Abstract
Precise regulation of flowering time is critical for plant reproductive success and, in cereals, to maximize grain yields. Seasonal cues including temperature and day length are integrated to regulate the timing of flowering. In temperate cereals, extended periods of cold (vernalization) release the repression of FLOWERING LOCUS T1 (FT1), which is upregulated in the leaves in response to inductive long-day photoperiods. FT1 is a homolog of rice HD3a, which encodes a protein transported from leaves to the shoot apical meristem to induce flowering. A rare FT-B1 allele from the wheat variety “Hope” has been previously shown to be associated with an early flowering phenotype under long-day photoperiods. Here, we demonstrate that the Hope FT-B1 allele accelerates flowering even under short days, and that it is epistatic to the VERNALIZATION 1 (VRN1) gene. On average, the introgression of Hope FT-B1 into 6 genetic backgrounds resulted in 2.6 days acceleration of flowering (P < 0.0001) and 4.1% increase in spike weight (P = 0.0093), although in one variety, it was associated with a decrease in spike weight. These results suggest that the Hope FT-B1 allele could be useful in wheat breeding programs to subtly accelerate floral development and increase adaptation to changing environments.
Subject areas: Gene action, regulation, transmission
Keywords: flowering, FT, Triticum, VRN3, wheat
In temperate cereals such as wheat (Triticum spp.), the precise control of flowering time is critical to ensure that the plant’s reproductive development coincides with optimal environmental conditions, thus maximizing grain yields. Several genes with important roles in regulating wheat flowering development in response to long exposures to cold temperatures (vernalization) or changes in day length (photoperiod) have been cloned and characterized, including VERNALIZATION 1 (VRN1) (Danyluk et al. 2003; Trevaskis et al. 2003; Yan et al. 2003), VRN2 (Yan et al. 2004b), VRN3 (Yan et al. 2006), and PHOTOPERIOD 1 (PPD1) (Turner et al. 2005; Beales et al. 2007).
VRN1 is a homologue of the Arabidopsis floral meristem identity gene APETALA1 (AP1) and is central to the vernalization response in temperate cereals. In varieties with a winter growth habit, vernalization induces the gradual increase of VRN1 transcript levels and results in competence to flower (Danyluk et al. 2003; Trevaskis et al. 2003; Yan et al. 2003). Large deletions and insertions in the regulatory regions of VRN1 reduce or eliminate vernalization requirement in wheat and barley and result in a spring growth habit (Yan et al. 2004a; Fu et al. 2005; Hemming et al. 2009).
It was recently demonstrated that a critical role of VRN1 is to repress the flowering repressor VRN2 in the leaves during the spring (Chen and Dubcovsky 2012). The VRN2 locus consists of duplicated genes encoding CCT-domain proteins (ZCCT1 and ZCCT2) and simultaneous deletion of, or nonfunctional mutation in, both copies is sufficient to confer a spring growth habit (Yan et al. 2004b). Under long days (LD), VRN2 suppresses the induction of VRN3, a gene encoding a phosphatidylethanolamine-binding protein (PEBP) structurally and functionally similar to FLOWERING LOCUS T (FT) in Arabidopsis (Yan et al. 2006). This gene, henceforth referred to as FT1, is one of at least 6 FT-like paralogs which have been identified in cereals (Faure et al. 2007; Lv et al. 2014). Of these, FT1 shows the most robust induction of flowering when transformed into rice and wheat (Yan et al. 2006; Kikuchi et al. 2009).
Before vernalization, the induction of FT1 by LD photo-period is blocked by VRN2. However, following the vernalization-induced upregulation of VRN1 and its subsequent repression of VRN2, FT1 is upregulated inducing flowering in the spring. Under LD, the induction of FT1 is mediated by the photoperiod gene PPD1 (Turner et al. 2005; Beales et al. 2007).
The FT1 ortholog in rice, HEADING DATE 3a (Hd3a), has been demonstrated to act as a graft-transmissible mobile signal, which is transported via the phloem to the shoot apical meristem (Tamaki et al. 2007). After transport to the apex, HD3a is incorporated into a floral activation complex which also comprises FD and 14-3-3 linker proteins. This complex binds to the promoters of target genes such as OsMADS15 (rice ortholog of Arabidopsis AP1), thus activating their expression and inducing the irreversible transition from vegetative to reproductive growth of the shoot apical meristem (Taoka et al. 2011). In wheat, the interaction between FT1 and FDL-2 and the binding of FDL-2 to the VRN1 promoter in vitro have also been demonstrated, suggesting that this mechanism is conserved among grass species (Li and Dubcovsky 2008).
A large number of natural VRN1 alleles have been described, and many have been incorporated into wheat breeding programs to alter both growth habit and flowering time (Yan et al. 2004a; Fu et al. 2005; Chu et al. 2011; Diaz et al. 2012). In contrast, limited natural variation has been described for the wheat FT1 gene. In barley, several haplotypes have been identified, and a dominant FT1 allele for early flowering has been shown to be associated with increased copy number (Nitcher et al. 2013). In wheat, sequence variation has been reported for FT-A1 (a SNP resulting in an amino acid substitution in exon 3) and FT-D1 (a single base pair insertion-deletion in the third exon), which results in small variations in flowering time for both loci (Bonnin et al. 2008). In FT-B1, an allele dominant for spring growth habit has been identified in the Triticum aestivum variety “Hope,” derived from a cross between the wild emmer “Yaraslov” (Triticum dicoccum) and the Canadian T. aestivum variety “Marquis” (Clark et al. 1926; McFadden 1930).
The Hope FT-B1 allele is defined by the insertion of a 5295 bp retrotransposon element in the promoter region and is associated with early flowering under LD photoperiods (Yan et al. 2006). This association was confirmed by the transformation of the winter hexaploid variety “Jagger” with the Hope FT-B1 allele, which resulted in greatly increased FT1 transcript levels and acceleration of flowering even in the absence of vernalization (Yan et al. 2006). The FT-B1 allele with the repetitive element insertion is not common among modern tetraploid durum and hexaploid bread wheat varieties. The narrow distribution of this allele and the lack of information regarding its effects on heading date and agronomic performance in different genetic backgrounds under field conditions have limited its utilization in wheat breeding programs.
In the current study, we demonstrate that the Hope FT-B1 allele is capable of accelerating flowering in a photoperiod-sensitive winter wheat variety even under short-day (SD) photoperiod, and that it is epistatic to VRN1 under both greenhouse and field environments. Using 6 pairs of near-isogenic sister lines with and without the Hope FT-B1 allele, we show that this allele significantly reduces days to heading (DTH) in the field in some varieties, and that in general, it has no significant negative effects on yield components, demonstrating its potential utility as a tool to adjust flowering time in wheat breeding programs.
Materials and Methods
Materials and Growing Conditions
Two chromosome substitution lines in the T. aestivum “Chinese Spring” (CS) background were used in this study: CS (Hope 7B) (Kuspira and Unrau 1957) and CS (Cheyenne 5D) (Morris et al. 1966). In CS (Hope 7B), CS chromosome 7B, carrying the wild-type FT-B1 allele, was replaced by chromosome 7B from “Hope,” which carries the highly expressed FT-B1 allele with a retroelement insertion in the promoter. In CS (Cheyenne 5D), CS chromosome 5D carrying the Vrn-D1 allele (spring growth habit) was replaced by Cheyenne chromosome 5D, which carries the vrn-D1 allele (winter growth habit). Since the Vrn-D1 allele is the only VRN1 allele for spring growth habit in CS, the CS (Cheyenne 5D) plants have a winter growth habit.
The substitution lines CS (Hope 7B) and CS (Cheyenne 5D) were crossed to produce a population segregating for Hope FT-B1 and Vrn-D1. Except for the segregating chromosomes 7B and 5D, the genetic background of these lines is expected to be identical. F2 individuals were genotyped for these 2 genes (see below), and those with different combinations of homozygous alleles were selected to produce F3 families for further evaluations. The first experiment included 29 F3 families (N = 109), representing the 4 genotypic classes expected from the segregation of these 2 genes, and the second experiment included derived F4 lines (N = 111). Plants were grown in a greenhouse under nonvernalizing, LD controlled conditions, and were periodically randomized. DTH was recorded as the number of days from germination to heading.
Transgenic plants obtained from the bombardment of the photoperiod-sensitive winter variety “Jagger” with a genomic copy of the Hope FT-B1 allele including its natural promoter (henceforth referred to as transgenic FT1HOPE lines) were described before (Yan et al. 2006). Transgenic and wild-type Jagger were compared under SD (8 h light/16 h dark) and LD (16 h light/8 h dark) photoperiods at 16 °C with 16% humidity in Conviron CMP3244 growth chambers (Conviron, Pembina, ND). Seeds were germinated in Petri dishes, incubated at 4 °C for 24 h, and then transferred to 25 °C for 48 h before transplanting into 6-inch pots in the growth chambers. These transgenic plants showed segregation for FT1 expression level, which is likely due to the segregation of multiple functional and nonfunctional transgene insertion events during bombardment. For this experiment, we selected 7 lines grown under LD and 11 lines grown under SD carrying the transgene (confirmed by PCR) and exhibiting FT1 expression greater than or equal to 2 times the average expression of the Jagger lines in each respective photoperiod.
To determine the effect of the Hope FT-B1 allele on heading time and yield components in the field, the CS (Hope 7B) chromosome substitution line was used as a donor to introgress the Hope FT-B1 allele into 1 tetraploid (UC1113) and 4 hexaploid varieties (UC1041, UC1110, Kern, and Clear White), all with a spring growth habit. The recurrent parents differ in the combinations of VRN1 alleles responsible for their spring growth habit. The tetraploid breeding line UC1113 (Chicaiza et al. 2006) carries the Vrn-A1c allele (first intron deletion), whereas the hexaploid breeding line UC1110 (Lowe et al. 2011) carries the Vrn-A1a allele (insertion and duplication in promoter region, Yan et al. 2004a). The bread wheat variety Clear White (PI 635044) carries Vrn-B1a and Vrn-D1a alleles for spring growth habit (both with first intron deletions, Fu et al. 2005), whereas an isogenic sister line designated as Clear White-vrn-B1 carries only the Vrn-D1a allele. The variety Kern (PI 612142) and the breeding line UC1041 (Uauy et al. 2009) also have Vrn-D1a as the only allele for spring growth habit (Fu et al. 2005). The Hope FT-B1 allele was introgressed into these 6 lines using marker-assisted backcrossing. After 4 generations of backcrossing, near-isogenic sister lines were selected, which are expected to be approximately 97% identical.
The BC4F2 lines were grown in the greenhouse and lines homozygous for the different VRN1 genotypes and for wild-type or Hope FT-B1 alleles were selected (see FT-B1 and VRN1 Genotyping section below). Selected BC4F3 (2011–2012 growing season) and BC4F4 (2012–2013 growing season) sister lines were grown in single rows at the University of California, Davis, CA (38°32′N, 121°46′W). Both experiments were planted in a Split-Plot Randomized Complete Block Design (10 blocks). These rows were cultivated following typical growing practices for the region as previously described (Brevis and Dubcovsky 2010). DTH was recorded as number of days from sowing to heading. We also recorded yield components including grains per spike (GpS), thousand kernel weight (TKW), spikelets per spike (SpS), and weight of the grain per spike (SW). The number of GpS was determined as the average of 5 spikes harvested randomly per row. SW was estimated from GpS and TKW values. Following the second field season, samples of each line (BC4F5 seed) were submitted to the National Small Grains Collection for use as germplasm. The PI numbers are as follows: UC1113 + Hope FT-B1, PI 671995; UC1113, PI 638741; UC1041 + Hope FT-B1, PI 671996; UC1041, PI 671997; UC1110 + Hope FT-B1, PI 671998; UC1110, PI 671999; Kern + Hope FT-B1, PI 672000; Kern, PI 672001; Clear White + vrn-B1 + Hope FT-B1, PI 672002; Clear White + vrn-B1, PI 672003; Clear White + Hope FT-B1, PI 672004; Clear White, PI 672005.
FT-B1 and VRN1 Genotyping
Wild-type and Hope FT-B1 alleles were assessed in field lines using the codominant marker described previously (Tsp509I assay, Yan et al. 2006). Transgenic FT1HOPE lines and CS populations were characterized using allele-specific (dominant) FT-B1 PCR markers (Yan et al. 2006), to detect the presence and absence of the retrotransposon insertion in the promoter region.
Allelic variants in the promoter of hexaploid Vrn-A1a were determined with a codominant marker, using a combination of primers developed in this and previous studies (Yan et al. 2004a) (Table 1). Two complementary sets of allele-specific primers were used to develop a codominant marker for the first intron deletion present in the Vrn-A1c allele. Specifically, the absence of the deletion in the wild-type allele was tested with primers described by Fu et al. (2005), while new primers were designed to detect the presence of the intron deletion (Table 1). Similarly, 2 pairs of complementary allele-specific primers were used to characterize the intron 1 deletions present in the Vrn-B1a and Vrn-D1a alleles (Fu et al. 2005) (Table 1).
Table 1.
Primers used for genotyping VRN1 and FT1 alleles
| Locus | Forward primer | Reverse primer | Product size (bp) spring/winter |
|---|---|---|---|
| Vrn-A1a promotera | TGAAAGGAAAAATTCTGCTCGT | GCAGGAAATCGAAATCGAAb | 945/713 |
| Vrn-A1c first intron (deletion) c | TTTGGTAGATTCCGTTTGCC | TGTTGGGAAAATAGAGTGACCC | 942/NA |
| vrn-A1 first intron (WT) d | GCACTCCTAACCCACTAACC | TCATCCATCATCAAGGCAAA | NA/1068 |
| Vrn-B1a first intron (deletion) d | CAAGTGGAACGGTTAGGACA | CTCATGCCAAAAATTGAAGATGA | 709/NA |
| vrn-B1 first intron (WT) d | CAAGTGGAACGGTTAGGACA | CAAATGAAAAGGAATGAGAGCA | NA/1149 |
| Vrn-D1a (deletion)d | GTTGTCTGCCTCATCAAATCC | GGTCACTGGTGGTCTGTGC | 1671/NA |
| vrn-D1 (WT)d | GTTGTCTGCCTCATCAAATCC | AAATGAAAAGGAACGAGAGCG | NA/997 |
| FT-B1 Hopee | CATAATGCCAAGCCGGTGAGTAC | ATGTCTGCCAATTAGCTAGC | 1200/NA |
| FT-B1 WTe | ATGCTTTCGCTTGCCATCC | CTATCCCTACCGGCCATTAG | NA/1140 |
| FT-B1e,f | TCCTAGCAAGCACACGAGTAA | CGTGCACATCATGATCAAAG | 120/154 |
NA, no amplification; WT, wild type.
Annealing temperature 60 °C.
Annealing temperature 56 °C.
Digest by Tsp509I.
Statistical Analysis
Data from the greenhouse experiments were analyzed with a 2-way factorial Anova including FT1 and VRN1 alleles, while data from field experiments were analyzed with a 3-way factorial Anova including FT1 alleles, variety, and year. Type III sum of squares and Least Squares means were used to compensate for the unequal number of plants in the different genotyping classes. Means were compared using Tukey’s test. Due to the presence of significant interactions in the field experiments, DTH was also tested separately by variety and yield components were tested separately by year and variety. All statistical analyses were performed with SAS version 9.1 (SAS Institute, Cary, NC).
Quantitative Reverse Transcription–PCR
Leaf tissue from Jagger and transgenic FT1HOPE plants was harvested (SD at 5 weeks and 14 weeks and LD at 5 weeks) and immediately frozen in liquid nitrogen before grinding into a fine powder. Tissue was harvested from the youngest, fully deployed leaf or from the flag leaf of flowering plants at 10 a.m. (4 h of light). Total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich, St Louis, MO) and 1μg of the RNA equivalent of cDNA was synthesized using High Capacity cDNA Reverse Transcription Kit with RNAse Inhibitor (Applied Biosystems, Foster City, CA). Quantitative reverse transcription–PCR (qRT – PCR) reactions were performed in 20 μl final volume including 1× USBR VeriQuest SYBRR Green qPCR Master Mix (Affymetrix, Santa Clara, CA) 0.5 μM of F and R primers and 10 ng of cDNA. qRT–PCR was carried out using an Applied Biosystems 7500 Fast Real-Time PCR System machine (Applied Biosystems). ACTIN was used as an endogenous control. Primers for FT1 (Shimada et al. 2009) and ACTIN (Fu et al. 2007) have been described previously. Transcript levels for all genes and experiments presented in this study are expressed as linearized fold-ACTIN levels calculated by the formula 2(ACTIN CT − TARGET CT) ± standard error. The resulting number indicates the ratio between the initial number of molecules of the target gene and the number of molecules of ACTIN and therefore, the Y scales are comparable across genes and experiments.
Results
Characterization of the Hope FT-B1 Allele Under Different Photoperiods
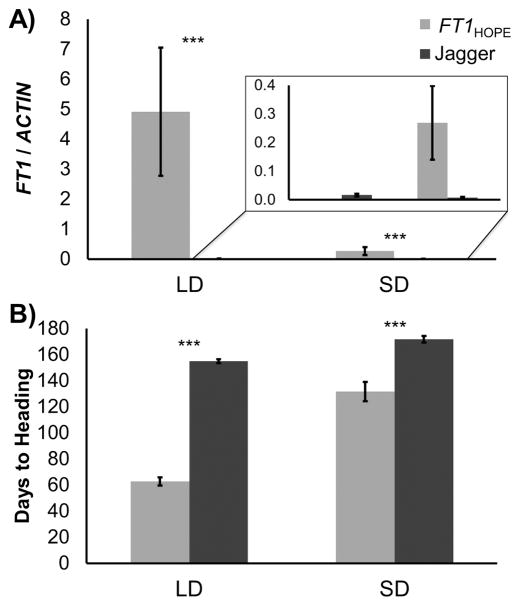
To analyze the effect of the Hope FT-B1 allele on heading time under different photoperiods, we compared wild-type Jagger with transgenic FT1HOPE lines in controlled growth chamber experiments under SD and LD photoperiods. Under LD conditions, FT1 transcript levels were low or undetectable in 5-week-old Jagger plants (average fold-ACTIN = 0.016 ± 0.004, Figure 1A). This is an expected result for unvernalized plants from a winter variety and is consistent with previous observations (Yan et al. 2006). By contrast, FT1 transcript levels were almost 5-fold higher than ACTIN (4.92 ± 2.14, Figure 1A) in the 7 transgenic plants selected for the presence of the active FT1HOPE transgene and grown under LD photoperiod. These plants headed significantly earlier (62.7 ± 3.2 days) than the wild-type Jagger plants (155 ± 1.5 days, P < 0.0001, Figure 1B). This result confirms the association between increased FT1 expression and the acceleration in flowering time under LD.
Figure 1.
(A) FT1 transcript levels (fold-ACTIN ± standard error) of 7–10 biological replicates at flowering (5 weeks LD, 14 weeks SD) of transgenic FT1HOPE and wild-type Jagger lines. (B) DTH in FT1HOPE transgenic plants and wild-type Jagger under LD and SD photoperiods. ***P ≤ 0.0001, comparison of FT1HOPE phenotype to wild-type Jagger phenotype in designated photoperiod.
Jagger plants flowered significantly later under SD (171 days) than they did under LD (155 days, P < 0.0001, Figure 1B). As expected for a photoperiod-sensitive line grown under SD, FT1 transcript levels of 5-week-old Jagger plants were almost undetectable. In transgenic Hope FT-B1 plants of the same age, we also found very low levels of FT1 expression, so we repeated the expression analysis in 14-week-old plants to match the developmental stage of the LD samples (initiation of heading). The 11 transgenic plants selected for the presence of the active FT1HOPE allele grown in a SD photoperiod showed FT1 transcript levels 38-fold higher (0.269 ± 0.129-fold-ACTIN) than those observed in the nontransgenic Jagger plants (0.007 ± 0.003-fold-ACTIN). The increased FT1 transcript levels in the transgenic plants was associated with a significantly earlier (P = 0.0003) flowering time (131.6 ± 7.5 days, Figure 1B) than that observed for the Jagger wild-type plants (171.75 ± 2.6 days). This result demonstrates that the Hope FT-B1 allele is able to accelerate flowering in a photoperiod-sensitive genotype under noninductive SD photoperiod. However, FT1 transcript levels detected in the transgenic plants grown under SD were lower (P = 0.073) than those observed in the transgenic plants grown under LD, which was also reflected in later flowering under SD than under LD (P < 0.0001, Figure 1B).
Epistatic Interactions Between VRN-D1 and FT-B1
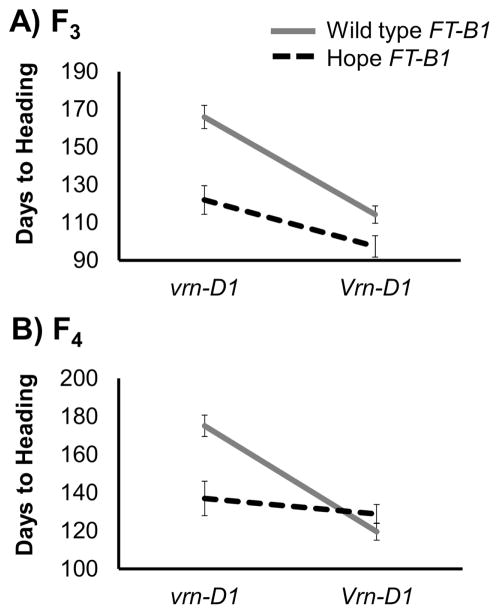
To better understand the effect of the Hope FT-B1 allele in spring and winter backgrounds, we analyzed the epi-static interactions between FT-B1 and VRN-D1. In F3 families derived from the cross between chromosome substitution lines CS (Hope 7B) and CS (Cheyenne 5D), which were homozygous for different VRN-D1/FT-B1 allelic combinations, we observed a significant interaction between these 2 loci (P = 0.028, Figure 2A). This interaction, validated in an independent experiment performed with derived F4 lines (P = 0.0003, Figure 2B), indicates that the effect of each gene is affected by the allele present in the other gene. Therefore, we describe the simple effects of each gene within each of the classes of the other gene. The Hope FT-B1 allele significantly accelerated flowering in the winter vrn-D1 background, both in the F3 (Hope FT-B1 44 days earlier, P < 0.0001) and F4 populations (Hope FT-B1 38.2 days earlier, P = 0.003). However, the effect was not significant within the plants carrying the Vrn-D1 allele (spring growth habit) either in the F3 (Hope FT-B1 16.9 days earlier, P = 0.104) or F4 populations (Hope FT-B1 9.3 days later, P = 0.508). The reciprocal analysis of VRN1 effects showed that the difference in heading time between the plants carrying the Vrn-D1 and vrn-D1 alleles was significant within the wild-type FT-B1 class (52 days F3, 56 da 4, P ysF < 0.0001) but not within the Hope FT-B1 class (25 days F3, 9 da 4, ysF P > 0.05, Figure 2). Taken together, these results indicate that the Hope FT-B1 and Vrn-D1 alleles for early flowering are epistatic to the alleles for late flowering in the other gene.
Figure 2.
Interaction graphs for heading time in the CS (Cheyenne 5D) × CS (Hope 7B) (A) F3 and (B) F4 populations segregating for both FT-B1 and VRN-D1. Nonparallel lines indicate a strong interaction (nonadditive effects).
Effect of the Hope FT-B1 Allele on Heading Time and Yield Components Under Field Conditions
Using 6 pairs of near-isogenic sister lines for the wild-type and Hope FT-B1 alleles, we detected significant differences in heading time that were consistent across years (Table 2) and small differences in yield components that were significant only for one of the 2 years (Table 3). Plants carrying the Hope FT-B1 allele flowered significantly earlier than the sister lines carrying the wild-type FT-B1 (P < 0.01) in both years. No significant interactions were detected between year and the other factors in the statistical model, so the results from both years were combined (Table 2). On average, plants carrying the Hope FT-B1 allele flowered 2.6 days earlier than those carrying the wild-type allele (P < 0.0001, Table 2). A significant interaction was detected between genotype and FT-B1 (P = 0.0003), so we also tested the effect of this locus on heading date separately for each genotype. Plants carrying the Hope FT-B1 allele flowered 4–5 days earlier than those carrying the wild-type allele and these differences were highly significant in UC1113, Clear White-vrn-B1, and Kern, and marginally nonsignificant in UC1110 (P = 0.057). The last 2 varieties, UC1041 and Clear White, showed less than 1 day difference in heading time between sib-lines with and without the Hope FT-B1 allele, and the differences were not significant (P > 0.56, Table 2).
Table 2.
Effect of FT-B1 alleles on average heading time in 6 pairs of near-isogenic sister lines
| FT-B1 | DTH | P value | ||
|---|---|---|---|---|
| FT-B1 Effect across line | WT | 139.72 | <0.0001 | |
| Hope | 137.13 | |||
|
| ||||
| Line by FT-B1 interaction | 0.0003 | |||
|
| ||||
| Line | VRN1 | FT-B1 | DTH | P value |
|
| ||||
| UC1113 (tetraploid) | Vrn-A1c | WT | 149.80 | 0.0005 |
| Hope | 145.80 | |||
| UC1041 (hexaploid) | Vrn-D1a | WT | 137.60 | 0.6970 |
| Hope | 138.05 | |||
| UC1110 (hexaploid) | Vrn-A1c (promoter) | WT | 137.15 | 0.0571 |
| Hope | 133.58 | |||
| Kern (hexaploid) | Vrn-D1a | WT | 138.10 | <0.0001 |
| Hope | 134.40 | |||
| Clear White-vrn-B1 (hexaploid) | Vrn-D1a | WT | 139.25 | <0.0001 |
| Hope | 134.65 | |||
| Clear White (hexaploid) | Vrn-B1a/D1a | WT | 135.56 | 0.5687 |
| Hope | 135.94 | |||
P values indicate the significance of the effects over 2 years of field experiments using a 3-way factorial Anova. Values are also presented separately by line due to the presence of a significant interaction between the FT-B1 locus and the line. WT, wild type.
Table 3.
Effect of FT-B1 alleles on yield components in 6 pairs of near-isogenic sister lines in 2 field experiments
| GpS
|
TKW
|
SpS
|
SW
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FT-B1 | 2012 | 2013 | 2012–2013 | 2012 | 2013 | 2012–2013 | 2012 | 2013 | 2012–2013 | 2012 | 2013 | 2012–2013 | |
| Across lines | |||||||||||||
| FT-B1 effect across lines | WT | 61.71 | 63.92 | 41.89 | 40.56 | 21.01 | 20.92 | 2.58 | 2.60 | ||||
| Hope | 64.41 | 66.92 | 41.75 | 40.80 | 21.32 | 21.24 | 2.66 | 2.73 | |||||
| P value | 0.1220 | 0.0017 | 0.0036 | 0.7945 | 0.6867 | 0.7611 | 0.2844 | 0.0865 | 0.0733 | 0.2002 | 0.0021 | 0.0093 | |
| Line by FT-B1 interaction | P value | 0.4069 | 0.0088 | 0.0236 | 0.2291 | 0.0424 | 0.0323 | 0.2973 | <0.0001 | <0.0001 | 0.3642 | 0.0256 | 0.0263 |
| Year by line interaction | P value | 0.0158 | 0.0001 | <0.0001 | 0.0033 | ||||||||
| Year by FT-B1 interaction | P value | 0.8178 | 0.8355 | 0.7275 | 0.5529 | ||||||||
| By line | |||||||||||||
| UC1113 (tetraploid) | WT | 65.56 | 70.00 | 47.11 | 49.77 | 20.26 | 20.58 | 3.48 | 3.09 | ||||
| Hope | 68.48 | 78.22 | 47.03 | 48.17 | 21.22 | 21.96 | 3.77 | 3.21 | |||||
| P value | 0.5690 | 0.0084 | 0.0584 | 0.9580 | 0.0428 | 0.3237 | 0.2100 | 0.0528 | 0.0232 | 0.6314 | 0.0409 | 0.1454 | |
| UC1041 (hexaploid) | WT | 58.16 | 57.14 | 35.15 | 35.45 | 21.98 | 19.52 | 2.02 | 2.05 | ||||
| Hope | 65.50 | 61.74 | 33.21 | 34.16 | 22.72 | 21.22 | 2.10 | 2.16 | |||||
| P value | 0.0394 | 0.0485 | 0.0043 | 0.1904 | 0.3917 | 0.1206 | 0.2167 | 0.0007 | 0.0015 | 0.2672 | 0.4370 | 0.1729 | |
| UC1110 (hexaploid) | WT | 52.91 | 55.18 | 46.39 | 43.14 | 20.29 | 20.14 | 2.40 | 2.47 | ||||
| Hope | 54.18 | 57.68 | 47.47 | 43.51 | 20.91 | 20.48 | 2.51 | 2.55 | |||||
| P value | 0.5358 | 0.2675 | 0.8267 | 0.6349 | 0.9757 | 0.6371 | 0.6537 | 0.3053 | 0.3218 | 0.6739 | 0.3672 | 0.7627 | |
| Kern (hexaploid) | WT | 56.04 | 63.22 | 45.05 | 41.25 | 20.44 | 21.04 | 2.61 | 2.52 | ||||
| Hope | 53.12 | 58.96 | 43.55 | 41.99 | 19.66 | 19.46 | 2.47 | 2.32 | |||||
| P value | 0.4130 | 0.0402 | 0.0778 | 0.0817 | 0.2254 | 0.4375 | 0.1923 | 0.0031 | 0.0027 | 0.2523 | 0.0934 | 0.0794 | |
| Clear White-vrn-B1 (hexaploid) | WT | 66.58 | 66.64 | 38.54 | 36.55 | 21.44 | 21.84 | 2.44 | 2.57 | ||||
| Hope | 70.52 | 71.10 | 39.88 | 39.37 | 21.48 | 22.28 | 2.80 | 2.81 | |||||
| P value | 0.3111 | 0.0784 | 0.0667 | 0.0606 | 0.0298 | 0.0040 | 0.9342 | 0.3730 | 0.4797 | 0.1293 | 0.0054 | 0.0029 | |
| Clear White (hexaploid) | WT | 75.77 | 71.34 | 38.03 | 36.89 | 21.93 | 22.4 | 2.63 | 2.90 | ||||
| Hope | 79.80 | 73.82 | 38.70 | 37.26 | 22.30 | 22.04 | 2.74 | 3.10 | |||||
| P value | 0.4668 | 0.3682 | 0.1997 | 0.2621 | 0.7116 | 0.1623 | 0.4702 | 0.4802 | 0.9600 | 0.4045 | 0.3145 | 0.1577 | |
Averages and P values are presented for the effects over 2 years of field experiments as determined by 3-way factorial Anova, as well as separately by year and line due to the presence of significant interactions. WT, wild type.
To assess the impact of FT-B1 alleles on yield components, we recorded GpS, TKW, and SpS, and we calculated the SW (Table 3). Overall, we observed a significant increase in GpS (P = 0.0036) and SW (P = 0.0093) and a marginally nonsignificant increase in SpS (P = 0.0733). Differences in TKW were not significant. Interestingly, the interaction between FT-B1 alleles and year was not significant for any of the traits (P > 0.55), indicating that the responses were consistent across both years.
For all 4 traits, we found significant interactions between lines and year (GpS: P = 0.0158; TKW: P = 0.0001; SpS: P < 0.0001; SW: P = 0.0033), as well as between line and FT-B1 (2013—GpS: P = 0.0088; TKW: P = 0.0424; SpS: P < 0.0001; SW: P = 0.0256). Therefore, comparisons between the wild-type and Hope FT-B1 alleles are presented separately by year and line (Table 3). An average increase of 4–5% in grain number per spike and of 3–5% in spike weight was observed across years, but when considered separately, the differences were significant only for the second year (2013 GpS: P = 0.0017; 2013 SW: P = 0.0021). Interestingly, this increase in grain number was not associated with a significant decrease in TKW (2012–2013, P = 0.7611; Table 3). The analysis by individual lines showed a heterogeneous response for all 4 traits (significant interactions line × FT-B1 allele; Table 3). Except in the variety Kern, the presence of the FT1 Hope allele was associated with slight increases in GpS, SpS, or SW, which were significant only in 6 of the 30 comparisons (P < 0.05, Table 3). By contrast, Kern showed slightly lower GpS, SpS, and SW values across both years in the sister lines carrying the FT1 Hope allele, a decrease that was significant only in 2 of the 6 comparisons (P < 0.05, Table 3). When the 2 years were combined, Kern showed a significant decrease in SpS (P = 0.0027), whereas UC1113 and UC1041 showed significant increases in this parameter (P = 0.0232 and P = 0.0015 respectively; Table 3). A similar trend was observed for GpS, which is correlated with SpS (Table 3). It is interesting to point out that in Clear White-vrn-B1, the presence of the Hope FT-B1 allele was associated with a highly significant increase in SW, as a result of simultaneous increases in GpS (P = 0.0667) and TKW (P = 0.0040).
Discussion
Hope FT-B1 and Photoperiod
The rare FT-B1 allele identified in the variety Hope offers the opportunity to expand the genetic tools available to breeders to modulate wheat flowering time. This allele was previously shown to be expressed at higher levels than the wild-type allele under LD conditions, resulting in an early flowering phenotype (Yan et al. 2006). In the current study, we demonstrate that the Hope FT-B1 allele is also expressed at higher levels than the wild-type allele under SD and that this is sufficient for the acceleration of flowering under SD. However, FT1 expression was detected at a much later stage of the plant’s development and to a lower level under SD (Figure 1A), and this was correlated with later flowering under SD than under LD (P < 0.0001, Figure 1B). This observation suggests that while the increased expression of the transgenic Hope FT-B1 allele is sufficient to accelerate flowering in wheat, the mechanisms which result in low FT1 transcription during SD are not fully eliminated by the insertion of the retroelement in the promoter region of the Hope FT-B1 allele.
Interaction Between FT-B1 and Other Flowering Loci
It was previously demonstrated in barley that increased FT1 copy number, and the associated earlier expression of FT1, is sufficient to induce flowering of winter barley varieties in the absence of vernalization (Nitcher et al. 2013). Likewise, previous studies have shown that the transgenic Hope FT-B1 allele is able to overcome the repressed state of the VRN1 locus in the winter wheat variety Jagger in the absence of vernalization. The natural Hope FT-B1 allele was also shown to increase FT1 transcript levels in spring lines (Yan et al. 2006) but has not been tested before in winter wheat varieties.
In this study, we found that the natural Hope FT-B1 allele significantly accelerates flowering of plants carrying the vrn-D1 allele for winter growth habit but had no significant effect in plants carrying the dominant Vrn-D1a allele for spring growth habit in the CS background (Figure 2). Similar epistatic interactions have also been reported recently between Hope FT-B1 and the VERNALIZATION 4 (VRN4) gene, where the dominant Vrn-D4 and Hope FT-B1 alleles for early flowering were epistatic to the wild-type alleles for late flowering (Kippes et al. 2014). A possible explanation for these epistatic interactions could be the existence of an upper threshold of FT1 or VRN1 expression beyond which further increases in expression do not result in additional acceleration of flowering.
Taken together, these results suggest that the increased expression of the natural Hope FT-B1 allele is sufficient to overcome the need for vernalization to induce the expression of vrn1 alleles, and that its effects are smaller in plants that have other dominant alleles for spring growth habit. The accelerated reproductive development associated with the presence of the Hope FT-B1 allele in winter varieties under LD and in photoperiod-sensitive varieties under SD may limit the utilization of this allele in winter wheat programs or in those that use photoperiod-sensitive varieties, particularly in regions with long and severe winters.
Effect of the Hope FT-B1 Allele Under Field Conditions
To determine the potential utility of the Hope FT-B1 allele to slightly accelerate flowering in spring wheat breeding programs, we evaluated its effect on heading time and yield components in near-isogenic sister lines in 5 spring hexa-ploid backgrounds and 1 spring tetraploid background under field conditions. Although the overall average effect of the Hope FT-B1 allele was to accelerate flowering by 2.6 days, the responses were heterogeneous among varieties (Table 2).
It is interesting to point out that the Hope FT-B1 allele accelerated flowering relative to the wild type by 4.6 days in the Clear White-vrn-B1 line (P < 0.0001) carrying only Vrn-D1a but had no effect in the Clear White variety carrying both Vrn-B1a and Vrn-D1a. Since these 2 lines are isogenic, the differential effect of the Hope FT-B1 allele on flowering time can be attributed to the additional Vrn-B1a allele. The presence of the additional Vrn-B1a allele in Clear White accelerates heading time by 3.7 days relative to Clear White-vrn-B1 (in FT-B1 wild-type allele background) and may have eliminated any residual vernalization requirement. Following the hypothesis presented above, the resulting VRN1 transcript levels may have exceeded the threshold beyond which further increases have no additional effect on heading time. As in our epistatic experiment, the Hope FT-B1 allele was associated with accelerated heading time only within the late heading class (in this case, Clear White-vrn-B1). In addition to Clear White-vrn-B1, Kern and UC1041 also carry Vrn-D1a as the only Vrn1 allele for spring growth habit. However, whereas Kern shows highly significant differences in heading time between FT-B1 alleles (3.6 days, P < 0.0001), UC1041 showed no significant differences (0.45 days, P = 0.6970). This result suggests that additional genetic factors other than VRN1 can modulate the effects of the FT-B1 alleles. As an example of the complexity of the regulation of this critical flowering gene, posttranscriptional regulation of FT1 by microRNAs has been recently reported in Brachypodium distachyon (Wu et al. 2013).
Taken together, our results indicate that the differences in heading time associated with the Hope FT-B1 allele are highly dependent on the genetic background. For example, the introgression of the Hope FT-B1 allele in winter wheat varieties can result in a spring growth habit and consequently, very large differences in heading time (in nonvernalized plants). Within the spring varieties, the differences in heading time between the FT-B1 alleles are expected to be smaller than those in the winter NILs. Even within the spring varieties, the Hope FT-B1 allele is expected to have a larger effect on heading time in those varieties that still have a residual vernalization requirement (e.g., those carrying only the Vrn-B1a or the Vrn-D1a alleles for early flowering). However, in lines with identical Vrn1 alleles, additional unknown genes can modulate the effect of the FT-B1 alleles, as shown in the Vrn-D1a varieties Kern, Clear White-vrn-B1, and UC1041 (Table 2). These results indicate that breeders interested in using the Hope FT-B1 allele to accelerate flowering would need to validate experimentally the effect of this allele in their selected genetic background and environment.
To provide additional information to wheat breeders interested in using the Hope FT-B1 allele to engineer earlier-flowering varieties, we evaluated the effect of this allele on GpS, TKW, SpS, and spike weight. We selected these traits based on their high heritability (Kisana et al. 1982; Ali et al. 2008; Al-Tabbal and Al-Fraihat 2012) and the possibility to evaluate them with limited amounts of seed. Although these traits are correlated with yield in common wheat and are useful predictors of total grain yield (Sokoto et al. 2012), yield trials with replicated small plots at multiple locations will be required to confirm the positive trends on yield components detected in some of the lines.
Previous studies have shown that FT1 transcript levels affect wheat spike development by upregulating gibberellin biosynthetic genes in the apical region (Pearce et al. 2013). It would be interesting to investigate if these changes in gibber-ellin biosynthetic genes are related to the increase in the number of SpS and GpS observed in many of the Hope FT-B1 lines. However, this general trend should be considered with caution since the yield component traits showed a variable response across lines. Although in most lines we observed a trend toward an increase in SpS, GpS, and SW, the variety Kern showed the opposite trend (Table 3).
The effects on TKW were more variable but interestingly were not always negatively correlated with the differences in grain number. For example, Clear White-vrn-B1 showed simultaneous increases in GpS and TKW, that when combined in the calculation of SW, resulted in significant increases in the combined Anova across years (P = 0.0029, Table 3). In summary, the absence of a consistent negative effect on yield components associated with the Hope FT-B1 allele is encouraging for the utilization of this allele to subtly accelerate flowering of commercial spring wheat varieties.
To help breeders test the effects of the Hope FT-B1 allele in their particular environments, the backcross lines developed in this study have been released as publicly available germplasm to the National Small Grains Collections (see Material and Methods for PI numbers). This might be particularly interesting for spring durum wheat breeding program, where the Hope FT-B1 allele has not been available before.
Acknowledgments
Funding
National Research Initiative Competitive Grants from the United States Department of Agriculture National Institute of Food and Agriculture (2011-67013-30077, 2011-68002-30029 Triticeae-CAP); Howard Hughes Medical Institute; Gordon and Betty Moore Foundation; Argentinean Ministry of Science and Technology (FONCYT-PICT 2011 – 509).
References
- Ali Y, Atta BM, Akhter J, Monneveux P, Lateef Z. Genetic variability, association and diversity studies in wheat (Triticum aestivum L.) germplasm. Pak J Bot. 2008;40:2087–2097. [Google Scholar]
- Al-Tabbal JA, Al-Fraihat AH. Heritability studies of yield and yield associated traits in wheat genotypes. J Agric Sci. 2012;4:11. [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Bonnin I, Rousset M, Madur D, Sourdille P, Dupuits C, Brunel D, Goldringer I. FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat. Theor Appl Genet. 2008;116:383–394. doi: 10.1007/s00122-007-0676-0. [DOI] [PubMed] [Google Scholar]
- Brevis JC, Dubcovsky J. Effects of the chromosome region including the Gpc-B1 locus on wheat grain and protein yield. Crop Sci. 2010;50:93–104. [Google Scholar]
- Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8:e1003134. doi: 10.1371/journal.pgen.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicaiza O, Khan IA, Zhang X, Brevis JC, Jackson L, Chen X, Dubcovsky J. Registration of five wheat isogenic lines for leaf rust and stripe rust resistance genes. Crop Sci. 2006;46:485–487. [Google Scholar]
- Chu C-G, Tan CT, Yu G-T, Zhong S, Xu SS, Yan L. A novel retrotransposon inserted in the dominant Vrn-B1 allele confers spring growth habit in the tetraploid wheat (Triticum turgidum L.) Genes Genomes Genetics. 2011;1:637–645. doi: 10.1534/g3.111.001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JA, Love HH, Parker JH. Registration of improved wheat varieties. Agron J. 1926;18:922–935. [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F. TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 2003;132:1849–1860. doi: 10.1104/pp.103.023523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Zikhali M, Turner AS, Isaac P, Laurie DA. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat( Triticum aestivum) PLoS One. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Higgins J, Turner A, Laurie DA. The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare) Genetics. 2007;176:599–609. doi: 10.1534/genetics.106.069500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Dunbar M, Dubcovsky J. Wheat VIN3-like PHD finger genes are up-regulated by vernalization. Mol Genet Genomics. 2007;277:301–313. doi: 10.1007/s00438-006-0189-6. [DOI] [PubMed] [Google Scholar]
- Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J. Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics. 2005;273:54–65. doi: 10.1007/s00438-004-1095-4. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Fieg S, Peacock WJ, Dennis ES, Trevaskis B. Regions associated with repression of the barley (Hordeum vulgare) VERNALIZATION1 gene are not required for cold induction. Mol Genet Genomics. 2009;282:107–117. doi: 10.1007/s00438-009-0449-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippes N, Zhu J, Chen A, Vanzetti L, Lukaszewski A, Nishida H, Kato K, Dvorak J, Dubcovsky J. Fine mapping and epistatic interactions of the vernalization gene VRN-D4 in hexaploid wheat. Mol Genet Genomics. 2014;289:47–62. doi: 10.1007/s00438-013-0788-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisana NS, Chaudhry AR, Mohammad T, Chaudhary MA. Heritability of some quantitative characters in five crosses of wheat (Triticum aestivum) Pak J Agr Res. 1982;3:211–214. [Google Scholar]
- Kuspira J, Unrau J. Genetic analyses of certain characters in common wheat using whole chromosome substitution lines. Can J Plant Sci. 1957;37:300–326. [Google Scholar]
- Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe I, Jankuloski L, Chao S, Chen X, See D, Dubcovsky J. Mapping and validation of QTL which confer partial resistance to broadly virulent post-2000 North American races of stripe rust in hexaploid wheat. Theor Appl Genet. 2011;123:143–157. doi: 10.1007/s00122-011-1573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B, Nitcher R, Han X, Wang S, Ni F, Li K, Pearce S, Wu J, Dubcovsky J, Fu D. Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS One. 2014;9:e94171. doi: 10.1371/journal.pone.0094171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden ES. A successful transfer of Emmer characters to Vulgare wheat. Agron J. 1930;22:1020–1034. [Google Scholar]
- Morris R, Schmidt JW, Mattern PJ, Johnson VA. Chromosomal location of genes for flour quality in the wheat variety ‘Cheyenne’ using substitu tion lines. Crop Sci. 1966;6:119–122. [Google Scholar]
- Nitcher R, Distelfeld A, Tan C, Yan L, Dubcovsky J. Increased copy number at the HvFT1 locus is associated with accelerated flowering time in barley. Mol Genet Genomics. 2013;288:261–275. doi: 10.1007/s00438-013-0746-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163:1433–1445. doi: 10.1104/pp.113.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, Suzuki T, Ikari C, Shitsukawa N, Abe T, Kawahigashi H, Kikuchi R, Handa H, et al. Agenetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 2009;58:668–681. doi: 10.1111/j.1365-313X.2009.03806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoto MB, Abubakar IU, Dikko AU. Correlation analysis of some growth, yield, yield components and grain quality of wheat (Triticum aestivum L.) Nig J Basic Appl Sci. 2012;20:349–356. [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Furuita K, Hayashi K, Yanase T, Yamaguchi M, Nakashima C, Purwestri YA, Tamaki S, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a. 2011 doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Liu D, Wu J, Zhang R, Qin Z, Liu D, Li A, Fu D, Zhai W, Mao L. Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon. Plant Cell. 2013;25:4363–4377. doi: 10.1105/tpc.113.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J. Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet. 2004a;109:1677–1686. doi: 10.1007/s00122-004-1796-4. [DOI] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004b;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]