Abstract
More than 100 trillion microbial cells that reside in the human gut heavily influence nutrition, metabolism and immune function of the host. Gut dysbiosis, seen commonly in patients with Chronic Kidney Disease (CKD), results from qualitative and quantitative changes in host microbiome profile and disruption of gut barrier function. Alterations in gut microbiota and a myriad of host responses have been implicated in progression of CKD, increased cardiovascular risk, uremic toxicity and inflammation. We present a discussion of dysbiosis, various uremic toxins produced from dysbiotic gut microbiome, and their roles in CKD progression and complications. We also review the gut microbiome in renal transplant, highlighting the role of commensal microbes in alteration of immune responses to transplantation, and conclude with therapeutic interventions that aim to restore intestinal dysbiosis.
Keywords: Gut microbiome, chronic kidney disease, uremic toxins
Introduction
Human gut is home to approximately 100 trillion microorganisms and has about 100 times the number of genes in our genome (1). In health, gut microbiota exists in “symbiosis” and provides the host with a wide range of metabolic capabilities such as breakdown of indigestible plant polysaccharides (2), synthesis of vitamins (3), biotransformation of conjugated bile acids and degradation of dietary oxalates (4). Postnatal colonization of the intestine by bacteria educates our immune system and reduces allergic responses to food and environmental antigens. Chronic Kidney Disease (CKD) is a global public health problem affecting up to 10% of the population (1). “Microbiome-centric theory of CKD progression” proposes that initial adaptive changes in gut microbiome become maladaptive in later stages of CKD, leading to CKD-related complications (5). Dysbiosis in patients with CKD is being increasingly recognized as a potential therapeutic target, but the science is in its nascent state. In this review we will discuss the recent advances in the field of gut microbiome and how it could revolutionize management of CKD-related complications.
Gut microbiome in health - symbiosis
Advances in sequencing technology and bio-informatics have revealed the complexity of human microbiome. The human microbial community (microbiota) includes bacteria, archaea, viruses, phages, fungi and other microbial eukarya. Findings from the Human Microbiome Project (HMP) suggest that each individual’s microbiome is unique, each niche is characterized by one or a few signature taxa, and diversity is greatest in the gut with little variation over time (6, 7). The predominant bacterial groups in the human gastrointestinal tract are Bacteroidetes, Firmicutes and Actinobacter (8).
Acquisition of the human microbiota begins at birth and it undergoes dynamic adaptive changes over time that are influenced by age, sex, race/ethnicity, geography, diet and host genetics. Findings from the Twins UK study indicate that abundance of many microbial taxa are influenced by host genetics (8). Thaiss et al. showed that intestinal microbiota exhibits diurnal oscillations that are influenced by feeding rhythms, leading to time-specific compositional changes (9). Metagenomic sequencing of fecal samples demonstrated that bacterial genes involved in energy metabolism and protein synthesis are abundant during the day, whereas those genes related to detoxification become abundant at night (9). Indeed, gut microbial ecosystem is considered as an organ by itself, whose metabolic capacity exceeds that of liver.
Diet has a significant impact on the gut microbiome profile. Muegge et al. studied gut microbiome profile in 33 mammalian species and 18 humans, and noted that there is clear separation of microbiome by host diet (10). They reported that the difference in microbiome profile stems from differing metabolic functions required to utilize the diet (10). When fecal microbiota of European children was compared to that of children from a rural African village who consume high fiber diet, it was evident that African children showed a significant enrichment in Bacteroidetes and depletion in Firmicutes, with a unique abundance of bacteria from the genus Prevotella and Xylanibacter, known to contain a set of bacterial genes for cellulose and xylan hydrolysis (11). It appears that gut microbiota in African children has coevolved with the polysaccharide-rich diet, allowing them to maximize energy intake from fibers.
Gut microbiome in disease - dysbiosis
Dysbiosis, a term first described by Elie Metchnikoff (12), refers to an imbalanced intestinal microbial community with quantitative and qualitative changes in the composition and metabolic activities of the gut microbiota. The “Hygiene hypothesis” and the “Old Friends hypothesis” propose that alterations in gut microbiome induced by hygienic practices may be related to the recent surge in autoimmune diseases and allergic diseases. Indeed, the declining incidence of infectious diseases over the past 50 years appears to be coinciding with the steady rise in the incidence of allergic and autoimmune diseases in developed countries (13). Dysbiosis of the gut microbiota has been implicated in the pathogenesis of both intestinal and extra-intestinal disorders such as allergy and asthma (14), obesity (15), diabetes (16), cardiovascular disease (CVD), and cancer (17). Emerging evidence indicate that dysbiosis may underlie the obesity epidemic. Chevalier et al. show that cold exposure induces marked increase in the ratio of Firmicutes to Bacteroidetes and almost a complete loss of Verrucomicrobia species, including Akkermansia muciniphila (18), which has been shown to promote energy harvest in a mouse model of obesity (19).
Gut microbiome as a potential source of uremic toxins
In 1965, Einheber and Carter showed that germfree anephric mice survived longer than anephric mice with intact gut microbiome (20). Yokoyama showed that sterilizing the intestine by antibiotics decreased the fecal and urinary excretion of phenolic and aromatic bacterial metabolites in weanling pigs with normal kidney function (21). Gut microbiota has been shown to contribute to the generation of several uremic toxins (Table 1). Using untargeted metabolomic mass spectrometry, Wikoff et al. reported that the presence of several protein-bound uremic toxins, such as indoxyl sulphate (IS), hippuric acid and phenylacetic acid are dependent on the presence of gut microflora (22). Aronov et al. studied the plasma samples from hemodialysis patients with and without colons and demonstrated that a number of solutes are absent or present only in low concentrations in subjects without colon, suggesting a colonic origin of these molecules (23). Impaired protein assimilation in uremia leads to influx of undigested proteins into the distal intestine, which favors the proliferation of proteolytic bacteria (24). Increased protein fermentation results in generation of potentially toxic metabolites such as, ammonia, phenols, amines, indoles, and thiols (25). Posen et al. studied the stool metabolomics in 20 hemodialysis patients, 20 healthy subjects, and 20 household contacts (26). Although a clear discrimination was noted between unrelated controls and end-stage renal disease (ESRD) patients, they noted similarity between hemodialysis patients and their household contacts, leading them to conclude that the CKD-related differences in the human colonic microbial metabolism can be attributed to a large extent to dietary restrictions and to a lesser extent to loss of renal function. However, they did not assess the dietary pattern in this study. Furthermore, in the same study they noted that the CKD and non-CKD rats that were fed with same diet had significantly different stool metabolomic profile (26).
Table 1.
Uremic toxins: source, mechanisms of injury and consequence
| Toxin | Source/ Mechanism of action | Consequences | Remarks |
|---|---|---|---|
| Ammonia and Urea | Ammonia, the end product of protein catabolism is converted to urea by Ornithine -Urea cycle. Gut bacteria expressing urease cleave urea into ammonia and carbon dioxide (1). |
Epithelial cell injury by increasing the pH of surrounding environment (2) |
Some ammonia is used for microbial synthesis of amino acids (3) |
| Creatinine, Guanidine and Uric acid |
Up to 60% of creatinine is excreted by routes other than urine in kidney failure (4). Colonic bacteria degrade creatinine in gut. Guanidine is produced by creatinine metabolism by Pseudomonas stutzeri (5). Intestinal organisms can degrade urate, generating allantoin, allantoic acid, urea and ammonia (6) |
Guanidine accumulation in CKD (7) increases mortality noticed in animal studies (8) |
|
| Indoles: Indoxyl sulphate (IS) |
Generated from metabolism of tryptophan (9), cleared by proximal tubules and increased in CKD (10, 11). Nephrotoxic via OAT mediated uptake by proximal tubule cells, activation of nuclear factor (NF)-Kb and plasminogen activator inhibitor type 1 (12–14) |
Increased expression of genes related to tubulointerstitial fibrosis (15), aortic and vascular calcification (16–18), endothelial cell damage (19, 20). Lowers erythropoietin production (21), bone turnover (22) |
Transport is mediated by organic anion transporter 1 and 3 (OAT1, OAT3) (69– 71). Not effectively removed by HD (23, 24) |
| Indole acetic acid (IAA) | Produced by intestinal bacteria form tryptophan. |
Glomerulosclerosis, interstitial fibrosis (3), oxidative stress (4) |
Partly removed by hemodialysis (25–27) |
| Phenols: Para (p) -Cresol |
Breakdown of tyrosine and phenylalanine by intestinal bacteria |
Renal fibrosis, oxidative stress, increased inflammatory cytokines (28), all- cause mortality and CVD . (29, 30)Inhibition of endothelial proliferation, increased endothelial permeability (20, 31) |
Uremic toxin excreted by tubular secretion through specific transporters (32–34), accumulates in CKD (35) |
| Phenylacetylglutamine (PAG) |
Microbe derived, accumulates in uremia (36–38). Phenyl acetic acid, a precursor of PAG is derived from phenylalanine (39). Impairs immunoregulation (40), increases oxidative stress (41) and osteoblast dysfunction (42) |
Tubular damage and progression of CKD (43) mediated by Phenyl acetic acid |
|
| Hippurate | Gut microbial-mammalian co metabolite (44, 45). Causes anion gap acidosis in CKD (46, 47) |
Interferes with erythropoiesis and platelet cyclooxygenase activity (48). Possible glucose intolerance. |
|
| Amines Polyamines | Play regulatory role in cell function and growth (49). Polyamine induced cellular downregulation play a role in lack of tissue responses to hormones in uremia (50) |
Erythropoietin inhibition, anemia of CKD (51) |
Many polyamines are generated by microbiota from precursor amino acids |
| D-amino Acids | Bacterial production in the intestinal tract |
Possible neurotoxicity (52) |
Plasma levels increase with declining kidney function (53) |
| Trimethylamine N Oxide (TMAO) |
Choline is catabolized by intestinal microbiota to trimethylamine gas, which is metabolized by liver to TMAO. Dietary carnitine is also a substrate for gut flora to produce TMAO (54). Promotes atherosclerosis by promoting foam cell formation and increasing scavenger receptors on macrophage (54–56) |
Associated with higher long term mortality (54–56). Levels are high in CKD (57). |
|
| Hydrogen sulfide (H2S) | Generated by sulfate reducing bacteria in colon. Inhibits mitochondrial cytochrome-c oxidase (58). Genotoxic, cytotoxic and inflammatory (59). May decrease inflammation and inhibiting renal fibrosis (60). |
Can be cardio protective (61). |
Levels reduced in hemodialysis patients (62) |
| Endotoxin | Binds to lipopolysaccharide-binding protein (LBP), forming a complex that interacts with the MD-2 part of the toll like receptor 4, anchored by CD14 (100). This stimulates production of inflammatory cytokines (35, 41) |
Endotoxin translocation causes inflammation in CKD (54, 63), possible role in progression of atherosclerosis (64, 65) |
Soluble CD14 associates with progression of CKD,CVD and mortality (66–68) |
Reference List
WALSER M, BODENLOS LJ. Urea metabolism in man. J.Clin.Invest 1959;38:1617-1626.
Vaziri ND, Dure-Smith B, Miller R et al. Pathology of gastrointestinal tract in chronic hemodialysis patients: an autopsy study of 78 cases. Am.J.Gastroenterol. 1985;80(8):608-611.
Stewart GS, Smith CP. Urea nitrogen salvage mechanisms and their relevance to ruminants, non-ruminants and man. Nutr.Res.Rev. 2005;18(1):49-62.
Jones JD, Burnett PC. Creatinine metabolism in humans with decreased renal function: creatinine deficit. Clin.Chem. 1974;20(9):1204-1212.
Eyk HG, van, Vermaat RJ et al. The conversion of creatinine by creatininase of bacterial origin. Enzymologia. 1968;34(3):198-202.
Yokozawa T, Mo ZL, Oura H. Comparison of toxic effects of methylguanidine, guanidinosuccinic acid and creatinine in rats with adenine-induced chronic renal failure. Nephron 1989;51(3):388-392.
OLSEN NS, BASSETT JW. Blood levels of urea nitrogen, phenol, guanidine and creatinine in uremia. Am.J.Med. 1951;10(1):52-59.
OLSEN NS, BASSETT JW. Blood levels of urea nitrogen, phenol, guanidine and creatinine in uremia. Am.J.Med. 1951;10(1):52-59.
Cummings JH. Fermentation in the human large intestine: evidence and implications for health. Lancet 1983;1(8335):1206-1209.
Lin CJ, Chen HH, Pan CF et al. p-Cresylsulfate and indoxyl sulfate level at different stages of chronic kidney disease. J.Clin.Lab Anal. 2011;25(3):191-197.
Wu IW, Hsu KH, Lee CC et al. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol.Dial.Transplant. 2011;26(3):938-947.
Lysaght MJ, Vonesh EF, Gotch F et al. The influence of dialysis treatment modality on the decline of remaining renal function. ASAIO Trans. 1991;37(4):598-604.
Motojima M, Hosokawa A, Yamato H et al. Uraemic toxins induce proximal tubular injury via organic anion transporter 1-mediated uptake. Br.J.Pharmacol. 2002;135(2):555-563.
Motojima M, Hosokawa A, Yamato H et al. Uremic toxins of organic anions up-regulate PAI-1 expression by induction of NF-kappaB and free radical in proximal tubular cells. Kidney Int. 2003;63(5):1671-1680.
Miyazaki T, Ise M, Seo H et al. Indoxyl sulfate increases the gene expressions of TGF-beta 1, TIMP-1 and pro-alpha 1(I) collagen in uremic rat kidneys. Kidney Int.Suppl 1997;62:S15-S22.
Adijiang A, Higuchi Y, Nishijima F et al. Indoxyl sulfate, a uremic toxin, promotes cell senescence in aorta of hypertensive rats. Biochem.Biophys.Res.Commun. 2010;399(4):637-641.
Barreto FC, Barreto DV, Liabeuf S et al. Serum indoxyl sulfate is associated with vascular disease and mortality in chronic kidney disease patients. Clin.J.Am.Soc.Nephrol. 2009;4(10):1551-1558.
Yamamoto H, Tsuruoka S, Ioka T et al. Indoxyl sulfate stimulates proliferation of rat vascular smooth muscle cells. Kidney Int. 2006;69(10):1780-1785.
Amabile N, Guerin AP, Leroyer A et al. Circulating endothelial microparticles are associated with vascular dysfunction in patients with end-stage renal failure. J.Am.Soc.Nephrol. 2005;16(11):3381-3388.
Dou L, Bertrand E, Cerini C et al. The uremic solutes p-cresol and indoxyl sulfate inhibit endothelial proliferation and wound repair. Kidney Int. 2004;65(2):442-451.
Chiang CK, Tanaka T, Inagi R et al. Indoxyl sulfate, a representative uremic toxin, suppresses erythropoietin production in a HIF-dependent manner. Lab Invest 2011;91(11):1564-1571.
Nii-Kono T, Iwasaki Y, Uchida M et al. Indoxyl sulfate induces skeletal resistance to parathyroid hormone in cultured osteoblastic cells. Kidney Int. 2007;71(8):738-743.
Niwa T, Takeda N, Tatematsu A et al. Accumulation of indoxyl sulfate, an inhibitor of drug-binding, in uremic serum as demonstrated by internal-surface reversed-phase liquid chromatography. Clin.Chem. 1988;34(11):2264-2267.
Martinez AW, Recht NS, Hostetter TH et al. Removal of P-cresol sulfate by hemodialysis. J.Am.Soc.Nephrol. 2005;16(11):3430-3436.
De SR, Dhondt A, Eloot S et al. Effect of the super-flux cellulose triacetate dialyser membrane on the removal of non-protein-bound and protein-bound uraemic solutes. Nephrol.Dial.Transplant. 2007;22(7):2006-2012.
Satoh M, Hayashi H, Watanabe M et al. Uremic toxins overload accelerates renal damage in a rat model of chronic renal failure. Nephron Exp.Nephrol. 2003;95(3):e111-e118.
Dou L, Sallee M, Cerini C et al. The cardiovascular effect of the uremic solute indole-3 acetic acid. J.Am.Soc.Nephrol. 2015;26(4):876-887.
Watanabe H, Miyamoto Y, Honda D et al. p-Cresyl sulfate causes renal tubular cell damage by inducing oxidative stress by activation of NADPH oxidase. Kidney Int. 2013;83(4):582-592.
Bammens B, Evenepoel P, Keuleers H et al. Free serum concentrations of the protein-bound retention solute p-cresol predict mortality in hemodialysis patients. Kidney Int. 2006;69(6):1081-1087.
Liabeuf S, Barreto DV, Barreto FC et al. Free p-cresylsulphate is a predictor of mortality in patients at different stages of chronic kidney disease. Nephrol.Dial.Transplant. 2010;25(4):1183-1191.
Cerini C, Dou L, Anfosso F et al. P-cresol, a uremic retention solute, alters the endothelial barrier function in vitro. Thromb.Haemost. 2004;92(1):140-150.
Meijers BK, Evenepoel P. The gut-kidney axis: indoxyl sulfate, p-cresyl sulfate and CKD progression. Nephrol.Dial.Transplant. 2011;26(3):759-761.
Niwa T. Indoxyl sulfate is a nephro-vascular toxin. J.Ren Nutr. 2010;20(5 Suppl):S2-S6.
Mutsaers HA, Wilmer MJ, van den Heuvel LP et al. Basolateral transport of the uraemic toxin p-cresyl sulfate: role for organic anion transporters? Nephrol.Dial.Transplant. 2011;26(12):4149.
Poesen R, Viaene L, Verbeke K et al. Renal clearance and intestinal generation of p-cresyl sulfate and indoxyl sulfate in CKD. Clin.J.Am.Soc.Nephrol. 2013;8(9):1508-1514.
Zimmerman L, Egestad B, Jornvall H et al. Identification and determination of phenylacetylglutamine, a major nitrogenous metabolite in plasma of uremic patients. Clin.Nephrol. 1989;32(3):124-128.
Smith EA, MacFarlane GT. Formation of Phenolic and Indolic Compounds by Anaerobic Bacteria in the Human Large Intestine. Microb.Ecol. 1997;33(3):180-188.
Seakins JW. The determination of urinary phenylacetylglutamine as phenylacetic acid. Studies on its origin in normal subjects and children with cystic fibrosis. Clin.Chim.Acta 1971;35(1):121-131.
Yang D, Beylot M, Agarwal KC et al. Assay of the human liver citric acid cycle probe phenylacetylglutamine and of phenylacetate in plasma by gas chromatography-mass spectrometry. Anal.Biochem. 1993;212(1):277-282.
Schmidt S, Westhoff TH, Krauser P et al. The uraemic toxin phenylacetic acid impairs macrophage function. Nephrol.Dial.Transplant. 2008;23(11):3485-3493.
Schmidt S, Westhoff TH, Krauser P et al. The uraemic toxin phenylacetic acid increases the formation of reactive oxygen species in vascular smooth muscle cells. Nephrol.Dial.Transplant. 2008;23(1):65-71.
Yano S, Yamaguchi T, Kanazawa I et al. The uraemic toxin phenylacetic acid inhibits osteoblastic proliferation and differentiation: an implication for the pathogenesis of low turnover bone in chronic renal failure. Nephrol.Dial.Transplant. 2007;22(11):3160-3165.
Yang D, Beylot M, Agarwal KC et al. Assay of the human liver citric acid cycle probe phenylacetylglutamine and of phenylacetate in plasma by gas chromatography-mass spectrometry. Anal.Biochem. 1993;212(1):277-282.
Remer T, Manz F. Paleolithic diet, sweet potato eaters, and potential renal acid load. Am.J.Clin.Nutr. 2003;78(4):802-803.
Li M, Wang B, Zhang M et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc.Natl.Acad.Sci.U.S.A 2008;105(6):2117-2122.
Cathcart-Rake W, Porter R, Whittier F et al. Effect of diet on serum accumulation and renal excretion of aryl acids and secretory activity in normal and uremic man. Am.J.Clin.Nutr. 1975;28(10):1110-1115.
Mitch WE, Brusilow S. Benzoate-induced changes in glycine and urea metabolism in patients with chronic renal failure. J.Pharmacol.Exp.Ther. 1982;222(3):572-575.
Uremic toxins. Proceedings of the Ghent Symposium. October 3-4, 1986, Ghent, Belgium. Adv.Exp.Med.Biol. 1987;223:1-296.
Igarashi K, Ueda S, Yoshida K et al. Polyamines in renal failure. Amino.Acids 2006;31(4):477-483.
Campbell RA, Grettie DP, Bartos F et al. Uremic polyamine dysmetabolism. Proc.Clin.Dial.Transplant.Forum 1978;8:194-198.
Kushner D, Beckman B, Nguyen L et al. Polyamines in the anemia of end-stage renal disease. Kidney Int. 1991;39(4):725-732.
Oh MS, Phelps KR, Traube M et al. D-lactic acidosis in a man with the short-bowel syndrome. N.Engl.J.Med. 1979;301(5):249-252.
Bruckner H, Hausch M. Gas chromatographic characterization of free D-amino acids in the blood serum of patients with renal disorders and of healthy volunteers. J.Chromatogr. 1993;614(1):7-17.
Koeth RA, Wang Z, Levison BS et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat.Med. 2013;19(5):576-585.
Tang WH, Wang Z, Levison BS et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N.Engl.J.Med. 2013;368(17):1575-1584.
Wang Z, Klipfell E, Bennett BJ et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472(7341):57-63.
Tang WH, Wang Z, Kennedy DJ et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ.Res. 2015;116(3):448-455.
Reiffenstein RJ, Hulbert WC, Roth SH. Toxicology of hydrogen sulfide. Annu.Rev.Pharmacol.Toxicol. 1992;32:109-134.
Aminzadeh MA, Vaziri ND. Downregulation of the renal and hepatic hydrogen sulfide (H2S)-producing enzymes and capacity in chronic kidney disease. Nephrol.Dial.Transplant. 2012;27(2):498-504.
Song K, Wang F, Li Q et al. Hydrogen sulfide inhibits the renal fibrosis of obstructive nephropathy. Kidney Int. 2014;85(6):1318-1329.
Perna AF, Lanza D, Sepe I et al. Hydrogen sulfide, a toxic gas with cardiovascular properties in uremia: how harmful is it? Blood Purif. 2011;31(1-3):102-106.
Song K, Wang F, Li Q et al. Hydrogen sulfide inhibits the renal fibrosis of obstructive nephropathy. Kidney Int. 2014;85(6):1318-1329.
Freudenberg MA, Tchaptchet S, Keck S et al. Lipopolysaccharide sensing an important factor in the innate immune response to Gram-negative bacterial infections: benefits and hazards of LPS hypersensitivity. Immunobiology 2008;213(3-4):193-203.
Eggesbo JB, Hjermann I, Ovstebo R et al. LPS induced procoagulant activity and plasminogen activator activity in mononuclear cells from persons with high or low levels of HDL lipoprotein. Thromb.Res. 1995;77(5):441-452.
Reidy MA, Bowyer DE. Distortion of endothelial repair. The effect of hypercholesterolaemia on regeneration of aortic endothelium following injury by endotoxin. A scanning electron microscope study. Atherosclerosis 1978;29(4):459-466.
Raj DS, Carrero JJ, Shah VO et al. Soluble CD14 levels, interleukin 6, and mortality among prevalent hemodialysis patients. Am.J.Kidney Dis. 2009;54(6):1072-1080.
Raj DS, Shah VO, Rambod M et al. Association of soluble endotoxin receptor CD14 and mortality among patients undergoing hemodialysis. Am.J.Kidney Dis. 2009;54(6):1062-1071.
Poesen R, Ramezani A, Claes K et al. Associations of Soluble CD14 and Endotoxin with Mortality, Cardiovascular Disease, and Progression of Kidney Disease among Patients with CKD. Clin.J.Am.Soc.Nephrol. 2015;10(9):1525-1533.
Prior studies by Vaziri et al. have demonstrated extensive change in the structure and function of the gut microbiome in humans and a animals with advanced CKD (27). Individuals with ESRD restrict fruits and vegetable intake, in an attempt to prevent hyperkalemia. These dietary changes have been shown to result in difference in bacterial taxa between ESRD and healthy individuals. Wong et.al have described significant expansion of intestinal bacteria that contain urease, uricase, and indole and p-cresol forming enzymes; and contraction of short chain fatty acid producing bacteria in subjects with ESRD. Indoxyl sulfate, p-cresol sulfate, and urea-derived ammonia have deleterious effects, whereas SCFAs are known to have beneficial effects, there changes in intestinal microbial metabolism, therefore may contribute to uremic toxicity and inflammation (28).
Uremia increases intestinal permeability, both in uremic rats and in patients with CKD allowing entry of gut derived uremic toxins in to the systemic circulation. Magnusson et al. showed that increased intestinal permeability to large-molecular-weight polyethylene glycols in the uremic animals and humans with CKD (29, 30). Ammonia and ammonium hydroxide are generated in abundance from hydrolysis of urea by microbial urease in uremic state. Ammonia generated this way, mediates breakdown of the gut epithelial tight junction and contributes to endotoxemia and systemic inflammation (31, 32).
In the presence of CKD, histologic changes, such as reduction of villous height, elongation of the crypts, and infiltration of lamina propria with inflammatory cells are evident in the intestine (33). Vaziri et al. observed marked reductions in tight junction protein abundance of claudin-1, occludin, and ZO-1 in colonic mucosa in animals with CKD (34). The disruption of colonic epithelial tight junction could allow translocation of bacteria and endotoxin across the intestinal wall contributing to systemic inflammation in CKD (35, 36). Thus disrupted gut barrier function allows translocation of uremic toxins and the impaired excretory function leads to accumulation of these toxins.
Immune modulation
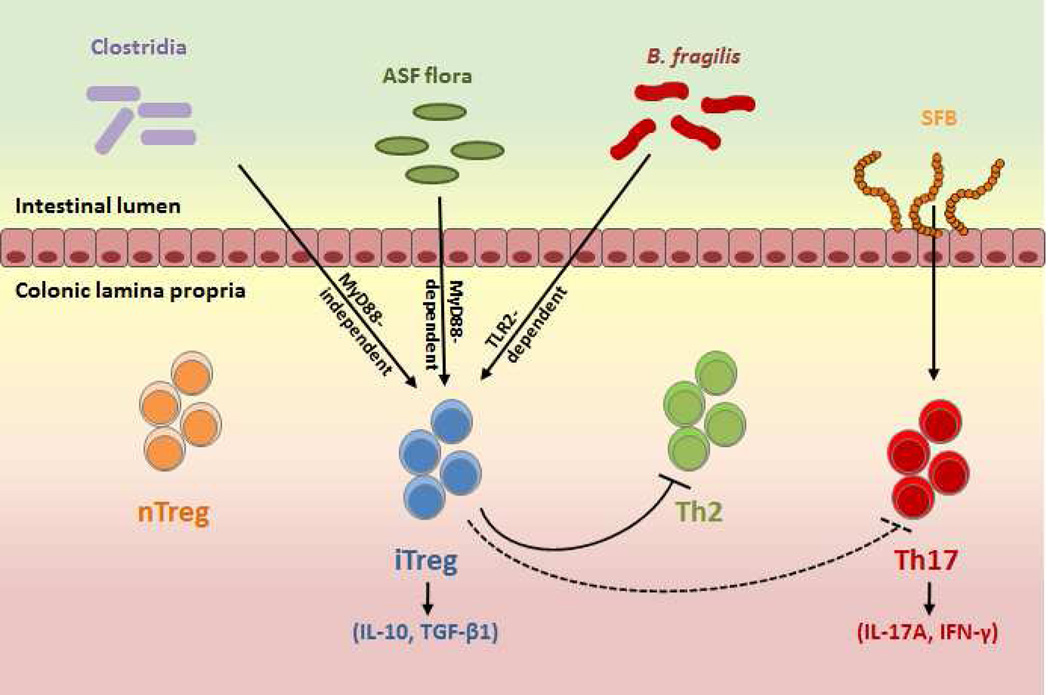
Microbiome immune modulation of host is best exemplified by gut microbial induction of host immune maturation. Various species of gut microbiota have been shown to promote expansion or differentiation of forkhead box protein 3 (Foxp-3) expressing regulatory T cells (Tregs). Some of these Tregs are implicated in recognition of microbial antigens (37, 38). Similarly, colonic Tregs are increased in germ free mice with a set of defined benign commensals (39). While some gut commensals such as Clostridium species have the potential to promote colonic Tregs , others can control the development and maturation of mucosal and systemic natural killer T cells (NKTs) (40), and lymphoid structures (41). Peptidoglycan, an essential component of bacterial cell wall is an important molecule that has the potential to modulate peripheral immune function (42). Gut microbiota are closely involved in regulation of intestinal CD4+ cell induction (Fig. 1). Segmented-filamentous bacteria (SFB) promote induction of Th17 responses, while the Bacteroides fragilis and Clostridia species induce IL-10-producing iTregs. A bacterial polysaccharide (PSA) produced by Bacteroides fragilis, directs the cellular and physical maturation of the developing immune system (43). Another class of bacterial metabolite, short chain fatty acids (SCFA), are known for their ability to promote T-cell differentiation into both effector and regulatory T cells, depending on immunological milieu of the host (44).
Figure 1.
Regulation of intestinal CD4+ T-cell by gut microbiota.
Since immune regulation is key to transplant survival, it would be most logical to assume that microbiome should influence transplant outcome. Generally, there is loss of diversity with the emergence of a new dominant bacterial population post-transplant. In kidney transplant recipients, an increase in the relative abundance of Proteobacteria in the stool samples is observed in the post transplantation specimens compared to pre-transplantation specimens (45). The microbiome profile post-transplant is strongly influenced by the intensity and type of immunosuppression, renal allograft function, infection, medication use including use of antibiotic and proton pump inhibitors. Gut microbiome is involved in metabolism of drugs and is relevant to dosing of medications in transplant patients. Indeed, fecal Faecalibacterium prausnitzii abundance in the first week of transplantation was positively correlated with future tacrolimus dosing at one month (46).
Gut microbiome and progression of CKD
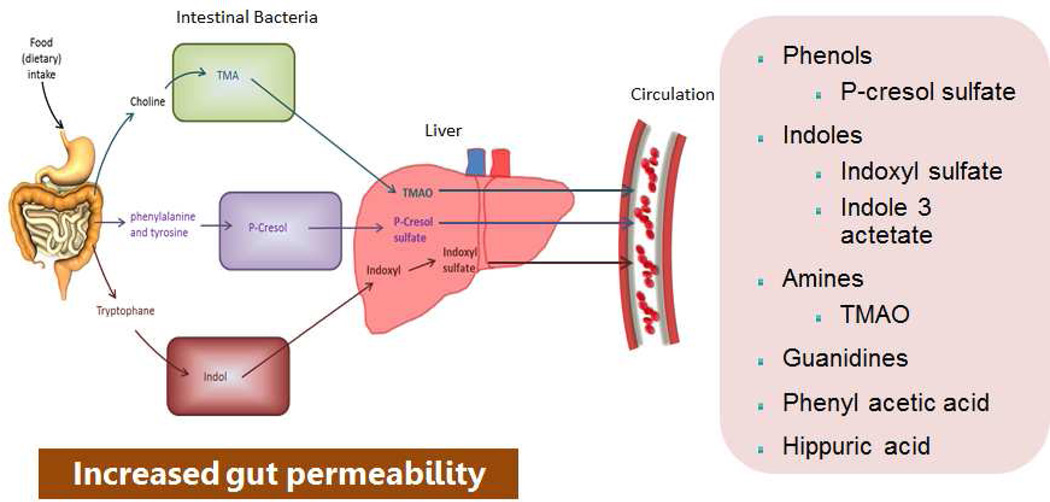
In 1997, Niwa et al. proposed the protein metabolite hypothesis, which claims that toxins generated from protein putrefaction by gut microbes are taken up by the organic anion transporters in the renal tubules. This in turn activates cellular pathways leading to cell death and interstitial fibrosis. We proposed that maladaptive dysbiosis contributes to progression of CKD through pathways beyond generation of uremic toxins (5). Dietary tryptophan is metabolized into indole by tryptophanase in intestinal bacteria such as Escherichia coli. Indole is absorbed into the blood from the intestine, and is metabolized to IS in the liver; it is normally excreted in urine (Fig. 2). IS is an aryl hydrocarbon receptor ligand and a transcriptional regulator. A prospective, observational study performed in 268 patients with CKD indicated that IS measured at baseline is a predictor of loss of kidney function (47). Evidence from animal models suggest that this uremic toxin may damage renal tubular cells (48). In uremic rats, administration of IS mediates the renal expression of genes related to tubulointerstitial fibrosis, such as TGF-β1 and tissue inhibitor of metalloproteinases (49). Osamu et al. showed that mouse podocytes exposed to IS exhibited a pro-inflammatory phenotype, decreased expression of podocyte-specific genes, and decreased cell viability (50). P-Cresol is a colonic fermentation product of the amino acid tyrosine and phenylalanine. Treatment of proximal tubular cells with IS and p-Cresol Sulfate (PCS) led to epithelial mesenchymal phenotypic transition in renal tubule with increased expression of transcription factor Snail, fibronectin and α-smooth muscle actin. Furthermore, in partial nephrectomized mice model, IS or PCS activated the intrarenal renin-angiotensin-aldosterone system (RAAS) system and interstitial fibrosis and glomerulosclerosis (51).
Figure 2.
Schematic representation of origin and synthesis of the major uremic toxins from dysbiotic gut microbiome, subsequent modification in the liver, and secretion into the circulation.
Trimethylamine-N-oxide (TMAO), a gut microbial-dependent metabolite of dietary choline, phosphatidylcholine (lecithin), and L-carnitine, is elevated in CKD. Tang et al. found that elevated TMAO concentrations in animal models were associated with corresponding increases in tubulointerstitial fibrosis. Animals with increased TMAO levels also had increased fibrosis and phosphorylation of Smad3, an important regulator of fibrosis (52). Further studies are needed to see whether TMAO plays a role in progression of CKD.
Gut microbiome in CKD related complications
Obesity
Obesity is associated with changes in the relative abundance of the two dominant bacterial divisions, the Bacteroidetes and the Firmicutes, as noted in comparisons of the distal gut microbiota of genetically obese mice and their lean littermates, as well as those of obese and lean human volunteers. Through metagenomic and biochemical analysis, Turnbaugh et al. (53) found that these changes affect the metabolic potential of the mouse gut microbiota indicating that the gut microbiota as an additional contributing factor to the pathophysiology of obesity
Type 2 diabetes mellitus
T2DM patients exhibited an altered intestinal microbiota, characterized by a decrease of Bacteroidetes/Firmicutes ratio and some functional bacteria (e.g. Bifidobacteria) with an increase of various opportunistic pathogens and some endotoxin-producing Gram-negative bacteria (54–56). It has also been proposed that altered microbiota in obesity modulates intestinal permeability and increases metabolic endotoxin secretion that lead to chronic low-level inflammation (57, 58), the pathogenesis of insulin resistance and onset of T2DM (59, 60).
CVD
Emerging evidence support the infectious theory of atherosclerosis. Using 454 pyrosequencing of 16S rRNA genes, Koren et al. (61) identified Chryseomonas, in all atherosclerotic plaque samples, and Veillonella and Streptococcus in atherosclerotic plaques in human subjects. Further study suggested that oral cavity and gut can be sources for atherosclerotic plaque-associated bacteria. A number of products from the microbiome are now known to be associated with CVD. The Bruneck study (62) showed that an elevated endotoxin level is a strong risk factor for the development of atherosclerosis in the general population. We showed that elevated plasma level of sCD14 (the receptor for endotoxin) is associated with CVD and mortality in patients with CKD and hemodialysis patients (63, 64).
Elevated IS level is associated with aortic calcification, increased vascular stiffness, and risk of cardiovascular mortality in patients with CKD(65). Similarly, elevated plasma PCS level is associated with all-cause mortality and CVD in CKD and ESRD patients (66). Using metabolomics approach, Hazen’s group showed that TMAO, choline and betaine to be associated with heart disease (67). In a follow-up study they showed that elevated TMAO level predicted an increased risk of major adverse cardiovascular events after adjustment for traditional risk factors in 4,007 patients undergoing elective coronary angiography (68). Stubbs et al. measured the TMAO levels in 200 patients with CKD, who underwent coronary angiography and showed that it is predictor of coronary atherosclerosis and mortality, after adjusting for traditional cardiovascular risk factors. Microbial taxa belonging to the Clostridiaceae and Peptostreptococcaceae families are positively associated with blood levels of TMAO in humans (69).
Bone mineral metabolism
Uremic toxins are prototypic protein-bound molecules which have a role in the development of CVD and mortality among patients with CKD (70). Recent research has implicated uremic toxins in the development of bone disease (71). Elimination of these toxins could be a noble therapeutic intervention in controlling low-turnover bone disease in patients with CKD (72).
Depression
The brain-gut-microbiota axis is a bidirectional signaling between the gut and the brain, which is regulated at neural, endocrine, and immune levels (73). The subtle alterations in microbiota acquisition or maintenance in early life (74–76) may act as a vulnerability factor, impacting on endocrine and immune signaling pathways of the brain-gut-microbiota axis, disruption of which may subsequently predispose to stress-related disorders in adulthood.
Microbiota impacts early in life during critical neurodevelopmental phases (77) and for hypothalamic pituitary adrenal axis (78). It’s an extension of the hygiene hypothesis (79) which has been reconceptualised as the “old friend’s hypothesis” (80), proposing that low microbial diversity leads to chronic inflammatory disorders including depression (81, 82).
Kwashiorkor
Kwashiorkor and Marasmus are two forms of severe under nutrition prevalent in regions confronting food insecurity and high burdens of infectious disease. A recent study implicates the gut microbiota as a central factor in the cause of kwashiorkor (83). To determine the causality of microbiota composition in kwashiorkor, investigators used fecal transplantation in gnotobiotic mice. The mice that received fecal transplants from human donors with kwashiorkor lost a significant amount of weight. Investigators identified potential targets for kwashiorkor that are rooted in the microbiota, and also found specific microbes, metabolic pathways, as well as signals that correlated with a lack of sustained improvement.
Application of meta-omics to the study of human gut microbiome
Until recently, studies of the gut microbiome were mostly based on traditional culture methods which allowed cultivation of only a fraction of gut microbiota (84, 85). Development of advanced next-generation sequencing technologies such as 16 S rDNA sequence analysis and metagenomics have facilitated the analysis of a much larger number of uncultivated gut microbial microorganisms (86–89). Both methods have their own unique advantages; the 16S method is more useful in identifying “who’s there?” by sequencing of the conserved 16S rDNA gene which is present in all bacteria (90), whereas the shotgun metagenomic sequencing aims at determining “what can they do” by random sequencing of all DNA extracted from the sample (91). Metagenomics have been successfully applied by both the American HMP (92) and the European project, MetaHIT (56) to facilitate the study of the human intestinal microbiome. Recently, Rossi and colleagues (93) reported the results of their clinical trial in which they evaluated whether synbiotic (pre- and probiotic) therapy alters the gut microbiota and reduces serum concentrations of microbiome–generated uremic toxins, IS and PCS, in patients with CKD. Using 16S gene sequencing method, these investigators determined the microbiome profiles of the study participants and found that synbiotics altered the stool microbiome, particularly with enrichment of Bifidobacterium and depletion of Ruminococcaceae. Furthermore, the results revealed a significant inverse correlation between changes in the relative abundance of Bifidobacterium spp. and free serum concentrations of both PCS and IS.
Despite its advantages, metagenomics is unable to determine microbial gene expression, and determining the functional activity of the gut microbiome requires application of other methods such as metaproteomics (94) and metabolomics (95). The two methods are very similar but with slightly different focuses; proteomics focuses on study of peptides and proteins, whereas metabolomics aims at identification of small metabolites (<1000 Daltons). Currently, metabolomics is widely applied to the study of human gut microbiome, particularly in context of intestinal disorders such as colorectal cancer, inflammatory bowel disease, and irritable bowel syndrome (IBS) (96–99). Using metabolite profiling of plasma samples from 1,434 Framingham study participants, Rhee et al. demonstrated that progression to CKD could be predicted using just 9 metabolites (100). Another metabolomic study applied NMR spectroscopy to determine differences in the plasma metabolic status of stage 3–4 CKD patients and healthy controls (101). This study revealed the elevation of 14 metabolites in the plasma of uremic patients, which included several previously identified uremic toxins, such as PCS, as well as two novel uremic retentions solutes, dimethyl sulphone and 2-hydroxyisobutyric acid (101). Ideally, to study uremic toxins, both proteomics and metabolomics approaches should be applied as these methods complement each other and each has a unique role in the discovery of uremic retention solutes.
Targeted interventions to treat intestinal dysbiosis
Recent advances in our understanding of the physiologic functions of gut microbiome and the pathologic consequences of dysbiosis have led to exploration of various ways of reestablishing symbiosis. Most therapies targeting the colonic microenvironment in CKD aim to modulate gut microbiota, or target adsorption of uremic toxin end products of microbial fermentation.
Modulation of gut microbiota
Probiotics
Probiotics are defined by the United Nations’ Food and Agriculture Organization and the World Health Organization as “live microorganisms that when administered in adequate amounts confer a health benefit on the host” (102). Probiotics consist of living bacteria, such as Bifidobacteria species, Lactobacilli, and Streptococci (103), that can alter gut microbiota and affect the inflammatory state (104, 105). Treatment with Bacillus pasteurii and Sporlac slowed the progression of kidney disease and prolonged the life span of fifth/sixth nephrectomized Sprague-Dawley rats (106). Hemodialysis patients treated with oral Lactobacillus acidophilus showed decreased serum dimethylamine, a potential uremic toxin. In another study, treatment with L. acidophilus ATCC-4356 reduced the atherosclerotic burden in ApoE 2/2 mice (107). However, the optimal dose of the bacteria needed to achieve engraftment and also whether they will survive in the uremic environment is unclear.
Smart bacteria
Prakash et al. (108) reduced BUN in uremic rats by orally administering microencapsulated, genetically engineered live cells that contained living urease-producing Escherichia coli–DH5. Scientist are envisioning the possibility of colonizing our gut with genetically modified “smart” bacteria that could potentially treat disease. Advancement in DNA technologies for manipulation of microbial genomes has made it possible to generate tailored bacteria for the treatment of human disorders. These bacteria could be altered to produce a continuous and inexpensive supply of required therapeutic molecules for treatment or scavengers that remove toxic molecules (109). These molecules may be designed to turn on and off the synthesis of molecules based on the biological need. Researchers have developed bacterial suicide strategies to mitigate possible risks and should permit the safe use of microbes in targeted therapy (110).
Rhabb et al. (111) showed that germfree mice have increased susceptibility to ischemia and reperfusion injury (IRI), which is reversed by colonization with commensal bacteria. This observation suggests that certain gut microbiota products might be protecting against IRI (112). In an elegant study, Andrade-Oliveira et al. (113) showed that treatment with SCFA reduces IRI–induced kidney injury by reducing inflammation, increase in autophagy, a reduction in apoptosis, and an improvement in mitochondrial biogenesis. SCFA are organic fatty acids with one to six carbons including acetate, propionate, and butyrate. They exert their systemic effects through G protein-coupled receptors and affect a variety of host processes, including energy utilization, host-microbe signaling and immune modulation. Interestingly, Pluznik et al. showed that SCFA may be involved in blood pressure regulation (114). Olfactory receptor 78 (a receptor for SCFA) expressed in the kidney, where it mediates renin secretion and increases blood pressure is counteracted by GPR43, which induces vasodilatation (114).
Prebiotics
A prebiotic is a nondigestible food ingredient that has a beneficial effect through its selective stimulation of the growth or activity of one or a limited number of bacteria in the colon (115, 116). The candidate prebiotics include inulin, fructo-oligosaccharides, galacto-oligo-saccharides, soya-oligosaccharides, xylo-oligosaccharides, and pyrodextrins. Prebiotics promote the growth of Bifidobacteria and Lactobacilli species at the expense of other groups of bacteria in the gut, such as Bacteroides species, Clostridia species, and Enterobacteria (117). Preliminary evidence indicates that prebiotic oligofructose-enriched inulin (p-inulin) promotes growth of Bifidobacteria species, mediates weight loss, reduces inflammation, and improves metabolic function (118–120). High dietary fiber intake is associated with lower risk of inflammation and reduced mortality in patients with CKD (121). Meijers et al. (122) reported that serum concentrations of p-cresol and IS are reduced by the oral intake of p-inulin in hemodialysis patients. Acarbose is an inhibitor of α-glucosidase enzymes in the intestinal brush-border that blocks the hydrolysis of poly- and oligosaccharides to glucose and other monosaccharides. The delivery of undigested oligo-saccharides to the colon promotes growth of saccarolytic bacteria over proteolytic bacteria leading to reduced generation of PCS (123). Evenepoel et al. showed that treatment with acarbose reduces the colonic generation of p-cresol in healthy persons (123).
Synbiotic is the combination of prebiotic and probiotic treatments. Recently completed, Synbiotics Easing Renal Failure by Improving Gut Microbiology (SYNERGY) is a randomized, double-blind, placebo-controlled, crossover trial. Patients with CKD, which showed six weeks of synbiotic treatment reduced PCS, and not IS (93).
High fiber (amylose) diet
Results from animal studies show favorable effects of a high fiber diet in restoring intestinal microbiome, and plasma, cecal, and urine metabolome (124). Similarly, rats with CKD fed high resistant starch diet had retardation in progression of CKD. These effects were mediated through partial restoration of intestinal epithelial tight junction, attenuated impairment of Nrf2 activity, lower oxidative stress, inflammation and fibrosis (125).
Novel therapies
Lubiprostone is a synthetic derivative of prostaglandin, which activates chloride channel in the gut and is used in the treatment of constipation. In a rat model of CKD, treatment with lubiprostone reduced BUN and histological evidence of fibrosis, which was associated with attenuated inflammation in the kidney and improvement in microbiome profile with proliferation of saccarolytic bacteria. They also noted that microbiome-derived uremic toxins such as indoxyl sulfate and TMAO decreased and the citric acid cycle pathway was activated indicating improved energy metabolism (126).
Trimethylamine (TMA) inhibitor: 3, 3-dimethyl-1-butanol (DMB) is a structural analogue of choline which inhibits microbial TMA formation through inhibition of microbial TMA lyases (127). DMB decreases plasma TMAO levels in mice placed chronically on choline-supplemented or carnitine-supplemented diet. Furthermore, DMB inhibited endogenous macrophage foam cell formation and atherosclerotic lesion development in mice. DMB was detected in some balsamic vinegars, in red wines, and in some olive oils and grape seed oils.
Conclusions and Future endeavors
Our understanding of the gut microbiome’s physiologic functions and pathologic consequences of dysbiosis have led to exploration of various ways of reestablishing symbiosis. Regulation of intestinal immune responses by the presence or absence of certain members of the microbiota opens the exciting possibility of directionally modulating these responses by either changing the composition of the gut bacteria or modifying relevant signaling pathways. Therefore, it will be crucial to elucidate the molecular mechanisms by which commensals influence mucosal responses. Until recently, a reliance on microbiological culture techniques to characterize flora composition limited the studies. Advances in sequencing techniques, bio-informatics and wide application of metabolomics have greatly expanded our understanding of the role of microbiome in health and disease in general. It is becoming evident that microbiome changes adaptively early in CKD, but later becomes maladaptive and becomes a source for number of uremic toxins (5).
A number of interventions have been proposed and tested in small studies. It is important to define the variation in microbiome profile and associate it with specific metabolome in well-defined studies in patients with CKD and ESRD, with specific attention to diet and other potential confounders. NIDDK is conducting two such studies under the auspicious of Hemodialysis Novel Therapies Consortium (ClinicalTrials.gov Identifier: NCT02572882) and CKD Pilot Studies consortium. Future studies should explore the interaction of microbiome with human genome and how the metabolic potential of the microbiome could be harnessed to treat CKD.
Acknowledgments
D.S.R. is supported by National Institutes of Health grants 1R01DK073665-01A1, 1U01DK099924-01, and 1U01DK099914-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflict of interest
All authors have read the journal’s policy on disclosure of potential conflicts of interest and have nothing to disclose. All authors have read the journal’s authorship agreement and that the manuscript has been reviewed by and approved by all named authors.
Reference List
- 1.Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu.Rev.Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 3.Hill MJ. Intestinal flora and endogenous vitamin synthesis. Eur.J.Cancer Prev. 1997;6(Suppl 1):S43–S45. doi: 10.1097/00008469-199703001-00009. [DOI] [PubMed] [Google Scholar]
- 4.Hylemon PB, Harder J. Biotransformation of monoterpenes, bile acids, and other isoprenoids in anaerobic ecosystems. FEMS Microbiol.Rev. 1998;22(5):475–488. doi: 10.1111/j.1574-6976.1998.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 5.Ramezani A, Massy ZA, Meijers B, et al. Role of the Gut Microbiome in Uremia: A Potential Therapeutic Target. Am.J.Kidney Dis. 2016;67(3):483–498. doi: 10.1053/j.ajkd.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thaiss CA, Zeevi D, Levy M, et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 10.Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De FC, Cavalieri D, Di PM, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc.Natl.Acad.Sci.U.S.A. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podolsky SH. Metchnikoff and the microbiome. Lancet. 2012;380(9856):1810–1811. doi: 10.1016/s0140-6736(12)62018-2. [DOI] [PubMed] [Google Scholar]
- 13.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N.Engl.J.Med. 2002;347(12):911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 14.Penders J, Thijs C, van den Brandt PA, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007;56(5):661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc.Natl.Acad.Sci.U.S.A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bollyky PL, Bice JB, Sweet IR, et al. The toll-like receptor signaling molecule Myd88 contributes to pancreatic beta-cell homeostasis in response to injury. PLoS.One. 2009;4(4):e5063. doi: 10.1371/journal.pone.0005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl.Environ.Microbiol. 1995;61(9):3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevalier C, Stojanovic O, Colin DJ, et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163(6):1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 20.Einheber A, Carter D. The role of the microbial flora in uremia I. Survival times of germfree, limited-flora, and conventionalized rats after bilateral nephrectomy and fasting. J.Exp.Med. 1966;123(2):239–250. doi: 10.1084/jem.123.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yokoyama MT, Tabori C, Miller ER, et al. The effects of antibiotics in the weanling pig diet on growth and the excretion of volatile phenolic and aromatic bacterial metabolites. Am.J.Clin.Nutr. 1982;35(6):1417–1424. doi: 10.1093/ajcn/35.6.1417. [DOI] [PubMed] [Google Scholar]
- 22.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc.Natl.Acad.Sci.U.S.A. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronov PA, Luo FJ, Plummer NS, et al. Colonic contribution to uremic solutes. J.Am.Soc.Nephrol. 2011;22(9):1769–1776. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorensen LB. Role of the intestinal tract in the elimination of uric acid. Arthritis Rheum. 1965;8(5):694–706. doi: 10.1002/art.1780080429. [DOI] [PubMed] [Google Scholar]
- 25.Gibson SA, McFarlan C, Hay S, et al. Significance of microflora in proteolysis in the colon. Appl.Environ.Microbiol. 1989;55(3):679–683. doi: 10.1128/aem.55.3.679-683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poesen R, Windey K, Neven E, et al. The Influence of CKD on Colonic Microbial Metabolism. J.Am.Soc.Nephrol. 2015 doi: 10.1681/ASN.2015030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaziri ND, Wong J, Pahl M, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83(2):308–315. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 28.Wong J, Piceno YM, Desantis TZ, et al. Expansion of urease- and uricase-containing, indole- and p-cresol-forming and contraction of short-chain fatty acid-producing intestinal microbiota in ESRD. Am.J.Nephrol. 2014;39(3):230–237. doi: 10.1159/000360010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnusson M, Magnusson KE, Sundqvist T, et al. Increased intestinal permeability to differently sized polyethylene glycols in uremic rats: effects of low- and high-protein diets. Nephron. 1990;56(3):306–311. doi: 10.1159/000186158. [DOI] [PubMed] [Google Scholar]
- 30.Magnusson M, Magnusson KE, Sundqvist T, et al. Impaired intestinal barrier function measured by differently sized polyethylene glycols in patients with chronic renal failure. Gut. 1991;32(7):754–759. doi: 10.1136/gut.32.7.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaziri ND, Goshtasbi N, Yuan J, et al. Uremic plasma impairs barrier function and depletes the tight junction protein constituents of intestinal epithelium. Am.J.Nephrol. 2012;36(5):438–443. doi: 10.1159/000343886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaziri ND, Yuan J, Norris K. Role of urea in intestinal barrier dysfunction and disruption of epithelial tight junction in chronic kidney disease. Am.J.Nephrol. 2013;37(1):1–6. doi: 10.1159/000345969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farhadi A, Banan A, Fields J, et al. Intestinal barrier: an interface between health and disease. J.Gastroenterol.Hepatol. 2003;18(5):479–497. doi: 10.1046/j.1440-1746.2003.03032.x. [DOI] [PubMed] [Google Scholar]
- 34.Vaziri ND, Yuan J, Rahimi A, et al. Disintegration of colonic epithelial tight junction in uremia: a likely cause of CKD-associated inflammation. Nephrol.Dial.Transplant. 2012;27(7):2686–2693. doi: 10.1093/ndt/gfr624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Almeida Duarte JB, de Aguilar-Nascimento JE, Nascimento M, et al. Bacterial translocation in experimental uremia. Urol.Res. 2004;32(4):266–270. doi: 10.1007/s00240-003-0381-7. [DOI] [PubMed] [Google Scholar]
- 36.Szeto CC, Kwan BC, Chow KM, et al. Endotoxemia is related to systemic inflammation and atherosclerosis in peritoneal dialysis patients. Clin.J.Am.Soc.Nephrol. 2008;3(2):431–436. doi: 10.2215/CJN.03600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cebula A, Seweryn M, Rempala GA, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497(7448):258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geuking MB, Cahenzli J, Lawson MA, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 40.Zeissig S, Blumberg RS. Commensal microbial regulation of natural killer T cells at the frontiers of the mucosal immune system. FEBS Lett. 2014;588(22):4188–4194. doi: 10.1016/j.febslet.2014.06.042. [DOI] [PubMed] [Google Scholar]
- 41.Maynard CL, Elson CO, Hatton RD, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clarke TB, Davis KM, Lysenko ES, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat.Med. 2010;16(2):228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 44.Park J, Kim M, Kang SG, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal.Immunol. 2015;8(1):80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation. 2014;98(7):697–705. doi: 10.1097/TP.0000000000000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS.One. 2015;10(3):e0122399. doi: 10.1371/journal.pone.0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu IW, Hsu KH, Lee CC, et al. p-Cresyl sulphate and indoxyl sulphate predict progression of chronic kidney disease. Nephrol.Dial.Transplant. 2011;26(3):938–947. doi: 10.1093/ndt/gfq580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satoh M, Hayashi H, Watanabe M, et al. Uremic toxins overload accelerates renal damage in a rat model of chronic renal failure. Nephron Exp.Nephrol. 2003;95(3):e111–e118. doi: 10.1159/000074327. [DOI] [PubMed] [Google Scholar]
- 49.Miyazaki T, Ise M, Seo H, et al. Indoxyl sulfate increases the gene expressions of TGF-beta 1, TIMP-1 and pro-alpha 1(I) collagen in uremic rat kidneys. Kidney Int.Suppl. 1997;62:S15–S22. [PubMed] [Google Scholar]
- 50.Ichii O, Otsuka-Kanazawa S, Nakamura T, et al. Podocyte injury caused by indoxyl sulfate, a uremic toxin and aryl-hydrocarbon receptor ligand. PLoS.One. 2014;9(9):e108448. doi: 10.1371/journal.pone.0108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun CY, Chang SC, Wu MS. Uremic toxins induce kidney fibrosis by activating intrarenal renin-angiotensin-aldosterone system associated epithelial-to-mesenchymal transition. PLoS.One. 2012;7(3):e34026. doi: 10.1371/journal.pone.0034026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang WH, Wang Z, Kennedy DJ, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ.Res. 2015;116(3):448–455. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Ma C, Han L, et al. Molecular characterisation of the faecal microbiota in patients with type II diabetes. Curr.Microbiol. 2010;61(1):69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- 55.Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS.One. 2010;5(2):e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 57.McArdle MA, Finucane OM, Connaughton RM, et al. Mechanisms of obesity-induced inflammation and insulin resistance: insights into the emerging role of nutritional strategies. Front Endocrinol.(Lausanne) 2013;4:52. doi: 10.3389/fendo.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J. Adipose tissue macrophages in the development of obesity-induced inflammation, insulin resistance and type 2 diabetes. Arch.Pharm.Res. 2013;36(2):208–222. doi: 10.1007/s12272-013-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diamant M, Blaak EE, de Vos WM. Do nutrient-gut-microbiota interactions play a role in human obesity, insulin resistance and type 2 diabetes? Obes.Rev. 2011;12(4):272–281. doi: 10.1111/j.1467-789X.2010.00797.x. [DOI] [PubMed] [Google Scholar]
- 60.Everard A, Cani PD. Diabetes, obesity and gut microbiota. Best.Pract.Res.Clin.Gastroenterol. 2013;27(1):73–83. doi: 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Koren O, Spor A, Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc.Natl.Acad.Sci.U.S.A. 2011;108(Suppl 1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiedermann CJ, Kiechl S, Dunzendorfer S, et al. Association of endotoxemia with carotid atherosclerosis and cardiovascular disease: prospective results from the Bruneck Study. J.Am.Coll.Cardiol. 1999;34(7):1975–1981. doi: 10.1016/s0735-1097(99)00448-9. [DOI] [PubMed] [Google Scholar]
- 63.Raj DS, Shah VO, Rambod M, et al. Association of soluble endotoxin receptor CD14 and mortality among patients undergoing hemodialysis. Am.J.Kidney Dis. 2009;54(6):1062–1071. doi: 10.1053/j.ajkd.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poesen R, Ramezani A, Claes K, et al. Associations of Soluble CD14 and Endotoxin with Mortality, Cardiovascular Disease, and Progression of Kidney Disease among Patients with CKD. Clin.J.Am.Soc.Nephrol. 2015;10(9):1525–1533. doi: 10.2215/CJN.03100315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin CJ, Wu V, Wu PC, et al. Meta-Analysis of the Associations of p-Cresyl Sulfate (PCS) and Indoxyl Sulfate (IS) with Cardiovascular Events and All-Cause Mortality in Patients with Chronic Renal Failure. PLoS.One. 2015;10(7):e0132589. doi: 10.1371/journal.pone.0132589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu IW, Hsu KH, Hsu HJ, et al. Serum free p-cresyl sulfate levels predict cardiovascular and all-cause mortality in elderly hemodialysis patients--a prospective cohort study. Nephrol.Dial.Transplant. 2012;27(3):1169–1175. doi: 10.1093/ndt/gfr453. [DOI] [PubMed] [Google Scholar]
- 67.Organ CL, Otsuka H, Bhushan S, et al. Choline Diet and Its Gut Microbe-Derived Metabolite, Trimethylamine N-Oxide, Exacerbate Pressure Overload-Induced Heart Failure. Circ.Heart Fail. 2016;9(1):e002314. doi: 10.1161/CIRCHEARTFAILURE.115.002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller PE, Haberlen SA, Brown TT, et al. Intestinal Microbiota-Produced Trimethylamine-N-oxide and its Association with Coronary Stenosis and HIV Serostatus. J.Acquir.Immune.Defic.Syndr. 2016 doi: 10.1097/QAI.0000000000000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Backhed F. Meat-metabolizing bacteria in atherosclerosis. Nat.Med. 2013;19(5):533–534. doi: 10.1038/nm.3178. [DOI] [PubMed] [Google Scholar]
- 70.Huang WH, Hung CC, Yang CW, et al. High correlation between clearance of renal protein-bound uremic toxins (indoxyl sulfate and p-cresyl sulfate) and renal water-soluble toxins in peritoneal dialysis patients. Ther.Apher.Dial. 2012;16(4):361–367. doi: 10.1111/j.1744-9987.2012.01068.x. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka H, Iwasaki Y, Yamato H, et al. p-Cresyl sulfate induces osteoblast dysfunction through activating JNK and p38 MAPK pathways. Bone. 2013;56(2):347–354. doi: 10.1016/j.bone.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 72.Kim YH, Kwak KA, Gil HW, et al. Indoxyl sulfate promotes apoptosis in cultured osteoblast cells. BMC.Pharmacol.Toxicol. 2013;14:60. doi: 10.1186/2050-6511-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grenham S, Clarke G, Cryan JF, et al. Brain-gut-microbe communication in health and disease. Front Physiol. 2011;2:94. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohland CL, Macnaughton WK. Probiotic bacteria and intestinal epithelial barrier function. Am.J.Physiol Gastrointest.Liver Physiol. 2010;298(6):G807–G819. doi: 10.1152/ajpgi.00243.2009. [DOI] [PubMed] [Google Scholar]
- 75.Swanson PA, Kumar A, Samarin S, et al. Enteric commensal bacteria potentiate epithelial restitution via reactive oxygen species-mediated inactivation of focal adhesion kinase phosphatases. Proc.Natl.Acad.Sci.U.S.A. 2011;108(21):8803–8808. doi: 10.1073/pnas.1010042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shifrin DA, Jr, McConnell RE, Nambiar R, et al. Enterocyte microvillus-derived vesicles detoxify bacterial products and regulate epithelial-microbial interactions. Curr.Biol. 2012;22(7):627–631. doi: 10.1016/j.cub.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Borre YE, O’Keeffe GW, Clarke G, et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol.Med. 2014;20(9):509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 78.Moloney RD, Desbonnet L, Clarke G, et al. The microbiome: stress, health and disease. Mamm.Genome. 2014;25(1–2):49–74. doi: 10.1007/s00335-013-9488-5. [DOI] [PubMed] [Google Scholar]
- 79.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rook GA, Martinelli R, Brunet LR. Innate immune responses to mycobacteria and the downregulation of atopic responses. Curr.Opin.Allergy Clin.Immunol. 2003;3(5):337–342. doi: 10.1097/00130832-200310000-00003. [DOI] [PubMed] [Google Scholar]
- 81.Rook GA, Lowry CA, Raison CL. Microbial ‘Old Friends’, immunoregulation and stress resilience. Evol.Med.Public Health. 2013;2013(1):46–64. doi: 10.1093/emph/eot004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rook GA, Raison CL, Lowry CA. Microbiota, immunoregulatory old friends and psychiatric disorders. Adv.Exp.Med.Biol. 2014;817:319–356. doi: 10.1007/978-1-4939-0897-4_15. [DOI] [PubMed] [Google Scholar]
- 83.Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl.Environ.Microbiol. 1999;65(11):4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tannock GW. Molecular assessment of intestinal microflora. Am.J.Clin.Nutr. 2001;73(2 Suppl):410S–414S. doi: 10.1093/ajcn/73.2.410s. [DOI] [PubMed] [Google Scholar]
- 86.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shendure J, Ji H. Next-generation DNA sequencing. Nat.Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 88.Tringe SG, Rubin EM. Metagenomics: DNA sequencing of environmental samples. Nat.Rev.Genet. 2005;6(11):805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 89.Tringe SG, von MC, Kobayashi A, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 90.Cole JR, Chai B, Farris RJ, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35(Database issue):D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lepage P, Leclerc MC, Joossens M, et al. A metagenomic insight into our gut’s microbiome. Gut. 2013;62(1):146–158. doi: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 92.A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rossi M, Johnson DW, Morrison M, et al. Synbiotics Easing Renal Failure by Improving Gut Microbiology (SYNERGY): A Randomized Trial. Clin.J.Am.Soc.Nephrol. 2016;11(2):223–231. doi: 10.2215/CJN.05240515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verberkmoes NC, Russell AL, Shah M, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME.J. 2009;3(2):179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 95.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat.Rev.Microbiol. 2005;3(5):431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 96.Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS.One. 2009;4(7):e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Le GG, Noor SO, Ridgway K, et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J.Proteome.Res. 2011;10(9):4208–4218. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 98.Marchesi JR, Holmes E, Khan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J.Proteome.Res. 2007;6(2):546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 99.Phua LC, Chue XP, Koh PK, et al. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol.Ther. 2014;15(4):389–397. doi: 10.4161/cbt.27625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rhee EP, Clish CB, Ghorbani A, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J.Am.Soc.Nephrol. 2013;24(8):1330–1338. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mutsaers HA, Engelke UF, Wilmer MJ, et al. Optimized metabolomic approach to identify uremic solutes in plasma of stage 3–4 chronic kidney disease patients. PLoS.One. 2013;8(8):e71199. doi: 10.1371/journal.pone.0071199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mutsaers HA, Engelke UF, Wilmer MJ, et al. Optimized metabolomic approach to identify uremic solutes in plasma of stage 3–4 chronic kidney disease patients. PLoS.One. 2013;8(8):e71199. doi: 10.1371/journal.pone.0071199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rastall RA, Gibson GR, Gill HS, et al. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol.Ecol. 2005;52(2):145–152. doi: 10.1016/j.femsec.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 104.Konstantinov SR, Smidt H, de Vos WM, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc.Natl.Acad.Sci.U.S.A. 2008;105(49):19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.van BP, Troost FJ, van HS, et al. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc.Natl.Acad.Sci.U.S.A. 2009;106(7):2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ranganathan N, Patel B, Ranganathan P, et al. Probiotic amelioration of azotemia in 5/6th nephrectomized Sprague-Dawley rats. ScientificWorldJournal. 2005;5:652–660. doi: 10.1100/tsw.2005.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen L, Liu W, Li Y, et al. Lactobacillus acidophilus ATCC 4356 attenuates the atherosclerotic progression through modulation of oxidative stress and inflammatory process. Int.Immunopharmacol. 2013;17(1):108–115. doi: 10.1016/j.intimp.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 108.Prakash S, Chang TM. Microencapsulated genetically engineered live E. coli DH5 cells administered orally to maintain normal plasma urea level in uremic rats. Nat.Med. 1996;2(8):883–887. doi: 10.1038/nm0896-883. [DOI] [PubMed] [Google Scholar]
- 109.Pinero-Lambea C, Ruano-Gallego D, Fernandez LA. Engineered bacteria as therapeutic agents. Curr.Opin.Biotechnol. 2015;35:94–102. doi: 10.1016/j.copbio.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 110.Mandell DJ, Lajoie MJ, Mee MT, et al. Corrigendum: Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;527(7577):264. doi: 10.1038/nature15536. [DOI] [PubMed] [Google Scholar]
- 111.Doi K, Rabb H. Impact of acute kidney injury on distant organ function: recent findings and potential therapeutic targets. Kidney Int. 2016;89(3):555–564. doi: 10.1016/j.kint.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 112.Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J.Am.Soc.Nephrol. 2014;25(4):657–670. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Andrade-Oliveira V, Amano MT, Correa-Costa M, et al. Gut Bacteria Products Prevent AKI Induced by Ischemia-Reperfusion. J.Am.Soc.Nephrol. 2015;26(8):1877–1888. doi: 10.1681/ASN.2014030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pluznick JL, Protzko RJ, Gevorgyan H, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc.Natl.Acad.Sci.U.S.A. 2013;110(11):4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J.Nutr. 1995;125(6):1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 116.Gibson GR, Probert HM, Loo JV, et al. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr.Res.Rev. 2004;17(2):259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 117.Silk DB, Davis A, Vulevic J, et al. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment.Pharmacol.Ther. 2009;29(5):508–518. doi: 10.1111/j.1365-2036.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- 118.Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 119.Gibson GR, Beatty ER, Wang X, et al. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology. 1995;108(4):975–982. doi: 10.1016/0016-5085(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 120.Pylkas AM, Juneja LR, Slavin JL. Comparison of different fibers for in vitro production of short chain fatty acids by intestinal microflora. J.Med.Food. 2005;8(1):113–116. doi: 10.1089/jmf.2005.8.113. [DOI] [PubMed] [Google Scholar]
- 121.Krishnamurthy VM, Wei G, Baird BC, et al. High dietary fiber intake is associated with decreased inflammation and all-cause mortality in patients with chronic kidney disease. Kidney Int. 2012;81(3):300–306. doi: 10.1038/ki.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meijers BK, De PV, Verbeke K, et al. p-Cresyl sulfate serum concentrations in haemodialysis patients are reduced by the prebiotic oligofructose-enriched inulin. Nephrol.Dial.Transplant. 2010;25(1):219–224. doi: 10.1093/ndt/gfp414. [DOI] [PubMed] [Google Scholar]
- 123.Evenepoel P, Bammens B, Verbeke K, et al. Acarbose treatment lowers generation and serum concentrations of the protein-bound solute p-cresol: a pilot study. Kidney Int. 2006;70(1):192–198. doi: 10.1038/sj.ki.5001523. [DOI] [PubMed] [Google Scholar]
- 124.Kieffer DA, Piccolo BD, Vaziri ND, et al. Resistant Starch Alters Gut Microbiome and Metabolomics Profiles Concurrent with Amelioration of Chronic Kidney Disease in Rats. Am.J.Physiol Renal Physiol. 2016 doi: 10.1152/ajprenal.00513.2015. ajprenal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vaziri ND, Liu SM, Lau WL, et al. High amylose resistant starch diet ameliorates oxidative stress, inflammation, and progression of chronic kidney disease. PLoS.One. 2014;9(12):e114881. doi: 10.1371/journal.pone.0114881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mishima E, Fukuda S, Shima H, et al. Alteration of the Intestinal Environment by Lubiprostone Is Associated with Amelioration of Adenine-Induced CKD. J.Am.Soc.Nephrol. 2015;26(8):1787–1794. doi: 10.1681/ASN.2014060530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang Z, Roberts AB, Buffa JA, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163(7):1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]