ABSTRACT
The omnivorous cockroach Periplaneta americana hosts a diverse hindgut microbiota encompassing hundreds of microbial species. In this study, we used 16S rRNA gene sequencing to examine the effect of diet on the composition of the P. americana hindgut microbial community. Results show that the hindgut microbiota of P. americana exhibit a highly stable core microbial community with low variance in compositions between individuals and minimal community change in response to dietary shifts. This core hindgut microbiome is shared between laboratory-hosted and wild-caught individuals, although wild-caught specimens exhibited a higher diversity of low-abundance microbes that were lost following extended cultivation under laboratory conditions. This taxonomic stability strongly contrasts with observations of the gut microbiota of mammals, which have been shown to be highly responsive to dietary change. A comparison of P. americana hindgut samples with human fecal samples indicated that the cockroach hindgut community exhibited higher alpha diversity but a substantially lower beta diversity than the human gut microbiome. This suggests that cockroaches have evolved unique mechanisms for establishing and maintaining a diverse and stable core microbiome.
IMPORTANCE The gut microbiome plays an important role in the overall health of its host. A healthy gut microbiota typically assists with defense against pathogens and the digestion and absorption of nutrients from food, while dysbiosis of the gut microbiota has been associated with reduced health. In this study, we examined the composition and stability of the gut microbiota from the omnivorous cockroach Periplaneta americana. We found that P. americana hosts a diverse core gut microbiome that remains stable after drastic long-term changes in diet. While other insects, notably ant and bee species, have evolved mechanisms for maintaining a stable association with specific gut microbiota, these insects typically host low-diversity gut microbiomes and consume specialized diets. In contrast, P. americana hosts a gut microbiota that is highly species rich and consumes a diverse solid diet, suggesting that cockroaches have evolved unique mechanisms for developing and maintaining a stable gut microbiota.
INTRODUCTION
Most insects host simple gut microbial communities, with only a few unique species represented; the reed beetle, honey bee, fruit fly, and gypsy moth all have fewer than 10 species of bacteria in their guts (1). The low complexity of these communities has been attributed to selective pressures dictated by host physiology (2) and the lack of extensive parental contact with offspring in many insects, which offers few opportunities for vertical and social transmission of gut microbes (1, 3). However, certain social and/or gregarious insect species, including cockroaches and their close relatives, the termites, host complex gut communities comprising hundreds of species (1, 4, 5).
The cockroach gut is composed of three compartments: the foregut, midgut, and hindgut. Of the three, the hindgut has the highest bacterial density and diversity (6). This hindgut microbial community breaks down recalcitrant dietary components from food that has passed through the fore- and midgut, supplying the cockroach with volatile fatty acids such as acetate (7). While this is not thought to be an obligate symbiosis, reducing the gut microbiota in Periplaneta americana slows development and results in lowered body weight and metabolic activity, suggesting that the gut microbiota plays an important role in the health and fitness of cockroaches (7–9). Recent work also suggested that the hindgut microbiota is responsible for producing pheromones, including volatile fatty acids, which promote social behavior among cockroaches (10).
While cockroach gut microbes are most closely related to microbes found in termites and other insects, they share many clades with those found in mammals, including humans (4, 11). Mammalian studies have found that diets can have strong impacts on the gut microbiome composition (12–14). As a result, we sought to determine the extent to which the response of the cockroach gut microbiota to dietary shifts resembles those identified in mammals.
Several studies were conducted that examined the effect of diet in various cockroach species (15–19). These studies found a variety of results, with Schauer et al. (18) reporting a highly stable core microbiome in Shelfordella lateralis, Bertino-Grimaldi et al. (17) reporting a small but significant response to dietary shifts in P. americana, and Pérez-Cobas et al. (19) reporting a strong response in Blattella germanica. However, these studies typically focused on responses to a limited range of substrates, particularly lignocellulosic materials, and all but that reported by Pérez-Cobas (19) (three replicate experiments per treatment) lack replication or characterization of the interindividual variability in microbiome composition. In this study, we utilized high-throughput 16S rRNA gene sequencing to characterize the hindgut microbiome of P. americana and its response to a wide range of dietary compositions, including high-fat, high-carbohydrate, and high-protein diets.
MATERIALS AND METHODS
Insects.
P. americana cockroaches were provided by the University of Georgia's entomology department from a colony that has been maintained in captivity for over 10 years. Cockroaches were maintained in mixed-age mixed-sex colonies in aquarium tanks at room temperature on a diet of dog food (Kroger nutritionally complete bite-sized adult dog food, composed of 21% protein, 8% fat, and 6% fiber) ad libitum. Each tank was provided with corn cob bedding, cardboard tubes for nesting, and a cellulose sponge saturated with water.
Adult cockroaches were selected, weighed, and marked for later identification. Initial 14-day experiments used 20 adult cockroaches (5 male, 15 female) per treatment. Later time-series experiments used either 43 (26 male, 17 female) or 20 (10 male, 10 female) adult cockroaches per treatment. Each dietary treatment group was housed in a single plastic tank that contained pebbles for bedding, polyvinyl chloride (PVC) tubes for nesting, and food and water in shallow plastic dishes. Food, water, and PVC tubes were changed daily, and any visible debris (or deceased cockroaches) was removed. Diet treatments included a diet of bran (Bob's Red Mill organic high fiber oat bran hot cereal), butter (Kroger unsalted butter sticks), filter paper (Whatman qualitative filter paper, grade 1), honey (Kroger pure clover grade A honey), tuna (StarKist Selects low-sodium chunk light tuna in water), white flour (King Arthur unbleached bread flour), whole-wheat flour (King Arthur 100% whole-grain whole-wheat flour), a mixed diet (calorie count of 25% tuna, 25% butter, 16.67% whole-wheat flour, 16.67% white flour, and 16.67% honey), and a starvation control (Table 1).
TABLE 1.
Nutrient information for 100-g servings of each diet treatmenta
| Diet treatmenta,b | Calories | Protein | Carbohydrate | Fat | Fiber |
|---|---|---|---|---|---|
| Bran (L) | 375 | 17.5 | 67.5 | 5 | 17.5 |
| Butter (S, L) | 714 | 0 | 0 | 79 | 0 |
| Filter paper (L) | Uc | U | U | U | U |
| Honey (S) | 286 | 0 | 81 | 0 | 0 |
| Tuna (S, L) | 107 | 27 | 0 | 1 | 0 |
| White flour (S, L) | 367 | 13 | 73 | 0 | 3 |
| Whole wheat flour (S) | 333 | 13 | 67 | 2 | 13 |
| Mixed (S) | 239 | 18 | 27 | 7 | 2 |
| Starvation (S, L) | NAd | NA | NA | NA | NA |
Nutritional facts are as stated by the manufacturers of each food product. The mixed diet was based on the general guidelines for a typical human diet. Thus, it was calorically composed of 25% tuna, 25% butter, 16.67% whole-wheat flour, 16.67% white flour, and 16.67% honey; the values shown assume a daily diet of 2,000 calories.
L, long-term dietary shift; S, short-term dietary shift.
U, unavailable.
NA, not applicable.
For studies of wild-caught cockroaches, insects were collected in traps placed outside on the University of Georgia's campus. The traps were glass jars containing glass wool saturated with beer as a lure and with petroleum jelly placed around the jar opening to prevent insects from escaping the jars after entering. Traps were checked daily, and any captured P. americana adults were either sacrificed immediately or placed in an aquarium tank under laboratory culture conditions (as described above) for 14 days before being sacrificed.
Sample collection and DNA extraction.
Hindgut samples were collected on day 14 of the short-term dietary shift and, as the treatment populations permitted, throughout the long-term dietary shift (see Data Sets S1A and S1B in the supplemental material). For comparisons with wild-caught cockroaches, hindgut sample collection occurred either within 24 h of collection or after 14 days under laboratory conditions (see Data Set S1C in the supplemental material). Cockroaches were removed from tanks, weighed, and placed on ice in sterile culture plates. After approximately 20 min, or when the cockroaches were sufficiently torpid, cockroaches were dissected and the entire gut was removed. Any visible debris, including fat bodies or exoskeleton, was removed with forceps. The hindgut was then separated from the rest of the gut using a scalpel and placed on Parafilm. The hindgut was submerged in 100 μl of RNAlater (Ambion, Austin, TX) and a pipette tip was used to break open the gut and disperse the contents into the RNAlater (Ambion). The suspended gut lumen was then removed from the hindgut wall and stored at −80°C.
DNA was extracted from an aliquot of the total preserved hindgut sample using a modified version of the EZNA Bacteria kit (Omega Bio-tek, Norcross, GA). Preserved frozen hindgut samples were thawed on ice. A 30-μl volume was removed for extraction while the rest was returned to storage at −80°C for future use. To each sample aliquot, 100 μl of balanced salt solution (2.5 g K2HPO4, 1 g KH2PO4, 1.6 g KCl, 1.4 g NaCl, and 10 ml of 1 M NaHCO3 per liter, pH 7.2) was added, and the sample was mixed and centrifuged for 10 min at 5,000 × g. After centrifugation, the supernatant was discarded and the pellet was resuspended in 100 μl TE buffer (10 nM Tris, 1 mM EDTA [pH 8]) and 10 μl lysozyme (as supplied by kit). The sample was incubated at 37°C for 30 min. Approximately 25 mg of glass beads (as supplied by kit) were added to the sample, which was bead beaten for 5 min at 3,000 rpm using a vortex mixer with a horizontal adaptor. To each sample, 100 μl BTL buffer and 20 μl proteinase K solution (as supplied by the kit) were added and the sample was incubated at 55°C while shaking at 600 rpm for 1 h. After this step, the manufacturer's protocol (June 2014 version) was followed beginning at step 11. Samples were eluted in 50 μl preheated elution buffer after a 5-min incubation at 65°C. The final DNA concentrations (typically between 5 to 50 ng/μl) and A260/A280 were measured using a NanoDrop Lite spectrophotometer (Thermo Scientific, Wilmington, DE).
Library preparation and sequencing.
The V4 region of the 16S rRNA gene from each gut sample was amplified in duplicate using a two-step PCR method on the basis of work by Caporaso et al. (20). The initial PCR used Q5 Hot Start high-fidelity DNA polymerase (New England BioLabs, Ipswich, MA) and 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT) primers in a 10-μl PCR mixture (1× Q5 reaction buffer, 200 μM deoxynucleoside triphosphates [dNTPs], 0.5 μM 515F, 0.5 μM 806R, 2 ng DNA, and 0.02 U/μl Q5 polymerase) under the following conditions: 98°C for 30 s, followed by 15 cycles at 98°C for 10 s, 52°C for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 2 min for the initial V4 region amplification.
Immediately following the initial amplification, the resulting product was reamplified using primers (see Table S1 in the supplemental material) that contained double Hamming barcodes (21). This two-step PCR scheme was used for ensuring high quality amplicons, as the initial replication occurred before the addition of Illumina-specific adaptors or sample-specific barcodes. The secondary amplification mixture contained 1× Q5 reaction buffer, 200 μM dNTPs, 0.5 μM 515F, 0.5 μM 806R, 2 ng DNA, and 0.02 U/μl Q5 polymerase. From this mixture, 21 μl was added to 9 μl of the initial reaction product. These reactions were then cycled under the following conditions: 98°C for 30 s, followed by 4 cycles at 98°C for 10 s, 52°C for 10 s, and 72°C for 30 s, followed by 6 cycles at 98°C for 10 s and 72°C for 1 min, concluding with a final extension at 72°C for 2 min.
Two independent PCRs with unique barcode combinations were generated for each sample. These technical replicates were pooled and purified using the EZ Cycle Pure kit (Omega Bio-tek) according to the manufacturer's protocol. Samples were eluted in 30 μl of elution buffer. Purified amplicons were quantified using a NanoDrop Lite spectrophotometer (Thermo Scientific). Amplicons from 12 guts obtained from cockroaches treated with the short-term dietary shift, all available guts excluding day 30 from cockroaches treated with the long-term dietary shift, and all available guts from the wild-caught cockroaches were normalized and pooled to a concentration of 10 nM on the basis of a predicted total product size of ∼400 bp. The quality of the prepared library was assessed using the Agilent 2100 Bioanalyzer DNA-HS assay (Agilent Technologies, Santa Clara, CA) before submission to the Georgia Genomics Facility for sequencing (Illumina MiSeq 250 × 250 bp; Illumina, Inc., San Diego, CA).
American Gut Project (AGP) data retrieval.
The American Gut Project (AGP) is a collaborative effort for characterizing the human gut microbiome through crowdsourcing fecal samples from the public for 16S rRNA gene analysis (22, 23). We used data from the AGP as a human comparison for our cockroach data, as the AGP uses the same 16S rRNA gene primers (515F/806R) and sequencing technology (Illumina MiSeq) that we used in our experiments (22). A file containing all demultiplexed full-length debloomed sequences from the AGP was downloaded (April 2015 version). From this file, a subset of 157 samples was randomly selected from individuals who provided their sex and were between 20 and 60 years of age. This subset of samples was analyzed using the method described below.
Data analysis.
The mothur software package was used for analyzing the sequences generated in this experiment (24). The MiSeq standard operating protocol was followed (25, 26) with the following modifications: after sequences were assembled, sequences that had any ambiguous bases or were longer than 275 bp were removed; sequences that passed this initial screening process were aligned to the Silva reference database (Release 123) (27–29); aligned sequences were again screened to remove sequences that contained homopolymers of 8 or more base pairs; UCHIME was used for identifying chimeras from the remaining sequences (30); after chimera removal, the Wang method was used for taxonomic classification of samples with the greengenes reference database (August 2013 version) (31–33); sequences that were unclassifiable or identified as chloroplasts, mitochondria, Eukaryota, or Blattabacterium (a cockroach endosymbiont found in fat body cells) were removed. The remaining sequences were clustered into operational taxonomic units (OTUs) on the basis of 97% or greater sequence identity.
To make an accurate comparison between data generated from this experiment and data provided by the AGP, sequences generated from this experiment were trimmed to match the length of samples provided by the AGP. All sequences were then analyzed using the same pipeline as described above. Figures containing only the unique data generated in this experiment used the original data set; figures containing comparisons to the AGP data used the trimmed data set.
Accession number(s).
The sequences generated from this experiment were submitted to the NCBI Sequence Read Archive and are available under the accession numbers SRP075213, SRP075102, and SRP075057.
RESULTS
Effect of diet on hindgut microbial community.
Laboratory-raised adult cockroaches were maintained for 14 days on a variety of diets, including tuna, butter, honey, whole-wheat flour, white flour, a mixture of the above, and a starvation diet. Over the course of the experiment, only the butter and starvation treatments were found to have significant effects on weight (paired t test, P < 0.001 and P < 0.05, respectively). After 14 days on each diet, cockroaches were sacrificed and hindgut lumen contents were used for microbial DNA extraction and 16S rRNA gene amplicon sequencing. A total of 28,742,658 16S rRNA gene sequences were obtained from 99 unique samples, of which 15,754,172 passed quality checks, resulting in an average of 1,294 OTUs per sample (see Data Set S1A in the supplemental material).
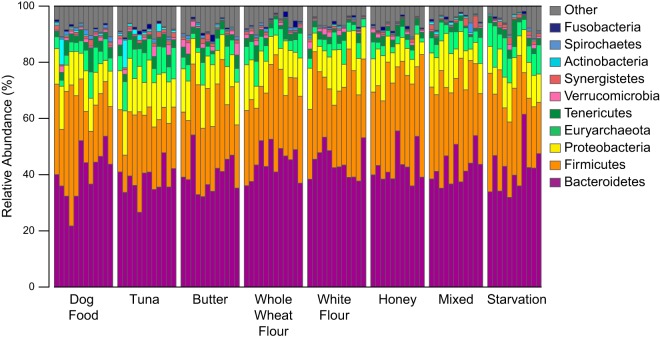
Bacteroidetes, Firmicutes, and Proteobacteria were the predominant phyla present in the gut microbiota of cockroaches receiving all treatments (Fig. 1; see also Fig. S1 in the supplemental material). Within the Bacteroidetes phylum, bacteria from the Porphyromonadaceae, Rikenellaceae, and Bacteroidaceae families were especially prevalent, accounting for over 40% of the total bacteria found in several cockroaches. Clostridia represented the majority of Firmicutes in the cockroach gut, though there were other classes present, such as Erysipelotrichia and Bacilli. In the Proteobacteria phyla, Desulfobacteraceae and Enterobacteriaceae were the major families represented. The predominant archaeal taxon was Methanomicrococcus blatticola, a methanogen associated most commonly with cockroaches (34). These results agree well with those from previously published studies of cockroaches (17–19). Overall, most identified microbes were typical of those found in the guts of omnivores, including the human gut (13, 35). However, many of the microbes found were unclassifiable above the class or family level, suggesting that they may belong to poorly characterized, insect-specific lineages.
FIG 1.
Relative abundances of microbial phyla across 14-day diet treatments (Table 1). Each bar represents an individual cockroach gut. The 10 most abundant phyla are shown.
Dietary shifts did not result in large changes in gut microbial community composition. No large differences in the relative abundances of major bacterial phyla or families were observed among dietary treatments (Fig. 1; see also Fig. S1 in the supplemental material). This is in contrast to results found in mammals, where dietary shifts have been found to change the ratio of Bacteroidetes to Firmicutes and the proportions of other members of the microbial community (13, 14). This stability in gut microbiome composition was apparent at all taxonomic resolutions. An ordination analysis did not identify a strong impact of diet on the microbial community composition at the 97% OTU level (see Fig. S2 in the supplemental material). Neither nonmetric multidimensional scaling nor principal component-based analyses detected clear separation between diet treatments, suggesting that diet does not have a strong impact on the microbial community composition. Permutational multivariate analysis of variance (PERMANOVA) found a significant effect for diet on community composition (P = 0.001). However, the biological significance of this difference is unclear, as the effect size was small (R2 = 0.21 overall, average R2 for 100 random permutations of data labels = 0.08). Similarly, pairwise comparisons of results from individual diets with those from dog food controls identified small (R2 = 0.11 to 0.23) but significant (P = 0.001 to 0.004) shifts in community composition (see Table S2 in the supplemental material). In addition, we did not observe large shifts in alpha or beta diversity following the treatments (see Fig. S3 in the supplemental material).
The initial short-term dietary perturbation was followed up with an extended time series. This long-term dietary shift included the two additional dietary treatments of bran and filter paper (Table 1) as well as more frequent sampling on days 1, 2, 3, 7, 14, 30, 60, and 90 (see Data Set S1B in the supplemental material). These experiments also showed minimal dietary effects on gut microbiota composition (see Fig. S4 through S7 in the supplemental material).
Individual-to-individual variation.
An initial hypothesis was that diet-driven changes in gut microbiome composition might have been obscured by high individual-to-individual variation. To test this, we compared the relative level of individual-to-individual variation observed for P. americana to that found in other animals with complex gut communities. For this comparison, we used 16S rRNA gene sequences from 157 randomly chosen human fecal samples obtained from the American Gut Project (AGP) (22). This data set was chosen because it represents an extensive examination of individual-to-individual variation in gut microbiome composition in an animal that shares many traits (an omnivorous diet and an anoxic, circumneutral hindgut lumen that is extensively colonized by microbes) with cockroaches. One potential caveat is that the degree to which fecal samples accurately reflect the microbial community composition of the gut lumen is poorly constrained. However, we feel that this comparison places our observations of cockroach gut microbial diversity in context. To minimize artifacts resulting from differences in the sequencing technologies used, we trimmed our cockroach data to match the read length for the human data and jointly reprocessed the combined human and cockroach data sets as described in Materials and Methods. After quality control measures, a total of 2,768,251 16S rRNA gene sequences remained from 138 unique human fecal samples, with an average richness of 1,075 OTUs per human fecal sample. The reprocessed cockroach data comprised 15,899,340 16S rRNA gene sequences with an average of 1,713 OTUs per sample.
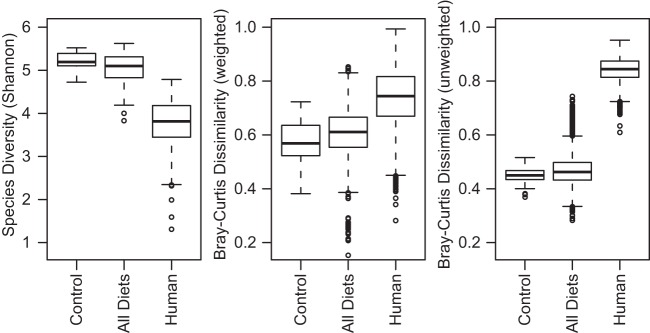
The comparison of P. americana hindgut community composition with the community composition of human fecal samples revealed that the cockroach gut community consistently exhibited higher alpha diversity at the 97% OTU level than did the human gut microbiota (Fig. 2). In contrast, comparisons of P. americana composition identified much lower beta diversity than that observed among human samples (Fig. 2). Similar trends were observed for comparisons of our data to data sets from studies of humans and humanized mice (36–38) (data not shown). Pairwise comparisons of individual cockroach gut samples found significantly lower average Bray-Curtis dissimilarities than a similar comparison with human fecal samples for abundance-weighted and unweighted measures (Fig. 2). This suggests that the cockroach population has less individual-to-individual variation than does the human population. Moreover, the lower unweighted (presence/absence-based) dissimilarity suggests that the cockroach population has a richer and more extensive core gut microbiota than does the human population (Fig. 2). A shared core community of 201 OTUs (see Table S3 in the supplemental material) was identified across all dietary treatment groups, averaging 67% of the sequences recovered from cockroaches from all dietary treatment groups. In contrast, only 5 OTUs were shared among all 138 human samples (see Table S4 in the supplemental material), accounting for an average of 31% of the sequences recovered from human fecal samples.
FIG 2.
Alpha and beta diversities among cockroach gut and human fecal samples. Boxplots show Shannon diversity indices (left) and weighted (middle) and unweighted (right) Bray-Curtis dissimilarities among the laboratory cockroaches raised on a dog food diet, all cockroach treatment groups, and human gut microbial communities at the OTU level (97% sequence identity). Human data were obtained from the American Gut Project (22). Cockroach data were trimmed to the same lengths and alignment positions as those from the human gut data prior to OTU calling, and all libraries were resampled to a depth of 4,000 sequences. For each group, the bars delineate the means, the hinges represent the lower and upper quartiles, the whiskers extend to the most extreme values (which are no more than 1.5 times the interquartile range from the box), and outliers are plotted, if present.
Comparison between gut microbiota of wild-caught and laboratory-raised insects.
The dietary perturbations resulted in a laboratory-raised P. americana host gut microbiota with very low individual-to-individual variability relative to that in human fecal samples. A comparison between the laboratory-raised and wild-caught P. americana microbiota was conducted to verify that this low diversity was a common property in this species and not an artifact of laboratory culture conditions. To do so, we examined the gut microbiota from freshly captured P. americana individuals immediately upon capture and following 14 days of culture under laboratory conditions.
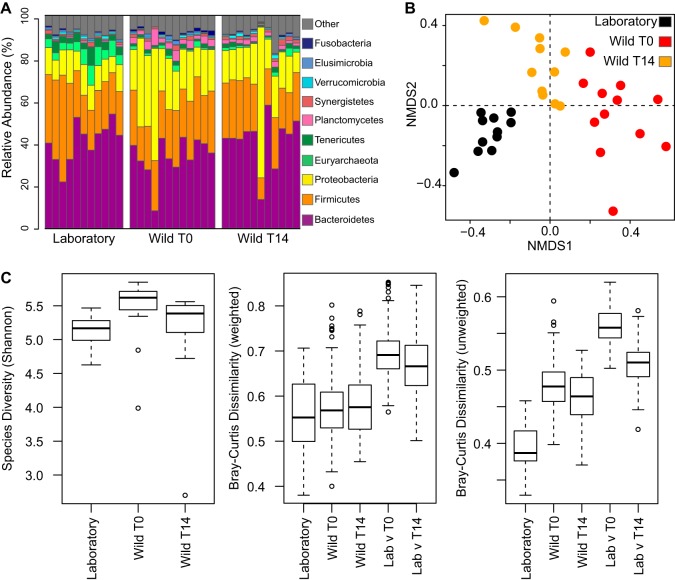
At the phylum level, the gut microbiota from wild-caught P. americana is similar to that from the laboratory cockroach population. Bacteroidetes, Firmicutes, and Proteobacteria were the predominant phyla present in the gut microbiota from all treatment groups (Fig. 3A). Wild-caught individuals exhibited a higher abundance of Proteobacteria and a relatively lower abundance of Bacteroidetes and Firmicutes (t test, P < 10−5 for the two time points), which became more similar in abundance following 14 days of cultivation under laboratory conditions.
FIG 3.
Comparison of laboratory-raised and wild-caught cockroach gut microbiota. (A) Relative abundances of the 10 most abundant phyla identified among laboratory-raised and wild-caught cockroach gut samples immediately following capture (T0) or after 14 days of culture under laboratory conditions (T14). Each bar represents an individual cockroach gut. (B) Nonmetric multidimensional scaling (NMDS) plot of laboratory-raised and wild-caught cockroaches. PERMANOVA based on dissimilarities was also conducted (R2 = 0.242; P = 0.001). (C) Boxplots comparing Shannon diversity (left) and weighted (middle) and unweighted (right) compositional dissimilarities among the three groups at the OTU level (97% sequence identity). For analyses presented in panels B and C, libraries were sampled to a constant depth of 4,000 sequences. For each group, the bars delineate the means, the hinges represent the lower and upper quartiles, the whiskers extend to the most extreme values (which are no more than 1.5 times the interquartile range from the box), and outliers are plotted, if present.
At the 97% identity OTU level, laboratory-raised and wild-caught populations were clustered independently by ordination analysis, with wild-caught cockroaches becoming more similar to the laboratory population following 14 days of housing under laboratory conditions (Fig. 3B). The microbiota of wild-caught cockroaches exhibited higher alpha diversity (Fig. 3C) and had increased individual-to-individual variation by unweighted Bray-Curtis dissimilarity metrics (Fig. 3C). However, the microbiota of wild-caught and laboratory-raised cockroaches had similar levels of individual-to-individual variation by abundance-weighted Bray-Curtis metrics (Fig. 3C), suggesting that much of the difference in beta diversity can be attributed to a greater representation of low-abundance, transiently hosted microbes in the guts of wild-caught cockroaches. This may have resulted from environmental exposure to a higher diversity of microbes. Consistent with this hypothesis, alpha diversity in the guts of wild-caught cockroaches decreased following 14 days of cultivation under laboratory conditions.
Direct comparisons between microbiota of laboratory-raised and wild-caught cockroaches identified significantly greater between-group than within-group dissimilarities (Fig. 3C). This suggests that there are differences in the specific microbial OTUs hosted by these two populations. However, these between-group dissimilarities are lower than those observed between individual human fecal samples, suggesting that these gut populations maintain a large degree of overlap after a decade of laboratory cultivation. Consistent with this, the three treatment groups, which had an average of 1,575 OTUs per sample, shared 199 microbial OTUs (see Table S5 in the supplemental material) that made up an average of 47% of the sequences in gut communities recovered from the initial wild-caught cockroaches, 55% from the wild-caught cockroaches after 14 days under laboratory conditions, and 54% from the laboratory-raised cockroaches. Interestingly, while alpha diversity within wild-caught populations decreased following 14 days in the laboratory, the level of dissimilarity between laboratory and wild-caught cockroach populations did not decrease substantially. This suggests that the core gut microbiome of wild-caught cockroaches was not replaced with laboratory-associated species during that time period.
DISCUSSION
Diet has a strong role in shaping the structure and function of the mammalian gut microbiome (12–14, 39). Our goal was to determine to what extent the microbiome in the omnivorous insect P. americana exhibits similar trends. Our results show that adult P. americana has a rich, extensive core gut microbial community with minimal variation between individuals. The cockroach core gut community (see Tables S3 and S4 in the supplemental material) is composed primarily of bacteria in the Bacteroidetes and Firmicutes phyla, although members of the Euryarchaeota, Actinobacteria, Proteobacteria, Synergistetes, Tenericutes, and Verrucomicrobia phyla are present along with multiple unclassified bacteria. This core was present in laboratory-raised and wild-caught cockroaches. These results contrast strongly with observations from human fecal samples, which exhibit substantial individual-to-individual variation and few, if any, shared microbial OTUs.
P. americana's stable extensive core microbial community appears to be a unique characteristic of the cockroach and is highly resilient to changes in host diet. Our results are in agreement with those from a study of the cockroach S. lateralis that found no observable differences among the gut microbiota of cockroaches fed a low- or high-fiber diet (11). Similar work in P. americana and the related cockroach species B. germanica identified significant changes in their gut communities in response to diet (17, 19). However, both studies used alternate sequencing technologies that resulted in smaller numbers of sequences (216 and 48,527, respectively) and examined fewer treatments (three and two treatments, respectively) (17, 19).
In mammals, different microbial groups are believed to specialize in the utilization of specific dietary substrates, in part because they tend to increase in abundance when these substrates are enriched in the host's diet. For example, Bacteroidetes are associated with high-protein diets, while Firmicutes are associated with high-fiber diets (35). This hypothesis is on the basis of two assumptions, (i) that not all gut microbes utilize all substrates equally well and that microbial abundance in the gut is dependent on their ability to obtain substrates for growth, and (ii) that a change in dietary composition translates into a change in substrate availability within the gut. The absence of diet-driven changes in the composition of the cockroach gut microbiome suggests that one of these assumptions is not true. One possibility is that cockroach-associated gut microbes are substantially more metabolically versatile than those in mammalian-associated species, and they can therefore survive equally well when presented with a wide range of dietary compositions. Similarly, the ability to utilize the dietary substrates tested may be widely distributed across cockroach gut microbial lineages, such that changes in substrate availability drive “hidden” changes in the microbial representation at a sub-OTU resolution. A final possibility is that cockroach gut microbes obtain growth substrates through an alternative pathway, such as metabolic cross-feeding between gut microbes or the provision of key substrates by the host. Future investigations of the metabolic capabilities of cockroach gut microbes should provide further insight into these questions.
Cockroaches are among the most diverse and abundant members of the animal kingdom and survive in a wide variety of habitats, from the tropical rainforest and mountainous caves to urban environments (40, 41). The American cockroach, P. americana, can be found throughout the world; however, it is best known as a common household pest that thrives in warm and moist environments, such as steam tunnels or boiler rooms (6, 42). Maturing to adulthood in as few as 6 months and living for up to 2 years, adult P. americana cockroaches are opportunistic feeders that can survive on a wide variety of food sources (40, 43) and frequently subsist on no or limited food for days at a time (5). Thus, a stable resident gut community provides a remarkable evolutionary advantage.
Insects have evolved diverse mechanisms for the maintenance of stable host-symbiont relationships with their gut microbiota. Heteropteran stinkbugs have developed highly species-specific associations with individual gut symbionts that are either maternally transmitted or acquired early in development (44–46). Other insects have established stable relationships with simple gut communities, including honey and bumble bees (47, 48) and ants (49). While the mechanisms by which bees regulate their gut microbiome have not been established, the Sonoran Desert turtle ant, Cephalotes rohweri, was recently found to have a mechanical filter that blocks any bacteria or particles larger than 0.2 μm from entering into the midgut and hindgut after an initial gut microbiome is established (50). However, stable host/gut symbiont associations have been found primarily in insects with specialized diets and low-diversity gut microbiota. Thus, it is unlikely that the same mechanisms are at work in P. americana, which consumes a wide-ranging, omnivorous diet and hosts a highly diverse gut microbiome that compositionally resembles that of mammalian omnivores (11).
Termites are known to have a symbiotic relationship with their gut microbial community, which, like the cockroach gut microbiota, is extensive and diverse (1, 51). The termite's more restricted herbivorous diet and social behavior are currently thought to be the key drivers that shape the development of their specialized gut microbiota (51). However, given that molecular analyses suggest that termites fall within the cockroach radiation (52), these results suggest an alternative hypothesis in which the ability to maintain a stable gut microbiome evolved prior to, and perhaps facilitated, the evolutionary shift to a lignocellulosic diet. Further work should provide insight into the mechanisms underlying this stability and its role in shaping cockroach (and termite) evolution and ecology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brian Forschler for providing the cockroaches used in this work, Vickie Trinh for her assistance in capturing and dissecting wild P. americana cockroaches, and Rob Knight and Daniel McDonald for providing access to data from the American Gut Project.
This work was supported by funds provided by the University of Georgia.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01837-16.
REFERENCES
- 1.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 2.Müller U, Vogel P, Alber G, Schaub GA. 2008. The innate immune system of mammals and insects. Contrib Microbiol 15:21–44. [DOI] [PubMed] [Google Scholar]
- 3.Dillon RJ, Dillon VM. 2004. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol 49:71–92. doi: 10.1146/annurev.ento.49.061802.123416. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich C, Kohler T, Brune A. 2014. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol 80:2261–2269. doi: 10.1128/AEM.04206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell WJ, Roth LM, Nalepa CA. 2007. Diets and Foraging, p 61–75. In Cockroaches: ecology, behavior, and natural history. The Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 6.Cruden DL, Markovetz AJ. 1987. Microbial ecology of the cockroach gut. Annu Rev Microbiol 41:617–643. doi: 10.1146/annurev.mi.41.100187.003153. [DOI] [PubMed] [Google Scholar]
- 7.Cruden DL, Markovetz AJ. 1984. Microbial aspects of the cockroach hindgut. Arch Microbiol 138:131–139. doi: 10.1007/BF00413013. [DOI] [PubMed] [Google Scholar]
- 8.Gijzen HJ, Barugahare M. 1992. Contribution of anaerobic protozoa and methanogens to hindgut metabolic activities of the American cockroach, Periplanta americana. Appl Environ Microbiol 58:2565–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bracke JW, Cruden DL, Markovetz AJ. 1978. Effect of metronidazole on the intestinal microflora of the American cockroach, Periplaneta americana l. Antimicrob Agents Chemother 13:115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wada-Katsumata A, Zurek L, Nalyanya G, Roelofs WL, Zhang A, Schal C. 2015. Gut bacteria mediate aggregation in the German cockroach. Proc Natl Acad Sci U S A 112:15678–15683. doi: 10.1073/pnas.1504031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schauer C, Thompson CL, Brune A. 2012. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol 78:2758–2767. doi: 10.1128/AEM.07788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Knight R. 2015. Dietary effects on human gut microbiome diversity. Br J Nutr 113(Suppl):S1–S5. doi: 10.1017/S0007114514004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane MD, Breznak JA. 1991. Effect of host diet on production of organic acids and methane by cockroach gut bacteria. Appl Environ Microbiol 57:2628–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gijzen HJ, van der Drift C, Barugahare M, Op den Camp HJ. 1994. Effect of host diet and hindgut microbial composition on cellulolytic activity in the hindgut of the American cockroach, Periplaneta americana. Appl Environ Microbiol 60:1822–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertino-Grimaldi D, Medeiros MN, Vieira RP, Cardoso AM, Turque AS, Silveira CB, Albano RM, Bressan-Nascimento S, Garcia ES, de Souza W, Martins OB, Machado EA. 2013. Bacterial community composition shifts in the gut of Periplaneta americana fed on different lignocellulosic materials. Springerplus 2:609. doi: 10.1186/2193-1801-2-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schauer C, Thompson C, Brune A. 2014. Pyrotag sequencing of the gut microbiota of the cockroach Shelfordella lateralis reveals a highly dynamic core but only limited effects of diet on community structure. PLoS One 9:e85861. doi: 10.1371/journal.pone.0085861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Cobas AE, Maiques E, Angelova A, Carrasco P, Moya A, Latorre A. 2015. Diet shapes the gut microbiota of the omnivorous cockroach Blattella germanica. FEMS Microbiol Ecol 91:pii:fiv022. doi: 10.1093/femsec/fiv022. [DOI] [PubMed] [Google Scholar]
- 20.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2010. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108(Suppl 1):S4516–S4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.American Gut Project. 2015. The American Gut. http://americangut.org/ Accessed December 2015.
- 23.McDonald D, Birmingham A, Knight R. 2015. Context and the human microbiome. Microbiome 3:52. doi: 10.1186/s40168-015-0117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL. 2014. MiSeq SOP. http://www.mothur.org/wiki/MiSeq_SOP Accessed March 2014.
- 27.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO. 2014. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprenger WW, Hackstein JH, Keltjens JT. 2007. The competitive success of Methanomicrococcus blatticola, a dominant methylotrophic methanogen in the cockroach hindgut, is supported by high substrate affinities and favorable thermodynamics. FEMS Microbiol Ecol 60:266–275. doi: 10.1111/j.1574-6941.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- 35.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. 2015. Role of the normal gut microbiota. World J Gastroenterol 21:8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. 2013. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schal C, Gautier JY, Bell WJ. 1984. Behavioural ecology of cockroaches. Biol Rev Camb Philos Soc 59:209–254. doi: 10.1111/j.1469-185X.1984.tb00408.x. [DOI] [Google Scholar]
- 41.Bell WJ, Roth LM, Nalepa CA. 2007. Habitats, p 37-60 In Cockroaches: ecology, behavior, and natural history. The Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- 42.Tee H-S, Saad AR, Lee C-Y. 2011. Population ecology and movement of the American cockroach (Dictyoptera: Blattidae) in sewers. J Med Entomol 48:797–805. doi: 10.1603/ME10255. [DOI] [PubMed] [Google Scholar]
- 43.Wharton DR, Lola JE, Wharton ML. 1967. Population density, survival, growth, and development of the American cockroach. J Insect Physiol 13:699–716. [DOI] [PubMed] [Google Scholar]
- 44.Fukatsu T, Hosokawa T. 2002. Capsule-transmitted gut symbiotic bacterium of the Japanese common plataspid stinkbug, Megacopta punctatissima. Appl Environ Microbiol 68:389–396. doi: 10.1128/AEM.68.1.389-396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. Specific developmental window for establishment of an insect-microbe gut symbiosis. Appl Environ Microbiol 77:4075–4081. doi: 10.1128/AEM.00358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohbayashi T, Takeshita K, Kitagawa W, Nikoh N, Koga R, Meng X, Tago K, Hori T, Hayatsu M, Asano K, Koamagata Y, Lee BL, Fukatsu T, Kikuchi Y. 2015. Insect's intestinal organ for symbiont sorting. Proc Natl Acad Sci U S A 112:E5179–E5188. doi: 10.1073/pnas.1511454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwong WK, Moran NA. 2015. Evolution of host specialization in gut microbes: the bee gut as a model. Gut Microbes 6:214–220. doi: 10.1080/19490976.2015.1047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwong WK, Engel P, Koch H, Moran NA. 2014. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 111:11509–11514. doi: 10.1073/pnas.1405838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kautz S, Rubin BE, Russell JA, Moreau CS. 2013. Surveying the microbiome of ants: comparing 454 pyrosequencing with traditional methods to uncover bacterial diversity. Appl Environ Microbiol 79:525–534. doi: 10.1128/AEM.03107-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanan MC, Rodrigues PA, Agellon A, Jansma P, Wheeler DE. 2016. A bacterial filter protects and structures the gut microbiome of an insect. ISME J 10:1866–1876. doi: 10.1038/ismej.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brune A, Dietrich C. 2015. The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu Rev Microbiol 69:145–166. doi: 10.1146/annurev-micro-092412-155715. [DOI] [PubMed] [Google Scholar]
- 52.Inward D, Beccaloni G, Eggleton P. 2007. Death of an order: a comprehensive molecular phylogenetic study confirms that termites are eusocial cockroaches. Biol Lett 3:331–335. doi: 10.1098/rsbl.2007.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.