Summary
Viruses in the Reoviridae, like the triple-shelled human rotavirus and the single-shelled insect cytoplasmic polyhedrosis virus (CPV), all package a genome of segmented dsRNAs inside the viral capsid and carry out endogenous mRNA synthesis through a transcriptional enzyme complex (TEC). By direct electron-counting cryoEM and asymmetric reconstruction, we have determined the organization of the dsRNA genome inside quiescent CPV (q-CPV) and the in situ atomic structures of TEC within CPV in both quiescent and transcribing (t-CPV) states. We show that the total 10 segmented dsRNAs in CPV are organized with 10 TECs in a specific, non-symmetric manner, with each dsRNA segment attached directly to a TEC. TEC consists of two extensively-interacting subunits: an RNA-dependent RNA polymerase (RdRP) and an NTPase VP4. We find that the bracelet domain of RdRP undergoes significant conformational change when converted from q-CPV to t-CPV, leading to formation of the RNA template entry channel and access to the polymerase active site. An N-terminal helix from each of two subunits of the capsid shell protein (CSP) interacts with VP4 and RdRP. These findings establish the link between sensing of environmental cues by the external proteins and activation of endogenous RNA transcription by the TEC inside the virus.
Each capsid of viruses in the Reoviridae contains 9-12 segmented dsRNAs and up to 12 transcriptional enzyme complexes (TECs). These RNA-containing viruses are fully capable of RNA transcribing and capping1. Crystal structures of the RNA-dependent RNA polymerase (RdRP) component of the TEC have been determined for rotavirus and mammalian reovirus (MRV)2,3, but no high-resolution in situ structure of the TEC is available. Moreover, the organization of TECs with the dsRNA genome and the mechanism of transcriptional activation have remained mysteries, in contrast to the well understood genome organization inside dsDNA viruses4,5.
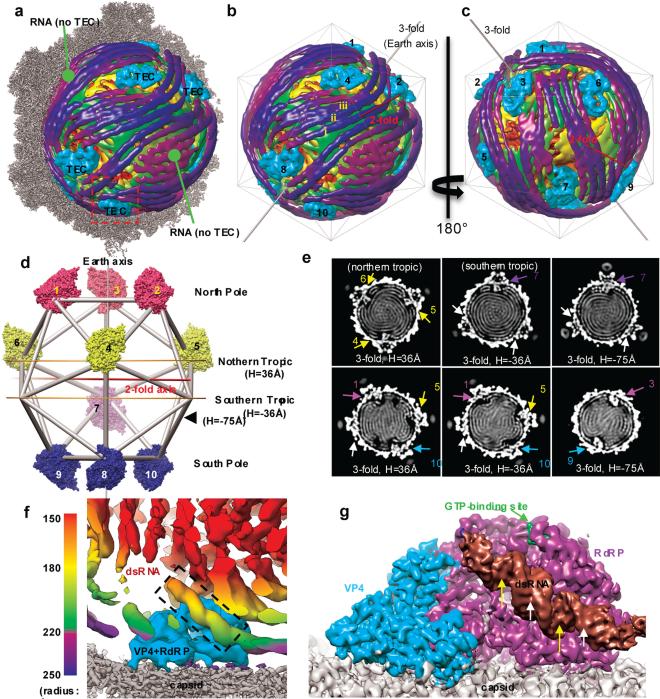
With only a single protein shell that encloses 10 different genome segments, CPV is one of the simplest dsRNA viruses6 and serves as a model system, as highlighted by its contribution to the discovery of RNA capping7. To gain insight into the organization of the TEC and segmented dsRNA genome, we have determined CPV structure in a quiescent (q-CPV) state at 5.1Å resolution (see Methods, Extended Data Figs. 1,2). The structure reveals that each CPV contains 10 TECs under 10 specific positions of the 12 icosahedral vertices (Fig. 1). The two vertices without TECs are occupied by rod-like densities (Fig. 1a-e, Supplementary Video 1, Extended Data Figs. 3-4). The previously ambiguous locations of TECs8,9 are now determined to be 10 specific positions in each CPV particle, related by pseudo-D3 symmetry, with only one on a “south tropic” position and three each around the “north tropic”, “north pole” and “south pole” positions (Fig. 1d, Supplementary Video 2).
Figure 1. Transcription enzyme complex (TEC) and dsRNA genome organization inside CPV.
a, Superposition of the high-resolution (3.9 Å) map of half a capsid (grey) and low-resolution (22 Å) map of dsRNA genome (radially colored as in f) and TECs (cyan). b-c, Front (b) and back (c) views of the dsRNA genome and TECs of (a). d, Earth-like representation, illustrating the locations of the ten TECs (surface-rendered) with pseudo-D3 symmetry: three on each pole and the northern tropic but only one on the southern tropic. e, Cross sections of the 22 Å-density map, perpendicular to either the “earth axis” in (d) (top row) or a pseudo-D3 2-fold axis (bottom row). Densities of TECs are numbered as in (d); and the two vertices without TEC but with RNA are indicated by white arrows. f, Boxed region in (a) containing RNA threads (radially colored as in the bar) and a TEC (cyan) with bound dsRNA (dashed box). g, Averaged TEC region, filtered to 4.5 Å and viewed as the southern-most TEC of (a). The RdRP-bound dsRNA has the same structure in all TECs and shows major (yellow arrows) and minor (white arrows) grooves.
Each TEC is surrounded by rod-like densities with lengths up to ~650Å (Fig. 1a-c, e-f, Extended Data Fig. 4a, Supplementary Video 1). In most regions, these rods form parallel striations with an inter-rod distance of ~27Å as suggested early (e.g., 10,11). Some of the rods exhibit the characteristic minor and major grooves typical of dsRNA duplex (Fig. 1g). We therefore interpret these rod-like densities as dsRNA duplexes. Unlike the model of each genome segment spiraling around one TEC (Ref 12), the duplexes do not spiral locally around TECs (Fig. 1a-c, Extended Data Fig. 4 and Supplementary Video 1); instead, many extend tangentially from one TEC to another (e.g., duplexes i-iii in Fig. 1b; Extended Data Fig. 4), indicating that each dsRNA segment is organized beyond one TEC. Indeed, the whole RNA genome is organized into 7 to 8 non-concentric layers with visible connections between adjacent layers (Fig. 1e and Extended Data Fig. 3). This extended organization of dsRNA is consistent with the rather long (~620 Å) persistence length of dsRNA13 and would reduce the energy needed for genome packaging and transcription. One RNA duplex (the brown one in Fig. 1g) binds to each of the 10 TECs at the same relative position and orientation, suggesting that this RNA duplex is a conserved feature among the 10 dsRNA segments. However, the organization of the remaining RNA duplex differs among the 10 TECs (Extended Data Fig. 4g). The two vertices without TECs are occupied only by roughly parallel dsRNA densities (Fig. 1a-c).
We also obtained a 3.9Å resolution asymmetric reconstruction directly from the raw images of q-CPV and subsequently used non-crystallographic averaging to improve the resolution to 3.3Å for the TEC-containing regions (see Methods and Extended Data Fig. 5). The averaged map retains a short (~35Å) RdRP-bound dsRNA density (Fig. 1g) and resolves the two protein components of the TEC: VP4 and RdRP (Fig. 2a). We built a backbone model of the RdRP-bound dsRNA and de novo atomic models of both VP4 and RdRP (Fig. 2c-f, Extended Data Fig. 6, Supplementary Videos 3-8). VP4 and RdRP interact extensively (Fig. 2a) with a buried interface area of ~2800Å2.
Figure 2. Averaged TEC map at 3.3Å resolution and de novo modelling of VP4.
a, Averaged map of the TEC region showing VP4 (cyan) and RdRP (purple), both anchored to the inner surface of the capsid (grey). b, Atomic model of VP4. c-f, The boxed regions in (b), showing density (meshes) superposed with atomic models of the GTP-binding site (c), a loop (d), a helix (e) and an RdRP-interacting loop (f).
VP4 appears “L”-shaped and consists of an N-terminal (aa1-252) and a C-terminal (aa 253-561) domain, with two unresolved/flexible segments (aa 23-40 and 86-131) (Fig. 2a,b, Supplementary Video 3). The N-terminal domain is formed by two small β-sheets and several α-helices and the main body of the C-terminal domain is a Walker-A α/β motif, a well-known NTP-binding motif found in the P-loop kinase family of proteins. Sequence analysis predicted an NTP binding site in VP4 (Refs 14,15). Indeed, the VP4 structure contains a GTP molecule at the predicted NTP binding site of the C-terminal domain (Fig. 2c, Supplementary Video 4). We thus rename the C-terminal domain as the NTPase domain (Fig. 2b,c). A similar fold was also observed in the N-terminal α/β domain of bluetongue virus VP4. But, remarkably, bluetongue virus VP4 is an RNA capping enzyme and its α/β domain does not bind GTP (Ref 16). CPV VP4 and its homologs in other dsRNA viruses have been speculated to function as an NTPase, as an RNA 5′-triphosphatase (RTPase) or as a helicase14,17,18. Our structure supports VP4 as an NTPase but shows no interaction with dsRNA, suggesting that VP4 is unlikely a helicase. Whether VP4 is the CPV RTPase or an RdRP regulatory factor remains to be determined.
Like other RdRP structures2,3,19, the CPV RdRP contains a polymerase core with finger (aa 349-515, 549-641), thumb (aa 730-863) and palm (aa 516-548, 642-729) subdomains (Fig. 3a). This polymerase core is sandwiched between the N-terminal (aa 1-348) and C-terminal bracelet (aa 864-1225) domains (Fig. 3 and Extended Data Fig. 6). A GTP is identified (Figs 1g, 3a) at the position equivalent to the cap-binding site observed in the MRV RdRP2. Interestingly, the bracelet domain of q-CPV RdRP differs from that of MRV significantly, despite close similarities between both their polymerase core and their N-terminal domains. Consequently, the crystal structures of MRV RdRP has an open RNA template entry channel and an accessible polymerase active site2; while in the q-CPV RdRP, the polymerase active site is covered by the bracelet domain and there is no recognizable channel for template entry (Figs 3a,4a, Supplementary Videos 5,8). Since q-CPV is incapable of mRNA transcription, we considered that these structural differences might be characteristic of conformational differences between bracelet-containing RdRPs in the quiescent and transcribing states.
Figure 3. Comparison of RdRP in quiescent and transcribing states.
a-b, Ribbon models of RdRP in quiescent (a) and transcribing (b) states. The latter contains fragments of RNA template (orange) and nascent mRNA (cyan) inside the active site (box). c-f, Superpositions of RdRP structures in quiescent (colour) and transcribing (grey) states shown in full (c) and as separate domains – N-terminal (d), polymerase (e), and bracelet (f) with Modules A (yellow) and B (magenta) further highlighted on its right panel. g-i, Densities (grey) and models (ribbons and sticks) of nucleic acids in the active site of RdRP. The fragments of the (−)RNA template and the nascent mRNA in the active site are modelled as a poly-G and poly-C, respectively. In (h), a CTP is placed in the NTP-binding site and in (i), the template and mRNA form RNA duplex in the active site of RdRP (surface-rendered model).
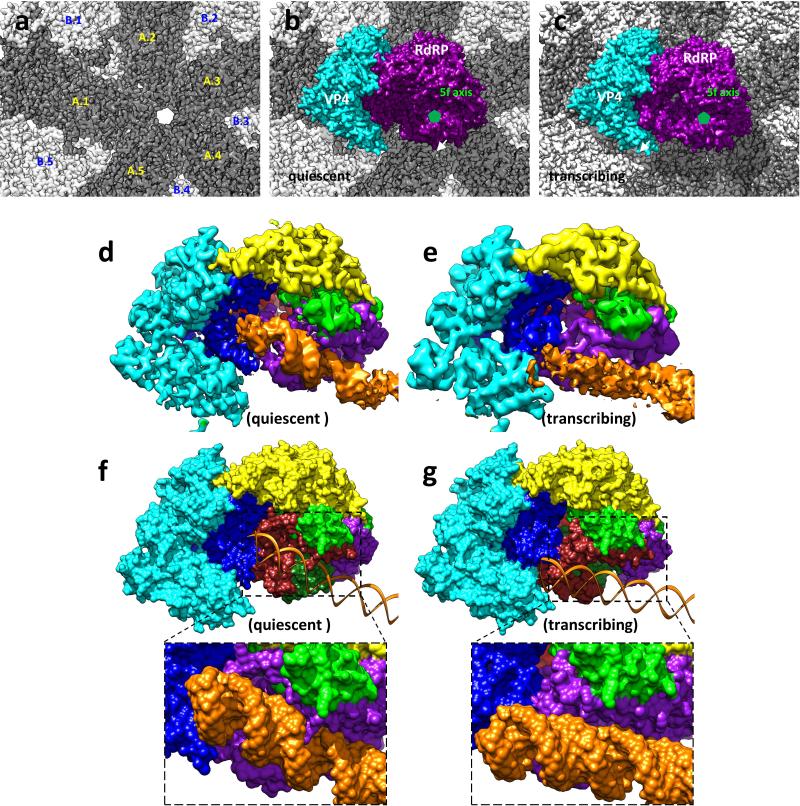
Figure 4. Interactions between TEC and capsid shell proteins (CSPs).
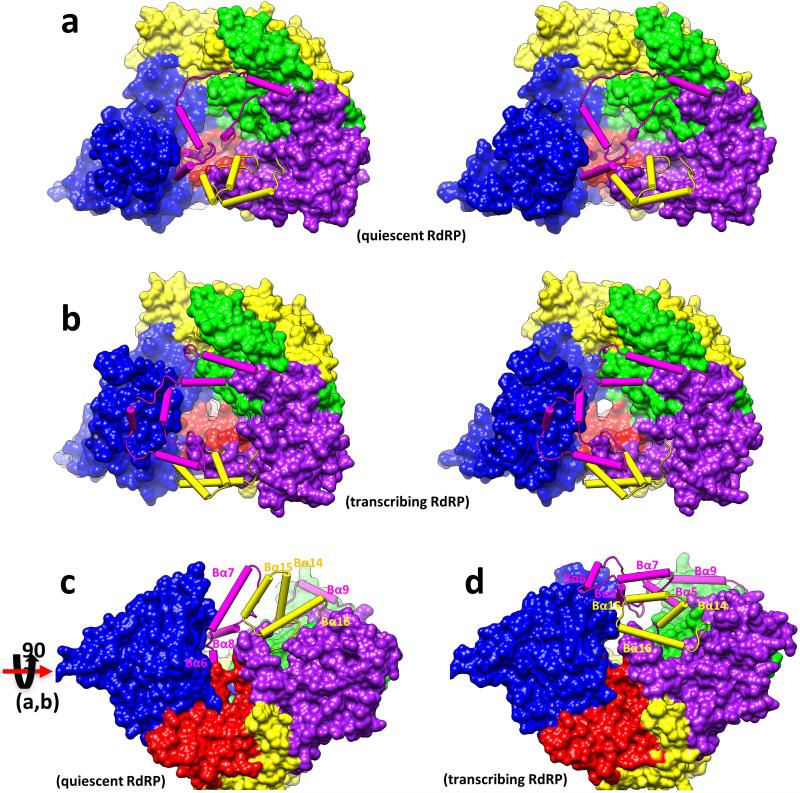
a-b, Conformational changes of modules A (yellow loops/helices as wires/cylinders) and B (magenta loops/helices as wires/cylinders) in quiescent (a) and transcribing (b) states. Module A interacts with the capsid shell, and the loop-Bα5 fragment of module B blocks the active site (inset) in the quiescent state (a) but retracts to expose the active site in the transcribing (b) state (see Extended Data Figure 7). c-d, The RdRP-bound dsRNA (ribbon) in the quiescent state (c) is detached from RdRP in the transcribing state (d). e-f, Interactions of CSPs (ribbons) with RdRP (purple and yellow) and VP4 (cyan). Residues of RdRP and VP4 within 4Å distance to the capsid shell are marked in red. An icosahedral 5-fold axis is indicated by a green line in (e) and a green pentagon in (f). Insets in (f) indicate two CSP N-terminal helices (white density with ribbon-and-stick models): one (upper) interacts only with RdRP while the other (lower) with both RdRP and VP4.
To test this hypothesis, we then determined the structure of actively transcribing CPV (t-CPV), obtained an averaged TEC map at 4.0 Å resolution, and built atomic models of VP4 and RdRP (Fig. 3b, Extended Data Figs 5, Supplementary Video 9). In t-CPV, the location of TECs remains the same, as do the structures of VP4 and those of the N-terminal and polymerase core domains of RdRP (Fig. 3a-f, Extended Data Figs 7-9, Supplementary Video 10). By contrast, the RdRP bracelet domain undergoes major conformational change (Fig. 3d,e). Consistent with the above hypothesis, the in situ structure of the t-CPV RdRP is quite similar to the crystal structure of MRV RdRP in its elongation state2 (Extended Data Fig. 10).
The most significant changes of the CPV RdRP between quiescent and transcribing states involve two neighboring structural modules in the bracelet domain, the capsid-proximal Module A (aa 1080-1140 containing helices Bα14-Bα16) and the VP4-proximal Module B (aa 912-1010 containing helices Bα5-Bα9) (Fig.4a-b, Extended Data Figs 9,10). Compared to that in q-CPV, Module A in t-CPV rotates ~40° towards the capsid shell (Fig. 3f, Extended Data Figs 9, 10f-k). Consistent with previous icosahedral reconstructions, our asymmetric reconstructions show that the capsid shell of t-CPV expands outwards from q-CPV, with the maximal (~10 Å) expansion occurring at the vertex region20,21, to which Module A of the bracelet domain is attached (Fig. 4e,f). Likewise, Module B refolds substantially from quiescent to transcribing state, such that a template entry channel is formed (Fig. 4a,b) and the blockage of the active site by the Bα5-loop-Bα6 fragment is removed (Fig. 3f, Fig. 4a-b, Extended Data Figs 9,10).
In the quiescent state, a helical dsRNA duplex is held inside a shallow cleft formed by Modules A and B (Figs.1g,4c, and Extended Data Fig. 7d,f) through interaction between a major groove of the RNA duplex and residue Arg979 of Module B (Fig. 4c inset). In the transcribing state, this RNA duplex becomes detached perhaps as a result of refolding the RdRP bracelet domain (Fig. 4d, and Extended Data Fig. 7e,g). We anticipate that detachment of the RNA duplex would permit RNA to slide towards the template entry channel for RNA synthesis in t-CPV. Indeed, in the catalytic center of the t-CPV RdRP, we observe weak densities (Fig. 3b,g-i) that match the RNA duplex in the crystal structure of the MRV RdRP elongation complex2. We are able to place a 5-basepair RNA backbone model in the active site and a CTP at the NTP binding site (Fig. 3g-i).
In addition to enclosing the viral genome and anchoring TECs, the capsid shell protein (CSP) also regulates polymerase activity in dsRNA viruses22-25. In particular, the CSP N-terminal fragment plays roles in genome replication, mRNA transcription and capping23,26,27. A CSP N-terminal fragment, unresolved in all previous structures21,28-30, is resolved here to form a helix in the two TEC-interacting CSP subunits in both q-CPV and t-CPV (Fig. 4e,f). The N-terminal helix of one CSP inserts into the interface between the NTPase domain of VP4 and the finger subdomain of RdRP (Fig. 4f lower inset) and that of the other CSP interacts with the bracelet domain of RdRP (Fig. 4f upper inset). Notably, the former is in proximity to the NTP-binding site of the VP4 NTPase, suggesting how the N-terminal fragment of CSP is positioned to affect TEC. In addition, the structures reveal that other regions (i.e., vertices area) of CSP also interact with Module A of the RdRP bracelet domain (Fig. 4e-f). From quiescent to transcribing state, Module A and the CSP regions involved in this interaction both undergo conformational changes. Taken together, these results point to a sequence of conformational changes that leads to activation of endogenous transcription. Specifically, environmental cues cause the capsid shell to expand21, which triggers refolding of the RdRP bracelet domain, leading to formation of the entry channel for a RNA template and exposure of the polymerase active site for RNA synthesis.
Methods
Sample preparation and cryoEM imaging
CPV particles were purified as described previously30. Purified polyhedra were treated at pH 10.8 with an alkaline solution (0.2 M Na2CO3-NaHCO3) for 1 hour, and then centrifuged at 10,000g for 40 minutes. The supernatant was collected and centrifuged at 80,000g for 60 minutes at 4°C to pellet the CPV virions. The resulting pellet was directly re-suspended in the quiescent buffer (70mM pH 8.0 Tris-Cl, 10 mM MgCl2, 100 mM NaCl and 2 mM GTP). In order to prepare the transcribing CPV (t-CPV) particles, 30μl purified CPV was incubated in a reaction buffer (70mM Tris, pH 8.0, 10 mM MgCl2, 100 mM NaCl, and 1 mM SAM+2mM GTP+2mM UTP+2mM CTP+4 mM ATP) at 31°C for 15 min, and then the reaction was stopped by quenching the reaction tubes on ice.
To prepare cryoEM grids, 2.5μl of purified CPV sample was applied to a Quantifoil grid (2/2), blotted for 15 seconds with an FEI vitrobot in 100% humidity, and then plunged into liquid ethane. CryoEM images of the quiescent CPV (q-CPV) were collected in an FEI Titan Krios cryo electron microscope, operated at 300kV with a nominal magnification of 49,000x (Extended Data Fig. 5g). The microscope was carefully aligned and electron beam tilt was minimized by a coma-free alignment procedure. Images were recorded on a Gatan K2 direct electron detection camera with the counting mode, and the pixel size was calibrated as 1.01Å/pixel on the specimen using catalase crystals. The dose rate of the electron beam was set to ~8e−/pixel/s, and the image stacks were recorded at 4 frames/sec for 3 seconds. The drift between frames in each image stack was corrected with the UCSF software31, and the total 12 frames of each stack were merged to generate a final image with a total dose of ~25e−/Å2. Contrast transfer function (CTF) parameters, including defocus values and astigmatism, were determined by CTFIND32 (Extended Data Fig. 5g).
Sample grid preparation, cryoEM imaging and drift correction of frames for the transcribing CPV (t-CPV) were performed using the same procedure described above for q-CPV with the exception of the camera used. The t-CPV cryoEM images were recorded on a new Gatan K2 direct electron detection camera attached to a Gatan imaging filter (GIF Quanta) with a pixel size of 1.36Å at the specimen scale (Extended Data Fig. 5g).
Asymmetric reconstruction based on original images
A total of 68,526 particles were selected for image processing using Frealign33 and Relion34. The 2x binned data set was first processed using icosahedral symmetry with Frealign33. The centers of all particles were then fixed and used for the asymmetrical global search with Frealign using 4x binned data set starting at 20 Å resolution.
To generate an initial model, we placed the crystal structure of the MRV RdRP2 under a previously obtained CPV capsid map35 at the location corresponding to that in MRV capsid as previously reported9 and imposed a tetrahedral symmetry (i.e., with 4 three-fold axes, 3 two-fold axes and 12 asymmetric units), resulting in a montage map with an empty CPV capsid containing 12 RdRPs but without any VP4. This montage map was filtered to 30Å resolution and used as the initial model for image processing with Frealign. After 9 iterations of global search and 2 iterations of refinement, the resolution of the density map was determined to be 3.9Å. In the final map, only 3 RdRPs (#8-10 in Fig.1d) remained at the same locations as in the initial model with the tetrahedral symmetry.
The final map was reconstructed using the top 47,968 (70%) particles of the original unbinned data set. Averaging all TEC densities under different vertices was performed following the procedure described previously36 to improve the density quality and the resolution. The effective resolution of the asymmetrical and averaged reconstructions were estimated to be 3.9 Å and 3.3 Å, respectively, based on the FSC (≥0.143) and the correlation coefficient (≥0.5) between the density map and atomic model calculated with Phenix (Extended Data Fig. 5g)37,38. These estimated resolutions are consistent with the observed structural features of the density maps (Fig. 2, Extended Data Fig. 5e, and Supplementary Videos 3-8). The averaged map was filtered to the spatial frequency of 1/(3.3 Å) and sharpened with a reverse B-factor of −120Å2. This B-factor was chosen with a trial-and-error method based on the optimization of noise level, backbone density continuity, and emergence of side-chain densities.
Since there were no densities in the initial montage model at the VP4 locations, the emergence of VP4 densities in the map and the match of side-chain densities to those expected from the VP4 amino acid sequence (Fig. 2) provide strong internal controls for the validity of the high resolution cryoEM map. Consistent with this assessment, the locations of the RdRP in the final reconstruction are not only different from those in the initial montage model, but also are related by D3 symmetry instead of the tetrahedral symmetry in the initial model. Most convincingly, the density features in the final map agree with the CPV RdRP amino acid sequence but differ from that of the MRV RdRP used in the initial model.
In addition, we also performed independent reconstruction without using the model of the 12 MRV RdRPs, and obtained a nearly identical structure from the same dataset. In this procedure, we first determined an icosahedral reconstruction without using any initial models. This icosahedral reconstruction was used to restrain refinement without symmetry (i.e., symmetry operator is C1) to search for orientation around the 60 icosahedral-symmetry-related locations with Relion34. This independent result further validate our TEC structures.
To obtain the 3D structure of the transcribing particles, we low-pass filtered the above 3D map of q-CPV to 30Å resolution and used it as the initial model. After 11 iterations of asymmetrical global search and 2 iterations of local refinement, the density map converged to a resolution of 4.8 Å, and the density quality of the TEC was further improved to ~4.0 Å resolution by aligning and averaging all TEC densities inside the asymmetric reconstruction (Extended Data Fig. 5d,f,g).
Asymmetric reconstruction using capsid-subtracted images
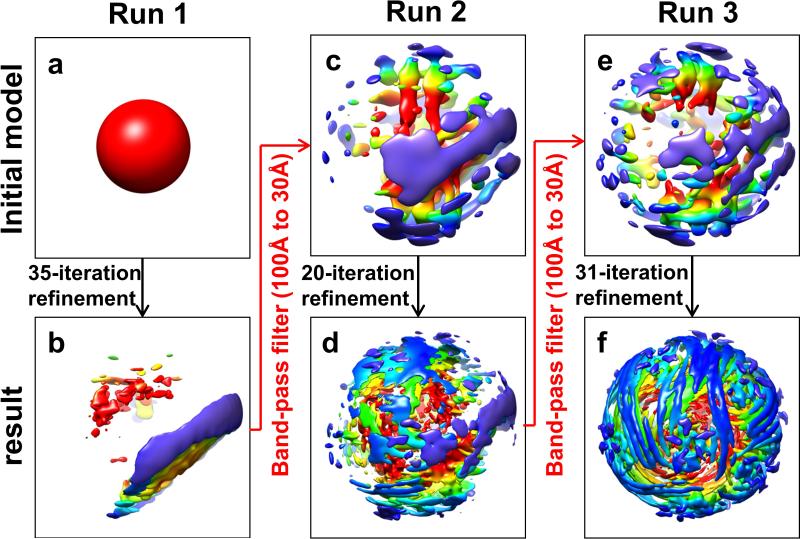
To further improve the genome structure, we used the following procedure to carry out asymmetric reconstruction of q-CPV with the same particle image dataset but with capsid contribution subtracted. As illustrated in Extended Data Fig. 1, this procedure includes four stages: 1, capsid subtraction in raw particle (orange); 2, initial model generation (green); 3, asymmetric feature emergence in Relion34 refinement (blue); 4, orientation selection (purple). In the first stage (orange in Extended Data Fig. 1), we determined the orientation and center parameters for each particle and obtained an icosahedral reconstruction with Frealign33 from raw particles with an inverse B-factor of −40 Å2 (a-b). Based on these parameters, a CTF-corrected projection (c) with empirical B-factor of 160 Å2 was generated. Next, the capsid contribution to the images was removed by subtracting the 2D projection corresponding to the icosahedral orientation of each image as done before39-42 with the following improvements. To accurately subtract the contribution from the capsid, we determined a scaling factor between capsid projection (c) and each raw particle image (a). The projection and raw images were both band-pass filtered between 1/400 Å−1 and 1/29 Å−1, then radially masked based on the inner and outer diameters of capsid to produce ring-shaped projections (d) and raw (e) images. The standard deviations of these ring-shaped images were calculated and used to normalize both the unmasked and masked (i.e., ring-shaped) projections. The cross-correlation coefficient (0 to1) between the ring-shaped raw image and the normalized ring-shaped projection was computed and used as the probability factor measuring the contribution of capsid signal in the raw particle image. Each raw image was then subtracted by the unmasked projection multiplied by this probability factor to generate a capsid-subtracted particle (f) for the following refinement. Particles with a probability factor less than 0.1 were not included in the subsequent analyses.
In the second stage (green in Extended Data Fig. 1), the map from the above Frealign asymmetric refinement (g) was low-pass filtered to 60 Å resolution, masked with a 260 Å radius (h), and used to refine the capsid-subtracted particle (f) with Relion version 1.2. The Tau2_fudge value (T-factor) in Relion was set to 0.5. T-factor is an ad hoc value in Relion to tune refinement speed, and a value of 0.5 slowed down the refinement progression thus ensuring the priority use of low resolution (up to 20Å, such as dsRNA) data in the refinement. This refinement led to a reconstruction without the capsid (i). This capsid-removed map has 12 TECs with D3 symmetry, which could be classified into two groups: the first group containing six better-resolved TECs close to the 3-fold axis (polar) and the other group containing six less-resolved TECs near the equator (tropical), suggesting potential smear of density due to orientation mis-assignments or TEC flexibility/lower occupancy near the equator.
In order to further eliminate potential orientation mis-assignments, we next conducted the third stage of data processing (blue in Extended Data Fig. 1). We first low-pass filtered the capsid-removed reference (i) to 32 Å resolution (j) and used it to drive Relion refinement with the capsid-subtracted particles (f). The T-factor used in this refinement is 0.1, only 2.5% of that used in Relion convention, thus ensuring slow progression of the refinement. Slower refinement provides time for asymmetrical feature to emerge. Relion global search was carried out with a 3.75 degrees angular interval, followed by local angular search with 1.875 degrees interval and highly-constrained translational search (0.7 pixel in range with 0.5 pixel interval). Asymmetrical RNA density feature with 10 TECs emerged after 10 iterations (k). In our procedure, one way to prevent trapping into local minima in orientation assignment due to symmetric structural elements is to filter the current refinement result back to ~32 Å resolution and refine with T-factor of 0.1 again to remove residual symmetric feature from the working reference. This process is carried out iteratively.
To further improve resolution of the 3D map, we carried out the fourth stage for particle orientation selection (purple in Extended Data Fig. 1). From the orientation of each particle determined in the high-resolution (~3Å) icosahedral reconstruction (b), we calculated 60 icosahedral-related orientation candidates. The task of the rest of the fourth stage of data processing is to select one out of these 60 orientation candidates to be the asymmetric orientation of the particle as done before4,5,43. To do this, we continued to run Relion refinement for 15 iterations using the above asymmetric map with 10 TECs (k) as initial model and the orientation determined by each iteration was recorded, giving rise to 15 Relion orientations for each particle. For each of these 15 Relion orientations, we calculated its angular distances to the 60 icosahedral-related orientation candidates; and the icosahedral-related orientation candidate with the smallest angular distance was selected as the working orientation for that iteration, resulting in a total of 15 working orientations for each particle. The particle would be retained if 14 or all of its 15 working orientations are the same (i.e., the selected orientation) and their averaged angular distance was less than 3 degrees. Otherwise, this particle will be discarded. This procedure yielded a total of 11,741 particles with selected orientation. The original raw images of these selected particles were combined to generate an asymmetric reconstruction using Frealign and the resolution was determined to be 5.1 Å.
As shown in Extended Data Figure 2, this procedure was repeated by using a Gaussian ball to replace the capsid+TEC model (g) in the initial model generation stage (green in Extended Data Fig. 1). The result is the same, confirming our procedure was not influenced by the choice of initial model.
Atomic modeling and visualization
The atomic models of both RdRP and VP4 in the quiescent state were built with Coot 44 and refined with Phenix38 as described previously45.
The atomic model of the VP4 structure was manually built with Coot. Because no homology models of VP4 previously existed, the Cα carbon backbone was constructed by matching the VP4 amino acid sequence to the density map. Once the correct placement of each residue was ensured, the backbone was converted to a purely alanine backbone by the function “Mainchain,” and mutated to the corresponding amino acids through the function “Mutate Residue Range.” With the initial model now completed, the “Density Fit Analysis” validation tool was used to screen for sequences of the model that did not fit the density. When identified, these sequences and the amino acids surrounding them were examined for any other possible conformations that would better fit the density. Due to the high resolution of this structure, this was completed through the refinement tool “Real Space Refine Zone,” which optimizes the fit of the model to the mass density while preserving stereochemistry. Additionally, refinement was also performed based on the Ramachandran plot, an important indicator of three-dimensional protein structure that validates the torsion angles of a protein chain. In the Ramachandran plot, any residues with disallowed values were selected, and the stereochemistry of that residue along with its surrounding residues was optimized with the refinement tool “Regularize Zone.” After ideal Ramachandran values were obtained (<1% outliers), the refinement function “Rotamers” was used to select a rotamer that best fit the density.
The atomic model of the polymerase structure was also manually built with Coot. However, since an atomic model for the MRV polymerase was available in the Protein Data Bank (accession number 1MUK), this model was used as a template to assist with model building through the identification of the N-terminus, C-terminus, and various secondary structures. Once the Cα carbon backbone was built by matching the polymerase amino acid sequence to the density map and mutated to the appropriate amino acids, the model was refined with “Regularize Zone,” “Rotamers,” and “Real Space Refine Zone.” The model was validated with the Ramachandran plot and the function “Density Fit Analysis.” The complex of VP4 and polymerase was then refined with Phenix, including the real space refinement38.
The atomic models of the transcribing state were built by fitting the atomic structures of RdRP and VP4 at quiescent state into the density, manually adjusting the changed residues with Coot44 , and refining the models with Phenix38.
Visualization, segmentation of density maps, and generation of videos were done with UCSF Chimera31.
Extended Data
Extended Data Figure 1.
Illustration of the asymmetric reconstruction procedure using particles with the capsid density subtracted.
Extended Data Figure 2.
Validation of asymmetric reconstruction from capsid-subtracted images using a Gaussian ball as the initial model.
Extended Data Figure 3. Sections of the q-CPV density map along the 3-fold (i.e., the earth axis) (a) and 2-fold (b) axes of the pseudo-D3 symmetry.
Note: the Lack of 3-fold and 2-fold symmetry in the RNA density in contrast to the perfect symmetry of the capsid shell proteins. Pixel size=4.04Å; Clipped map size=166*166*120 pixels.
Extended Data Figure 4. dsRNA density maps in the quiescent state.
a, View of TEC+RNA densities with the same orientation of Figure 1d. b-c, The same view as in (a) but rotated by +90° (b) or −90° (c) along x axis in (a) to view from either north (b) or south (c) poles. d-f, Three views from three 2-fold axes on the equator, each is rotated by 120 ° along the y axis from each other. g. dsRNA density maps at the twelve vertices. TECs are arranged and numbered according to Figure 1d. First row: TECs 1, 2, 3; Second row: TECs 4, 5, 6; Third row: TEC 7 and two unoccupied positions; Fourth row: TECs 8, 9, 10. All TECs have a dsRNA segment bonded at the flange, each marked with a black arrow. Compared with polar TECs, all tropical TECs (4-7) have extra piece of dense rods, with locations indicated with an opened black arrow.
Extended Data Figure 5. CryoEM reconstructions of CPV in the quiescent and transcribing states.
a-b, CryoEM images of CPV particles in quiescent (a) and transcribing (b) states. These images were obtained by aligning and averaging frames in direct electron counting image stacks. Fiber-like nascent mRNAs are visible over background in (b) (marked by green arrows), while the background in (a) is clean. c-d, Fourier shell correlation coefficients (FSCs) as a function of spatial frequency between two half maps for reconstructions in the quiescent (c) and transcribing (d) states. The black and red lines represent FSCs for the asymmetrical reconstructions of capsid + genome and the locally averaged TEC densities, respectively. The effective resolutions of the local averaged maps are ~3.3 Å (c) and ~4.0 Å (d) resolution (FSC≥0.143) for maps in the quiescent and transcribing, respectively. e-f, CryoEM densities (grey surface representations) superimposed with atomic models (ribbons and sticks) for the quiescent (e) and transcribing (f) states. The α-helix (Pα12) and the four-stranded β-sheet (P4, P7-8 & P11) in (e) and (f) are both from the palm subdomain of the polymerase domain at 3.3 Å (e) and ~4.0 Å (f) resolutions. g, Statistics of CPV reconstructions and atomic model refinement.
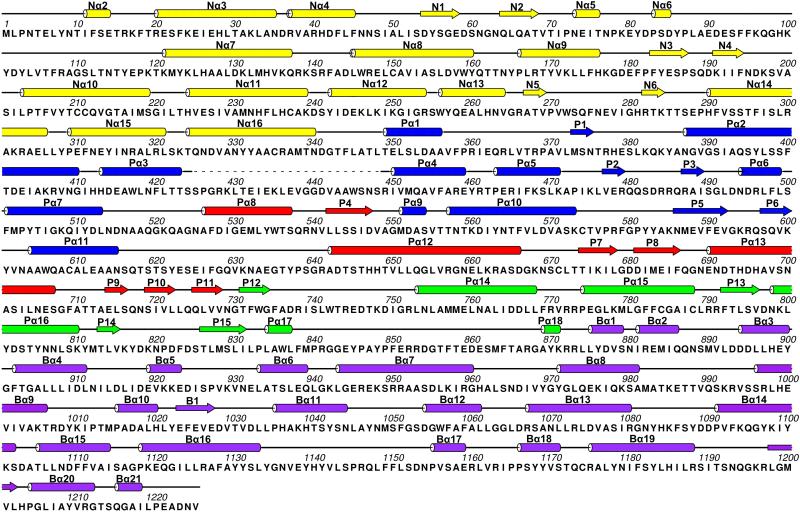
Extended Data Figure 6. Sequence and secondary structure assignment of CPV RdRP in the quiescent state.
α-helices were marked by cylinders, β-strands by arrows, loops by thin lines, and the flexible tip domain by dashed lines. The colour scheme is the same as Figure 3a.
Extended Data Figure 7. The RdRP-bound dsRNA in the quiescent and transcribing states.
a-c, Location of a TEC on the inner surface of the capsid shell in the quiescent and transcribing states. The inner surface of the CPV capsid (a) with 10 CSPs labelled (CSP-A.1/B.1 ~ CSP-A.5/B.5). Position of a TEC on the inner surface of capsid in the quiescent (b) and transcribing (c) states. VP4 and RdRP are colored cyan and purple, respectively. An icosahedral 5-fold axis is indicated with a small green pentagon. d-e, CryoEM densities of TEC and dsRNA (orange) in the quiescent (d) and transcribing (e) states. f-g, Models of TEC (surface representation) and dsRNA (ribbons) in the quiescent (f) and transcribing (g) states. Close-up views show the bound dsRNA (surface representation) on RdRP in the quiescent state (f) and its detachment in the transcribing state (g). VP4 is coloured cyan and the RdRP is coloured as in Figure 3a. Note: All surfaces displayed in this figure were rendered from models, except for the density maps of RdRP+dsRNA in (d-e).
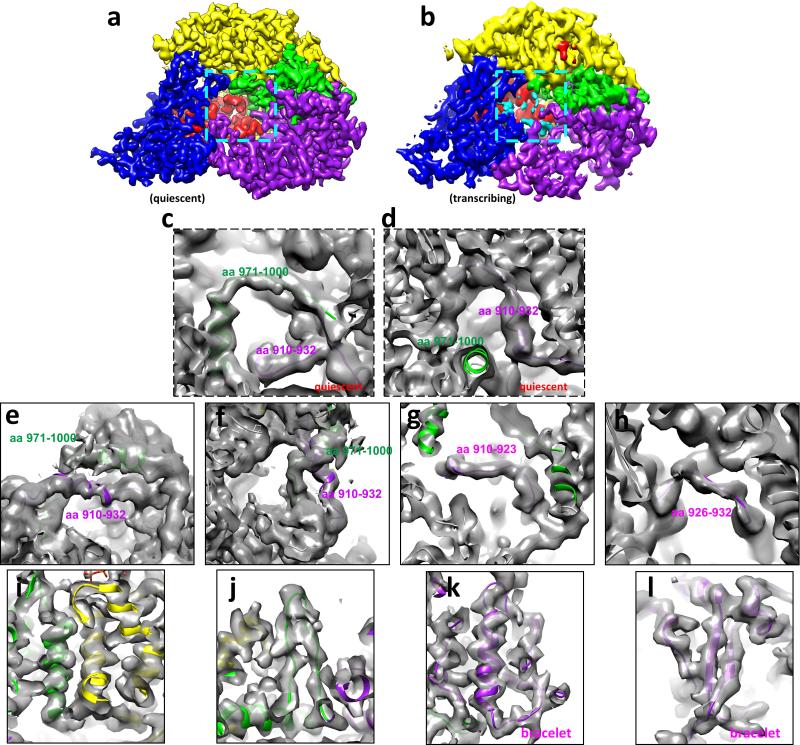
Extended Data Figure 8. Tracing amino acid residues 910-932 and 971-1000 of module B of the bracelet domain of RdRP in the quiescent and transcribing states.
a-b, CryoEM densities of RdRP in the quiescent (a) and transcribing (b) states. The locations of the residues 910-932 and 971-1000 are indicated with cyan boxes in (a) and (b). Due to their flexibility, these residues are not readily visible when displayed as in (a) and (b) but become visible when the maps are filtered to a lower resolution (e.g., 4.5Å resolution) as in (c-f). The colour scheme of domains/subdomains is the same as in Figure 3a. c, Trace of the residues 971-1000 (green) and 910-932 (purple) of module B of the bracelet domain of RdRP in the quiescent state. d, The same as (c) but in a different view. e, Trace of the residues 971-1000 (green) and 910-932 (purple) of module B of the bracelet domain of RdRP in the transcribing state. f, The same as (e) but in a different view to show the unambiguous trace of the two peptide fragments. g-h, Trace of the residues 910-923 (g) (purple) and 926-932 (h) (purple) of the bracelet domain of RdRP in the transcribing state, showing the unambiguous trace of the two peptide fragments. i-j, CryoEM densities (grey) and model (ribbon) of RdRP in the transcribing state, showing α-helices (i) and a β-hairpin (j). The colour scheme of domains/subdomains is the same as in Figure 3a. k-l, Trace of the residues of the bracelet domain of RdRP in the transcribing state, showing a α-helix (k) and a β-sheet (l).
Extended Data Figure 9. Stereo and rotated views of Figure 4a and 4b.
a-b, Stereo views of modules A (yellow cylinders and loops) and B (purple cylinders and loops) of the bracelet domain of RdRP in the quiescent (a) and transcribing (b) states. c-d, Same as in (a-b) but rotated around the X-axis by 90°. Note: All surfaces displayed in this figure were rendered from models.
Extended Data Figure 10. Comparisons of RdRPs from CPV and MRV.
a-b, CryoEM In situ structure of the RdRP in t-CPV (a) and crystal structure of the MRV RdRP (b), both containing a RNA duplex in the active site. c-e, Superposition of domains of RdRPs from t-CPV (colour) and MRV (grey): N-terminal (c), polymerase (d) and bracelet (e) domains. f-h, Comparisons of modules A (yellow) and B (magenta) of the bracelet domain of RdRPs from q-CPV (f) t-CPV (g) and MRV (h). i-k, The same as in (f-h), but with helices shown as cylinders, as in Figure 4 a,b.
Supplementary Material
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (AI094386 and GM071940 to Z.H.Z.), NSFC (31172263 to J.S.) and NSFGD (S2013010016750 to J.S.). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057, 1S10OD018111) and NSF (DBI-1338135). We thank Peng Ge for carrying out a Relion reconstruction without an initial model as an independent verification step, Stan Schein and Laurie Wang for proof-reading the paper, and Pavel Afonine for model refinement.
Footnotes
Author Contributions Z.H.Z. supervised research; X.Z., X.Y. and Z.H.Z. designed and performed the experiments, analyzed and interpreted data and wrote the paper; K.D. wrote programs, analyzed data and prepared figures; W.C. built models; JS prepared reagents. All authors reviewed and finalized the paper.
Author Information 3D cryoEM density maps have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-6408 (3.3Å averaged TEC in q-CPV), EMD-6404 (4.0Å averaged TEC in t-CPV), EMD-6407 (3.9Å full q-CPV), EMD-6405 (4.8Å full t-CPV), EMD-6406 (5.1Å asymmetric reconstruction of q-CPV by capsid-subtraction method) and EMD-6409 (filtered 22 Å q-CPV asymmetric reconstruction). The coordinates of atomic models of the TEC in q-CPV and t-CPV have been deposited in the Protein Data Bank under the accession number 3JB6 and 3JB7, respectively.
The authors declare no competing financial interests.
Online Content Any additional Methods and Extended Data display items are available in the online version of the paper; references unique to these sections appear only in the online paper.
REFERENCES
- 1.Mertens PPC, Rao S, Zhou ZH. In: Virus Taxonomy, VIIIth Report of the ICTV. Fauquet CM, et al., editors. Elsevier/Academic Press; 2004. pp. 522–533. [Google Scholar]
- 2.Tao Y, Farsetta DL, Nibert ML, Harrison SC. RNA synthesis in a cage— structural studies of reovirus polymerase λ3. Cell. 2002;111:733–745. doi: 10.1016/s0092-8674(02)01110-8. [DOI] [PubMed] [Google Scholar]
- 3.Lu X, et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 6.Zhou ZH. In: Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology. Patton John T., editor. Caister Academic Press; 2008. pp. 27–43. [Google Scholar]
- 7.Furuichi Y. “Methylation-coupled” transcription by virus-associated transcriptase of cytoplasmic polyhedrosis virus containing double-stranded RNA. Nucleic Acids Res. 1974;1:809–822. doi: 10.1093/nar/1.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estrozi LF, et al. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. Journal of molecular biology. 2013;425:124–132. doi: 10.1016/j.jmb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Walker SB, Chipman PR, Nibert ML, Baker TS. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nature Structural & Molecular Biology. 2003;10:1011–1018. doi: 10.1038/nsb1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nason EL, et al. Interactions between the inner and outer capsids of bluetongue virus. Journal of virology. 2004;78:8059–8067. doi: 10.1128/JVI.78.15.8059-8067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Q, Jakana J, Zhang J-Q, Zhou ZH. Structural Comparisons of Empty and Full Cytoplasmic Polyhedrosis Virus PROTEIN-RNA INTERACTIONS AND IMPLICATIONS FOR ENDOGENOUS RNA TRANSCRIPTION MECHANISM. Journal of Biological Chemistry. 2003;278:1094–1100. doi: 10.1074/jbc.M205964200. [DOI] [PubMed] [Google Scholar]
- 12.Gouet P, et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell. 1999;97:481–490. doi: 10.1016/s0092-8674(00)80758-8. [DOI] [PubMed] [Google Scholar]
- 13.Abels J, Moreno-Herrero F, Van der Heijden T, Dekker C, Dekker N. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys J. 2005;88:2737–2744. doi: 10.1529/biophysj.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nibert ML, Kim J. Conserved sequence motifs for nucleoside triphosphate binding unique to turreted Reoviridae members and coltiviruses. Journal of virology. 2004;78:5528–5530. doi: 10.1128/JVI.78.10.5528-5530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao S, Liang C, Hong J, Peng H. Genomic sequence analyses of segments 1 to 6 of Dendrolimus punctatus cytoplasmic polyhedrosis virus. Arch Virol. 2003;148:1357–1368. doi: 10.1007/s00705-003-0103-z. [DOI] [PubMed] [Google Scholar]
- 16.Sutton G, Grimes JM, Stuart DI, Roy P. Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol. 2007;14:449–451. doi: 10.1038/nsmb1225. [DOI] [PubMed] [Google Scholar]
- 17.Stauber N, Martinez-Costas J, Sutton G, Monastyrskaya K, Roy P. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J Virol. 1997;71:7220–7226. doi: 10.1128/jvi.71.10.7220-7226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Parker JS, Murray KE, Nibert ML. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. Journal of Biological Chemistry. 2004;279:4394–4403. doi: 10.1074/jbc.M308637200. [DOI] [PubMed] [Google Scholar]
- 19.Choi KH, Rossmann MG. RNA-dependent RNA polymerases from Flaviviridae. Current opinion in structural biology. 2009;19:746–751. doi: 10.1016/j.sbi.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Yang C, et al. Cryo-EM structure of a transcribing cypovirus. Proceedings of the National Academy of Sciences. 2012;109:6118–6123. doi: 10.1073/pnas.1200206109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Jiang J, Sun J, Zhou ZH. A putative ATPase mediates RNA transcription and capping in a dsRNA virus. Elife. 2015 doi: 10.7554/eLife.07901. eLife 2015.07901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luongo CL, et al. Loss of activities for mRNA synthesis accompanies loss of λ2 spikes from reovirus cores: an effect of λ2 on λ1 shell structure. Virology. 2002;296:24–38. doi: 10.1006/viro.2001.1258. [DOI] [PubMed] [Google Scholar]
- 23.Patton JT, Jones MT, Kalbach AN, He Y-W, Xiaobo J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. Journal of virology. 1997;71:9618–9626. doi: 10.1128/jvi.71.12.9618-9626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansell EA, Patton JT. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. Journal of virology. 1990;64:4988–4996. doi: 10.1128/jvi.64.10.4988-4996.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gridley CL, Patton JT. Regulation of rotavirus polymerase activity by inner capsid proteins. Curr Opin Virol. 2014;9:31–38. doi: 10.1016/j.coviro.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald SM, Patton JT. Rotavirus VP2 core shell regions critical for viral polymerase activation. Journal of virology. 2011;85:3095–3105. doi: 10.1128/JVI.02360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starnes MC, Joklik WK. Reovirus protein λ3 is a poly (C)-dependent poly (G) polymerase. Virology. 1993;193:356–366. doi: 10.1006/viro.1993.1132. [DOI] [PubMed] [Google Scholar]
- 28.Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- 29.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 30.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 32.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 33.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. Journal of structural biology. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X, Ge P, Jiang J, Atanasov I, Zhou ZH. Atomic Model of CPV Reveals the Mechanism Used by This Single-Shelled Virus to Economically Carry Out Functions Conserved in Multishelled Reoviruses. Structure. 2011;19:652–661. doi: 10.1016/j.str.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huiskonen JT, Jaalinoja HT, Briggs JA, Fuller SD, Butcher SJ. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J Struct Biol. 2007;158:156–164. doi: 10.1016/j.jsb.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Briggs JA, et al. Classification and three-dimensional reconstruction of unevenly distributed or symmetry mismatched features of icosahedral particles. Journal of structural biology. 2005;150:332–339. doi: 10.1016/j.jsb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 41.Booy F, et al. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Kostyuchenko VA, Rossmann MG. Structural analysis of viral nucleocapsids by subtraction of partial projections. Journal of structural biology. 2007;157:356–364. doi: 10.1016/j.jsb.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao Y, et al. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, et al. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 Å resolution. Elife. 2013;2 doi: 10.7554/eLife.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.