ABSTRACT
Besides being part of anti-Helicobacter pylori treatment regimens, proton pump inhibitors (PPIs) are increasingly being used to treat dyspepsia. However, little is known about the effects of PPIs on the human gastric microbiota, especially those related to H. pylori infection. The goal of this study was to characterize the stomach microbial communities in patients with dyspepsia and to investigate their relationships with PPI use and H. pylori status. Using 16S rRNA gene pyrosequencing, we analyzed the mucosa-associated microbial populations of 24 patients, of whom 12 were treated with the PPI omeprazole and 9 (5 treated and 4 untreated) were positive for H. pylori infection. The Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria phyla accounted for 98% of all of the sequences, with Helicobacter, Streptococcus, and Prevotella ranking among the 10 most abundant genera. H. pylori infection or PPI treatment did not significantly influence gastric microbial species composition in dyspeptic patients. Principal-coordinate analysis of weighted UniFrac distances in these communities revealed clear but significant separation according to H. pylori status only. However, in PPI-treated patients, Firmicutes, particularly Streptococcaceae, were significantly increased in relative abundance compared to those in untreated patients. Consistently, Streptococcus was also found to significantly increase in relation to PPI treatment, and this increase seemed to occur independently of H. pylori infection. Our results suggest that Streptococcus may be a key indicator of PPI-induced gastric microbial composition changes in dyspeptic patients. Whether the gastric microbiota alteration contributes to dyspepsia needs further investigation.
IMPORTANCE Although PPIs have become a popular treatment choice, a growing number of dyspeptic patients may be treated unnecessarily. We found that patients treated with omeprazole showed gastric microbial communities that were different from those of untreated patients. These differences regarded the abundances of specific taxa. By understanding the relationships between PPIs and members of the gastric microbiota, it will be possible to envisage new strategies for better managing patients with dyspepsia.
INTRODUCTION
Specifically designed to shut down the H+/K+-ATPase of gastric parietal cells, through the blockage of acid transport (1), proton pump inhibitors (PPIs) are increasingly being used to treat gastroesophageal reflux disease (GERD) and other acid-related gastroduodenal disorders (2). Additionally, PPIs are included in standard 1-week triple or sequential therapies which are currently recommended to eradicate Helicobacter pylori from the human stomach (3).
While it is still unclear whether the presence of H. pylori influences the composition of the gastric microbial community, PPI administration is thought to alter gastric microbiota toward a more carcinogenic microbiota (i.e., dominated by bacteria that predispose to inflammation and cancer) (4), suggesting that H. pylori may be just a marker of these alterations (5, 6). It was also hypothesized that PPIs may affect the microbiota directly by targeting P-type ATPase enzymes of naturally occurring bacteria like H. pylori and Streptococcus pneumoniae (7) or indirectly by reducing the acidity of the gastric environment, which in turn leads to gastric bacterial overgrowth (8). Indeed, whatever the mechanisms by which PPIs affect microbes (9), gastric acid suppression proved to substantially increase the number of cultivable non-H. pylori bacteria in either the gastric mucosa or the stomach lumen; notably, this effect was largely influenced by the infection with H. pylori and the duration of acid suppression, which occurred through both histamine2-receptor antagonists (H2RA) and PPIs (10). By means of nonculturing methods (i.e., quantitative PCR and 16S rRNA gene pyrosequencing), a recent study by Tsuda et al. revealed very similar bacterial numbers in the gastric fluid microbiota between PPI users and PPI nonusers (11). However, the PPI administration induced a small but significant increase in the intersubject diversity (11), which was consistent with previous findings by Amir et al. showing an increase in the beta diversity of the gastric fluid microbiota of subjects after 8 weeks of PPI treatment (12). Furthermore, H. pylori was found to be a minor bacterium in the gastric luminal samples in the Tsuda and coworkers' study (11), whereas, as expected, the organism was identified as a dominant bacterium in gastric mucosal samples from H. pylori-infected patients (13).
Therefore, while the relationship between PPIs and gastric mucosa microbiota remains not fully understood, it is also plausible that PPIs affect the microbiota structure through the H. pylori interaction. So, understanding the gastric microbiota-H. pylori-PPI axis might be important in view of the clinical repercussions that would arise from this, for example, by clarifying the medical usefulness of PPI treatments (14). Dyspepsia is a highly prevalent condition characterized by symptoms (e.g., heartburn) originating from the gastroduodenal region (15). In western populations, 25% report having heartburn at least once a month and 5% describe daily symptoms, explaining the large demand for PPIs (16). However, especially in patients with functional dyspepsia, which is defined as the presence of symptoms with no evidence of pathologically based gastroduodenal disorders (i.e., GERD, hiatal hernia, peptic ulcer) (17), the efficacy of PPIs appears to be limited (18, 19). Nevertheless, there is growing perception of indiscriminate use of PPIs worldwide (20), which would arise from either medical hyperprescription (21, 22) or self-diagnosis and treatment due to the over-the-counter availability of these medications (23).
In this study, we investigated the effects of PPIs on the gastric microflora by profiling the mucosa-associated bacterial communities of patients with dyspepsia. Biopsy samples from 24 patients (12 on and 12 off PPIs) were thus analyzed using 16S rRNA gene pyrosequencing in order to (i) provide a comprehensive survey of dyspeptic stomach microbiotas and (ii) clarify the association between PPI administration, H. pylori infection, and gastric community diversity.
MATERIALS AND METHODS
Experimental samples and DNA isolation.
Gastric mucosal biopsy specimens were collected, in strict compliance with the Università Cattolica del Sacro Cuore (UCSC) Ethics Committee requirements (UCSC EC no. 4138/15), from adult patients who underwent upper gastrointestinal endoscopy for symptoms of dyspepsia (i.e., heartburn, nausea, epigastric pain and discomfort, bloating, and regurgitation) at the Department of Gastroenterology and Hepatology of the Agostino Gemelli Hospital (UCSC) in Rome, Italy. Informed consent was obtained from all patients included in the study. Twelve patients were currently on PPI therapy (i.e., omeprazole, 40 mg/day) and 12 were either PPI naive (n = 10) or had discontinued PPI therapy at least 6 months before sample collection (n = 2). All patients on PPIs were treated for at least 12 months prior to endoscopy (Table 1). Patients were not enrolled if they were taking PPIs for fewer than 12 months or were taking antibiotics in the past 3 months prior to endoscopy or if they were having or had a history of peptic ulcer disease, previous gastric surgery, or chronic use of nonsteroidal anti-inflammatory drugs. Patients who were on or had been treated with any other acid-reducing drugs like H2RA (e.g., ranitidine) or antacids (e.g., alginate rafts) were also excluded. After enrollment, patients were determined to be positive for H. pylori if both histology and rapid urease tests provided a positive result (24) and physical and clinical examinations did not reveal comorbidities, and all the patients also reported normal dietary habits. Details about demographic and clinical characteristics of the 24 patients are shown in Table S1 in the supplemental material.
TABLE 1.
Characteristics of the patient groups studieda
| Characteristic | PPI-treated (n = 12) | Untreated (n = 12) |
|---|---|---|
| Age (mean ± SD) (yr) | 56 ± 16.6 | 44.7 ± 9.6 |
| Gender (men/women) | 3/9 | 4/8 |
| Body mass index (mean ± SD) (kg/m2) | 23.0 ± 1.5 | 22.0 ± 1.8 |
| Helicobacter pylori status (positive/negative) | 4/8 | 5/7 |
| Gastric mucosa inflammation degree (mild/moderate) | 4/8 | 6/5 |
| PPI treatment before upper endoscopy (mean ± SD) (mo) | 16 ± 3 |
None of the comparisons were significantly different (P > 0.05, for all comparisons). Only one of the patients on proton-pump inhibitor (PPI) therapy had histological findings showing severe gastric inflammation.
Patients followed a 12-h fasting period before mucosal biopsy specimens were recovered from the stomach antrum using a Pentax EG2990i gastroscope (Pentax, Tokyo, Japan) and single-use, sterile biopsy forceps. All biopsy specimens were repeatedly washed in sterile phosphate-buffered saline (PBS) immediately after their recovery to ensure that the microbiota from the biofilm associated with the gastric mucosa (i.e., those bacteria attached to the upper mucous layer or the juxtaepithelial mucus) could be analyzed and/or to significantly decrease the number of bacteria from the luminal fluid (25). Gastric biopsy samples were snap-frozen on dry ice and stored at −80°C until processed. A total of 10 negative controls without samples (i.e., obtained by inserting the forceps through the gastroscope, bringing them into the stomach, and pulling back and inserting them into an empty vial) were included and processed identically to the biopsy samples. Samples were homogenized using a mini-bead beater (BioSpec Products, Bartlesville, OK) at maximum speed for 1 min (three pulses of 20 s each) to enhance the yield of material to be subjected to PCR amplification (see below), particularly from robust bacteria such as Clostridium, Veillonella, or Streptococcus (25). Then, DNA was extracted from the samples by using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany), and DNA concentrations were measured by a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Minneapolis, MN).
16S rRNA gene pyrosequencing and analysis.
Multitag (“multiplex”) pyrosequencing was performed using GS titanium technology (Roche 454 Life Sciences, Branford, CT, USA), mainly according to a well-established protocol (26), to survey the V1-to-V3 variable region corresponding to positions 28 to 519 of the Escherichia coli 16S rRNA gene. Among the targeted gene regions that afford variable levels of taxonomic and phylogenetic informativeness (27), the V1-to-V3 region was shown to provide the greatest classification rate when different samples of the same mock community are tested (i.e., “defined mixture of microbial cells created in vitro to simulate the composition of a microbiome sample”) (25). To this end, the 454 adaptor-linked 28F (5′-TTTGATCNTGGCTCAG) and 519R (5′-GTNTTACNGCGGCKGCTG) primers were designed to carry, on the reverse primer, sample-specific multiple identifier barcodes between the sequences of the 5′-adaptor A and the primer and, on the forward primer, only the 5′-adaptor B sequence (28). The amplification reaction was prepared in a 50-μl final volume containing 0.4 μM each primer, 200 μM each deoxynucleoside triphosphate (dNTP), 1.8 μM MgCl2, 5 μl of 10× PCR buffer, 50 ng of template DNA, and 2.5 U of FastStart high-fidelity enzyme blend (Roche). The PCR conditions were as follows: 95°C for 2 min, followed by 35 cycles of 95°C for 20 s, 56°C for 45 s, and 72°C for 5 min, with final extension step at 72°C for 7 min. For each PCR, one additional PCR negative control without the DNA template was added. PCR products were purified with the Agencourt AMPure XP kit (Beckman Coulter, Milan, Italy), visually inspected on agarose gels to observe bands of the specific size (approximately 600 bp), and quantified with the Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Life Technologies; Monza MB, Italy). After cleanup and quality control, amplicons from all of the 24 biopsy samples were normalized, pooled, purified, and then unidirectionally sequenced on the 454 GS Junior platform using a GS titanium sequencing kit according to the manufacturer's instructions (Roche). A negative-control sample that yielded any visible band (this occurred for 3 of the 10 controls without sample included) was sequenced, and sequence reads in the biopsy samples also present in the negative-control sample were excluded from further analysis (see below). Sequence data were processed with the QIIME (Quantitative Insights Into Microbiology Ecology) 1.8.0 pipeline (29). Sequence reads were demultiplexed and quality-filtered according to default parameters, and pyrosequencing errors were removed using the PyroNoise algorithm (30). The reads were then sorted and grouped into operational taxonomic units (OTUs) using the UCLUST algorithm (http://drive5.com/usearch/manual/uclust_algo.html) at a distance-based similarity of 97% (31). Potentially chimeric sequences were removed using the UCHIME algorithm (32). OTUs from the 3 controls mentioned above were filtered from the analysis. The OTU (phylotype) representative sequences were aligned with PyNAST (33) and assigned to different taxonomic levels (from phylum to genus) using the UCLUST consensus taxonomy classifier (34) and the Greengenes taxonomy reference database (version 13.8.0), with a sequence identity threshold of 97%. Sequences that did not match with the existing sequences in the database were pooled as “unassigned” (Table 2). A phylogenetic tree was built with FastTree (35), whereas a table showing the counts of each assigned OTU in each sample was created, using the QIIME pipeline. To control for differences in coverage and to limit the effects of uneven sampling, the OTU table was rarefied to a depth of 863 sequences, as the sample with the fewest sequences contained 872 sequences, and was used in downstream analyses. Alpha diversity (diversity of microbial communities found within individual samples) was estimated using the Chao1 index, Shannon index, Simpson index, Shannon's index of evenness, and observed species. Also, Good's method was used (36) to estimate what percentage of the total species was represented in a sample (i.e., the percentage of coverage). Beta diversity (diversity of microbial communities found between different samples) was estimated using weighted UniFrac distances (37).
TABLE 2.
Sequence diversity, coverage, and taxonomic complexity by groups of patients
| Measurement |
H. pylori statusa |
Proton pump inhibitor status |
|||
|---|---|---|---|---|---|
| Positive (n = 9) | Negative (n = 15) | Treated (n = 12) | Untreated (n = 12) | Combined (n = 24) | |
| No. of sequences | |||||
| Total | 95,949 | 95,709 | 103,912 | 87,746 | 191,658 |
| Assigned | 95,758 | 93,854 | 103,519 | 86,093 | 189,612 |
| Unassigned | 191 | 1,855 | 393 | 1,653 | 2,046 |
| Helicobacter sequences | 45,739 | 994 | 15,834 | 30,899 | 46,733 |
| No. of OTUsb | 304 | 479 | 419 | 378 | 519 |
| Chao1 estimator of species richness | 313.9 | 479.6 | 423.0 | 387.5 | 519.0 |
| Shannon's index for diversity | 3.5 | 5.7 | 4.9 | 4.6 | 4.9 |
| Simpson's index for diversity | 0.7 | 0.9 | 0.9 | 0.8 | 0.9 |
| Evennessc | 0.4 | 0.6 | 0.6 | 0.5 | 0.5 |
| Good's estimator of coverage (%) | 99.9 | 99.9 | 99.9 | 99.9 | 100 |
| Classification success (% [no. of identified taxa]) | |||||
| Phylum | 70.6 (12) | 100 (17) | 94.1 (16) | 88.2 (15) | 100 (17) |
| Family | 77.1 (64) | 94.0 (78) | 85.5 (71) | 91.6 (76) | 100 (83) |
| Genus | 64.3 (74) | 94.8 (109) | 81.7 (94) | 82.6 (95) | 100 (115) |
As determined by conventional testing.
OTU, operational taxonomic unit.
The Shannon's index of evenness was calculated using the formula E = eD/N, where D is the Shannon diversity index.
Statistics.
Statistical analyses were performed using the R (v. 3.2.5) and GraphPad Prism (v. 6.07) software packages. Prior to statistical analysis, the relative abundance was calculated as the number of sequence reads for each taxon and normalized for sample, such that the total relative abundance for each sample sums to 100%. The normality of the data was examined by the Shapiro-Wilk test. Statistical comparison of alpha diversity between samples was performed using a t test, analysis of variance (ANOVA), or a nonparametric test, depending on either the distribution of the variable (normal versus nonnormal) or the number of groups (2 versus >2). The weighted UniFrac-based distance matrix was exported to the R package vegan (v. 2.3-0), and principal-coordinate analysis (PCoA) plots were generated using the first two principal coordinates (PCs). Relations between the PCoA scores of patient categories (i.e., PPI-treated/untreated, H. pylori-positive/H. pylori-negative, etc.) were assessed by means of the R Adonis function (2015-06-09 r68498), which implements a permutational multivariate analysis of variance (PERMANOVA) using distance matrices.
Differentially abundant taxa in the gastric microbiota between the groups of patients categorized according to medical (with/without PPI treatment) or biological (presence/absence of H. pylori infection) conditions (or classes) were analyzed using the Prism multiple t test analysis that performs many t tests at once. Additionally, differences in taxonomic (i.e., genus level) abundances were estimated using the linear discriminant analysis (LDA) effect size (LEfSe) method (38). In particular, the LEfSe algorithm (https://huttenhower.sph.harvard.edu/galaxy/) identifies features (e.g., bacterial genera) that are statistically different with respect to the class of interest by coupling standard tests for statistical significance (i.e., the nonparametric Kruskal-Wallis rank sum test, or the pairwise Wilcoxon rank sum test) with tests of biological consistency and effect size estimation (LDA). The LEfSe alpha parameter for pairwise tests was set to 0.05, and the threshold on the logarithmic score of LDA was set to 2.0. As was required, all the pairwise comparison rejected the null hypothesis for detecting the biomarker; thus, no multiple testing corrections were needed (38).
Following statistical analyses with multiple comparisons, P values were corrected using the Benjamini-Hochberg method to control the false discovery rate (FDR). An FDR value (Q) of 0.05 was used as a statistically significant cutoff. When interpreted individually without respect to the others, each P value was considered to be statistically significant if it was less than the significance level, alpha, which was set to 0.05.
Accession number(s).
The 16S rRNA gene sequences from this study are available through the NCBI Sequence Read Archive under the accession number SRP060417.
RESULTS
Bacterial genera found in the gastric mucosal microbiota of dyspeptic patients.
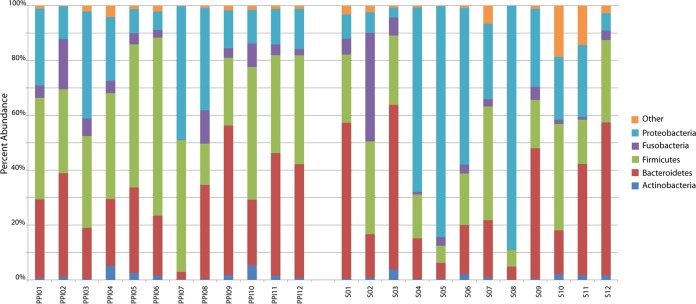
To investigate the gastric microbial community in human dyspepsia, DNA was extracted from individual stomach mucosa biopsy specimens of 24 dyspeptic patients (71% women; aged from 27 to 85 years), 12 of whom were treated with PPIs (Table 1; also see Table S1 in the supplemental material). As shown, although the average age did not differ significantly according to the PPI status, the only two subjects aged older than 80 were in the group of patients on PPIs. By means of PCR amplification and 454 pyrosequencing of the bacterial 16S rRNA gene, we obtained a total of 191,658 high-quality sequence reads, with a length of 424 ± 84 bp (mean ± SD). Excluding three samples that yielded highest numbers of reads (35,873, 27,624, and 21,610, respectively) and one sample that yielded the lowest number of reads (872), there were 5,284 ± 2,265 sequences (mean ± SD) per sample. Rarefaction curves showing the number of OTUs at a 3% genetic distance (see Fig. S1 in the supplemental material) suggested that enough sequencing effort was achieved for all 24 samples. Sequence clustering yielded a total of 519 OTUs (phylotypes), representing an overall Good's coverage of 99.9 (Table 2; see also Table S2 in the supplemental material). The OTUs were classified into 17 bacterial phyla, with some phyla being undetectable in some samples. Accounting for 98% of all sequence reads, Proteobacteria, Firmicutes, Bacteroidetes, Fusobacteria, and Actinobacteria represented the five most abundant phyla. Among them, Proteobacteria (3.56% to 88.97%), Firmicutes (6.10% to 64.92%), and Bacteroidetes (2.68% to 60.05%) were dominant (Fig. 1; see also Table S3 in the supplemental material). Rare phyla, defined as those with a relative abundance of less than 1%, were grouped in the “other” category (Fig. 1). The OTUs were classified into 83 different families, of which the most abundant (i.e., being ≥1% in relative abundance) were Pseudomonadaceae, Comamonadaceae, and Neisseriaceae (phylum Proteobacteria), Streptococcaceae, Gemellaceae, and Veillonellaceae (phylum Firmicutes), and Prevotellaceae and Bacteroidaceae (phylum Bacteroidetes). At finer taxonomic levels, we found that, among 115 genera identified in total, Helicobacter (24.6%), Prevotella (18.6%), Streptococcus (17.9%), Veillonella (6.7%), Neisseria (4.6%), Porphyromonas (4.4%), Fusobacterium (3.7%), Gemella (3.2%), Haemophilus (2.4%), and Leptotrichia (1.4%) represented the 10 most abundant genera. Of these, all taxa except Helicobacter and Leptotrichia were present among all samples. Sequences classified as Helicobacter were found in all of the 9 samples from H. pylori-positive patients (abundance varied from <1.0% to 88.0%) and in 5 of 15 samples from H. pylori-negative patients (abundance varied from <1.0% to 4.5%) (see Table S4 in the supplemental material). As mentioned above, the positive or negative H. pylori status was defined based on the results of conventional tests for all 24 patients; of these, 12 patients (including 4 H. pylori-positive and 8 H. pylori-negative) were treated and 12 patients (including 5 H. pylori-positive and 7 H. pylori-negative) were not treated with PPIs (see Table S1 in the supplemental material).
FIG 1.
Relative abundance at the phylum level (97% similarity) of most dominant bacteria found in the gastric mucosal biopsy specimens of 24 patients with dyspepsia. Biopsy specimens were named as PPI01 to PPI12 or as S01 to S12, depending on whether the patients were (PPI) or were not (S) treated with a proton pump inhibitor (PPI) drug at the time of sampling.
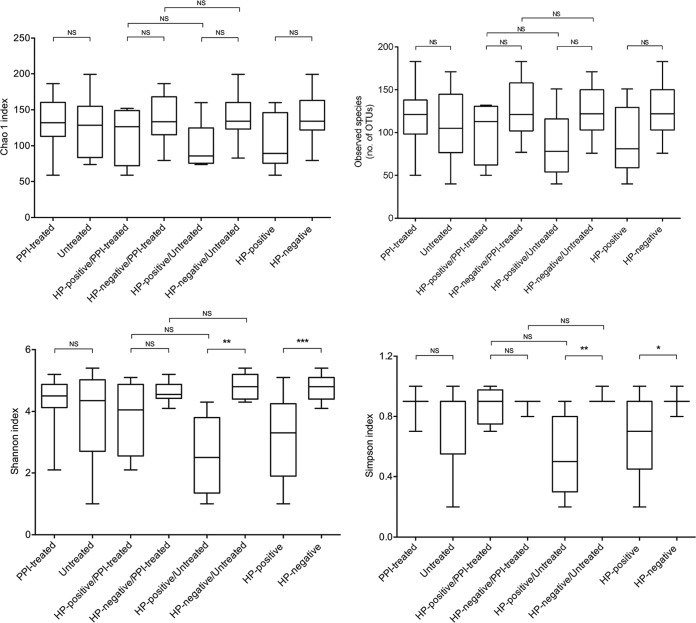
Species richness, evenness, and diversity indices were calculated for the entire sample set according to the H. pylori and PPI treatment status (Table 2). As shown in Fig. 2, Shannon's (ANOVA, P < 0.001) and Simpson's (ANOVA, P < 0.05) indices were significantly higher in samples from H. pylori-negative patients than in samples from H. pylori-positive patients. These results remained significant also when the comparisons were performed including only the samples from patients who did not receive PPIs (Fig. 2). Overall, no significant differences across the sample groups were found with respect to the Chao1 richness estimator and the observed species (ANOVA, P > 0.05, for all comparisons), indicating that neither the presence of H. pylori nor the treatment with PPIs may influence the number of species which compose the gastric mucosa-associated microbiota in patients with dyspepsia (Fig. 2). As expected, when Helicobacter sequences were left out of the analysis, species evenness and diversity among samples in which Helicobacter sequences were found (samples were from both patients positive and negative for H. pylori by conventional testing) were higher (albeit not significantly; P > 0.05) than those of samples without Helicobacter sequences (all samples were from patients negative for H. pylori by conventional testing) (data not shown).
FIG 2.
Alpha diversity plots of Chao1 estimator richness, observed species (number of OTUs), Shannon index, and Simpson index measures for the 24 patient samples grouped by the positive or negative H. pylori (HP) status, or by the presence (treated) or absence (untreated) of proton pump inhibitor (PPI) treatment. The HP-positive and HP-negative patient groups were subdivided by PPI treatment as indicated. The middle line in the box plot represents the median value, and the box is drawn from 25% to 75% quartiles. Whiskers show minimum and maximum values and the ends of the whiskers represent the nonoutlier range. P values of <0.05 (*), <0.01 (**), and <0.001 (***), by an ANOVA and Tukey's post hoc test, were used to determine the statistical significance of differences between groups. A P value of >0.05 indicated the absence of statistically significant differences. NS, not significant.
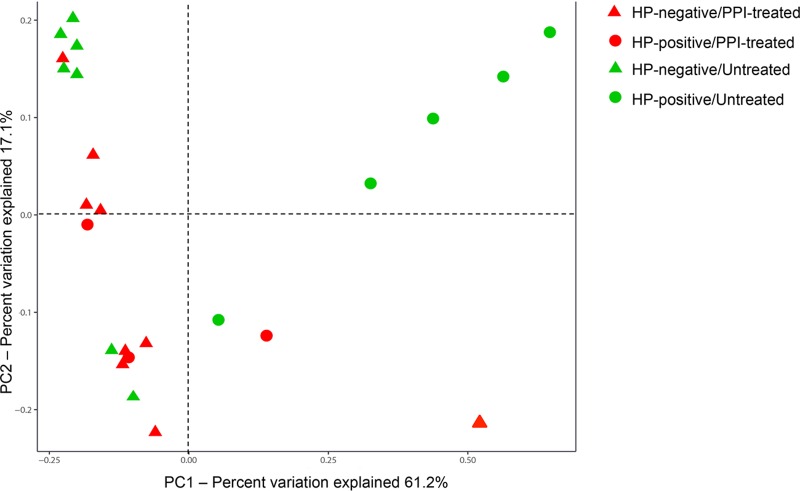
Using the UniFrac analysis, we then examined the relative relatedness of the gastric bacterial communities of 24 dyspeptic patients. The PCoA visualization of weighted UniFrac distances and PERMANOVA analysis of this comparison are shown in Fig. 3. Notably, samples from H. pylori-positive and H. pylori-negative patients and samples from PPI-treated and untreated patients were seen to separate along the axes of two coordinates (i.e., PC1 and PC2, which accounted for 61.2% and 17.1% of the total variation), respectively. However, a significant difference in the overall bacterial community structure was observed comparing only H. pylori-positive and H. pylori-negative patients (P = 0.001). When Helicobacter sequences were left out of the analysis, similar PCoA results were obtained (data not shown). Consistent with the PCoA separation (Fig. 3), additional pairwise comparisons showed that the bacterial community structures within different patient groups might be distinguished in a manner that related microbial composition with both of the patient conditions (see Table S5 in the supplemental material). Interestingly, among untreated patients, a significant distinction was observed when H. pylori-positive patients were compared with H. pylori-negative patients (P = 0.001); in contrast, among H. pylori-negative patients, distinction did not reach statistical significance when PPI-treated patients were compared with untreated patients (P = 0.159) (see Table S5 in the supplemental material).
FIG 3.
Principal-coordinate analysis (PCoA) of weighted UniFrac distances highlighting differences in gastric mucosal biopsy samples of 24 patients with dyspepsia. PC1 and PC2 represent the first two highest discriminating axes. The percent variation explained by each PC axis is indicated. Circle and triangle symbols represent the patients with a positive or negative H. pylori (HP) status (which was determined using conventional testing methods, as specified in the text), respectively. Differently colored symbols represent the patients treated or not treated with proton pump inhibitors (PPIs), respectively.
Influences of H. pylori and PPIs on the gastric microbiota composition of dyspeptic patients.
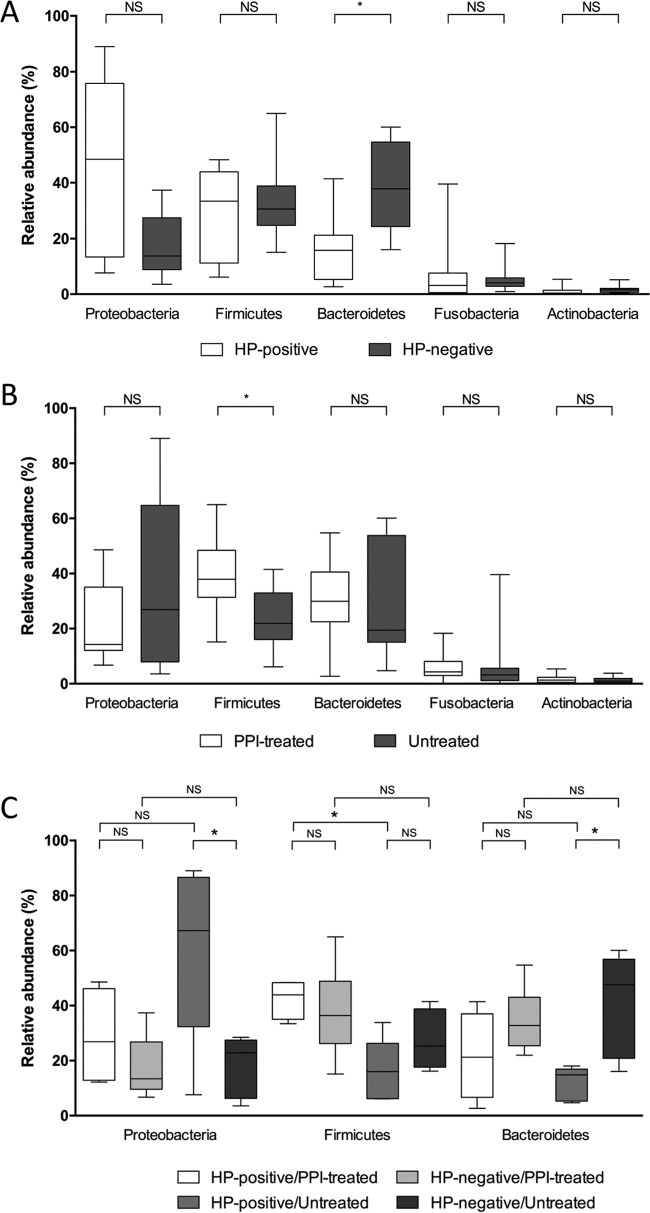
At the phylum level, H. pylori-positive patients displayed lower relative abundance of Bacteroidetes (FDR-adjusted Q = 0.022) than H. pylori-negative patients (Fig. 4A). In contrast, PPI-treated patients tended to have a higher relative abundance of Firmicutes (FDR-adjusted Q = 0.047) compared with that of untreated patients (Fig. 4B). Among the H. pylori-positive patients stratified by PPI treatment, we found that Firmicutes were significantly more abundant in PPI-treated patients than in untreated patients (FDR-adjusted Q = 0.048); in contrast, among the untreated patients stratified by H. pylori status, we found that Proteobacteria were significantly more abundant and Bacteroidetes less abundant (FDR-adjusted Q = 0.049, for both comparisons) in H. pylori-positive patients than in H. pylori-negative patients (Fig. 4C). These results did not remain significant (all FDR-adjusted Q = >0.05) when H. pylori sequences were left out of the analysis; otherwise, among untreated patients, Firmicutes became significantly more abundant (FDR-adjusted Q = 0.029) in H. pylori-positive patients than in H. pylori-negative patients (see Fig. S2 in the supplemental material).
FIG 4.
Differences in relative abundances of bacterial phyla composing the gastric mucosa-associated microbiota of 24 patients, who were stratified according to the positive or negative H. pylori (HP) status (which was determined using conventional testing methods, as specified in the text) (A) and the presence or absence of treatment with proton pump inhibitor (PPI) drugs (B). (C) Comparison of phylum abundances among patient groups, which were defined according to both of the above-mentioned conditions. The middle line in the box plot represents the median value, and the box is drawn from 25% to 75% quartiles. Whiskers show minimum and maximum values, and the ends of the whiskers represent the nonoutlier range. An asterisk indicates a statistically significant difference after the P value of each comparison by t test analysis was corrected for multiple testing (FDR-adjusted Q = <0.05); a nonsignificant (NS) difference (FDR-adjusted Q = >0.05) between each comparison is also indicated.
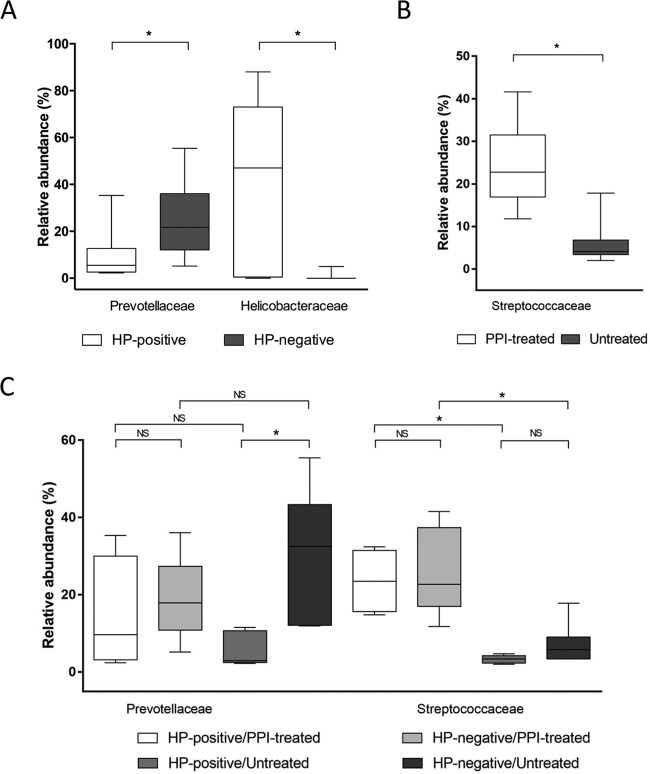
At the family level, across the taxa significantly affected by H. pylori status, we found that Helicobacteraceae (phylum Proteobacteria) were relatively more abundant (FDR-adjusted Q = 0.001), whereas Prevotellaceae (phylum Bacteroidetes) were relatively less abundant (FDR-adjusted Q = 0.013) in H. pylori-positive patients than in H. pylori-negative patients (Fig. 5A). In contrast, across the taxa significantly affected by PPI treatment, we found that Streptococcaceae (phylum Firmicutes) were relatively more abundant (FDR-adjusted Q = 0.0001) in PPI-treated patients than in untreated patients (Fig. 5B). Interestingly, we found that in PPI-treated patients, the relative Streptococcaceae abundance was significantly increased, compared to that in untreated patients, either only considering H. pylori-positive patients (FDR-adjusted Q = 0.048) or H. pylori-negative patients (FDR-adjusted Q = 0.043) (Fig. 5C). Among the untreated patients, H. pylori-positive patients displayed significantly decreased relative Prevotellaceae abundance (FDR-adjusted Q = 0.043) compared to that of H. pylori-negative patients (Fig. 5C). These results remained significant (all FDR-adjusted Q = <0.05) only for the Streptococcaceae family when H. pylori sequences were left out of the analysis (see Fig. S3 in the supplemental material).
FIG 5.
Significant differences in relative abundances of bacterial families composing the gastric mucosa-associated microbiota of 24 patients, who were stratified according to positive or negative H. pylori (HP) status (which was determined based on the results of conventional testing methods, as specified in the text) (A) and the presence or absence of treatment with proton pump inhibitor (PPI) drugs (B). (C) Comparison of family abundances among patient groups, which were defined according to both of the above-mentioned conditions. The middle line in the box plot represents the median value, and the box is drawn from 25% to 75% quartiles. Whiskers show minimum and maximum values, and the ends of the whiskers represent the nonoutlier range. An asterisk indicates a statistically significant difference after the P value of each comparison by t test analysis was corrected for multiple testing (FDR-adjusted Q = <0.05). In panel C, a nonsignificant (NS) differences (FDR-adjusted Q = >0.05) between each comparison is also indicated.
To confirm the identification of such differently abundant family taxa (Fig. 4 and 5) by identifying the genera whose relative abundance might change in relation to the above-mentioned patient conditions, we applied a recently developed biomarker discovery method, LEfSe (38). Using this method, across the bacterial genera whose relative abundances were significantly changed in relation to H. pylori status, we found that 9 genera, including Helicobacter (family Helicobacteraceae), among others, were increased, whereas 6 genera, including Tannerella (family Porphyromonadaceae), Enhydrobacter (family Moraxellaceae), and Mogibacterium (family Mogibacteriaceae), among others, were decreased using H. pylori-negative patients as the reference group (Table 3). Across the bacterial genera whose relative abundances were significantly changed in relation to PPI treatment, we found that Capnocytophaga (family Flavobacteriaceae), Granulicatella (family Carnobacteriaceae), and Streptococcus (family Streptococcaceae) were increased using untreated patients as the reference group (Table 3). Interestingly, when H. pylori sequences were left out of the analysis, these results were unchanged only for Tannerella, Enhydrobacter, Mogibacterium, and Streptococcus; in contrast, Veillonella (the fourth-ranked among most abundant genera of the gastric microbiota in this study) was seen to significantly increase in relation to PPI treatment (Table 3).
TABLE 3.
Genus level taxa that were significantly changed in relation to the patients' conditions in gastric mucosal samplesa
| Taxon/biomarker | Comparison of patients positive vs patients negative to the indicated condition, performed with: |
|||
|---|---|---|---|---|
|
Helicobacter sequences included in the analysis |
Helicobacter sequences excluded from the analysis |
|||
| Trend | LEfSe P value | Trend | LEfSe P value | |
| H. pylori infectionb | ||||
| p_Actinobacteria, g_Propionibacterium | Increased | 0.003 | No | |
| p_Bacteroidetes, g_Tannerella | Decreased | 0.001 | Decreased | 0.009 |
| p_Proteobacteria, g_Campylobacter | Increased | 0.006 | No | |
| p_Proteobacteria, g_Enhydrobacter | Decreased | 0.030 | Decreased | 0.043 |
| p_Proteobacteria, g_Haemophilus | Increased | 0.029 | No | |
| p_Bacteroidetes, g_Capnocytophaga | Increased | 0.044 | No | |
| p_Bacteroidetes, g_Prevotella | Increased | 0.013 | No | |
| p_Firmicutes, g_Bulleidia | Increased | 0.011 | No | |
| p_Proteobacteria, g_Pseudomonas | Increased | 0.049 | No | |
| p_Proteobacteria, g_Helicobacter | Increased | 0.0001 | No | |
| p_Proteobacteria, g_Acinetobacter | Decreased | 0.041 | No | |
| p_Firmicutes, g_Staphylococcus | Increased | 0.042 | No | |
| p_Actinobacteria, g_Corynebacterium | Decreased | 0.014 | No | |
| p_Firmicutes, g_Mogibacterium | Decreased | 0.021 | Decreased | 0.040 |
| p_Firmicutes, g_Dialister | Decreased | 0.012 | No | |
| PPI treatmentc | ||||
| p_Bacteroidetes, g_Capnocytophaga | Increased | 0.048 | No | |
| p_Proteobacteria, g_Actinobacillus | Increased | 0.028 | No | |
| p_Firmicutes, g_Granulicatella | Increased | 0.003 | No | |
| p_Firmicutes, g_Veillonella | No | Increased | 0.024 | |
| p_Firmicutes, g_Streptococcus | Increased | 0.003 | Increased | 0.037 |
Bold denotes genera whose relative abundance was found to change significantly when the linear discriminant analysis (LDA) effect size (LEfSe) method was applied in order to detect differentially abundant taxa among the patients' stomach bacterial communities, regardless of whether Helicobacter sequences were included or not included in the analysis. p_, phylum; g_, genus.
Patients were categorized as positive or negative for the indicated condition, i.e., H. pylori (HP) infection, according to the results of conventional (nonsequencing) HP testing methods as specified in the text. Trends of increase or decrease for each taxon were calculated using the HP-negative patients as the reference group. Of the genera found to significantly differ in relative abundance among these patients, two genera, Mogibacterium (family Mogibacteriaceae) and Dialister (family Veillonellaceae), are members of the order Clostridiales.
Patients were categorized as positive or negative for the indicated condition, i.e., proton-pump inhibitor (PPI) treatment, depending on whether they were, respectively, PPI users and PPI nonusers as specified in the text. Trends of increase or decrease for each taxon were calculated using the untreated patients (PPI nonusers) as the reference group. Of the genera found to significantly differ in relative abundance among these patients, two genera, Granulicatella (family Carnobacteriaceae) and Streptococcus (family Streptococcaceae), are members of the order Lactobacillales.
DISCUSSION
To date, the small number of studies exploring the human stomach microbiota using advanced, culture-independent, molecular methods (i.e., 16S rRNA gene sequencing) showed that the gastric mucosa-associated microbial community is, in the apparently “normal” acidic condition, strongly dominated by Proteobacteria (to which H. pylori belongs), Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria (39–41; for review, see reference 42). It is now clear that the accuracy of classification increases with the length of the sequenced 16S rRNA gene region (27). However, using an earlier version of 454 pyrosequencing technology (i.e., based on shorter 16S rRNA gene sequence reads) to explore the differences in microbial communities along the digestive tract, Andersson et al. identified 177 phylotypes in stomach but not in throat, with the majority of them belonging to the Proteobacteria (40). Later, a core set of 19 bacterial genera was identified in all of the gastric biopsy specimens analyzed by Delgado et al. (43), whereas 8 bacterial genera were uniquely present in the gastric and lung fluid but not in the oropharyngeal communities of 116 children studied (44).
Our first goal was to provide a detailed characterization of the stomach-resident microbial community in patients with dyspepsia. Here, 454 pyrosequencing analysis of the gastric mucosa community underscores the prominence of genera such as Streptococcus, Prevotella, Veillonella, Porphyromonas, and Haemophilus. In the pioneer studies by Bik et al. (39) and Li et al. (41) on the gastric mucosa-associated microbiota, Streptococcus and Prevotella were detected as the two major abundant genera, although these studies analyzed two geographically and ethnically divergent populations and two medically different populations. Surprisingly, Streptococcus and Prevotella were also detected as the top two genera in the gastric luminal microbiota, suggesting similarities of the bacterial compositions between the two intragastric compartments (11). Thus, the question of whether specific, stomach-resident microbial communities or bacterial groups/organisms may contribute to or be associated with, the development of gastric disease becomes intriguing (43, 45, 46). In the aforementioned study, Delgado et al. included subjects who had dyspepsia without detectable gastric pathology (i.e., functional dyspepsia) (43). In the present study, all dyspeptic patients had histologically documented gastric mucosa inflammation (Table 1; see also Table S1 in the supplemental material). A comparison of the two studies revealed differences in the array of bacteria dominating the gastric mucosal samples. Thus, Gram-positive organisms like Streptococcus, Propionibacterium, Lactobacillus, and Enterococcus dominated in that study (43), whereas a mixing of Gram-positive (only Streptococcus) and Gram-negative organisms was found by us. However, it is worth noting that differences in DNA extraction methods and PCR amplification protocols as well as in the sampling procedures used in the two studies might have biased the detection of some (e.g., Gram-positive) bacterial groups/organisms.
Previous studies on humans (39, 40, 47) reported significant changes in the gastric mucosa-associated microbiota in response to H. pylori. In one study, eight bacterial phyla (128 phylotypes) were identified in 23 North American patients, with no differences in species richness by H. pylori status (39); in another study, 13 bacterial phyla were identified in six Swedish patients, but the species diversity in H. pylori-negative stomachs (262 phylotypes) was higher than in H. pylori-positive stomachs (33 phylotypes) (40). Furthermore, using a high-density 16S rRNA gene microarray, Maldonado-Contreras et al. showed that 28% of total variation in the gastric microbiota of 12 patients (10 Amerindians and 2 non-Amerindians) was explained by H. pylori status (47). In the present study, Bacteroidetes were found to be the primary driver of the distinction in microbial composition between patients with positive or negative H. pylori status (Fig. 3 and 4A), although differences in the gastric mucosa communities between the two patient groups could be explained by relative abundance differences of at least 15 specific biomarkers (Table 3). These included not only Prevotella but also low-abundance genera, such as Bacteroidetes other than Prevotella (Tannerella and Capnocytophaga), Proteobacteria (Campylobacter, Enhydrobacter, Pseudomonas, and Acinetobacter), Firmicutes (Bulleidia, Staphylococcus, Mogibacterium, and Dialister), and Actinobacteria (Propionibacterium and Corynebacterium) organisms. Whether these microbial alterations might have a role in the pathogenesis of gastritis needs to be elucidated in future studies. Unless infected subjects display symptoms, H. pylori infections are not treated, implying that firmly established infections persist in the stomach (48). It is plausible that H. pylori infections lead to changes in the microbiota over time (49) and that drug-induced variations in specific microbiota members affect the immune response to H. pylori, thereby contributing to H. pylori-associated upper gastrointestinal diseases (50).
The acidity of the human stomach can be greatly compromised in the case of H. pylori infection (and H. pylori-associated atrophic gastritis) and also in the case of pharmacological interventions, for example, with PPIs (6). Therefore, we looked at the role of PPI use on the gastric mucosa-associated microbiota in our patients, and we showed that PPI treatment was associated with increased abundance of Firmicutes, particularly Streptococcaceae (Fig. 4B and 5B). A closer inspection of the microbial composition in PPI-treated patients revealed a few genera that were relatively abundant in all gastric samples from these patients and thus considered biomarkers of these samples (Table 3). So, Streptococcus and Granulicatella (both from the phylum Firmicutes) together with Capnocytophaga (phylum Bacteroidetes) and Actinobacillus (phylum Proteobacteria) were differentiated by relative abundance. The Streptococcus genus in particular was detected by LEfSe with a very high LDA score (data not shown), reflecting marked abundance in PPI-treated patients and consistently low abundance in untreated patients. Using Metastasts analysis, the only recent method that is an alternative to LEfSe analysis (51), Rosen et al. also found that Streptococcus was relatively more abundant in the gastric fluid of children treated with PPIs (44). Similarly, PPI use was shown to be associated with profound changes in the gut microbiota, involving the genera Streptococcus, Enterococcus, and Staphylococcus, and the species Escherichia coli (52). In another study, significant differences in the abundance of stool-associated bacterial taxa between PPI users and non-PPI users were seen for the Holdemania, Streptococcus, and Blautia genera (53). Also, in the largest study published to date, 16S rRNA gene profiling of fecal samples collected from 1,827 healthy twins found that PPI use was associated with significant increases in the abundances of oral and upper gastrointestinal tract bacteria and in particular of the Streptococcaceae family (54). Remarkably, these associations were replicated in a small interventional data set indicating causality (54). Because of their presumed overgrowth in the upper gastrointestinal tract (4), streptococci can thrive under PPI-induced hypochlorhydria conditions, triggering increased host susceptibility to disease and infection (55, 56). As aciduric bacteria, Streptococcus (and Lactobacillus) may directly contribute to the onset of dyspepsia by producing acid within the esophagus and oral cavity, whereas at least one Streptococcus species is known to have its own H+/K+-ATPase, which may be targeted by PPIs (7). In this context, it is likely that the increased Streptococcus abundance in the stomach is an almost exclusive indicator of PPI use in dyspeptic patients and that this abundance is ultimately responsible for exacerbation and/or persistence of symptoms of dyspepsia during PPI therapy.
Finally, we tried to elucidate whether the interaction of PPIs with the gastric mucosa-associated microbiota is mediated by H. pylori infection. The PPIs are believed to interact directly with H. pylori by inhibiting the urease activity of the bacterium or by exerting a bacteriostatic effect against the bacterium (7). Thus, PPIs might mask the influence of H. pylori on the gastric microbiota via the overgrowth of specific taxa among the indigenous non-H. pylori bacteria (8). When searching for significant differences in the relative taxon abundance among the patient subgroups, we noticed that the significant increase in the relative Streptococcaceae abundance (Fig. 5B) was independent of whether the PPI-treated patients had a positive or negative H. pylori status (Fig. 5C). Importantly, this effect was not blunted when the statistical analyses were performed with our data set excluding the Helicobacter sequences (see Fig. S3 in the supplemental material). On the other hand, we noticed that Prevotellaceae were significantly decreased in patients with positive H. pylori status (Fig. 5A) and that this decrease was also significant in the untreated but not in the PPI-treated patient subgroup (Fig. 5C).
Taken together, our findings are strongly suggestive of a concomitant role of H. pylori and PPIs on the gastric mucosa-associated microbiota, although concomitant H. pylori and non-H. pylori bacterial infections may be implied in the pathogenesis of gastric inflammation/disease. It is biologically plausible that PPIs, if given after H. pylori is eradicated, contribute to gastric carcinogenesis by causing non-H. pylori gastric dysbiosis that perpetuates Correa's stepwise inflammatory process in the human stomach (20). It should be recalled that eradication of H. pylori significantly decreases the development of gastric cancer only in patients without H. pylori-induced precancerous gastric lesions such as atrophy and intestinal metaplasia (57). Once again, increased abundance of Streptococcus might be regarded as a biomarker of PPI-related modifications toward a less healthy gastric microbiota (46). If these modifications are linked with altered abundance and location of gastric H. pylori needs to be explored in the future.
In conclusion, the present study shows the potential of using 16S rRNA gene sequence data analyses to better understand the impact of xenobiotics such as PPIs on the stomach microbiota and to identify specific organisms as indicators of microbial composition changes under foreign stimuli. Although a popular treatment choice for dyspepsia, which remains one of most common gastrointestinal disorders among western people, PPIs exhibit substantial intersubject variability and commonly fail to provide a complete cure for the disorder (2). Thus, a patient-tailored use of these drugs could be derived by careful monitoring of the gastric microbiota changes that occur during PPI therapy. Future studies will help to reveal whether the way taken is the right one.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by UCSC-Linea D1 grants to B.P. and M.S, by a UCSC-Linea D.3.2-2013 grant to M.S., and by an independent group leader starting grant from the Technische Universität Dresden to C.V.C.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01437-16.
REFERENCES
- 1.Shin JM, Munson K, Sachs G. 2011. Gastric H+,K+-ATPase. Compr Physiol 1:2141–2153. doi: 10.1002/cphy.c110010. [DOI] [PubMed] [Google Scholar]
- 2.Yang YX, Metz DC. 2010. Safety of proton pump inhibitor exposure. Gastroenterology 139:1115–1127. doi: 10.1053/j.gastro.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 3.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, El-Omar EM, Kuipers EJ; European Helicobacter Study Group . 2012. Management of Helicobacter pylori infection—-the Maastricht IV/ Florence Consensus Report. Gut 61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
- 4.Williams C, McColl KE. 2006. Review article: proton pump inhibitors and bacterial overgrowth. Aliment Pharmacol Ther 23:3–10. doi: 10.1111/j.1365-2036.2006.02707.x. [DOI] [PubMed] [Google Scholar]
- 5.Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ. 2014. Participation of microbiota in the development of gastric cancer. World J Gastroenterol 20:4948–4952. doi: 10.3748/wjg.v20.i17.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker MM, Talley NJ. 2014. Review article: bacteria and pathogenesis of disease in the upper gastrointestinal tract—beyond the era of Helicobacter pylori. Aliment Pharmacol Ther 39:767–779. doi: 10.1111/apt.12666. [DOI] [PubMed] [Google Scholar]
- 7.Vesper BJ, Jawdi A, Altman KW, Haines GK III, Tao L, Radosevich JA. 2009. The effect of proton pump inhibitors on the human microbiota. Curr Drug Metab 10:84–89. doi: 10.2174/138920009787048392. [DOI] [PubMed] [Google Scholar]
- 8.Del Piano M, Pagliarulo M, Tari R, Carmagnola S, Balzarini M, Lorenzini P, Pane M. 2014. Correlation between chronic treatment with proton pump inhibitors and bacterial overgrowth in the stomach: any possible beneficial role for selected lactobacilli? J Clin Gastroenterol 48(Suppl 1):S40–S46. doi: 10.1097/MCG.0000000000000256. [DOI] [PubMed] [Google Scholar]
- 9.Bengmark S. 2013. Gut microbiota, immune development and function. Pharmacol Res 69:87–113. doi: 10.1016/j.phrs.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Sanduleanu S, Jonkers D, De Bruine A, Hameeteman W, Stockbrügger RW. 2001. Non-Helicobacter pylori bacterial flora during acid-suppressive therapy: differential findings in gastric juice and gastric mucosa. Aliment Pharmacol Ther 15:379–388. doi: 10.1046/j.1365-2036.2001.00888.x. [DOI] [PubMed] [Google Scholar]
- 11.Tsuda A, Suda W, Morita H, Takanashi K, Takagi A, Koga Y, Hattori M. 2015. Influence of proton-pump inhibitors on the luminal microbiota in the gastrointestinal tract. Clin Transl Gastroenterol 6:e89. doi: 10.1038/ctg.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. 2014. Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environ Microbiol 16:2905–2914. doi: 10.1111/1462-2920.12285. [DOI] [PubMed] [Google Scholar]
- 13.Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF. 2014. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 19:407–416. doi: 10.1111/hel.12145. [DOI] [PubMed] [Google Scholar]
- 14.Talley NJ, Holtmann G, Walker MM. 2015. Therapeutic strategies for functional dyspepsia and irritable bowel syndrome based on pathophysiology. J Gastroenterol 50:601–613. doi: 10.1007/s00535-015-1076-x. [DOI] [PubMed] [Google Scholar]
- 15.Ford AC, Moayyedi P. 2013. Dyspepsia. Curr Opin Gastroenterol 29:662–668. doi: 10.1097/MOG.0b013e328365d45d. [DOI] [PubMed] [Google Scholar]
- 16.Moayyedi P, Talley NJ. 2006. Gastro-oesophageal reflux disease. Lancet 367:2086–2100. doi: 10.1016/S0140-6736(06)68932-0. [DOI] [PubMed] [Google Scholar]
- 17.Karamanolis GP, Tack J. 2012. Current management of functional dyspepsia: impact of Rome III subdivision. Ann Gastroenterol 25:96–99. [PMC free article] [PubMed] [Google Scholar]
- 18.Zullo A, Hassan C, De Francesco V, Repici A, Manta R, Tomao S, Annibale B, Vaira D. 2014. Helicobacter pylori and functional dyspepsia: an unsolved issue? World J Gastroenterol 20:8957–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacy BE, Talley NJ, Locke GR III, Bouras EP, DiBaise JK, El-Serag HB, Abraham BP, Howden CW, Moayyedi P, Prather C. 2012. Review article: current treatment options and management of functional dyspepsia. Aliment Pharmacol Ther 36:3–15. doi: 10.1111/j.1365-2036.2012.05128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freedberg DE, Lebwohl B, Abrams JA. 2014. The impact of proton pump inhibitors on the human gastrointestinal microbiome. Clin Lab Med 34:771–785. doi: 10.1016/j.cll.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald EG, Milligan J, Frenette C, Lee TC. 2015. Continuous proton pump inhibitor therapy and the associated risk of recurrent Clostridium difficile infection. JAMA Intern Med 175:784–791. doi: 10.1001/jamainternmed.2015.42. [DOI] [PubMed] [Google Scholar]
- 22.Kelly OB, Dillane C, Patchett SE, Harewood GC, Murray FE. 2015. The inappropriate prescription of oral proton pump inhibitors in the hospital setting: a prospective cross-sectional study. Dig Dis Sci 60:2280–2286. doi: 10.1007/s10620-015-3642-8. [DOI] [PubMed] [Google Scholar]
- 23.Boardman HF, Delaney BC, Haag S. 2015. Partnership in optimizing management of reflux symptoms: a treatment algorithm for over-the-counter proton-pump inhibitors. Curr Med Res Opin 31:1309–1318. doi: 10.1185/03007995.2015.1047745. [DOI] [PubMed] [Google Scholar]
- 24.Mégraud F, Bessède E, Lehours P. 2014. Diagnosis of Helicobacter pylori infection. Helicobacter 19(Suppl 1):S6–S10. doi: 10.1111/hel.12161. [DOI] [PubMed] [Google Scholar]
- 25.Yang I, Nell S, Suerbaum S. 2013. Survival in hostile territory: the microbiota of the stomach. FEMS Microbiol Rev 37:736–761. doi: 10.1111/1574-6976.12027. [DOI] [PubMed] [Google Scholar]
- 26.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soergel DA, Dey N, Knight R, Brenner SE. 2012. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J 6:1440–1444. doi: 10.1038/ismej.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binladen J, Gilbert MT, Bollback JP, Panitz F, Bendixen C, Nielsen R, Willerslev E. 2007. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One 2:e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, Swings J. 2005. Opinion: Re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 32.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Good IJ. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264. doi: 10.1093/biomet/40.3-4.237. [DOI] [Google Scholar]
- 37.Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. 2008. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One 3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XX, Wong GL, To KF, Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C. 2009. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engstrand L, Lindberg M. 2013. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol 27:39–45. doi: 10.1016/j.bpg.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Delgado S, Cabrera-Rubio R, Mira A, Suárez A, Mayo B. 2013. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol 65:763–772. doi: 10.1007/s00248-013-0192-5. [DOI] [PubMed] [Google Scholar]
- 44.Rosen R, Hu L, Amirault J, Khatwa U, Ward DV, Onderdonk A. 2015. 16S community profiling identifies proton pump inhibitor related differences in gastric, lung, and oropharyngeal microflora. J Pediatr 166:917–923. doi: 10.1016/j.jpeds.2014.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stearns JC, Lynch MD, Senadheera DB, Tenenbaum HC, Goldberg MB, Cvitkovitch DG, Croitoru K, Moreno-Hagelsieb G, Neufeld JD. 2011. Bacterial biogeography of the human digestive tract. Sci Rep 1:170. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. 2009. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol 58:509–516. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 47.Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG. 2011. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J 5:574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salama NR, Hartung ML, Müller A. 2013. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol 11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J Clin Invest 119:2475–2487. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolig AS, Cech C, Ahler E, Carter JE, Ottemann KM. 2013. The degree of Helicobacter pylori-triggered inflammation is manipulated by preinfection host microbiota. Infect Immun 81:1382–1389. doi: 10.1128/IAI.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJ, Dijkstra G, Franke L, Xavier RJ, Jonkers D, Wijmenga C, Weersma RK, Zhernakova A. 2016. Proton pump inhibitors affect the gut microbiome. Gut 65:740–748. doi: 10.1136/gutjnl-2015-310376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clooney AG, Bernstein CN, Leslie WD, Vagianos K, Sargent M, Laserna-Mendieta EJ, Claesson MJ, Targownik LE. 2016. A comparison of the gut microbiome between long-term users and non-users of proton pump inhibitors. Aliment Pharmacol Ther 43:974–984. doi: 10.1111/apt.13568. [DOI] [PubMed] [Google Scholar]
- 54.Jackson MA, Goodrich JK, Maxan ME, Freedberg DE, Abrams JA, Poole AC, Sutter JL, Welter D, Ley RE, Bell JT, Spector TD, Steves CJ. 2016. Proton pump inhibitors alter the composition of the gut microbiota. Gut 65:749–756. doi: 10.1136/gutjnl-2015-310861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canani RB, Cirillo P, Roggero P, Romano C, Malamisura B, Terrin G, Passariello A, Manguso F, Morelli L, Guarino A, Working Group on Intestinal Infections of the Italian Society of Pediatric Gastroenterology Hepatology and Nutrition (SIGENP) . 2006. Therapy with gastric acidity inhibitors increases the risk of acute gastroenteritis and community-acquired pneumonia in children. Pediatrics 117:e817–e820. doi: 10.1542/peds.2005-1655. [DOI] [PubMed] [Google Scholar]
- 56.Gulmez SE, Holm A, Frederiksen H, Jensen TG, Pedersen C, Hallas J. 2007. Use of proton pump inhibitors and the risk of community-acquired pneumonia: a population-based case-control study. Arch Intern Med 167:950–955. doi: 10.1001/archinte.167.9.950. [DOI] [PubMed] [Google Scholar]
- 57.Wong BC, Lam SK, Wong WM, Chen JS, Zheng TT, Feng RE, Lai KC, Hu WH, Yuen ST, Leung SY, Fong DY, Ho J, Ching CK, Chen JS, China Gastric Cancer Study Group . 2004. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA 291:187–194. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.