Abstract
Doxorubicin (Adriamycin) is an anthracycline chemotherapy agent effective in treating a wide range of malignancies with a well–established dose–response cardiotoxic side effect that can lead to heart failure. At present, it is not possible to predict which patients will be affected by doxorubicin-induced cardiotoxicity (DIC). Here we demonstrate that patient–specific human induced pluripotent stem cell–derived cardiomyocytes (hiPSC–CMs) can recapitulate individual patients’ predilection to DIC at the single cell level. hiPSC–CMs derived from breast cancer patients who suffered clinical DIC are consistently more sensitive to doxorubicin toxicity, demonstrating decreased cell viability, mitochondrial and metabolic function, calcium handling, and antioxidant pathway activity, along with increased reactive oxygen species (ROS) production compared to hiPSC–CMs from patients who did not experience DIC. Together, our data indicate that hiPSC–CMs are a suitable platform for identifying and verifying the genetic basis and molecular mechanisms of DIC.
Introduction
The anthracycline doxorubicin, one of the first chemotherapeutic agents to be introduced, is still one of the the most common and effective antineoplastic drugs currently in use1. Despite the advent of targeted tyrosine kinase- and monoclonal antibody-based therapies, anthracyclines are still used for 40–50% of breast cancer patients2, commonly alongside the alkylating agent cyclophosphamide (Cytoxan) or the antimicrotuble taxanes paclitaxel (Taxol) or docetaxel (Taxotere). The cardiotoxic side effects of doxorubicin are well established3, 4. Even at relatively low cumulative concentrations of 200–250 mg m−2, the risk of cardiotoxicity is estimated at 7.8% to 8.8%5, 6. The cardiotoxic side effects experienced with doxorubicin can range from asymptomatic reductions in left ventricular ejection fraction (LVEF), to tachycardia and arrhythmias, cardiomyopathy, myocardial infarction, and highly symptomatic congestive heart failure (Class III to IV)7, 8. At present, it is not possible to predict which patients will be affected by DIC or adequately protect patients who are at risk for suffering this devastating side effect9.
The anti-cancer effects of doxorubicin are thought to be threefold: via the stabilization of the toposimerase II α (TOP2A)–DNA cleavage complex, preventing DNA religation and double stranded cut repair; the intercalation of doxorubicin with double-stranded DNA directly resulting in transcriptome and epigenome modulation; and the generation of free radicals. The cardiotoxic effects of doxorubicin are thought to be more complex, but can be grouped into three interrelated subsets, generation of reactive oxygen species (ROS), toposimerase II β (TOP2B)10 and TOP1MT11 inhibition, and calcium release12.
The generation of reactive oxygen species (ROS) by redox cycling within cardiomyocytes, both dependent and independent of iron, causes mitochondrial dysregulation, lipid peroxidation, DNA damage, and protein carbonylation. ROS can be deactivated by endogenous antioxidants such as glutathione peroxidase, catalase and superoxide dismutase, yet doxorubicin also directly reduces the activity of these antioxidants, further increasing oxidative stress7. The mitochondria are the major site of doxorubicin-induced ROS generation due to the localization of the major redox cycling enzymes such as NAD(P)H and that doxorubicin becomes nearly irreversibly bound to cardiolipin on the mitochondrial membrane13. Doxorubicin also increases mitochondrial iron accumulation which further increases ROS production in the mitochondria14. Topoismerase inhibition (including TOP2B and TOP1MT) causes transcriptional modulation of the cell and mitochondrial genomes, DNA damage-induced apoptosis, and specifically, TOP2B has been associated with PPARGC1A and PPARGC1B (peroxisome proliferator-activated receptor gamma coactivator 1-α and -β) induced reduction in mitochondrial biogenesis10. Finally, doxorubicin and its metabolite doxorubicinol can induce Ca2+ release from the sarcoplasmic reticulum (SR) causing Ca2+ overload that leads to sarcomeric disarray and myofibril deterioration12, 15.
Here we demonstrate hiPSC–CMs derived from breast cancer patients who experienced doxorubicin-induced cardiotoxicity successfully recapitulate this phenotype. As the patient-specific human gene expression response to doxorubicin is essentially unknown, we go on extensively probe the transcriptomes associated with patient–specific cardiotoxicity responses. Finally, we show that hiPSC-CMs from patients with toxicity have significant reduction in basal metabolism and mitochondrial content.
Results
Generation of patient–specific hiPSC-derived cardiomyocytes
We recruited twelve female patients: eight breast cancer patients who had been treated at Stanford University Hospital with 240 mg m−2 doxorubicin or equivalent (Supplementary Table 1), including four patients (referred to as ‘DOX’) who did not experience clinical cardiotoxicity (as documented by minimum post–treatment LVEF > 56%); four patients (‘DOXTOX’) who did experience clinical cardiotoxicity (post–treatment LVEF = 10–45%); and four age– and gender–matched control volunteers who had never been treated with any chemotherapeutic agent (‘Healthy’) (Supplementary Table 2). We derived hiPSCs from these patients16 and all lines passed common assessments for pluripotency17 and genomic stability18 (Supplementary Fig. 1 and Supplementary Fig. 2). We differentiated hiPSCs into cardiomyocytes19, with modifications to the differentiation protocol to enhance cardiomyocyte purity20, mitochondrial metabolism21, and maturation22, resulting in cardiomyocyte purities of >85% (Supplementary Fig. 3). We based suitable in vitro hiPSC–CM doxorubicin treatments on the terminal plasma half–life of doxorubicin of 20–48 h, and peak plasma concentration (Cmax) of 2–6 µg ml−1 (average 6.9 µM) for a single concentration of 60 mg m−2 23, 24. Therefore, we primarily used concentrations in this range (0.1 µM to 10 µM). We selected time points of 24 h and 72 h based on prior reports on primary neonatal rat ventricular cardiomyocytes (NRVMs)25–27.
Characterization of in vitro doxorubicin-induced cardiotoxicity
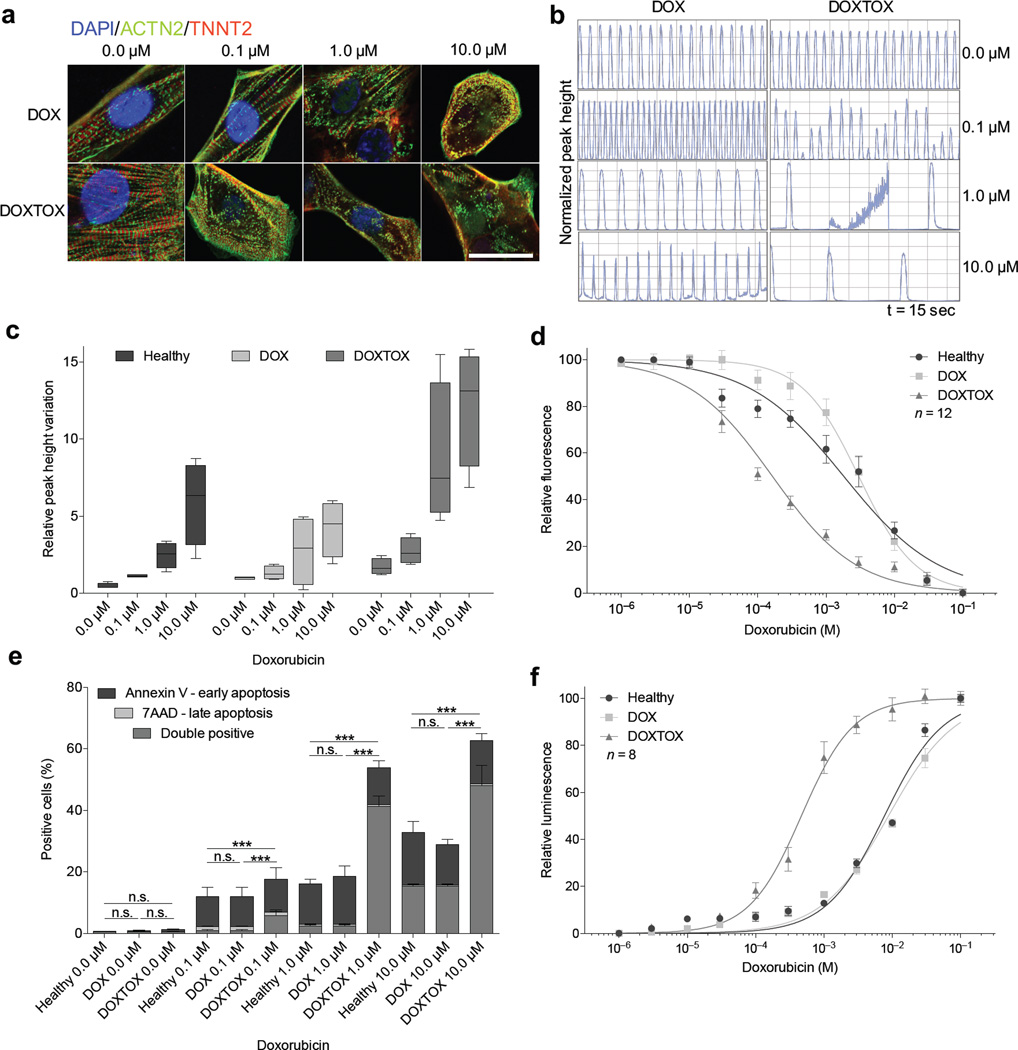
We began by establishing whether a differential response to doxorubicin existed between Healthy, DOX, and DOXTOX hiPSC–CM groups. Immunofluorescent imaging demonstrated a concentration–response increase in sarcomeric disarray, a well–established effect of doxorubicin on NRVMs28, 29, which was consistently seen at 0.1 µM in the DOXTOX cells but not in DOX cells (Fig. 1a). Cardiomyocyte contraction assays demonstrated increased beat rates in DOX cells at 1 µM and increasing arrhythmia at higher concentrations, which was more severe in DOXTOX cells (Fig. 1b, c). After 72 h treatment with doxorubicin, the four DOXTOX hiPSC–CM lines consistently showed significantly reduced cell viability compared to the four DOX and four Healthy hiPSC–CM lines at all concentrations tested (F–test statistic = 170.5, P < 0.0001), with an LD50 of 0.1643 µM for the DOXTOX cells and 3.015 µM for the DOX cells (Fig. 1d). This increase in doxorubicin cardiotoxicity susceptibility could be further validated using a wide range of assays: membrane integrity and maintenance of a reducing environment, ATP quantitation, adenylate kinase release, and lactate dehydrogenase release (Supplementary Fig 4a–e). Flow cytometry demonstrated a concentration–dependent increase in cells double–positive for annexin V+ and 7-aminoactinomycin D (7–AAD), suggesting programmed cell death as the principal mechanism of cell loss. A significant (P < 0.005) increase in annexin V+ and 7–AAD+ cells was also noted in DOXTOX hiPSC–CMs compared to DOX hiPSC–CMs at higher doxorubicin concentrations (Fig. 1e). A second assay for apoptosis, the measurement of caspase 3 and 7, further confirmed this result (Fig. 1f).
Figure 1.
Assessment of in vitro doxorubicin–induced cardiotoxicity in patient–specific hiPSC–CMs. (a) Immunofluorescent staining for α–actinin (ACTN2) and cardiac troponin T (TNNT2) to demonstrate sarcomeric organization in hiPSC–CMs derived from patients who did not experience doxorubicin–induced cardiotoxicity (DOX) versus those who did experience doxorubicin-induced cardiotoxicity (DOXTOX) after 24 h treatment with doxorubicin. Scale bar, 20 µm. (b) Representative camera–capture contraction assay demonstrating variation in beat frequency in response to doxorubicin after 24 h treatment with doxorubicin (hiPSC–CMs from lines DOX1 and DOXTOX4 shown). (c) Camera–capture contraction assay demonstrating variation on relative peak high in response to doxorubicin in Heathy, DOX, and DOXTOX hiPSC–CMs after 24 h treatment with doxorubicin. Each data point represents four hiPSC–CM lines repeated three times (n = 12). (d) Effect of doxorubicin (72 h) on hiPSC–CM viability (n = 12). LD50: Healthy, 1.82 µM; DOX, 3.015 µM; DOXTOX, 0.1643 µM. (e) Detection of early and late apoptosis in hiPSC–CMs after 72 h treatment with doxorubicin (n = 4). Unpaired two–tailed t–test with *P < 0.05, **P < 0.01, ***P < 0.005, n.s. = not significant. (f) Effect of doxorubicin (72 h) on caspase 3 and 7 expression (n = 8). Error bars represent standard error of the mean (s.e.m.)
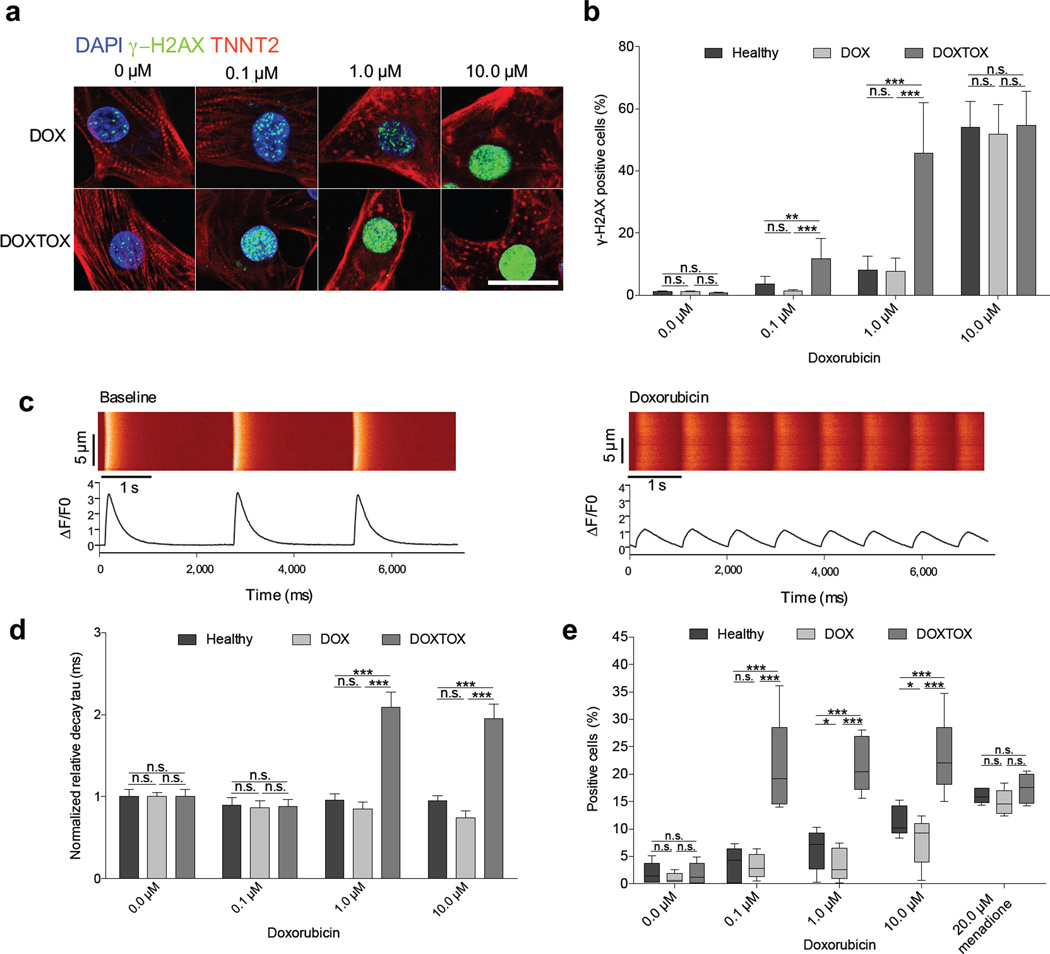
Given that the therapeutic mechanism of doxorubicin in tumor cells is via induction of DNA damage, we next assessed the level of double–stranded DNA damage by staining for phosphorylated H2A histone family member X (γ–H2AX). Similarly, we observed a concentration–dependent increase in DNA damage (Fig. 2a), which was significantly (P < 0.005) higher in DOXTOX hiPSC–CMs at the 0.1 µM and 1 µM doxorubicin concentrations (Fig. 2b). To probe the potential mechanisms for the observed differences in response to doxorubicin, we next assessed spontaneous Ca2+ transients by loading single cells with Fluo–4 (Fig. 2c). In DOXTOX hiPSC–CM samples treated with 1 µM or 10 µM doxorubicin, there was a significant (P < 0.005) increase in the transient decay time (decay tau) in the DOXTOX hiPSC–CM over the DOX hiPSC–CM group (Fig. 2d), likely a function of the reduced transient amplitude and time to peak (Supplementary Fig. 4f, g). No significant effect on beat rate between groups was detected (Supplementary Fig 4h).
Figure 2.
Assessment of the effect of doxorubicin on DNA damage, calcium handling, and whole–cell oxidative stress in patient–specific hiPSC–CMs. (a) Detection of DNA double–stranded breaks using immunofluorescent staining for γ–H2AX after 24 h doxorubicin. Scale bar, 20 µm. (b) Quantification of γ–H2AX staining by flow cytometry (n = 4). (c) Representative recording of spontaneous calcium activity of patient–derived hiPSC–CMs at baseline or treated with doxorubicin, DOXTOX1 shown. (d) Normalized relative decay tau of calcium imaging. Each data point represents an average of four hiPSC–CM lines (n > 35 cells cells per line). Groups were normalized to 0 µM healthy group. Comparisons were conducted via Fisher’s exact test, one way–ANOVA test followed by All Pairwise Multiple Comparison Procedures (Holm–Sidak method). (e) Assessment of the effect of 24 h doxorubicin treatment (before cell death) on whole cell reactive oxygen species (ROS) levels using CellROX. Unpaired two–tailed t–test with *P < 0.05, **P < 0.01, ***P < 0.005, n.s. = not significant. Error bars represent s.e.m.
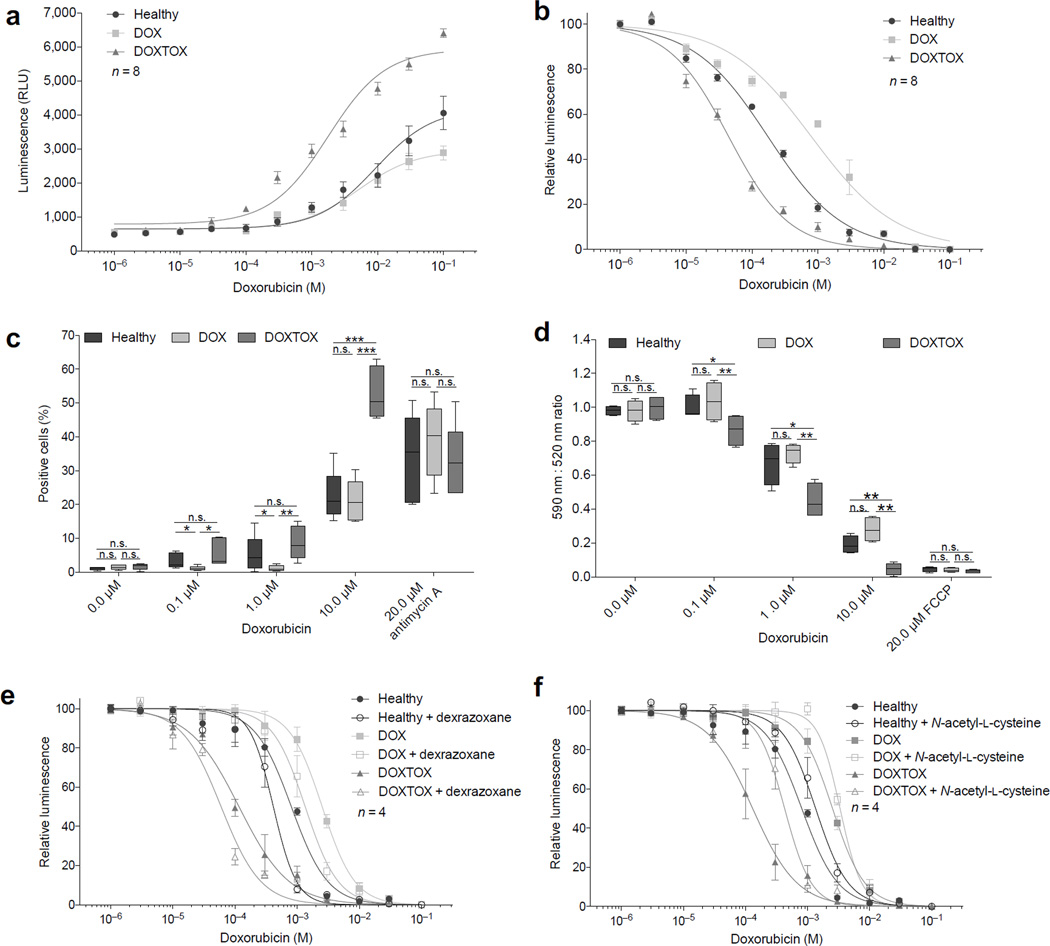
Several studies have implicated oxidative stress, as evidenced by increased ROS, as the central mechanism underlying the cardiotoxic effects of doxorubicin30–33. We next assessed cellular ROS production by flow cytometry using the fluorescent CellROX probe. After 24 h of doxorubicin, ROS was significantly (P < 0.005) higher in the DOXTOX hiPSC–CMs compared to the DOX hiPSC–CMs by an average of >2–fold after 0.1 to 10 µM (Fig. 2e). A second assay measuring whole cell hydrogen peroxide (H2O2) confirmed this >2–fold increase in ROS from concentrations as low as 0.01 µM doxorubicin (Fig. 3a). We next assayed the antioxidant glutathione (GSH) as a marker of cellular oxidative stress response and found a significantly (P < 0.005) greater drop in GSH levels in DOXTOX hiPSC–CMs at lower levels of doxorubicin than DOX hiPSC–CMs (EC50 of 0.04 µM vs. 0.8 µM) (Fig. 3b). Mitochondria are one of the major sources of increased ROS production and also a target for doxorubicin toxicity. Using a mitochondrial superoxide probe (MitoSOX), we demonstrated significantly (P < 0.005) higher levels of mitochondrial ROS in the DOXTOX hiPSC–CMs compared to DOX hiPSC–CMs (>2.5–fold at 10 µM) (Fig. 3c). Finally, we demonstrated that there was a consistent concentration–dependent decrease in mitochondrial membrane potential to doxorubicin using the JC–10 probe, which was significantly (P < 0.01) greater in the DOXTOX hiPSC–CMs vs. DOX hiPSC–CMs at 0.1 to 10 µM (Fig. 3d).
Figure 3.
Assessment of the effect of doxorubicin on oxidative stress in patient–specific hiPSC–CMs. (a) Assessment of the effect of 24 h doxorubicin treatment on hydrogen peroxide (H2O2) levels. (b) Assessment of the effect of 24 h doxorubicin treatment on the presence of the antioxidant glutathione (GSH) levels. (c) Assessment of the effect of 24 h doxorubicin treatment on mitochondrial superoxide (but not other reactive oxygen or nitrogen species) levels. (d) Assessment of the effect of 24 h doxorubicin treatment on mitochondrial membrane potential. (e) ATP–based cell viability assay demonstrating the effect of 72 h treatment of doxorubicin in the presence of iron chelator dexrazoxane (DRZ). (f) ATP–based cell viability assay demonstrating the effect of 72 h treatment of doxorubicin in the presence of antioxidant N–acetyl–L–cysteine (NAC) in response to doxorubicin. Unpaired two–tailed t–test with *P < 0.05, **P < 0.01, ***P < 0.005, n.s. = not significant. Error bars represent s.e.m.
To help further understand the mechanisms of action, we pre– and co–treated hiPSC–CMs with the cardioprotectant dexrazoxane (DRZ)34, a catalytic topoisomerase inhibitor and iron chelator, used clinically to try to reduce doxorubicin cardiotoxicity35. Unexpectedly, we found that DRZ significantly (P < 0.005) increased toxicity, specifically in DOXTOX hiPSC–CMs at 0.1 µM and DOX hiPSC–CMs at 3 µM (Fig. 3e). To determine whether this effect was a problem with our model or limited to this one agent, we next assessed the effects of a well–established antioxidant, N–acetyl–L–cysteine (NAC)36. NAC significantly (P < 0.005) decreased doxorubicin–induced toxicity in DOXTOX hiPSC–CMs (Fig. 3f), confirming that ROS–based toxicity is a major component of reduced viability
Effect on doxorubicin on patient-specific gene expression
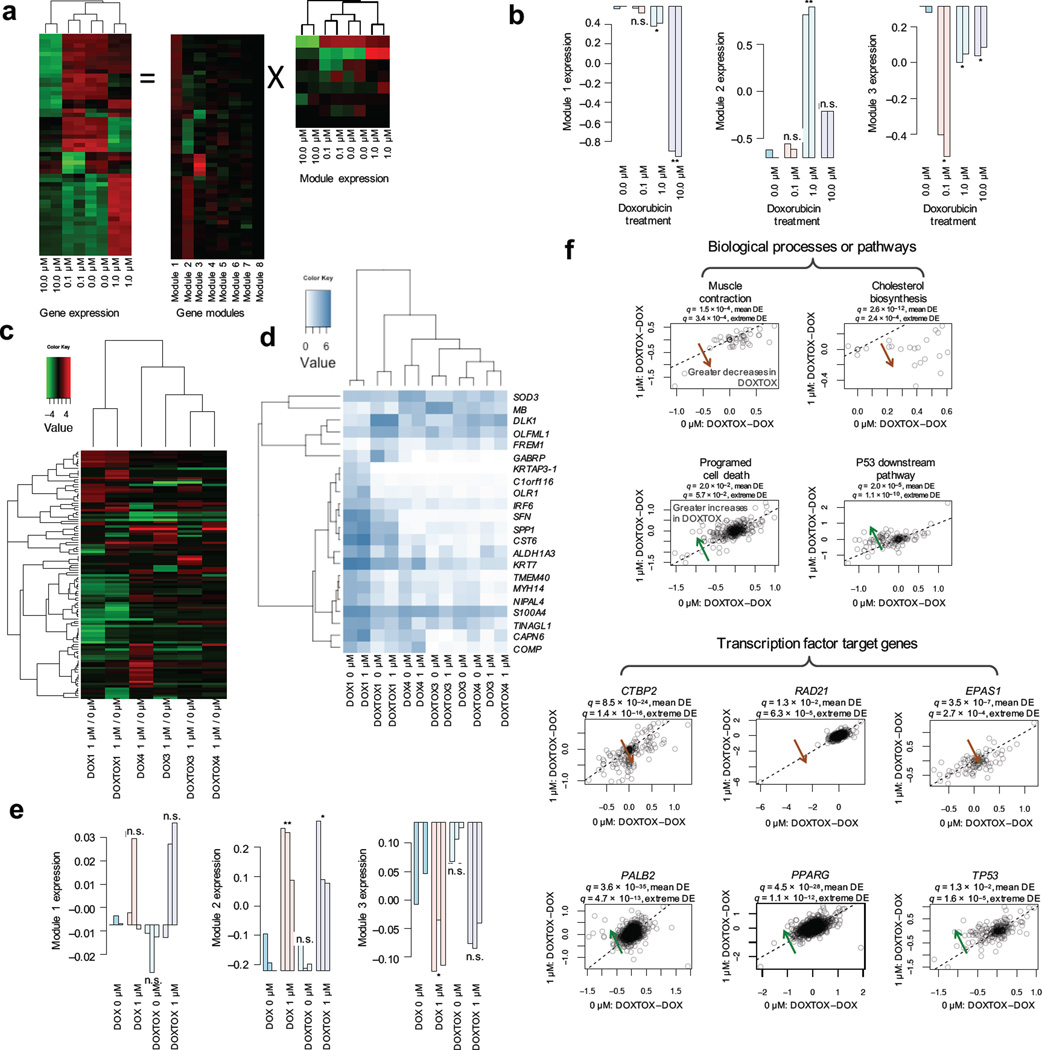
We hypothesized that the mechanism of DIC may vary depending on the concentration of doxorubicin. To probe this question, we began by performing microarray analysis on control hiPSC–CMs exposed to increasing concentrations of doxorubicin (Fig. 4a). After 24 h doxorubicin treatment, minimal changes were seen with 0.1 µM concentration. By contrast, numerous cardiac development-related transcription factors were significantly down-regulated at 1 µM concentration compared to the untreated group, including NKX2–5 (homeobox protein Nkx-2.5), MEF2A (myocyte-specific enhancer factor 2), TBX5 (T-box transcription factor TBX5), and MYOCD (myocardin) (Supplementary Table 3). Using Ingenuity Toxicogenomics analysis, we found that TGF–β signaling, p53, cardiac hypertrophy, G1/S checkpoint regulation, and retinoic acid receptor activation were differentially regulated between the 1 µM and 10 µM groups (Supplementary Table 4). We next applied the unsupervised machine learning approach Independent Component Analysis37 to discover 8 modules of gene expression related to the different concentrations of doxorubicin, identifying key transcription regulators such as TP53 (tumor protein p53), RELA (v-rel avian reticuloendotheliosis viral oncogene homolog A), NFKB1 (nuclear factor of kappa light polypeptide gene enhancer in B-cells 1), and EP300 (E1A binding protein p300) that have previously been classified as involved in doxorubicin pharmacodynamics and cardiotoxicity pathways by PharmGKB38 (Fig. 4b and Supplementary Fig. 5a,b).
Figure 4.
Modulation of gene expression in hiPSC–CMs by doxorubicin. (a) Functional gene modules of transcriptional responses following doxorubicin treatment of cardiomyocytes. Illustration of the ICA decomposition of gene expression data (showing 51 most varied genes) into the linear combination of multiple statistically independent gene modules. (b) The first three gene modules are significantly associated with doxorubicin dosages. Module 1, oxidation and hypoxia stress response and enrichment of genes related to heart morphogenesis. Module 2, inflammation and DNA damage repair. Module 3, cell cycle arrest and apoptosis. (c) Differential gene expression in DOX and DOXTOX hiPSC lines after treatment with 1 µM doxorubicin for 24 h normalized to baseline expression (12 samples total). (d) Heatmap of top genes significantly and differentially expressed between DOX and DOXTOX hiPSC–CMs after doxorubicin treatment. (e) Relationship between DOX and DOXTOX with 1 µM doxorubicin results and gene expression modules, demonstrating strong correlation between modules 2 and 3. (f) Scatter plots of the DOXTOX to DOX patient differences at 1 µM of doxorubicin (y–axis) vs. the differences at 0 µM of doxorubicin (x–axis) for selected biological processes, pathways, and transcription factors. A systematic deviation from the diagonal (dashed line) suggests patient–specific responses upon doxorubicin treatment. Significance of a given gene set (e.g., biological process, pathway or TF target gene set) is evaluated in two ways, namely mean differential expression and extreme differential expression. For mean differential expression (DE), we test if delta–delta–expression (y–x) has identical means for the member genes of the gene set against other genes using t–test. For extreme differential expression, we test if extreme delta–delta–expression values are enriched for member genes of the gene set using Fisher’s exact test. Significant: multi–test corrected P–value < 0.05.
To identify a genetic rationale for the observed differences in response to doxorubicin in the DOX and DOXTOX cohorts, we next performed RNA–seq on hiPSC–CMs derived from three DOX and three DOXTOX lines, both with and without 1 µM doxorubicin exposure for 24 h, and after normalizing for baseline expression in untreated cells (Fig. 4c) and identified the most differentially regulated genes between the DOX and DOXTOX hiPSC–CM populations after doxorubicin treatment (Fig. 4d and Supplementary Fig. 5c). The relative activity of gene modules in the concentration–response microarray experiment is nicely recapitulated (Fig. 4e and Supplementary Figure 6a, b). Modules 1–3 explain 41%, 39%, and 7.8% of the total expression variances, respectively, whereas in the patient-response study, the three modules explain 0.7%, 73%, and 16%, respectively.
An independent pairwise comparison of the DOX vs. DOXTOX at baseline and DOX vs. DOXTOX at 1 µM again identified RELA, NFKB1, and RARA (retinoic acid receptor alpha) with significantly (P < 1.8 × 10−4) altered target gene expression, and signal transducer and activator of transcription 3 as significant signaling pathway, which were previously described39, 40 (Supplementary Table 5). In addition, we identified significantly (q < 1.3 × 10−2) increased expression of Programed Cell Death, p53 downstream pathway, and target genes of transcription factors TP53, BRCA1 (breast cancer 1), PALB2 (partner and localizer of BRCA2), STAT3, CEBPA (CCAAT/enhancer binding protein alpha), and PPARG (peroxisome proliferator activated receptor gamma), and significant (q < 1.3 × 10−2) decreases of Muscle Contraction, Cholesterol Biosynthesis, Anatomical Structure Development, and target genes of SUZ12 (SUZ12 polycomb repressive complex 2 subunit), CTBP2 (C-terminal binding protein 2), EPAS1 (endothelial PAS domain protein 1), and RAD21 (RAD21 cohesin complex component) in DOXTOX hiPSC–CMs treated with 1 µM doxorubicin compared to the untreated group (Fig. 4f, Supplementary Fig. 6c and Supplementary Table 6). Based on the RNA–seq data, we determined the coding sequence variant for BRCA1, BRCA2 (breast cancer 2) and PALB2 and found that homozygous non–synonymous variants in BRCA1 might be associated with DOXTOX subject cardiotoxicity (data not shown).
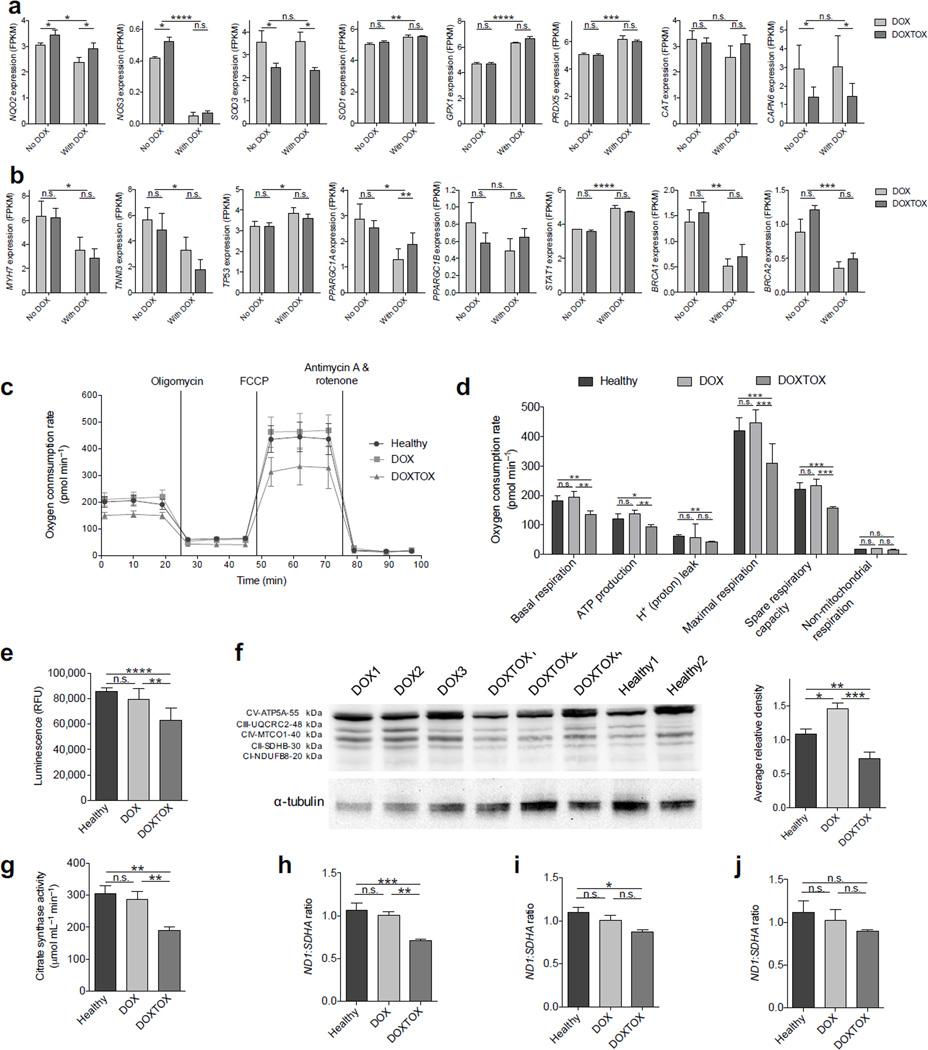
We next examined the expression of genes identified by PharmGKB38 as associated with DIC. Genes differentially expressed between the DOX and DOXTOX populations include those associated with redox cycling of doxorubicin and production of ROS NQO2 (NAD(P)H dehydrogenase, quinone 2), the generation of reactive nitrogen species NOS3 (nitric oxide synthase 3 (endothelial cell), and those associated with antioxidants such as SOD3 (superoxide dismutase 3), although no variance was found in CAT (catalase)41 (Fig. 5a and Supplementary Fig. 7a, b). We found significant (P < 0.05) differences in CAPN6 (calpain 6), which is involved in the Ca2+ overload response28, although we noted no differences in other calcium handling–related genes (Supplementary Fig. 7c). The down–regulation of cardiac structural genes in response to doxorubicin is well established26, 42, and we confirmed a consistent down regulation of such transcripts as MYH7 (β–myosin heavy chain) and TNNI3 (cardiac troponin I), among others, after doxorubicin treatment in both populations (Fig. 5b, Supplementary Fig. 7d). Additionally, we found significant (P < 0.05) differences in genes involved with iron transport and storage32 (Supplementary Fig. 7e), chromatin remodeling and chemotherapy resistance-related gene SMARCA4 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4)43 (Supplementary Fig. 7f), and apoptosis signaling40 (Fig. 5b, Supplementary Fig. 7g), as well as genes previously identified by DIC targeted SNP studies44 (Supplementary Fig. 7h). PPARGC1A was less down-regulated in DOXTOX cells than DOX cells after doxorubicin treatment and PPARGC1B was upregulated in DOXTOX cells and up-regulated in DOX cells after doxorubicin treatment (Fig. 5b). Finally, significant (P < 0.01) differences were also noted in STAT1 and STAT3, and BRCA1 and BRCA2 (Fig. 5b and Supplementary Fig. 7i).
Figure 5.
Assessment of the baseline mitochondrial function in patient–specific hiPSC–CMs. (a) Expression of ROS– ands calcium overload–related genes differentially expressed in DOX and DOXTOX hiPSC–CMs after exposure to doxorubicin. (b) Expression of sarcomeric proteins, apoptosis markers, TOP2B related PGC1–α and PGC1–β, STAT, and BRCA–related genes differentially expressed in DOX and DOXTOX hiPSC–CMs after exposure to doxorubicin. No doxorubicin vs. with doxorubicin, two–way ANOVA. DOX vs. DOXTOX, t–test. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001, n.s. = not significant. Error bars represent s.e.m. (c) Representative Seahorse extracellular flux assay measuring oxygen consumption rate (OCR). (d) Analysis of Seahorse extracellular flux assay demonstrating lower levels of oxygen consumption in DOXTOX hiPSC–CMs at baseline (i.e., without doxorubicin treatment) in comparison to DOX hiPSC–CMs (n = 12). (e) Quantification of ATP levels in 100,000 hiPSC–CMs (n = 8 hiPSC–CM samples). (f) Western blotting for select proteins involved in oxidative phosphorylation and graph with quantification of average releative density of Western blot data. (g) Citrate synthase assay to measure the baseline presence of intact mitochondria in 5 million iPSC-CMs (n = 8, each data point is an average of four cell lines with two experimental replicates). (h) Quantification of mitochondria encoded Complex I ND1 DNA ratio to nuclear encoded complex II SDHA DNA in healthy control patient, DOX, and DOXTOX hiPSC–CMs (n = 4). (i) ND1:SDHA expression ratio in DOX and DOXTOX patient fibroblasts and healthy (non–doxorubicin treated) patient fibroblasts (n = 4). (j) Average ND1:SDHA expression ratio in four Healthy, four DOX, and four DOXTOX hiPSC lines (n = 3). Paired two–tailed t–test with *P < 0.05, **P < 0.01, ***P < 0.005, n.s. = not significant. Error bars represent s.e.m.
Patient-specific baseline metabolic variance
As mitochondrial function has been identified as a key component of DIC45, we next performed a baseline mitochondrial stress test in DOX and DOXTOX hiPSC–CMs using a Seahorse XF24 extracellular flux analyzer (Fig. 5c). More extreme reductions in basal and maximal respiration in DOXTOX cells were also noted after doxorubicin treatments, particularly with 1 and 10 µM treatment (Supplementary Fig. 8a–d). Surprisingly, even without doxorubicin treatment, variance was noted in the basal respiration, maximal mitochondrial respiration, and spare respiratory capacity, as assessed by subjecting cells to FCCP (carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone), which promotes a dissipation of the membrane potential and leads to an increase in oxygen consumption that is accompanied by a decrease in ATP synthesis (Fig. 5d). To further probe this result, we assessed baseline ATP activity and found this to be significantly (P < 0.01) lower in DOXTOX hiPSC–CMs than that of healthy and DOX hiPSC–CMs (Fig. 5e). Western blot for select proteins involved in oxidative phosphorylation showed a similar trend (Fig. 5f and Supplementary Fig. 8e), as did quantification of citrate synthase activity, a marker for mitochondrial content (Fig. 5g). We then compared nuclear vs. mitochondrial DNA content as a surrogate to estimate the relative cellular mitochondrial content. DOXTOX hiPSC–CMs contained ~30.1±2.4% fewer mitochondria than DOX hiPSC–CMs (Fig. 5h). To determine whether this was generalizable to other cell types, we also assessed mitochondrial content directly in patient fibroblasts and undifferentiated hiPSCs, but did not find a significant difference (Fig. 5i, j). To further study whether patient–derived fibroblasts or hiPSCs could be used as a surrogate for predicting DIC, we performed viability assays with 5–log range concentrations of doxorubicin. In contrast to our results with hiPSC–CMs, we found that these cell types could not recapitulate patient response to doxorubicin (Supplementary Fig. 9).
Discussion
Although the effects of doxorubicin on pure populations of cardiomyocytes in vitro may not mimic all processes that occur in vivo, our results suggest that, patient–specific hiPSC–CMs can recapitulate patients’ predilection to DIC and enable the prediction of clinical susceptibilities of high–risk populations to drug–induced cardiotoxicity. The differences between DOX and DOXTOX hiPSC–CMs manifest at multiple points in the DIC mechanism (summarized in Fig. 6) and these differences are not detectable in other patient-derived cell types such as fibroblasts or hiPSCs. In particular, we confirm numerous DIC phenotypes shown in animal models, particularly concentrating on the ROS pathways and mitochondrial dysregulation.
Figure 6.
Schematic of findings in relationship to the established DIC pathways. Numbered cyan boxes demonstrate our findings. Doxorubicin (DOX), doxorubinol (DOX–ol), doxorubicin–semiquinone (DOX–semiquinone), C7 centered radical aglycone (C7 radical), nitric oxide synthase 3 (NOS3), NADH dehydrogenases (collectively NAD(P)H oxidoreductases), P450 (cytochrome) oxidoreductase (POR), xanthine oxidase (XDH) superoxide radical (O2–•), hydrogen peroxide (H2O2), hydroxyl radical (OH•), nitric oxide (NO•), peroxynitrite (ONOO–), superoxide dismutase (SOD), catalase (CAT), glutathione (GSH), glutathione peroxide (GSH), glutathione disulfide (GSSG), peroxiredoxin (PRDX), myoglobin (MB), ferrous iron (Fe2+), ferric iron (Fe3+), dexrazoxane (DRZ), N–acetyl–L–cysteine (NAC), topoisomerase (DNA) 1 mitochondrial (TOP1MT), BCL2–associated X protein (BAX), cytochrome C (CYCS) tumor protein p53 (TP53), topoisomerase 2B (TOP2B), ryanodine receptor 2 (RYR2), ATPase, Ca2+ transporting, cardiac muscle slow twitch 2 (ATP2A2), myosin light chain (MYL), cardiac troponin T (TNNT), α–actinin (ACTA), peroxisome proliferator-activated receptor gamma, coactivator 1–α (PPARGC1A) and peroxisome proliferator-activated receptor gamma, coactivator 1–β (PPARGC1B)
The hiPSC-CM model has weaknesses, such as a lack of suitable maturity46 and sub-type specificity47 and a lack of co-localization with other cell types found in the heart such as fibroblasts and endothelial cells48. Yet this is also one of the model’s great strengths as immature cells are easier to dissociate, pure cells are more suited to plate assays and high–content imaging, and RNA-seq analysis is more accurate due to the lack of contamination with other cell types. The demonstration here that this human surrogate system is suitable for assessing patient–specific DIC will allow the future validation of gene variants identified through genome–wide association studies (GWAS)49. This platform also holds considerable promise for the discovery of novel DIC cardioprotectants, although our data on the lack of efficacy of the iron-chelator dexrazoxane but success of the antioxidant NAC may highlight the differences between a whole-animal model and an in vitro isolated cardiomyocyte-model. In particular, it is thought that the heart is uniquely at risk to DIC due to high proportion of cardiomyocyte mitochondria, making up 35% of the total cell volume50. In hiPSC-CMs, however, mitochondrial numbers are lower51, and the confirmation of this ‘mitochondrial content’ risk effect will require further analysis. Finally, our work on variation of basal metabolism needs to take into account that all cells (fibroblasts, hiPSC, and hiPSC-CMs) were derived after patients had received doxorubicin, and any systemic damage to the genomic or mitochondrial DNA in the original patient fibroblasts used for reprogramming may be carried over in these cell types. Therefore a better model in the future would be to derive hiPSCs prior to doxorubicin treatment and perform similar sets of experiments outlined here. Although this would require significantly larger patient subjects, it would move us toward precision medicine for cardio-oncology.
ONLINE METHODS
Human induced pluripotent cell derivation
All pluripotent and reprogramming cultures were maintained at 37 °C in a New Brunswick Galaxy 170R humidified incubator (Eppendorf) with 5% CO2 and 5% O2. Primary cell and differentiation cultures were maintained at 5% CO2 and 21% O2. Protocols were approved by the Stanford University Human Subjects Research Institutional Review Board. With informed written consent, two 2–mm skin punch biopsies were taken from each volunteer, diced with a scalpel, digested with 1 mg ml−1 collagenase IV (Life Technologies) for 2 h at 37 °C. Fibroblasts were then grown in DMEM with GlutaMAX (Life Technologies) supplemented with 10% fetal bovine serum (FBS, US origin, Life Technologies) on 6–well plates (Greiner) coated with a 1:200 dilution of growth–factor reduced Matrigel (9 µg cm−2, Corning). Media was changed every other day. When confluent (~1 week), fibroblasts were passaged with TrypLE Express (Life Technologies) onto Matrigel–coated T225 flasks (Thermo Scientific Nunc). For Sendai virus reprogramming, early passage (p2–p3) fibroblasts were seeded at 40,000 cells per well on Synthemax II–SC (625 ng cm−2, Corning)–coated52 6–well plates in E853. E8 was made in–house and consisted of DMEM/F12 (10–092–CM, Corning), 20 µg ml−1 E. coli–derived recombinant human insulin (Life Technologies), 64 µg ml−1 L–ascorbic acid 2–phosphate sesquimagnesium salt hydrate (Sigma–Aldrich), 10.7 µg ml−1 Oryza sativa–derived recombinant human transferrin (Optiferrin, InVitria/Sigma–Aldrich), 14 ng ml−1 sodium selenite (Sigma–Aldrich), 100 ng ml−1 recombinant human FGF2 (154 amino acid, E. coli–derived, Peprotech), 2 ng ml−1 recombinant human TGFβ1 (112 amino acid, HEK293–derived, Peprotech), and 100 ng ml−1 heparin sodium salt (>180 U mg−1, Sigma–Aldrich). After 24 h, media was changed to fresh E8 and supplemented with four CytoTune–iPS Sendai Reprogramming Kit viral particle factors (Life Technologies)54 diluted to 20% of the manufacturer’s recommendations (3 × 105 cell infectious units [CIU] of each particle per well, multiplicity of infection [MOI] = 7.5). Media was changed after 24 h and thereafter once every day. For the first 7 days, cultures were maintained in E8 supplemented with 100 nM hydrocortisone (Sigma–Aldrich) and 200 µM sodium butyrate (Sigma–Aldrich) to enhance reprogramming efficiency55. At day 7, cells were passaged with TrypLE Express and seeded onto Synthemax II–SC–coated 6–well plate in E7N media (E8 minus TGFβ1; supplemented with 200 µM sodium butyrate). 10 µM Y27632 (Biorbyt) was added for the first 24 h after passage. Media was changed every day, and switched to E8 at day 20. Individual colonies with hESC–like morphology were picked onto a 12–well plate (1 colony per well) at day 17–25 in E8 with 10 µM Y27632 for 24 h after picking. Subsequently, cells were expanded in 6–well plates by passaging 1:1, 1:4, 1:6, 1:8, and finally 1:12 using 0.5 mM EDTA (Life Technologies) in DPBS without Ca2+ or Mg2+ (Life Technologies) for 6 min at RT. Specific hiPSC clones used for this study were: 59FSDNC3 (DOX1), 60FSDNC1 (DOX2), 64FSDNC1 (DOX3), 65FSDNC2 (DOX4), 31FSDNC14 (DOXTOX1), 40FSDNC15 (DOXTOX2), 51FSDNC18 (DOXTOX3), and 79FSDNC1 (DOXTOX4). For hESC experiments, the line H7 (WA07)56 was used (WiCell Research Institute).
Human induced pluripotent stem cell culture
Cells were routinely maintained in E8 (made as above) on 1:200 growth factor–reduced Matrigel (9 µg cm−2) and passaged every 3–4 days using 0.5 mM EDTA (as above). Cell lines were used between passages 20 and 85. All cultures (primary, pluripotent, and differentiation) were maintained with 2 ml media per 10 cm2 of surface area or equivalent. All cultures were routinely tested for mycoplasma using a MycoAlert Plus Kit (Lonza).
Immunofluorescence staining for pluripotency markers
For assessment of pluripotency, hiPSCs were passaged with EDTA and plated onto Synthemax II–SC–coated (625 ng cm−2) Lab–Tek II 8–chamber glass slides (Thermo Scientific Nunc) in E8 with 10 µM Y27632 (for 24 h) and allowed to grow for 3 days. Cells were fixed with 4% paraformaldehyde (Electron Microscopy Services) in DPBS (Life Technologies) for 10 min at RT, permeabilized with 0.1% saponin (Sigma–Aldrich) in DPBS for 20 min at RT, blocked in 3% bovine serum albumin (BSA, Sigma–Aldrich) in DPBS for 15 min at RT, and stained with 1:200 monoclonal mouse IgM TRA–1–60 (sc–21705), 1:200 monoclonal mouse IgM TRA–1–81 (sc–21706), 1:200 monoclonal mouse IgG3 SSEA4 (sc–21704), 1:200 monoclonal mouse IgG2b POU5F1 (sc–5279), 1:200 monoclonal mouse IgG1 NANOG (sc–33759), or 1:200 monoclonal mouse IgG1 SOX2 (sc–365823) in 0.1% saponin in DPBS overnight at 4 °C. Cells were washed and then stained with secondary antibodies 1:250 Alexa Fluor 488 goat anti–mouse IgM, or Alexa Fluor 488 goat anti–mouse IgG3, and 1:250 Alexa Fluor 594 goat anti–mouse IgG2b or Alexa Fluor 594 goat anti–mouse IgG1 (all Life Technologies) in 0.1% saponin in DPBS for 1 h at RT in the dark. Cells were washed three times and mounted with ProLong Diamond Antifade Mountant with DAPI (Life Technologies). Slides were imaged with a DM IL LED inverted fluorescent microscope (Leica Microsystems) at 40× and with a DFC550 camera (Leica Microsystems) and LAS X software and processed using Volocity 6.0 (PerkinElmer).
Flow cytometry to assess pluripotency
For assessment of pluripotency, hiPSCs at three days after passage were dissociated with TrypLE Express for 3 min at 37 °C and 1 million cells were transferred to flow cytometry tubes (BD Biosciences). Cells were then fixed with 4% PFA in DPBS for 10 min, permeabilized with 0.1% saponin (Sigma–Aldrich) in DPBS for 20 min, and stained using 1:50 mouse IgM TRA–1–81-488 (BD Biosciences, 560173), mouse IgG3 SSEA4-488 (BD Biosciences 560308), POU5F1-488 (BD Biosciences, 560217), or SOX2 (BD Biosciences, 245610) for 30 min at RT. Isotype controls FITC mouse IgM κ (BD Biosciences, 555583) and Alexa Fluor 488 Mouse IgG3, κ (BD Biosciences, 563536), Alexa Fluor 488 Mouse IgG1, κ (BD Biosciences, 557702), Alexa Fluor 647 Mouse IgG2a, κ (BD Biosciences, 557715) were used to establish gating. Cells were analyzed using a FACSAria II (BD Biosciences) with a 100 µM nozzle and FACSDiva software. Data was analyzed using FlowJo (Tree Star).
Quantitative Real–time PCR
To analyze pluripotent gene expression, cells were dissociated with TrypLE Express for 5 min at 37 °C, triturated and diluted in E8 and centrifuged at 200 g for 4 min. Media was aspirated and pellets of cells were snap frozen in liquid nitrogen and stored at −80 °C. RNA was isolated using an RNeasy Plus kit (QIAGEN), cDNA was produced using a High Capacity RNA–to–cDNA kit (Life Technologies), and real–time PCR was performed using TaqMan Gene Expression Assays 18S (Hs99999901_s1), NANOG (Hs02387400_g1), POU5F1 (Hs00999632_g1), SOX2 (Hs01053049_s1), KLF4 (Hs00358836_m1), LIN28 (Hs00702808_s1), MYC (Hs00153408_m1), UTF1 (Hs00747497_g1), ABCG2 (Hs01053790_m1), DMNT3B (Hs01002405_m1), TERT (Hs99999022_m1), TP53 (Hs99999147_m1), TaqMan Gene Expression Master Mix, and a 7900HT Real–Time PCR System (Life Technologies). All PCR reactions were performed in quadruplicate, normalized to 18S endogenous control gene, and assessed using the comparative Ct method5.
Teratoma analysis
Three confluent wells of pluripotent cells were dissociated with 0.5 mM EDTA, centrifuged at 200 g for 4 min, resuspended in 100 µl of growth factor–reduced Matrigel, and injected into the kidney capsule of female NOD–SCID mice (NOD.CB17–Prkdcscid/NcrCrl Strain code 394, Charles River). After 4–6 weeks, teratomas were removed, fixed in 4% PFA, embedded in paraffin wax, sectioned, and hematoxylin– and eosin–stained (H&E) by the Stanford Tissue Bank. Slides were imaged and analyzed by a qualified clinical pathologist.
Single nucleotide polymorphism karyotyping
A single well of >p20 pluripotent cells was dissociated with 0.5 mM EDTA, centrifuged at 200 g for 4 min, and pellet was snap frozen in liquid nitrogen and stored at −80 °C. Genomic DNA was extracted from the cell pellets using a Blood and Tissue DNA extraction kit (QIAGEN) following the manufacturer’s directions. SNP karyotyping was performed using a Genome–Wide CytoScan HD Array (Affymetrix) covering 2.7 million markers and 750,000 SNPs, and was analyzed using Chromosome Analysis Suite (ChAS, Affymetrix).
Cardiac differentiation of hiPSCs
hiPSCs (>p20) were split at 1:12 to 1:15 ratios using EDTA as above and grown for 3–4 days at which time they reached ~75% confluence. Media was changed to CDM36, consisting of RPMI 1640 (10–040–CM, Corning), 500 µg ml−1 Oryza sativa–derived recombinant human albumin (Oryzogen Sciencell), and 213 µg ml−1 L–ascorbic acid 2–phosphate (Sigma–Aldrich). Media was changed every other day (48 h). For days 0–2, media was supplemented with 6 µM CHIR99021 (MedChem Express)19, 20. On day 2, media was changed to CDM3 supplemented with 2 µM Wnt–C59 (Biorbyt). Media was changed on day 4 and every other day for CDM3. Contracting cells were noted from day 7. At day 10, media was changed to CDM3L made with using RPMI 1640 no glucose (11879–020, Life Technologies), 500 µg ml−1 recombinant human albumin, and 213 µg ml−1 L–ascorbic acid 2–phosphate supplemented with 4 mM L–lactic acid (Sigma–Aldrich). At day 15, media was changed to CDM3M consisting of RPMI 1640 no glucose, 500 µg ml−1 recombinant human albumin, 213 µg ml−1 L–ascorbic acid 2–phosphate supplemented with 10 mM D–galactose (Sigma–Aldrich)21, 4 mM L–lactic acid, 1 mM sodium pyruvate (Life Technologies), 20 µg ml−1 insulin (Life technologies), 1 × chemically defined lipid concentrate (Life Technologies), and 200 ng ml−1 tri–iodo–L–thyronine (Sigma–Aldrich)22.
Immunofluorescence staining for cardiomyocyte markers
Cells at differentiation day 15 were dissociated using TrypLE Express for 10 min at 37 °C, triturated, centrifuged at 200 g for 4 min, plated onto Synthemax II–SC–coated (625 ng cm−2) coverslips in CDM3 and allowed to adhere for 3–5 days. Cells were then processed as above and stained with 1:200 monoclonal mouse IgG1 TNNT2 (13–11, Thermo Scientific, MA5-12960), 1:200 polyclonal rabbit IgG ACTN2 (α–actinin, H–300, Santa Cruz Biotechnology, sc-15336), 1:50 monoclonal rabbit IgG P4HB (Abcam, ab137110), 1:400 polyclonal rabbit IgG VWF (Abcam, ab201336), 1:500 polyclonal rabbit IgG MKI67 (Ki67, Thermo Scientific, PA5-16785), and 1:200 monoclonal mouse γ–H2AX (Millipore, 05-636). Cells were washed 4 times, for 10 min, with 1% BSA in DPBS–T and then incubated for 1 h at RT in the dark with 1:400 Alexa Fluor secondary antibodies (Life Technologies) diluted in 2% BSA. Cells were washed again as above and mounted with ProLong Diamond Antifade Mountant with DAPI (Life Technologies) onto Superfrost Plus (Thermo Scientific) slides and imaged with an LSM510Meta Confocal Microscope (Zeiss).
Flow cytometry cardiac differentiation efficiency
For assessment of cardiac differentiation efficiency, cells on day 15 of differentiation were dissociated with TrypLE Express for 5 min at 37 °C and transferred to flow cytometry tubes (BD Biosciences). Cells were then fixed with 4% PFA for 10 min, permeabilized with 0.1% saponin for 20 min, and stained using 1:50 mouse monoclonal IgG1 TNNT2-647 (13–11, BD Biosciences, 565744) or 1:5 mouse monoclonal IgG2b MYH14-PE (MF20, BD Biosciences, 564408) for 30 min at RT. These conditions were shown to stain negative on human skin fibroblasts. Isotype controls Alexa Fluor 647 mouse IgG1 κ (BD Biosciences, 557714) and Alexa Fluor 488 Mouse IgG2b, κ (BD Biosciences, 555743) were used to establish gating. Cells were analyzed using a FACSAria II (BD Biosciences) with a 100 µM nozzle and FACSDiva software. Data were analyzed using FlowJo X (TreeStar).
Cardiomyocyte plating and doxorubicin treatment
Cells at differentiation day 20 were dissociated using TrypLE Express freshly supplemented with liberase TH (50:1) for 15 min at 37 °C, centrifuged at 300 g for 5 min, filtered through a 100 µM cell strainer (Partec CellTrics), counted for live and dead cells using acridine orange and propidium iodide (AO/PI) and a LUNA–FL Dual Fluorescence cell counter (Logos Biosystems) and plated onto Synthemax–II–coated coverslip chamber sides (Nunc) in 24–well cell culture (750,000 cells per well), 96–well black–sided (fluorescence) or white–sided (luminescence) plates (100,000 cells per well, all Greiner), or 24–well Seahorse plates (250,000 cells per well, Seahorse Bioscience) in CDM3 supplemented with 20% dislyzed FBS (Corning). Doxorubicin hydrochloride (HY–15142, MedChem Express) was resuspended to 10 mM in WFI water (Corning). For treatments on day 30 hiPSC-CMs, doxorubicin (0.01–10 µM) was diluted in RPMI 1640 no glucose without phenol red supplemented with 500 µg ml−1 recombinant human albumin (Sigma–Aldrich) and cells were treated for 24 h to 96 h. For dexrazoxane treatment, hiPSC-CMs were treated with 100 mM of dexrazoxane HCl (HY–76201, MedChem Express) 12 h prior to doxorubicin administration and then a second concentration was co–administered with the doxorubicin concentrations. For N–acetyl–L–cysteine, hiPSC-CMs were co–treated with 1 mM N–acetyl–L–cysteine (A9165, Sigma–Aldrich) with doxorubicin treatment.
Phase contrast imaging and contraction assays
Phase contrast images were captured using a Leica DMIR–LED microscope with a Leica DFC550 camera and Leica Application Suite 4.1 software. Contraction data were extracted using a Cellogy Pulse video microscope and image analysis platform57.
Plate–based cellular viability, caspase 3 and 7, H2O2, GSH and mitochondrial membrane potential assays
After doxorubicin treatment, PrestoBlue (Life Technologies), 10 µl was added directly to each well of the 96–well plates, incubated at 37 °C for 2 h and fluorescence was read using a Cytation 5 Cell Imaging Multi–Mode Reader (BioTek Instruments) with monochromators set to 560 nm excitation and 590 nm emission. CCK–8 (Dojindo), 10 µl was added directly to each well of the 96–well plates, incubated at 37 °C for 4 h and absorbance was read at 450 nm. CellTiter–Glo 2.0 (Promega), 100 µl was added directly to each well of the 96–well plates, incubated at RT on an orbital shaker (Thermo) for 10 min and read using fiber–optic luminescence with an integration time of 0.25 sec. For all cell viability assays, 10 µM staurosporine was used as a positive control. Caspase 3/7–Glo, ROS–Glo H2O2, and GSH–Glo (Promega) were performed following manufacturer’s instructions and read using fiber–optic luminescence with an integration time of 1 sec. Lactate dehydrogenase (LDH) (Clonetech) and ToxiLight (adenylate kinase) (Lonza) were performed following manufacturer’s instructions. JC–10 (Enzo Life Sciences) was diluted to 2 µM in RPMI 1640 and stained at 37 °C for 30 min, cells were washed and read with monochromators set to 490 nm excitation and 520 nm (green) or 590 nm (red) emission. 20 µM FCCP was used a positive control. Data were analyzed using Prism 6.0f (GraphPad) using standard concentration–response guidelines.
Flow cytometry–based apoptosis, reactive oxygen species, mitochondrial membrane potential, and DNA damage assays
After doxorubicin treatment, cells were dissociated with TrypLE Express and stained in suspension. For apoptosis detection, cells were stained with 7–AAD (7–aminoactinomycin D) and Annexin V (both BD Biosciences) following the manufacturer’s instructions. 6 µM of camptothecin (Sigma–Aldrich) was used as a positive control to set the flow cytometry gates. For DNA damage detection, cells were stained with Anti–phospho–histone γ–H2AX (Millipore) and propidium iodide (Life Technologies) following standard protocols58. For whole cell ROS detection, CellROX Green (Life Technologies) was used at a 1 µM concentration, incubating for 30 min, excluding dead cells using SYTOX Red and using 20 µM menadione (Sigma–Aldrich) as a positive control. For mitochondrial superoxide detection, MitoSOX Red (Life Technologies) was used at a 5 µM concentration, incubating for 30 min, excluding dead cells using SYTOX Green and using 20 µM antimycin A (Sigma–Aldrich) as a positive control59. Cells were analyzed by flow cytometry on a BD Biosciences FACS Aria II using FACSDiva software. Data analysis was performed using FlowJo X (TreeStar).
RNA–seq gene expression analysis
hiPSC–CMs were treated with doxorubicin for 24 h. Cells were dissociated with TrypLE Express and Liberase TH, centrifuged, and cell pellets were flash frozen in liquid nitrogen and stored at −80 °C then shipped to Macrogen who performed RNA extraction, cDNA synthesis, library preparation and sequencing on a HiSeq 2000 (Illumina) at 40 million reads (20 million in each direction). The probe level data was preprocessed with the Robust Multiarray Analysis (RMA) algorithm60, quantile normalized61, and summarized to the gene level.
RNA–seq gene expression analysis and coding sequence variant calling
RNA–seq analyses were performed on the hiPSC–CMs from 6 patients (3 DOX without cardiotoxicity responses and 3 DOXTOX with cardiotoxicity responses), each treated with 0 or 1 µM doxorubicin. Reads were mapped to human genome (hg19) using tophat2 (version v2.0.13)62, assembled into transcripts using cufflinks (version 2.2.1) and cuffmerge with known transcripts as guides63, and quantified using cuffdiff (version 2.2.1). Only transcripts mapped to unique genes were retained. The read–counts of transcripts were aggregated to the gene level by summation. Gene expression were then log2 transformed with pseudo–count of 1, and quantile normalized61. Only known protein coding genes were retained for subsequent analysis. Gene variants calling from the RNA–seq data are performed using samtools (version 1.2) and bcftools (version 1.2)64 and annotated with Genome Analysis Toolkit (version v3.4–46)14. The mutation effects of the variants are predicted using snpEff65. The variant calling results are visualized and inspected by using the Integrative Genome Viewer66.
Independent component analysis (ICA)
ICA is an unsupervised machine learning method for solving the blind source separation problem67. Given a g×s matrix X for the log–transformed expression values of g genes in s samples, ICA estimates a g×m source matrix S for m independent gene modules, and an m× s matrix A of mixing coefficients of the m modules in the s samples, such that X≈S·A. ICA has been shown to identify more functionally coherent gene clusters than previous methods such as K–mean and PCA37, 68. We applied ICA to derive the statistically independent gene modules (pathways) in the doxorubicin drug treatment data. Gene ontology enrichment analysis was performed using Fisher’s exact test algorithm69. To compare the patient–response study (RNA–seq analysis of DOX vs. DOXTOX) and the concentration–response study (microarray analysis of doxorubicin 0–10 µM), we projected patient–response data Y onto the ICA modules S derived from the concentration–response data X to obtain the estimated expression of these modules, i.e., AY = St · Y, where St is the transpose of S.
Transcription factor activity inference
To identify the transcription factors involved in a biological process, we applied Fisher’s exact test to evaluate if the target genes of each transcription factor are significantly over–represented in the up– or down–regulated genes. We defined the up–regulated, down–regulated, and unchanged genes for each of the m gene modules as those with standard deviation–normalized expression values z ≥ q0.95, ≤ z ≥ q0.95, and −q < z < q0.95, where q0.95, is the 95% quantile of standard normal distribution. The transcription factor target gene database was compiled from literature reported regulatory relationships from low–throughput experiments as well as ENCODE70 and additional ChIP–seq data retrieved from Gene Expression Omnibus71. Two–tailed Fisher’s exact tests were performed. For the transcriptional regulatory network visualization, we used a linear model per gene module to estimate the activity values of each TF in our transcription factor target gene database. The P–values of TF activities are obtained from Wald test of the estimated activities, and q–values were then estimated from the P–values.
Estimation of gene expression changes associated with patient–specific doxorubicin toxicity responses
Body mass index (BMI) is known to affect doxorubicin dosing and responses. To reduce the effect from BMI, we required it to be in the range of 25 to 34 when selecting patients. In addition, we included BMI as a control variable in the linear model when estimating patient specific doxorubicin responses. Specifically, to extract the differential response between DOX and DOXTOX patients upon doxorubicin treatment, the following linear model for observed gene expression level xid for patient i (1–6) and doxorubicin concentrations d (0, 1uM) is applied to each gene:
where bi and ti are the BMI and response type (DOX or DOXTOX), respectively, βb, βt, and βd are the effects of BMI, patient type, and doxorubicin treatment on the gene expression, whereas βbd and βtd are the interaction effects of BMI and doxorubicin treatment, and patient response type and doxorubicin treatment, respectively. We did not strictly control for age in patient selection, and there is a spurious inverse correlation between patient age and toxicity response in our dataset (R = −0.81, P–value = 0.052). However, as we found that including age as control variable did not change the major findings, we present the results based on the model that does not include age variable. In addition, age is not a known risk factor for doxorubicin cardiotoxicity. Despite these, it is possible that the identified patient specific drug responses and regulatory mechanisms could be partly due to unknown age differences between the patients.
Seahorse extracellular metabolic flux assay
hiPSC–CMs were plated at 250,000 cells per well in Synthemax–II–coated Seahorse 24–well plates in CDM3M. The bioenergetics response of hiPSC–CMs was measured with the Seahorse Bioscience XF24 Flux Analyzer following directions in the XF Cell Mito Stress Test Kit User Guide. Media was aspirated and replaced with 525 µL assay medium (Seahorse Bioscience) and pre–equilibrated for 1 h at 37 °C. Baseline OCR measurements were performed followed by injection of 1 µM of oligomycin with three OCR measurements, then injection of 0.5 µM FCCP with three OCR measurements, and finally injection of 0.5 µM of rotenone and antimycin A followed by three OCR measurements. Four hiPSC lines were used for each sample group and contained 3 technical replicates. The 0 µM well from each set of samples was stained with NucBlu Live Ready Probes reagent and number of cells per well were counted with a high–content imager (Biotek Cytation 5) to confirm plating homogeneity. (confirm dose is correct)
Mitochondrial copy number quantification
Cells were dissociated with TrypLE Express and pellets flash frozen in liquid nitrogen. Genomic DNA (gDNA) extraction was performed using DNeasy Blood and Tissue Kit (QIAGEN), according to manufacturer’s instructions, with RNase treatment. Following determination of gDNA concentration using a UV–Vis spectrophotometer, all samples were adjusted to have equal starting concentrations. The mitochondrial DNA (mtDNA) content of cells is expressed as a relative mtDNA : nuclear DNA (nDNA) ratio with the nDNA copy number per cell being considered constant and alteration of the ratio is attributed to changes in mtDNA content. Real–time amplification for a nuclear gene Succinate dehydrogenase complex, subunit A, flavoprotein variant (SDHA), and a mitochondrial gene NADH dehydrogenase subunit I (ND1) was performed using an ABI Prism 7000 Sequence Detection System.
Total oxidative phosphorylation protein analysis
Western blots were generated using standard Bio–Rad wet–blot protocols and antibodies for Total OXPHOS Antibody Cocktail (Abcam, ab110413), containing an optimized premixed cocktail 5 mAbs, one each against: CI subunit NDUFB8 (20 kDa, a subunit of NADH dehydrogenase, ab110242), CII subunit SDHB– (30 kDa, the iron–sulfur protein (IP) subunit of succinate dehydrogenase (SDH), ab14714), CIII–subunit UQCRC2 (48 kDA, Core protein 2 – a component of the ubiquinol–cytochrome c reductase complex, ab14745), CIV subunit MTCO1 (40 kDa, a subunit of cytochrome c oxidase, ab14705), and CV ATP5A (55 kDa, a subunit of ATP synthase, ab14748). α–tubilin (ab7291) was used as a loading control.
Calcium imaging
Dissociated hiPSC–CMs were reseeded in Matrigel–coated eight–well Lab Tek II coverglass chambers (Thermo Scientific Nunc) and were treated with 5 µM Fluo–4 AM (Life Technologies) and 0.02% Pluronic F–127 (Life Technologies) in Tyrode’s solution [140 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM glucose, 1.8 mM CaCl2, 10 mM HEPES, pH 7.4 with NaOH at 25 °C] for 15 min at 37 °C. Cells were washed with Tyrode’s solution afterwards. Ca2+ imaging was conducted using a Zeiss LSM 510Meta confocal microscope (Carl Zeiss AG) with a 63× objective and analyzed using Zen imaging software. Spontaneous Ca2+ transients were obtained at 37 °C using a single–cell line scan mode.
Statistical methods
Data were analyzed in Excel or R and graphed in Prism (GraphPad). Data are presented as mean ± SEM. Comparisons were conducted via Fisher’s exact test, one way–ANOVA test followed by All Pairwise Multiple Comparison Procedures (Holm–Sidak method), or an unpaired, two–tailed Student’s t–test with significant differences defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***). No blinding or randomization was used. Our sample size (4 patients in each category) was based on the feasibility of handling of 12 hiPSC lines. Patient exclusion criterial are included in Supplementary Table 1.
Supplementary Material
Acknowledgments
We thank J. Odegaard for analysis of teratoma slides. This work was supported by the National Institutes of Health (NIH) K99/R00 HL121177, American Heart Association (AHA) Beginning 14BGIA20480329, Dixon Translational Research Grant Young Investigator Award (P.W.B.), AHA 13POST14480004 (A.C.C.), Muscular Dystrophy Association grant 4320 (H.M.B.), NIH R21 HL123655 (D.B.), NIH LM05652 GM102365 and GM61374 (R.B.A.), and AHA 13EIA14420025, Burroughs Wellcome Fund, NIH R01 HL123968, NIH R01 HL126527, NIH R01 HL128170, and NIH R01 HL130020 (J.C.W).
Footnotes
Accession codes: RNA–seq data have been deposited in the Gene Expression Omnibus (GEO) database, accession code GSE76314.
Author contributions: P.W.B. performed project planning, experimental design, iPSC reprogramming, characterization, differentiation, cardiotoxicity analysis, flow cytometry, data analysis, and wrote the manuscript; Y.F.L. performed computational analyses of microarray and RNA–seq data, gene enrichment analysis, and wrote part of the manuscript; E.M. and A.H. performed cell culture. E.M. and A.S. performed immunohistochemistry; H.W. performed Ca2+ imaging; P.W.B., S.–G.O., A.C.C., M.J.C., and A.D.E. performed Seahorse analysis; S.–G.O. performed mitochondrial analysis. J.W.K., M.L.T., and R.M.W. performed patient recruitment; H.M.B., D.B., R.B.A., and J.C.W. aided in manuscript preparation; J.C.W. and P.W.B. provided project planning, conceptual design of study, and funding support.
Competing financial interests: P.W.B. is a shareholder in Stem Cell Theranostics. J.C.W. is a founder and shareholder in Stem Cell Theranostics.
REFERENCES
- 1.Lipshultz SE, Franco VI, Miller TL, Colan SD, Sallan SE. Cardiovascular disease in adult survivors of childhood cancer. Annu. Rev. Med. 2015;66:161–176. doi: 10.1146/annurev-med-070213-054849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giordano SH, Lin Y-L, Kuo YF, Hortobagyi GN, Goodwin JS. Decline in the use of anthracyclines for breast cancer. J. Clin. Oncol. 2012;30:2232–2239. doi: 10.1200/JCO.2011.40.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefrak EA, Pitha J, Rosenheim S, Gottlieb JA. A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer. 1973;32:302–314. doi: 10.1002/1097-0142(197308)32:2<302::aid-cncr2820320205>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Hoff, Von DD, et al. Risk factors for doxorubicin-induced congestive heart failure. Ann. Intern. Med. 1979;91:710–717. doi: 10.7326/0003-4819-91-5-710. [DOI] [PubMed] [Google Scholar]
- 5.Swain SM, Whaley FS, Ewer MS. Congestive heart failure in patients treated with doxorubicin: a retrospective analysis of three trials. Cancer. 2003;97:2869–2879. doi: 10.1002/cncr.11407. [DOI] [PubMed] [Google Scholar]
- 6.Kremer LCM, van der Pal HJH, Offringa M, van Dalen EC, Voûte PA. Frequency and risk factors of subclinical cardiotoxicity after anthracycline therapy in children: a systematic review. Ann. Oncol. 2002;13:819–829. doi: 10.1093/annonc/mdf167. [DOI] [PubMed] [Google Scholar]
- 7.Shakir DK, Rasul KI. Chemotherapy induced cardiomyopathy: pathogenesis, monitoring and management. J. Clin. Med. Res. 2009;1:8–12. doi: 10.4021/jocmr2009.02.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein D, Burridge P. Patient-specific pluripotent stem cells in doxorubicin cardiotoxicity: a new window into personalized medicine. Prog. Pediatr. Cardiol. 2014;37:23–27. doi: 10.1016/j.ppedcard.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granger CB. Prediction and prevention of chemotherapy-induced cardiomyopathy: can it be done? Circulation. 2006;114:2432–2433. doi: 10.1161/CIRCULATIONAHA.106.666248. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S, et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 11.Khiati S, et al. Mitochondrial topoisomerase I (top1mt) is a novel limiting factor of doxorubicin cardiotoxicity. Clin. Cancer Res. 2014;20:4873–4881. doi: 10.1158/1078-0432.CCR-13-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna AD, Lam A, Tham S, Dulhunty AF, Beard NA. Adverse effects of doxorubicin and its metabolic product on cardiac RyR2 and SERCA2A. Mol. Pharmacol. 2014;86:438–449. doi: 10.1124/mol.114.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goormaghtigh E, Brasseur R, Huart P, Ruysschaert JM. Study of the adriamycin-cardiolipin complex structure using attenuated total reflection infrared spectroscopy. Biochemistry. 1987;26:1789–1794. doi: 10.1021/bi00380a043. [DOI] [PubMed] [Google Scholar]
- 14.Ichikawa Y, et al. Cardiotoxicity of doxorubicin is mediated through mitochondrial iron accumulation. J. Clin. Invest. 2014;124:617–630. doi: 10.1172/JCI72931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmberg SR, Williams AJ. Patterns of interaction between anthraquinone drugs and the calcium-release channel from cardiac sarcoplasmic reticulum. Circ. Res. 1990;67:272–283. doi: 10.1161/01.res.67.2.272. [DOI] [PubMed] [Google Scholar]
- 16.Burridge PW, et al. Modeling cardiovascular diseases with patient-specific human pluripotent stem cell-derived cardiomyocytes. Methods Mol. Biol. 2015;1353:1–12. doi: 10.1007/7651_2015_196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson SE, Loring JF. Genomic instability in pluripotent stem cells: implications for clinical applications. J. Biol. Chem. 2014;289:4578–4584. doi: 10.1074/jbc.R113.516419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burridge PW, et al. Chemically defined generation of human cardiomyocytes. Nat. Meth. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burridge PW, Holmström A, Wu JC. Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Curr. Protoc. Hum. Genet. 2015;87:21.3.1–21.3.15. doi: 10.1002/0471142905.hg2103s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rana P, Anson B, Engle S, Will Y. Characterization of human-induced pluripotent stem cell-derived cardiomyocytes: bioenergetics and utilization in safety screening. Toxicol. Sci. 2012;130:117–131. doi: 10.1093/toxsci/kfs233. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J. Mol. Cell Cardiol. 2014;72:296–304. doi: 10.1016/j.yjmcc.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert J, et al. Comparative pharmacokinetics and metabolism of doxorubicin and epirubicin in patients with metastatic breast cancer. Cancer Treat Rep. 1985;69:633–640. [PubMed] [Google Scholar]
- 24.Bramwell VHC, et al. Safety and efficacy of the multidrug-resistance inhibitor biricodar (VX-710) with concurrent doxorubicin in patients with anthracycline-resistant advanced soft tissue sarcoma. Clin. Cancer Res. 2002;8:383–393. [PubMed] [Google Scholar]
- 25.Berdichevski A, et al. TVP1022 protects neonatal rat ventricular myocytes against doxorubicin-induced functional derangements. J. Pharmacol. Exp. Ther. 2010;332:413–420. doi: 10.1124/jpet.109.161158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito H, et al. Doxorubicin selectively inhibits muscle gene expression in cardiac muscle cells in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4275–4279. doi: 10.1073/pnas.87.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruan Y, et al. SIRT1 suppresses doxorubicin-induced cardiotoxicity by regulating the oxidative stress and p38MAPK pathways. Cell Physiol. Biochem. 2015;35:1116–1124. doi: 10.1159/000373937. [DOI] [PubMed] [Google Scholar]
- 28.Lim CC, et al. Anthracyclines induce calpain-dependent titin proteolysis and necrosis in cardiomyocytes. J. Biol. Chem. 2004;279:8290–8299. doi: 10.1074/jbc.M308033200. [DOI] [PubMed] [Google Scholar]
- 29.Chen B, et al. Disruption of a GATA4/Ankrd1 signaling axis in cardiomyocytes leads to sarcomere disarray: implications for anthracycline cardiomyopathy. PLoS One. 2012;7:e35743. doi: 10.1371/journal.pone.0035743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doroshow JH. Effect of anthracycline antibiotics on oxygen radical formation in rat heart. Cancer Res. 1983;43:460–472. [PubMed] [Google Scholar]
- 31.Shi Y, Moon M, Dawood S, McManus B, Liu PP. Mechanisms and management of doxorubicin cardiotoxicity. Herz. 2011;36:296–305. doi: 10.1007/s00059-011-3470-3. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Persson HL, Richardson DR. Molecular pharmacology of the interaction of anthracyclines with iron. Mol. Pharmacol. 2005;68:261–271. doi: 10.1124/mol.105.013383. [DOI] [PubMed] [Google Scholar]
- 33.Venditti P. Free radical involvement in doxorubicin-induced electrophysiological alterations in rat papillary muscle fibres. Cardiovasc. Res. 1998;38:695–702. doi: 10.1016/s0008-6363(98)00034-0. [DOI] [PubMed] [Google Scholar]
- 34.Swain SM, et al. Cardioprotection with dexrazoxane for doxorubicin-containing therapy in advanced breast cancer. J. Clin. Oncol. 1997;15:1318–1332. doi: 10.1200/JCO.1997.15.4.1318. [DOI] [PubMed] [Google Scholar]
- 35.Deng S, et al. The catalytic topoisomerase II inhibitor dexrazoxane induces DNA breaks, ATF3 and the DNA damage response in cancer cells. Br. J. Pharmacol. 2015;172:2246–2257. doi: 10.1111/bph.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farshid AA, et al. Effects of histidine and N-acetylcysteine on doxorubicin-induced cardiomyopathy in rats. Cardiovasc. Toxicol. 2014;14:153–161. doi: 10.1007/s12012-013-9239-6. [DOI] [PubMed] [Google Scholar]
- 37.Engreitz JM, Daigle BJ, Marshall JJ, Altman RB. Independent component analysis: mining microarray data for fundamental human gene expression modules. J. Biomed. Inform. 2010;43:932–944. doi: 10.1016/j.jbi.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorn CF, et al. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet. Genomics. 2011;21:440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hussner J, et al. Regulation of interferon-inducible proteins by doxorubicin via interferon γ-Janus tyrosine kinase-signal transducer and activator of transcription signaling in tumor cells. Mol. Pharmacol. 2012;81:679–688. doi: 10.1124/mol.111.075994. [DOI] [PubMed] [Google Scholar]
- 40.Zhu W, Zhang W, Shou W, Field LJ. P53 inhibition exacerbates late-stage anthracycline cardiotoxicity. Cardiovasc. Res. 2014;103:81–89. doi: 10.1093/cvr/cvu118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doroshow JH, Locker GY, Myers CE. Enzymatic defenses of the mouse heart against reactive oxygen metabolites: alterations produced by doxorubicin. J. Clin. Invest. 1980;65:128–135. doi: 10.1172/JCI109642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torti SV, Akimoto H, Lin K, Billingham ME, Torti FM. Selective inhibition of muscle gene expression by oxidative stress in cardiac cells. J. Mol. Cell Cardiol. 1998;30:1173–1180. doi: 10.1006/jmcc.1998.0681. [DOI] [PubMed] [Google Scholar]
- 43.Naidu SR, Love IM, Imbalzano AN, Grossman SR, Androphy EJ. The SWI/SNF chromatin remodeling subunit BRG1 is a critical regulator of p53 necessary for proliferation of malignant cells. Oncogene. 2009;28:2492–2501. doi: 10.1038/onc.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visscher H, et al. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr. Blood Cancer. 2013;60:1375–1381. doi: 10.1002/pbc.24505. [DOI] [PubMed] [Google Scholar]
- 45.Lebrecht D, Kokkori A, Ketelsen U-P, Setzer B, Walker UA. Tissue-specific mtDNA lesions and radical-associated mitochondrial dysfunction in human hearts exposed to doxorubicin. J. Pathol. 2005;207:436–444. doi: 10.1002/path.1863. [DOI] [PubMed] [Google Scholar]
- 46.Zhu R, et al. Physical developmental cues for the maturation of human pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther. 2014;5:117. doi: 10.1186/scrt507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.David R, Franz WM. From Pluripotency to Distinct Cardiomyocyte Subtypes. Physiology. 2012;27:119–129. doi: 10.1152/physiol.00044.2011. [DOI] [PubMed] [Google Scholar]
- 48.Mathur A, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aminkeng F, et al. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat. Genet. 2015;47:1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HD, Kim CH, Rah BJ, Chung HI, Shim TS. Quantitative study on the relation between structural and functional properties of the hearts from three different mammals. Anat. Rec. 1994;238:199–206. doi: 10.1002/ar.1092380206. [DOI] [PubMed] [Google Scholar]
- 51.Hattori F, et al. Nongenetic method for purifying stem cell–derived cardiomyocytes. Nat. Methods. 2009;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
ONLINE REFERENCES
- 52.Melkoumian Z, et al. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat. Biotech. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 53.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad., Ser. B. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mali P, et al. Butyrate Greatly Enhances Derivation of Human Induced Pluripotent Stem Cells by Promoting Epigenetic Remodeling and the Expression of Pluripotency-Associated Genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomson JA. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 57.Maddah M, et al. A non-invasive platform for functional characterization of stem-cell-derived cardiomyocytes with applications in cardiotoxicity testing. Stem Cell Reports. 2015;4:621–631. doi: 10.1016/j.stemcr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang X, Darzynkiewicz Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol. Biol. 2006;314:73–80. doi: 10.1385/1-59259-973-7:073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukhopadhyay P, et al. Simultaneous detection of apoptosis and mitochondrial superoxide production in live cells by flow cytometry and confocal microscopy. Nat. Protoc. 2007;2:2295–2301. doi: 10.1038/nprot.2007.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 61.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 62.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hyvärinen A, Oja E. Independent component analysis: algorithms and applications. Neural Netw. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 68.Lee S-I, Batzoglou S. Application of independent component analysis to microarrays. Genome Biol. 2003;4:R76. doi: 10.1186/gb-2003-4-11-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 70.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.