ABSTRACT
Partially acetylated chitosan oligosaccharides (paCOS) are potent biologics with many potential applications, and their bioactivities are believed to be dependent on their structure, i.e., their degrees of polymerization and acetylation, as well as their pattern of acetylation. However, paCOS generated via chemical N-acetylation or de-N-acetylation of GlcN or GlcNAc oligomers, respectively, typically display random patterns of acetylation, making it difficult to control and predict their bioactivities. In contrast, paCOS produced from chitin deacetylases (CDAs) acting on chitin oligomer substrates may have specific patterns of acetylation, as shown for some bacterial CDAs. However, compared to what we know about bacterial CDAs, we know little about the ability of fungal CDAs to produce defined paCOS with known patterns of acetylation. Therefore, we optimized the expression of a chitin deacetylase from the fungus Puccinia graminis f. sp. tritici in Escherichia coli. The best yield of functional enzyme was obtained as a fusion protein with the maltose-binding protein (MBP) secreted into the periplasmic space of the bacterial host. We characterized the MBP fusion protein from P. graminis (PgtCDA) and tested its activity on different chitinous substrates. Mass spectrometric sequencing of the products obtained by enzymatic deacetylation of chitin oligomers, i.e., tetramers to hexamers, revealed that PgtCDA generated paCOS with specific acetylation patterns of A-A-D-D, A-A-D-D-D, and A-A-D-D-D-D, respectively (A, GlcNAc; D, GlcN), indicating that PgtCDA cannot deacetylate the two GlcNAc units closest to the oligomer's nonreducing end. This unique property of PgtCDA significantly expands the so far very limited library of well-defined paCOS available to test their bioactivities for a wide variety of potential applications.
IMPORTANCE We successfully achieved heterologous expression of a fungal chitin deacetylase gene from the basidiomycete Puccinia graminis f. sp. tritici in the periplasm of E. coli as a fusion protein with the maltose-binding protein; this strategy allows the production of these difficult-to-express enzymes in sufficient quantities for them to be characterized and optimized through protein engineering. Here, the recombinant enzyme was used to produce partially acetylated chitosan oligosaccharides from chitin oligomers, whereby the pronounced regioselectivity of the enzyme led to the production of defined products with novel patterns of acetylation. This approach widens the scope for both the production and functional analysis of chitosan oligomers and thus will eventually allow the detailed molecular structure-function relationships of biologically active chitosans to be studied, which is essential for developing applications for these functional biopolymers for a circular bioeconomy, e.g., in agriculture, medicine, cosmetics, and food sciences.
INTRODUCTION
Chitosans are a family of functional biopolymers derived from the partial de-N-acetylation of chitin, one of the most abundant biopolymers in the world. Whereas chitin, a linear homopolymer of β-1,4-glycosidically linked N-acetylglucosamine (GlcNAc) residues, forms crystalline fibers and is insoluble in aqueous solvents, chitosans are soluble at slightly acidic pH values, due to the positive charges conveyed by their de-N-acetylated glucosamine (GlcN) residues. The unique polycationic nature of chitosans is at least partially responsible for the many bioactivities of chitosans, such as their antimicrobial, plant-strengthening, and wound-healing activities (1–6). Additionally, though, their biological functionalities are dependent on the degree of polymerization and acetylation of the chitosan, and possibly also on its pattern of acetylation (7–9). However, detailed structure-function relationships are not yet fully understood, in particular regarding the postulated role of the pattern of acetylation. This is mostly because all chitosans available commercially today are prepared from chitin by partial de-N-acetylation, or by full de-N-acetylation followed by partial re-N-acetylation, and the chemical methods used for these conversions invariably lead to chitosans with random patterns of acetylation (10, 11). In contrast, chitosans are also generated naturally by the action of chitin de-N-acetylases (CDAs; E.C. 3.5.1.41) (9, 12, 13), but the patterns of acetylation found in natural chitosans are presently unknown.
It has recently been proposed that some if not most of the biological activities of chitosan polymers are attributable to the oligomeric degradation products generated by chitosan hydrolases present in the target tissues, such as bacterial chitinases and chitosanases, plant chitinases, or human chitotriosidase (9). As these hydrolytic enzymes have more or less pronounced sequence specificities, e.g., cleaving exclusively between two acetylated GlcNAc residues, the pattern of acetylation of the chitosan polymer will define the quantity and quality of the resulting oligomers (14). Importantly, even if the polymeric substrate had a random pattern of acetylation, the acetylation pattern of the enzymatically produced oligomers is expected not to be completely random, as the cleavage specificity of the hydrolase will define the nature of the monosaccharide units present at the oligomeric products' reducing and nonreducing ends (15). As a consequence, understanding the structure-function relationships of partially acetylated chitosan oligosaccharides (paCOS) will be key to developing reliable applications of chitosans in food sciences, agriculture, and biomedicine.
Although it is critical to produce paCOS with fully defined structures, the paCOS available today are typically mixtures of chitosan oligomers that differ in their degrees of polymerization and acetylation as well as in their patterns of acetylation (16). The paCOS in these mixtures can be separated according to their degrees of polymerization and degrees of acetylation, which allows one to experimentally study how these parameters influence the biological activities of paCOS (17–20), but it is not yet possible to separate paCOS with identical degrees of polymerization and acetylation but that differ in their patterns of acetylation.
Thus, to study how the acetylation pattern affects paCOS bioactivities, we must be able to produce paCOS with defined nonrandom patterns of acetylation. This can best be achieved using a CDA that acts on fully acetylated chitin oligomers of a defined degree of polymerization. However, to date, only a few CDAs have been analyzed in detail to determine their products' pattern of acetylation. The best known examples of CDAs yielding paCOS of fully defined architecture are the bacterial NodB (21) and COD (22, 23) from Rhizobium and Vibrio species, respectively. NodB acts specifically on the GlcNAc unit at the nonreducing end of chitin oligomers, while COD specifically deacetylates the penultimate GlcNAc unit next to the nonreducing end. Thus, when acting on a chitin oligomer with a degree of polymerization “n” (GlcNAcn), these enzymes will generate the monodeacetylated paCOS GlcN-GlcNAc(n−1) and GlcNAc-GlcN-GlcNAc(n−2), respectively. We have recently shown that these two CDAs can be combined to yield the dideacetylated paCOS GlcN2-GlcNAc(n−2) (24). Because regioselective CDAs such as these are emerging as a powerful tool for producing paCOS of fully defined architecture (12, 13), more CDAs with different and known regioselectivities are urgently required. However, most of the bacterial and fungal CDAs analyzed to date yield mixtures of paCOS and often eventually fully de-N-acetylate chitin oligomers, even if the sequence of deacetylation varies between enzymes (20, 25–27). Hence, it would be highly advantageous to identify CDAs that are both regioselective and can lead to specific deacetylation patterns.
One problem hampering progress in this field is the difficulty in expressing eukaryotic CDA genes (such as those from fungi) in prokaryotic hosts, such as Escherichia coli. Issues arise because these hosts use different codons and cannot perform posttranslational modifications, such as glycosylation or disulfide bond formation, both of which occur in fungal CDAs (13), possibly leading to incorrect protein folding in E. coli. While this problem can be overcome by using yeast expression systems, such as Pichia pastoris (28, 29), these systems are less suitable than E. coli for comprehensive large-scale mutational studies that would be required to understand the regioselectivity of CDA on a molecular level, a prerequisite for the targeted engineering of CDAs with novel or improved specificities.
So far, a few fungal CDA genes, such as Saccharomyces cerevisiae CDA1 and CDA2 (30–32), ClCDA from Colletotrichum lindemuthianum UPS9 (33), Flammulina velutipes CDA (34), and Rhizopus circinans CDA (35), have been expressed successfully in eukaryotic hosts, namely, S. cerevisiae and Pichia pastoris. Even fewer fungal CDA genes have been expressed successfully in E. coli, namely, ClCDA (ATCC 56676) (36), CDA2 from Saccharomyces cerevisiae (37), as well as Aspergillus nidulans CDA (38).
In this study, we systematically optimized the expression of a fungal CDA gene from the plant-pathogenic basidiomycete Puccinia graminis f. sp. tritici, a natural chitosan producer (39), in E. coli, and we then characterized the recombinant enzyme in terms of its regioselectivity.
MATERIALS AND METHODS
Materials.
N-Acetylglucosamine (GlcNAc) was purchased from Sigma-Aldrich (Munich, Germany). N-Acetyl-chito-oligosaccharides, namely, dimers to hexamers (A2 to A6; A, GlcNAc), were procured from Seikagaku Corporation (Tokyo, Japan), and a chito-oligosaccharide mixture (COS mixture, A5/A4 of ca. 85% A5 and 15% A4, with traces of A4D1, [D, GlcN]) was obtained from the Bio Base Europe Pilot Plant (BBEPP, Ghent, Belgium). Chitosan polymers and the colloidal chitin polymer were produced and characterized as previously described (40, 41). The α-chitin and β-chitin polymers were bought from Mahtani Chitosan Pvt. Ltd. (Veraval, India), and glycol chitin was prepared from glycol chitosan (Sigma-Aldrich, St. Louis, MO) (42). The plasmid purification and gel extraction kits were acquired from Analytik Jena (Biometra, Jena, Germany). All the FastDigest restriction enzymes were from Fermentas (St. Leon Rot, Germany), and the T4 DNA ligase kit for PCR ligations was from Fermentas (Thermo Scientific, Waltham, MA). The PCR for gene amplification and cloning was done using the Phusion II Hot Start high-fidelity DNA polymerase from Finnzymes (New England BioLabs GmbH, Frankfurt, Germany). All reagents were of analytical grade, unless otherwise stated (Sigma-Aldrich, Munich, Germany). Acetate release was measured by using the acetate kit from R-Biopharm (Darmstadt, Germany).
Three-dimensional modeling.
The crystallographic structure of ClCDA from Colletotrichum lindemuthianum (PDB 2IWO) served as a template for generating the model of PgtCDA. Sequence alignment between the target and template was carried out using Clustal Omega (43). The online structure prediction program SWISS-MODEL was applied for structure modeling. The model obtained was refined by the online tool KoBaMIN (44). The quality of the model was assessed by MolProbity (45). Both PyMOL and VMD (46, 47) were used for structure visualization and image generation. The protein topology graph was generated by Pro-origami, available online (48).
CDA cloning.
To assess the PgtCDA expression in a variety of different bacterial strains, we first had to clone the gene (NCBI accession no. XP_003323413.1) in different vectors. The gene for P. graminis CDA (49) (see Fig. S1 in the supplemental material) was codon optimized by GenScript (NJ, USA) and was obtained in plasmid pJET1.2. The vector pET-22b(+) was purchased from Novagen (Darmstadt, Germany) and used as a template to include a Strep II tag (WSHPQFEK) encoding a sequence downstream of the multiple-cloning site (50). This was accomplished by PCR using the 5′-phosphorylated primer pair pET22b_Strep II_for (Pho-CAATTTGAAAAATAGGCTTGCGGCCGCACTC) and pET22b_Strep II_rev (Pho-AGGATGTGACCATTGTCGACGGAGCTCG), resulting in pET-22b(+)::Strep IIC. The PgtCDA gene was inserted into the vector pET-22b(+)::Strep IIC by digestion with NdeI and SacI, yielding pET-22b(+)::PgtCDA-Strep IIC. To enhance solubility, the maltose-binding protein (MBP) was fused to the N terminus of the PgtCDA protein. To this end, the MBP gene was amplified from the pMAL-p5x vector obtained from New England BioLabs (Ipswich, MA, USA) using the primer pair MalE_for (GCTCTAGAAATAATTTTGTTTAAGAAGGAGATATAATTATGAAAATCGAAGAAGGTAAAC), with an XbaI restriction site followed by a ribosomal binding site; and MalE_rev (GAGATTCCATATGCCTTCCCTCGATCCCG), with an NdeI restriction site. The amplified MBP gene was ligated into the pET22b(+)::PgtCDA::Strep IIC plasmid, yielding pET22b(+)::MBP-PgtCDA-Strep IIC. For periplasmic localization of the MBP-PgtCDA fusion protein, we cloned the PgtCDA-Strep IIC gene into the pMAL-p5x vector, which contained an N-terminal signal sequence encoded in its malE gene designed to direct the protein into the periplasm. Thus, we created a set of different bacterial plasmids coding for different versions of PgtCDA to check for cytoplasmic and periplasmic expression in different bacterial expression strains. The strains Rosetta2(DE3)/pLysSRARE from Merck (Darmstadt, Germany), BL21(DE3) from Novagen (Merck, Germany), E. coli K-12 ER2508 from New England BioLabs (NEB), and Rosetta2(DE3)/pLysSRARE/pCDFDuet-1::DsbC/TrxA were used. The plasmid pCDFDisul, namely, pCDFDuet-1 (Novagen) containing DsbC/TrxA, encodes a cytoplasm-localized version of the disulfide bridge isomerase DsbC and thioredoxin A, thus generating a more-oxic cytoplasm to help in disulfide bridge formation. pCDFDisul was constructed by cloning the optimized open reading frames (ORFs) (GenScript, Piscataway, NJ, USA) of the disulfide bridge isomerase DsbC from E. coli K-12 without the leader peptide-encoding sequence (amino acids [aa] 13 to 228; GenBank accession no. AMH34137.1) via NcoI/HindIII and thioredoxin A (trxA) in its Grx-like form from E. coli K-12 (G34P, P35Y; GenBank accession no. BAE77517.1) via NdeI/XhoI in pCDFDuet-1 (Novagen, Madison, WI).
Regardless of the plasmid used, the recombinant protein PgtCDA-Strep II had an expected molecular mass of 29.21 kDa and a theoretical isoelectric point of 7.31, and the same construct with MBP had an expected molecular mass of 71.81 kDa with a theoretical isoelectric point of 5.69, as deduced by the in silico tool Sci-Ed software clone manager.
Protein purification from E. coli strains.
Transformation of competent bacterial strains with PgtCDA plasmids (51) was followed by the induction of protein production and purification. LB medium (1 liter) was inoculated with the corresponding bacterial strains and the requisite antibiotics (100 μg · ml−1 ampicillin, 34 μg · ml−1 chloramphenicol, 25 μg · ml−1 kanamycin, and 50 μg · ml−1 streptomycin final concentrations). The final culture was incubated for 48 h at 120 rpm and 26°C. Autoinduction medium was used as described by Studier (52), followed by a second induction using 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) 3 h before harvesting the cells. The cells were centrifuged at 4,000 × g and 4°C for 30 min, and the supernatant was discarded. The pellet was dissolved in 50 ml of 20 mM triethanolamine (TEA) buffer (pH 8.0). During the freeze-thaw lysis, the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF, 1 mM; Sigma-Aldrich, Munich, Germany), was added. The cell pellet was sonicated after the addition of 4 μl (100 U) of Benzonase nuclease (Merck KGaA, Darmstadt, Germany) dissolved in 10 mM MgCl2, which reduced the turbidity and sticky texture of the lysate. The crude extract was obtained by centrifuging the lysate at 40,000 × g and 4°C for 30 min. The supernatant was then loaded onto the affinity column for purification. The Strep-Tactin affinity chromatography was carried out using a 1-ml Strep-Tactin superflow Plus cartridge (Qiagen, Helden, Germany) with the ÄKTAprime plus fast protein liquid chromatography (FPLC) system (GE Healthcare, Freiburg, Germany). The fractions with CDA activity were pooled and concentrated using Amicon Ultra 15-ml centrifugal filters (10 kDa; Merck Millipore, Billerica, MA) and dialyzed against 20 mM TEA buffer (pH 8.0). The purified proteins were concentrated to a 1-ml volume and stored at 4°C after adding 10% glycerol.
Zymography by seminative activity PAGE.
The protein purity and activity on glycol chitin were determined by zymography. A seminative polyacrylamide gel containing 1% glycol chitin as the substrate was run at 4°C at 20 mA after loading 2 μg of recombinant enzyme per lane. The gel was washed with 20 mM TEA buffer (pH 8.0) containing 10% Triton X-100 and incubated overnight in 20 mM TEA buffer (pH 8.0). The protein activity was observed under UV light after staining the gel with calcofluor white M2R (Sigma) before and after treatment with sodium nitrite and sulfuric acid that depolymerized glycol chitosan produced by the action of CDA (53).
CDA activity assay.
After protein purification, the CDA activity assay was used for all subsequent optimization and characterization experiments. CDA activity was estimated by measuring the amount of acetate released using a coupled enzymatic assay. The reactions were stopped by adding 10 μl of 5% formic acid. The acetate assay kit from R-Biopharm (Darmstadt, Germany) was diluted (by a factor of 10) for use in a microtiter plate in a total reaction volume of 400 μl, and the increase in NADH + H+ was measured spectrophotometrically at a wavelength of 340 nm using a Multiskan GO microplate spectrophotometer (Thermo Scientific, Germany). All experiments reported in the study were carried out a minimum of three times and in triplicate, using three separate batches of purified proteins.
Comparison of expression strains.
We then determined which recombinant protein had the highest specific activity (nanokatals per nanomole). A total reaction volume of 400 μl of 50 mM TEA buffer (pH 8) containing 210 nmol chitin hexamer (A6) or 250 μg of COS mixture was incubated with 0.21 nmol of each enzyme for 4 h at 37°C, and then the acetate assay was performed. The total protein yield (in nanomoles per milliliter and nanokatals per milliliter) from each expression strain was also determined. However, for all subsequent characterization and mode-of-action experiments, we used the periplasmic MBP-PgtCDA-Strep II enzyme that displayed the maximum specific activity, here referred to as recombinant PgtCDA.
Optimal pH and temperature.
To determine the optimum pH, recombinant PgtCDA (0.207 nmol) was incubated for 1 h with the COS mixture (250 μg) at 37°C in 400 μl of the universal buffer (40 mM NH4HCO3, 20 mM Na2HPO4, 20 mM KH2PO4, 20 mM TEA) adjusted to pH values in the range of pH 3 to 10.
The optimal temperature was determined by incubating the same recombinant enzyme (0.207 nmol) at temperatures ranging from 10°C to 60°C for 1 h with the COS mixture (250 μg) in 400 μl of 50 mM TEA buffer (pH 8.0).
Effect of metal cofactors and EDTA.
Since most CDAs are metalloenzymes, we tested if adding metal cofactors or EDTA affected enzyme activity. Recombinant PgtCDA (0.207 nmol) was preincubated for 1 h at room temperature with 1 mM EDTA and 1 mM metal ions in the form of chloride salts (MgCl2, KCl, CaCl2 MnCl2, CoCl2, ZnCl2, FeCl2, and CuCl2) in 400 μl of 50 mM TEA buffer (pH 8.0). This was followed by addition of substrate (250 μg of COS mixture) and further incubation for 1 h at 37°C for monitoring the activity of the enzyme.
Activity on substrates: chitin polymers, chitosan polymers, and chitin oligomers.
To assess CDA activity on different substrates, recombinant PgtCDA (0.207 nmol) was incubated with 0.5 mg · ml−1 different chitin polymers (α-chitin, β-chitin, and colloidal chitin) and chitosan polymers with different degrees of acetylation (DAs, 32%, 40%, 50%, and 61%) in 400 μl of 50 mM TEA buffer (pH 8.0) for 5 h at 37°C.
To further assess CDA activity on smaller substrates, chitin oligomers (1 mg · ml−1) ranging from degree of polymerization (DP) 1 to 6, namely, chitin monomers to hexamers (A1 to A6), were incubated with periplasmic recombinant PgtCDA (0.207 nmol) for 30 min each at 37°C in 400 μl of 50 mM TEA buffer (pH 8.0).
HP-TLC.
Recombinant CDAs were tested for activity against chitin oligomers ranging from chitin dimers (A2) to hexamers (A6) using high-performance thin-layer chromatography (HP-TLC) (see the supplemental material). The chitin oligomers (1 mg · ml−1) were incubated at 37°C with 0.34 nmol recombinant enzyme overnight in a 20-μl reaction mixture in 20 mM TEA buffer (pH 8.0). The enzyme was removed from the samples using centrifugal filters (modified PES 3K; VWR, Germany) with a cutoff of 3 kDa, and the flowthrough was used for HP-TLC analysis. From the flowthrough, 10 μl of the sample was applied via Camag automatic TLC sampler 4 (Camag, Berlin, Germany) to the silica plates (60 F254 plates from Merck, Darmstadt, Germany). A mixture of butanol-methanol-ammonia-water (5:4:2:1) was used as the mobile phase. The bands were visualized using a protocol modified from that of Gal (54). After solvent evaporation, the silica plate was dipped in 30% ammonium bisulfate solution. Finally, a stream of hot air was used to visualize the bands on the plate.
Ultraperformance liquid chromatography–evaporative light-scattering detection–electrospray ionization–mass spectrometry in MSn mode.
To analyze the gradual formation of differentially deacetylated products over time, chitin oligomers (1 mg · ml−1) with DP 4 to 6 (A4, A5, and A6) were incubated in a master mix containing 0.6 nmol recombinant PgtCDA in a total reaction volume of 200 μl of 50 mM TEA buffer (pH 8.0), from which 30-μl aliquots were evaluated over time (at 3, 6, 12, 24, and 48 h, respectively). The reactions were stopped by adding 6 μl of 5% formic acid, and the protein was removed by centrifuging the samples for 30 min at 13,000 × g in centrifugal filters (modified PES 3K; VWR, Germany), with a cutoff of 3 kDa. These samples were analyzed with mass spectrometry (24) after previous re-N-acetylation with [2H6]acetic anhydride (MS1) and 18O-labeling (MS2), using a modified protocol based on Lavertu et al. (55) and Remoroza et al. (56) (S. Cord-Landwehr, P. Ihmor, H. Luftmann, B. M. Moerschbacher, and M. Mormann, unpublished data), allowing us to quantify the partially deacetylated intermediates and final products and also to determine the sequence of deacetylation caused by the enzyme.
RESULTS AND DISCUSSION
Bioinformatic analysis of the chitin deacetylase PgtCDA gene.
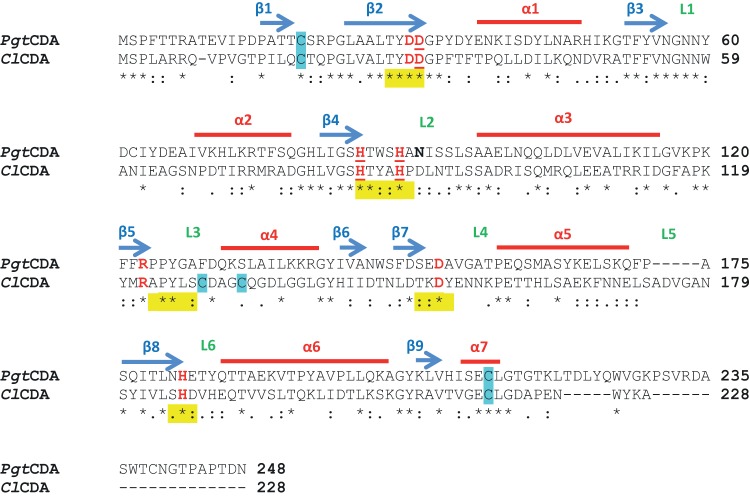
The deduced protein from P. graminis f. sp. tritici, PgtCDA (49), was analyzed by a conserved domain search on NCBI (CDD) (57), revealing the presence of the polysaccharide deacetylase NodB homology domain, characteristic of chitin deacetylases (EC 3.5.1.41), containing the five distinct sequence motifs (MT 1 to 5, also known as CE4 motif) (Fig. 1). No other domains, such as the substrate-binding domains, were identified. The amino acid sequence alignment (Clustal Omega 2.1) (43, 58) of PgtCDA compared to ClCDA from C. lindemuthianum, which has been characterized extensively (25, 33), showed an amino acid identity of 33.63%. Analysis of PgtCDA by I-TASSER (59–61) and SWISS-MODEL (62) predicted that ClCDA's crystal structure (PDB 2IWO) was the closest template for PgtCDA modeling, which was done using PyMOL (Molecular Graphics System, version 1.8) (see Fig. S2A in the supplemental material) (46). After KoBaMIN refinement, MolProbity analysis showed that >94% of the residues were present in the Ramachandran-favored region, representing a good-quality model for further analysis. The model shows good agreement among the secondary structure elements. Like ClCDA, the modeled structure of PgtCDA contains seven alpha-helices and nine beta-sheets, forming an inner-core beta-sheet barrel surrounded by outer-core alpha-helices. Also, the connections between helices and sheets as shown in the Pro-origami diagram (see Fig. S2B in the supplemental material) are similar to the ClCDA topological diagram (25).
FIG 1.
Clustal Omega amino acid sequence alignment of PgtCDA from P. graminis and ClCDA (PDB 2IWO) from C. lindemuthianum. The sequence analysis shows comparison of amino acid sequences of ClCDA and PgtCDA without signal peptide removed using SignalP 3.0 server. Alpha-helices and beta-sheets are marked by red lines and blue arrows, respectively, and the positions of loops (L) decorating the active site are indicated in green. The five conserved motifs of CE4 enzymes are marked in yellow. The catalytic and metal ligand binding (underlined) residues are marked in red. The cysteines predicted in disulfide bonds are marked in blue. One potential N-glycosylation site is marked in bold. The consensus symbols have the usual meanings, i.e., asterisks (*) indicate positions which have a single, fully conserved residue, colons (:) indicate conservation between groups of strongly similar properties, and periods (.) indicate conservation between groups of weakly similar properties.
Since some fungal CDAs are known to be glycosylated (13), the deduced PgtCDA protein was analyzed for potential N-glycosylation sites (NetNGlyc1.0 server) (63); one such site was identified to be Asn91 (marked on Fig. 1). However, after visually inspecting PgtCDA on the PyMOL viewer (PyMOL Molecular Graphics System, version 1.8) (46), Asn91 was found to be located on the surface of the protein. Since ClCDA, as PgtCDA's most closely related CDA, is not glycosylated, even when expressed in P. pastoris (33), we assumed that PgtCDA is also not a glycoprotein. In addition to glycosylation, we assessed if and where disulfide bonds might form in PgtCDA. The DiANNA server (64) predicted that a single disulfide bond forms between Cys19 and Cys213. On homology modeling using the ClCDA structure as a template, we found that these conserved cysteine residues in PgtCDA correspond to the first disulfide bridge in ClCDA (Cys18-Cys217). PgtCDA does not contain the corresponding cysteines that form ClCDA's second disulfide bond.
Determining which recombinant PgtCDA had the highest specific activity.
Expressing fungal enzymes in bacterial systems is often difficult, but several factors, such as codon optimization (65), solubility-enhancing fusion proteins (66), or varying subcellular localization (67), can be helpful for enhancing protein expression and folding. So far, only a very few fungal CDAs have been successfully expressed in E. coli. A. nidulans CDA was obtained by cytoplasmic expression in E. coli, leading to the formation of inclusion bodies from which the active enzyme had to be refolded (38). CDA2 from S. cerevisiae was obtained as an active enzyme in the periplasmic space of E. coli (37), while ClCDA was secreted from the bacterial cells into the medium with the help of a signal sequence from Streptomyces lividans (36).
To systematically study the recombinant expression of PgtCDA in E. coli, the codon-optimized gene was cloned in vectors containing a Strep II tag for purification at the C terminus but not containing the N-terminal 22-amino-acid signal sequence as predicted by the SignalP 3.0 server (68). Additionally, the vectors either did or did not have the N-terminal addition of the maltose-binding protein (MBP), and they either did or did not have the signal peptide of MBP for periplasmic secretion.
When the recombinant enzymes resulting from the different constructs were purified and their activities toward chitin hexamer (A6) were compared (Table 1 and Fig. 2), we found that the constructs producing cytoplasmic PgtCDA without the MBP (CDA1 to CDA3) showed the lowest yields and lowest specific activities. Cytoplasmic production of MBP-PgtCDA fusion proteins (CDA4 to CDA6) showed higher yields and higher specific activities, and secretion of the fusion proteins into the periplasm (CDA7 and CDA8) gave even higher specific activities, although with slightly lower yields. Expressing the genes in E. coli Rosetta2 containing the pLysSRARE plasmid for better handling of rare codons or in a strain containing pLysSRARE in addition the pCDFDisul plasmid (which allowed for better formation of disulfide bonds), negatively affected production of the active enzyme. The secretion of MBP-PgtCDA fusion protein into the periplasm (CDA8) gave the highest specific activity and very good yields, possibly caused by the positive effect of an oxidizing milieu in the periplasm favoring the formation of the disulfide bond predicted in PgtCDA, and the presence of MBP, which can aid protein folding (69). This enzyme was therefore used for further characterization.
TABLE 1.
Different bacterial expression strains and plasmids used in the expression study to determine the strain yielding recombinant PgtCDA with the highest yield and highest specific activity
| Construct no. | Expression strain | Plasmid | Protein yield (nmol/ml) | Enzyme activity (nkat/ml) | Specific activity (nkat/nmol) |
|---|---|---|---|---|---|
| CDA1 | Rosetta disulfide/pLysSRARE/pCDFDuet1-DsbC/TrxA | pET22b::PgtCDA-Strep IIC | 3.08 | 2 | 1 |
| CDA2 | Rosetta2(DE3)/pLysSRARE | pET22b::PgtCDA-Strep IIC | 2.26 | 58 | 25 |
| CDA3 | BL21(DE3) | pET22b::PgtCDA-Strep IIC | 2.45 | 202 | 82 |
| CDA4 | Rosetta disulfide/pLysSRARE/pCDFDuet1-DsbC/TrxA | pET22b::MBP-PgtCDA-Strep IIC | 4.38 | 719 | 164 |
| CDA5 | Rosetta2(DE3)/pLysSRARE | pET22b::MBP- PgtCDA-Strep IIC | 4.85 | 1,001 | 206 |
| CDA6 | BL21(DE3) | pET22b::MBP-PgtCDA-Strep IIC | 6.57 | 1,700 | 258 |
| CDA7 | ER2508 | pMAL-p5x::MBP-PgtCDA-Strep IIC | 1.70 | 697 | 410 |
| CDA8 | Rosetta2(DE3)/pLysSRARE | pMAL-p5x::MBP-PgtCDA-Strep IIC | 3.30 | 1,485 | 450 |
FIG 2.
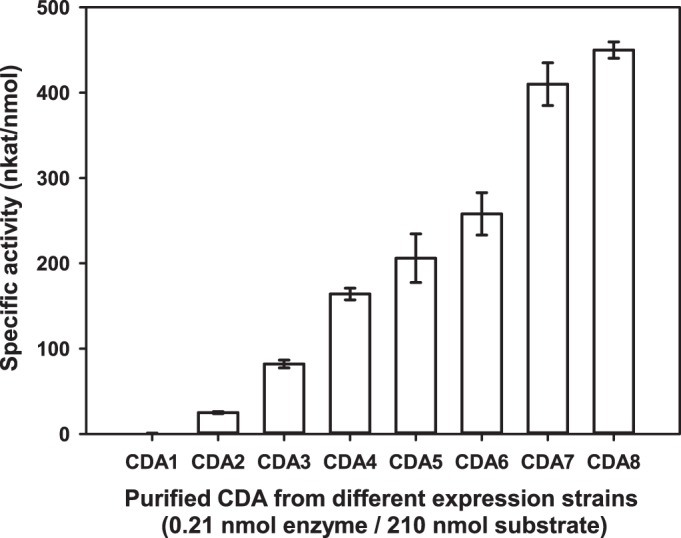
Specific enzyme activities of recombinant PgtCDA produced in different bacterial expression strains. PgtCDA (0.21 nmol) from each construct was incubated with chitin hexamer as the substrate (210 nmol) at 37°C for 4 h in 50 mM TEA buffer (pH 8.0) to determine specific activity of the enzyme. Strain numbers refer to those shown in Table 1. Data given represent mean ± standard deviation (SD) values of the results from three independent experiments.
How temperature, pH, metal ions, and EDTA affected enzyme activity.
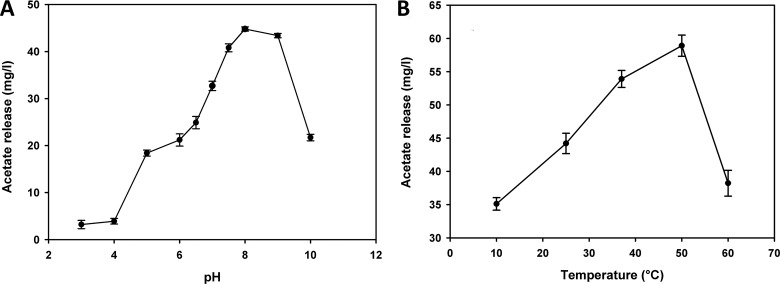
The recombinant PgtCDA showed an optimum pH of 8 to 9 (Fig. 3A) and an optimum temperature of 50°C (Fig. 3B) toward a mixture of chitin pentamers and tetramers (A5/A4). As the PgtCDA gene has a signal peptide (which was removed for expression in E. coli), the native enzyme is expected be secreted into the medium. Most extracellular fungal CDAs, such as ClCDA ATCC 56676, ClCDA DSM 16344, ClCDA UPS9, A. nidulans CDA, and S. cerevisiae CDA2p, have been reported to have an optimum pH ranging between pH 7 and 12 and an optimum temperature in the range of 50°C to 60°C (13, 26, 31, 33, 36, 70, 71).
FIG 3.
Effects of pH (A) and temperature (B) on enzyme activity. (A) The optimal pH was determined at 37°C by incubating the enzyme (0.207 nmol) at different pH values in universal buffer (40 mM NH4HCO3, 20 mM Na2HPO4, 20 mM KH2PO4, 20 mM TEA) with the A5-A4 chitin oligomer mixture (250 μg) as a substrate for 1 h. (B) For optimum temperature, the enzyme (0.207 nmol) was subjected to incubation at different temperatures in 50 mM TEA buffer (pH 8.0) with the chitin oligomer mixture (250 μg) as a substrate for 1 h. Data given represent the mean ± SD values of the results from three independent experiments.
Chitin deacetylases are metalloenzymes requiring divalent cations, such as Zn2+, in their active site (25), and the addition of metal ions, such as Co2+ in Mortierella sp. DY52 CDA and in F. velutipes CDA, or, to a minor extent, Ca2+ in these two CDAs as well as in A. nidulans CDA, have been reported to enhance activity (13, 34, 38, 71). However, for the recombinant PgtCDA, the presence of 1 mM EDTA did not inhibit the enzyme's activity, and the presence of 1 mM metal ions (Mg2+, K+, Ca2+, Mn2+, Co2+, Zn2+, Fe2+, and Cu2+ as chloride salts) did not increase its activity (see Fig. S3 in the supplemental material). Possibly, the metal ion is tightly bound in the catalytic site of the enzyme and is thus unable to equilibrate with the solution, as was also observed for ClCDA (25).
How chitin deacetylase affected different chitinous substrates.
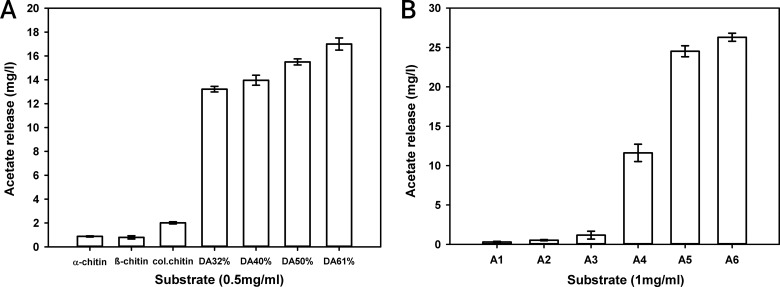
Recombinant PgtCDA was almost inactive toward insoluble α- and β-chitin and showed only slight activity toward colloidal chitin (Fig. 4A), but it was active toward the soluble chitin derivative glycol-chitin in a seminative zymography assay (see Fig. S4 in the supplemental material). The enzyme was also active on soluble partially acetylated chitosan polymers, and activity increased with increasing degree of acetylation. These properties resemble results that have been reported for CDAs from Mucor rouxii, A. nidulans, and F. velutipes (26, 34, 72).
FIG 4.
Enzyme activity on chitin and chitosan polymers (A) and chitin oligomers (B). (A) Different substrates: α-chitin, β-chitin, colloidal chitin (col.chitin), and chitosan polymers, with degree of acetylation (DA) of 32 to 61%, with a final concentration of 0.5 mg · ml−1 were incubated with PgtCDA (0.207 nmol) at 37°C for 5 h. (B) Chitin oligomers (final concentration, 1 mg · ml−1) with DP 1 to 6, i.e., monomers to hexamers, were treated with PgtCDA (0.207 nmol) for 30 min at 37°C. Data given represent the mean ± SD values of the results from three independent experiments.
PgtCDA was inactive toward very small chitin oligomers of DP 1 to 3 (A1 to A3), but its activity increased with increasing DP from 4 to 6 (A4 to A6) (Fig. 4B). HP-TLC analysis showed the formation of multiple deacetylation products from DP 4 to 6 (see Fig. S5 in the supplemental material). This is a significant difference compared to ClCDA, which has shown activity toward chitin dimers and trimers (73). Thus, recombinant PgtCDA has a more limited oligomer substrate range than ClCDA and CDAs from A. nidulans, Mortierella spp., and F. velutipes, all of which are active on dimers and larger oligomers (26, 34, 73, 74). It is also slightly more limited than the CDA from M. rouxii, which is inactive on GlcNAc monomers and dimers but can deacetylate chitin oligomers that are trimers or larger (27).
Sequential de-N-acetylation of chitin oligomers by recombinant PgtCDA.
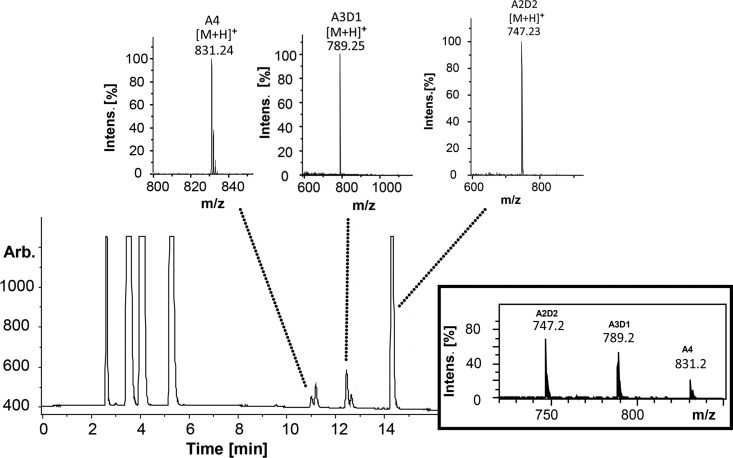
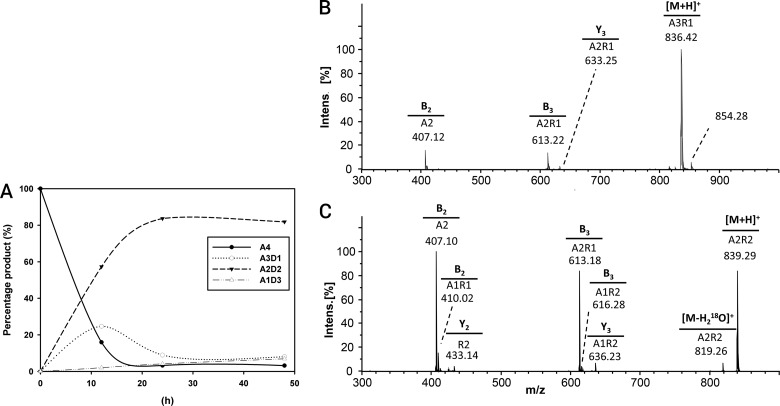
By re-N-acetylating the enzymatic products of chitin oligomers using [2H6]acetic anhydride, labeling their reducing ends using 18O, and then applying ultraperformance liquid chromatography–evaporative light-scattering detection–electrospray ionization–mass spectrometry in MSn mode (UHPLC-ELSD-ESI-MSn) analysis, we were able to quantify recombinant PgtCDA's partially deacetylated intermediate and final products and determine the products' sequence of deacetylation. The MS1 spectra for chitin tetramer deacetylation are shown in Fig. 5; the time course of deacetylation is given in Fig. 6A (for chitin pentamers and hexamers, see the supplemental material). Invariably, the final products of recombinant PgtCDA digestion of chitin oligomers (An with n = 4 to 6) were paCOS, with two remaining acetylated units [A2D(n−2)]. Only upon very long incubation times (>24 h) or when excessive enzyme was added was further and eventually complete deacetylation observed. This is in stark contrast to the results reported for ClCDA, which readily proceeds to complete deacetylation yielding fully deacetylated glucosamine oligomers (Dn).
FIG 5.
UHPLC-ELSD-ESI-MS analysis of chitin tetramer (A4) (1 mg · ml−1) treated with PgtCDA (0.6 nmol). The ELSD chromatogram shows the elution of initial buffer peaks (2 to 6 min) followed by target peaks. The m/z ratios in the MS spectra correspond to the mass of the substrate (A4; m/z 831.24), which after incubation with enzyme gives rise to single- and double-deacetylated products A3D1 (m/z 789.25) and A2D2 (m/z 747.23), respectively. A combined spectrum is also attached for an overview. See the supplemental material for the same analysis of chitin pentamer (A5) and hexamer (A6) treated with PgtCDA. intens., intensity; Arb., arbitrary units.
FIG 6.
(A to C) Time curve of deacetylation (A) and hydrophilic interaction liquid chromatography-tandem mass spectrometry (HILIC-MS2) chromatogram of the deacetylation products from PgtCDA (0.6 nmol) incubated with chitin tetramer (A4) (1 mg · ml−1) as the substrate, giving single deacetylation A-A-D-A (B) and then double deacetylation A-A-D-D (C). After incubation with the enzyme for different lengths of time, the products were re-N-acetylated with [2H6]acetic anhydride, 18O-labeled at the reducing ends, followed by UHPLC-ELSD-ESI-MSn. (B and C) The precursor ion m/z values and the MS2 spectra for sequencing the precursor ion are shown. See the supplemental material for the same analysis of chitin pentamer (A5) and hexamer (A6) treated with PgtCDA.
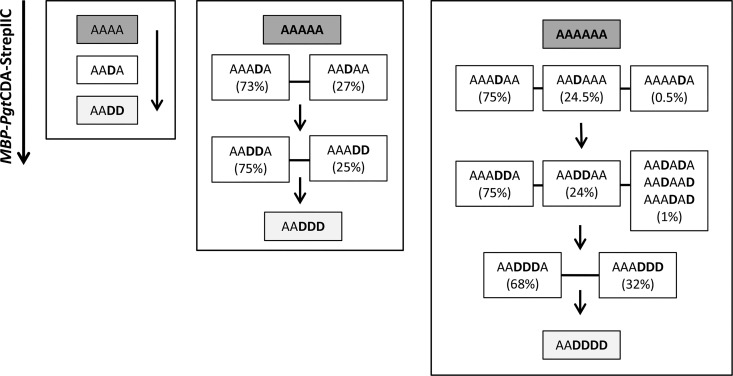
In a comparison of the stepwise deacetylation of chitin tetramers, pentamers, and hexamers, we found that the first deacetylation never occurred at the two units closest to the nonreducing end, nor at the reducing end unit (Fig. 7). Thus, the chitin tetramer (A4 [A-A-A-A]) was first deacetylated to A-A-D-A (Fig. 6B), followed by deacetylation at the reducing end, yielding the final product A-A-D-D (Fig. 6C). The first deacetylation in the chitin pentamer and hexamer (An with n = 5 or 6) occurred at the fourth or third unit from the nonreducing end in a ratio of 3:1, yielding A3-D-A(n−4) (i.e., A-A-A-D-A and A-A-A-D-A-A) as a major product and A2-D-A(n−3) (i.e., A-A-D-A-A and A-A-D-A-A-A) as a minor product. The monodeacetylated hexamer A4-D-A (i.e., A-A-A-A-D-A) occurred in trace amounts only. The second deacetylation of the pentamer yielded the products A-A-D-D-A and A-A-A-D-D in a 3:1 ratio, showing that deacetylation of the reducing end unit is disfavored, and the third deacetylation then yielded A-A-D-D-D as the final product. The second deacetylation of the hexamer yielded the products A-A-A-D-D-A and A-A-D-D-A-A in a 3:1 ratio (and traces of other patterns); the third deacetylation yielded more than twice as much A-A-D-D-D-A than A-A-A-D-D-D (and traces of other patterns), again showing that deacetylation at the reducing end is less favored. Finally, the fourth deacetylation then yielded A-A-D-D-D-D as the final product. A similar analysis has been reported for C. lindemuthianum CDA but only up to a DP of 4 where, like for recombinant PgtCDA, the first deacetylation is two units from the nonreducing end, giving rise to A-A-D-A. However, subsequent steps lead to a complete deacetylation with intermediate products (such as D-A-D-A and A-D-D-A, and finally D-D-D-D) (73).
FIG 7.
Sequence of deacetylation of chitin oligomers by PgtCDA. Chitin tetramers to hexamers were treated with PgtCDA, and at different time points, the products were analyzed as described in the Fig. 6 legend.
This prominent difference in the products formed by the CDAs from P. graminis and C. lindemuthianum is very interesting, especially given that both enzymes are rather closely related, as detailed above. The product patterns of CDAs have recently been explained by a subsite capping model based on the crystal structure of the bacterial CDA COD from Vibrio compared to those of other CDAs. The subsite capping model states that the position and movement of loops play a potent role in substrate preference and regioselectivity of the deacetylation, thus determining the pattern of acetylation of the chitosan oligomers produced (75). Such a detailed structural analysis and comparison with other CDAs, however, will have to wait until the crystal structure of PgtCDA has been solved.
In conclusion, this study reports the bacterial expression and characterization of a fungal chitin deacetylase, PgtCDA, from the basidiomycete P. graminis. It elucidates the mode of action of recombinant PgtCDA on a range of substrates, such as chitin and chitosan polymers, as well as on chitin oligomers. Recombinant PgtCDA produces paCOS with a fully defined structure, namely, A-A-Dn (with n > 1), a novel pattern that has not been reported so far for CDAs that produce specific deacetylations. CDAs, like PgtCDA, thus emerge as a powerful tool for the production of defined paCOS that cannot currently be produced in any other way. These paCOS can be further analyzed for their structure-function relationships and used in various applications, such as plant protection or biomedicine, based on their biological activities (4, 76).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to André Nordhues (IBBP, University of Münster) and Subhanarayan Das (University of Hyderabad) for scientific suggestions and discussions. We thank Victoria Schneider for her technical assistance and Florian Raths for inputs on the graphical data. S.N. gratefully acknowledges Celeste R. Brennecka for editing the manuscript.
We thank the European Union for funding via the European Union Sixth Framework Programme (grant FP6/2002-2006) in the framework of the European Research Area-Industrial Biotechnology (ERA-IB) project “ChitoBioEngineering” under grant agreement EIB.10.042. S.N. also gratefully acknowledges the doctoral fellowship from Deutscher Akademischer Austauschdienst (DAAD).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01961-16.
REFERENCES
- 1.Dutta J, Tripathi S, Dutta PK. 2012. Progress in antimicrobial activities of chitin, chitosan and its oligosaccharides: a systematic study needs for food applications. Food Sci Technol Int 18:3–34. doi: 10.1177/1082013211399195. [DOI] [PubMed] [Google Scholar]
- 2.Kong M, Chen XG, Xing K, Park HJ. 2010. Antimicrobial properties of chitosan and mode of action: a state of the art review. Int J Food Microbiol 144:51–63. doi: 10.1016/j.ijfoodmicro.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 3.El Hadrami A, Adam LR, El Hadrami I, Daayf F. 2010. Chitosan in plant protection. Mar Drugs 8:968–987. doi: 10.3390/md8040968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das SN, Madhuprakash J, Sarma PVSRN, Purushotham P, Suma K, Manjeet K, Rambabu S, Gueddari El NE, Moerschbacher BM, Podile AR. 2015. Biotechnological approaches for field applications of chitooligosaccharides (COS) to induce innate immunity in plants. Crit Rev Biotechnol 35:29–43. doi: 10.3109/07388551.2013.798255. [DOI] [PubMed] [Google Scholar]
- 5.Aam BB, Heggset EB, Norberg AL, Sørlie M, Vårum KM, Eijsink VGH. 2010. Production of chitooligosaccharides and their potential applications in medicine. Mar Drugs 8:1482–1517. doi: 10.3390/md8051482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altiok D, Altiok E, Tihminlioglu F. 2010. Physical, antibacterial and antioxidant properties of chitosan films incorporated with thyme oil for potential wound healing applications. J Mater Sci Mater Med 21:2227–2236. doi: 10.1007/s10856-010-4065-x. [DOI] [PubMed] [Google Scholar]
- 7.Vander P, Vårum K, Domard A, Eddine El Gueddari N, Moerschbacher BM. 1998. Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol 118:1353–1359. doi: 10.1104/pp.118.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Gueddari NE, Schaaf A, Kohlhoff M, Gorzelanny C, Schneider SW, Moerschbacher BM. 2007. Substrates and products of chitin and chitosan modifying enzymes. Adv Chitin Sci 10:119–126. [Google Scholar]
- 9.El Gueddari NE, Kolkenbrock S, Schaaf A, Chilukoti A, Brunel F, Gorzelanny C, Fehser S, Chachra S, Bernard F, Nampally M, Kalagara T, Ihmor P, Moerschbacher BM. 2014. Chitin and chitosan modifying enzymes: versatile novel tools for the analysis of structure-function relationships of partially acetylated chitosans. Adv Chitin Sci 14:40–47. [Google Scholar]
- 10.Vårum KM, Ottoy MH, Smidsrod O. 1994. Water-solubility of partially N-acetylated chitosans as a function of pH: effect of chemical composition and depolymerisation. Carbohydr Polym 25:65–70. doi: 10.1016/0144-8617(94)90140-6. [DOI] [Google Scholar]
- 11.Trombotto S, Ladavière C, Delolme F, Domard A. 2008. Chemical preparation and structural characterization of a homogeneous series of chitin/chitosan oligomers. Biomacromolecules 9:1731–1738. doi: 10.1021/bm800157x. [DOI] [PubMed] [Google Scholar]
- 12.Tsigos I, Martinou A, Kafetzopoulos D, Bouriotis V. 2000. Chitin deacetylases: new, versatile tools in biotechnology. Trends Biotechnol 18:305–312. doi: 10.1016/S0167-7799(00)01462-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y, Park R-D, Muzzarelli RAA. 2010. Chitin deacetylases: properties and applications. Mar Drugs 8:24–46. doi: 10.3390/md8010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duo-Chuan L. 2006. Review of fungal chitinases. Mycopathologia 161:345–360. doi: 10.1007/s11046-006-0024-y. [DOI] [PubMed] [Google Scholar]
- 15.Jung WJ, Park RD. 2014. Bioproduction of chitooligosaccharides: present and perspectives. Mar Drugs 5328–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinhold MX, Sauvageau JCM, Kumirska J, Thöming J. 2009. Studies on acetylation patterns of different chitosan preparations. Carbohydr Polym 78:678–684. doi: 10.1016/j.carbpol.2009.06.001. [DOI] [Google Scholar]
- 17.Domard A, Cartier N. 1989. Glucosamine oligomers: 1. Preparation and characterization. Int J Biol Macromol 11:297–302. doi: 10.1016/0141-8130(89)90023-8. [DOI] [PubMed] [Google Scholar]
- 18.Huang G-L, Zhang D-W, Zhao H-J, Zhang H-C, Wang P-G. 2006. Chemo-enzymatic synthesis of allyl penta-N-acetyl-chitopentaose. Bioorg Med Chem Lett 16:2042–2043. doi: 10.1016/j.bmcl.2005.12.055. [DOI] [PubMed] [Google Scholar]
- 19.Samain E, Drouillard S, Heyraud A, Driguez H, Geremia RA. 1997. Gram-scale synthesis of recombinant chitooligosaccharides in Escherichia coli. Carbohydr Res 302:35–42. doi: 10.1016/S0008-6215(97)00107-9. [DOI] [PubMed] [Google Scholar]
- 20.Tang M-C, Nisole A, Dupont C, Pelletier JN, Waldron KC. 2011. Chemical profiling of the deacetylase activity of acetyl xylan esterase A (AxeA) variants on chitooligosaccharides using hydrophilic interaction chromatography-mass spectrometry. J Biotechnol 155:257–265. doi: 10.1016/j.jbiotec.2011.06.041. [DOI] [PubMed] [Google Scholar]
- 21.John M, Röhrig H, Schmidt J, Wieneke U, Schell J. 1993. Rhizobium NodB protein involved in nodulation signal synthesis is a chitooligosaccharide deacetylase. Proc Natl Acad Sci U S A 90:625–629. doi: 10.1073/pnas.90.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadokura K, Sakamoto Y, Saito K, Ikegami T, Hirano T, Hakamata W, Oku T, Nishio T. 2007. Production of a recombinant chitin oligosaccharide deacetylase from Vibrio parahaemolyticus in the culture medium of Escherichia coli cells. Biotechnol Lett 29:1209–1215. doi: 10.1007/s10529-007-9386-6. [DOI] [PubMed] [Google Scholar]
- 23.Kadokura K, Rokutani A, Yamamoto M, Ikegami T, Sugita H, Itoi S, Hakamata W, Oku T, Nishio T. 2007. Purification and characterization of Vibrio parahaemolyticus extracellular chitinase and chitin oligosaccharide deacetylase involved in the production of heterodisaccharide from chitin. Appl Microbiol Biotechnol 75:357–365. doi: 10.1007/s00253-006-0831-6. [DOI] [PubMed] [Google Scholar]
- 24.Hamer SN, Cord-Landwehr S, Biarnés X, Planas A, Waegeman H, Moerschbacher BM, Kolkenbrock S. 2015. Enzymatic production of defined chitosan oligomers with a specific pattern of acetylation using a combination of chitin oligosaccharide deacetylases. Sci Rep 5:8716. doi: 10.1038/srep08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blair D, Hekmat O, Schüttelkopf AW, Shrestha B, Tokuyasu K, Withers SG, van Aalten DM. 2006. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 45:9416–9426. doi: 10.1021/bi0606694. [DOI] [PubMed] [Google Scholar]
- 26.Alfonso C, Nuero OM, Santamarfa F, Reyes F. 1995. Purification of a heat-stable chitin deacetylase from Aspergillus nidulans and its role in cell wall degradation. Curr Microbiol 30:49–54. doi: 10.1007/BF00294524. [DOI] [PubMed] [Google Scholar]
- 27.Tsigos I, Zydowicz N, Martinou A, Domard A, Bouriotis V. 1999. Mode of action of chitin deacetylase from Mucor rouxii on N-acetylchitooligosaccharides. Eur J Biochem 261:698–705. [DOI] [PubMed] [Google Scholar]
- 28.Higgins DR. 2001. Overview of protein expression in Pichia pastoris. Curr Protoc Protein Sci Chapter 5:Unit 5.7. doi: 10.1002/0471140864.ps0507s02. [DOI] [PubMed] [Google Scholar]
- 29.Cregg JM, Cereghino JL, Shi J, Higgins DR. 2000. Recombinant protein expression in Pichia pastoris. Mol Biotechnol 16:23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 30.Mishra C, Semino CE, McCreath KJ, de la Vega H, Jones BJ, Specht CA, Robbins PW. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327–336. doi:. [DOI] [PubMed] [Google Scholar]
- 31.Martinou A, Koutsioulis D, Bouriotis V. 2002. Expression, purification, and characterization of a cobalt-activated chitin deacetylase (Cda2p) from Saccharomyces cerevisiae. Protein Expr Purif 24:111–116. doi: 10.1006/prep.2001.1547. [DOI] [PubMed] [Google Scholar]
- 32.Christodoulidou A, Bouriotis V, Thireos G. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J Biol Chem 271:31420–31425. doi: 10.1074/jbc.271.49.31420. [DOI] [PubMed] [Google Scholar]
- 33.Shrestha B, Blondeau K, Stevens WF, Hegarat FL. 2004. Expression of chitin deacetylase from Colletotrichum lindemuthianum in Pichia pastoris: purification and characterization. Protein Expr Purif 38:196–204. doi: 10.1016/j.pep.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 34.Yamada M, Kurano M, Inatomi S, Taguchi G, Okazaki M, Shimosaka M. 2008. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol Lett 289:130–137. doi: 10.1111/j.1574-6968.2008.01361.x. [DOI] [PubMed] [Google Scholar]
- 35.Gauthier C, Clerisse F, Dommes J, Jaspar-Versali MF. 2008. Characterization and cloning of chitin deacetylases from Rhizopus circinans. Protein Expr Purif 59:127–137. doi: 10.1016/j.pep.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Tokuyasu K, Kaneko S, Hayashi K, Mori Y. 1999. Production of a recombinant chitin deacetylase in the culture medium of Escherichia coli cells. FEBS Lett 458:23–26. doi: 10.1016/S0014-5793(99)01113-8. [DOI] [PubMed] [Google Scholar]
- 37.Martinou A, Koutsioulis D, Bouriotis V. 2003. Cloning and expression of a chitin deacetylase gene (CDA2) from Saccharomyces cerevisiae in Escherichia coli: purification and characterization of the cobalt-dependent recombinant enzyme. Enzyme Microb Technol 32:757–763. doi: 10.1016/S0141-0229(03)00048-6. [DOI] [Google Scholar]
- 38.Wang Y, Song J-Z, Yang Q, Liu Z-H, Huang X-M, Chen Y. 2010. Cloning of a heat-stable chitin deacetylase gene from Aspergillus nidulans and its functional expression in Escherichia coli. Appl Biochem Biotechnol 162:843–854. doi: 10.1007/s12010-009-8772-z. [DOI] [PubMed] [Google Scholar]
- 39.El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB. 2002. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytol 156:103–112. doi: 10.1046/j.1469-8137.2002.00487.x. [DOI] [Google Scholar]
- 40.Lamarque G, Lucas JM, Viton C, Domard A. 2005. Physicochemical behavior of homogeneous series of acetylated chitosans in aqueous solution: role of various structural parameters. Biomacromolecules 6:131–142. doi: 10.1021/bm0496357. [DOI] [PubMed] [Google Scholar]
- 41.Hsu SC, Lockwood JL. 1975. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl Microbiol 29:422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Araki Y, Ito E. 1975. A pathway of chitosan formation in Mucor rouxii. Enzymatic deacetylation of chitin. Eur J Biochem 55:71–78. [DOI] [PubMed] [Google Scholar]
- 43.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodrigues J, Levitt M, Chopra G. 2012. KoBaMIN: a knowledge based minimization Web server for protein structure refinement. Nucleic Acids Res 40:W323–W328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson RC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schrödinger LLC. 2010. The AxPyMOL molecular graphics plugin for Microsoft PowerPoint, version-1.8. Schrödinger, LLC, New York, NY. [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. Mol Graph 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Stivala A, Wybrow M, Wirth A, Whisstock JC, Stuckey PJ. 2011. Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics 27:3315–3316. doi: 10.1093/bioinformatics/btr575. [DOI] [PubMed] [Google Scholar]
- 49.Jaszczuk BK, Moerschbacher BM, Schaaf A. June 2014. Nucleic acid molecules coding for a protein with deacetylase activity, said protein, and method for the production of chitosan. European patent EP2148924 B1.
- 50.Schmidt TGM, Skerra A. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc 2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 52.Studier FW. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 53.Trudel J, Asselin A. 1989. Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem 178:362–366. doi: 10.1016/0003-2697(89)90653-2. [DOI] [PubMed] [Google Scholar]
- 54.Gal AE. 1968. Separation and identification of monosaccharides from materials by thin-layer chromatography. Anal Biochem 24:452–461. doi: 10.1016/0003-2697(68)90152-8. [DOI] [PubMed] [Google Scholar]
- 55.Lavertu M, Darras V, Buschmann MD. 2012. Kinetics and efficiency of chitosan reacetylation. Carbohydr Polym 87:1192–1198. doi: 10.1016/j.carbpol.2011.08.096. [DOI] [Google Scholar]
- 56.Remoroza C, Cord-Landwehr S, Leijdekkers AGM, Moerschbacher BM, Schols HA, Gruppen H. 2012. Combined HILIC-ELSD/ESI-MSn enables the separation, identification and quantification of sugar beet pectin derived oligomers. Carbohydr Polym 90:41–48. doi: 10.1016/j.carbpol.2012.04.058. [DOI] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer CR, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwede T, Kopp J, Guex N, Peitsch MC. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pacific Symp Biocomput 7:310–322. [PubMed] [Google Scholar]
- 64.Ferrè F, Clote P. 2006. DiANNA 1.1: an extension of the DiANNA Web server for ternary cysteine classification. Nucleic Acids Res 34:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novy R, Drott D, Yaeger K, Mierendorf R. 2001. Overcoming the codon bias of E. coli for enhanced protein expression. Innov News Lett 12:1–3. [Google Scholar]
- 66.Esposito D, Chatterjee DK. 2006. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol 17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 67.Bassford PJ., Jr 1990. Export of the periplasmic maltose-binding protein of Escherichia coli. J Bioenerg Biomembr 22:401–439. doi: 10.1007/BF00763175. [DOI] [PubMed] [Google Scholar]
- 68.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 69.Kapust RB, Waugh DS. 1999. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci 8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsigos I, Bouriotis V. 1995. Purification and characterization of chitin deacetylase from Colletotrichum lindemuthianum. J Biol Chem 270:26286–26291. doi: 10.1074/jbc.270.44.26286. [DOI] [PubMed] [Google Scholar]
- 71.Tokuyasu K, Ohnishi-Kameyama M, Hiyashi K. 1996. Purification and characterization of extracellular chitin deacetylase from Colletotrichum lindemuthianum. Biosci Biotech Biochem 60:1598–1603. doi: 10.1271/bbb.60.1598. [DOI] [PubMed] [Google Scholar]
- 72.Davis LL, Bartnicki-Garcia S. 1984. The coordination of chitosan and chitin synthesis in Mucor rouxii. J Gen Microbiol 130:2095–2102. [DOI] [PubMed] [Google Scholar]
- 73.Hekmat O, Tokuyasu K, Withers S. 2003. Subsite structure of the endo-type chitin deacetylase from a deuteromycete, Colletotrichum lindemuthianum: an investigation using steady-state kinetic analysis and MS. Biochem J 374:369–380. doi: 10.1042/bj20030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim YJ, Zhao Y, Oh KT, Nguyen VN, Park RD. 2008. Enzymatic deacetylation of chitin by extracellular chitin deacetylase from a newly screened Mortierella sp. DY-52. J Microbiol Biotechnol 18:759–766. [PubMed] [Google Scholar]
- 75.Andrés E, Albesa-Jové D, Biarnés X, Moerschbacher BM, Guerin ME, Planas A. 2014. Structural basis of chitin oligosaccharide deacetylation. Angew Chem Int Ed Engl 53:6882–6887. doi: 10.1002/anie.201400220. [DOI] [PubMed] [Google Scholar]
- 76.Liu H-T, Li W-M, Huang P, Chen W-J, Liu Q-S, Bai X-F, Yu C, Du Y-G. 2010. Chitosan oligosaccharides inhibit TNF-α-induced VCAM-1 and ICAM-1 expression in human umbilical vein endothelial cells by blocking p38 and ERK1/2 signaling pathways. Carbohydr Polym 81:49–56. doi: 10.1016/j.carbpol.2010.01.054. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.