Abstract
Current seasonal influenza virus vaccines are effective against infection but they have to be reformulated on a regular basis to counter antigenic variations. The majority of the antibodies induced in response to seasonal vaccination are strain-specific. However, antibodies targeting conserved epitopes on the hemagglutinin protein have been identified and they offer broad protection. Most of these antibodies bind the hemagglutinin stalk domain and are generated from preexisting memory B cells. Broadly protective stalk-biased responses induced by antigenically divergent influenza strains, in concert with prior immunity, are sufficient to eradicate seasonally circulating strains. Future vaccine trials should aim to harness and maintain such a response with the realistic goal of developing a universal influenza vaccine.
Introduction
Influenza virus epidemics contribute to 250,000 to 500,000 deaths per year worldwide [1]. Current seasonal influenza virus vaccines are effective against infection with some limitations, such as the need to be reformulated most years to counter antigenic variations, also called antigenic drift [2]. Due to the timely production of the vaccine, the strains composing the seasonal vaccine have to be determined based on prediction and surveillance; mismatches between vaccine and circulating strains occasionally occur [3]. Furthermore such vaccines do not protect against novel pandemic strains, which are occasionally introduced into the human population, typically due to antigenic shift [4]. Seasonal vaccination generally induces a narrow, strain-specific response against the highly variable head domain of hemagglutinin (HA) and thus antibodies targeting the globular head quickly lose efficacy against drifted strains [5,6]. The stalk domain, in contrast, is more conserved among influenza A (group 1 and 2) and B viruses allowing antibodies that target this region to neutralize a wide spectrum of influenza virus subtypes [7–9]. Such antibodies are relatively rare in the human population but novel approaches to enhance these antibodies are currently being developed [10,11]. Importantly, it is believed that targeting such conserved epitopes is the key to the elimination of seasonal influenza strains. Broadly neutralizing stalk-reactive antibodies are emerging therapeutic tools against influenza virus infections and are a promising prospect for the development of a universal influenza virus vaccine. A key issue in the field is whether or not an antibody response to HA stalk epitopes could sufficiently protect and sustain for permanent immunity to all, or most, circulating influenza strains. We argue herein that indeed a properly designed stalk-based vaccine could provide broad immunity.
Antibody responses to influenza virus
The influenza virus has two main surface glycoproteins: HA and neuraminidase (NA) [12]. HA is a trimeric protein with an immunodominant head domain that is preferentially mutated during immune evasion [4,13,14]. There is a receptor-binding site within the head domain that binds to sialic acid moieties on the surface of host cells to facilitate viral infection [15]. Antibodies blocking this binding site are characterized by their ability to prevent influenza virus mediated agglutination; in vitro these antibodies can be identified using a hemagglutination-inhibition assay (HAI) [12]. The HA stalk domain is composed of three helical bundles and is functionally required for the pH induced conformational changes involved in membrane fusion during viral entry and exit from the host cell [8,14,16,17]. Antibodies specific for this region can be identified by their ability to block viral cell infection independently of HAI activity, using in vitro microneutralization or plaque assay. NA, on the other hand, is required for cleaving the HA-sialic acid tethering to release new virions, allowing for viral spread [18,19]. Potentially protective NA-reactive antibodies are identified by their ability to block NA cleavage [20,21].
Influenza A viruses are subtyped based on the sequence and antigenic divergence of the HA and NA surface proteins. A total of 18 HA and 11 NA subtypes have been identified so far, with the type of HA expressed splitting influenza A viruses into two phylogenetic groups (Group 1: H1, H2, H5, H6, H8, H9, H11, H13, H16, H17, H18; and Group 2: H3, H4, H7, H10, H14, H15) [22–25]. Influenza B viruses are divided into two antigenically different lineages (Victoria and Yamagata) [26]. The majority of protective antibodies generated in response to influenza target the HA protein [27]. Less is known about how the antibody response to NA alters the course of an influenza infection, although NA-inhibitors such as Oseltamivir (Tamiflu), Zanamivir (Relenza), Laninamivir (Inavir), and Peramivir (Rapivab) have some efficacy in reducing severity if used early during the course of infection [28,29]. This review focuses on the antibody response to HA.
Conserved protective epitopes on HA
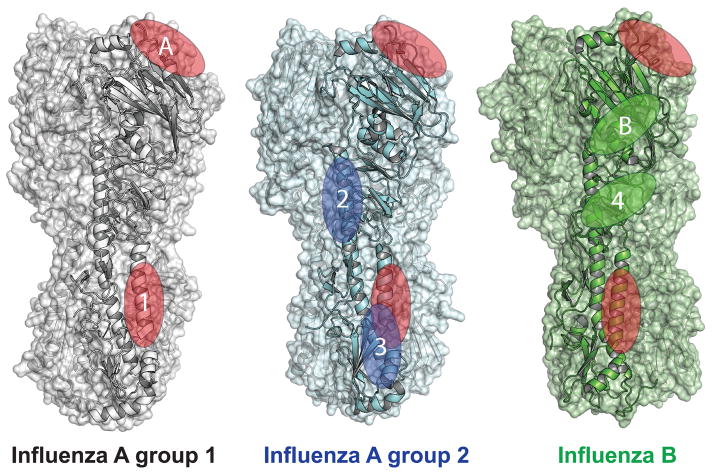
Despite the fact that the majority of the protective antibodies targeting HA recognize the head domain and display a high level of strain specificity [6], a number of head specific antibodies have been identified with varying levels of cross-reactivity between influenza strains [30–42]. All of these antibodies identified thus far, target one of two cross-protective head epitopes (Figure 1). Antibodies that target epitope A must overcome the extreme variability of the HA head, by forming key interactions within the highly conserved receptor-binding site [30–39,42]. An extensive study of antibodies binding to this epitope revealed that they are fairly common in a vaccine response, have diverse V(D)J gene usage and utilize sialic acid mimicry on their HCDR3 loops to directly engage the receptor binding site [42]. These interactions are sufficient to overcome the extreme strain-to-strain variation present in the surrounding contact residues [15,42]. Epitope B is protective in both B strain lineages and includes the vestigial esterase domain at the base of the HA head [31].
Figure 1. Conserved protective hemagglutinin (HA) epitopes.
Epitopes shared between all three influenza groups are indicated by red ovals, influenza A group 2 are blue and influenza B epitopes are green. Two conserved epitopes have been identified within the HA head domain. Epitope A includes the receptor-binding site, which is conserved across all influenza subtypes. Epitope B contains the vestigial esterase domain and is conserved across both B strain lineages. Four conserved epitopes have been identified within the HA stalk domain. Epitope 1, which includes the A α helix of HA2, is conserved across influenza A group 1, group 2 and influenza B strains. Stalk epitope 2 consists of the CD α helix in HA2, while epitope 3 encompasses regions of at the base of the HA2 stalk. Both of these epitopes are conserved within influenza A group 2. Epitope 4 is conserved between both B lineages and is located in the C terminal portion of HA1. Each epitope is only indicated on a single monomer within the HA trimer. Accession numbers: Group 1 H5N1 (2FK0), Group 3 H3 (3ZTJ) and influenza B/Brisbane/60/2008 (4FQM).
In contrast to the HA-head variability, the stalk domain displays a much higher level of conservation across influenza strains with some central residues being identical across all subtypes [7–9]. Three protective epitopes, with varying levels of cross-reactivity between group 1 and 2 influenza strains, have been identified within the stalk portion of influenza A HA (Figure 1) [8,9,31,43–47]. Epitope 1 is centered on the A α-helix of the HA2 region of HA [8,9,31,44,48]. Targeting this epitope is also protective against B strains, but the antibody must have unique properties to accommodate key modifications helping to obscure the epitope surface [31]. Epitopes 2 and 3 are protective across group 2 influenza A subtypes. Epitope 2 includes the upper portion of the long alpha helix CD in HA2 [49], whereas epitope 3 is located at the base of the HA2 stalk spanning regions of the fusion peptide and helix-capping loops [43]. The fourth protective stalk epitope is located in the C terminal portion of HA1 and offers broad protection across both B strain lineages [50]. Generating a strong antibody response against any of these conserved epitopes can offer broader and more durable protection against influenza by circumventing reliance on epitopes prone to antigenic drift.
The antibodies specific for the conserved epitopes within the head and stalk are protective due to their neutralizing capacity. Recently we, and others, discovered a novel group of non-neutralizing protective antibodies. We identified these antibodies following H7N9 vaccination in humans. They are broadly cross-reactive, harbor high level of somatic mutations and target new epitopes on the HA protein conserved between group 1 and 2 [51]. Such antibodies have also been isolated after H7 immunization in mice [52]. Both studies showed that protection by these non-neutralization antibodies is partially dependent on Fc-FcγR interactions. The discovery of such antibodies is reinforced by serum studies showing that cross-reactive antibodies were found in influenza seropositive humans in the absence of neutralization [53] and in the absence of HAI activity [54]. Moreover passive transfer of both neutralizing and non-neutralizing antibodies from vaccinated individual sera improved virus clearance in a mouse model [55]. Targeting antibodies against any of these conserved protective epitopes would offer significantly improved protection against circulating influenza strains. Most hope is currently being placed on antibodies specific for the stalk domain because it is so widely conserved and can be independently targeted as an immunogen.
Occurrence of broadly neutralizing stalk-reactive antibodies
Broadly neutralizing stalk-reactive responses have been characterized following natural infection with seasonal H1N1 and H3N2 strains, but are more rare after seasonal influenza vaccination [56,57]. However, plasmablasts with HA stalk specificity have been reported after vaccination with the seasonal trivalent vaccine [58] and more recently, we found that group 2 cross-reactive stalk antibodies induced by seasonal vaccination were not uncommon [45]. Studies are now revealing that broadly neutralizing stalk-reactive antibodies are boosted more efficiently in humans upon exposure to antigenically divergent head HA domains, which was the case with the 2009 pandemic H1N1 strain [36,59–63]. Interestingly, this pandemic H1N1 virus led to the disappearance of all pre-pandemic H1N1 strains in the human population. In nature, the replacement of existing seasonal viruses by novel pandemic strains in the human population is a recurrent phenomenon. Over the past 60 years, multiple instances of seasonal circulating viruses being eradicated by the emergence of pandemic influenza viruses have occurred. In 1957, the H2N2 pandemic virus replaced the seasonal H1N1 strains that were previously circulating in humans [64]. In 1968, a boost in antibodies against conserved epitopes on neuraminidase after the introduction of the H3N2 strain might have caused the extinction of human H2N2 strains [65]. Both the 1957 and 2009 pandemic H1N1 virus expressed divergent head domains compared to the previous circulating strains but had conserved stalk domains. These observations lead to the hypothesis that antibodies against conserved epitopes, regulated by the immune status of the general population, could be responsible for the extinction of circulating seasonal influenza viruses after the emergence of novel pandemic strains [66]. In support of this, an elegant study using chimeric hemagglutinin proteins revealed that anti-stalk antibodies generated after the pandemic H1N1 infection played a substantial role in the disappearance of the existing seasonal H1 viruses [67].
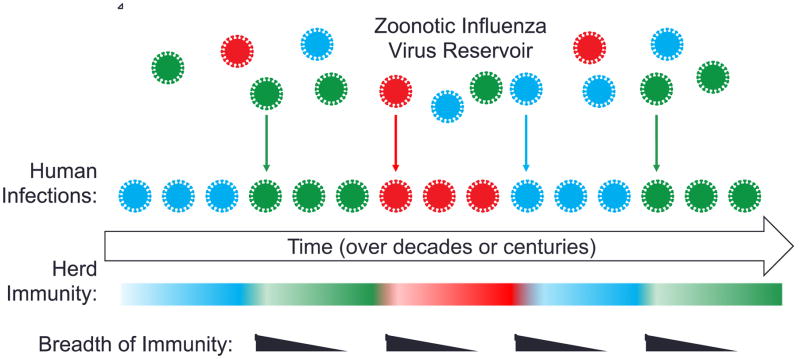
Zoonotic reservoirs are the source of pandemic influenza strains [68]. Our documented history of influenza infections only spans 100 years and it is likely that many more subtypes than are currently appreciated have existed throughout human history. We discussed above how broad protection induced from exposure to novel strains in conjunction with existing partial immunity to antigenically drifted epitopes could cause circulating strains to be eliminated. The current evidence suggests that targeted vaccines that induce broad immunity on a wide scale could eliminate all currently circulating influenza strains. However, the progeny of past strains, or genetic reassortants thereof, are always available within these zoonotic reservoirs to re-enter human populations as reoccurring seasonal strains (Figure 2). People who have been exposed to similar antigens in the past will meet these strains with broad protective immunity, but the majority of the population will be vulnerable due to loss of antigen breadth and herd immunity over time. This cycle will only be broken by removal of zoonotic reservoirs, which is likely not possible, or by widespread and durable immunization against conserved epitopes. This could come in the form of a broadly protective vaccination, which eliminates currently circulating strains, and is reformulated upon cyclic re-entry from zoonotic reservoirs. These issues will need to be addressed in the design of a universal influenza vaccine.
Figure 2. Model for cyclic re-entry of zoonotic influenza strains to humans.
Diverse strains of influenza persist in zoonotic reservoirs and can re-enter the human population. Introduction of novel zoonotic strains, most often due to genetic reassortment, boosts immunity preferentially to conserved epitopes. The breadth of this response decreases over time with subsequent exposures. Herd immunity to past circulating strains is lost over generations and they can again become infectious to humans. Both sources of novel influenza strains result in a never-ending cycle of re-entry. Inducing broad immunity on a wide scale, and maintaining it indefinitely at the population level, could eradicate influenza infections of humans.
Memory origin of stalk-reactive antibodies
Humans have an extensive immune history and upon antigen re-exposure, antigen-specific memory B cells are recalled in the immune response. The antibody producing plasmablast population bursts after vaccination, or infection, and is mostly comprised of antigen-specific cells [6]. It is now well appreciated that the adult influenza vaccine response is driven by activation of preexisting memory B cells, which can be identified by extensively mutated variable region genes [6,36,48]. By analyzing the B cell response to vaccination in adult subjects who received the trivalent influenza vaccine over consecutive years, we showed that memory B cells are the predominant precursors to the plasmablast influenza response [69]. Based on their substantial mutation load and their binding affinity, stalk-reactive antibodies appear to be pre-existent within the memory compartment [36,48,69]. Protective H7N9 stalk-reactive antibodies have even been identified in people who have never been exposed to this virus, due to presence of group 2 cross-reactive memory B cells [45]. Additionally, an H7 vaccine study showed the generation of a vigorous, high-affinity, stalk-specific antibody response with a consistent increase in circulating memory B-cell frequencies [70]. Finally, geriatric populations with extensive memory B cell compartments but limited naïve B cells, showed a stalk-biased serum antibody response following seasonal vaccination [71]. These studies suggest that the key to inducing broad protection against influenza is to activate cross-reactive memory B cells.
How to boost stalk-reactive antibody responses
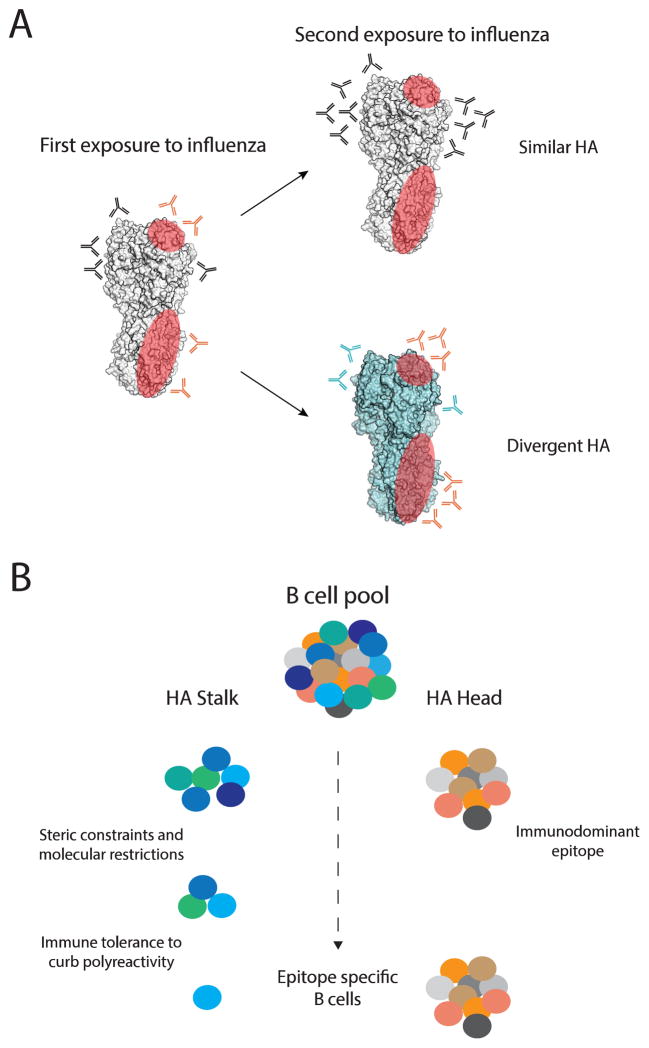
It is known that immunological memory acquired against influenza strains alters the response to subsequent viral encounters [72], but how sequential exposure to antigenically distinct influenza strains shapes the humoral immune response remains poorly understood. Two recent longitudinal studies provide important new insights. The first study is an analysis of antibody titers against various pandemic and seasonal influenza strains spanning a 20-year period, before the pandemic 2009 H1N1. HAI neutralizing titers specific to pandemic viruses in human circulation between 1957 and 2008 (H2N2, H3N2, and H1N1) exhibited sustained increases over the course of study. Interestingly group 1 and 2 stalk-reactive antibodies also rose modestly over the same period of time, even in the absence of major antigenic shift. However, group 1 HA stalk-reactive antibody titers were greatest in individuals who were exposed to the most diverse group 1 viruses [73]. An antigenically more stable virus, human cytomegalovirus, did not induced sustained increase in neutralizing antibody titers, suggesting that antigenic variation of influenza A viruses play a role in shaping the humoral response. The second study analyzed the B cell response to the pandemic 2009 H1N1 strain at the monoclonal antibody level upon first or second exposure [69]. Only individuals with low preexisting serological levels of pandemic H1N1-specific antibodies generated a broadly neutralizing plasmablast response directed toward the HA stalk. This observation confirmed that in the context of exposure to divergent influenza strains, immune history directly determines the likelihood of generating a broadly protective response (Figure 3A). Moreover maintaining a sustainable broadly neutralizing stalk-biased response upon subsequent exposures is a challenge, as re-exposure to the pandemic 2009 H1N1 strain by vaccination induced an HA head–biased response [69].
Figure 3. Broadly protective antibodies.
(A) Antibodies against conserved epitopes, shown in red on a single monomer of the HA trimer, are generated at a low frequency during a primary exposure. The B cells generating these antibodies are recalled upon secondary exposure to an antigenically distinct influenza strain, and contribute significantly when only these protective epitopes are conserved. Unfortunately, due to the immuno-dominant nature of the HA-head, when secondary exposure is against an antigenically similar influenza strain the stalk antibodies will be lost in the crowd of strain specific HA-head antibodies. (B) There are multiple factors proposed to be responsible for the relative scarcity of broadly protective stalk antibodies. First, the subdominant immunogenicity of the stalk domain has been attributed to steric constrains, which impose harsh molecular restrictions on the antibodies recognizing these epitopes. Second, immune tolerance to curb polyreactivity may also select against these cells.
The effects of immune history on induction of these broadly neutralizing antibodies result from characteristics of the virus and the antibodies themselves (Figure 3B). Firstly, there is an immuno-dominance towards epitopes located on the globular head. One reason proposed for sub-dominance of the stalk HA epitopes is a limited access due to steric shielding by the HA globular head and/or because of their proximity to the viral envelope [74,75]. However, structural studies have demonstrated that the HA stalk epitopes are accessible for antibody binding [76]. Although the epitopes are accessible, we found that stalk-reactive antibodies have reduced affinity to whole virus but a similar affinity to soluble HA protein compared to head-reactive antibodies. [69]. The structure of HA on whole virions must be the limiting factor to antibody binding. In addition to these steric restrictions there are also molecular/biochemical constrains imposed by the epitopes themselves, with most antibodies specific for the conserved HA stalk epitope 1 preferring the VH1-69 and VH1-18 genes [8,9,43,69,77]. This is attributed to three conserved hydrophobic residues located within the HCDR2 & HCDR3 loops of this V gene, which are required for the heavy chain mediated interaction [8,9,43,78]. Less common antibodies specific for this epitope have been identified without gene restriction and they utilize the more canonical antigen binding mediated by both the heavy and the light chain [44]. Broad immunoglobulin variable gene usage has also been identified for the other stalk epitopes [43,79]. These observations suggest that the limited accessibility of the HA stalk epitopes imposes molecular constraints on antibodies, leading to restricted VH usage of neutralizing HA stalk–reactive antibodies.
Anti-HIV antibodies that bind the gp140 glycoprotein have been shown to be polyreactive. The antibodies have one high-affinity binding site on gp140 and one low-affinity binding site on another molecule at the surface of HIV virus. This mechanism, referred to as heteroligation, demonstrably increases the apparent affinity of polyreactive antibodies to HIV and improves viral neutralization [80,81]. Interestingly, broadly neutralizing stalk-reactive antibodies have also been reported to have higher levels of polyreactivity [69]. We found that polyreactivity is a specific characteristic of antibodies capable of binding broadly protective epitopes on the HA stalk, independently of the VH usage. Therefore, immune checkpoints that curb possible self-reactivity, including polyreactivity [82] may also contribute to the scarcity of these broadly protective cells, further contributing to HA head immuno-dominance (Figure 3B).
New approaches towards a universal influenza virus vaccine
Two main avenues are currently being explored to modify the seasonal influenza vaccine to induce a more protective stalk-biased response. The first approach utilizes immunizations with recombinant HA proteins; either a stabilized headless version or a chimeric HA with a conserved stalk region combined with a diverse HA head [74,83–85]. The second focuses on modifying the current vaccine to include an adjuvant or to incorporate a live attenuated influenza vaccine boost prior to the inactivated vaccine [70,86,87]. These approaches have been successful at biasing the antibody response against the conserved stalk domains in animal models, and coincide with the literature on recalling stalk antibodies from within the pre-existing memory compartment. The chimeric-HA immunization strategy is in preparation for clinical trials. It will be quite interesting to see the results of this trial vaccine when placed in the context of a diverse immune memory/history [88,89].
Summary and outlook
We have learned from nature that the eradication of particular influenza viruses is possible. Further, the discovery of stalk-reactive antibodies has been a catalyst for the goal of a universal influenza virus vaccine. Understanding the impact of immune memory to conserved influenza virus epitopes in humans is critical for the induction of a broadly stalk-reactive antibody response and its sustainability over time. Future clinical trials with vaccine candidates targeting such a response in humans will demonstrate how realistic a universal influenza vaccine is.
Highlights.
Conserved epitopes on the influenza HA protein offer broad protection
Antibodies specific for these conserved epitopes are induced by exposure to antigenically distinct strains
Immune history directly determines the likelihood of generating a broadly protective response
Current vaccine trials are underway to harness this response with the realistic goal of eradicating seasonal influenza strains
Acknowledgments
We thank Charles L. Dulberger for critical help with the figures. This project was funded in parts from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under CEIRS contract HHSN272201400006C, and grant numbers: U19AI109946-01, U19AI082724, P01AI097092-03, and U19AI057266-11.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
• of special interest
•• of outstanding interest
- 1.World Health Organization. Influenza (seasonal) fact sheet. 2014;2014 [Google Scholar]
- 2.Palese MLSaP. Orthomyxoviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Lippincott Williams and Wilkins; Philadelphia: 2013. [Google Scholar]
- 3.de Jong JC, Beyer WE, Palache AM, Rimmelzwaan GF, Osterhaus AD. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J Med Virol. 2000;61:94–99. [PubMed] [Google Scholar]
- 4.Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–736. doi: 10.1126/science.1178258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krystal M, Elliott RM, Benz EW, Jr, Young JF, Palese P. Evolution of influenza A and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci USA. 1982;79:4800–4804. doi: 10.1073/pnas.79.15.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 11.Krammer F. The Quest for a Universal Flu Vaccine: Headless HA 2. 0. Cell Host Microbe. 2015;18:395–397. doi: 10.1016/j.chom.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Laver WG, Valentine RC. Morphology of the isolated hemagglutinin and neuraminidase subunits of influenza virus. Virology. 1969;38:105–119. doi: 10.1016/0042-6822(69)90132-9. [DOI] [PubMed] [Google Scholar]
- 13.Temoltzin-Palacios F, Thomas DB. Modulation of immunodominant sites in influenza hemagglutinin compromise antigenic variation and select receptor-binding variant viruses. J Exp Med. 1994;179:1719–1724. doi: 10.1084/jem.179.5.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 15.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 16.Skehel JJ, Bayley PM, Brown EB, Martin SR, Waterfield MD, White JM, Wilson IA, Wiley DC. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 18.Seto JT, Rott R. Functional significance of sialidose during influenza virus multiplication. Virology. 1966;30:731–737. doi: 10.1016/0042-6822(66)90178-4. [DOI] [PubMed] [Google Scholar]
- 19.Marcelin G, Sandbulte MR, Webby RJ. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol. 2012;22:267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen HT, Sheu TG, Mishin VP, Klimov AI, Gubareva LV. Assessment of pandemic and seasonal influenza A (H1N1) virus susceptibility to neuraminidase inhibitors in three enzyme activity inhibition assays. Antimicrob Agents Chemother. 2010;54:3671–3677. doi: 10.1128/AAC.00581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westgeest KB, Bestebroer TM, Spronken MI, Gao J, Couzens L, Osterhaus AD, Eichelberger M, Fouchier RA, de Graaf M. Optimization of an enzyme-linked lectin assay suitable for rapid antigenic characterization of the neuraminidase of human influenza A(H3N2) viruses. J Virol Methods. 2015;217:55–63. doi: 10.1016/j.jviromet.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Air GM. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci USA. 1981;78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, Tong S, Paulson JC, Wilson IA. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci USA. 2013;110:1458–1463. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology. 1990;175:59–68. doi: 10.1016/0042-6822(90)90186-u. [DOI] [PubMed] [Google Scholar]
- 27.Kaur K, Sullivan M, Wilson PC. Targeting B cell responses in universal influenza vaccine design. Trends Immunol. 2011;32:524–531. doi: 10.1016/j.it.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 29.Air GM. Influenza neuraminidase. Influenza Other Respir Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, Wakiyama M, Shirouzu M, Okada J, Okuno Y, et al. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol. 2014;88:7130–7144. doi: 10.1128/JVI.00420-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE, Jr, Wilson IA, Basler CF. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PloS Pathog. 2012;8:e1003067. doi: 10.1371/journal.ppat.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE, Jr, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181–193. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci USA. 2012;109:17040–17045. doi: 10.1073/pnas.1212371109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 2009;5:e1000350. doi: 10.1371/journal.ppat.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE., Jr A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol. 2011;85:10905–10908. doi: 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. Viral receptor-binding site antibodies with diverse germline origins. Cell. 2015;161:1026–1034. doi: 10.1016/j.cell.2015.04.028. Schmidt et al., identify the signature motif for the receptor-binding site-specific antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 45•.Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, Huang M, Qu X, Huang Y, Salgado-Ferrer M, et al. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest. 2015;125:1255–1268. doi: 10.1172/JCI74374. The authors show that preexisting cross-reactive immunity offers protection against novel emerging influenza strains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci USA. 2014;111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, et al. Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci USA. 2012;109:9047–9052. doi: 10.1073/pnas.1118979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, Palese P. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasugi M, Kubota-Koketsu R, Yamashita A, Kawashita N, Du A, Sasaki T, Nishimura M, Misaki R, Kuhara M, Boonsathorn N, et al. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog. 2013;9:e1003150. doi: 10.1371/journal.ppat.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, et al. Neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies offer protection in vivo. Cell Host Microbe. 2016 doi: 10.1016/j.chom.2016.05.014. In press. This paper identifies protective non-neutralizing antibodies from vaccinated individuals. These antibodies target conserved epitopes on the HA protein and rely on Fc-FcγR interactions to mediate protection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Tan GS, Leon PE, Albrecht RA, Margine I, Hirsh A, Bahl J, Krammer F. Broadly-reactive neutralizing and non-neutralizing antibodies directed against 2 the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 2016 doi: 10.1371/journal.ppat.1005578. In press. Tan et al., identify protective non-neutralizing antibodies in mice following H7 HA immunization that bind at the interface between the head and the stalk domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, et al. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol. 2013;190:1837–1848. doi: 10.4049/jimmunol.1201574. [DOI] [PubMed] [Google Scholar]
- 54.Terajima M, Co MD, Cruz J, Ennis FA. High Antibody-Dependent Cellular Cytotoxicity Antibody Titers to H5N1 and H7N9 Avian Influenza A Viruses in Healthy US Adults and Older Children. J Infect Dis. 2015;212:1052–1060. doi: 10.1093/infdis/jiv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krammer F, Jul-Larsen A, Margine I, Hirsh A, Sjursen H, Zambon M, Cox RJ. An H7N1 influenza virus vaccine induces broadly reactive antibody responses against H7N9 in humans. Clin Vaccine Immunol. 2014;21:1153–1163. doi: 10.1128/CVI.00272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, et al. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One. 2011;6:e25797. doi: 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, Palucka K, Garcia-Sastre A, Palese P, Treanor JJ, et al. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J Virol. 2013;87:4728–4737. doi: 10.1128/JVI.03509-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller MS, Tsibane T, Krammer F, Hai R, Rahmat S, Basler CF, Palese P. 1976 and 2009 H1N1 influenza virus vaccines boost anti-hemagglutinin stalk antibodies in humans. J Infect Dis. 2013;207:98–105. doi: 10.1093/infdis/jis652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sui J, Sheehan J, Hwang WC, Bankston LA, Burchett SK, Huang CY, Liddington RC, Beigel JH, Marasco WA. Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis. 2011;52:1003–1009. doi: 10.1093/cid/cir121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sangster MY, Baer J, Santiago FW, Fitzgerald T, Ilyushina NA, Sundararajan A, Henn AD, Krammer F, Yang H, Luke CJ, et al. B cell response and hemagglutinin stalk-reactive antibody production in different age cohorts following 2009 H1N1 influenza virus vaccination. Clin Vaccine Immunol. 2013;20:867–876. doi: 10.1128/CVI.00735-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomson CA, Wang Y, Jackson LM, Olson M, Wang W, Liavonchanka A, Keleta L, Silva V, Diederich S, Jones RB, et al. Pandemic H1N1 Influenza Infection and Vaccination in Humans Induces Cross-Protective Antibodies that Target the Hemagglutinin Stem. Front Immunol. 2012;3:87. doi: 10.3389/fimmu.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, et al. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci USA. 2014;111:13133–13138. doi: 10.1073/pnas.1414070111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guan Y, Vijaykrishna D, Bahl J, Zhu H, Wang J, Smith GJ. The emergence of pandemic influenza viruses. Protein Cell. 2010;1:9–13. doi: 10.1007/s13238-010-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kilbourne ED, Cerini CP, Khan MW, Mitchell JW, Jr, Ogra PL. Immunologic response to the influenza virus neuraminidase is influenced by prior experience with the associated viral hemagglutinin. I. Studies in human vaccinees. J Immunol. 1987;138:3010–3013. [PubMed] [Google Scholar]
- 66.Palese P, Wang TT. Why do influenza virus subtypes die out? A hypothesis. MBio. 2011:2. doi: 10.1128/mBio.00150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pica N, Hai R, Krammer F, Wang TT, Maamary J, Eggink D, Tan GS, Krause JC, Moran T, Stein CR, et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci USA. 2012;109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Reperant LA, Kuiken T, Osterhaus AD. Adaptive pathways of zoonotic influenza viruses: from exposure to establishment in humans. Vaccine. 2012;30:4419–4434. doi: 10.1016/j.vaccine.2012.04.049. Ellebedy et al., report that antibodies and memory B cells directed against the HA stalk domain are prevalent in humans and that H5N1 vaccination induces broadly-reactive stalk antibodies. [DOI] [PubMed] [Google Scholar]
- 69••.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med. 2015;7:316ra192. doi: 10.1126/scitranslmed.aad0522. The study shows that preexisting head antibodies are immunodominant, preventing clear access to the stalk and that stalk-reactive antibodies are couterselected due to polyreactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halliley JL, Khurana S, Krammer F, Fitzgerald T, Coyle EM, Chung KY, Baker SF, Yang H, Martinez-Sobrido L, Treanor JJ, et al. High-Affinity H7 Head and Stalk Domain-Specific Antibody Responses to an Inactivated Influenza H7N7 Vaccine After Priming With Live Attenuated Influenza Vaccine. J Infect Dis. 2015 doi: 10.1093/infdis/jiv210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nachbagauer R, Choi A, Izikson R, Cox MM, Palese P, Krammer F. Age Dependence and Isotype Specificity of Influenza Virus Hemagglutinin Stalk-Reactive Antibodies in Humans. MBio. 2016:7. doi: 10.1128/mBio.01996-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med. 2013;210:1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller MS, Gardner TJ, Krammer F, Aguado LC, Tortorella D, Basler CF, Palese P. Neutralizing antibodies against previously encountered influenza virus strains increase over time: a longitudinal analysis. Sci Transl Med. 2013;5:198ra107. doi: 10.1126/scitranslmed.3006637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graves PN, Schulman JL, Young JF, Palese P. Preparation of influenza virus subviral particles lacking the HA1 subunit of hemagglutinin: unmasking of cross-reactive HA2 determinants. Virology. 1983;126:106–116. doi: 10.1016/0042-6822(83)90465-8. [DOI] [PubMed] [Google Scholar]
- 75.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, Palese P. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio. 2010:1. doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, et al. Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci USA. 2013;110:4592–4597. doi: 10.1073/pnas.1214913110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henry Dunand CJ, Wilson PC. Restricted, canonical, stereotyped and convergent immunoglobulin responses. Philos Trans R Soc Lond B Biol Sci. 2015:370. doi: 10.1098/rstb.2014.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78••.Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang CY, Cadwell G, et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PloS Pathog. 2014;10:e1004103. doi: 10.1371/journal.ppat.1004103. The authors identify the key residues used by VH1-69 stalk-reactive antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whittle JR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J Virol. 2014;88:4047–4057. doi: 10.1128/JVI.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mouquet H, Warncke M, Scheid JF, Seaman MS, Nussenzweig MC. Enhanced HIV-1 neutralization by antibody heteroligation. Proc Natl Acad Sci USA. 2012;109:875–880. doi: 10.1073/pnas.1120059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 83.Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol. 2013;3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol. 2013;87:6542–6550. doi: 10.1128/JVI.00641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, Garcia-Sastre A, et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J Virol. 2013;87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Babu TM, Levine M, Fitzgerald T, Luke C, Sangster MY, Jin H, Topham D, Katz J, Treanor J, Subbarao K. Live attenuated H7N7 influenza vaccine primes for a vigorous antibody response to inactivated H7N7 influenza vaccine. Vaccine. 2014;32:6798–6804. doi: 10.1016/j.vaccine.2014.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goff PH, Eggink D, Seibert CW, Hai R, Martinez-Gil L, Krammer F, Palese P. Adjuvants and immunization strategies to induce influenza virus hemagglutinin stalk antibodies. PLoS One. 2013;8:e79194. doi: 10.1371/journal.pone.0079194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krammer F. Novel universal influenza virus vaccine approaches. Curr Opin Virol. 2016;17:95–103. doi: 10.1016/j.coviro.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89••.Tran EE, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, et al. Cryo-electron Microscopy Structures of Chimeric Hemagglutinin Displayed on a Universal Influenza Vaccine Candidate. MBio. 2016:7. doi: 10.1128/mBio.00257-16. This paper reports the first three-dimensional (3D) visualization of chimeric hemagglutinin proteins displayed on the surface of the influenza virus, in the context of a candidate universal influenza vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]