Abstract
The SAGA complex contains two enzymatic modules, which house histone acetyltransferase (HAT) and deubiquitinase (DUB) activities. USP22 is the catalytic subunit of the DUB module, but two adaptor proteins, ATXN7L3 and ENY2, are necessary for DUB activity toward histone H2Bub1 and other substrates. ATXN7L3B shares 74% identity with the N-terminal region of ATXN7L3, but the functions of ATXN7L3B are not known. Here we report that ATXN7L3B interacts with ENY2 but not other SAGA components. Even though ATXN7L3B localizes in the cytoplasm, ATXN7L3B overexpression increases H2Bub1 levels, while overexpression of ATXN7L3 decreases H2Bub1 levels. In vitro, ATXN7L3B competes with ATXN7L3 to bind ENY2, and in vivo, knockdown of ATXN7L3B leads to concomitant loss of ENY2. Unlike the ATXN7L3 DUB complex, a USP22-ATXN7L3B-ENY2 complex cannot deubiquitinate H2Bub1 efficiently in vitro. Moreover, ATXN7L3B knockdown inhibits migration of breast cancer cells in vitro and limits expression of ER target genes. Collectively, our studies suggest that ATXN7L3B regulates H2Bub1 levels and SAGA DUB activity through competition for ENY2 binding.
INTRODUCTION
USP22 and the SAGA deubiquitinase (DUB) module are important for normal embryonic development (1), and alterations in the expression or structure of component proteins are linked to neurodegenerative disease and cancer (2, 3). The DUB module consists of a catalytic subunit, USP22, and two adaptor proteins, ATXN7L3 and ENY2, required for deubiquitinase activity and stability of the DUB module (4–6). Another protein, ATXN7, functions as a bridge to integrate the core DUB module into the greater SAGA complex (7, 8) and has also been reported to affect DUB activity (6, 9, 10).
ATXN7L3 contains a Sus1/ENY2-binding region in its N-terminal region, a ZnF-Sgf11 domain, and a SCA7 domain in its C-terminal region (6). The presence of ATXN7L3 is essential for the deubiquitinase activity of the DUB module. No stable complex could be formed in the absence of ATXN7L3 (6). Moreover, the ZnF-Sgf11 domain of ATXN7L3 plays a pivotal role in the enzymatic activity, but is dispensable for the assembly, of the DUB module (6). The ZnF-Sgf11 domain of ATXN7L3 is essential for DUB activity toward H2Bub1 in vitro (6). The ZnF-Sgf11 domain is required for ATXN7L3 binding to nucleosomal DNA (11), and the crystal structure of the DUB module reveals that an arginine cluster in the ZnF-Sgf11 domain directly interacts with ubiquitinated nucleosomes and H2A/H2B heterodimer (12).
Loss of ATXN7L3B, a paralog of ATXN7L3, may also be associated with neurodegenerative disease (13), as three family members exhibiting loss of chromosome region 12q21, where ATXN7L3B lies, exhibited motor and cognitive deficiencies, as well as learning difficulties and cerebellar ataxia. ATXN7L3B and ATXN7L3 share 74% identity within their N-terminal ∼60 amino acid residues (Fig. 1A), including the Sus1/ENY2-binding region (Fig. 1A, in red) (6, 14). Interestingly, a truncated form of ATXN7L3 that only contains amino acids 3 to 76 was shown by others to interact with USP22 and ENY2 in vitro, but the complex containing the truncated protein could not remove ubiquitin from H2Bub1, although it still bound ubiquitin vinyl sulfone (Ub-VS), an irreversible and specific inhibitor that binds only to active deubiqitinating enzymes (6). Since the native structure of ATXN7L3B resembles this artificial form of ATXN7L3, we reasoned that it might also interact with USP22 and affect DUB activity in vivo.
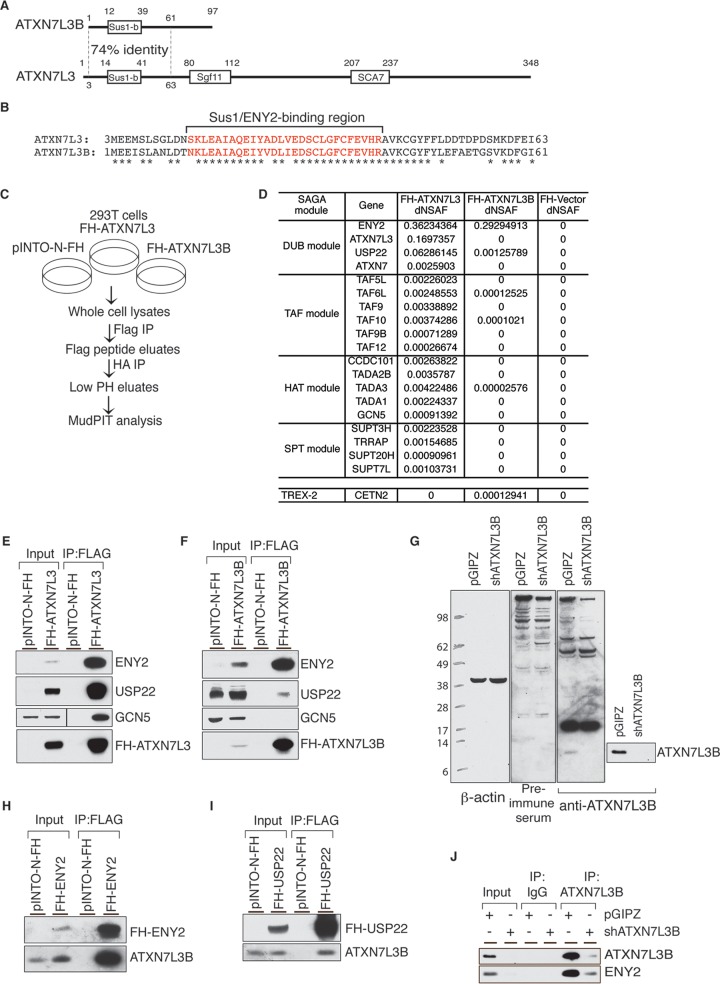
FIG 1.
ATXN7L3B interacts with DUB module components but not SAGA. (A) Comparison of the protein structures of ATXN7L3 and ATXN7L3B. The N termini share 74% identity, but ATXN7L3B lacks the Sgf11 and SCA7 domains that are present in ATXN7L3. (B) Comparison of the protein sequences of ATXN7L3 and ATXN7L3B N termini. Identical amino acid residues are indicated by asterisks. Red lettering indicates the Sus1/ENY2-binding region. (C) Schematic of tandem Flag and HA affinity purification using 293T stable cells expressing pINTO-N-FH, FH-ATXN7L3, or FH-ATXN7L3B. (D) Distributed normalized spectral abundance factor (dNSAF) of SAGA components in different modules and TREX-2 components immunoprecipitated by FH-ATXN7L3 or FH-ATXN7L3B. (E and F) Flag-IP from 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ATXN7L3 (E), or pINTO-N-FH-ATXN7L3B (F). Bound proteins were resolved by electrophoresis and detected by immunoblotting with the indicated antibodies. (G) Whole-cell lysates from MCF7T cells stably expressing nontargeting shRNA (pGIPZ) or shATXN7L3B were resolved by SDS-PAGE. Proteins were transferred onto membranes, which were probed with preimmune serum and ATXN7L3B antibody. (H and I) Flag-IP from 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ENY2 (H), or pINTO-N-FH-USP22 (I). Bound proteins were resolved by electrophoresis and detected by immunoblotting with the indicated antibodies. (J) ATXN7L3B-IP using MCF7T cells stably expressing nontargeting shRNA (pGIPZ) or shATXN7L3B. Bound proteins were resolved by electrophoresis and detected by immunoblotting with the indicated antibodies.
Here we report that ATXN7L3B interacts with ENY2, and weakly with USP22, but not with other SAGA components. These interactions limit USP22 activity toward H2Bub1 by sequestering ENY2 and destabilizing ATXN7L3. Our data provide new insights into ATXN7L3B functions in regulating USP22 activity and localization. Since USP22 overexpression is observed in several highly aggressive cancers (15, 16), we also asked whether ATXN7L3B has functions in breast cancer. We found that ATXN7L3B is highly expressed in estrogen receptor-positive (ER+) breast cancer cell lines. Knockdown of ATXN7L3B inhibits breast cancer cell migration and limits ER functions in vitro. Overall, our findings suggest that ATXN7L3B may provide a novel target for therapy development for human cancers.
MATERIALS AND METHODS
Antibodies.
Antibodies used in this study include anti-ATXN7L3B (4331-1; homemade), anti-ATXN7L3 (catalog number A301-800A; Bethyl Labs), anti-ENY2 (catalog number GTX128034; GeneTex), anti-USP22 (3933-1; homemade [17]), anti-GCN5 (catalog number 3305; Cell Signaling), anti-β-actin (catalog number Sc-47778; Santa Cruz), anti-β-tubulin (catalog number 2146S; Millipore), anti-H2Bub1 (catalog number 05-1312; Millipore), anti-H2B (catalog number 07-371; Millipore), anti-Flag (M2; catalog number F3165; Sigma), antihemagglutinin (anti-HA; catalog number 3724; Cell Signaling), and anti-V5 (catalog number 46-0705; Invitrogen).
Cell culture.
293T cells were grown in high-glucose Dulbecco's modified Eagle medium (DMEM; Thermo Scientific) supplemented with 10% fetal bovine serum (FBS) (catalog number SH30910.03; HyClone) and 1% penicillin-streptomycin (Thermo Scientific). MCF7T cells were provided by Khandan Keyomarsi and were grown in alpha medium (minimum essential medium alpha [catalog number 15-012-CV; Corning] supplemented with 10% FBS, 10 mM HEPES, 1% nonessential amino acids, 2 mM glutamine, 1% sodium pyruvate, 1 μg/ml of insulin, 1 μg/ml of hydrocortisone, and 12.5 ng/ml of epidermal growth factor). Sf21 insect cells were grown in Sf-900 II SFM (catalog number 10902-096; Life Technologies) supplemented with 1% penicillin-streptomycin. Cells were maintained in flasks (250 ml [catalog number 431144] or 500 ml [catalog number 431145]; Corning) on an orbital shaker at 26°C and 120 rpm. Sf21 cells were passaged every 3 days.
Protein lysates.
Adherent cells were trypsinized and neutralized in culture medium and then pelleted by centrifugation at 200 × g for 5 min at room temperature (RT). Sf21 cells were harvested by centrifugation at 200 × g for 5 min. In both cases, cell pellets were then washed twice in ice-cold phosphate-buffered saline (PBS) containing protease inhibitors (protease inhibitor cocktail; catalog number P8340; Sigma). Washed cell pellets were lysed in buffer C (20 mM Tris-HCl [pH 7.9], 20% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.1% NP-40, 0.2 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitors) by pipetting vigorously several times, vortexed briefly, and incubated on ice for 20 min. An equal volume of 10 mM HEPES [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 1 mM PMSF, and protease inhibitors was then added to the lysates. Cell lysates were sonicated for 5 min using Biorupter Twin (Diagenode, UCD-400) and then centrifuged at 12,000 rpm for 10 min at 4°C. The cleared supernatants were taken as whole-cell lysates.
Immunoprecipitation.
After obtaining whole-cell lysates, protein concentrations were measured by Bradford assays (catalog number 500-0006; Bio-Rad). A total of 0.8 to 1 mg of total proteins was used for each immunoprecipitation (IP). Thirty microliters of anti-Flag M2 (catalog number A2220; Sigma) beads was added to the lysates and incubated for 4 h on a rocking platform at 4°C. After incubation, beads were centrifuged at 1,000 rpm for 1 min and washed in wash buffer 150 (10 mM Tris-HCl [pH 7.9], 10% glycerol, 150 mM NaCl, 1.5 mM MgCl2, 0.1% NP-40, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, protease inhibitors) once and wash buffer 350 (10 mM Tris-HCl [pH 7.9], 350 mM NaCl, 1.5 mM MgCl2, 0.1% NP-40, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, protease inhibitors) once. In each wash, beads were incubated at 4°C for 5 min on a rocking platform and then centrifuged at 1,000 rpm for 1 min. Beads with precipitated complexes were boiled in equal volume of 2× SDS sample buffer at 95°C for 10 min.
Immunoblotting.
Twenty to 40 μg of whole-cell lysates or IP proteins was resolved on 4 to 12% NuPAGE gels (catalog number NW04122BOX; Life Technologies). After electrophoresis, proteins were transferred to 0.2-μm nitrocellulose in transfer buffer (25 mM Tris, 190 mM glycine, 10% methanol) for 1 h at a constant 300 mA and 4°C. After blocking in 5% nonfat milk–Tris-buffered saline with Tween 20 (TBST) for 1 h at RT, nitrocellulose membranes were incubated with primary antibodies overnight at 4°C. After three washes in TBST for 5 min, membranes were incubated with secondary horseradish peroxidase (HRP)-conjugated antibodies (catalog number NA934V for rabbit and catalog number NA931V for mouse; GE Healthcare) for 45 min. After three washes in TBST for 5 min, membranes were incubated with ECL Prime Western blotting detection reagent (RPN2232; GE Healthcare Life Sciences) for 2 min and exposed.
Tandem IP sample preparation for MudPIT analyses.
Sample preparation for multidimensional protein identification technology (MudPIT) analyses was performed as described previously (17). Briefly, 293T cells, stably expressing pINTO-N-FH-ATXN7L3, pINTO-N-FH-ATXN7L3B, or empty vector, were harvested by trypsinization, washed twice in ice-cold PBS, pelleted by centrifugation, and resuspended in 5 pellet volumes of buffer C (20 mM Tris-HCl [pH 7.9], 20% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.1% NP-40, 0.2 mM EDTA, 1 mM DTT, 1 mM PMSF, protease inhibitors). Lysates were incubated on ice for 30 min and further homogenized using a glass homogenizer with 15 strokes using a tight pestle. An equal volume of ice-cold 10 mM HEPES [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 1 mM PMSF, and protease inhibitors was added to homogenates. After centrifugation, the clear supernatants were collected and a small fraction was saved as input. Equal amounts of total protein for all samples were subjected to Flag IP and subsequent HA IP, after which precipitated complexes were eluted by 150 μg/ml of 3× Flag peptide (catalog number F3290; Sigma) and 100 mM glycine (pH 2.0), respectively. Ten percent of the eluates (100 μl) was saved for silver staining and immunoblotting following gel electrophoresis. For MudPIT samples, the final eluates were treated with Benzonase (catalog number E8263-5KU; Sigma) and precipitated by trichloroacetic acid (TCA; catalog number T6399; Sigma). Eluate-TCA mixtures were incubated at 4°C overnight. After this incubation, precipitated proteins were pelleted and washed twice with ice-cold acetone. Pellets were air dried in a fume hood and submitted for MudPIT analysis as described previously (18). Original mass spectrometry data can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/libpb-1053.
Subcellular fractionation.
Cells were resuspended in 200 μl of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 1 mM PMSF, protease inhibitor) to separate nuclei from a crude cytoplasmic fraction, which was further clarified by high-speed centrifugation to remove cell debris and insoluble aggregates. The supernatant was designated the cytoplasmic fraction and mixed with 1/4 volume of 5× SDS loading buffer and boiled. Nuclei were washed with buffer A and resuspended in 200 μl of 2× SDS loading buffer, boiled for 10 min, and sonicated for 15 s at 25% amplitude using an EpiShear probe sonicator (model CL-18; Active Motif).
Transfection and viral infection.
When 293T cells reached 60% confluence, pINTO-N-FH empty vector, pINTO-N-FH-ATXN7L3, or pINTO-N-FH-ATXN7L3B was transfected using Lipofectamine 2000 (catalog number 11668019; Life Technologies) by following the manufacturer's instructions. Six to 8 h after transfection, the medium was changed to normal culture medium as described above. Viral medium was produced using 293T cells cotransfected with lentiviral or retroviral vectors and psPAX.2 and pMD2.G (Addgene) for lentiviral production or pCL-Ampho (Addgene) for retroviral production. Forty-eight hours posttransfection, the culture medium containing the viral particles (viral medium) was collected, filtered through 0.45-μm filters, and used for infection. For infection of cells, the viral medium was diluted 1:3 with the culture medium containing 8 μg/ml of Polybrene (catalog number H9268; Sigma).
Immunofluorescence.
Appropriate amounts of cells were seeded onto sterile coverslips placed in 6-well plates 24 h before staining. Cells were rinsed with PBS once and fixed with 3% formaldehyde in PBS for 15 min at RT, followed by three rinses with PBS. Cells were then permeabilized with 0.5% Triton X-100 for 5 min at RT and rinsed again with PBS three times. After blocking with 10% FBS in PBS for 45 min at RT, cells were incubated with anti-Flag antibody for 1 h at RT, followed by three rinses with PBS. Then cells were incubated with a secondary anti-mouse antibody for 1 h at RT and rinsed three times with PBS. One milliliter of 100% ethanol was added onto the cells for 1 min and then removed. Slides were dried for 5 min at RT, and coverslips were removed from the 6-well plate and mounted onto a microslide using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; catalog number H-1200; Vector Laboratories). Images were acquired using a laser scanning spectral confocal microscope (Leica STP6000).
Protein purification from the baculovirus system.
Large-scale protein purification from the baculovirus system was performed as previously described (2). Briefly, 4 × 107 Sf21 cells were infected and maintained in flasks in an orbital shaker for 3 days at 27°C and 120 rpm. Cells were harvested and lysed in protein purification buffer C (50 mM HEPES [pH 7.9], 20% glycerol, 300 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 1 mM DTT, 1 mM PMSF, protease inhibitors). The supernatant was recovered by centrifugation and then incubated with 80 μl of anti-Flag M2, anti-HA matrix (catalog number 1181506001; Roche), or anti-V5 agarose affinity gel (catalog number A7345; Sigma) overnight at 4°C. After being washed with wash buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.1% Triton X-100, 300 mM NaCl, 1 mM DTT, 1 mM PMSF, protease inhibitor) 3 times, bound proteins were eluted with 200 μg/ml of Flag peptide (catalog number F3290; Sigma) at 4°C or 400 μg/ml of HA peptide (catalog number 11666975001; Sigma and Roche) at RT or 700 μg/ml of V5 peptide (catalog number V7754; Sigma) at RT for 2 h. The eluted complexes or recombinant proteins were further purified through a gel filtration Superdex 200 column (GE Healthcare).
In vitro deubiquitination assay.
An in vitro deubiquitination assay was performed as previously described (2). Total free histones were extracted from 293T cells using a histone purification minikit (catalog number 40026; Active Motif) per the manufacturer's instruction. Free histones were incubated with purified recombinant USP22 or deubiquitinase complexes in reaction buffer (100 mM Tris-HCl [pH 8.0], 5% glycerol, 1 mM EDTA, 3 mM DTT) for 2 h at 37°C. A total of ¼ volume of 5× SDS loading buffer was added and the mixture was boiled to stop the reaction. Immunoblots were performed to assess ubiquitination levels of the histones.
In vitro competition assay.
Individual recombinant proteins were purified from Sf21 cells. Equal amounts of V5-ATXN7L3 and Flag-ENY2 and increasing amounts of V5-ATXN7L3B were added into 1 ml of wash buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.1% Triton X-100, 300 mM NaCl, 1 mM DTT, 1 mM PMSF, protease inhibitor). Reaction mixtures were incubated in 4°C overnight on a rocking platform. After addition of anti-Flag M2 beads, reaction mixtures were incubated in 4°C for another hour. Bead-protein complexes were washed three times with wash buffer and boiled in equal volume of 1× SDS loading buffer. The supernatants were analyzed by immunoblotting.
RNA extraction, reverse transcription, and quantitative real-time PCR.
Total RNA was isolated using an RNeasy minikit (catalog number 74104; Qiagen) by following the manufacturer's recommended procedure. One microgram of RNA was reverse-transcribed to cDNA using a SuperScript VILO cDNA synthesis kit (catalog number 11754050; ThermoFisher Scientific) by following the manufacturer's procedure. Ten nanograms of cDNA was used for one quantitative real-time PCR with three technical replicates.
Transwell migration assay.
MCF7T cells were used for Transwell migration assays. Transwell chambers with transparent polyethylene terephthalate (PET) membranes (catalog number 353097; Falcon) were placed in 24-well plates with normal culture medium, alpha medium. Then 80,000 cells suspended in alpha medium containing 0.1% bovine serum albumin (BSA) were placed on the upper layer of the chambers. After 24 h of incubation at 37°C with 5% CO2, the membranes at the bottom of chambers were fixed with methanol for 5 min and stained with 0.5% crystal violet for 10 min. Then the cells above the membranes were removed with cotton swabs. Membranes were dried, and cells attached below the membranes were imaged using microscope (Axiovert 40 CFL; Zeiss).
Colony formation assays.
Stable MCF7T pGIPZ-, shATXN7L3-, and shATXN7L3B-expressing cells were trypsinized thoroughly, neutralized with alpha medium, and counted 3 times for each sample. Three hundred cells for each sample were put into single wells of a 6-well plate, with three replicates, and incubated at 37°C with 5% CO2. Twenty to 30 days later, cells were washed with PBS once and fixed with fixation buffer (acetic acid-methanol, 1:7 [vol/vol]) for 5 min. Then cells were stained with 0.5% crystal violet for 2 h to overnight, washed, dried, and imaged.
RESULTS
ATXN7L3B interacts with DUB module components but not SAGA.
ATXN7L3B and ATXN7L3 share 74% identity within their N-terminal ∼60 amino acid residues (Fig. 1A), including a region that interacts with ENY2 (Fig. 1B, in red) (6) called the Sus1-binding region for the yeast ENY2 ortholog (14). Interestingly, others have reported that a truncated form of ATXN7L3 that contains only amino acids 3 to 76 interacts with USP22 and ENY2 in vitro, but the complex containing the truncated protein cannot remove monoubiquitination from H2Bub1 (6). Since the native structure of ATXN7L3B resembles this artificial form of ATXN7L3, we reasoned that it might also interact with USP22 or other SAGA components and affect DUB activity in vivo.
We first determined whether ATXN7L3B is incorporated into the SAGA complex. Using tandem IP, we affinity purified ATXN7L3- or ATXN7L3B-interacting proteins from 293T cells stably expressing Flag- and HA-tagged ATXN7L3 (FH-ATXN7L3) or ATXN7L3B (FH-ATXN7L3B) (Fig. 1C). Multidimensional Protein Identification Technology (MudPIT) indicated that FH-ATXN7L3 interacted with most known components of the SAGA complex, with ENY2 and USP22 as the top interacting partners, as expected (Fig. 1D). ATXN7L3B also bound strongly to ENY2 and less strongly to USP22, but it did not bind ATXN7 (Fig. 1D). The interaction between ATXN7L3B and ENY2 was comparable with that between ATXN7L3 and ENY2, but the interaction between ATXN7L3B and USP22 was much weaker than with ATXN7L3 (∼60-fold difference). Except for very low levels of TAF10, TAF6L, and TADA3, no other SAGA components were detected among FH-ATXN7L3B-associated proteins (Fig. 1D). Interactions between USP22, ENY2, and FH-ATXN7L3 or FH-ATXN7L3B were confirmed by coimmunoprecipitations (co-IPs) followed by immunoblotting (Fig. 1E and F). These results indicate that ATXN7L3B, even though it interacts with ENY2, does not associate with SAGA.
ENY2 is also an important component of the transcription and export complex 2 (TREX-2) complex, which is involved in RNA transport (19). Examination of our MudPIT results indicates only a weak interaction of FH-ATXN7L3B with CETN2 (Fig. 1D), with no evidence for interaction with other TREX-2 components, including PCID2, DSS1, GANP, and CETN3 (20). We were not able to confirm interaction between ATXN7L3B and CETN2 by immunoprecipitation, further suggesting that ATXN7L3B does not associate with TREX-2.
To investigate the functions of endogenous ATXN7L3B, we generated a polyclonal antibody targeting amino acids 75 to 97 in the ATXN7L3B C-terminal region, which are not homologous with ATXN7L3. Although this antibody recognized multiple nonspecific bands, it also recognized a protein of the expected size for endogenous ATXN7L3B (∼11 kDa) that is depleted after short hairpin RNA (shRNA)-mediated knockdown of ATXN7L3B (Fig. 1G). This 11-kDa protein is not recognized by the preimmune sera. Unfortunately, a very strong nonspecific band recognized by this antibody corresponds to the size of FH-ATXN7L3B (∼17 kDa), precluding use of this antibody for detection of the tagged protein. Therefore, we used anti-Flag or anti-HA antibodies to detect FH-ATXN7L3B throughout our study. Nonetheless, we were able to confirm interaction between endogenous ATXN7L3B and ENY2 by probing immunoblots of FH-ENY2 immunoprecipitates with our anti-ATXN7L3B antibody (Fig. 1H). We also confirmed interaction of FH-USP22 with endogenous ATXN7L3B (Fig. 1I). Moreover, immunoprecipitations of endogenous ATXN7L3B using our antibody confirmed interactions with endogenous ENY2 (Fig. 1J). Depletion of endogenous ATXN7L3B resulted in less ENY2 in the immunoprecipitate, further confirming the specificity of the interaction. Together, these data demonstrate that ATXN7L3B binds strongly to ENY2 and more weakly with USP22.
ATXN7L3B mainly localizes to the cytoplasm, whereas ATXN7L3 is nuclear.
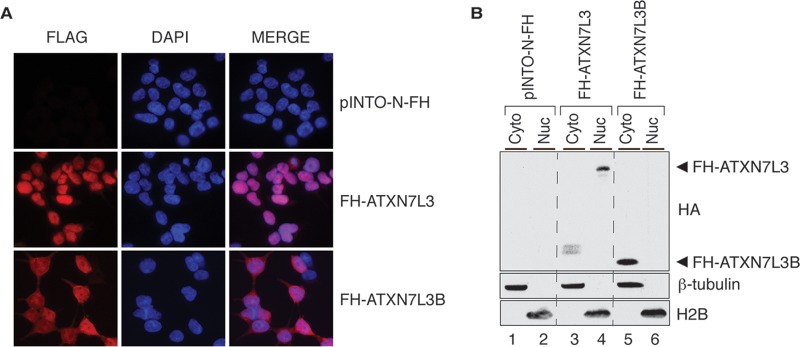
SAGA is best characterized as a transcriptional coactivator functioning in the cell nucleus (5). To determine the subcellular distribution of ATXN7L3B, we performed immunofluorescence assays using 293T cells that stably express FH-ATXN7L3 or FH-ATXN7L3B. Staining with anti-Flag antibodies and DAPI revealed that ATXN7L3B is mainly localized outside the nucleus, in the cytoplasm (Fig. 2A). In contrast, as expected, ATXN7L3 mainly localized to the nucleus (Fig. 2A). We confirmed the different distributions of these proteins by separating cells into cytoplasmic and nuclear fractions followed by immunoblotting (Fig. 2B). While FH-ATXN7L3 localized in the nuclear fraction with histone H2B, FH-ATXN7L3B localized in the cytoplasmic fraction with β-tubulin. The distinct subcellular locations of ATXN7L3 and ATXN7L3B suggest that despite their similarities in sequence, these two proteins likely perform different functions in cells.
FIG 2.
ATXN7L3B mainly localizes to the cytoplasm, whereas ATXN7L3 is nuclear. (A) Immunofluorescence using Flag antibody (red) on 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ATXN7L3, or pINTO-N-FH-ATXN7L3B. Nuclei were counterstained with DAPI (blue). (B) Cytoplasmic and nuclear fractions were isolated from 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ATXN7L3, or pINTO-N-FH-ATXN7L3B. Proteins were resolved by SDS-PAGE and detected by immunoblotting with the indicated antibodies. FH-ATXN7L3 and FH-ATXN7L3B proteins bands are indicated with arrowheads. H2B marks the nuclear fraction, while β-tubulin marks the cytoplasmic fraction.
ATXN7L3 and ATXN7L3B affect global levels and subcellular distributions of H2Bub1, ENY2, and USP22.
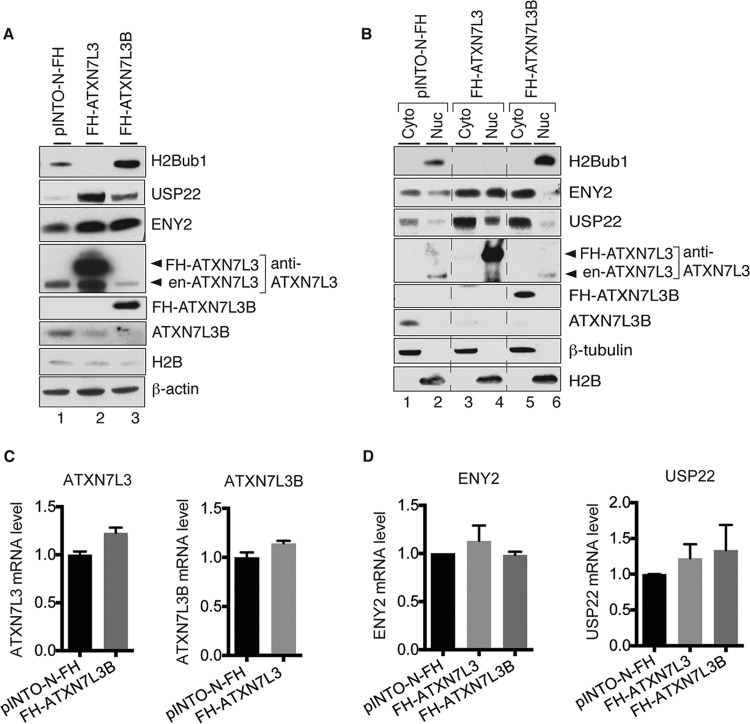
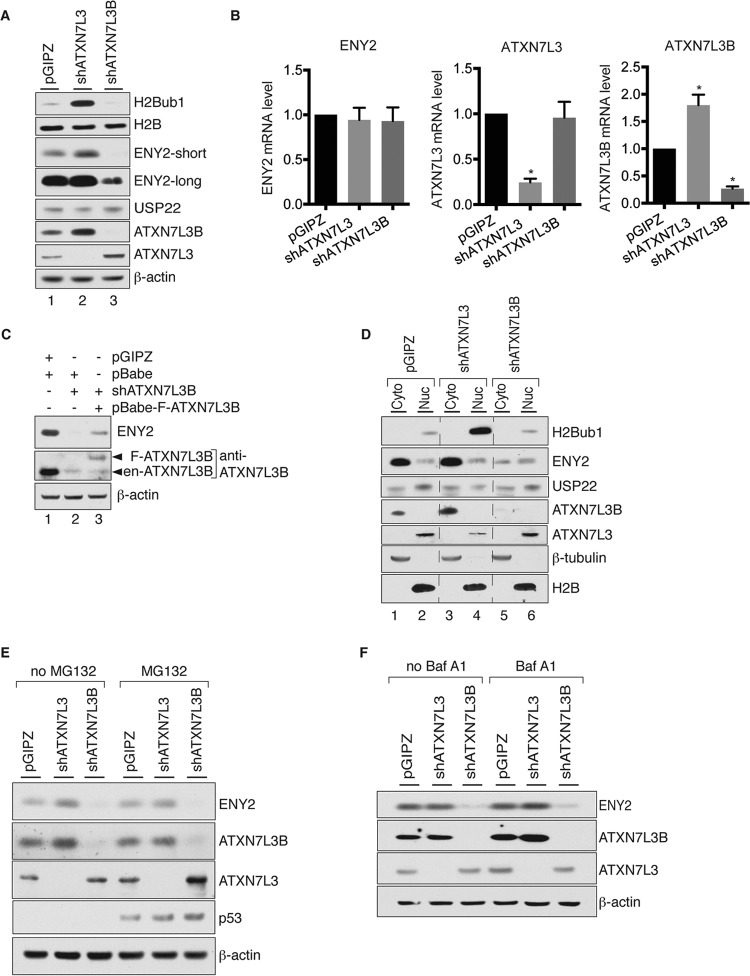
The best-characterized substrate of the SAGA DUB module is histone H2Bub1. Fluctuations in ENY2 protein levels affect DUB module activity and overall levels of H2Bub1 (5, 17, 21). Since ATXN7L3B interacts with ENY2, we determined how changes in ATXN7L3B levels affect H2Bub1. As expected, based on previously published data (17), overexpression of ATXN7L3 greatly diminished H2Bub1 (Fig. 3A, lanes 1 and 2), consistent with increased DUB activity. Surprisingly, overexpression of ATXN7L3B had an opposite effect, dramatically increasing H2Bub1 levels (Fig. 3A, lanes 1 and 3), consistent with loss of DUB activity. Examination of ENY2 and USP22 levels by immunoblotting revealed that overexpression of ATXN7L3 led to increased levels of both ENY2 and USP22 proteins (Fig. 3A, lanes 1 and 2), consistent with the increased DUB activity and decreased H2Bub1 levels observed. Interestingly, ATXN7L3B overexpression also led to increased of levels of ENY2 and USP22 (Fig. 3A, lanes 1 and 3), although the extent of increase in USP22 levels was lower than that caused by ATXN7L3 overexpression. These results were puzzling, as previous work has shown that overexpression of USP22 leads to decreased H2Bub1 (17). Examination of the effects of overexpression of ATXN7L3 or ATXN7L3B on the endogenous levels of each other provided a possible explanation. Overexpression of ATXN7L3B resulted in significantly decreased levels of endogenous ATXN7L3 (Fig. 3A, lanes 1 and 3), consistent with the increased H2Bub1 levels observed. Conversely, overexpression of ATXN7L3 led to decreased levels of endogenous ATXN7L3B (Fig. 3A, lanes 1 and 2). Unexpectedly, we also observed a significant decrease in endogenous ATXN7L3B after ectopic overexpression of FH-ALTXNL3B (Fig. 3A, lanes 1 and 3), suggesting self-regulation of ATXN7L3B protein levels.
FIG 3.
ATXN7L3 and ATXN7L3B affect global levels and subcellular distributions of H2Bub1, ENY2, and USP22. (A) Whole-cell lysates from 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ATXN7L3, or pINTO-N-FH-ATXN7L3B were resolved by SDS-PAGE. Proteins were transferred onto membranes and detected by immunoblotting with the indicated antibodies. Arrowheads indicate endogenous (en-) and exogenous (FH-) ATXN7L3 proteins. (B) Cytoplasmic (Cyto) and nuclear (Nuc) fractions were isolated from 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ATXN7L3, or pINTO-N-FH-ATXN7L3B. Proteins were transferred onto membranes and detected by immunoblotting with the indicated antibodies. Arrowheads indicate endogenous and exogenous ATXN7L3 proteins. (C and D) Effects of overexpression of FH-ATXN7L3 or FH-ATXN7L3B on the levels of the indicated proteins occur posttranscription. RNAs were extracted from 293T cells stably expressing pINTO-N-FH vector, pINTO-N-FH-ATXN7L3, or pINTO-N-FH-ATXN7L3B and used as the templates for cDNA generation and quantitative real-time PCR. The mRNA levels of ATXN7L3 and ATXN7L3B (C) and ENY2 and USP22 (D) were examined.
Since ATXN7L3 and ATXN7L3B are located in different subcellular compartments (Fig. 2), we isolated cytoplasmic and nuclear fractions from FH-ATXN7L3- or FH-ATXN7L3B-expressing cells in order to better understand effects on H2Bub1, ENY2, and USP22 levels. H2Bub1 levels in the nuclear fraction faithfully reflected levels observed in whole-cell lysates (Fig. 3B, compare lanes 2, 4, and 6). Interestingly, overexpression of FH-ATXN7L3 led to increased levels of ENY2 and USP22 in both the cytoplasmic and nuclear fractions (Fig. 3B, compare lanes 1 and 2 to lanes 3 and 4), whereas overexpression of FH-ATXN7L3B led to increased levels of ENY2 and USP22 only in the cytoplasmic fraction and decreased levels of ENY2 in the nuclear fraction (Fig. 3B, compare lanes 1 and 2 to lanes 5 and 6). These results help to explain why the increased global levels of USP22 and ENY2 observed upon overexpression of FH-ATXN7L3B did not correlate to decreased levels of H2Bub1 (Fig. 3A), since the majority of H2Bub1 is associated with chromatin in the nucleus. In contrast, upon overexpression of ATXN7L3B, ATXN7L3 levels were decreased in the nuclear fraction (Fig. 3B, lanes 2 and 6), consistent with the increased H2Bub1 levels observed. Overexpression of either FH-ATXN7L3 or FH-ATXN7L3B decreased the cytoplasmic levels of ATXN7L3B (Fig. 3B, lanes 1, 3, and 5), consistent with the global changes observed (Fig. 3A). Interestingly, the changes in the protein levels observed upon overexpression of FH-ATXN7L3 or FH-ATXN7L3B were not reflected by changes in corresponding RNA levels (Fig. 3C and D), indicating that these effects occur posttranscriptionally.
ATXN7L3B competes with ATXN7L3 for ENY2 binding in vitro.
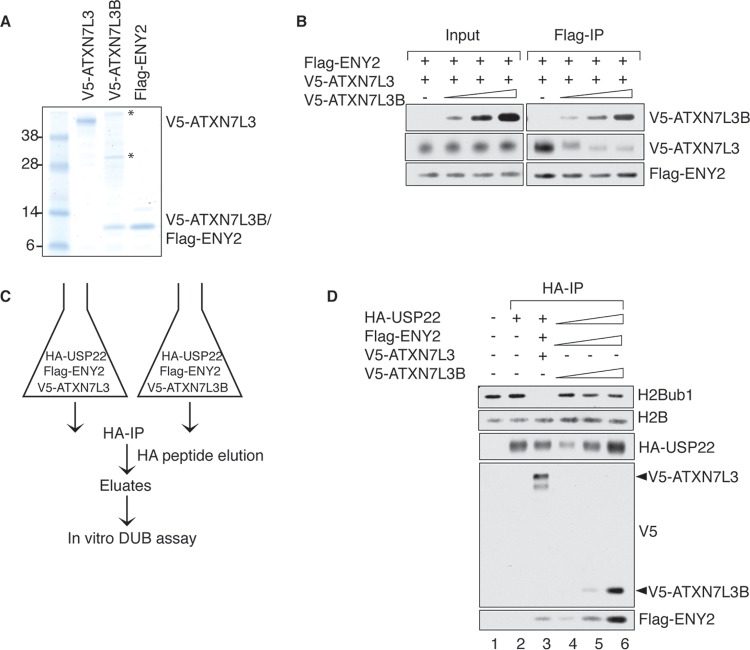
Since both ATXN7L3 and ATXN7L3B physically interact with ENY2, we determined whether ATXN7L3B competes with ATXN7L3 for ENY2 binding in vitro. We purified recombinant Flag-ENY2, V5-ATXN7L3, and V5-ATXN7L3B separately from insect cells (Fig. 4A) and added increasing amounts of V5-ATXN7L3B into reaction mixtures containing a constant amount of Flag-ENY2 and V5-ATXN7L3 (Fig. 4B). Using Flag immunoprecipitations, we observed increasing amounts of ATXN7L3B but decreasing amounts of ATXN7L3, associated with Flag-ENY2, indicating that ATXN7L3B is able to compete with ATXN7L3 for ENY2 binding. This result provides a plausible explanation for the decreased levels of ATXN7L3 protein level observed after overexpression of ATXN7L3B (Fig. 3A and B), as previous studies have shown that ENY2 is necessary for stabilization of ATXN7L3 (17, 20). Overexpression of ATXN7L3B likely sequesters ENY2, resulting in less ENY2-ATXN7L3 association and destabilization of ATXN7L3.
FIG 4.
ATXN7L3B competes with ATXN7L3 for ENY2 binding in vitro. (A) Recombinant proteins were purified from Sf21 cells and resolved by electrophoresis, followed by colloidal blue staining. Asterisks indicate background protein bands. (B) The indicated recombinant proteins were mixed in an in vitro competition reaction, and the formed complexes were precipitated with anti-Flag resin. The resin-bound proteins were eluted, resolved by SDS-PAGE, and detected by immunoblotting with the indicated antibodies. Five percent of the in vitro reactions was used as input. (C) Schematic of HA affinity purification using Sf21 cells coinfected with baculovirus containing expressing vector of HA-USP22, Flag-ENY2, and V5-ATXN7L3 or V5-ATXN7L3B. (D) The indicated immunoprecipitated complexes from panel C were used in in vitro deubiquitination reactions in which purified histones served as the substrates. Proteins in reactions were resolved by electrophoresis and detected by immunoblotting with the indicated antibodies.
To test whether USP22, ATXN7L3B, and ENY2 can form a stable complex in vitro, we coinfected cells with baculovirus containing expression vectors for HA-USP22, Flag-ENY2, and V5-ATXN7L3 or V5-ATXN7L3B and used the lysates to perform HA immunoprecipitations (Fig. 4C). As expected, HA-USP22 pulled down V5-ATXN7L3 and Flag-ENY2 (Fig. 4D, lane 3). HA-USP22 also pulled down V5-ATXN7L3B and Flag-ENY2 (Fig. 4D, lanes 4 to 6), suggesting that USP22, ATXN7L3B, and ENY2 can form a stable complex in vitro. To determine whether the ATXN7L3B-containing complex has deubiquitinase activity, we performed in vitro deubiquitination assays using histone H2Bub1 as the substrate. Consistent with previous results (6), the complex that contained USP22, ATXN7L3, and ENY2 deubiquitinated H2Bub1 completely after 1 h (Fig. 4D, lane 3). However, the complex containing ATXN7L3B instead of ATXN7L3 had only weak, if any, deubiquitinase activity toward H2Bub1, even though there was more USP22 in the reaction (Fig. 4D, lanes 3 and 6). These results indicate that if ATXN7L3B-containing complexes function as DUBs in vivo, they likely target nonhistone proteins, consistent with the cytoplasmic location of ATXN7L3B.
ATXN7L3B regulates the protein levels of ENY2.
To complement the overexpression experiments (Fig. 3), we next compared the effects of depletion of ATXN7L3B or ATXN7L3 on levels of H2Bub1 and DUB module components. We used MCF7T cells for these experiments, as endogenous ATXN7L3B is easily detectable in these cells (Fig. 5A, lane 1). As expected, depletion of ATXN7L3 dramatically increased the levels of H2Bub1 (Fig. 5A, lanes 1 and 2). However, knockdown of ATXN7L3B slightly decreased H2Bub1 levels in MCF7T cells (Fig. 5A, lanes 1 and 3). Interestingly, knockdown of ATXN7L3B caused a significant reduction in ENY2 levels, whereas knockdown of ATXN7L3 increased levels of ENY2 (Fig. 5A). In addition, knockdown of ATXN7L3 caused an increase in ATXN7L3B protein levels, and conversely, depletion of ATXN7L3B led to increased levels of ATXN7L3 (Fig. 5A). Taken together with our observation that overexpression of ATXN7L3 or ATXN7L3B led to decreased expression of the other protein (Fig. 3A), these results further suggest cross regulation of ATXN7L3 and ATXN7L3B. Knockdown of ATXN7L3 or ATXN7L3B did not significantly affect ENY2 mRNA levels, indicating that regulation of ENY2 by ATXN7L3 or ATXN7L3B occurred posttranscriptionally (Fig. 5B). Knockdown of ATXN7L3B also did not alter mRNA levels of ATXN7L3, whereas knockdown of ATXN7L3 increased ATXN7L3B mRNA (Fig. 5B). Additionally, the reduction of ENY2 upon knockdown of ATXN7L3B was restored by expression of exogenous ATXN7L3B (Fig. 5C), indicating that the effect is not due to an off-target effect of the knockdown reagents.
FIG 5.
ATXN7L3B regulates the protein levels of ENY2. (A) Whole-cell lysates from MCF7T cells stably expressing shATXN7L3, shATXN7L3B, or nontargeting shRNA (pGIPZ) were resolved by SDS-PAGE. Proteins were transferred onto membranes and detected by immunoblotting with the indicated antibodies. (B) RNAs from ATXN7L3 depleted, ATXN7L3B depleted, or control (pGIPZ) MCF7T cells were isolated. The mRNA levels of ENY2, ATXN7L3, and ATXN7L3B were examined using quantitative real-time PCR. P values were calculated using Student's t test (*, P < 0.05). (C) Ectopic expression of shRNA immune ATXN7L3B restored the ENY2 levels in ATXN7L3B-depleted cells. Whole-cell lysates from ATXN7L3B-depleted MCF7T cells were used to monitor the ENY2 protein levels before (lane 2) and after (lane 3) ectopic expression of Flag-ATXN7L3B. Arrowheads indicate endogenous and exogenous ATXN7L3B proteins. (D) Cytoplasmic and nuclear fractions were isolated from the cells used for panel A, resolved by electrophoresis. Proteins were transferred onto membranes and detected by immunoblotting with the indicated antibodies. (E and F) ATXN7L3-depleted, ATXN7L3B-depleted, or control (pGIPZ) MCF7T cells were treated with MG132 (20 μM, 5 h) or BafA1 (100 nM, 2 h). Whole-cell lysates were extracted and resolved by electrophoresis. Proteins were transferred onto membranes and detected by immunoblotting with the indicated antibodies.
We next determined how ATXN7L3B or ATXN7L3 depletion affects the subcellular distribution of ENY2. Knockdown of ATXN7L3B significantly decreased levels of ENY2 in the cytoplasm (Fig. 5D, compare lanes 1 and 2 to lanes 5 and 6). Conversely, depletion of ATXN7L3 led to an increase in the cytoplasmic ENY2 levels (Fig. 5D, compare lanes 1 and 2 to lanes 3 and 4), in which the mechanism is not clear.
The dramatic decrease of ENY2 protein levels upon ATXN7L3B knockdown triggered our curiosity as to whether ATXN7L3B depletion also affects the functions of the TREX-2 complex. ENY2 is crucial for protein stability of GANP, another critical component in TREX-2 complex (20). Silencing of ENY2 results in defects in mRNA export and mRNA nuclear accumulation in HeLa cells (20). However, in MCF7T cells, silencing of either ATXN7L3B or ENY2 had no effects on both protein levels of GANP or mRNA export (data not shown). These results indicate that ATXN7L3B does not affect the functions of TREX-2 in MCF7T cells, consistent with our finding that AXN7L3B does not interact with TREX-2 components (Fig. 1D).
To examine whether ENY2 levels are regulated through the proteasome or through lysosome-dependent protein degradation, we treated control, ATXN7L3-depleted, or ATXN7L3B-depleted MCF7T cells with MG132, a proteasome inhibitor, or bafilomycin A1 (BafA1), a lysosome inhibitor. Neither MG132 nor BafA1 treatment rescued reductions in ENY2 protein levels caused by ATXN7L3B knockdown (Fig. 5E and F). These data indicate that ATXN7L3B likely regulates ENY2 levels at the level of RNA processing, RNA transport, or protein translation.
ATXN7L3B regulates migration of ER+ breast cancer cells.
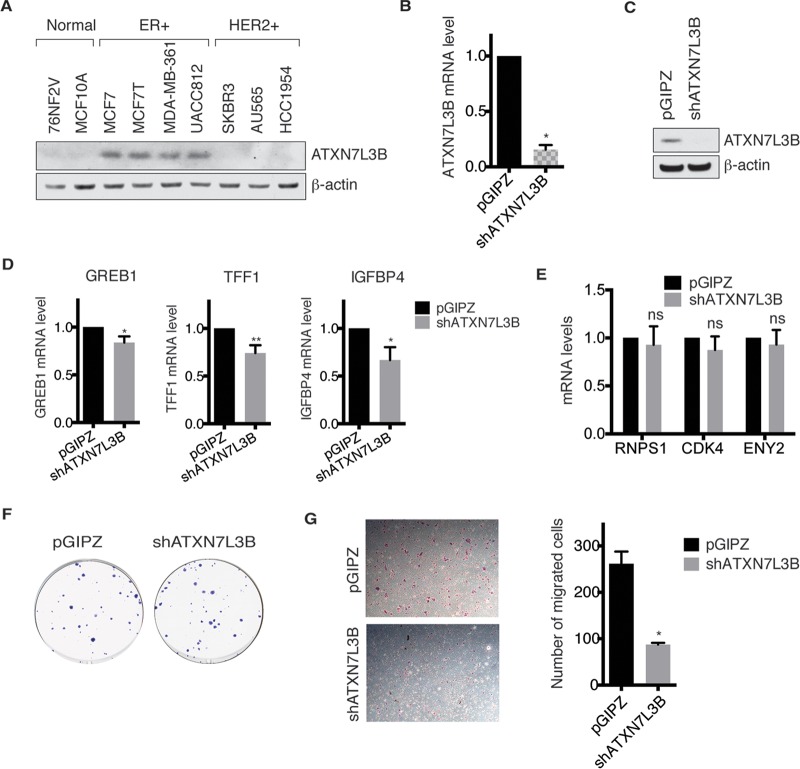
Given that USP22 overexpression is correlated with several aggressive cancers (3, 16, 22), we examined ATXN7L3B protein levels in normal mammary, estrogen receptor-positive (ER+), and HER2-positive (HER2+) breast cancer cell lines (Fig. 6A). Interestingly, higher ATXN7L3B protein levels were observed only in the ER+ breast cancer cell lines (Fig. 6A). We depleted ATXN7L3B in MCF7T cells (Fig. 6B and C) to determine whether ATXN7L3B loss affects ER functions, cell migration, or colony formation. ATXN7L3B depletion slightly, but significantly, decreased expression of three well-characterized ER target genes, those for growth regulation by estrogen in breast cancer 1 (GREB1), trefoil factor 1 (TFF1) (23), and insulin-like growth factor binding protein 4 (IGFBP4) (24) (Fig. 6D). Expression of non-ER target genes, including the genes for RNA-binding protein with serine-rich domain 1 (RNPS1), cyclin-dependent kinase 4 (CDK4), and ENY2, was not affected by ATXN7L3B depletion (Fig. 6E). ATXN7L3B depletion did not affect colony formation (Fig. 6F), but it significantly decreased migration of MCF7T cells (Fig. 6G). Therefore, ATXN7L3B appears to function specifically in the context of ER-positive breast cancers, in which it affects cell motility.
FIG 6.
Depletion of ATXN7L3B inhibits migration of ER-positive breast cancer cells. (A) Protein levels of ATXN7L3B in two normal mammary, four ER+, and three HER2+ cell lines were detected by immunoblotting. (B and C) Efficient silencing of ATXN7L3B in MCF7T cells stably expressing shATXN7L3B. The mRNA levels of ATXN7L3B were examined using quantitative real-time PCR (B), and protein levels of ATXN7L3B were examined by immunoblotting (C). (D) Expression levels of ER target genes, the GREB1, TFF1, and IGFBP4 genes, in AXTN7L3B-depleted and control (pGIPZ) MCF7T cells. mRNA levels was examined using quantitative real-time PCR. (E) Expression levels of non-ER target genes in AXTN7L3B-depleted and control (pGIPZ) MCF7T cells. mRNA levels was examined using quantitative real-time PCR. (F) ATXN7L3B-depleted or control (pGIPZ) cells were used to perform colony formation assay. After 20 days of incubation, colonies were stained and imaged. (G) ATXN7L3B ablation impairs MCF7T cell migration. AXTN7L3B-depleted or control (pGIPZ) cells were used to perform a Transwell migration assay. Migrated cells were stained with crystal violet, imaged, and counted.
DISCUSSION
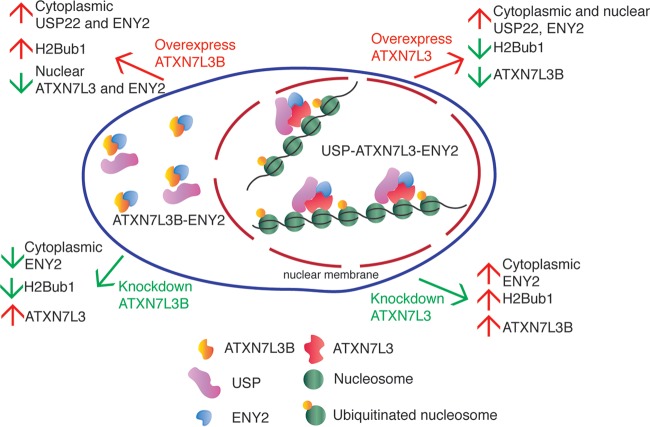
Our data indicate that USP22 DUB activity toward histones is regulated by ATXN7L3B through cytoplasmic sequestration of ENY2, resulting in destabilization of ATXN7L3 (Fig. 7). Unlike ATXN7L3, ATXN7L3B does not interact with most SAGA components, but it exhibits strong interaction with ENY2. Interactions of ENY2 with ATXN7L3 or ATXN7L3B appear to be mutually exclusive. Moreover, manipulation of ATXN7L3B levels by overexpression or shRNA-mediated depletion leads to effects on H2Bub1 that are opposite to those caused by changes in ATXN7L3 levels. These effects are consistent with changes in ATXN7L3 protein levels observed upon overexpression or knockdown of ATXN7L3B. While overexpression of ATXN7L3 leads to increased DUB activity and decreased levels of H2Bub1, overexpression of ATXN7L3B leads to reduction of ATXN7L3 and increased levels of H2Bub1, consistent with a loss of DUB activity directed toward this histone modification. In contrast to the loss of ATXN7L3, overexpression of ATXN7L3B increases cytoplasmic levels of ENY2 but decreases nuclear levels of ENY2. Conversely, depletion of ATXN7L3B leads to decreased H2Bub1 levels and increased ATXN7L3 protein levels but decreased cytoplasmic levels of ENY2. As ENY2 is required for stabilization of ATXN7L3 within the DUB module (20), ATXN7L3B likely affects H2Bub1 levels indirectly, by changing the subcellular distribution of ENY2 and altering ENY2 availability for ATXN7L3 interaction.
FIG 7.
Regulation of protein levels of H2Bub1 and DUB components by ATXN7L3 and ATXN7L3B. ATXN7L3 mainly resides in the nucleus, while ATXN7L3B mainly localizes in the cytoplasm. They both interact strongly with ENY2. Overexpression of ATXN7L3 leads to increases of cytoplasmic and nuclear USP22 and ENY2 but decreases of H2Bub1 and ATXN7L3B. Overexpression of ATXN7L3B leads to increases in H2Bub1 and cytoplasmic USP22 and ENY2 but decreases in nuclear ENY2 and ATXN7L3. Depletion of ATXN7L3 results in upregulation of ENY2, H2Bub1, and ATXN7L3B in the cytoplasm. Depletion of ATXN7L3B decreases ENY2 and H2Bub1 in the cytoplasm but increases nuclear ATXN7L3 protein levels.
In a previous study, Tan and colleagues identified ATXN7L3B, also named lnc-SCA7, as a long noncoding RNA (25). Our work, using an antibody targeting the C terminus of ATXN7L3B, indicates that the ATXN7L3B gene encodes a 97-amino-acid protein of ∼11 kDa. Although ATXN7L3B contains the Sus1/ENY2-binding region, it lacks the ZnF-Sgf11 and SCA7 domains that reside in C-terminal region of ATXN7L3. A previous report by another group demonstrated that the ZnF-Sgf11 domain of ATXN7L3 is essential for DUB activity toward H2Bub1 in vitro (6). The ZnF-Sgf11 domain is also required for ATXN7L3 binding to nucleosomal DNA (11), and the crystal structure of the DUB module reveals that an arginine cluster in the ZnF-Sgf11 domain directly interacts with ubiquitinated nucleosomes and H2A/H2B heterodimer (12). Therefore, the absence of this domain in ATXN7L3B indicates that it is unlikely to interact with histones, consistent with our findings that the ATXN7L3B-DUB module cannot efficiently deubiquitinate histones in vitro and that ATXN7L3B is largely localized to the cytoplasm in vivo. Future work will determine whether an ATXN7L3B-DUB module targets nonhistone, cytoplasmic proteins.
The effects of ATXN7L3B on USP22 activity in vivo indicate that it might also impact USP22 functions in cancer. As early as 2005, USP22 was identified as a member of an 11-gene “death-from-cancer” signature for highly aggressive tumors (15). Elevated expression of USP22 has since been reported to be a prognostic factor for poor survival in patients with colorectal cancer (16), breast cancer (22), pancreatic cancer (26, 27), cervical cancer (28), oral squamous cell carcinoma (29), hepatocellular carcinoma (30), and gastric carcinoma (31).
The functions of USP22 in cancer are not yet well defined. They might reflect its functions in deubiquitination of histones H2B and H2A (4, 5, 32) or of nonhistone substrates, such as telomeric repeat-binding factor 1 (TRF1) (33), far upstream element (FUSE)-binding protein 1 (FBP1) (34), and sirtuin 1 (SIRT1) (1, 35), which have all been implicated in cancer (36–38). USP22 can also act as a coactivator for oncoproteins, such as Myc (4). Expression of ENY2 has also been correlated with breast cancer progression (17, 39). Our finding that ATXN7L3B is specifically expressed in ER+ breast cancer subtypes raises the possibility that ATXN7L3B impacts ER functions, perhaps indirectly by regulating SAGA DUB module functions in the nucleus or through direct effects on yet-to-be defined proteins in the cytoplasm. Going forward, it will be important to determine whether overexpression of USP22, ENY2, or ATXN7L3B can directly promote cancer progression or metastasis. If so, these proteins may provide unique targets for small-molecule inhibition as cancer therapies.
ACKNOWLEDGMENTS
We thank Khandan Keyomarsi for the breast cancer cell lines used in this study. We also acknowledge the Science Park Molecular Biology Core, supported in part by The Tobacco Fund and the Center for Environmental and Molecular Carcinogenesis, for DNA sequencing. We also acknowledge support from the Stowers Institute for Medical Research to S.K.S., R.D.M., L.F., M.P.W., and J.L.W.
This work was supported by NIH grant R01 GM096472 (USP22) and CPRIT RP110417 (Lonestar Project) to S.Y.R.D. and NIH grant R01 GM099945 to J.L.W. and M.P.W.
REFERENCES
- 1.Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, Fang D. 2012. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell 46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 2.Lan X, Koutelou E, Schibler AC, Chen YC, Grant PA, Dent SY. 2015. Poly(Q) expansions in ATXN7 affect solubility but not activity of the SAGA deubiquitinating module. Mol Cell Biol 35:1777–1787. doi: 10.1128/MCB.01454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glinsky GV. 2006. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle 5:1208–1216. doi: 10.4161/cc.5.11.2796. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köhler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. 2008. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol 10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Dent SY. 2014. Functions of SAGA in development and disease. Epigenomics 6:329–339. doi: 10.2217/epi.14.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Köhler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koehler C, Bonnet J, Stierle M, Romier C, Devys D, Kieffer B. 2014. DNA binding by Sgf11 protein affects histone H2B deubiquitination by Spt-Ada-Gcn5-acetyltransferase (SAGA). J Biol Chem 289:8989–8999. doi: 10.1074/jbc.M113.500868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan MT, Haj-Yahya M, Ringel AE, Bandi P, Brik A, Wolberger C. 2016. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351:725–728. doi: 10.1126/science.aac5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajakulendran S, Roberts J, Koltzenburg M, Hanna MG, Stewart H. 2013. Deletion of chromosome 12q21 affecting KCNC2 and ATXN7L3B in a family with neurodevelopmental delay and ataxia. J Neurol Neurosurg Psychiatry 84:1255–1257. doi: 10.1136/jnnp-2012-304555. [DOI] [PubMed] [Google Scholar]
- 14.Ellisdon AM, Jani D, Kohler A, Hurt E, Stewart M. 2010. Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1. J Biol Chem 285:3850–3856. doi: 10.1074/jbc.M109.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glinsky GV, Berezovska O, Glinskii AB. 2005. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest 115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YL, Yang YM, Xu H, Dong XS. 2011. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J Surg Oncol 103:283–289. doi: 10.1002/jso.21802. [DOI] [PubMed] [Google Scholar]
- 17.Atanassov BS, Mohan RD, Lan X, Kuang X, Lu Y, Lin K, Mclvor E, Li W, Zhang Y, Florens L, Byrum SD, Mackintosh SG, Davis T, Koutelou E, Wang L, Tang D, Tackett AJ, Washburn MP, Workman JL, Dent SY. 2016. ATXN7L3 and ENY2 coordinate activity of multiple H2B deubiquitinases important for cellular proliferation and tumor growth. Mol Cell 62:558–571. doi: 10.1016/j.molcel.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Florens L, Washburn MP. 2006. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol 328:159–175. [DOI] [PubMed] [Google Scholar]
- 19.Jani D, Lutz S, Hurt E, Laskey RA, Stewart M, Wickramasinghe VO. 2012. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res 40:4562–4573. doi: 10.1093/nar/gks059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. 2013. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 126:2656–2667. doi: 10.1242/jcs.118000. [DOI] [PubMed] [Google Scholar]
- 21.Köhler A, Pascual-Garcia P, Llopis A, Zapater M, Posas F, Hurt E, Rodriguez-Navarro S. 2006. The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11. Mol Biol Cell 17:4228–4236. doi: 10.1091/mbc.E06-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. 2011. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J Cancer Res Clin Oncol 137:1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown AM, Jeltsch JM, Roberts M, Chambon P. 1984. Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc Natl Acad Sci U S A 81:6344–6348. doi: 10.1073/pnas.81.20.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin CY, Strom A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, Vergara LA, Tang S, Chong A, Bajic VB, Miller LD, Gustafsson JA, Liu ET. 2004. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol 5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan JY, Vance KW, Varela MA, Sirey T, Watson LM, Curtis HJ, Marinello M, Alves S, Steinkraus BR, Cooper S, Nesterova T, Brockdorff N, Fulga TA, Brice A, Sittler A, Oliver PL, Wood MJ, Ponting CP, Marques AC. 2014. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat Struct Mol Biol 21:955–961. doi: 10.1038/nsmb.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ning Z, Wang A, Liang J, Xie Y, Liu J, Feng L, Yan Q, Wang Z. 2014. USP22 promotes the G1/S phase transition by upregulating FoxM1 expression via beta-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinoma. Int J Oncol 45:1594–1608. [DOI] [PubMed] [Google Scholar]
- 27.Liang J, Zhang X, Xie S, Zhou X, Shi Q, Hu J, Wang W, Qi W, Yu R. 2014. Ubiquitin-specific protease 22: a novel molecular biomarker in glioma prognosis and therapeutics. Med Oncol 31:899. doi: 10.1007/s12032-014-0899-2. [DOI] [PubMed] [Google Scholar]
- 28.Yang M, Liu YD, Wang YY, Liu TB, Ge TT, Lou G. 2014. Ubiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeutics. Tumour Biol 35:929–934. doi: 10.1007/s13277-013-1121-4. [DOI] [PubMed] [Google Scholar]
- 29.Piao S, Liu Y, Hu J, Guo F, Ma J, Sun Y, Zhang B. 2012. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS One 7:e42540. doi: 10.1371/journal.pone.0042540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang B, Liang X, Tang F, Zhang J, Zeng S, Jin S, Zhou L, Kudo Y, Qi G. 2015. Expression of USP22 and Survivin is an indicator of malignant behavior in hepatocellular carcinoma. Int J Oncol 47:2208–2216. [DOI] [PubMed] [Google Scholar]
- 31.He Y, Jin YJ, Zhang YH, Meng HX, Zhao BS, Jiang Y, Zhu JW, Liang GY, Kong D, Jin XM. 2015. Ubiquitin-specific peptidase 22 overexpression may promote cancer progression and poor prognosis in human gastric carcinoma. Transl Res 165:407–416. doi: 10.1016/j.trsl.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. 2008. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle 7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atanassov BS, Dent SY. 2011. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep 12:924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ao N, Liu Y, Feng H, Bian X, Li Z, Gu B, Zhao X. 2014. Ubiquitin-specific peptidase USP22 negatively regulates the STAT signaling pathway by deubiquitinating SIRT1. Cell Physiol Biochem 33:1863–1875. doi: 10.1159/000362964. [DOI] [PubMed] [Google Scholar]
- 36.Kishi S, Wulf G, Nakamura M, Lu KP. 2001. Telomeric protein Pin2/TRF1 induces mitotic entry and apoptosis in cells with short telomeres and is down-regulated in human breast tumors. Oncogene 20:1497–1508. doi: 10.1038/sj.onc.1204229. [DOI] [PubMed] [Google Scholar]
- 37.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T, Lorkiewicz P, St Clair D, Hung MC, Evers BM, Zhou BP. 2013. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell 23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan H, Su L, Chen WY. 2013. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther 6:1399–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Armakolas A, Stathopoulos GP, Nezos A, Theos A, Stathaki M, Koutsilieris M. 2012. Subdivision of molecularly-classified groups by new gene signatures in breast cancer patients. Oncol Rep 28:2255–2263. [DOI] [PubMed] [Google Scholar]