Abstract
As regulators of bioenergetics in the cell and the primary source of endogenous reactive oxygen species (ROS), dysfunctional mitochondria have been implicated for decades in the process of aging and age-related diseases. Mitochondrial DNA (mtDNA) is replicated and repaired by nuclear-encoded mtDNA polymerase γ (Pol γ) and several other associated proteins, which compose the mtDNA replication machinery. Here, we review evidence that errors caused by this replication machinery and failure to repair these mtDNA errors results in mtDNA mutations. Clonal expansion of mtDNA mutations results in mitochondrial dysfunction, such as decreased electron transport chain (ETC) enzyme activity and impaired cellular respiration. We address the literature that mitochondrial dysfunction, in conjunction with altered mitochondrial dynamics, is a major driving force behind aging and age-related diseases. Additionally, interventions to improve mitochondrial function and attenuate the symptoms of aging are examined.
Keywords: Mitochondrial DNA mutations, MtDNA replication, DNA polymerase gamma, POLG, Aging, Age-related diseases
1. Introduction
Aging is a highly complex, progressive process characterized by impairment in physiological function eventually leading to disease and death. This impairment is the result of accumulated molecular damage associated with metabolic activity and regulated by genetic and environmental effects (Kirkwood, 2005). In the 1950’s, Harmon first proposed that ROS generated by normal metabolism directly regulate the aging process (Harman, 1956). He later refined his theory to include mitochondria as both the primary source and target of ROS, leading to the accumulation of defective mitochondria and the hindrance of functional replacements (Harman, 1972). Others specifically advocated that accumulation of ROS-driven somatic mtDNA mutations, secondary to a lack of adequate repair mechanisms, is the main driving force behind aging (Fleming et al., 1982; Linnane et al., 1989). The mtDNA mutations lead to disruption of the electron transport chain (ETC), which generates more ROS followed by additional mtDNA mutations, thus creating a “vicious” cycle of damage culminating in apoptosis (Mandavilli et al., 2002).
Support for the classical Mitochondrial Free Radical Theory of Aging (MFRTA) waned as new evidence linked ROS to both toxicity and signaling pathways associated with longevity. Newer theories associate aging and mitochondrial dysfunction with clonally expanded mtDNA mutations caused by replication errors and altered mitochondrial dynamics. In particular, much of the mtDNA damage is linked with the nuclear-encoded proteins responsible for its replication. Here, we focus on aspects of the mitochondrial replication machinery that contribute to mtDNA mutagenesis and aging.
2. Mitochondrial biology and genetics
Mitochondria are intracellular organelles that produce the majority of adenosine triphosphate (ATP) in cells via the ETC and oxidative phosphorylation (OXPHOS). Mitochondria are also involved in buffering cytosolic calcium, regulating metabolic and signaling pathways and controlling apoptosis through the mitochondrial permeability transition pore (Wallace, 2005).
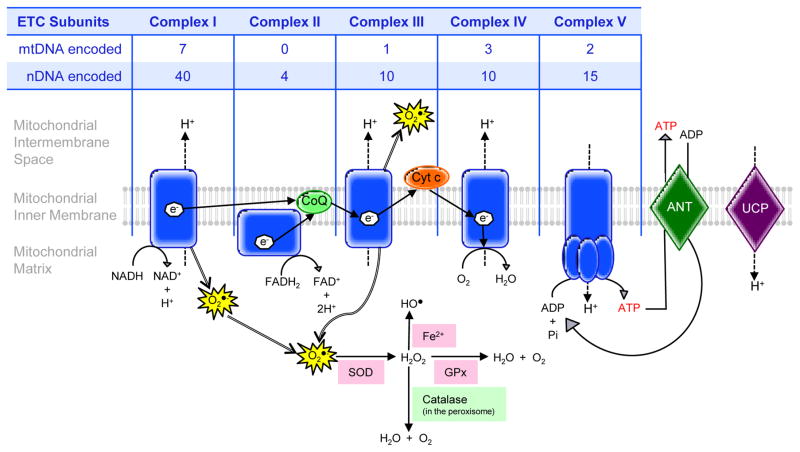
The ETC consists of four complexes located in the inner mitochondrial membrane (Figure 1). Energy from dietary reducing equivalents, nicotinamide adenine dinucleotide (NADH) and flavine adenine dinucleotide (FADH2), flow down the ETC as electrons that are used by Complexes I, III and IV to pump protons across the mitochondrial inner membrane, generating a proton electrochemical gradient. Protons allowed back into the matrix along this gradient are used by ATP synthase (Complex V) to drive synthesis of ATP from ADP and inorganic phosphate (Pi). ATP is then translocated into the intermembrane space by adenine nucleotide translocases (ANT1-4), which in conjunction with the voltage-dependent anion channel (VDAC) allow ATP into the cytosol (Wallace, 1997). Greater than 90% of the oxygen taken up into the cell is consumed by the ETC. Premature electron leakage from Complexes I and III convert 1–2% of the oxygen to ROS as superoxide (O2•−). The superoxide anions are converted to hydrogen peroxide (H2O2) by the superoxide dismutases (SODs). H2O2 is then detoxified either by peroxisomal catalase or the glutathione system. If not detoxified, H2O2 is transformed into the hydroxyl radical (HO•) via the Fenton reaction (Murphy, 2009).
Figure 1.
The four complexes of the ETC and ATP synthase are located in the inner mitochondrial membrane. Electrons from NADH and FADH2 are transferred from Complex I (NADH dehydrogenase) and Complex II (succinate dehydrogenase) to coenzyme Q (CoQ) before being transported to Complex III (cytochrome c reductase). From Complex III, cytochrome c (Cyt c) delivers the electrons to Complex IV (cytochrome c oxidase), which ultimately reduces oxygen to water. This flow of electrons down the ETC generates a proton electrochemical gradient, which is used by Complex V (ATP synthase) to synthesize ATP. ATP is then transported back into the intermembrane space by the ANT1-4 and beyond to the cytosol with a combination of ANT and VDAC. Protons can also enter the matrix through the uncoupling protein (UCP), which results in the uncoupling or separation of electron transport through the ETC from ATP synthesis. Premature electron leakage from Complexes I and II form ROS as superoxide (O2•−) which can be transformed into H2O2 or the hydroxyl radical (HO•). Subunits of each of the five complexes of the ETC, with the exception of Complex II, are encoded by both mtDNA and nDNA as recounted in the table.
Human mtDNA is maternally inherited (Giles et al., 1980) and exists as a circular, double-stranded genome organized into 16,569 base pairs. It encodes 37 mitochondrial genes: 22 transfer RNAs, 2 mitochondrial ribosomal RNAs and 13 proteins subunits of ETC complexes, with the exception of Complex II which is entirely nuclear encoded. There are several mtDNA copies per mitochondrion and hundreds of mitochondria per cell (Miller et al., 2003). MtDNA can exist as a pure population of wild type (homoplasmy) or a mixture of wild type and mutant mtDNA, known as heteroplasmy. Proteins, such as transcription factor A of mitochondria (TFAM), mtDNA helicase Twinkle and mitochondrial single-stranded DNA binding protein (mtSSB) can colocalize with mtDNA forming distinct, dynamic structures known as nucleoids (Garrido et al., 2003). Depletion of mtDNA leads to dysfunctional mitochondria, which can contribute to aging, disease and ultimately cell death. Additionally, mtDNA point mutations and deletions can disrupt mitochondrial function if the percentage of mutant mtDNA per cell reaches a threshold where not enough functional mitochondria remain to maintain proper energetic function (Wallace, 1999). Therefore, effective and faithful mtDNA replication is necessary for cell survival.
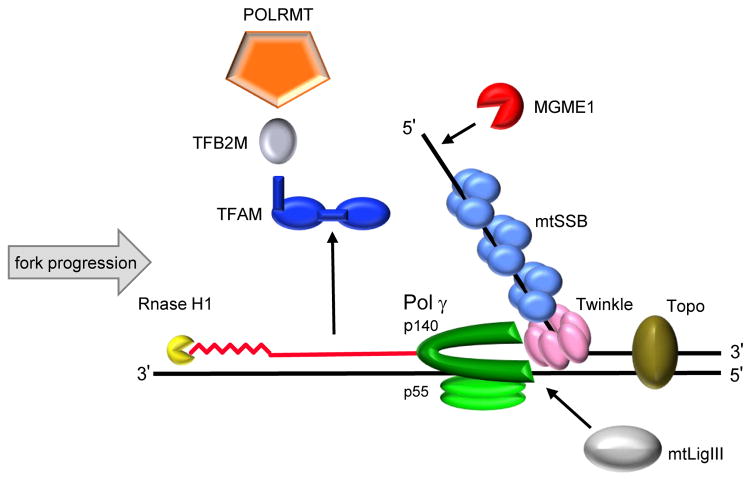
Of the 17 human cellular DNA polymerases, polymerase gamma (Pol γ) is the only replicative polymerase known to function in mitochondrial DNA replication and repair (Bebenek and Kunkel, 2004; Ropp and Copeland, 1996; Sweasy et al., 2006). The Pol γ holoenzyme is a heterotrimer consisting of a single 140 kDa catalytic subunit (nuclear encoded by POLG at chromosomal locus 15q25) and a 55 kDa accessory subunit that forms a dimer (nuclear encoded by POLG2 at chromosomal locus 17q24.1). The catalytic subunit has DNA polymerase, 3′→5′ exonuclease and 5′-dRP lyase activities (Graziewicz et al., 2006; Kaguni, 2004). The accessory subunit is required for enhanced DNA processivity by augmenting the affinity of the catalytic subunit for DNA (Lim et al., 1999; Young et al., 2015). The Pol γ holoenzyme functions in conjunction with Twinkle, mtSSB, topoisomerase, mitochondrial RNA polymerase (POLRMT), RNase H1 and mitochondrial DNA ligase III (mtLigIII) to faithfully replicate mtDNA (Figure 2). Other proteins involved in maintenance of the mitochondrial genome include: (1) TFAM, which enhances mtDNA packaging and compaction (Ngo et al., 2014); (2) transcription factor B of mitochondria (TFB2M); (3) Optic Atrophy 1 (OPA1), which plays a key role in mitochondrial fusion and mitophagy (Chen and Chan, 2005); (4) the RecB-type mitochondrial genome maintenance 5′→3′ exonuclease 1 (MGME1); (5) the RNA and DNA 5′ flap endonuclease (FEN1) and (6) the helicase/nuclease DNA2. MGME1 may participate in DNA repair, initiation of mtDNA replication, RNA primer removal or even the destructive elimination of improper mtDNA molecules, while the latter two proteins are involved in mtDNA base excision repair (BER) (Copeland and Longley, 2014).
Figure 2.
A schematic diagram of the mtDNA replication machinery. POLRMT forms the RNA primer (jagged red line) needed to initiate DNA replication in conjunction with TFAM (royal blue), TFB2M (gray). After initiation, POLRMT, TFAM and TFB2M separately leave the DNA and the RNA primer is degraded by RNase H1 (yellow). In a 5′ to 3′ direction, the Twinkle helicase (pink) unwinds dsDNA at the replication fork. The ssDNA is stabilized by mtSSB (light blue), while MGME1 (red) degrades ssDNA. MtLigIII (white) seals the mtDNA nick. Nascent DNA (solid red line) is synthesized by Pol γ (green). Topoisomerases (brown) relieve the torsional tension in the DNA caused by unwinding.
3. MtDNA damage in aging: ROS versus replication errors
Accumulations of mtDNA damage in the form of point mutations, deletions and depletions are correlated with aging and age-related diseases as observed in human skeletal muscle (Bua et al., 2006; Fayet et al., 2002), liver (Yen et al., 1991), brain (Corral-Debrinski et al., 1992a; Corral-Debrinski et al., 1994; Lin et al., 2002), heart (Corral-Debrinski et al., 1992b; Cortopassi and Arnheim, 1990), skin fibroblasts (Michikawa et al., 1999) and colonic crypt stem cells (Taylor et al., 2003). Early reports comparing nucleotide substitutions in mtDNA from somatic tissues of different primates revealed a 10-fold higher rate of evolution for mtDNA relative to the nuclear genome (Brown et al., 1979; Brown et al., 1982), implying a relatively high mutation rate for mtDNA. More recently, estimates have increased the mutation rate from 10- to 17-fold higher in mtDNA than nuclear genomic (nDNA) (Tuppen et al., 2010). Therefore, it is essential to understand the origins of mutations in mtDNA.
It was proposed that the higher mutation rate in mtDNA is caused by ROS because the mitochondrial genome is located near the ETC in the inner mitochondrial membrane. These mutations impact the ETC complexes, which leads to disruption of bioenergetics and production of more ROS in a feedback loop (Kwong and Sohal, 2000). Decreased protection of the mitochondrial genome due to both the lack of true histones and limited DNA repair mechanisms may also contribute to the higher mutation rate in mtDNA. Since high levels of ROS cause oxidative damage to cellular components such as DNA, RNA, proteins and lipids, these observations garnered support for the MFRTA (Chakravarti and Chakravarti, 2007; Cooke et al., 2003).
The most common oxidative lesions in mtDNA are the G:C to T:A transversion mutations caused by adenine pairing with the guanine adduct 7,8-dihydro-8-oxo-deoxyguanosine (8-oxo-dG) (Wang et al., 1998). Correlative evidence between increased levels of 8-oxo-dG and age-related deletions in mtDNA from humans (Capel et al., 2005; Hayakawa et al., 1992; Hayakawa et al., 1991), mice (Vermulst et al., 2007), rats (Sawada and Carlson, 1987) and houseflies (Sohal and Sohal, 1991) has been reported. Conversely, G:C to T:A transversions are only a small fraction of the mutations found in aged Drosophila (Itsara et al., 2014) or humans (Kennedy et al., 2013; Zheng et al., 2006). The enzyme 8-oxoguanine DNA glycosylase (OGG1) functions in the mitochondria to initiate removal of the 8-oxo-dG from mtDNA, while 8-oxoGTPase (MTH1) eliminates oxidized dGTP precursors to reduce their incorporation into mtDNA. The ogg1 or ogg1/mth1 knock-out mice have 20-fold higher levels of 8-oxoguanine in mtDNA (de Souza-Pinto et al., 2001) and half the life expectancy relative to wild-type (WT) mice (Xie et al., 2004), but no increase in mtDNA mutations (Halsne et al., 2012). Taken together, this suggests that ROS and 8-oxo-dG are not major contributors to mtDNA mutations in WT organisms.
Antioxidant systems eliminate ROS. Therefore, if the MFRTA were sustainable then over-expressed antioxidant systems should lead to greater longevity and fewer mtDNA mutations. This is the exact scenario found in transgenic mice over-expressing mitochondrially-targeted human catalase, which decreases H2O2 (Schriner et al., 2005). These same mice are also protected from mitochondrial dysfunction, decreased energy metabolism and lipid-induced insulin resistance in skeletal muscle (Lee et al., 2010). Interestingly, when human catalase is localized to peroxisomes or targeted to the nucleus in mice, no detectable reduction in mtDNA damage or significant extension of lifespan are observed (Schriner and Linford, 2006; Schriner et al., 2000). Over-expression of genes involved in the maintenance of thiol redox homeostasis or co-overexpression of Cu-Zn superoxide dismutase (Cu-ZnSOD) and catalase increase the lifespan of Drosophila melanogaster by 33–48% (Orr et al., 2013; Orr and Sohal, 1994). When either Cu-ZnSOD or catalase is over-expressed individually, only Cu-ZnSOD extends lifespan up to 48% while catalase has neutral or slightly negative effects (Sun and Tower, 1999). However, when manganese superoxide dismutase (MnSOD) and catalase are ectopically targeted to the mitochondrial matrix of transgenic flies, lifespan is decreased up to 43% (Bayne et al., 2005). If overexpressing antioxidant systems reduces ROS and leads to increased longevity, then the opposite should hold true when these systems are knocked-out or deficient. For example, mice deficient in either Cu-ZnSOD or MnSOD have widespread oxidative damage and hepatocarcinogenesis or neurodegeneration with reduced lifespan (Elchuri et al., 2005; Lebovitz et al., 1996). On the other hand, inactivation of any of the five isoforms of SOD in Caenorhabditis elegans fails to decrease longevity, in spite of increased oxidative stress, and actually prolongs lifespan (Van Raamsdonk and Hekimi, 2012). Similarly, while SOD knock-out mice (Sod2−/−) die within the first 10 days of life (Li et al., 1995), heterozygous mice (Sod2+/−) are viable with longevity and OXPHOS activity comparable to WT mice (Mansouri et al., 2006). This occurs despite presenting with a premature aging (progeroid) phenotype as evidenced by increased mitochondrial oxidative damage, cataracts and an impaired immune response (Van Remmen et al., 2003).
The mixed results for whether ROS contributes to decreased or increased longevity may be explained by mitohormesis. In this theory, high levels of ROS promote mtDNA damage and the aged phenotype, while low levels of ROS are signaling molecules that regulate stress response pathways and activate cellular processes that enhance lifespan (Hekimi et al., 2011). As ROS gradually increase over time, so do the cumulative levels of mtDNA damages, with ROS eventually reaching unstoppable toxic concentrations resulting in further mitochondrial dysfunction and aging.
Mutations can also arise as spontaneous errors of replication during either DNA replication or repair events. As the only replicative DNA polymerase known to exist in mammalian mitochondria, Pol γ is likely to produce these spontaneous errors. Comparison of mutation spectrum in mtDNA derived from in vivo sources with DNA copied in vitro by the highly purified human Pol γ reveals that over 85% of mutation detected in vivo could be recapitulated in vitro by Pol γ (Zheng et al., 2006). This suggests that spontaneous replication errors by Pol γ, and not ROS-induced damage, account for the majority of base pair substitution mutations in mtDNA. Thus, understanding the fidelity of Pol γ is critical.
4. Fidelity of DNA synthesis by pol γ
The human catalytic subunit of Pol γ has high base substitution fidelity over short repeat sequences (< 3 nucleotides), as a misinsertion event only occurs approximately once in every 500,000 base pairs synthesized (Longley et al., 2001). High fidelity is due to high nucleotide selectivity and exonucleolytic proofreading. The intrinsic 3′ → 5′ exonuclease activity of Pol γ improves replication fidelity at least 20-fold (Longley et al., 2001). Biochemical characterization of the human catalytic subunit found that 3′ → 5′ exonuclease function could be eliminated by substituting D198 and E200 with alanine in the exonuclease domain (Longley et al., 1998). To examine whether exonuclease function is critical to replication fidelity in vivo, several experiments were conducted. For example, expression of an exonuclease-deficient Pol γ fusion protein in cultured human cells resulted in the accumulation of mtDNA point mutations (Spelbrink et al., 2000). Exonuclease-deficient Pol γ pancreatic islet cells caused impaired glucose tolerance, diabetes, and increased apoptosis in β-cells (Bensch et al., 2007; Bensch et al., 2009). In mice, transgenic overexpression of exonuclease-deficient Pol γ in cardiac tissue caused a greater than 23-fold increase in mtDNA point mutations and detectable levels of large mtDNA deletions (Zhang et al., 2000). These mice also presented with secondary effects, such as increased apoptosis and cardiomyopathy (Zhang et al., 2005; Zhang et al., 2003). However, the first strong experimental evidence demonstrating a causal relationship between aging and the accumulation of mtDNA mutations/deletions secondary to an exonuclease-deficient Pol γ was obtained with the creation of the mtDNA mutator mouse.
5. Exonuclease-deficient pol γ mouse links mtDNA mutations and deletions to aging
Independently developed by two labs, the mtDNA mutator mouse has a homozygous knock-in that expresses a proofreading-deficient version of Pol γ, resulting in elevated levels of mtDNA mutations (Kujoth et al., 2005; Trifunovic et al., 2004). The mice appear normal through adolescence but develop a progeroid phenotype including alopecia (hair loss) and graying, kyphosis (curvature of the spine), osteoporosis (loss of bone density), sarcopenia (loss of muscle mass), weight loss, reduced subcutaneous fat, presbycusis (hearing loss), reduced fertility, anemia, cardiac hypertrophy (heart enlargement), and a reduced lifespan. Trifunovic et al. (2004) also describe impaired OXPHOS with a reduction in ETC enzyme activities and decreased ATP production.
The progeroid phenotype and accumulated mutations of mtDNA mutator mice are not due to increased ROS production and oxidative stress (Trifunovic et al., 2005). Kujoth et al. (2005) found no biomarkers of ROS induced damage to DNA, proteins or lipids, but report that tissue dysfunction is the result of apoptosis. In a follow-up study to experimentally address the “vicious” cycle aspect of the MFRTA, mouse embryonic fibroblasts from mtDNA mutator mice are shown to produce normal amounts of ROS and demonstrate no increased sensitivity to oxidative stress (Trifunovic et al., 2005). There are also no changes in the levels of antioxidants, protein carbonyls or aconitase enzyme activity in their tissues.
Despite only normal levels of ROS, mtDNA mutator mice do accumulate mutations of different types: (1) a 3- to 8-fold increase in progressive, yet random, somatic point mutations; (2) circular mtDNA with large deletions; and (3) multiple linear deleted mtDNA fragments approximately 12kb in length that are localized to the arc between the two origins of replication (Kujoth et al., 2005; Trifunovic et al., 2004). It remains highly contested as to which of these is the cause of the premature aging phenotype.
Unlike the homozygous mtDNA mutator mice, mice heterozygous for the Pol γ mutation have no progeroid phenotype or reduced lifespan (Kujoth et al., 2005; Trifunovic et al., 2004). The mutation load of these mice was originally reported to be indistinguishable from WT animals (Trifunovic et al., 2004) as determined by PCR cloning and sequencing, a method that is limited by the error frequency of the PCR polymerases, which is 0.13 mutations per kbp (Kujoth et al., 2005). However, using an adaptation of the more sensitive “random capture method,” which relies on PCR amplification of single molecules for mutation detection but is not limited by polymerase fidelity, the heterozygous mice are found to have a 500-fold higher mutational burden than age-matched wild-type controls and a 30-fold increase over the oldest wild-type animals (Vermulst et al., 2007). Thus, at least at the level observed in the heterozygous mice, mtDNA point mutations do not limit natural lifespan. However, the higher mutation load observed by the random capture method (2000-fold higher in the homozygous mtDNA mutator mouse than WT) showed that the heteroplasmic level of mutations clearly exceeded the bioenergetic threshold to elicit the premature aging phenotype.
WT and heterozygous mice mainly accumulate deletions at one hotspot between two 15-bp direct repeats, whereas homozygous mutant mice accumulate deletions regardless of the presence of direct repeats (Vermulst et al., 2008). Excluding this hotspot, homozygous mutant mice showed a 90-fold increase in circular mtDNA with large deletions, as compared to WT controls. Thus, the data suggest that deletions are the driving force behind the progeroid phenotype of the mtDNA mutator mouse (Vermulst et al., 2008). However, these findings were contested because when employing Southern blots and long-range PCR, these circular mtDNA molecules with large deletions are remarkably absent or present at extremely low levels, and as such, cannot be causal for the advanced aging phenotype (Edgar et al., 2009). In fact, the overall fraction of deletions in tissues from the mtDNA mutator mouse was only measured to be ~1.0% (Kraytsberg et al., 2009). But according Vermulst et al. (2009), the random nature of the deletions makes these methods insufficient for quantifying deletions. Thus, the number of deletions in the mtDNA mutator mouse continues to be debated.
Edgar et al. (2009) reported frequent point mutations in ETC genes in homozygous mtDNA mutator mice, while heterozygous mice were not examined. The resulting amino acid substitutions impair the stability of the ETC complexes, resulting in OXPHOS deficiency. Therefore, they conclude that point mutations, particularly in protein coding genes, are ultimately responsible for causing the progeroid phenotype in mtDNA mutator mice (Edgar et al., 2009). Lastly, a third type of mutation found in the mtDNA mutator mouse is the linear deleted mtDNA fragment. The deleted mtDNA was comparable in all tissues and did not change over time. Therefore, these fragments are surmised to arise from stalled replication due to Pol γ exonuclease deficiency and do not contribute to the progeroid phenotype (Kukat and Trifunovic, 2009). Hence, although it is agreed that mtDNA mutations contribute to the aging phenotype, it remains controversial as to whether these mutations are in the form of point mutations, deletions or both.
6. Age-related mitochondrial diseases associated with defective mtDNA replication machinery
6.1. DNA Polymerase γ
Several mitochondrial diseases have been linked to ineffective mtDNA replication by Pol γ. To date, there are over 300 disease-associated mutations in POLG listed in the Human DNA Polymerase Gamma Mutation Database (http://tools.niehs.nih.gov/polg). Some associated diseases manifest in adulthood and include symptoms of aging, namely autosomal dominant and recessive forms of Progressive External Ophthalmoplegia (adPEO/arPEO) and Parkinson’s disease.
PEO is associated with mtDNA point mutations and deletions (Hirano et al., 2001; Van Goethem et al., 2001; Van Goethem et al., 2003b) first identified in an Italian family with heritable autosomal dominant PEO (adPEO) (Zeviani et al., 1989). PEO manifests during adulthood and is characterized by bilateral ptosis and progressive weakening of the external eye muscles, proximal muscle weakness and wasting, skeletal muscle with ragged red fibers, impaired ETC enzyme activity, exercise intolerance, sensory ataxic neuropathy and respiratory failure due to muscle fatigue.
In 2001, a heterozygous mutation in Pol γ that substituted a tyrosine with cysteine at codon 955 (Y955C) was first identified as a disease locus for adPEO while three more missense mutations were associated with arPEO (Van Goethem et al., 2001). The following year, several more Pol γ mutations were identified that cause both adPEO and arPEO (Lamantea et al., 2002). Four adPEO mutations (G923D, R943H, Y955C and A957S) were biochemically characterized (Graziewicz et al., 2004). Both R943H and Y955C alter direct interactions with incoming dNTPs resulting in severely decreased processivity and a 99% decrease in polymerase activity. The reduced catalytic efficiency and the profound stalling of DNA synthesis are predictive for the in vivo dysfunction presented by PEO patients (Graziewicz et al., 2004). The Y955C mutation has a wild-type catalytic rate, but a 45-fold decrease in binding affinity for the incoming dNTP. The proofreading deficient exonuclease domain causes a 10- to 100-fold increase in nucleotide misinsertion rates (Ponamarev et al., 2002). In a transgenic mouse model, human Pol γ encoding Y955C was targeted to the heart. These mice develop a molecular signature of PEO in the heart with cardiomyopathy, loss of mtDNA, increased 8-OHdG and premature death (Lewis et al., 2007). The G923D and A957S mutations have 21% and 23%, respectively, of the catalytic activity of the wild-type enzyme, which seems to correlate with the presentation of a milder clinical phenotype (Graziewicz et al., 2004).
Most of the mutations in Pol γ are associated with arPEO. They are compound heterozygotes and often occur with common Pol γ mutations, such as A467T (Van Goethem et al., 2003a), W748S (Van Goethem et al., 2004), R309L (Lamantea et al., 2002), G848S (Lamantea et al., 2002), R627W (Van Goethem et al., 2003a), N846S (Van Goethem et al., 2003c) or T251I in cis with P587L (Lamantea et al., 2002; Scuderi et al., 2015), to name a few. Patients with arPEO have the same clinical features as with adPEO, but with greater variation in symptoms and time of onset.
Parkinson’s disease (PD) is a progressive neurological disorder affecting greater than 1% of people over 60 years of age and 4% of those over 80 years (Luo et al., 2015). It is characterized by the loss of dopaminergic neurons in the substantia nigra and the formation of protein inclusions called Lewy bodies (Duvoisin, 1992). However, the absence of Lewy bodies in the substantia nigra has been found postmortem in patients with Pol γ associated PD (Moslemi et al., 1999). Clinically, PD presents with motor and non-motor symptoms. Motor dysfunctions include tremor, rigidity, bradykinesia, postural instability and uncontrollable movements while non-motor symptoms include sleep disturbances, depression, cognitive impairment and autonomic dysfunction (Jankovic, 2008). Aberrant mitochondrial functioning has been implicated in the pathogenesis of idiopathic PD with genetic analysis focusing on age-related mtDNA mutations (Bender et al., 2006; Coxhead et al., 2015; Hudson et al., 2014; Keogh and Chinnery, 2015; Kraytsberg et al., 2006).
Seven PEO families of various ethnic origins were reported to have Pol γ mutations with multiple mtDNA deletions (Luoma et al., 2004) and reduced mtDNA copy number (Gui et al., 2015). The dominant Y955C mutation and two new missense mutations, N468D and A1105T, were discovered in these families (Luoma et al., 2004). Most of the women had early menopause or primary amenorrhea. Similar results were obtained in a family with premature ovarian failure associated with PEO and PD in which the diseases segregated with Y955C (Pagnamenta et al., 2006). A novel missense mutation in Pol γ (A2492G) was identified in another family with PEO, PD, hypogonadism, hearing loss, muscle weakness and psychiatric problems (Mancuso et al., 2004). However, it was later determined that the substitution is probably a neutral polymorphism as it has a 1.1% frequency in controls (Luoma et al., 2007; Tiangyou et al., 2006). Two sisters with early-onset PD, peripheral neuropathy and without PEO had a 37% decrease in mtDNA content with multiple deletions and were found to be compound heterozygous for two missense mutations (G737R and R853W) in Pol γ (Davidzon et al., 2006). Several other point mutations in Pol γ have been identified in patients diagnosed with a combination of PEO and PD, such as heterozygous S511N with L463F (Hudson et al., 2007), homozygous W748S (Remes et al., 2008), heterozygous F943H (Brandon et al., 2013), heterozygous P587L and W748S (Ylonen et al., 2013) and heterozygous H945L (Delgado-Alvarado et al., 2015). Lastly, Pol γ contains a polyglutamine tract in the N-terminal region encoded by a CAG-repeat and rare variants in the CAG-repeat were found in patients with idiopathic PD (Luoma et al., 2007). However, a similar analysis revealed no changes (Taanman and Schapira, 2005). In support of the latter study, another group reported that there is no contribution of common genetic variants of Pol γ contributing to the etiology of idiopathic PD (Tiangyou et al., 2006). Despite these two confounding studies, the role of Pol γ in contributing to mtDNA mutations and reduced copy number thus impacting the pathogenesis of PEO and PD patients appears convincing.
6.2. PolG2
PolG2 is a 110 kDa homodimeric accessory subunit for the catalytic subunit PolG (Carrodeguas et al., 2001) that functions to enhance processivity of DNA replication by augmenting the binding of PolG to DNA (Lim et al., 1999). Of the handful of heterozygous mutations identified in POLG2, G451E (Longley et al., 2006) and R369G (Craig et al., 2012; Young et al., 2011) are linked to PEO. Homodimeric mutants have problems stimulating PolG processivity, with G451E being more severely defective than R396G (Craig et al., 2012; Longley et al., 2006; Young et al., 2011). G451E homodimers also fail to enhance binding of PolG to DNA, while R369G has weaker binding to PolG. Only heterozygotes have been clinically identified, and recent biochemical characterization of heterodimers has found that the WT/G451E heterodimer has significantly reduced stimulation of PolG processivity due to weaker binding to both PolG and DNA (Young et al., 2015). Thus G451E PolG2 is a dominant negative mutation causing adPEO. The WT/R369G heterodimer functions like the wild-type enzyme in vitro. Though WT/G451E and WT/R369G localize to mtDNA like WT PolG2, bioenergetic analysis of transfected HEK-293 cells finds that both mutant cell lines have significantly reduced reserve capacity (Young et al., 2015). Skeletal muscle with reduced reserve capacity has muscle weakness and exercise intolerance, hallmarks of PEO. A complete POLG2 knock-out is embryonic lethal in mice, but the heterozygous knockout (+/−) mouse has a normal phenotype (Humble et al., 2013). Currently, there are no mouse models available for conditional knock-out or knockdown of POLG2. Knockdown of porcine oocyte PolG2 causes an aging-like phenotype as evidenced by decreased production of ATP and maturation promoting factor (MPF), reducing the fraction of oocytes that reach MII stage (Lee et al., 2015).
6.3. Twinkle
Twinkle is a 72 kDA hexamer/heptamer that functions as a 5′ → 3′ helicase for mtDNA replication (Korhonen et al., 2003). It was classified as a helicase based on sequence similarity to bacteriophage T7 gene 4 primase-helicase (T7 gp4) (Spelbrink et al., 2001). Twinkle has two domains, a primase-like N-terminal domain and a C-terminal helicase domain (Shutt and Gray, 2006). The N-terminal domain is important for ssDNA binding and stimulation of the C-terminal helicase activity (Farge et al., 2007). Twinkle is essential for mtDNA maintenance and copy number (Tyynismaa et al., 2004). It is involved in D-loop formation and localizes to the nucleoid (Jemt et al., 2015; Milenkovic et al., 2013; Spelbrink et al., 2001). Helicase activity along with strand annealing activity has been confirmed (Sen et al., 2012), but no primase activity has been detected to date.
Twinkle, encoded at chromosomal locus 10q24, was originally identified through sequencing analysis of a collection of PEO patients with unidentified disease causality (Spelbrink et al., 2001). In all, 31 dominant and 3 recessive mutations have been recognized, the majority of which cause age-related disease (Sanchez-Martinez et al., 2012; Spelbrink et al., 2001). Most of the mutations cluster in a region that is predicted to be the linker region (Sanchez-Martinez et al., 2012). In vitro analysis on some of these mutations has had mixed results concerning helicase activity despite some mutants displaying altered DNA binding affinity, nucleotide hydrolysis or thermal stability (Holmlund et al., 2009; Korhonen et al., 2008; Longley et al., 2010; Matsushima et al., 2008).
Overexpression of adPEO Twinkle mutations in HEK293 cells cause an accumulation of replication intermediates suggesting that these mutations are triggering replication stalling (Goffart et al., 2009; Wanrooij et al., 2007). These mutants have decreased mtDNA copy number and enlarged nucleoids. Taken together, these data support Twinkle’s role in mtDNA replication.
A homozygous Twinkle knock-out mouse is embryonic lethal at E8.5, but a conditional tissue-specific knockout in heart and skeletal muscle is viable. This mouse has progressive mtDNA depletion leading to severely impaired mtDNA expression and respiratory chain deficiency (Milenkovic et al., 2013). Conversely, systemic Twinkle overexpression in a transgenic mouse causes a significant increase in mtDNA copy number (Tyynismaa et al., 2004); however, the mice have enlarged nucleoid structures with impaired mtDNA replication and transcription (Ylikallio et al., 2010). Depleting Twinkle in Drosophila Schneider cells induces slow growth, limits viability and reduces mtDNA copy number 5-fold (Matsushima and Kaguni, 2007). Overexpressing Twinkle in these same cells causes a modest increase in mtDNA copy number (Matsushima and Kaguni, 2007). Taken together, these data further indicate that Twinkle is essential for mtDNA replication.
MtDNA typically exists as monomeric double-stranded, covalently closed circles. In adult human hearts however, mtDNA has been shown to exist as dimers and is organized into highly complex, branched molecules (Pohjoismaki et al., 2009). This form of mtDNA has not been detected in human infants (Pohjoismaki et al., 2010) or found endogenously in other species (Pohjoismaki et al., 2009). Systemic overexpression of Twinkle in mouse hearts causes an increase in complex DNA forms and recombination junctions similar to adult human hearts (Pohjoismaki et al., 2009). Heart tissue samples from patients harboring a Twinkle duplication mutation (amino acids 352–364) that causes adPEO have mtDNA that is lacking in these complex higher order structures (Pohjoismaki et al., 2010). This suggests Twinkle has an important role in maintenance of mtDNA higher order structures in adult human heart.
Mice overexpressing Twinkle are protected from left ventricular remodeling in the heart (Tanaka et al., 2013). This mouse attenuates VO-induced eccentric hypertrophy and improves cardiac function while suppressing the production of mitochondrial ROS and subsequent redox-sensitive signals (Ikeda et al., 2015). The Twinkle overexpression mouse was crossed with a manganese superoxide dismutase heterozygous (MnSOD+/−) knockout mouse (Pohjoismaki et al., 2013). The MnSOD+/− mouse has increased oxidative damage leading to replication stalling and the formation of unicircular dimers in the heart (Li et al., 1995). Crossing the MnSOD+/− mouse with the systemic Twinkle overexpression mouse abolishes the MnSOD+/− phenotype (Pohjoismaki et al., 2013). Additionally, a transgenic knock-in mouse was created expressing a dominant-negative K320E Twinkle in the heart (Baris et al., 2015). These mice develop ventricular arrhythmia with aging due to dysfunctional ETC enzyme activity. This suggests Twinkle provides cardioprotection and could be a potential therapy for heart failure patients.
Transgenic mice overexpressing dominant PEO Twinkle mutations (either a point mutant A360T or an in-frame duplication of amino acids 353–365) were created (Tyynismaa et al., 2005). The Twinkle duplication mouse, herein referred to as the deletor mouse, presents with mitochondrial myopathy around 12 months of age (Tyynismaa et al., 2005). The phenotype of the deletor mouse is strikingly similar to PEO patients, thus creating a PEO mouse model (Tyynismaa et al., 2010). The deletor mouse phenotypes can be attenuated with a lipid-lowering agent, such as bezafibrate, however this treatment causes severe hepatic side effects (Yatsuga and Suomalainen, 2012). The hepatic side effects are not expected in humans as the dosage amounts are much lower. Thus, bezafibrate has the potential to be a therapy for PEO patients (Yatsuga and Suomalainen, 2012).
A conditional transgenic mouse overexpressing the PEO Twinkle duplication targeted to neurons has been generated (Song et al., 2012). The mouse has neuron loss in the substantia nigra with significant mtDNA deletions and reduction of the protein Parkin resulting in motor abnormality (Song et al., 2012). This mouse is used to study substantia nigra specific accumulation of mtDNA deletions in Parkinson’s disease.
Moderate overexpression of Twinkle adPEO disease mutations, I334T or A442P, in Schneider cells allows for normal growth despite a 2-fold reduction in copy number, while other adPEO disease mutations do not (Matsushima and Kaguni, 2007). However, overexpression of A442P in Drosophila melanogaster is lethal at the pupal stage (Matsushima and Kaguni, 2007). MtDNA copy number is severely depleted along with mtDNA-encoded transcripts leading to reduced ETC Complex IV activity.
6.4. Transcription Factor A of Mitochondria (TFAM)
TFAM, a member of the high mobility group (HMG) proteins (Parisi and Clayton, 1991), is a 25 kDa protein that binds upstream of both the heavy- and light-strand promoters of mtDNA (Fisher and Clayton, 1985; Fisher et al., 1987). TFAM was originally identified as a regulator of mtDNA transcription (Fisher and Clayton, 1985). More recently TFAM has been identified as a core protein of the mtDNA nucleoid (Kanki et al., 2004), a regulator of mtDNA copy number (Ekstrand et al., 2004) and potentially a key factor in mtDNA replication (Campbell et al., 2012).
Homozygous and heterozygous knock-outs of TFAM have been created in mice (Larsson et al., 1998). The homozygous knock-out is embryonic lethal at E8.5, whereas the heterozygous mouse remains viable with reduced mtDNA copy number, reduced mitochondrial transcription and impaired ETC activity in the heart (Larsson et al., 1998). Several conditional knock-out mice have been created targeting cardiomyocytes (Li et al., 2000; Wang et al., 1999), skeletal muscle cells (Wang et al., 1999; Wredenberg et al., 2002), pancreatic β-cells (Silva et al., 2000), pyramidal neurons (Sorensen et al., 2001) and midbrain dopamine (DA) neurons (Ekstrand et al., 2007). For all the targeted cell types, TFAM depletion leads to mtDNA depletion (not reported for midbrain DA neurons), mitochondrial transcript depletion (not reported for midbrain DA neurons) and respiratory chain deficiency, suggesting that TFAM is essential for mtDNA maintenance in vivo (Ekstrand et al., 2007; Li et al., 2000; Silva et al., 2000; Sorensen et al., 2001; Wang et al., 1999; Wredenberg et al., 2002). Both heart- and muscle-specific conditional TFAM knock-out mice have cardiomyopathy with greater severity in the heart-specific knock-out (Li et al., 2000; Wang et al., 1999). Skeletal muscle-specific TFAM knock-out mice have myopathy with an abundance of COX-deficient red ragged fibers similar to mitochondrial disease patients (Wredenberg et al., 2002). TFAM deletion in pancreatic β-cells leads to diabetic mice (Silva et al., 2000). TFAM deletion in mouse pyramidal neurons causes mitochondrial late-onset neurodegeneration (MILON) (Sorensen et al., 2001) whereas deletion in mouse midbrain DA neurons leads to PD (Ekstrand et al., 2007). Heart failure, myopathy, diabetes, neurodegeneration and PD are all age-related diseases strongly implicating TFAM in aging.
Despite the potential for TFAM to play a role in PD, separate studies of 250 and 301 patients, respectively, identified no TFAM mutations (Alvarez et al., 2008; Bertram et al., 2007). To date, there are no confirmed TFAM clinical mutations, although the genetic variant rs2306604 may be linked to Alzheimer’s disease (Belin et al., 2007) pending more comprehensive clinical and biochemical analyses. However, TFAM has shown promise for being an anti-aging therapeutic agent. Studies of cardiomyocytes (Suarez et al., 2008), SH-SY5Y neuroblastomas (Xu et al., 2009), and rat pancreatic islet cell lines (Gauthier et al., 2009) have been performed. TFAM overexpression in cardiomyocytes improves mitochondrial function when cells are exposed to hyperglycemic conditions (Suarez et al., 2008). Exogenous introduction of recombinant TFAM (rhTFAM) to cardiomyocytes increases mtDNA copy number and attenuates hypertrophy, suggesting TFAM as a potential therapeutic agent to combat heart failure.
Mice heterozygous for Pdx1 (Pdx+/−), a transcription factor needed for pancreatic development and βcell maturation/survival, show diabetic symptoms and significantly reduced TFAM transcript levels (Oliver-Krasinski and Stoffers, 2008). Rat pancreatic islet cells transduced with a dominant-negative Pdx1 variant have the same TFAM transcript reduction (Gauthier et al., 2009). TFAM overexpression in rat pancreatic islet cells normalized Nd1 mRNA levels and restored glucose-induced ATP generation along with insulin secretion (Gauthier et al., 2009). This suggests TFAM could similarly be a therapeutic agent for diabetes.
SH-SY5Y neuroblastoma cells were used to study TFAM’s role in neurodegenerative disease. Overexpression of TFAM in these cells provides protection against Aβ1-42 induced mtDNA oxidative damage and MPP+ induced damage (Xu et al., 2009) suggesting TFAM could also be used as a therapeutic agent in neurodegenerative disorders. To test this premise, SH-SY5Y neuroblastoma cybrids were created that mimic sporadic PD (Xu et al., 2009). These cells were then treated with protofection technology using a modified rhTFAM that has an N-terminal 11 Arginine transduction domain followed by a mitochondrial localization signal (Khan and Bennett, 2004). Uptake and mitochondrial localization of rhTFAM restored mitochondrial bioenergetics to impaired cells without altering normal cells, further supporting the use of rhTFAM as a therapeutic agent (Iyer et al., 2009).
Mice overexpressing human TFAM (hTFAM) are protected from memory impairment (Hayashi et al., 2008) and cardiac damage after myocardial infarction (Ikeuchi et al., 2005). Overexpression increases mtDNA copy number, attenuates volume overload induced eccentric hypertrophy, improves cardiac function and suppresses the production of mitoROS (Ikeda et al., 2015). However, no increases in ETC enzyme activities suggest a disassociation between mtDNA copy number and the ETC (Ikeda et al., 2015; Ikeuchi et al., 2005). Another mouse model system crossed mice overexpressing TFAM with the mito-mice that have mutant mtDNA carrying a deletion of 4696 bp leading to an overall increase in mtDNA content and copy number without a decrease in the amount of deleted mtDNA (Nishiyama et al., 2010). TFAM overexpression improved the symptoms caused by mitochondrial deficiency signifying that WT mtDNA is important for maintaining mitochondrial function irrespective of mutant mtDNA proportions (Nishiyama et al., 2010). However, Ylikallio et al. (2010) suggest caution should be taken when considering the use of either recombinant TFAM and/or mtDNA as therapeutic agents since increased mtDNA copy number causes nucleoid enlargement and defective replication and transcription.
6.5. Mitochondrial Genome Maintenance Exonuclease 1 (MGME1)
MGME1 is a 37 kDa monomeric protein that is a mitochondrially-located, metal-dependent exonuclease (Szczesny et al., 2013). MGME1 belongs to the PD-(D/E)XK nuclease superfamily and is a close homologue of the bacterial RecB nuclease (Kornblum et al., 2013; Szczesny et al., 2013). MGME1 selectively degrades ssDNA in both 3′ → 5′ and 5′ → 3′ directions, preferring the latter (Kornblum et al., 2013; Szczesny et al., 2013). It can degrade DNA flap and RNA-DNA flaps (Kornblum et al., 2013; Szczesny et al., 2013), but not circular DNA (Kornblum et al., 2013). HeLa cells and patient fibroblasts with MGME1 knocked-down have increased amounts and lengths of 7S mtDNA (Nicholls et al., 2014). Conversely, patient fibroblasts that were transfected with MGME1 have 7S mtDNA decreased in both amount and length, especially the 11 kb linear dsDNA fragments whose ends map to the two classically derived replication origins, OH and OL. MGME1 interacts directly with PolG suggesting a role in mtDNA replication, likely providing 5′ → 3′ exonuclease activity (Nicholls et al., 2014).
Three different families presenting with PEO, skeletal muscle wasting/weakness, emaciation and respiratory distress bear MGME1 mutations (Kornblum et al., 2013). Two of these families (five affected individuals) have the same homozygous nonsense mutation (c.456G>A, P.W152*) while a different sporadic case presents with homozygous c.698A>G, P.W233C point mutation. The W152* nonsense mutation truncates MGME1 upstream of the nuclease motif. W152* fibroblasts have no detectable full length or truncated MGME1. Protein folding appears to be impaired in W233C substituted MGME1.
6.6. Ribonuclease H1 (RNase H1)
RNase H1, first identified in 1969 (Stein and Hausen, 1969), is a 29 kDa monomeric protein that functions in both the nucleus and the mitochondria (Cerritelli et al., 2003). Alternative start codons permit translation of the mitochondrial (first AUG) and nuclear (second AUG) forms of RNase H1 (Cerritelli and Crouch, 2009). RNase H1 cleaves RNA/DNA hybrids containing at least four ribonucleotides (Ohtani et al., 1999). RNase H1 associates with the protein P32 suggesting RNase H1 involvement in mitochondrial pre-rRNA processing (Wu et al., 2013). Mutations in the catalytic domain have been identified in PEO patients (Reyes et al., 2015).
In mice, an RNase H1 knockout is embryonic lethal at E8.5 when mtDNA replication begins suggesting the mitochondrially located RNase H1 is essential for development (Cerritelli et al., 2003). This is similar to the RNaseH2 null embryos that show lethality and a large number of ribonucleotides in the genomic DNA (Reijns et al., 2012). The RNaseH1 KO embryos also have a loss of mtDNA indicating that RNase H1 has a vital role in mtDNA replication. In Drosophila melanogaster, RNase H1 knockout is also lethal during the last larval instar stage, when rapid mtDNA replication is important for survival (Filippov et al., 2001). Knockdown of RNase H1 in human bone osteosarcoma cells (143B TK-) led to lower mtDNA copy number and complete stalling of replication (Ruhanen et al., 2011), suggesting RNase H1 is vital for mtDNA replication. Mouse embryonic fibroblasts lacking RNaseH1 revealed retention of primers at OriH and OriL, which causes an obstacle to the replication machinery upon the next round of replication (Holmes et al., 2015).
7. The contribution of clonal expansion of mtDNA mutations to mitochondrial dysfunction
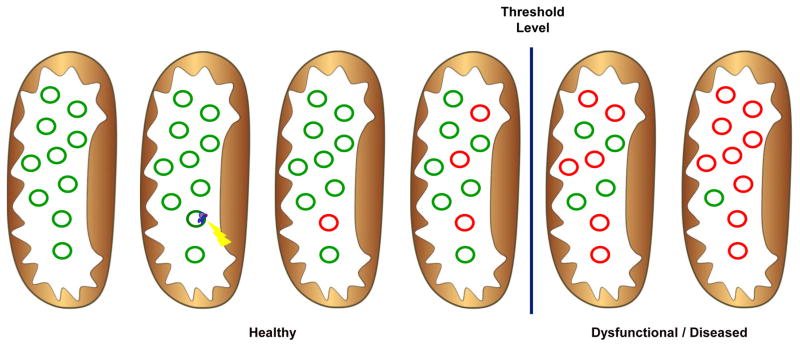
If aging is caused by mtDNA mutations, then how do these mutations lead to dysfunction? The percentage of mutated mtDNA must be between 60% and 90% in a given cell before dysfunction is measurable (Edgar and Trifunovic, 2009) (Figure 3). Elevated heteroplasmy can occur by clonal expansion of mutated mtDNA, mitotic segregation, positive selection and/or random genetic drift. Genetic drift has been proposed to be the more likely situation due to the relaxed replication of mtDNA, which is untethered to the cell cycle (Elson et al., 2001). However, reduced size gives deleted mtDNA a selective replication advantage over wild-type mtDNA, supporting positive selection (Diaz et al., 2002; Hayashi et al., 1991; Kowald and Kirkwood, 2011).
Figure 3.
Clonal expansion and heteroplasmy. Individual mitochondria contain many copies of mtDNA. During replication, a mutation and/or deletion can occur. This damage accumulates by clonal expansion. When the fraction of mutated mtDNA relative to total mtDNA exceeds 60% depending on the tissue affected, dysfunction and disease are measurable. The mitochondria are represented as the brown ovals, green circles are healthy mtDNA and red circles are damaged mtDNA. The yellow lightening bolt represents damage occurring during replication and the blue circles represent the replication machinery.
Clonal expansion of mtDNA mutations has been described in numerous tissues from aged individuals, such as skeletal muscle (Fayet et al., 2002; Muller-Hocker et al., 1993), substantia nigra neurons (Cottrell et al., 2001; Kraytsberg et al., 2006), heart (Muller-Hocker, 1989) and colonic crypts (Taylor et al., 2003). Typically, the clonal expansion of a mtDNA mutation results in highly focal ETC defects, such as cytochrome c oxidase deficiency. For example, the pattern of mosaic respiratory chain deficiency in the heart of the mtDNA mutator mice is thought to indicate clonal expansion (Trifunovic et al., 2004). A similar pattern of focal ETC defects has also been reported in human brain (Taylor et al., 2014), colorectal epithelium (Greaves et al., 2014) and colonic crypt stem cells (Taylor et al., 2003) and deemed to be the result of clonal expansion of pre-existing mutations.
The timing of when mtDNA mutations occur remains debated (Khrapko, 2011; Popadin et al., 2014). Somatic mtDNA mutations were originally thought to occur late in life when the mtDNA is more prone to chemical damage and thereafter accumulate exponentially (Khrapko, 2011). However, in order to reveal a pathological effect caused by clonal expansion, it was argued that these mutations would have to occur early in life (Elson et al., 2001). For instance, the linear accumulation of mutations from mid-gestation to late adult life as seen in the mtDNA mutator mice, along with the same mutations between tissues, implies that the mutations occurred extremely early in development (Trifunovic et al., 2004). This hypothesis was later confirmed with ultra-deep sequencing to study the genome-wide mtDNA mutation load in the mtDNA mutator mouse (Ameur et al., 2011) and in human tissues (He et al., 2010). Using Digital Deletion Detection (3D), a newer technology that allows for the study of rare, low occurring mtDNA deletions, Taylor et al. (2014) report that although the total deletion load increases in human brain with aging, the number and diversity of deletions remains constant. Therefore, the authors agree that mtDNA mutations associated with premature aging occur from early, pre-existing deletions and not de novo deletions (Taylor et al., 2014). Mutations that occur in early development also support the stem cell hypothesis in which mtDNA mutagenesis modulates somatic stem cell dysfunction. For instance, mtDNA mutator mice were found to have impaired neural and hematopoietic progenitor cells during embryogenesis, which resulted in decreased self-renewal, quality and amount of quiescent cells and may contribute to the progeroid phenotype (Ahlqvist et al., 2012).
Conversely, the spatial distribution of expanded mutations, within or between tissues, implicates mutations later in life (Khrapko et al., 2004). Should mutations occur during embryogenesis, one would expect specific mutations to be more widely distributed among tissues as seen in the mtDNA mutator mice. However, mutations limited to a local area, such as a single cell or a particular tissue, would be an indication of mutagenesis later in life. For example, Nicholas et al. (2009) describe that mtDNA deletions from two distinct but proximal areas of muscle from an 82-year-old individual are completely different with no mutations in common. Within each area, the mutations are clonally expanded but are of late origin since the mutations are not shared between the same tissue (Nicholas et al., 2009). More recently, a combined scenario for mutations has been proposed in which mtDNA mutations seen in aging tissue are a mixture of pre-existing and de novo mutations that expand at different rates depending on cell type and/or conditions placed upon the cells (Popadin et al., 2014).
8. Mitochondrial dynamics and aging
Mitochondria do not exist as solitary, rigidly structured organelles, but rather are continuously remodeled through the dynamic processes of biogenesis, fusion, fission and mitophagy (Chan, 2006, 2012). Mitochondrial biogenesis is needed to meet the energy demands of the cell and to replace the damaged mitochondria removed by mitophagy. Biogenesis is mediated by the transcriptional co-activator, peroxisome-proliferator-activated receptor-γ coactivator 1α (PGC-1α) (Ventura-Clapier et al., 2008). Age-related symptoms, particularly sarcopenia, have been linked to decreased expression of PGC-1α and hence, decreased mitochondrial biogenesis (Joseph et al., 2013; Wenz, 2011).
Fusion and fission are opposing actions that work together to maintain the number, size, shape and function of mitochondria (Chan, 2012). Fusion forms interconnected mitochondrial networks that allow for the exchange of metabolites, proteins and DNA between mitochondria. There are three GTPases involved in fusion: the mitofusins, Mfn1 and Mfn2, for the outer mitochondrial membrane and OPA1 for the inner mitochondrial membrane. Depletion of any of one these proteins results in severe dysfunction in respiratory capacity and fragmentation of mitochondria (Chen et al., 2005; Chen et al., 2010; Song et al., 2009), while homozygous knock-outs of either Mfn1 or Mfn2 are embryonic lethal (Chen et al., 2003). Additionally, conditional deletion of Mfn1 and Mfn2 in mouse skeletal muscle correlates with increased accumulation of mtDNA point mutations and deletions (Chen et al., 2010). Fission segregates the mitochondria into daughter cells during cell division and allows for the selective elimination of damaged mitochondria by mitophagy (Chen and Chan, 2009; Twig et al., 2008). The dynamin-related protein Drp1 and its four potential receptors: Fis1, Mff, MiD49 and MiD51/MIEF1 that recruit Drp1 to the outer mitochondrial membrane are critical for fission (Chan, 2012). Loss of function in these proteins results in severe fission defects (Lee et al., 2004). Similar to fusion proteins, silencing Drp1 and Fis1 in human cells that are heteroplasmic for the pathological A3243G mtDNA mutation results in increased mtDNA mutation loads as compared with untransformed cells (Malena et al., 2009). As expected, ageing is associated with decreases in both fission and fusion (Bereiter-Hahn, 2014; Chauhan et al., 2014; Jendrach et al., 2005; Seo et al., 2010).
As previously discussed, mtDNA mutations increase with age, but heteroplasmy must exceed ~60% in a given cell to have measurable dysfunction. Clonal expansion to this level may be delayed via fusion-directed complementation, resulting in dilution of the mutated mtDNA through content mixing and, ultimately, protection of mitochondrial function (Nakada et al., 2009; Seo et al., 2010). Support for inter-mitochondrial complementation as a protectant of respiratory function has been shown in human cells and in mice. In the first study, fusion with WT mtDNA rescued respiratory enzyme activity in two cell lines rendered respiration-deficient by pathogenic mitochondrial tRNA mutations (Ono et al., 2001). The same group studied the pathogenesis of mutant mtDNA carrying a 4696-basepair deletion in mice. Mixing with WT mtDNA preserved respiratory function until the mutated mtDNA exceeded 85% (Nakada et al., 2001). In another study, Chen et al. (2010) knocked out Mfn1 in the mtDNA mutator mouse to demonstrate the protective effects of mitochondrial fusion on existing mtDNA mutations. Although the double knock-out leads to a synthetic neonatal lethality, mouse embryonic fibroblasts recover and show significantly reduced production of ATP due to severe disruption in ETC complexes. Mitochondrial fusion apparently allows the mtDNA mutator mouse to carry a higher mutational load without affecting respiratory function (Chen et al., 2010).
When fusion-directed complementation cannot decrease the mtDNA mutational load, severely damaged mitochondria may be eliminated. In mammalian cells, Twig et al. (2008) describe fission of individual mitochondria to form daughter organelles with unequal mitochondrial membrane potentials (ΔΨMs). Daughter mitochondria with high ΔΨM undergo subsequent fusion. Daughter mitochondria with low ΔΨM (depolarized) have reduced levels of OPA1. Selective fusion segregates them from the population, after which they may be removed by mitophagy, thus maintaining a healthy mitochondrial population and upholding the bioenergetic efficiency of the cell (Twig et al., 2008). As with the fission and fusion processes, insufficient mitophagy and autophagy contribute to the accumulation of mutant mtDNA and are a factor in aging and age-related diseases (Gaziev et al., 2014; Green et al., 2011; Hubbard et al., 2012; Rubinsztein et al., 2011).
9. Interventions to attenuate the symptoms of aging
Although there is potential for the genetic manipulation of the proteins of the mtDNA replication machinery to be used as future therapies for aging and age-related diseases, currently, the best-known strategies for reducing the symptoms of aging caused by mitochondrial dysfunction appear to be caloric restriction (CR) and exercise. CR is the reduction in caloric intake by 10 to 40% while providing essential nutrients and vitamins, thereby eschewing malnutrition (Testa et al., 2014). CR has been shown to extend both medium and maximum lifespan and delay the onset of age-related diseases in a variety of species, including yeast (Kaeberlein et al., 2007), C. elegans (Lee et al., 2006), rodents (Masoro, 2009) and Rhesus monkeys (Colman et al., 2014).
Several mechanisms have been proposed to explain the effects of CR on extending longevity. Originally, it was shown that CR decreases ROS production in rodents, particularly at Complex I, thereby preventing oxidative damage in mitochondria from heart (Gredilla et al., 2002), liver (Lopez-Torres et al., 2002), skeletal muscle (Lass et al., 1998) and brain (Sanz et al., 2005). However, mitohormesis predicts low levels of ROS, possibly due to increasing the respiration rate, can activate systemic defense mechanisms enhancing longevity (Hekimi et al., 2011). CR apparently increases oxidative metabolism, respiration and biogenesis through multiple pathways. For example, CR induces mitochondrial biogenesis and increases respiration through activation of sirtuin 1 (SIRT1), an NAD+-dependent histone deacetylase, and its downstream induction of PGC-1α (Chistiakov et al., 2014; Martin-Montalvo and de Cabo, 2013). CR likewise activates nutrient- or energy-sensing pathways such as 5′-adenosine monophosphate-activated protein kinase (AMPK), which can then both activate PGC-1α and inactivate the mechanistic target of rapamycin (mTOR) (Chistiakov et al., 2014; Ruetenik and Barrientos, 2015). For example, CR induced mitochondrial biogenesis in this manner and improved bioenergetic efficiency in a variety of cell lines (Lopez-Lluch et al., 2006). CR-increased mitochondrial biogenesis improved mitochondrial function and rescued some progeroid phenotypes in both humans and mtDNA mutator mice (Civitarese et al., 2007; Dillon et al., 2012). CR has also been reported to enhance genomic stability by reversing mtDNA damage (Cassano et al., 2004; Civitarese et al., 2007) and/or promoting mtDNA repair (Cabelof et al., 2003; Heydari et al., 2007).
Various types of exercise training verifiably improve mitochondrial oxidative function in aging and mitochondrial myopathic muscles (Irving et al., 2015; Jeppesen et al., 2006; Short et al., 2004; Short et al., 2003; Wenz et al., 2009). As with CR, exercise triggers mitochondrial biogenesis and increases ETC enzyme levels through upregulation of PGC-1α and/or TFAM in aged skeletal muscle (Barbieri et al., 2015; Kang et al., 2013). Exercise also influences mitochondrial dynamics by changing fusion and fission protein expression and by promoting mitochondrial turnover through mitophagy (Drake et al., 2016). For instance, increased levels of Drp1 were measured in skeletal muscle of mice after 60 minutes of endurance treadmill running, while Mnf1 and OPA1 did not significantly change (Pagano et al., 2014). However, acute exercise in rats increased mRNA expression of Mnf1 and Mfn2 24 hours post-exercise (Cartoni et al., 2005). In two human studies, Fis1 expression was higher in regularly active individuals as compared to their sedentary age-matched controls (Bori et al., 2012) and Fis1, Mfn1 and Mfn2 proteins were upregulated after twelve weeks of aerobic exercise training in older men (Konopka et al., 2014). Furthermore, mitophagy/autophagy markers were increased in normal mice following short-term intense treadmill exercise (Grumati et al., 2011; Saleem et al., 2014). Similarly, resistance and endurance exercise training resulted in increased autophagy activity and reduced apoptosis in aged skeletal muscle (Kim et al., 2013; Luo et al., 2013). Even five months of endurance exercise in the mtDNA mutator mouse moderated systemic apoptosis, thus helping to rescue the progeroid phenotype in these mice (Safdar et al., 2011).
Of great importance is whether exercise can alter the augmented mtDNA mutation load observed in aging patients and/or patients with mitochondrial myopathies. In the mtDNA mutator mouse, endurance exercise improves oxidative capacity, prevents or repairs mtDNA depletion and mutations through tumor suppressor protein p53 and increases lifespan (Safdar et al., 2011; Safdar et al., 2015). However, others have reported that although oxidative capacity is improved, there are no changes in the mtDNA mutation load (Jeppesen et al., 2009; Taivassalo et al., 2006). A promising alternative is gene shifting, the activation of satellite muscles cells or dormant myofibers that do not harbor any mtDNA mutations to reenter the cell cycle and fuse with existing mature myofibers and shift the heteroplasmy ratio in favor of WT over mutated mtDNA (Taivassalo et al., 1999). This activation is accomplished with either eccentric or concentric resistance exercise, as shown in a patient with the G12315A tRNAleu(CUN) point mutation, where resistance training significantly improved heteroplasmic ratios and increased the number of respiratory-competent fibers in muscle fibers (Taivassalo et al., 1999). Resistance training in patients with a large mtDNA deletion led to increased muscle strength, myofiber damage and regeneration, improved oxidative capacity, increased satellite cell portion (Murphy et al., 2008) and apparent mtDNA shifting (Spendiff et al., 2013). Preliminary data suggests that mtDNA shifting also occurs in aging skeletal muscle following resistance exercise training (Tarnopolsky, 2009).
Adherence to a strict CR regime and/or exercise training has proven difficult in humans. Therefore, the use of CR mimetics, which target the same metabolic and stress pathways as CR and/or exercise, is now being examined. Examples include: SIRT-1 activators, such as resveratrol and green tea polyphenols; glycolytic inhibitors, such as 2-deoxy-D-glucose; insulin/AMPK activators, such as metformin; and mTOR inhibitors, such as rapamycin, which have been reviewed elsewhere (Ingram and Roth, 2015; Testa et al., 2014).
10. Conclusion
Aging is a multifactorial, progressive, biological process with mitochondria playing a central role. The MFRTA, once at the forefront of aging theory, has lost favor since its fundamental notion of a “vicious” cycle of ROS generating ETC disruption, and hence more ROS, is based on anecdotal evidence. This does not imply that high levels of ROS do not contribute to the aging phenotype through oxidative damage to cellular components, such as proteins and lipids. However, ROS does not appear to be the initiating or critical factor. The theory of mitohormesis states that low levels of ROS are important signaling molecules that regulate stress response pathways and enhance longevity. Therefore, clonally expanded mtDNA mutations caused by replication errors leads to mitochondrial dysfunction. It is this process, in conjunction with altered mitochondrial dynamics, that emerges as the driving force behind aging (Figure 4). Here, we showed that deficiency and/or dysfunction in many of the nuclear-encoded mtDNA replication proteins, including Pol γ, PolG2, Twinkle, TFAM, MGME1 and RNase H1, results in age-related diseases and/or phenotypes both in vitro and in vivo. Future studies focused on understanding the mechanisms of action are needed to effectively delay aging and prevent age-related diseases.
Figure 4.
Factors causal to aging. Spontaneous errors in the mitochondrial replication machinery cause mtDNA damage. Accumulations of this mtDNA damage occur via clonal expansion until tissue specific heteroplasmy is reached and mitochondrial dysfunction is measurable. Increased mitochondrial dysfunction along with decreased mitochondrial dynamics leads to aging and age-related diseases. Mitochondrial dysfunction may also cause increased apoptosis, which is associated with aging. At low levels, ROS triggers stress response and biogenesis pathways and increases mitochondrial dynamics, which are protective and prevent aging. Caloric restriction and exercise functions similarly. Lastly, high levels of ROS can cause damage to cellular components, such as DNA, RNA, protein and lipids, which may also contribute to aging.
Highlights.
Mitochondrial dysfunction in conjunction with altered mitochondrial dynamics drives aging
Spontaneous errors of replication cause mtDNA point mutations, deletions and depletions
Clonally expanded mtDNA mutations lead to mitochondrial dysfunction
ROS does not appear to be a critical factor in aging
Acknowledgments
The authors would like to thank Drs. Scott Lujan, Matthew Longley and Rachel Krasich for critically reading and editing this manuscript and Ms. Maggie Humble for reviewing. We also thank Dr. Scott Lujan for his expertise with 3-D graphics.
Funding
This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES 065078).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, Forsstrom S, Salven P, Angers-Loustau A, Kopra OH, Tyynismaa H, Larsson NG, Wartiovaara K, Prolla T, Trifunovic A, Suomalainen A. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Alvarez V, Corao AI, Sanchez-Ferrero E, De Mena L, Alonso-Montes C, Huerta C, Blazquez M, Ribacoba R, Guisasola LM, Salvador C, Garcia-Castro M, Coto E. Mitochondrial transcription factor A (TFAM) gene variation in Parkinson’s disease. Neurosci Lett. 2008;432:79–82. doi: 10.1016/j.neulet.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Ameur A, Stewart JB, Freyer C, Hagstrom E, Ingman M, Larsson NG, Gyllensten U. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 2011;7:e1002028. doi: 10.1371/journal.pgen.1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri E, Agostini D, Polidori E, Potenza L, Guescini M, Lucertini F, Annibalini G, Stocchi L, De Santi M, Stocchi V. The pleiotropic effect of physical exercise on mitochondrial dynamics in aging skeletal muscle. Oxidative medicine and cellular longevity. 2015;2015:917085. doi: 10.1155/2015/917085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baris OR, Ederer S, Neuhaus JF, von Kleist-Retzow JC, Wunderlich CM, Pal M, Wunderlich FT, Peeva V, Zsurka G, Kunz WS, Hickethier T, Bunck AC, Stockigt F, Schrickel JW, Wiesner RJ. Mosaic Deficiency in Mitochondrial Oxidative Metabolism Promotes Cardiac Arrhythmia during Aging. Cell Metab. 2015;21:667–677. doi: 10.1016/j.cmet.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Bayne AC, Mockett RJ, Orr WC, Sohal RS. Enhanced catabolism of mitochondrial superoxide/hydrogen peroxide and aging in transgenic Drosophila. Biochem J. 2005;391:277–284. doi: 10.1042/BJ20041872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Functions of DNA polymerases. Adv Protein Chem. 2004;69:137–165. doi: 10.1016/S0065-3233(04)69005-X. [DOI] [PubMed] [Google Scholar]
- Belin AC, Bjork BF, Westerlund M, Galter D, Sydow O, Lind C, Pernold K, Rosvall L, Hakansson A, Winblad B, Nissbrandt H, Graff C, Olson L. Association study of two genetic variants in mitochondrial transcription factor A (TFAM) in Alzheimer’s and Parkinson’s disease. Neurosci Lett. 2007;420:257–262. doi: 10.1016/j.neulet.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- Bensch KG, Degraaf W, Hansen PA, Zassenhaus HP, Corbett JA. A transgenic model to study the pathogenesis of somatic mtDNA mutation accumulation in beta-cells. Diabetes, obesity & metabolism. 2007;9(Suppl 2):74–80. doi: 10.1111/j.1463-1326.2007.00776.x. [DOI] [PubMed] [Google Scholar]
- Bensch KG, Mott JL, Chang SW, Hansen PA, Moxley MA, Chambers KT, de Graaf W, Zassenhaus HP, Corbett JA. Selective mtDNA mutation accumulation results in beta-cell apoptosis and diabetes development. American journal of physiology. 2009;296:E672–680. doi: 10.1152/ajpendo.90839.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Mitochondrial dynamics in aging and disease. Progress in molecular biology and translational science. 2014;127:93–131. doi: 10.1016/B978-0-12-394625-6.00004-0. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bori Z, Zhao Z, Koltai E, Fatouros IG, Jamurtas AZ, Douroudos II, Terzis G, Chatzinikolaou A, Sovatzidis A, Draganidis D, Boldogh I, Radak Z. The effects of aging, physical training, and a single bout of exercise on mitochondrial protein expression in human skeletal muscle. Exp Gerontol. 2012;47:417–424. doi: 10.1016/j.exger.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon BR, Diederich NJ, Soni M, Witte K, Weinhold M, Krause M, Jackson S. Autosomal dominant mutations in POLG and C10orf2: association with late onset chronic progressive external ophthalmoplegia and Parkinsonism in two patients. J Neurol. 2013;260:1931–1933. doi: 10.1007/s00415-013-6975-2. [DOI] [PubMed] [Google Scholar]
- Brown WM, George MJ, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, Prager EM, Wang A, Wilson AC. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18:225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am J Hum Genet. 2006;79:469–480. doi: 10.1086/507132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof DC, Yanamadala S, Raffoul JJ, Guo Z, Soofi A, Heydari AR. Caloric restriction promotes genomic stability by induction of base excision repair and reversal of its age-related decline. DNA Repair (Amst) 2003;2:295–307. doi: 10.1016/s1568-7864(02)00219-7. [DOI] [PubMed] [Google Scholar]
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Capel F, Rimbert V, Lioger D, Diot A, Rousset P, Mirand PP, Boirie Y, Morio B, Mosoni L. Due to reverse electron transfer, mitochondrial H2O2 release increases with age in human vastus lateralis muscle although oxidative capacity is preserved. Mech Ageing Dev. 2005;126:505–511. doi: 10.1016/j.mad.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C. Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell. 2001;7:43–54. doi: 10.1016/s1097-2765(01)00153-8. [DOI] [PubMed] [Google Scholar]
- Cartoni R, Leger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Deriaz O, Zorzano A, Gobelet C, Kralli A, Russell AP. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. The Journal of physiology. 2005;567:349–358. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano P, Lezza AM, Leeuwenburgh C, Cantatore P, Gadaleta MN. Measurement of the 4,834-bp mitochondrial DNA deletion level in aging rat liver and brain subjected or not to caloric restriction diet. Ann N Y Acad Sci. 2004;1019:269–273. doi: 10.1196/annals.1297.045. [DOI] [PubMed] [Google Scholar]
- Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Chakravarti B, Chakravarti DN. Oxidative modification of proteins: age-related changes. Gerontology. 2007;53:128–139. doi: 10.1159/000097865. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Vera J, Wolkenhauer O. The systems biology of mitochondrial fission and fusion and implications for disease and aging. Biogerontology. 2014;15:1–12. doi: 10.1007/s10522-013-9474-z. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14(Spec No 2):R283–289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Sobenin IA, Revin VV, Orekhov AN, Bobryshev YV. Mitochondrial aging and age-related dysfunction of mitochondria. BioMed research international. 2014;2014:238463. doi: 10.1155/2014/238463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E, Team CP. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS medicine. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun. 2014;5:3557. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- Copeland WC, Longley MJ. Mitochondrial genome maintenance in health and disease. DNA Repair (Amst) 2014;19:190–198. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992a;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, McKee AC, Beal MF, Graham BH, Wallace DC. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23:471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat Res. 1992b;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell DA, Blakely EL, Johnson MA, Ince PG, Borthwick GM, Turnbull DM. Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol Aging. 2001;22:265–272. doi: 10.1016/s0197-4580(00)00234-7. [DOI] [PubMed] [Google Scholar]
- Coxhead J, Kurzawa-Akanbi M, Hussain R, Pyle A, Chinnery P, Hudson G. Somatic mtDNA variation is an important component of Parkinson’s disease. Neurobiol Aging. 2015 doi: 10.1016/j.neurobiolaging.2015.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig K, Young MJ, Blakely EL, Longley MJ, Turnbull DM, Copeland WC, Taylor RW. A p.R369G POLG2 mutation associated with adPEO and multiple mtDNA deletions causes decreased affinity between polymerase gamma subunits. Mitochondrion. 2012;12:313–319. doi: 10.1016/j.mito.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidzon G, Greene P, Mancuso M, Klos KJ, Ahlskog JE, Hirano M, Dimauro S. Early-onset familial parkinsonism due to POLG mutations. Ann Neurol. 2006;59:859–862. doi: 10.1002/ana.20831. [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- Delgado-Alvarado M, de la Riva P, Jimenez-Urbieta H, Gago B, Gabilondo A, Bornstein B, Rodriguez-Oroz MC. Parkinsonism, cognitive deficit and behavioural disturbance caused by a novel mutation in the polymerase gamma gene. J Neurol Sci. 2015;350:93–97. doi: 10.1016/j.jns.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, Moraes CT. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 2002;30:4626–4633. doi: 10.1093/nar/gkf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon LM, Williams SL, Hida A, Peacock JD, Prolla TA, Lincoln J, Moraes CT. Increased mitochondrial biogenesis in muscle improves aging phenotypes in the mtDNA mutator mouse. Hum Mol Genet. 2012;21:2288–2297. doi: 10.1093/hmg/dds049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JC, Wilson RJ, Yan Z. Molecular mechanisms for mitochondrial adaptation to exercise training in skeletal muscle. FASEB J. 2016;30:13–22. doi: 10.1096/fj.15-276337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvoisin RC. Overview of Parkinson’s disease. Ann N Y Acad Sci. 1992;648:187–193. doi: 10.1111/j.1749-6632.1992.tb24537.x. [DOI] [PubMed] [Google Scholar]
- Edgar D, Shabalina I, Camara Y, Wredenberg A, Calvaruso MA, Nijtmans L, Nedergaard J, Cannon B, Larsson NG, Trifunovic A. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Edgar D, Trifunovic A. The mtDNA mutator mouse: Dissecting mitochondrial involvement in aging. Aging. 2009;1:1028–1032. doi: 10.18632/aging.100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson FS, Trifunovic A, Hoffer B, Cullheim S, Mohammed AH, Olson L, Larsson NG. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc Natl Acad Sci U S A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- Elson JL, Samuels DC, Turnbull DM, Chinnery PF. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am J Hum Genet. 2001;68:802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge G, Pham XH, Holmlund T, Khorostov I, Falkenberg M. The accessory subunit B of DNA polymerase {gamma} is required for mitochondrial replisome function. Nucleic Acids Res. 2007;35:902–911. doi: 10.1093/nar/gkl1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet G, Jansson M, Sternberg D, Moslemi AR, Blondy P, Lombes A, Fardeau M, Oldfors A. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul Disord. 2002;12:484–493. doi: 10.1016/s0960-8966(01)00332-7. [DOI] [PubMed] [Google Scholar]
- Filippov V, Filippov M, Gill SS. Drosophila RNase H1 is essential for development but not for proliferation. Mol Genet Genomics. 2001;265:771–777. doi: 10.1007/s004380100483. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA. A transcription factor required for promoter recognition by human mitochondrial RNA polymerase. Accurate initiation at the heavy- and light-strand promoters dissected and reconstituted in vitro. J Biol Chem. 1985;260:11330–11338. [PubMed] [Google Scholar]
- Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- Fleming JE, Miquel J, Cottrell SF, Yengoyan LS, Economos AC. Is cell aging caused by respiration-dependent injury to the mitochondrial genome? Gerontology. 1982;28:44–53. doi: 10.1159/000212510. [DOI] [PubMed] [Google Scholar]
- Garrido N, Griparic L, Jokitalo E, Wartiovaara J, Van Der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier BR, Wiederkehr A, Baquie M, Dai C, Powers AC, Kerr-Conte J, Pattou F, MacDonald RJ, Ferrer J, Wollheim CB. PDX1 deficiency causes mitochondrial dysfunction and defective insulin secretion through TFAM suppression. Cell Metab. 2009;10:110–118. doi: 10.1016/j.cmet.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziev AI, Abdullaev S, Podlutsky A. Mitochondrial function and mitochondrial DNA maintenance with advancing age. Biogerontology. 2014;15:417–438. doi: 10.1007/s10522-014-9515-2. [DOI] [PubMed] [Google Scholar]
- Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffart S, Cooper HM, Tyynismaa H, Wanrooij S, Suomalainen A, Spelbrink JN. Twinkle mutations associated with autosomal dominant progressive external ophthalmoplegia lead to impaired helicase function and in vivo mtDNA replication stalling. Hum Mol Genet. 2009;18:328–340. doi: 10.1093/hmg/ddn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat Struct Mol Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in Mitochondrial DNA Replication and Repair. Chemical Reviews. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Greaves LC, Nooteboom M, Elson JL, Tuppen HA, Taylor GA, Commane DM, Arasaradnam RP, Khrapko K, Taylor RW, Kirkwood TB, Mathers JC, Turnbull DM. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 2014;10:e1004620. doi: 10.1371/journal.pgen.1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R, Lopez-Torres M, Barja G. Effect of time of restriction on the decrease in mitochondrial H2O2 production and oxidative DNA damage in the heart of food-restricted rats. Microscopy research and technique. 2002;59:273–277. doi: 10.1002/jemt.10204. [DOI] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Schiavinato A, Castagnaro S, Bertaggia E, Sandri M, Bonaldo P. Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy. 2011;7:1415–1423. doi: 10.4161/auto.7.12.17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui YX, Xu ZP, Lv W, Zhao JJ, Hu XY. Evidence for polymerase gamma, POLG1 variation in reduced mitochondrial DNA copy number in Parkinson’s disease. Parkinsonism & related disorders. 2015;21:282–286. doi: 10.1016/j.parkreldis.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Halsne R, Esbensen Y, Wang W, Scheffler K, Suganthan R, Bjoras M, Eide L. Lack of the DNA glycosylases MYH and OGG1 in the cancer prone double mutant mouse does not increase mitochondrial DNA mutagenesis. DNA Repair (Amst) 2012;11:278–285. doi: 10.1016/j.dnarep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? Journal of the American Geriatrics Society. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Hattori K, Sugiyama S, Ozawa T. Age-associated oxygen damage and mutations in mitochondrial DNA in human hearts. Biochem Biophys Res Commun. 1992;189:979–985. doi: 10.1016/0006-291x(92)92300-m. [DOI] [PubMed] [Google Scholar]
- Hayakawa M, Torii K, Sugiyama S, Tanaka M, Ozawa T. Age-associated accumulation of 8-hydroxydeoxyguanosine in mitochondrial DNA of human diaphragm. Biochem Biophys Res Commun. 1991;179:1023–1029. doi: 10.1016/0006-291x(91)91921-x. [DOI] [PubMed] [Google Scholar]
- Hayashi J, Ohta S, Kikuchi A, Takemitsu M, Goto Y, Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc Natl Acad Sci U S A. 1991;88:10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Yoshida M, Yamato M, Ide T, Wu Z, Ochi-Shindou M, Kanki T, Kang D, Sunagawa K, Tsutsui H, Nakanishi H. Reverse of age-dependent memory impairment and mitochondrial DNA damage in microglia by an overexpression of human mitochondrial transcription factor a in mice. J Neurosci. 2008;28:8624–8634. doi: 10.1523/JNEUROSCI.1957-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekimi S, Lapointe J, Wen Y. Taking a “good” look at free radicals in the aging process. Trends in cell biology. 2011;21:569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydari AR, Unnikrishnan A, Lucente LV, Richardson A. Caloric restriction and genomic stability. Nucleic Acids Res. 2007;35:7485–7496. doi: 10.1093/nar/gkm860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Marti R, Ferreiro-Barros C, Vila MR, Tadesse S, Nishigaki Y, Nishino I, Vu TH. Defects of intergenomic communication: autosomal disorders that cause multiple deletions and depletion of mitochondrial DNA. Semin Cell Dev Biol. 2001;12:417–427. doi: 10.1006/scdb.2001.0279. [DOI] [PubMed] [Google Scholar]
- Holmes JB, Akman G, Wood SR, Sakhuja K, Cerritelli SM, Moss C, Bowmaker MR, Jacobs HT, Crouch RJ, Holt IJ. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication. Proc Natl Acad Sci U S A. 2015;112:9334–9339. doi: 10.1073/pnas.1503653112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmlund T, Farge G, Pande V, Korhonen J, Nilsson L, Falkenberg M. Structure-function defects of the twinkle amino-terminal region in progressive external ophthalmoplegia. Biochim Biophys Acta. 2009;1792:132–139. doi: 10.1016/j.bbadis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Hubbard VM, Valdor R, Macian F, Cuervo AM. Selective autophagy in the maintenance of cellular homeostasis in aging organisms. Biogerontology. 2012;13:21–35. doi: 10.1007/s10522-011-9331-x. [DOI] [PubMed] [Google Scholar]
- Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 2014;10:e1004369. doi: 10.1371/journal.pgen.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Schaefer AM, Taylor RW, Tiangyou W, Gibson A, Venables G, Griffiths P, Burn DJ, Turnbull DM, Chinnery PF. Mutation of the linker region of the polymerase gamma-1 (POLG1) gene associated with progressive external ophthalmoplegia and Parkinsonism. Arch Neurol. 2007;64:553–557. doi: 10.1001/archneur.64.4.553. [DOI] [PubMed] [Google Scholar]
- Humble MM, Young MJ, Foley JF, Pandiri AR, Travlos GS, Copeland WC. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum Mol Genet. 2013;22:1017–1025. doi: 10.1093/hmg/dds506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Ide T, Fujino T, Arai S, Saku K, Kakino T, Tyynismaa H, Yamasaki T, Yamada K, Kang D, Suomalainen A, Sunagawa K. Overexpression of TFAM or twinkle increases mtDNA copy number and facilitates cardioprotection associated with limited mitochondrial oxidative stress. PLoS One. 2015;10:e0119687. doi: 10.1371/journal.pone.0119687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- Ingram DK, Roth GS. Calorie restriction mimetics: can you have your cake and eat it, too? Ageing research reviews. 2015;20:46–62. doi: 10.1016/j.arr.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Irving BA, Lanza IR, Henderson GC, Rao RR, Spiegelman BM, Nair KS. Combined training enhances skeletal muscle mitochondrial oxidative capacity independent of age. J Clin Endocrinol Metab. 2015;100:1654–1663. doi: 10.1210/jc.2014-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara LS, Kennedy SR, Fox EJ, Yu S, Hewitt JJ, Sanchez-Contreras M, Cardozo-Pelaez F, Pallanck LJ. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014;10:e1003974. doi: 10.1371/journal.pgen.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Thomas RR, Portell FR, Dunham LD, Quigley CK, Bennett JP., Jr Recombinant mitochondrial transcription factor A with N-terminal mitochondrial transduction domain increases respiration and mitochondrial gene expression. Mitochondrion. 2009;9:196–203. doi: 10.1016/j.mito.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic J. Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry. 2008;79:368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- Jemt E, Persson O, Shi Y, Mehmedovic M, Uhler JP, Davila Lopez M, Freyer C, Gustafsson CM, Samuelsson T, Falkenberg M. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015;43:9262–9275. doi: 10.1093/nar/gkv804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendrach M, Pohl S, Voth M, Kowald A, Hammerstein P, Bereiter-Hahn J. Morpho-dynamic changes of mitochondria during ageing of human endothelial cells. Mech Ageing Dev. 2005;126:813–821. doi: 10.1016/j.mad.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Jeppesen TD, Duno M, Schwartz M, Krag T, Rafiq J, Wibrand F, Vissing J. Short- and long-term effects of endurance training in patients with mitochondrial myopathy. Eur J Neurol. 2009;16:1336–1339. doi: 10.1111/j.1468-1331.2009.02660.x. [DOI] [PubMed] [Google Scholar]
- Jeppesen TD, Schwartz M, Olsen DB, Wibrand F, Krag T, Duno M, Hauerslev S, Vissing J. Aerobic training is safe and improves exercise capacity in patients with mitochondrial myopathy. Brain. 2006;129:3402–3412. doi: 10.1093/brain/awl149. [DOI] [PubMed] [Google Scholar]
- Joseph AM, Adhihetty PJ, Wawrzyniak NR, Wohlgemuth SE, Picca A, Kujoth GC, Prolla TA, Leeuwenburgh C. Dysregulation of mitochondrial quality control processes contribute to sarcopenia in a mouse model of premature aging. PLoS One. 2013;8:e69327. doi: 10.1371/journal.pone.0069327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kang C, Chung E, Diffee G, Ji LL. Exercise training attenuates aging-associated mitochondrial dysfunction in rat skeletal muscle: role of PGC-1alpha. Exp Gerontol. 2013;48:1343–1350. doi: 10.1016/j.exger.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Kanki T, Nakayama H, Sasaki N, Takio K, Alam TI, Hamasaki N, Kang D. Mitochondrial nucleoid and transcription factor A. Ann N Y Acad Sci. 2004;1011:61–68. doi: 10.1007/978-3-662-41088-2_7. [DOI] [PubMed] [Google Scholar]
- Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9:e1003794. doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MJ, Chinnery PF. Mitochondrial DNA mutations in neurodegeneration. Biochim Biophys Acta. 2015;1847:1401–1411. doi: 10.1016/j.bbabio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Khan SM, Bennett JP., Jr Development of mitochondrial gene replacement therapy. J Bioenerg Biomembr. 2004;36:387–393. doi: 10.1023/B:JOBB.0000041773.20072.9e. [DOI] [PubMed] [Google Scholar]
- Khrapko K. The timing of mitochondrial DNA mutations in aging. Nat Genet. 2011;43:726–727. doi: 10.1038/ng.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K, Ebralidse K, Kraytsberg Y. Where and when do somatic mtDNA mutations occur? Ann N Y Acad Sci. 2004;1019:240–244. doi: 10.1196/annals.1297.040. [DOI] [PubMed] [Google Scholar]
- Kim YA, Kim YS, Oh SL, Kim HJ, Song W. Autophagic response to exercise training in skeletal muscle with age. Journal of physiology and biochemistry. 2013;69:697–705. doi: 10.1007/s13105-013-0246-7. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Konopka AR, Suer MK, Wolff CA, Harber MP. Markers of human skeletal muscle mitochondrial biogenesis and quality control: effects of age and aerobic exercise training. The journals of gerontology Series A, Biological sciences and medical sciences. 2014;69:371–378. doi: 10.1093/gerona/glt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5′ -> 3′ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- Korhonen JA, Pande V, Holmlund T, Farge G, Pham XH, Nilsson L, Falkenberg M. Structure-function defects of the TWINKLE linker region in progressive external ophthalmoplegia. J Mol Biol. 2008;377:691–705. doi: 10.1016/j.jmb.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Kornblum C, Nicholls TJ, Haack TB, Scholer S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Iuso A, Wieland T, Sciacco M, Ronchi D, Comi GP, Moggio M, Quinzii CM, DiMauro S, Calvo SE, Mootha VK, Klopstock T, Strom TM, Meitinger T, Minczuk M, Kunz WS, Prokisch H. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowald A, Kirkwood TB. Evolution of the mitochondrial fusion-fission cycle and its role in aging. Proc Natl Acad Sci U S A. 2011;108:10237–10242. doi: 10.1073/pnas.1101604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y, Simon DK, Turnbull DM, Khrapko K. Do mtDNA deletions drive premature aging in mtDNA mutator mice? Aging Cell. 2009;8:502–506. doi: 10.1111/j.1474-9726.2009.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Kukat A, Trifunovic A. Somatic mtDNA mutations and aging--facts and fancies. Exp Gerontol. 2009;44:101–105. doi: 10.1016/j.exger.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Kwong LK, Sohal RS. Age-related changes in activities of mitochondrial electron transport complexes in various tissues of the mouse. Arch Biochem Biophys. 2000;373:16–22. doi: 10.1006/abbi.1999.1495. [DOI] [PubMed] [Google Scholar]
- Lamantea E, Tiranti V, Bordoni A, Toscano A, Bono F, Servidei S, Papadimitriou A, Spelbrink H, Silvestri L, Casari G, Comi G, Zeviani M. Mutations of mitochondrial DNA polymerase gamma are a frequent cause of autosomal dominant or recessive Progressive External Ophthalmoplegia. Ann Neurol. 2002;52:211–219. doi: 10.1002/ana.10278. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lass A, Sohal BH, Weindruch R, Forster MJ, Sohal RS. Caloric restriction prevents age-associated accrual of oxidative damage to mouse skeletal muscle mitochondria. Free Radic Biol Med. 1998;25:1089–1097. doi: 10.1016/s0891-5849(98)00144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci U S A. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, Ingram DK, Zou S. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Choi CS, Birkenfeld AL, Alves TC, Jornayvaz FR, Jurczak MJ, Zhang D, Woo DK, Shadel GS, Ladiges W, Rabinovitch PS, Santos JH, Petersen KF, Samuel VT, Shulman GI. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab. 2010;12:668–674. doi: 10.1016/j.cmet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Zhao MH, Zheng Z, Kwon JW, Liang S, Kim SH, Kim NH, Cui XS. Polymerase subunit gamma 2 affects porcine oocyte maturation and subsequent embryonic development. Theriogenology. 2015;83:121–130. doi: 10.1016/j.theriogenology.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis W, Day BJ, Kohler JJ, Hosseini SH, Chan SSL, Green E, Haase CP, Keebaugh E, Long R, Ludaway T, Russ R, Steltzer J, Tioleco N, Santoianni R, Copeland WC. MtDNA depletion, oxidative stress, cardiomyopathy, and death from transgenic cardiac targeted human mutant polymerase gamma. Lab Invest. 2007;87:326–335. doi: 10.1038/labinvest.3700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Wilhelmsson H, Hansson A, Thoren P, Duffy J, Rustin P, Larsson NG. Genetic modification of survival in tissue-specific knockout mice with mitochondrial cardiomyopathy. Proc Natl Acad Sci U S A. 2000;97:3467–3472. doi: 10.1073/pnas.97.7.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, Olson JL, Noble LJ, Yoshimura MP, Berger C, Chan PH, Wallace DC, Epstein CJ. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- Lin MT, Simon DK, Ahn CH, Kim LM, Beal MF. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum Mol Genet. 2002;11:133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Clark S, Yu Wai Man C, Hudson G, Durham SE, Taylor RW, Nightingale S, Turnbull DM, Copeland WC, Chinnery PF. Mutant POLG2 Disrupts DNA Polymerase gamma Subunits and Causes Progressive External Ophthalmoplegia. Am J Hum Genet. 2006;78:1026–1034. doi: 10.1086/504303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Humble MM, Sharief FS, Copeland WC. Disease variants of the human mitochondrial DNA helicase encoded by C10orf2 differentially alter protein stability, nucleotide hydrolysis and helicase activity. J Biol Chem. 2010;285:29690–29702. doi: 10.1074/jbc.M110.151795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley MJ, Nguyen D, Kunkel TA, Copeland WC. The Fidelity of Human DNA Polymerase gamma with and without Exonucleolytic Proofreading and the p55 Accessory Subunit. J Biol Chem. 2001;276:38555–38562. doi: 10.1074/jbc.M105230200. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Prasad R, Srivastava DK, Wilson SH, Copeland WC. Identification of 5′-deoxyribose phosphate lyase activity in human DNA polymerase gamma and its role in mitochondrial base excision repair in vitro. Proc Natl Acad Sci U S A. 1998;95:12244–12248. doi: 10.1073/pnas.95.21.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Torres M, Gredilla R, Sanz A, Barja G. Influence of aging and long-term caloric restriction on oxygen radical generation and oxidative DNA damage in rat liver mitochondria. Free Radic Biol Med. 2002;32:882–889. doi: 10.1016/s0891-5849(02)00773-6. [DOI] [PubMed] [Google Scholar]
- Luo L, Lu AM, Wang Y, Hong A, Chen Y, Hu J, Li X, Qin ZH. Chronic resistance training activates autophagy and reduces apoptosis of muscle cells by modulating IGF-1 and its receptors, Akt/mTOR and Akt/FOXO3a signaling in aged rats. Exp Gerontol. 2013;48:427–436. doi: 10.1016/j.exger.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Luo Y, Hoffer A, Hoffer B, Qi X. Mitochondria: A Therapeutic Target for Parkinson’s Disease? International journal of molecular sciences. 2015;16:20704–20730. doi: 10.3390/ijms160920704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoma P, Melberg A, Rinne JO, Kaukonen JA, Nupponen NN, Chalmers RM, Oldfors PA, Rautakorpi I, Peltonen PL, Majamaa PK, Somer H, Suomalainen A. Parkinsonism, premature menopause, and mitochondrial DNA polymerase gamma mutations: clinical and molecular genetic study. Lancet. 2004;364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- Luoma PT, Eerola J, Ahola S, Hakonen AH, Hellstrom O, Kivisto KT, Tienari PJ, Suomalainen A. Mitochondrial DNA polymerase gamma variants in idiopathic sporadic Parkinson disease. Neurology. 2007;69:1152–1159. doi: 10.1212/01.wnl.0000276955.23735.eb. [DOI] [PubMed] [Google Scholar]
- Malena A, Loro E, Di Re M, Holt IJ, Vergani L. Inhibition of mitochondrial fission favours mutant over wild-type mitochondrial DNA. Hum Mol Genet. 2009;18:3407–3416. doi: 10.1093/hmg/ddp281. [DOI] [PubMed] [Google Scholar]
- Mancuso M, Filosto M, Oh SJ, DiMauro S. A novel polymerase gamma mutation in a family with ophthalmoplegia, neuropathy, and Parkinsonism. Arch Neurol. 2004;61:1777–1779. doi: 10.1001/archneur.61.11.1777. [DOI] [PubMed] [Google Scholar]
- Mandavilli BS, Santos JH, Van Houten B. Mitochondrial DNA repair and aging. Mutat Res. 2002;509:127–151. doi: 10.1016/s0027-5107(02)00220-8. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Muller FL, Liu Y, Ng R, Faulkner J, Hamilton M, Richardson A, Huang TT, Epstein CJ, Van Remmen H. Alterations in mitochondrial function, hydrogen peroxide release and oxidative damage in mouse hind-limb skeletal muscle during aging. Mech Ageing Dev. 2006;127:298–306. doi: 10.1016/j.mad.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Martin-Montalvo A, de Cabo R. Mitochondrial metabolic reprogramming induced by calorie restriction. Antioxid Redox Signal. 2013;19:310–320. doi: 10.1089/ars.2012.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Caloric restriction-induced life extension of rats and mice: a critique of proposed mechanisms. Biochim Biophys Acta. 2009;1790:1040–1048. doi: 10.1016/j.bbagen.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Farr CL, Fan L, Kaguni LS. Physiological and biochemical defects in carboxyl-terminal mutants of mitochondrial DNA helicase. J Biol Chem. 2008;283:23964–23971. doi: 10.1074/jbc.M803674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima Y, Kaguni LS. Differential phenotypes of active site and human adPEO mutations in drosophila mitochondrial DNA helicase expressed in schneider cells. J Biol Chem. 2007;282:9436–9444. doi: 10.1074/jbc.M610550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Matic S, Kuhl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet. 2013;22:1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FJ, Rosenfeldt FL, Zhang C, Linnane AW, Nagley P. Precise determination of mitochondrial DNA copy number in human skeletal and cardiac muscle by a PCR-based assay: lack of change of copy number with age. Nucleic Acids Res. 2003;31:e61. doi: 10.1093/nar/gng060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moslemi AR, Melberg A, Holme E, Oldfors A. Autosomal dominant progressive external ophthalmoplegia: distribution of multiple mitochondrial DNA deletions. Neurology. 1999;53:79–84. doi: 10.1212/wnl.53.1.79. [DOI] [PubMed] [Google Scholar]
- Muller-Hocker J. Cytochrome-c-oxidase deficient cardiomyocytes in the human heart--an age-related phenomenon. A histochemical ultracytochemical study. Am J Pathol. 1989;134:1167–1173. [PMC free article] [PubMed] [Google Scholar]
- Muller-Hocker J, Seibel P, Schneiderbanger K, Kadenbach B. Different in situ hybridization patterns of mitochondrial DNA in cytochrome c oxidase-deficient extraocular muscle fibres in the elderly. Virchows Archiv A, Pathological anatomy and histopathology. 1993;422:7–15. doi: 10.1007/BF01605127. [DOI] [PubMed] [Google Scholar]
- Murphy JL, Blakely EL, Schaefer AM, He L, Wyrick P, Haller RG, Taylor RW, Turnbull DM, Taivassalo T. Resistance training in patients with single, large-scale deletions of mitochondrial DNA. Brain. 2008;131:2832–2840. doi: 10.1093/brain/awn252. [DOI] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada K, Inoue K, Ono T, Isobe K, Ogura A, Goto YI, Nonaka I, Hayashi JI. Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat Med. 2001;7:934–940. doi: 10.1038/90976. [DOI] [PubMed] [Google Scholar]
- Nakada K, Sato A, Hayashi J. Mitochondrial functional complementation in mitochondrial DNA-based diseases. Int J Biochem Cell Biol. 2009;41:1907–1913. doi: 10.1016/j.biocel.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Ngo HB, Lovely GA, Phillips R, Chan DC. Distinct structural features of TFAM drive mitochondrial DNA packaging versus transcriptional activation. Nat Commun. 2014;5:3077. doi: 10.1038/ncomms4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A, Kraytsberg Y, Guo X, Khrapko K. On the timing and the extent of clonal expansion of mtDNA deletions: evidence from single-molecule PCR. Experimental neurology. 2009;218:316–319. doi: 10.1016/j.expneurol.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls TJ, Zsurka G, Peeva V, Scholer S, Szczesny RJ, Cysewski D, Reyes A, Kornblum C, Sciacco M, Moggio M, Dziembowski A, Kunz WS, Minczuk M. Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease. Hum Mol Genet. 2014;23:6147–6162. doi: 10.1093/hmg/ddu336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S, Shitara H, Nakada K, Ono T, Sato A, Suzuki H, Ogawa T, Masaki H, Hayashi J, Yonekawa H. Over-expression of Tfam improves the mitochondrial disease phenotypes in a mouse model system. Biochem Biophys Res Commun. 2010;401:26–31. doi: 10.1016/j.bbrc.2010.08.143. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry. 1999;38:605–618. doi: 10.1021/bi982207z. [DOI] [PubMed] [Google Scholar]
- Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–275. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- Orr WC, Radyuk SN, Sohal RS. Involvement of redox state in the aging of Drosophila melanogaster. Antioxid Redox Signal. 2013;19:788–803. doi: 10.1089/ars.2012.5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- Pagano AF, Py G, Bernardi H, Candau RB, Sanchez AM. Autophagy and protein turnover signaling in slow-twitch muscle during exercise. Medicine and science in sports and exercise. 2014;46:1314–1325. doi: 10.1249/MSS.0000000000000237. [DOI] [PubMed] [Google Scholar]
- Pagnamenta AT, Taanman JW, Wilson CJ, Anderson NE, Marotta R, Duncan AJ, Bitner-Glindzicz M, Taylor RW, Laskowski A, Thorburn DR, Rahman S. Dominant inheritance of premature ovarian failure associated with mutant mitochondrial DNA polymerase gamma. Hum Reprod. 2006;21:2467–2473. doi: 10.1093/humrep/del076. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Taylor RW, Turnbull DM, Suomalainen A, Jacobs HT, Karhunen PJ. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS One. 2010;5:e10426. doi: 10.1371/journal.pone.0010426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, Jacobs HT. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Williams SL, Boettger T, Goffart S, Kim J, Suomalainen A, Moraes CT, Braun T. Overexpression of Twinkle-helicase protects cardiomyocytes from genotoxic stress caused by reactive oxygen species. Proc Natl Acad Sci U S A. 2013;110:19408–19413. doi: 10.1073/pnas.1303046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponamarev MV, Longley MJ, Nguyen D, Kunkel TA, Copeland WC. Active Site Mutation in DNA Polymerase gamma Associated with Progressive External Ophthalmoplegia Causes Error-prone DNA Synthesis. J Biol Chem. 2002;277:15225–15228. doi: 10.1074/jbc.C200100200. [DOI] [PubMed] [Google Scholar]
- Popadin K, Safdar A, Kraytsberg Y, Khrapko K. When man got his mtDNA deletions? Aging Cell. 2014;13:579–582. doi: 10.1111/acel.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, Devenney PS, Sexton D, Grimes G, Holt IJ, Hill RE, Taylor MS, Lawson KA, Dorin JR, Jackson AP. Enzymatic removal of ribonucleotides from DNA is essential for Mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remes AM, Hinttala R, Karppa M, Soini H, Takalo R, Uusimaa J, Majamaa K. Parkinsonism associated with the homozygous W748S mutation in the POLG1 gene. Parkinsonism & related disorders. 2008;14:652–654. doi: 10.1016/j.parkreldis.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Reyes A, Melchionda L, Nasca A, Carrara F, Lamantea E, Zanolini A, Lamperti C, Fang M, Zhang J, Ronchi D, Bonato S, Fagiolari G, Moggio M, Ghezzi D, Zeviani M. RNASEH1 Mutations Impair mtDNA Replication and Cause Adult-Onset Mitochondrial Encephalomyopathy. Am J Hum Genet. 2015;97:186–193. doi: 10.1016/j.ajhg.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropp PA, Copeland WC. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase gamma. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Ruetenik A, Barrientos A. Dietary restriction, mitochondrial function and aging: from yeast to humans. Biochim Biophys Acta. 2015;1847:1434–1447. doi: 10.1016/j.bbabio.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhanen H, Ushakov K, Yasukawa T. Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim Biophys Acta. 2011;1813:2000–2007. doi: 10.1016/j.bbamcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, Prolla TA, Tarnopolsky MA. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Safdar A, Khrapko K, Flynn JM, Saleem A, De Lisio M, Johnston AP, Kratysberg Y, Samjoo IA, Kitaoka Y, Ogborn DI, Little JP, Raha S, Parise G, Akhtar M, Hettinga BP, Rowe GC, Arany Z, Prolla TA, Tarnopolsky MA. Exercise-induced mitochondrial p53 repairs mtDNA mutations in mutator mice. Skeletal muscle. 2015;6:7. doi: 10.1186/s13395-016-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Saleem A, Carter HN, Hood DA. p53 is necessary for the adaptive changes in cellular milieu subsequent to an acute bout of endurance exercise. Am J Physiol Cell Physiol. 2014;306:C241–249. doi: 10.1152/ajpcell.00270.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martinez A, Calleja M, Peralta S, Matsushima Y, Hernandez-Sierra R, Whitworth AJ, Kaguni LS, Garesse R. Modeling pathogenic mutations of human twinkle in Drosophila suggests an apoptosis role in response to mitochondrial defects. PLoS One. 2012;7:e43954. doi: 10.1371/journal.pone.0043954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A, Caro P, Ibanez J, Gomez J, Gredilla R, Barja G. Dietary restriction at old age lowers mitochondrial oxygen radical production and leak at complex I and oxidative DNA damage in rat brain. J Bioenerg Biomembr. 2005;37:83–90. doi: 10.1007/s10863-005-4131-0. [DOI] [PubMed] [Google Scholar]
- Sawada M, Carlson JC. Changes in superoxide radical and lipid peroxide formation in the brain, heart and liver during the lifetime of the rat. Mech Ageing Dev. 1987;41:125–137. doi: 10.1016/0047-6374(87)90057-1. [DOI] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ. Extension of mouse lifespan by overexpression of catalase. Age. 2006;28:209–218. doi: 10.1007/s11357-006-9010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Schriner SE, Ogburn CE, Smith AC, Newcomb TG, Ladiges WC, Dolle ME, Vijg J, Fukuchi K, Martin GM. Levels of DNA damage are unaltered in mice overexpressing human catalase in nuclei. Free Radic Biol Med. 2000;29:664–673. doi: 10.1016/s0891-5849(00)00352-x. [DOI] [PubMed] [Google Scholar]
- Scuderi C, Borgione E, Castello F, Lo Giudice M, Santa Paola S, Giambirtone M, Di Blasi FD, Elia M, Amato C, Citta S, Gagliano C, Barbarino G, Vitello GA, Musumeci SA. The in cis T251I and P587L POLG1 base changes: description of a new family and literature review. Neuromuscul Disord. 2015;25:333–339. doi: 10.1016/j.nmd.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Sen D, Nandakumar D, Tang GQ, Patel SS. Human mitochondrial DNA helicase TWINKLE is both an unwinding and annealing helicase. J Biol Chem. 2012;287:14545–14556. doi: 10.1074/jbc.M111.309468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123:2533–2542. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KR, Vittone JL, Bigelow ML, Proctor DN, Nair KS. Age and aerobic exercise training effects on whole body and muscle protein metabolism. American journal of physiology. 2004;286:E92–101. doi: 10.1152/ajpendo.00366.2003. [DOI] [PubMed] [Google Scholar]
- Short KR, Vittone JL, Bigelow ML, Proctor DN, Rizza RA, Coenen-Schimke JM, Nair KS. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes. 2003;52:1888–1896. doi: 10.2337/diabetes.52.8.1888. [DOI] [PubMed] [Google Scholar]
- Shutt TE, Gray MW. Twinkle, the mitochondrial replicative DNA helicase, is widespread in the eukaryotic radiation and may also be the mitochondrial DNA primase in most eukaryotes. J Mol Evol. 2006;62:588–599. doi: 10.1007/s00239-005-0162-8. [DOI] [PubMed] [Google Scholar]
- Silva JP, Kohler M, Graff C, Oldfors A, Magnuson MA, Berggren PO, Larsson NG. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet. 2000;26:336–340. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- Sohal RS, Sohal BH. Hydrogen peroxide release by mitochondria increases during aging. Mech Ageing Dev. 1991;57:187–202. doi: 10.1016/0047-6374(91)90034-w. [DOI] [PubMed] [Google Scholar]
- Song L, Shan Y, Lloyd KC, Cortopassi GA. Mutant Twinkle increases dopaminergic neurodegeneration, mtDNA deletions and modulates Parkin expression. Hum Mol Genet. 2012;21:5147–5158. doi: 10.1093/hmg/dds365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen L, Ekstrand M, Silva JP, Lindqvist E, Xu B, Rustin P, Olson L, Larsson NG. Late-onset corticohippocampal neurodepletion attributable to catastrophic failure of oxidative phosphorylation in MILON mice. J Neurosci. 2001;21:8082–8090. doi: 10.1523/JNEUROSCI.21-20-08082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, Toivonen JM, Hakkaart GA, Kurkela JM, Cooper HM, Lehtinen SK, Lecrenier N, Back JW, Speijer D, Foury F, Jacobs HT. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- Spendiff S, Reza M, Murphy JL, Gorman G, Blakely EL, Taylor RW, Horvath R, Campbell G, Newman J, Lochmuller H, Turnbull DM. Mitochondrial DNA deletions in muscle satellite cells: implications for therapies. Hum Mol Genet. 2013;22:4739–4747. doi: 10.1093/hmg/ddt327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- Suarez J, Hu Y, Makino A, Fricovsky E, Wang H, Dillmann WH. Alterations in mitochondrial function and cytosolic calcium induced by hyperglycemia are restored by mitochondrial transcription factor A in cardiomyocytes. Am J Physiol Cell Physiol. 2008;295:C1561–1568. doi: 10.1152/ajpcell.00076.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Tower J. FLP recombinase-mediated induction of Cu/Zn-superoxide dismutase transgene expression can extend the life span of adult Drosophila melanogaster flies. Mol Cell Biol. 1999;19:216–228. doi: 10.1128/mcb.19.1.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- Szczesny RJ, Hejnowicz MS, Steczkiewicz K, Muszewska A, Borowski LS, Ginalski K, Dziembowski A. Identification of a novel human mitochondrial endo-/exonuclease Ddk1/c20orf72 necessary for maintenance of proper 7S DNA levels. Nucleic Acids Res. 2013;41:3144–3161. doi: 10.1093/nar/gkt029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taanman JW, Schapira AH. Analysis of the trinucleotide CAG repeat from the DNA polymerase gamma gene (POLG) in patients with Parkinson’s disease. Neurosci Lett. 2005;376:56–59. doi: 10.1016/j.neulet.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Taivassalo T, Fu K, Johns T, Arnold D, Karpati G, Shoubridge EA. Gene shifting: a novel therapy for mitochondrial myopathy. Hum Mol Genet. 1999;8:1047–1052. doi: 10.1093/hmg/8.6.1047. [DOI] [PubMed] [Google Scholar]
- Taivassalo T, Gardner JL, Taylor RW, Schaefer AM, Newman J, Barron MJ, Haller RG, Turnbull DM. Endurance training and detraining in mitochondrial myopathies due to single large-scale mtDNA deletions. Brain. 2006;129:3391–3401. doi: 10.1093/brain/awl282. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Ide T, Fujino T, Onitsuka K, Ikeda M, Takehara T, Hata Y, Ylikallio E, Tyynismaa H, Suomalainen A, Sunagawa K. The overexpression of Twinkle helicase ameliorates the progression of cardiac fibrosis and heart failure in pressure overload model in mice. PLoS One. 2013;8:e67642. doi: 10.1371/journal.pone.0067642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnopolsky MA. Mitochondrial DNA shifting in older adults following resistance exercise training. Applied physiology, nutrition, and metabolism = Physiologie appliquee, nutrition et metabolisme. 2009;34:348–354. doi: 10.1139/H09-022. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Barron MJ, Borthwick GM, Gospel A, Chinnery PF, Samuels DC, Taylor GA, Plusa SM, Needham SJ, Greaves LC, Kirkwood TB, Turnbull DM. Mitochondrial DNA mutations in human colonic crypt stem cells. J Clin Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SD, Ericson NG, Burton JN, Prolla TA, Silber JR, Shendure J, Bielas JH. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell. 2014;13:29–38. doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G, Biasi F, Poli G, Chiarpotto E. Calorie restriction and dietary restriction mimetics: a strategy for improving healthy aging and longevity. Current pharmaceutical design. 2014;20:2950–2977. doi: 10.2174/13816128113196660699. [DOI] [PubMed] [Google Scholar]
- Tiangyou W, Hudson G, Ghezzi D, Ferrari G, Zeviani M, Burn DJ, Chinnery PF. POLG1 in idiopathic Parkinson disease. Neurology. 2006;67:1698–1700. doi: 10.1212/01.wnl.0000238963.07425.d5. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci U S A. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, Oresic M, Moraes CT, Pietilainen K, Hovatta I, Suomalainen A. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19:3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum Mol Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Mutation of POLG is associated with progressive external ophthalmoplegia characterized by mtDNA deletions. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Luoma P, Rantamaki M, Al Memar A, Kaakkola S, Hackman P, Krahe R, Lofgren A, Martin JJ, De Jonghe P, Suomalainen A, Udd B, Van Broeckhoven C. POLG mutations in neurodegenerative disorders with ataxia but no muscle involvement. Neurology. 2004;63:1251–1257. doi: 10.1212/01.wnl.0000140494.58732.83. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Martin JJ, Dermaut B, Lofgren A, Wibail A, Ververken D, Tack P, Dehaene I, Van Zandijcke M, Moonen M, Ceuterick C, De Jonghe P, Van Broeckhoven C. Recessive POLG mutations presenting with sensory and ataxic neuropathy in compound heterozygote patients with progressive external ophthalmoplegia. Neuromuscul Disord. 2003a;13:133–142. doi: 10.1016/s0960-8966(02)00216-x. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Martin JJ, Van Broeckhoven C. Progressive external ophthalmoplegia characterized by multiple deletions of mitochondrial DNA: unraveling the pathogenesis of human mitochondrial DNA instability and the initiation of a genetic classification. Neuromolecular medicine. 2003b;3:129–146. doi: 10.1385/NMM:3:3:129. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Schwartz M, Lofgren A, Dermaut B, Van Broeckhoven C, Vissing J. Novel POLG mutations in progressive external ophthalmoplegia mimicking mitochondrial neurogastrointestinal encephalomyopathy. Eur J Hum Genet. 2003c;11:547–549. doi: 10.1038/sj.ejhg.5201002. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Superoxide dismutase is dispensable for normal animal lifespan. Proc Natl Acad Sci U S A. 2012;109:5785–5790. doi: 10.1073/pnas.1116158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiological genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, Loeb LA. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Loeb LA. On mitochondria, mutations, and methodology. Cell Metab. 2009;10:437. doi: 10.1016/j.cmet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA in aging and disease. Sci Am. 1997;277:40–47. doi: 10.1038/scientificamerican0897-40. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat Res. 1998;400:99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- Wang J, Wilhelmsson H, Graff C, Li H, Oldfors A, Rustin P, Bruning JC, Kahn CR, Clayton DA, Barsh GS, Thoren P, Larsson NG. Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat Genet. 1999;21:133–137. doi: 10.1038/5089. [DOI] [PubMed] [Google Scholar]
- Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic Acids Res. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T. Mitochondria and PGC-1alpha in Aging and Age-Associated Diseases. Journal of aging research. 2011;2011:810619. doi: 10.4061/2011/810619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Diaz F, Hernandez D, Moraes CT. Endurance exercise is protective for mice with mitochondrial myopathy. Journal of applied physiology. 2009;106:1712–1719. doi: 10.1152/japplphysiol.91571.2008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wredenberg A, Wibom R, Wilhelmsson H, Graff C, Wiener HH, Burden SJ, Oldfors A, Westerblad H, Larsson NG. Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci U S A. 2002;99:15066–15071. doi: 10.1073/pnas.232591499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun H, Liang X, Lima WF, Crooke ST. Human RNase H1 is associated with protein P32 and is involved in mitochondrial pre-rRNA processing. PLoS One. 2013;8:e71006. doi: 10.1371/journal.pone.0071006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, Miller JH. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- Xu S, Zhong M, Zhang L, Wang Y, Zhou Z, Hao Y, Zhang W, Yang X, Wei A, Pei L, Yu Z. Overexpression of Tfam protects mitochondria against beta-amyloid-induced oxidative damage in SH-SY5Y cells. FEBS J. 2009;276:3800–3809. doi: 10.1111/j.1742-4658.2009.07094.x. [DOI] [PubMed] [Google Scholar]
- Yatsuga S, Suomalainen A. Effect of bezafibrate treatment on late-onset mitochondrial myopathy in mice. Hum Mol Genet. 2012;21:526–535. doi: 10.1093/hmg/ddr482. [DOI] [PubMed] [Google Scholar]
- Yen TC, Su JH, King KL, Wei YH. Ageing-associated 5 kb deletion in human liver mitochondrial DNA. Biochem Biophys Res Commun. 1991;178:124–131. doi: 10.1016/0006-291x(91)91788-e. [DOI] [PubMed] [Google Scholar]
- Ylikallio E, Tyynismaa H, Tsutsui H, Ide T, Suomalainen A. High mitochondrial DNA copy number has detrimental effects in mice. Hum Mol Genet. 2010;19:2695–2705. doi: 10.1093/hmg/ddq163. [DOI] [PubMed] [Google Scholar]
- Ylonen S, Ylikotila P, Siitonen A, Finnila S, Autere J, Majamaa K. Variations of mitochondrial DNA polymerase gamma in patients with Parkinson’s disease. J Neurol. 2013;260:3144–3149. doi: 10.1007/s00415-013-7132-7. [DOI] [PubMed] [Google Scholar]
- Young MJ, Humble MM, DeBalsi KL, Sun KY, Copeland WC. POLG2 disease variants: analyses reveal a dominant negative heterodimer, altered mitochondrial localization and impaired respiratory capacity. Hum Mol Genet. 2015;24:5184–5197. doi: 10.1093/hmg/ddv240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MJ, Longley MJ, Li FY, Kasiviswanathan R, Wong LJ, Copeland WC. Biochemical analysis of human POLG2 variants associated with mitochondrial disease. Hum Mol Genet. 2011;20:3052–3066. doi: 10.1093/hmg/ddr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- Zhang D, Mott JL, Chang SW, Denniger G, Feng Z, Zassenhaus HP. Construction of transgenic mice with tissue-specific acceleration of mitochondrial DNA mutagenesis. Genomics. 2000;69:151–161. doi: 10.1006/geno.2000.6333. [DOI] [PubMed] [Google Scholar]
- Zhang D, Mott JL, Chang SW, Stevens M, Mikolajczak P, Zassenhaus HP. Mitochondrial DNA mutations activate programmed cell survival in the mouse heart. Am J Physiol Heart Circ Physiol. 2005;288:H2476–2483. doi: 10.1152/ajpheart.00670.2004. [DOI] [PubMed] [Google Scholar]
- Zhang D, Mott JL, Farrar P, Ryerse JS, Chang SW, Stevens M, Denniger G, Zassenhaus HP. Mitochondrial DNA mutations activate the mitochondrial apoptotic pathway and cause dilated cardiomyopathy. Cardiovasc Res. 2003;57:147–157. doi: 10.1016/s0008-6363(02)00695-8. [DOI] [PubMed] [Google Scholar]
- Zheng W, Khrapko K, Coller H, Thilly WG, Copeland WC. Origins of human mitochondrial point mutations as DNA polymerase gamma-mediated errors. Mutation Research. 2006;599:11–20. doi: 10.1016/j.mrfmmm.2005.12.012. [DOI] [PubMed] [Google Scholar]