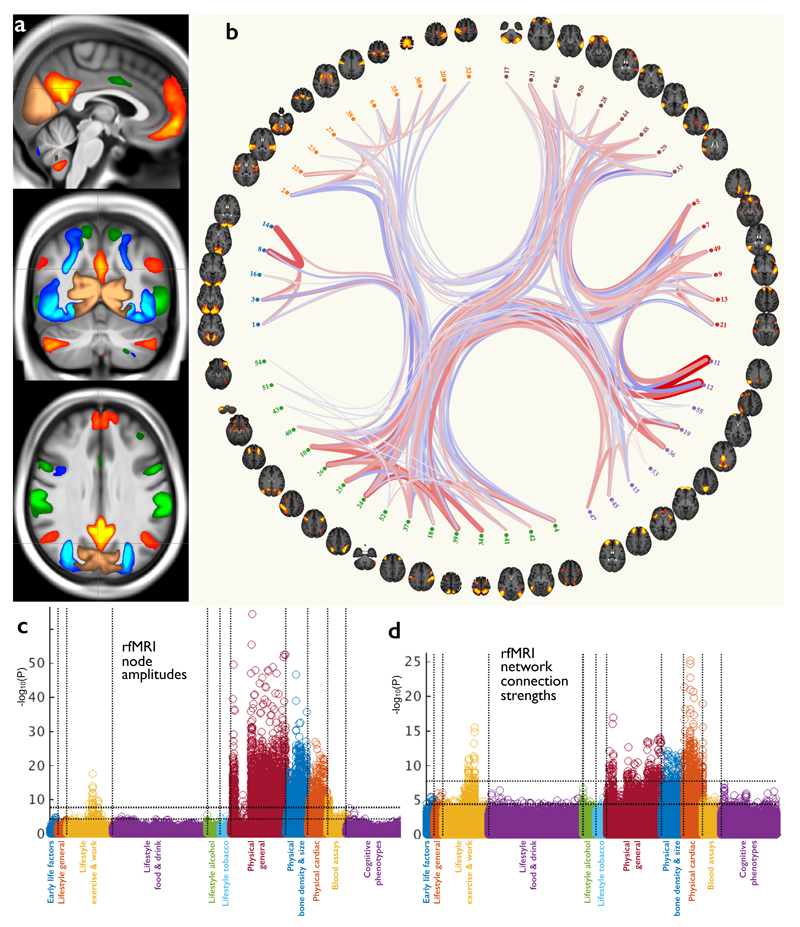
fMRI reflects neural activity indirectly, based on dynamic changes in blood oxygenation and flow resulting from changes in neural metabolic demand20. The task deployed in tfMRI (Fig 3) involves matching shapes and emotionally negative faces21 and was chosen to engage a range of neural systems, from low-level sensory and motor to perceptual (e.g., fusiform) and emotional (e.g., amygdala) areas. The 16 tfMRI IDPs quantitate the strength of brain activity changes for specific aspects of the task, in regions defined using the group-averaged activation maps shown, in three task conditions. Resting-state fMRI (Fig 4) identifies connected brain regions based on common fluctuations in activity over time in the absence of an explicit task22. Sets of voxels that co-fluctuate most strongly correspond to brain regions, referred to as network “nodes”; different nodes may have weaker co-fluctuations indicating a connection between them, a network “edge”. The group analysis of the rfMRI data generated two atlases of these functional networks - a “low-dimensional” decomposition of the brain into 21 functional sub-divisions and a higher-dimensional parcellation into 55 sub-divisions. IDPs represent a) edge connectivity strengths and b) node fluctuation amplitudes (Fig 4).
Figure 3. The task fMRI data in UK Biobank.
(a) The task paradigm temporal model (time running vertically) depicting the periods of the two task types (shapes and faces); for more information on this paradigm view, see http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FEAT/UserGuide. (b) Example fitted activation regression model vs. timeseries data (time running horizontally), for the voxel most strongly responding to the “faces > shapes” contrast in a single subject (Z=12.3). (c) Percentage of subjects passing simple voxel-wise activation thresholding (Z>1.96) for the same contrast. Note reliable focal activation in left and right amygdala. The underlying image is the group-averaged raw fMRI image. (d) Group-averaged activation for the 3 contrasts of most interest, overlaid on the group-average T1 atlas (fixed-effects group average, Z>100, voxelwise Pcorrected<10-30). (e) Plot relating the 16 tfMRI IDPs to 1100 non-imaging variables (see Fig 1g for details). Maximum r2 = 0.018, minimum r2 (passing Bonferroni) = 0.0062. Dotted horizontal lines (multiple comparison thresholds) are the same as in Fig 1g.
Figure 4. The resting-state fMRI data in UK Biobank.
(a) Example group-average resting-state network (RSN) atlases from the low-dimensional group-average decomposition, showing four out of 21 estimated functional brain networks, including the default mode network (red-yellow), dorsal attention network (green), primary visual (copper), and higher level visual (dorsal and ventral streams, blue). The three slices shown are (top to bottom) sagittal, coronal and axial. (b) The 55 non-artefact components from a higher-dimensional parcelation of the brain (axial views). These are shown as displayed by the connectome browser (www.fmrib.ox.ac.uk/analysis/techrep/ukb/netjs_d100), which allows interactive investigation of individual connections in the group-averaged functional network modeling. The 55 brain regions (network nodes) are clustered into groups according to their average population connectivity, and the strongest individual connections are shown (positive in red, anticorrelations in blue). (c) Plot relating the 76 rfMRI “node amplitude” IDPs to 1100 non-imaging variables (see Fig 1g for details). Maximum r2 = 0.065, minimum r2 (passing Bonferroni) = 0.0059. (d) Plot relating the 1695 rfMRI “functional connectivity” IDPs to 1100 non-imaging variables. Maximum r2 = 0.032, minimum r2 = 0.0059. Dotted horizontal lines (multiple comparison thresholds) in (c) and (d) are the same as in Fig 1g.
Voxel-wise associations with aging
IDPs reduce raw data into a compact set of biologically-meaningful measures, with current measures condensing ~2GB raw data per subject into 2501 IDPs, but such summary measures can lose valuable information. For example, once aligned to common coordinate systems, images can be analysed for cross-subject variation at the voxel level to provide a more spatially-detailed exploration than can be achieved via IDPs. However, this requires greater imaging expertise and computational resources, as well as often leading to lower statistical power (due to the greatly increased number of multiple comparisons and the higher noise in voxel-wise measures compared with regional averages). Below, we present example voxel-wise associations with aging.
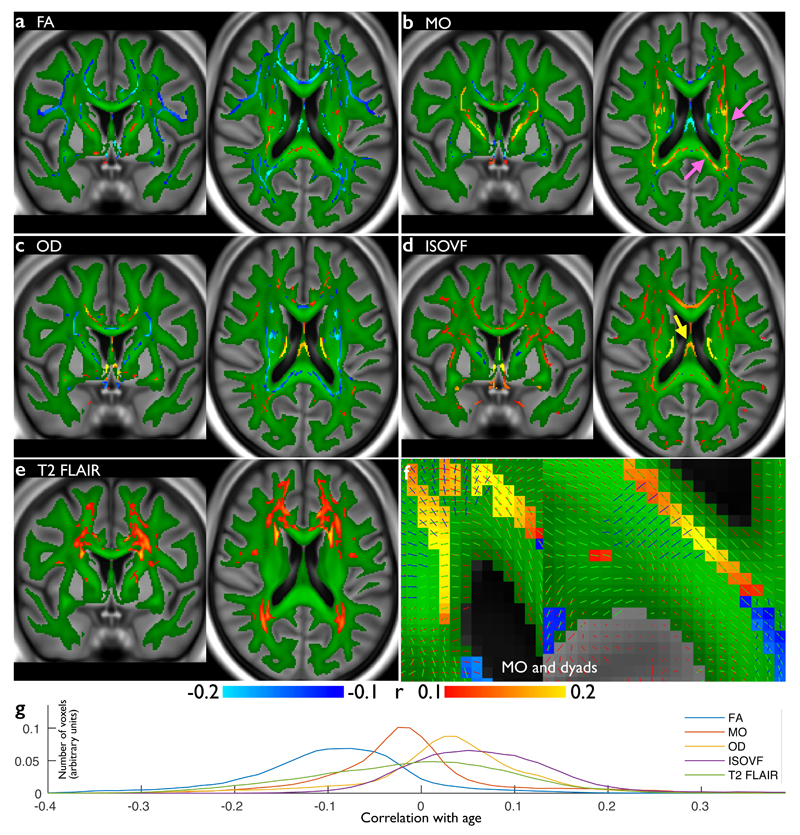
Fig 5 presents voxel-wise correlations of age with several parameters modeled from the dMRI data (along the centers of the main white matter tracts), as well as normalized T2 FLAIR intensity in the white matter. Fractional anisotropy (FA), a sensitive but non-specific marker of white matter integrity, predominantly demonstrates the established reduction of FA with aging (Fig 5a,g). However, some voxels exhibit the opposite, of FA increasing with aging, which may reflect degradation of secondary fibers or reduced fiber dispersion23; notably, none of the FA-based IDPs exhibit this significant positive correlation, demonstrating that averaging across tracts can sacrifice richness of information. The tensor mode24 (Fig 5b), which primarily describes whether a voxel contains one vs. multiple tracts, was even more sensitive with highly significant positive correlations in certain association fiber areas and posterior corpus callosum (arrows), likely the same effect seen as FA increases23. We further observed an increase in free water with aging (Fig 5d); the strongest increase, in the fornix (arrow), is likely due to an increase of the fraction of CSF within voxels spanning this thin tract as it atrophies. Finally, we calculated voxel-wise cross-subject correlation of age with T2 images. This analysis identified peri-ventricular areas, which are most susceptible to white matter hyperintensities known to be associated with aging (Fig 5e).
Figure 5. Voxel-wise correlations of participants’ age against several white matter measures from the dMRI and T2 FLAIR data.
(a) Voxel-wise (cross-subject) correlation of FA (fractional anisotropy) vs. age. Group-average FA in white matter is shown in green, overlaid onto the group-average T1. (b) Correlation of MO (tensor mode) vs. age, using the same color scheme. Nearby areas of MO increase are shown in greater detail in (f), which also shows the distinct primary fiber directions. (c) Correlation of OD (orientation dispersion) vs. age, including a reduction in dispersion in posterior corpus callosum. (d) Correlation of ISOVF (isotropic or free water volume fraction) vs. age, showing increases in “free water” with age in a broad range of tracts. (e) Voxel-wise correlation of T2 FLAIR intensity, showing increased intensity with aging in white matter. For (a-e), blue and red-yellow show negative and positive Pearson correlation with age, respectively (Pcorrected<0.05, with Bonferroni correction across voxels resulting in significance at r=0.1 (dMRI n=3722; T2 FLAIR n=3781). (g) Histograms (across voxels) of the voxel-wise age correlation of the correlation maps shown above, with correlation value on the x axis. FA and MO largely decrease with age, while OD and ISOVF largely increase.
A further example voxelwise analysis is shown in Supplementary Fig 1, where the rfMRI data was used to investigate aging effects in the default-mode resting-state network25. This also provides a demonstration that group analyses do not degrade with increasingly-large subject numbers (e.g., due to alignment issues), using group sizes from 15 to 5000. With increasing subject numbers, background noise is suppressed without increase in spatial blurring, and localized estimates of age-dependence stabilize, with statistical significance rising indefinitely.
Pairwise associations between brain IDPs and other measures
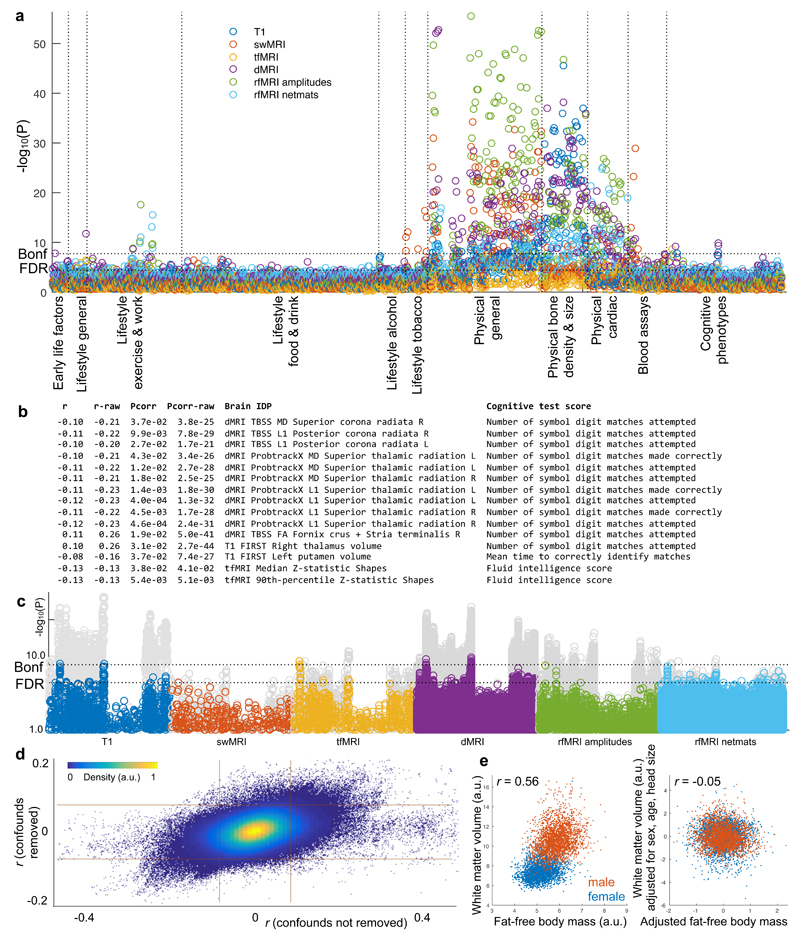
We conducted simple univariate association analyses to illustrate the richness of relationships between IDPs and other available variables, as well as the statistical power afforded by ~5000 subjects. We individually correlated all 2501 brain IDPs with 1100 other Biobank variables, where the latter were broadly grouped into 11 categories (Figs 1-4,6, Supplementary Fig 2). Even after false discovery rate (FDR) multiple comparison correction for these 2.8 million correlations, 57 out of the 66 combinations of brain modalities and non-brain-imaging categories show significant associations. Some variable categories exhibited large numbers of associations with IDPs (e.g., height and weight), whereas others (e.g. cognitive measures and alcohol/tobacco intake) had more focused associations.
Figure 6. Visualisation of 2.8 million univariate cross-subject association tests between 2501 IDPs and 1100 other variables in the UK Biobank database.
(a) Manhattan plot showing, for each of the 1100 non-brain-imaging variables, the statistically strongest association of that variable with each distinct imaging sub-modality’s IDPs. (i.e., 6 results plotted for each x axis position, each with a color indicating a brain imaging modality; this plot differs from the other Manhattan plots, which show correlations with all IDPs). Whereas the Manhattan plots in Figs 1-4 indicated associations for each brain imaging modality separately, here we depict all associations in a single plot. (b) List of all IDP-cognitive score associations passing Bonferroni correction for multiple comparisons (Pcorrected<0.05; Puncorrected<1.8x10-8). The first column lists the age-adjusted correlation coefficient, and the second shows the unadjusted correlation, both being correlations between a specific brain IDP (fifth column) and a cognitive test score (seventh column). UK Biobank cognitive tests carried out include Fluid Intelligence, Prospective Memory, Reaction Time (Shape Pairs Matching), Memorised Pairs Matching, Trail Making (Symbol Ordering), Symbol Digit Substitution, and Numeric Memory. (c) IDP associations with the cognitive phenotype variables (the full set of 174 cognitive variables, repeated for each brain imaging modality). Shown behind, in gray, are the same associations without adjustment for age, with a large number of stronger associations. Dotted horizontal lines (multiple comparison thresholds) in (a) and (c) are the same as in Fig 1g. (d) Scatterplot showing the relationship between adjusted correlations and those obtained without first regressing out the confound variables (each point is a pairing of one IDP with one non-brain-imaging variable, 2.8 million points). The grid lines indicate Bonferroni-corrected significance level (as described in Fig 1). (e) Example association between unadjusted white matter volume and fat-free body mass is high (r=0.56) when pooling across the sexes. After adjusting for several variables (including sex), the correlation falls almost to zero.
The above associations were estimated after adjusting all variables for age, sex, age-sex interaction, head motion and head size (“de-confounding”). Some factors can be unambiguously considered a confound to be removed (e.g., head motion, which can corrupt IDPs but also correlates with disease/aging26). For other factors (e.g., age), the appropriateness of de-confounding depends on the question being asked and needs to be taken into consideration when interpreting associations (see Discussion). The relationship between the correlations estimated with vs. without de-confounding (Fig 6d) demonstrates that in almost all cases the strength of association was reduced by de-confounding, in some cases being almost entirely removed (horizontal cloud around y=0).
We considered associations between cognitive tests and brain IDPs, including potential age interactions, in greater detail. Sex, head motion, and head size were regressed out of all data before computing correlations (see Online Methods). Figure 6b shows Bonferroni-significant (Puncorrected<1.8x10-8) associations with brain IDPs, both with and without adjusting for age. The task-fMRI vs. Fluid Intelligence associations are unchanged by adjusting for age, whereas all other cognition-IDP correlations are approximately doubled, being significantly stronger (Pcorrected<0.005) without age adjustment.
In the Symbol Digit Substitution test, participants replace symbols with numbers using a substitution key. Strong IDP associations were found with two scores: Number of symbol digit matches made correctly and Number of symbol digit matches attempted in the time allowed (because subjects rarely make mistakes, these two scores are highly correlated, r=0.97). These scores correlated negatively with measures of water diffusivity in the corona radiata and superior thalamic radiation, and with FA in the posterior fornix (consistent with literature27, which as discussed above may reflect variations in tract thickness28). Finally, there was significant association with thalamus volume (right thalamic volume significant, left thalamic volume close to significance with r=0.10), consistent with literature29. These negative associations likely reflect lower cognitive performance with aging and pathology (increased diffusivity and atrophy).
In the Reaction Time test, subjects confirm whether two abstract symbols match as quickly as possible. Mean time to correctly identify matches was found to correlate inversely with left putamen volume (right putamen had similar correlation, r=-0.06, but was below significance). These negative associations are consistent with literature30 and indicate increased volume correlating positively with cognitive speed (and negatively with reaction time).
The Fluid Intelligence score reports how many numerical, logic and syntactic questions subjects were able to answer in two minutes. This was negatively correlated with strength of gray matter activation in the simple shapes matching task in tfMRI, with no age interaction. The shapes matching task incurs low cognitive demand, and it is plausible that higher intelligence requires less neural activity for this task, a mechanism that has previously been ascribed to “minimization of cognitive workload”31.
All cognitive scores reported above involve processing speed as a significant factor, consistent with previous studies27. However, the observation that different test scores do not all correlate identically with each other or with the same brain IDPs suggests that there is not a single (speed-related) cognitive factor at play. The increases in association strengths when not controlling for age suggest that age-related cognitive decline is a major source of cross-subject variability for these IDP-cognition associations28. Plotting all IDP-cognitive associations (Fig 6c) demonstrated that a large number of non-age-adjusted associations (gray circles) were stronger than the results after age adjustment (colored circles); below, we show how interpretation of such results can be aided further through multivariate analyses. These age interactions provide an early indication that UK Biobank should provide cognitive biomarkers of clinical significance as health outcomes accumulate.
Multivariate associations: modes of population variation
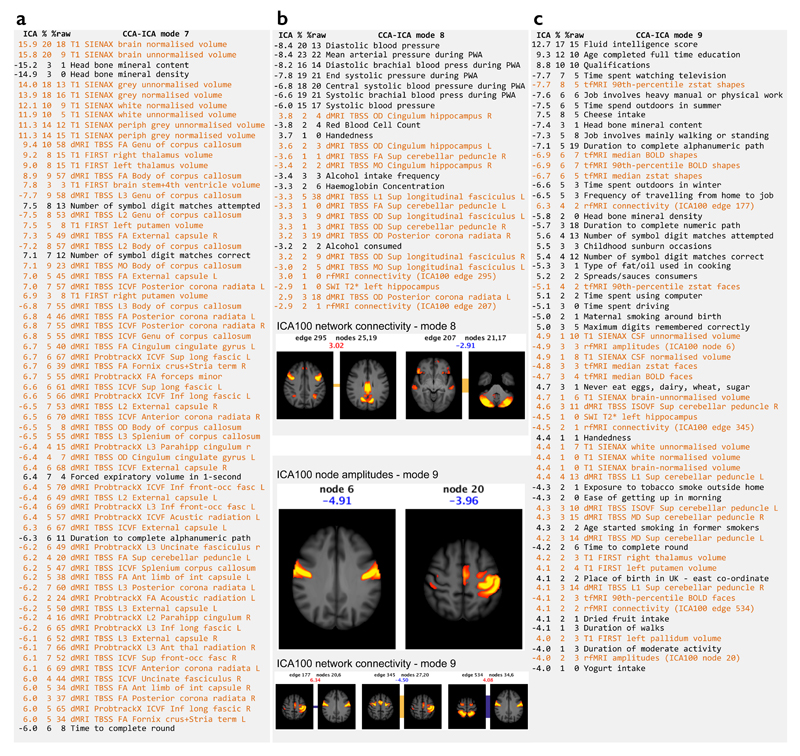
We conducted multivariate analyses using canonical correlation analysis (CCA32) combined with independent component analysis (ICA33) (see Figs 7-8, Supplementary Figs 3-7 and Online Methods). This analysis identifies “modes” of population covariation linking IDPs to non-imaging measures. Each mode consists of one linear combination of IDPs and a separate combination of non-imaging measures that have a highly similar variation across subjects. The strength of involvement of a variable in a given mode is dictated by the variable weight (Fig 7). Multiple population modes may be identified, provided they describe different (independent) cross-subject variation, meaning that the implied association between a given pair of variables can vary from mode to mode.
Figure 7. Details of three modes from the doubly-multivariate CCA-ICA analyses across all IDPs and non-brain-imaging variables.
IDPs are listed in orange and non-brain-imaging variables in black. The text lists show the variables most strongly associated with each mode; where multiple very similar (and highly correlated) non-imaging variables are found, only the most significant is listed here for brevity. The first column shows the weight (strength and sign) of a given variable in the ICA mode, the second shows the (cross-subject) percentage variance of the data explained by this mode, and the third column shows the percentage variance explained in the data without the confounds first regressed out. Mode 7 links measures of bone density, brain structure/tissue volumes and cognitive tests. Mode 8 links measures of blood pressure and alcohol intake to IDPs from the diffusion and functional connectivity data; two functional network connections strongly involved are displayed, with the population mean connection indicated by the bar connecting the two nodes forming the connection (red indicates positive mean correlation, blue negative, and the width of the bar indicates the connection strength). The group-ICA maps are thresholded at Z>5, and the colored text is the ICA weight shown in the table list. Mode 9 includes a wide range of imaging and non-imaging variables (see main text for details); as well as showing 3 strong functional network connections, we also show two functional nodes whose resting fluctuation amplitude is associated with this mode.
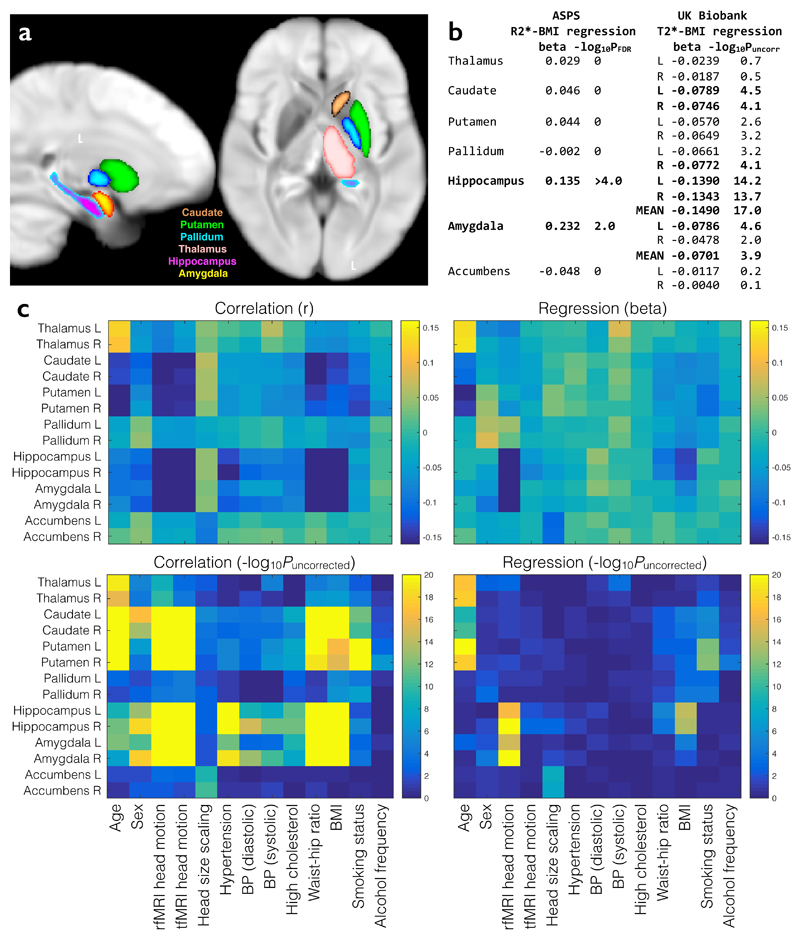
Figure 8. Hypothesis-driven study of age, BMI and smoking associations with subcortical T2*.
(a) UK Biobank population-average map of T2*, overlaid with the main subcortical structures being investigated. The T2* IDPs reflect individuals’ median T2* values within these regions. The relatively low T2* in putamen and pallidum likely reflects greater iron content. (b) BMI regression betas from multiple regressions of R2* (from the ASPS study) and T2* (from UK Biobank) against relevant covariates (see (c)). All variables are standardized so that beta values can be interpreted as (partial) correlation coefficients. R2* significance is reported as FDR-corrected P. T2* significance is reported as –log10Puncorrected with the more conservative Bonferroni correction (for Pcorrected=0.05) resulting in a threshold here of 3.6. (c) Full set of univariate and multiple regression betas and significance values for all brain regions tested and all model covariates. The regression results are much sparser, reflecting the higher associational specificity obtained by reporting unique variance explained.
From the current UK Biobank release, we identified nine modes that were highly significant (Pcorrected<0.002, no further modes significant at Pcorrected<0.05). Similar methodology using Human Connectome Project (HCP) data previously identified a single statistically-significant mode of population covariation in 461 young healthy adults8, 34. Our ability to identify more modes than in the HCP dataset could be due to the tenfold increase in the number of subjects, the larger range of imaging modalities and non-imaging variables, and the older subject ages.
While these modes are not guaranteed to reflect biological processes, in practice ICA often produces such interpretability35. Of the nine modes, some reflected dominant physical factors (e.g., body size or heart rate), while others linked rich subsets of non-imaging measures to IDPs. Modes 7-9 are displayed in Fig 7, with modes 7 and 9 discussed in detail below, continuing our focus on the relationship between IDPs and cognitive tests. Modes 1-6 are overviewed in Supplementary Figs 3-7. The relationship of these multivariate associations to potential confounds and variables of interest, including some clinical outcomes, is explored in Supplementary Fig 8. Modes 1,2,4,5,7,8 were strongly associated with aging, whereas 3,6,9 were not.
Mode 7 primarily links bone density measures and cognitive scores to brain structure and dMRI measures. There is extensive literature linking volume and diffusivity measures to cognition, but a relationship between these measures and bone density has not to our knowledge been reported. This link could reflect variations in physical properties of non-interest that are not fully accounted for by de-confounding. However, correlations between low bone density and accelerated cognitive decline have been reported36 including association of bone density with Alzheimer’s disease37.
Mode 9 exhibited the most complex population pattern (Fig 7c). The most strongly-involved non-imaging measures were intelligence, education levels and occupational factors; additionally, some physical and dietary measures are involved, that may reflect socio-economic status as a latent factor (e.g., cheese intake or time spent outdoors in winter). Associated brain IDPs include task fMRI (with a negative weight, consistent with the sign of univariate associations, discussed above), followed by a range of functional and structural IDPs.
There is some overlap between modes 7 and 9 in terms of cognitive tasks (e.g., symbol digit matches), bone density and T1-based brain volumes. However, the fact that CCA-ICA separated modes 7 and 9 indicates that they constitute distinct biophysical patterns of variation across subjects; for example, mode 7 correlates with age, while mode 9 does not. The broader range of non-imaging measures involved in mode 9, and the ability to interpret many of them in terms of positive or negative life factors, is reminiscent of the single mode previously reported from HCP data8, 34. That mode resembled the well-established observation of strong correlations in subject performance across a broad range of cognitive and behavioral tests (the “general intelligence” g-factor), but also included demographic and life factors. However, there is not perfect correspondence between mode 9 reported here and the previous HCP mode. This may be due to key differences in the HCP and Biobank datasets, including different non-imaging measures, the use of only rfMRI in the HCP analysis, the different cohort profiles (e.g., age range) and the ability to separate more modes from the larger Biobank cohort.
Illustrative hypothesis-driven study
The Austrian Stroke Prevention Study (ASPS) recently reported associations between aging, smoking and body mass index (BMI) with gray matter T2* in 314 participants (38-82 years)38, likely reflecting iron accumulation in local tissue13. We sought to replicate several of their key findings as a demonstration of a hypothesis-led investigation. The ASPS study reported R2*, which is the reciprocal of the T2* value we estimate in UK Biobank; therefore we expected T2* associations with opposite sign to those reported by ASPS. The main results from ASPS in deep gray matter structures were as follows: BMI was the strongest determinant of R2* generally, and was significantly related to R2* in amygdala (beta=0.23, PFDR=0.009) and hippocampus (beta=0.14, PFDR<0.0001). Further associations with R2* (averaged across subcortical structures) were found for age (beta=0.03, PFDR =0.027) and recent smoking level (beta=0.02, PFDR=0.001). No equivalent associations were found for sex or hypertension. The ASPS conducted univariate correlations and multiple regressions to identify both shared and unique variance in the associations, using FDR correction. Based on these results, we hypothesize negative association between T2* in subcortical structures with BMI, age and smoking.
We conducted similar analyses, applying univariate correlations and multiple regressions against a similar set of covariates to ASPS (see Fig 8). The regressions used the n=4,891 subjects with complete data in all IDPs and covariates. In order to maximize the complementarity of information content between the univariate correlations and multiple regressions, no adjustments for factors such as age and sex were applied in correlations, whereas these factors were included as confound covariates in the multivariate regressions. We applied Bonferroni multiple comparisons correction across covariates and brain regions, resulting (for Pcorrected<0.05) in a -log10Puncorrected threshold of 3.6.
Our results were highly concordant with ASPS. BMI was significantly associated with T2* in amygdala (averaged across left and right: beta=-0.07, -log10Puncorrected=3.9) and hippocampus (beta=-0.15, -log10Puncorrected=17.0; for comparison, FDR correction would result in PFDR<10-10). Individual subcortical BMI associations are shown in Fig 8b. In accordance with our hypothesis, the signs of regression betas are universally negative with T2* from UK Biobank data. Associations with T2* were found for age in thalamus, caudate and putamen (see Fig 8c) and for smoking status in caudate, putamen and right pallidum (beta ranging from -0.03 to -0.1). Association of T2* with sex was only found in right amygdala, and no association was found for hypertension.
The increased specificity of multiple regression is notable for many of the tests – for example, a significant univariate association of T2* with cholesterol disappears after controlling for the other covariates. Similarly, for T2* in hippocampus and amygdala, many of the associations with age, sex, BMI and other factors become much weaker after controlling for all variables, in particular amount of head motion. Despite the fact that this motion was recorded from the functional data (not the T2* data), it is likely a general indicator of head motion, and these results illustrate why interpretation of imaging associations requires care. For example, BMI could be predictive of head motion (e.g., comfort in the scanner) while also potentially relating to biophysical parameters of deeper interest.
The BMI and smoking associations with T2* are found in distinct subcortical structures. Interestingly, this distinction is reflected in the CCA-ICA results, where these associations appear in separate population modes. The association of T2* in caudate and putamen with smoking (and more weakly with alcohol, Fig 8c) is highly concordant with CCA-ICA mode 5 (Supplementary Fig 4b), and is associated with aging (Fig 8c and Supplementary Fig 8). The association of T2* in hippocampus and amygdala with BMI is highly concordant with CCA-ICA mode 3 (Supplementary Fig 3c), a distinct mode of population covariation that is not associated with aging (in either analysis). Neither mode includes cognitive test scores, suggesting that while these associations clearly relate to biological processes, they may be only indirectly linked to cognitive health.
Discussion
Challenges of population imaging
UK Biobank data is openly available to researchers including non-imaging experts. However, imaging data is considerably more complex than most of the existing UK Biobank measures. Extensive post-processing is required to align images across subjects and remove artefacts. Moreover, information is usually encoded across multiple voxels, requiring further processing to extract relevant features. Even with carefully-prepared IDPs, meaningful interpretation requires care because MRI is generally an indirect measure of the biology of interest. Apparent structural atrophy can be susceptible to misinterpretation39; functional MRI signals can reflect vascular properties rather than neural activity40; and diffusion MRI is sensitive to many aspects of tissue microstructure14. A final challenge is that data sizes have become extremely large, requiring “big data” techniques; the brain imaging data in UK Biobank will ultimately surpass 0.2PB even without data inflation during post-processing.
Large cohorts face the further challenge that statistically significant associations are identified even when their explanatory power is small. In the present dataset, significance is reached at a correlation of just r≈0.1, i.e., 1% of population variance explained41 even with multiple comparison correction. Large genome-wide association studies (GWAS) face this challenge, where it is accepted that small effect sizes can be meaningful, particularly where multiple factors combine to create a large effect. However, in GWAS, genetic variants can be interpreted as causal factors (whether direct or indirect42), whereas apparent associations across IDPs and non-imaging phenotypes could result from a shared latent (non-measured) cause. For example, education level could result in a dietary factor associating with a brain IDP, despite no direct causal connection between diet and IDP. This danger is inflated with larger subject numbers, but may here be mitigated by the rich life factor and biological variables that can be controlled for or used to match sub-groups. Population variances explained in the pairwise associations reached maxima of around 5% (Supplementary Fig 2), but these are higher with the multivariate analyses (up to 20-50% variance explained in the most highly involved variables within population modes), partly reflecting increased sensitivity gained when appropriately combining across related variables.
The importance of accounting for relevant confounds is exemplified in Fig 6e, which shows a strong apparent association between total white matter volume and fat-free body mass (one scatter point per subject) without de-confounding. In fact, this association is largely driven by the average differences in body mass and head size between sexes (see color coding) and disappears after adjusting for sex, age and head size. This is an example of Simpson’s paradox43, in which suboptimal pooling across variables (here, sex) results in a misleading association. Other pitfalls include failing to consider study population selection bias44 and inappropriate de-confounding of variables that are caused by (and not feeding into) the variables of interest45. While there is no guarantee that UK Biobank is an unbiased sample of the full population, that does not imply that studies using subsets of the data have to retain any biases (though again it is still possible for bias to arise44); one important aspect of study design will be the method of sub-selection of Biobank subjects to feed into an analysis. In the case of focused hypothesis testing, it is likely that carefully selected sub-groups of subjects should be utilized. For example, once a group of subjects is identified with a clinical diagnosis, it is likely that optimal sensitivity and interpretability will require a control sub-group that is matched over many relevant factors (e.g. sex, age, relevant life factors not appearing in the predictive model).
Future studies might seek to find causalities between variables, for example using structural equation modeling, Bayes Nets or non-linear/non-Gaussian methods46. The dangers of inferring causalities from observational datasets like UK Biobank are well-known; the inclusion of genotype and other “instrumental” measures enable analyses such as Mendelian randomization, although important caveats must be considered42. The safest way to confirm causal results discovered from such observational datasets is to use such results to form hypotheses for new focused interventional studies.
The mapping of disease associations and population patterns (e.g., learned from UK Biobank data) onto individuals will be an important long-term goal. For example, population distributions of imaging measures and health outcomes can be learned, and used to form patient-specific prior distributions to combine with measures from a new patient. While this might not provide statistical certainty for a diagnosis or interventional recommendation, it should allow for single-patient imaging to be used in a similar way to current state-of-the-art patient-tuned genetic testing.
Data analysis in population imaging
The analyses presented here demonstrate some of the possibilities offered by the UK Biobank resource. Focused association studies may select just two variables to investigate, for example, one IDP correlated against one life factor, genetic marker, physical assay or health outcome. More complex analyses could model a larger number of variables simultaneously, for example, looking to predict health outcome from multiple linear regression against several predictor variables. Nonlinear methods (e.g. penalized regression or data-driven feature selection)22 could enable use of much larger number of predictor variables. A further extension could identify nonlinear interactions between predictor variables, for example considering an imaging measure, a life factor and an interaction term between the two as three distinct predictors. An even more complex analysis might predict multiple outcome variables, looking for “doubly-multivariate” associations between two or more sets of variables; the CCA-ICA analyses presented above are an example of this. Finally, imaging measures may in some cases be more sensitive or specific than clinical symptoms47, thereby providing proxies for healthcare outcomes and/or enabling clustering of patients that is more predictive of prognosis or therapeutic response48.
Pairwise correlation analyses result in simple outcomes that require an understanding of the caveats in imaging-derived measures. Data-driven multivariate analyses identifying associations between sets of variables have complementary benefits, including improved sensitivity to biological processes and a streamlined set of results compared with millions of univariate associations. Further, multivariate analyses can separate distinct biological processes with opposing relationships between variables. For example, our CCA-ICA analysis revealed one aging-related process that involved changes in heart rate and fMRI measures (mode 4) while another aging-related process related blood pressure and white matter microstructure (mode 8). A simple correlational analysis would show associations between all of these factors, including even those that appeared in separate modes (e.g. fMRI and white matter changes). Additionally, as with multiple regression, simultaneous identification of multiple modes of association reduces the unexplained residual variance (effectively data “de-noising”).
Multivariate analyses of multi-modal data like UK Biobank enable discovery of (potentially complex) clinical phenotypes. This is a powerful alternative to diagnostic categories that rely on clinical symptoms which do not map cleanly onto underlying disease mechanisms. For many complex diseases, the discovery of distinct mechanisms/sub-diseases that are currently conflated may be unlikely to occur solely through symptom-based investigations. Discovering relevant population axes and sub-groups based on imaging, genetics and other objective markers may therefore be expected to increase our understanding of the etiology and pathogenesis of a wide variety of diseases. For example, this concept is at the heart of the recently-proposed Research Domain Criteria (RDoC) in psychiatry48.
Population imaging landscape
In the early 2000s, several ambitious studies built cohorts numbering thousands of subjects. Several recent brain imaging studies are aiming to image tens of thousands of subjects, including the Maastricht Study (n=10,000)49, the German National Cohort (n=30,000)50 and the Rhineland Study (n=30,000). In addition to having even larger numbers, UK Biobank will benefit from the breadth of organ systems imaged, the highly multi-modal brain protocol, and the existing rich phenotyping. A longitudinal component is planned for a subset of the UK Biobank imaging participants (n=10,000), as in the Rhineland and GNC studies. Most of these studies use identical MRI scanners at a small number of dedicated sites, with the goal of maximizing data homogeneity within study. A future challenge to further leveraging these large datasets is to develop analysis tools that can harmonize data across these studies for combined analyses, where there could be considerable impact in focusing on harmonization of a few very large cohorts.
Even with just 5% of the eventual cohort size, the results presented here demonstrate the statistical benefits that are conferred by large numbers. However, the primary rationale for the size of the study is not to boost statistical power across 100,000 subjects, but rather to provide prospective imaging data suitable for discovering early markers and risk factors for as broad a set of diseases as possible. For some rare diseases with few established risk factors, this approach is uniquely suited to discovery of pre-symptomatic markers; for example, 50-100 imaging participants are expected to develop sporadic amyotrophic lateral sclerosis (ALS) by 2027. This rich imaging addition to the ongoing UK Biobank study will provide scientists with novel insights into the causes of brain disease, provide markers with predictive power for therapeutic interventions, and advance non-invasive imaging-based screening for preventative healthcare.
Online Methods
Protocol Overview
Imaging protocols were designed by the UK Biobank Imaging Working Group (www.ukbiobank.ac.uk/expert-working-groups), in consultation with a large number of brain imaging experts (listed in the acknowledgments). MRI provides many imaging modalities offering complementary information. As part of this consultancy, a number of modalities were determined to be infeasible or lower priority for a range of reasons. Considerations included time constraints, generalizability, feasibility of automated analysis, and existence of robust, well-tested acquisition methods. The advisory network therefore decided not to include quantitative relaxometry, MR spectroscopy or angiography. Arterial spin labeling is currently being piloted, as described below.
To maximize data compatibility, three dedicated imaging centers will have identical scanners with fixed platforms (i.e., no major software or hardware updates throughout the study). Each center is equipped with a 3T Siemens Skyra (software platform VD13), 1.5T Siemens Aera (VD13), carotid ultrasound and dual energy X-ray absorptiometry (DEXA). Brain imaging is being conducted on the 3T system using a 32-channel receive head coil.
Key acquisition parameters for each modality are summarized in Supplementary Table 1, grouped according to primary modality categories (structural, diffusion and functional MRI). Order of acquisition was optimized in consideration of subject compliance, assuming subject motion might increase over the scan (favoring early acquisition of the T1 due to its central importance – for example the processing pipeline cannot run without the T1) and subject wakefulness might decrease (favoring early acquisition of fMRI). The order is: (1) T1, (2) resting fMRI, (3) task fMRI, (4) T2 FLAIR, (5) dMRI, (6) swMRI.
Further protocol details are available at: http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=2367 and further description of post-processing pipelines and data outputs included in the first data release are available at: http://biobank.ctsu.ox.ac.uk/crystal/docs/brain_mri.pdf. All software used in these pipelines is freely available51, 52 and full pipeline processing scripts will shortly be publicly available.
The processing pipeline utilized for the initial data release was primarily based on tools from FSL (the FMRIB Software Library51), but it will be gradually expanded to utilize a broader range of methods and software, where this will increase the quality, robustness and scope of IDPs generated. For example, one high priority is to adapt the Human Connectome Project pipelines53 to provide cortical surface modeling.
The intention is that non-imaging experts will be able to use the IDPs directly without having to become expert in the complexities of data processing, although we encourage engagement with imaging experts in light of the numerous and subtle caveats and confounds associated with interpreting these data.
Data access requests from all academic or commercial researchers (with no exclusive or preferential access) are processed by the UK Biobank’s Research Access Administration Team and approved relatively rapidly provided that they fulfill UK Biobank’s aims of supporting health research in the public interest. Researchers’ institutions then sign a Material Transfer Agreement agreeing not to attempt to identify any participant, and to return any derived data (e.g., new IDPs) to UK Biobank, to be made available to other approved researchers after an agreed “embargo” period (to allow findings to be published or IP protected by the researchers). Thus, while the first set of IDPs described here from internally commissioned research is being made available immediately, the range of IDPs is expected to grow rapidly as additional contributions from the wider user community are added.
Protocol Considerations
Design of the brain imaging protocol was conducted through broad consultation with neuroimaging experts and required careful balance of a range of considerations, often specifically relating to the high throughput nature of UK Biobank. In setting up the pilot protocol, the primary challenge was to achieve the target of one participant scanned every 36 minutes without serious compromise to data quality compared to research protocols that might conventionally require up to an hour of scan time. Despite these tight time constraints, we aimed to include as many MRI modalities as possible, to take advantage of the full richness of information that can be provided by MRI. Here, we highlight the primary considerations that required a different approach from more conventional imaging studies.
With each additional minute of scanning per subject effectively costing an additional ~£1million, there is enormous value associated with seemingly small efficiency savings. We recovered several minutes of scan time by systematically minimizing the overheads associated with subject placement, scan prescription, and calibration measurements. For example, corrections to the static magnetic field homogeneity (shimming) and strict enforcement of a single shim calibration harvested 2 minutes (changing system defaults to improve and accelerate shimming), which is equivalent to the scan time associated with some of the included modalities.
Tight imaging FOVs (fields of view – the physical size of the imaged volume) are in general favorable to reduce scan time; however, these restrictions exclude subjects with larger heads or brains. For UK Biobank, even a “conservative” FOV that includes 99% of the population will exclude 1000 participants. As detailed statistics on brain size (as distinct from head size) were not available in the literature, we conducted a study of population brain size54 that (in conjunction with optimal slice angling) enables our FOVs to target 99.9%.
It is critically important that all analyses are automated. This translates to an additional role for certain imaging modalities beyond their intrinsic information content. Thus, although we considered methods for reducing scan time for T1-weighted structural scans while retaining coverage and resolution (e.g., elliptical sampling with consequent image blurring), this was deemed an unacceptable risk given the central role of the T1 to cross-subject and cross-modal alignment for most processing pipelines, including that implemented here for the initial data release.
The EPI (echo-planar imaging) acquisitions for fMRI and dMRI result in significant image distortion that creates local misalignment in certain brain regions. Correction of this requires measurement of the magnetic field inhomogeneities that cause distortion. Two types of measurements are possible: a non-EPI gradient-echo acquisition with two echoes (conventional fieldmap) or two EPI-based spin echo acquisitions with opposite phase encode direction55. We chose the latter, which can be incorporated into the dMRI protocol as additional b=0 scans to reduce acquisition time (total acquisition time ~30s).
To provide data with as rich and broad a range of applications as possible, we include imaging modalities that are not yet widely used in clinical practice, such as fMRI and dMRI. These modalities have demonstrated mechanistic and biological insights, and will hopefully see greater clinical take-up in the future, in part because of projects such as UK Biobank. We took advantage of recent advances in acquisition, largely developed as part of the Human Connectome Project, to obtain research quality data in limited time. Specifically, simultaneous multi-slice (or multiband, MB) acquisitions56–59 that enable rapid fMRI and dMRI without sacrificing statistical robustness or directions/b-values60, respectively. Without these accelerations, a seven-minute dMRI scan of the same spatial resolution would have been limited to ~32 directions and a single shell, precluding NODDI16 and other more advanced biological modeling.
After early piloting, a clinical T2/PD-weighted acquisition was removed from the protocol. This decision reflected the limited relevance to UK Biobank goals (given the inclusion of the higher-quality and more biologically informative T2 FLAIR) and the value in recovering this scan time (just over 1 minute).
One shortcoming of the current protocol is the lack of a direct measure of neurovascular health. We are piloting a protocol change to include a 2-minute perfusion scan (using arterial spin labeling). This would require reducing task fMRI to 2 minutes; while this is an extremely short task, early analyses (using truncated copies of existing initial tfMRI datasets) predict that it will be sufficiently robust.
A major ethical question in studies of this nature relates to identification and handling of incidental findings of previously unknown pathology. The procedure to be followed in UK Biobank has been considered in great depth with major external ethical, legal and clinical radiology bodies, and with the funders and their external review group. An assessment of different approaches to the identification of incidental findings and the impact of their feedback on participants and the health service has been conducted as part of the pilot phase of UK Biobank’s imaging project, and will be published separately. Based on its results and the deliberative process undertaken with external experts, the UK Biobank protocol for dealing with incidental findings does not involve the routine review of all scans for potential pathology by radiologists. Instead, if a radiographer incidentally identifies evidence of potentially serious pathology (i.e., likely to threaten life span, quality of life or major body functions) during the imaging process then a formal radiologist review is undertaken and, if it is confirmed as potentially serious, feedback is given to the participant and their doctor.
Informed consent is obtained from all UK Biobank participants; ethical procedures are controlled by a dedicated Ethics and Guidance Council (http://www.ukbiobank.ac.uk/ethics) that has developed with UK Biobank an Ethics and Governance Framework (given in full at http://www.ukbiobank.ac.uk/wp-content/uploads/2011/05/EGF20082.pdf), with IRB approval also obtained from the North West Multi-centre Research Ethics Committee.
Subjects are excluded from scanning according to fairly standard MRI safety/quality criteria, such as exclusions for metal implants, recent surgery, or health conditions directly problematic for MRI scanning, such as problems hearing, breathing or extreme claustrophobia.
Once the second and third imaging centers are complete and running, UK Biobank will use non-human phantom objects and traveling volunteers to confirm quality and consistency across sites.
Structural Imaging
The T1 structural protocol is acquired at 1mm isotropic resolution using a 3D MPRAGE acquisition, with inversion and repetition times optimized for maximal contrast. The superior-inferior field-of-view is large (256mm), at little cost, in order to include reasonable amounts of neck/mouth, as those areas will be of interest to some researchers (for example, in the study of sleep apnea). Pre-processing of this modality included removal of the face (which was deemed important to subject anonymisation for the standard data dissemination), brain extraction (removal of non-brain tissues from the image), linear alignment to the standard MNI152 brain template61 and non-linear warping to this template62 to maximize correspondence across individuals in light of significant cross-subject variation in brain structure. These alignments are used throughout the majority of the processing pipeline for other modalities.
T1 images are further analyzed to estimate volumes of a range of tissues and structures in each subject, which may reflect atrophy due to age and disease, as well as normal variation due to (e.g.) use-dependent plasticity. Images are segmented into tissue types (gray matter, white matter and cerebro-spinal fluid)63. Cortical gray matter volume is estimated, comparing the segmented gray matter to an atlas reference (where the external skull surface is used to normalize for head size)64. Sub-cortical volumes are estimated65, utilizing population priors on shape and intensity variation across subjects. T1-based IDPs are generated for the volumes of major tissue types of the whole brain and for specific structures (primarily sub-cortically).
Too much reliance on spatial registration could limit the usefulness or accuracy of some IDPs. This is in part why many of the IDPs are in fact generated from within-subject analyses that do not depend on exact voxelwise spatial alignment to standard space (or between subjects): for example, 283 of the 715 structural and diffusion IDPs do not rely on exact spatial alignment and are carried out in the original space of each subjects’ data.
The T2 protocol uses a fluid-attenuated inversion recovery (FLAIR) contrast with the 3D SPACE optimized readout66. This shows strong contrast for white matter hyperintensities. An automated pipeline for delineating these hyperintensities is currently being developed and future data releases will include IDPs reflecting the lesion “load”.
The swMRI scan uses a 3D gradient echo acquisition at 0.8x0.8x3mm resolution, acquiring two echo times (TE=9.4 and 20 ms). Anisotropic voxels can enhance certain contrast mechanisms, particularly for vascular conspicuity due to through-plane dephasing effects, but are less ideal for other susceptibility-based processing. Ultimately, however, this choice was motivated by the desire for whole brain coverage in the face of very limited scan time (2.5 minutes). Signal decay times (T2*) are estimated from the magnitude images at the two TEs, and the generated IDPs are the median T2* estimated within the various subcortical regions delineated from the T1 processing. Venograms are generated through non-linear filtering of the magnitude and phase images12, which produces enhanced conspicuity of medium and large veins. Automated segmentation of microbleeds and venograms would provide significant value, but to our knowledge robust tools for this are not yet available; future pipeline versions can hope to include such analyses. Future work will also consider whether this data will support quantitative susceptibility mapping, which would provide further information on tissue constituents as discussed in the main text.
Diffusion Imaging
Diffusion data is acquired with two b-values (b=1000 and 2000 s/mm2) at 2mm spatial resolution, with multiband acceleration factor of 3 (three slices are acquired simultaneously instead of just one). For each diffusion-weighted shell, 50 distinct diffusion-encoding directions were acquired (covering 100 distinct directions over the two b-values). The diffusion preparation is a standard (“monopolar”) Stejskal-Tanner pulse sequence. This enables higher SNR due to a shorter echo time (TE=92ms) than a twice-refocused (“bipolar”) sequence at the expense of stronger eddy current distortions, which are removed using the Eddy tool67 (which also corrects for static field distortion and motion68).
Both diffusion tensor and NODDI models are fit voxel-wise, and IDPs of the various model outputs are extracted from a set of white matter tracts. Tensor fits utilize the b=1000 s/mm2 data, producing maps including fractional anisotropy, tensor mode and mean diffusivity. The NODDI16 model is fit using the AMICO (Accelerated Microstructure Imaging via Convex Optimization) tool52, with outputs including intra-cellular volume fraction (which is often interpreted to reflect neurite density) and orientation dispersion (a measure of within-voxel disorganization). For tractography, a parametric approach is first used to estimate fibre orientations. The generalised ball & stick model is fit to the multi-shell data, estimating up to 3 crossing fibre orientations per voxel.17, 69 Tractography is then performed in a probabilistic manner to estimate white matter pathways using the voxel-wise orientations.
Cross-subject alignment of white matter pathways is critical for extracting meaningful IDPs; here, two complementary approaches are used. The first used tract-based spatial statistics (TBSS18, 70), in which a standard-space white matter skeleton is mapped to each subject using a high-dimensional warp, after which ROIs are defined as the intersection of the skeleton with standard-space masks for 48 tracts71 (see the “JHU ICBM-DTI-81 white-matter labels atlas” described at fsl.fmrib.ox.ac.uk/fsl/fslwiki/Atlases for definitions of the tract regions and names). The second approach utilizes subject-specific probabilistic diffusion tractography run using standard-space protocols to identify identify 27 tracts18; in this case, the output IDPs are weighted by the tractography output to emphasize values in regions that can most confidently be attributed to the tract of interest. Currently, no structural connectivity estimates from the diffusion tractography are provided as IDPs, but the probabilistic maps are available and future work will generate measures similar to those provided for resting-state fMRI.
Functional MRI
Task and resting-state fMRI use the same acquisition parameters, with 2.4mm spatial resolution and TR=0.735s, with multiband acceleration factor of 8. A “single-band” reference image (without the multiband excitation, exciting each slice independently) is acquired that has higher tissue-type image contrast; this is used as the target for motion correction and alignment. For both data sets, the raw data are corrected for motion72 and distortion55 and high-pass filtered to remove temporal drift.
The task scan used the Hariri faces/shapes “emotion” task21, 73, as implemented in the HCP22,, but with shorter overall duration and hence fewer total stimulus block repeats. The participants are presented with blocks of face or shape trials and asked to decide which of two faces (or shapes) presented on the bottom of the screen match the face (or shape) at the top of the screen. The faces have either angry or fearful expressions. The ePrime stimulus script is available for download (http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=1462). Task-induced activation is modeled with FEAT, including auto-correlation correction74, using 5 activation contrasts. Of these, the three activation contrasts of most interest (shapes, faces and faces>shapes) are used to generate output measures, including two IDPs for the faces-shapes task (one including all voxels above a group-level fixed-effects Z>120, and one including only the amygdala regions above threshold). IDPs corresponding to both percent signal change and statistical significance (Z statistics) are generated.
During resting-state scans, subjects are instructed to keep their eyes fixated on a crosshair, relax and “think of nothing in particular”. Resting-state are identified using ICA (independent component analysis33, 75), which identifies components within the data that are spatially independent (where a component comprises a spatial map and a single associated time course). Following the pre-processing described above, resting-state fMRI data for each subject is further “cleaned” using an ICA-based algorithm for automatically identifying and removing structured artefacts76. This data is fed into group-level ICA (including an initial group-level dimensionality reduction77), which is used to parcellate the data set into sets of 25 and (separately) 100 spatially-independent components. Where a small (<30) number of components is estimated78, it is common to consider each component as a separate “network” in its own right; each component will often include several non-contiguous regions, all having the same timecourse (according to the model). If a higher number of components is estimated79, these are more likely to be smaller regions (parcels), which can then be considered as nodes for use in network analysis80, where the spatial maps are used to define subject-specific time courses (the first stage of dual regression1). These time courses are used to estimate the size of signal fluctuation in each node, as well as to estimate connectivity between pairs of nodes using L2-regularised partial correlation81. The connectivity estimates are provided as IDPs at both parcellation dimensionalities (25 and 100 nodes); after removal of group-ICA components considered to be artefactual (i.e., relating either to scanning artefacts, or to non-neuronal biophysical processes such as cardiac fluctuations and head motion), this results in 21 and (respectively) 55 nodes left for forming the IDPs such as network matrices (functional connectivities between pairs of nodes).
Quality control
To date, raw data and pipeline outputs have been manually checked for gross problems of quality and robustness, with problematic data tagged and removed from pipeline outputs; see main text for results on proportions of usable data in the different modalities. However several quality-related IDPs are automatically generated by the pipeline (for example, number of outlier slices in the dMRI data, and measures of signal-to-noise ratio in the various modalities), and these can be used to help automatically identify problematic data. An expanded set of such quality measures is being produced, in addition to an automated machine learning system for flagging problematic data on the basis of the many IDPs and quality measures; future versions of the pipeline and data releases will benefit from the results of these ongoing developments.
Statistics
The two sections below describe the statistical analysis carried out using IDPs and non-brain-imaging measures. As described below, univariate statistics were primarily carried out using Pearson correlation (though see details below regarding Gaussian-distribution normalization and linear removal of confound effects) and multivariate statistics were carried out using a combination of canonical correlation analysis and independent component analysis (with permutation testing used to identify the significant number of components estimable). As discussed in the main text, the primary rationale for the size of the study is not to boost statistical power across 100,000 subjects, but rather to provide prospective imaging data suitable for discovering early markers and risk factors for as broad a set of diseases as possible, both rare and highly prevalent. Hence while calculations have been made to estimate the expected numbers of subjects developing different diseases over coming years (see introductory section of main text), no statistical methods were used to pre-determine sample sizes for any one specific disease, given that individual disease sample sizes are not prospectively controlled, and given the very broad expected set of future tests between different imaging measures and different diseases that will be ultimately applied from this prospective long-term resource. Details on significance testing and multiple comparison corrections are included in the two sections below. A Supplementary Methods Checklist is available.
Simple associations between brain IDPs and other measures
We report simple correlation analyses between each of the 2501 brain IDPs and each of 1100 other variables extracted from the UK Biobank database (these other variables are mostly not derived from imaging, though some do come from the non-brain imaging modalities); for the list of general classes of these variables, see Fig. 6a, and for many examples of individual variables, see the lists associated with the CCA-ICA modes presented in Fig. 7 and Supplementary Figs. The initial set of variables extracted from the UK Biobank database was automatically reduced to those (1100 variables) containing sufficient numbers of valid (non-missing) data entries, using very similar selection rules to those applied in the recent CCA-based analysis of Human Connectome Project data34. Some variables are defined (in the UK Biobank database) such that the numerical encoding is the inverse of what one might naturally assume - for example in the variable “Qualifications”, higher numbers refer to lower levels of educational qualifications. In such cases we have inverted the sign of the ICA weightings printed in the figures, for ease of interpretation. Further, some variables are categorical, with no clear quantitative meaning to the values (e.g., “Transport type to work”); where we find an apparent association, this can be considered to be indicative of a real association (one might think of the analysis therefore as an over-conservative poor implementation of an ANOVA), but interpretation of the sign of the association clearly needs care. The analysis used data from the first 5430 subjects scanned and having usable imaging data: age range 44-78y (IQR 56-68y); 53% of subjects were female.
Eight confound variables are generated: age, age2, sex, age×sex, age2×sex, average head motion during tfMRI, average head motion during rfMRI and head size. To enforce Gaussianity, all confound variables, IDPs and non-IDP variables are first passed through a rank-based inverse Gaussian transformation; this improves the robustness of correlations (for example, to avoid undue influence of potential outlier values). The confounds are then regressed out of all IDPs and non-IDP variables to reduce the risk of finding non-meaningful associations. For example, head motion corrupts imaging data in complex ways26, and also correlates with some diseases and with aging (r=0.15 in this data); hence, if not adjusted for, uninteresting associations would likely arise. However, some measures may have both biologically interesting associations with IDPs, and also act as imaging confounds. For example, abnormal heart rate or blood pressure could alter the fMRI signal through disrupted cerebral auto-regulation (independent of any changes to neural activity)40, but cardiovascular pathology could also be related to neurological pathology. Similarly, overall brain size and gray matter thickness IDPs are sensitive simple markers of aging and disease; however, these properties can also affect other IDPs by changing the mixture of tissue types in an imaging voxel, creating an apparent age/disease dependence that is driven by the volume of tissue rather than the properties of a given tissue type (such as fMRI activation or white matter microstructural properties). It is therefore important to interpret apparent associations carefully.
The full set of 2.8 million (2501×1100) Pearson correlations is then estimated and corrected for multiple comparisons. Bonferroni correction, which is likely to be somewhat conservative in such situations, due to non-independence across variables tested, resulted in pcorrected<0.05 being equivalent to requiring puncorrected<1.8×10-8. An alternative popular approach for multiple comparison correction is false discovery rate (FDR);82 we use the more conservative FDR option (making no assumption of variable dependencies83), resulting here in requiring puncorrected<3.8×10-5. These two threshold levels are shown with dotted lines in all Manhattan plots in the main figures.
Multivariate associations between brain IDPs and other measures
In the example multivariate analyses shown in Figs 7-8, canonical correlation analysis (CCA32) combined with independent component analysis (ICA33) is used to identify several “modes” of population covariation which link multiple brain IDPs to sets of other Biobank variables. This is very similar to the methodology used recently to identify a single mode of population covariation between imaging measures and many behavioral and lifestyle measures in data from 461 subjects in the Human Connectome Project8, 34.
IDP and non-IDP variables are prepared as for the univariate correlation analyses described above, resulting in a brain-IDP matrix of size 5034×2501 (subjects × IDPs) and a non-IDP matrix of size 5034×1100 (subjects × non-IDP variables). The intention is to feed these into CCA in order to identify population modes linking multiple variables from both matrices. However, in order to avoid an over-determined (rank deficient) CCA solution, we first compress both matrices along the respective phenotype dimension to 200 columns (i.e., much smaller than the numbers of subjects). This was done by separately reducing each matrix to the top 200 subject-eigenvectors using PCA. To achieve this while avoiding the problem of missing data, we use the approach detailed recently34 of estimating first a pseudo-covariance matrix ignoring missing data, projecting this onto the nearest valid (positive definite) covariance matrix, and then carry out an eigenvalue decomposition. The two resulting (IDP and non-IDP) matrices of size 5034×200 are then fed into standard CCA (“canoncorr” in Matlab), resulting in 200 CCA modes being estimated. The CCA aims to identify symmetric linear relations between the two sets of variables. Each significant CCA mode identifies a linear combination of IDPs and a linear combination of non-IDPs, where the variation in mode strength across subjects is maximally correlated. That is, CCA finds modes that relate sets of brain measures to sets of subjects’ non-brain-imaging measures; for a graphical illustration of this approach, see Smith et al34 (Supplementary information).
Permutation testing is then applied to estimate (family-wise-error, multiple-comparison-corrected) p-values for the CCA modes estimated. Nine modes are found to be significant (Pcorrected<0.002, with all later modes having Pcorrected>0.05). Because CCA can in general only unambiguously estimate distinct modes up to an orthogonal rotation amongst them (by direct analogy to PCA), we identify a non-ambiguous unmixing of the modes using ICA to optimize the final set of modes reported. Because we expect meaningful population modes to be much more structured (for example, sparser) in the cross-variable dimension than in the cross-subject dimension, we calculate ICA components that are statistically independent from each other in the cross-variable dimension. In order to take full advantage of the numbers of variables originally prepared, we first multiply the nine CCA subject-weight vectors into the original IDP and non-IDP data matrices (after concatenating these across variables), resulting in nine CCA variable-weight vectors of length 2501+1100=3601. These nine vectors are then fed into FastICA33 in order to estimate nine population data sources having maximal statistical independence. This general approach (CCA, followed by concatenation of CCA weight vectors, followed by ICA) is similar to that proposed by Sui84, except that we return to the full feature space (as described above) for the ICA stage, rather than staying in the PCA-reduced space. The ICA result is extremely robust, with split-half (cross-subjects) reproducibility across the 9 ICA components of r>0.89. Interestingly, 5 of these ICA modes (including modes 7, 8 and 9, shown in Fig. 7) are virtually unchanged if the de-confounding step was omitted (correlation of variable-weights vectors: r>0.8).
Data, code and results availability
As described above, all source data (including raw and processed brain imaging data, derived IDPs, and non-imaging measures) is available from UK Biobank via their standard data access procedure (see www.ukbiobank.ac.uk/register-apply).
The image processing pipeline will be made publicly available in early 2017 from www.fmrib.ox.ac.uk/ukbiobank - this is the pipeline used to process the raw imaging data and generate IDPs, and hence is not needed in order to replicate the results of this paper, which could be achieved by accessing IDPs as described above, and then using the IDP analysis code described below.
The Matlab code for the univariate and multivariate tests described in this paper, and the results of those tests (all univariate correlations and multivariate weight vectors) are available from www.fmrib.ox.ac.uk/ukbiobank - this online resource will be updated as more subjects’ data and more IDPs become available.
Higher resolution supplementary figures are available as a supplementary note.
Supplementary Material
Acknowledgements
We would like to acknowledge the valuable contributions of members of the UK Biobank Imaging Working Group and the UK Biobank coordinating centre. UK Biobank (including the imaging enhancement) has been generously supported by the UK Medical Research Council and the Wellcome Trust. KLM and SMS receive further support from the Wellcome Trust. PMM acknowledges support from the Edmund J Safra Foundation and Lily Safra, the Imperial College Healthcare Trust Biomedical Research Centre and the Medical Research Council. We are very grateful for additional input into the imaging protocol and image processing pipelines from the following: Michael Chappell, Stuart Clare, Eugene Duff, David Flitney, Moises Hernandez Fernandez, Heidi Johansen-Berg, Paul McCarthy, John Miller, Duncan Mortimer, Jonathan Price, Gholamreza Salimi-Khorshidi, Emmanuel Vallee, Diego Vidaurre, Matthew Webster, Anderson Winkler, Alan Young, Eddie Auerbach, Steen Moeller, Kamil Ugurbil, Daniel Alexander, Nick Fox, Enrico Kaden, Sebastien Ourselin, Gary Zhang, Alessandro Daducci, Tony Stoecker, Deanna Barch, Nick Bloom, Greg Burgess, Matt Glasser, Michael Harms, Dan Nolan, Bruce Fischl, Doug Greve, Jonathan Polimeni, Tom Nichols, Anna Murphy, Geoff Parker, Fred Barkhof, Christian Beckmann, Maarten Mennes, Meike Vernooij, Nikolaus Weiskopf, Chris Rorden, Joanna Wardlaw. We are grateful for the provision of simultaneous multi-slice (multiband) pulse sequence and reconstruction algorithms from the Center for Magnetic Resonance Research, University of Minnesota. Finally, we are extremely grateful to all UK Biobank study participants, who have generously donated their time to make this resource possible.
Footnotes
Contributions
KLM, RC, PMM and SMS provided the overall scientific strategy for UK Biobank brain imaging; KLM, NKB, DLT, EY, JX, AJB, SJ, SNS, JLRA, MJ, PMM and SMS developed acquisition protocols; NKB, KLM, TWO, PW, ID, SG and SH implemented the imaging protocol at the dedicated imaging centre; FAA, KLM, SJ, SNS, JLRA, LG, GD, MJ and SMS developed post-processing pipelines and IDP calculation; KLM and SMS carried out the univariate/multivariate analyses and prepared figures; KLM and SMS wrote the manuscript, which was edited by all authors.
Competing financial interests
PW and ID are employees of Siemens Healthcare UK, the vendor of MRI scanners for UK Biobank selected under a competitive bidding process. Other authors declare no competing financial interests.
References
- 1.Filippini N, et al. Distinct patterns of brain activity in young carriers of the APOE-e4 allele. Proc National Academy of Sciences USA. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douaud G, et al. Brain Microstructure Reveals Early Abnormalities more than Two Years prior to Clinical Progression from Mild Cognitive Impairment to Alzheimer’s Disease. Journal of Neuroscience. 2013;33:2147–2155. doi: 10.1523/JNEUROSCI.4437-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudlow C, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Medicine. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen N, et al. UK Biobank: Current status and what it means for epidemiology. Health Policy and Technology. 2012;1:123–126. [Google Scholar]
- 5.Wick J. Understanding frailty in the geriatric population. Consult Pharm. 2011;26:634–645. doi: 10.4140/TCP.n.2011.634. [DOI] [PubMed] [Google Scholar]
- 6.Stanek K, et al. Obesity is associated with reduced white matter integrity in otherwise healthy adults. Obesity. 2011;19:500–504. doi: 10.1038/oby.2010.312. [DOI] [PubMed] [Google Scholar]
- 7.Petersen S, et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. J Cardio Magn Reson. 2013;15:46. doi: 10.1186/1532-429X-15-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Essen DC, et al. The WU-Minn Human Connectome Project: An overview. NeuroImage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good CD, et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. NeuroImage. 2001;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 10.Karas G, et al. A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel-based morphometry. NeuroImage. 2003;18:895–907. doi: 10.1016/s1053-8119(03)00041-7. [DOI] [PubMed] [Google Scholar]
- 11.Debette S, Markus H. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. British Medical Journal. 2010;341:c3666. doi: 10.1136/bmj.c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haacke EM, Xu Y, Cheng Y-CN, Reichenbach J. Susceptibility weighted imaging (SWI) Magn Reson Med. 2004;52:612–618. doi: 10.1002/mrm.20198. [DOI] [PubMed] [Google Scholar]
- 13.Duyn JH. MR susceptibility imaging. J Magn Reson. 2013;229:198–207. doi: 10.1016/j.jmr.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002;15:435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- 15.Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson B. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Schneider T, Wheeler-Kingshott C, Alexander D. NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. NeuroImage. 2012;61:1000–1016. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- 17.Jbabdi S, Sotiropoulos S, Savio A, Grana M, Behrens T. Model-based analysis of multishell diffusion MR data for tractography: How to get over fitting problems. Magn Reson Med. 2012;68:1846–1855. doi: 10.1002/mrm.24204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Groot M, et al. Improving alignment in Tract-based spatial statistics: Evaluation and optimization of image registration. NeuroImage. 2013;76:400–411. doi: 10.1016/j.neuroimage.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakana S, et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage. 2007;36:630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453:869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 21.Hariri A, Tessitore A, Mattay V, Fera F, Weinberger D. The amygdala response to emotional stimuli: a comparison of faces and scenes. NeuroImage. 2002;17:317–323. doi: 10.1006/nimg.2002.1179. [DOI] [PubMed] [Google Scholar]
- 22.Smith S, et al. Functional connectomics from resting-state fMRI. Trends in Cognitive Sciences. 2013;17:666–682. doi: 10.1016/j.tics.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douaud G, et al. DTI measures in crossing-fibre areas: Increased diffusion anisotropy reveals early white matter alteration in MCI and mild Alzheimer's disease. NeuroImage. 2011;55:880–890. doi: 10.1016/j.neuroimage.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ennis D, Kindlmann G. Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magn Reson Med. 2006;55:136–146. doi: 10.1002/mrm.20741. [DOI] [PubMed] [Google Scholar]
- 25.Raichle ME, et al. A default mode of brain function. Proc Natl Acad Sci. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satterthwaite T, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. NeuroImage. 2013;64:240–256. doi: 10.1016/j.neuroimage.2012.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznetsova K, et al. Brain white matter structure and information processing speed in healthy older age. Brain Structure and Function. 2015:1–13. doi: 10.1007/s00429-015-1097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madden DJ, et al. Diffusion Tensor Imaging of Cerebral White Matter Integrity in Cognitive Aging. Biochim Biophys Acta. 2012;1822:386–400. doi: 10.1016/j.bbadis.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Werf Y, et al. Thalamic volume predicts performance on tests of cognitive speed and decreases in healthy aging. A magnetic resonance imaging-based volumetric analysis. Cognitive Brain Research. 2001;11:377–385. doi: 10.1016/s0926-6410(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 30.Fjell A, Walhovd KB. Structural Brain Changes in Aging: Courses, Causes and Cognitive Consequences. Reviews in the Neurosciences. 2010;21:187–221. doi: 10.1515/revneuro.2010.21.3.187. [DOI] [PubMed] [Google Scholar]
- 31.Reichle E, Carpenter P, Just M. The Neural Bases of Strategy and Skill in Sentence–Picture Verification. Cognitive Psychology. 2000;40:261–295. doi: 10.1006/cogp.2000.0733. [DOI] [PubMed] [Google Scholar]
- 32.Hotelling H. Relations between two sets of variates. Biometrika. 1936;28:321–377. [Google Scholar]
- 33.Hyvärinen A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Transactions on Neural Networks. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
- 34.Smith S, et al. A positive-negative mode of population co-variation links brain connectivity, demographics and behavior. Nature Neuroscience. 2015;18:1565–1567. doi: 10.1038/nn.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith SM, et al. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaffe K, Browner W, Cauley J, Launer L, Harris T. Association between bone mineral density and cognitive decline in older women. J Am Geriatr Soc. 1999;47:1176–1182. doi: 10.1111/j.1532-5415.1999.tb05196.x. [DOI] [PubMed] [Google Scholar]
- 37.Tan A, et al. Bone mineral density and the risk of Alzheimer disease. Arch Neurol. 2005;62:107–111. doi: 10.1001/archneur.62.1.107. [DOI] [PubMed] [Google Scholar]
- 38.Pirpamer L, et al. Determinants of iron accumulation in the normal aging brain. Neurobiol Aging. 2016;43:149–155. doi: 10.1016/j.neurobiolaging.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Hamidi M, Drevets WC, Price JL. Glial reduction in amygdala in major depressive disorder is due to oligodendrocytes. Biological Psychiatry. 2004;55:563–569. doi: 10.1016/j.biopsych.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 40.D'Esposito M, Deouell LY, Gazzaley A. Alterations in the BOLD fMRI signal with ageing and disease: a challenge for neuroimaging. Nature Reviews Neuroscience. 2003;4:863–872. doi: 10.1038/nrn1246. [DOI] [PubMed] [Google Scholar]
- 41.Kirk R. Practical significance: A concept whose time has come. Educ Psychol Meas. 1996;56:746–759. [Google Scholar]
- 42.Gage S, Smith G, Ware J, Flint J, Munafo M. G = E: What GWAS Can Tell Us about the Environment. PLoS Genetics. 2016;12:e1005765. doi: 10.1371/journal.pgen.1005765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson E. The Interpretation of Interaction in Contingency Tables. J Roy Stat Soc Ser B. 1951;13:238–241. [Google Scholar]
- 44.Swanson J. The UK Biobank and selection bias. The Lancet. 2012;380:110. doi: 10.1016/S0140-6736(12)61179-9. [DOI] [PubMed] [Google Scholar]
- 45.Berkson J. Limitations of the Application of Fourfold Table Analysis to Hospital Data. Biometrics Bulletin. 1946;2:47–53. [PubMed] [Google Scholar]
- 46.Pearl J. Causality: Models, Reasoning and Inference. Cambridge University Press; 2009. [Google Scholar]
- 47.Duff E, et al. Learning to identify CNS drug action and efficacy using multistudy fMRI data. Sci Transl Med. 2015;7:274ra216. doi: 10.1126/scitranslmed.3008438. [DOI] [PubMed] [Google Scholar]
- 48.Insel T, et al. Research Domain Criteria (RDoC): Toward a New Classification Framework for Research on Mental Disorders. Am J Psychiatry. 2010;167:748–752. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 49.Schram M, et al. The Maastricht Study: an extensive phenotyping study on determinants of type 2 diabetes, its complications and its comorbidities. Eur J Epidemiol. 2014;29:439–451. doi: 10.1007/s10654-014-9889-0. [DOI] [PubMed] [Google Scholar]
- 50.German National Cohort (GNC) Consortium. The German National Cohort: aims, study design and organization. Eur J Epidemiol. 2014;29:371–382. doi: 10.1007/s10654-014-9890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkinson M, Beckmann CF, Behrens T, Woolrich M, Smith S. FSL. NeuroImage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 52.Daducci A, et al. Accelerated microstructure imaging via convex optimization (AMICO) from diffusion MRI data. NeuroImage. 2015;105:32–44. doi: 10.1016/j.neuroimage.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 53.Glasser M, et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mennes M, et al. Optimizing full-brain coverage in human brain MRI through population distributions of brain size. NeuroImage. 2014;98:513. doi: 10.1016/j.neuroimage.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 55.Andersson J, Skare S, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- 56.Ugurbil K, et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. NeuroImage. 2013;80:80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larkman DJ, et al. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. JMRI. 2001;13:313–317. doi: 10.1002/1522-2586(200102)13:2<313::aid-jmri1045>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 58.Moeller S, et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Setsompop K, et al. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med. 2012;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feinberg D, et al. Multiplexed Echo Planar Imaging for Sub-Second Whole Brain FMRI and Fast Diffusion Imaging. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenkinson M, Bannister PR, Brady M, Smith S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain. NeuroImage. 2002 doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 62.Andersson J, Jenkinson M, Smith S. Technical report: Non-linear registration aka spatial normalisation. FMRIB Centre, Oxford University; Oxford, UK: 2007. [Google Scholar]
- 63.Zhang Y, Brady M, Smith S. Segmentation of Brain MR Images through a Hidden Markov Random Field Model and the Expectation Maximization Algorithm. IEEE Trans on Medical Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 64.Smith SM, et al. Longitudinal and Cross-Sectional Analysis of Atrophy in Alzheimer's Disease: Cross-Validation of BSI, SIENA and SIENAX. NeuroImage. 2007;36:1200–1206. doi: 10.1016/j.neuroimage.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 65.Patenaude B, Smith S, Kennedy D, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mugler J. Optimized Three-Dimensional Fast-Spin-Echo MRI. J Magn Reson Imag. 2014;39:745–767. doi: 10.1002/jmri.24542. [DOI] [PubMed] [Google Scholar]
- 67.Andersson JL, Sotiropoulos SN. Non-parametric representation and prediction of single- and multi-shell diffusion-weighted MRI data using Gaussian processes. NeuroImage. 2016 doi: 10.1016/j.neuroimage.2015.07.067. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersson JL, Sotiropoulos SN. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage. 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hernandez M, et al. Accelerating Fibre Orientation Estimation from Diffusion Weighted Magnetic Resonance Imaging Using GPUs. PLoS ONE. 2013;8:e61892. doi: 10.1371/journal.pone.0061892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith SM, et al. Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1506. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 71.Wakana S, Jiang H, Nagae-Poetscher L, van Zijl PCM, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- 72.Bannister PR, Brady JM, Jenkinson M. Integrating Temporal Information with a Non-rigid Method of Motion Correction for Functional Magnetic Resonance Images. Image and Vision Computing. 2007;25:311–320. [Google Scholar]
- 73.Barch D, et al. Function in the Human Connectome: Task-fMRI and individual differences in behavior. NeuroImage. 2013;80:169–189. doi: 10.1016/j.neuroimage.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woolrich MW, Ripley BD, Brady JM, Smith SM. Temporal Autocorrelation in Univariate Linear Modelling of FMRI Data. NeuroImage. 2001;14:1370–1386. doi: 10.1006/nimg.2001.0931. [DOI] [PubMed] [Google Scholar]
- 75.Beckmann CF, Smith SM. Probabilistic Independent Component Analysis for Functional Magnetic Resonance Imaging. IEEE Trans on Medical Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- 76.Salimi-Khorshidi G, et al. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. NeuroImage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith S, Hyvarinen A, Varoquaux G, Miller K, Beckmann C. Group-PCA for very large fMRI datasets. NeuroImage. 2014;101:738–749. doi: 10.1016/j.neuroimage.2014.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kiviniemi V, Kantola J-H, Jauhiainen J, Hyvärinen A, Tervonen O. Independent Component Analysis of nondeterministic fMRI signal sources. NeuroImage. 2003;19:253–260. doi: 10.1016/s1053-8119(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 79.Kiviniemi V, et al. Functional segmentation of the brain cortex using high model order group PICA. Human Brain Mapping. 2009;30:3865–3886. doi: 10.1002/hbm.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith S. The future of FMRI connectivity. NeuroImage. 2012;62:1257–1266. doi: 10.1016/j.neuroimage.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 81.Smith SM, et al. Network modelling methods for FMRI. NeuroImage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 82.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 83.Genovese CR, Lazar N, Nichols T. Thresholding of Statistical Maps in Functional Neuroimaging Using the False Discovery Rate. NeuroImage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 84.Sui J, et al. A CCA + ICA based model for multi-task brain imaging data fusion and its application to schizophrenia. NeuroImage. 2010;51:123–134. doi: 10.1016/j.neuroimage.2010.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.