ABSTRACT
The development of resistance in foodborne pathogens to food preservation techniques is an issue of increasing concern, especially in minimally processed foods where safety relies on hurdle technology. In this context, mild heat can be used in combination with so-called nonthermal processes, such as high hydrostatic pressure (HHP), at lower individual intensities to better retain the quality of the food. However, mild stresses may increase the risk of (cross-)resistance development in the surviving population, which in turn might compromise food safety. In this investigation, we examined the evolution of Escherichia coli O157:H7 strain ATCC 43888 after recurrent exposure to progressively intensifying mild heat shocks (from 54.0°C to 60.0°C in 0.5°C increments) with intermittent resuscitation and growth of survivors. As such, mutant strains were obtained after 10 cycles of selection with ca. 106-fold higher heat resistance than that for the parental strain at 58.0°C, although this resistance did not extend to temperatures exceeding 60.0°C. Moreover, these mutant strains typically displayed cross-resistance against HHP shock and displayed signs of enhanced RpoS and RpoH activity. Interestingly, additional cycles of selection maintaining the intensity of the heat shock constant (58.5°C) selected for mutant strains in which resuscitation speed, rather than resistance, appeared to be increased. Therefore, it seems that resistance and resuscitation speed are rapidly evolvable traits in E. coli ATCC 43888 that can compromise food safety.
IMPORTANCE In this investigation, we demonstrated that Escherichia coli O157:H7 ATCC 43888 rapidly acquires resistance to mild heat exposure, with this resistance yielding cross-protection to high hydrostatic pressure treatment. In addition, mutants of E. coli ATCC 43888 in which resuscitation speed, rather than resistance, appeared to be improved were selected. As such, both resistance and resuscitation speed seem to be rapidly evolvable traits that can compromise the control of foodborne pathogens in minimal processing strategies, which rely on the efficacy of combined mild preservation stresses for food safety.
INTRODUCTION
Minimal processing of foods is based on the combination of mild preservation methods (or hurdles) for maximizing retention of the sensorial and nutritional properties of the food while maintaining the appropriate level of food safety and shelf life (1, 2). Methods such as mild heating, acidification, and the use of natural antimicrobial compounds, high hydrostatic pressure (HHP), pulsed electric fields, ultrasound, and irradiation are some of the techniques commonly used in hurdle approaches (2–4). While the efficacy of minimal processing relies on the additive, or even synergistic, lethal or growth-inhibitory effects of such mild hurdles, the mild intensity might nevertheless pose the risk of increasing resistance to the corresponding treatments. More specifically, rare mutant strains with an increased stress resistance that can spontaneously emerge in a population might survive milder stress conditions and thus become enriched within the population (5, 6). In fact, several studies found that increased resistance emerged in a variety of bacteria after exposure to certain stresses (7–11).
The notorious foodborne pathogen Escherichia coli strain O157:H7 is an important concern in this context, as it combines a low infectious dose with the capacity to cause hemorrhagic colitis, which in humans can potentially be aggravated by the development of hemolytic uremic syndrome (12). While the stress response pathways of this and other E. coli strains are fairly well understood, the evolutionary adaptability of E. coli O157:H7 in terms of its stress resistance remains poorly documented. Nevertheless, the large phenotypic differences typically observed between various E. coli O157:H7 isolates are likely indicative of this strain's adaptive potential. In fact, much of this phenotypic variation can be traced back to polymorphisms in the gene encoding the sigma factor RpoS, which controls expression of the general stress response regulon (13–15).
Interestingly, while growth on poor carbon sources, such as succinate, was previously shown to select for compromised rpoS alleles in E. coli O157:H7 EDL933 at the expense of its pathogenic potential (16), we recently demonstrated that a limited number of exposures to mild HHP were sufficient to select for HHP-resistant mutants of E. coli O157:H7 ATCC 43888 that display signs of increased RpoS activity (17). Moreover, likely the result of the general stress response triggered by this alternative sigma factor, these mutant strains also exhibited cross-resistance against heat (17). This tradeoff between resistance or self preservation on the one hand and nutritional competence on the other hand has been referred to as the SPANC balance (18).
To further improve our understanding of the adaptive potential of E. coli O157:H7 ATCC 43888, this study focused on its evolution under mild heat stress. As such, we demonstrate that recurrent exposure to heat can readily select for mutants displaying increased heat and HHP (cross-)resistance, although distinct mechanisms that differ in the upregulated stress response pathways and the level of incurred sublethal injury after stress were discriminated. Interestingly, mutant strains displaying shorter resuscitation times were also selected, indicating that not only resistance but also the speed of recovery are evolvable traits.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Stationary phase-grown cultures of E. coli O157:H7 ATCC 43888 (obtained from the American Type Culture Collection), its evolved heat-resistant derivatives, and its previously constructed rssB knockout mutant with a transposon (mini-Tn5Km2) insertion (19) were used throughout this study. Strain ATCC 43888 was originally isolated from human feces and lacks the genes for Shiga-like toxins I and II (see https://www.lgcstandards-atcc.org/products/all/43888.aspx) and was chosen as a laboratory surrogate for enterohemorrhagic strains because of its attenuated virulence. Where indicated, the wild-type E. coli ATCC 43888 and its derivatives were transformed with pFPV-PdnaK-gfp (previously designated pAA212 and encoding the E. coli MG1655 dnaK promoter upstream of gfp) (20) and pFPV-PbolA-gfp (encoding the E. coli MG1655 bolA promoter upstream of gfp [see description of its construction below]) or pACYC184-rssB (harboring the E. coli ATCC 43888 rssB gene under the control of its native promoter) (19) and the corresponding backbone control plasmid (pACYC184) (21) by electroporation.
Bacterial cultures were obtained by inoculating test tubes containing 4 ml of tryptone soy broth (TSB) (Oxoid, Basingstoke, United Kingdom) with a single colony grown on a tryptone soy agar (TSA) plate and then incubating the bacteria aerobically with shaking (300 rpm) for 18 h at 37°C. When necessary, a final concentration of (i) 100 μg/ml ampicillin (AppliChem, Darmstadt, Germany) was added to select for the presence of bacteria with pFPV-PdnaK-gfp and pFPV-PbolA-gfp (20) or (ii) 30 μg/ml chloramphenicol (Sigma-Aldrich, Geel, Belgium) to select for the presence of bacteria with pACYC184-based complementation plasmids (19, 21).
Heat and HHP treatment.
Cells from a stationary phase-grown culture were harvested by centrifugation (4,000 × g for 5 min) and resuspended in an equal volume of 0.85% KCl (Sigma-Aldrich, St. Louis, MO). For thermal treatment, 3 sterile PCR tubes were aseptically filled with 75 μl of resuspended cells and subjected to heat (54.0°C to 60.0°C) for 15 min using a PCR apparatus (TPersonal 48; Biometra GmbH, Gottingen, Germany). For HHP treatment, 300 μl of the suspension was heat sealed in a sterile polyethylene bag after exclusion of air bubbles and subjected to pressure (500 MPa) for 15 min in an 8-ml pressure vessel (HPIU-10000, 95/1994; Resato, Roden, The Netherlands) that was held at 20°C with an external water jacket connected to a cryostat. Please note that the slow pressure increase (100 MPa/min) and the external water jacket attenuated adiabatic heating during pressure buildup, and conservative estimates indicated only a transient increase in the sample temperature of 13°C at 800 MPa. Finally, decompression was almost instantaneous. After heat or HHP treatment, samples were aseptically retrieved from the PCR tubes or polyethylene bags, respectively, and survival was determined as described below.
Please note that for reasons that are currently unclear, we observed that the apparent heat and HHP resistance of the parental ATCC 43888 strain was slightly increased over that in our previous study (19), although this did not affect the conclusions drawn in this study.
Selection of heat-resistant mutants.
To obtain mutants of E. coli ATCC 43888 with enhanced heat resistance, several independent bacterial cultures were reiteratively exposed to heat shocks (15 min) either by progressively increasing the treatment temperature (0.5°C each cycle) or by maintaining a constant temperature. After each heat shock, an aliquot of the treated sample was diluted 1/100 into fresh prewarmed TSB and incubated for 23 h at 37°C prior to the next round of treatment. After every five cycles of selection, a number of survivors from each evolved culture were purified and challenged with the last heat shock to find single clones that represented the behavior of the pool against heat. Finally, the stability of the isolated phenotypes was assessed by daily passages in the growth medium without intermittent exposure to heat, which corresponded to ca. 6.6 (log2[100]) generations per day.
Determination of viability.
Samples were diluted in 0.85% KCl supplemented with 0.1% bacteriological peptone (Oxoid) and subsequently spot plated (5 μl) or spread plated (20 μl) on TSA. Where indicated, cells were also recovered from violet red bile glucose agar (VRBGA) (Oxoid) to determine the extent of sublethal injury. After 24 h of incubation at 37°C, the colonies were counted and the logarithmic reduction factor was calculated as log10(N0/N), in which N0 and N represent the number of CFU/ml before and after treatment, respectively. Thus, the detection limits were 200 and 50 CFU/ml for spot- and spread-plated samples, respectively.
Construction of pFPV-PbolA-gfp.
The pFPV-PbolA-gfp reporter was constructed following the same procedure described for pFPV-PdnaK-gfp (previously designated pAA212) (20). Briefly, the promoter region of bolA was amplified from E. coli MG1655 by PCR (Phusion DNA polymerase; Thermo Scientific, Waltham, MA) using the primers 5′-TGTTGGATCCTGTTTGGTAAAAATTCCC-3′ and 5′-TGGTTCTAGATTATTCTTCTATCCGCTCACG-3′. Subsequently, this amplicon was digested with BamHI and XbaI and directionally cloned upstream of the promoterless gfp gene in pFPV25 (22), which was digested with the same enzymes.
Measurement of reporter gene activity.
To determine the fluorescence derived from strains equipped with pFPV-PdnaK-gfp (20) or pFPV-PbolA-gfp (this study), 200 μl of the corresponding stationary phase-grown culture was transferred to microplate wells and placed in a Fluoroskan Ascent FL fluorimeter (Thermo Labsystems, Brussels, Belgium). The basal green fluorescent protein (GFP) fluorescence was measured at an excitation wavelength of 480 nm and an emission wavelength of 520 nm. The obtained fluorescence values were subsequently divided by the optical density of the same sample at 600 nm (OD600) to obtain the relative fluorescence units. Differences in RpoH and RpoS activities are expressed as fold change with respect to the parental strain.
Determination of resuscitating-culture lag times.
The apparent lag times of resuscitating cultures were monitored by optical density (OD600) using a Bioscreen C plate-reader system (Thermo Labsystems Oy, Helsinki, Finland). For each experiment, heat-treated samples and untreated populations were serially diluted to 1/100,000 in TSB. Then, 300 μl of the different dilutions was placed in a microtiter plate and incubated in the Bioscreen C system for a 24-h period at 37°C with regular shaking and automatic measurement of OD600 every 10 min. The data obtained for each well were defined by the growth model described by Baranyi and Roberts (23) using the DMFit 3.0 software (Institute of Food Research, Norwich Research Park, Norwich, United Kingdom), which generates best-fit growth parameters, including the apparent lag time and growth rate (μmax), with a determination coefficient (R2) of >0.990. For each strain, the apparent lag times estimated from control and treated samples were plotted against the initial viable-bacterium counts obtained by regular plating.
Statistical analysis.
Statistical analyses (analysis of variance and t tests) were carried out using the software GraphPad Prism 5.0 (GraphPad Software, Inc., San Diego, CA), and differences were regarded to be significant when P values were ≤0.05. All microbial inactivation outcomes shown in the figures correspond to averages and standard deviations calculated from at least three replicate experiments performed on different days. Data gathered from OD600 monitoring were obtained in quadruplicate.
RESULTS
Development of heat resistance in E. coli O157:H7 ATCC 43888.
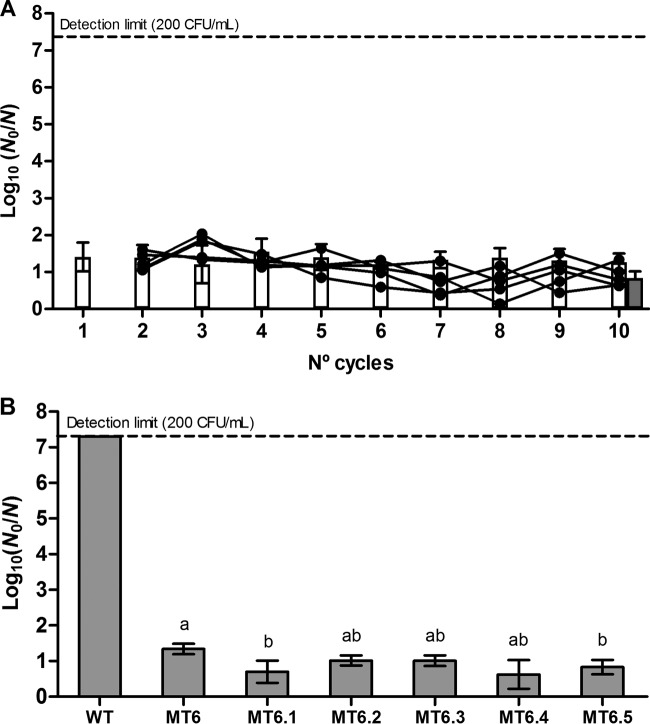
To examine the impact of heat stress on the evolution of E. coli ATCC 43888, 10 independent cultures were individually subjected to repeated cycles of increasingly severe heat treatment with intermittent recovery and outgrowth of survivors. The selection process started with a heat shock that inactivated approximately 1-log10 cycle of the parental strain (54.0°C for 15 min) (Fig. 1A), after which the treatment temperature was progressively increased by 0.5°C each cycle until bacterial growth was no longer observed after treatment (which occurred at 60.0°C for 15 min). The heat resistance evolution of the 10 lineages during this selection regimen is depicted in Fig. 1A. For comparison, the corresponding inactivation of the parental strain (without previous exposure to thermal stress) for each heat treatment is included. As observed, the number of survivors of the parental strain decreased exponentially with increasing treatment temperature. Overall, the inactivation of cultures repetitively challenged by heat shock followed the same profile as that of the parental strain during the first five cycles (i.e., heat shocks from 54.0°C to 56.0°C), although at this point, individual clones isolated from 4 of 10 lineages already showed significantly higher heat resistance (1.6-log10 cycles at 56.0°C on average; data not shown). After further temperature increments, however, heat resistance development occurred in all the lineages. It should be noted that three control lineages subjected to 10 similar cycles without intermittent heat shock did not acquire increased heat resistance. In fact, heat resistance even diminished ca. 17-fold at 56.0°C in these lineages, indicating that the serial passaging of cultures itself was not selecting for heat resistance (data not shown).
FIG 1.
Selection of E. coli ATCC 43888 toward heat resistance. (A) Evolution of the heat resistance of 10 independent axenic cultures iteratively exposed to progressively intensifying heat treatments (15 min), with intermittent resuscitation and growth of survivors. The black lines indicate the inactivation [i.e., logarithmic reduction factor, log10(N0/N)] of the evolving lineages during the stepwise selection regimen with 0.5°C increments, while white bars represent the resistance of the (unevolved) parental strain. Gray bars correspond to the average inactivation of the 10 individual clones that were isolated from each of the 10 evolved lineages after either five (reaching a 56.0°C exposure [light-gray bar]) or 10 (reaching a 58.5°C exposure [dark-gray bar]) cycles. (B) Logarithmic reduction factors of the 10 individual E. coli ATCC 43888 clones (designated MT1 to MT10) that were isolated from each of the 10 evolved lineages after 10 cycles (i.e., breakdown of the data for the 10 clones represented by the dark-gray bar plotted in panel A) and the wild-type (WT) strain by a heat shock at 58.5°C for 15 min. Survivors were recovered from plates with nonselective medium (TSA). Letters indicate statistically significant differences (P ≤ 0.05) in inactivation among different isolates.
The heat resistance of one individual clone (designated MT1 to MT10) from each of the evolved populations after 10 cycles of progressively intensifying heat treatment (i.e., after exposure to a heat shock at 58.5°C) is illustrated in Fig. 1B. Please note that each of the three clones randomly picked from each lineage exhibited approximately the same heat resistance as that shown by the evolved culture from which it was isolated, indicating that resistant variants constituted the majority of the population. The viability of the 10 isolated clones after a 58.5°C heat shock was reduced by only 2.6-log10 cycles on average, whereas the survival of the parental strain fell below the detection limit (≥7.3-log10 reductions). Overall, inactivation among MT1 to MT10 populations varied more than 1.5-log10 cycles.
When three independent lineages of clones displaying high heat resistance (i.e., MT3, MT6, and MT9) were subcultured for ca. 66 generations (i.e., 10 cycles) without heat exposure, their heat resistance levels declined by 1.2-log10 cycles on average at 58.5°C (data not shown), in accordance with the decline mentioned earlier for the parental strain after serial passaging. Nevertheless, their remaining resistance still outperformed that of the parental strain at ca. >104-fold, suggesting that the acquired heat resistance was mutationally fixed.
Examination of heat resistance and HHP cross-resistance in selected E. coli O157:H7 ATCC 43888 mutants.
Subsequently, the levels of heat resistance in MT3, MT6, and MT9 were more thoroughly examined. Figure 2 shows the reduction factors of the three mutants and the parental strain subjected to heat treatments of various intensities. Please note that survivors were recovered from nonselective (TSA) and selective (VRBGA) media to determine the extent of sublethal injury. The three mutants showed markedly enhanced resistance to heat compared to the resistance of the parental strain at all temperatures tested. The survival of all heat-resistant mutants on the nonselective medium was only slightly affected after 56.0°C heat shock (0.6-log10 reduction on average on TSA), while the viability of the parental strain was reduced by >99.9%. Furthermore, the inactivation of the three variants was <2-log10 cycles at 58.0°C, whereas the inactivation of the wild-type strain at that temperature was 7.1-log10 cycles. At the two temperatures, the heat resistance levels of MT3, MT6, and MT9 did not differ widely in the nonselective medium. Interestingly, however, counts on VRBGA plates showed that large proportions of the surviving populations of MT3 and MT9 were sublethally injured, whereas this did not appear to be the case for MT6 survivors. For instance, although the viable population of MT6 recovered from TSA from a heat shock at 58.0°C was slightly larger than that of MT3 (8.4- and 7.6-log10 cycles, respectively), more than 99.99% of the surviving MT3 cells were sublethally injured against only 65.81% of damaged cells scored in MT6. After exposure to 59.0°C, the viability of MT6 in the nonselective medium was 43- and 9-fold higher than that of MT3 and MT9, respectively, and more than 3 × 104-fold higher than that of the parental strain. Importantly, the increased heat resistance of the mutants did not extend to higher temperatures such as 60.0°C (data not shown).
FIG 2.

Logarithmic reduction factors of E. coli ATCC 43888 MT3, MT6, and MT9 isolated by 10 cycles of selection enrichment with progressively intensifying heat shocks and the wild-type (WT) strain by heat treatment at 56.0°C (A), 58.0°C (B), or 59.0°C (C) for 15 min. Survivors were recovered from TSA (gray bars) or VRBGA (white bars) plates. Letters indicate statistically significant differences (P ≤ 0.05) in inactivation among the wild-type and mutant strains recovered from TSA plates. Asterisks indicate statistically significant differences between the numbers of survivors recovered from TSA and VRBGA plates for each of the strains.
Although selected solely on the basis of heat resistance, the three mutants also exhibited increased cross-resistance to HHP compared to that of the parental strain (Fig. 3). In fact, MT3, MT6, and MT9 were 2.3-, 2.9-, and 2.1-log10 cycles more piezotolerant, respectively, than the parental strain after a 500-MPa exposure for 15 min. In addition, the extents of sublethal injury stemming from HHP exposure were larger in the surviving populations of MT3 and MT9 (2.9- and 3.5-log10 cycles, respectively) than in MT6 (1.4-log10 cycles) but lower than in the parental strain.
FIG 3.

Logarithmic reduction factors of E. coli ATCC 43888 wild-type (WT) strain and heat-resistant mutants MT3, MT6, and MT9 by an HHP treatment at 500 MPa for 15 min. Survivors were recovered from TSA (gray bars) or VRBGA (white bars) plates. Letters indicate statistically significant differences (P ≤ 0.05) in inactivation among the wild-type and mutant strains recovered from TSA plates. Asterisks indicate statistically significant differences between the numbers of survivors recovered from TSA and VRBGA plates for each of the strains.
Increased RpoH and RpoS activities govern development of heat resistance in selected E. coli O157:H7 ATCC 43888 mutants.
In an attempt to mechanistically infer the origin of resistance development in MT3, MT6, and MT9, the basal activities of two important stress-related sigma factors (RpoH and RpoS) were examined. More specifically, the activity of RpoH (directing expression of the heat shock response) was assayed by quantifying expression of the dnaK promoter (PdnaK, using the pFPV-PdnaK-gfp construct) (20, 24), while the activity of RpoS (directing expression of the general stress response) was assayed by quantifying expression of the bolA promoter (PbolA, using the pFPV-PbolA-gfp construct) (25). Please note that while dnaK and bolA promoter sequences originated from E. coli MG1655, their sequence identities with the corresponding sequences of E. coli ATCC 43888 are 99.6% and 99.0%, respectively, and both reporters proved to be functional in the ATCC 43888 background (reference 17 and Fig. 4A).
FIG 4.

(A) Fluorescence stemming from pFPV-PdnaK-gfp (encoding the E. coli MG1655 dnaK promoter upstream of gfp [gray bars]) and pFPV-PbolA-gfp (encoding the E. coli MG1655 bolA promoter upstream of gfp [white bars]) in the E. coli ATCC 43888 wild-type (WT) strain, rssB::mini-Tn5Km2 transposon mutant, and MT3, MT6, and MT9. Values are expressed as fold change with respect to the average value of the parental strain. Lowercase and capital letters indicate statistically significant differences (P ≤ 0.05) in the fluorescence values derived from the PdnaK-gfp and PbolA-gfp promoters, respectively, in the wild-type and mutant strains. (B) Logarithmic reduction factors of the E. coli ATCC 43888 wild-type (WT) strain, MT3, MT6, and MT9 equipped with pACYC184 (control vector [gray bars]) or pACYC184-rssB (white bars) by heat treatment (58.0°C for 15 min). Survivors were recovered from TSA (solid bars) or VRBGA (striped bars) plates. Asterisks indicate statistically significant differences (P ≤ 0.05) in inactivation between strains equipped with pACYC184-rssB and the control vector recovered from TSA or VRBGA plates.
Using these reporters, it could be inferred that MT3 and MT9 exhibited significantly increased PdnaK and PbolA expression, indicative of constitutively increased RpoH and RpoS activity (Fig. 4A). In fact, PbolA-gfp expression was similar (for MT3) or even higher (for MT9) than that of the rssB-compromised ATCC 43888 mutant that was attenuated in quenching its RpoS activity (19). In contrast, PdnaK-gfp expression did not seem to be affected in MT6, and this mutant only displayed increased PbolA-gfp expression.
Moreover, while MT3 and MT9 suffered attenuation of heat resistance after quenching of their RpoS activity (using pACYC-rssB), such quenching did not affect the inactivation or sublethal injury of MT6 (Fig. 4B), further suggesting that the resistance development of MT6 differs from that of MT3 and MT9. Furthermore, none of these mutants exhibited any sequence alterations at their rpoS or rssB loci (data not shown).
Prolonged selection at 58.5°C allows for the emergence of E. coli O157:H7 ATCC 43888 mutants with faster resuscitation.
Further focusing on MT6 as the most heat-resistant mutant, we wondered whether its heat resistance could be further improved. Since Fig. 1A demonstrates that a regimen with further temperature increments proved futile, we decided to impose a regimen with repetitive exposures at the same challenge temperature of 58.5°C to see whether the remaining ca. 1.6-log10 cycles of inactivation of MT6 at 58.5°C (Fig. 1B) could be further reduced. However, when five individual lineages of MT6 were challenged with 10 cycles of a 58.5°C heat shock and intermittent outgrowth of survivors, the resistance levels of their respective isolated clones (designated MT6.1 to MT6.5) increased only 0.5-log10 cycles on average with regard to the parental MT6 clone (Fig. 5), suggesting that the mechanism and/or population distribution of heat resistance did not evolve to protect the entirety or even majority of the population.
FIG 5.
Selection of E. coli ATCC 43888 MT6 toward faster resuscitation. (A) Evolution of the heat resistance of five independent axenic cultures iteratively exposed to heat shock at 58.5°C (15 min), with intermittent resuscitation and growth of survivors. The black lines indicate the logarithmic reduction factors of the evolving lineages during 10 cycles of selection, while white bars represent the resistance of the ancestral MT6. The gray bar corresponds to the average inactivation of the five individual clones that were isolated from each of the five evolved lineages after exposure to 10 cycles of selection by heat treatment at 58.5°C (15 min). (B) Logarithmic reduction factors of the five individual E. coli ATCC 43888 clones (designated MT6.1 to MT6.5) that were isolated from each of the five evolved lineages after 10 cycles (i.e., breakdown of the dark-gray bar plotted in panel A), the ancestral MT6, and the wild-type (WT) strain by heat shock at 58.5°C for 15 min. Survivors were recovered from plates with nonselective medium (TSA). Letters indicate statistically significant differences (P ≤ 0.05) in inactivation among different isolates.
In an effort to look for subtler differences, however, the inactivation kinetics at 58.5°C of one of these mutants (i.e., MT6.5) was compared in more detail to its corresponding ancestral MT6 and the wild-type strain (Fig. 6). While the inactivation of the parental strain followed log-linear kinetics, the survival curves of MT6 and MT6.5 displayed prominent shoulders prior to a gentler exponential inactivation phase (decimal reduction time [D] = 1.10, 4.64, and 4.80 min for the WT, MT6, and MT6.5, respectively). The survival of MT6.5 on the nonselective medium remained higher (0.46-log10 cycles; P < 0.05) than that of MT6 throughout the log-linear decay, although equal numbers of intact MT6.5 and MT6 cells were recovered from VRBGA plates.
FIG 6.
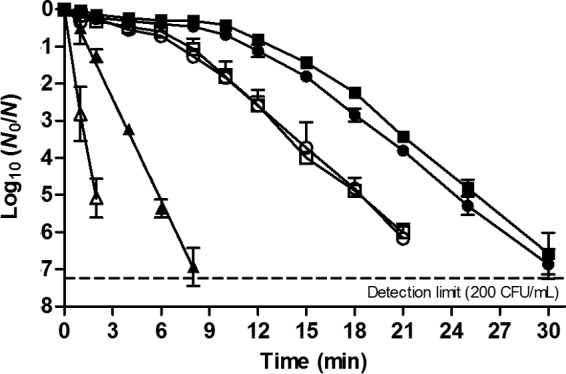
Inactivation curves of E. coli ATCC 43888 wild type (▲,△), MT6 (●,○), and MT6.5 (■,□) exposed to 58.5°C for the indicated times. Survivors were recovered from TSA (solid symbols) or VRBGA (open symbols) plates.
Since heat survival of MT6.5 was improved only modestly compared to that of its MT6 parent (Fig. 6), we wondered whether the isothermic selection regimen instead might have selected for the enrichment of variants displaying faster recovery. To more closely examine and compare the resuscitation times of MT6 and MT6.5, the durations of their respective apparent lag phases after recovery from a heat shock that inactivated ca. 1-log10 cycle of each strain using TSA as the plating medium (i.e., 58.5°C for 12 and 14 min for MT6 and MT6.5, respectively; Fig. 6) were monitored spectrophotometrically. Figure 7 illustrates the relationship between the lag time and the logarithm of initial viable-bacterium counts present in recovering cultures of MT6 and MT6.5 exposed to heat and of the corresponding control cultures. Interestingly, while there was no difference in lag phases (or growth rate − μmax = 0.157 ± 0.103 h−1 and 0.155 ± 0.138 h−1 for MT6 and MT6.5, respectively) between the two strains in the absence of stress, the lag phase of the resuscitating culture of MT6.5 (although it incurred a higher degree of injury) was, on average, 0.84 h shorter than that of MT6 regardless of variations in inoculum size (Fig. 7).
FIG 7.

Relationship between apparent lag times determined by OD600 monitoring and initial viable-bacterium counts of E. coli ATCC 43888 MT6 (●,○) and MT6.5 (■,□) after heat exposure (58.5°C for 12 and 14 min, respectively; open symbols) or those left untreated (solid symbols). The means and standard deviations of four replicate experiments for the inactivation of MT6 and MT6.5 were 1.10 ± 0.12 and 1.05 ± 0.16 log10 cycles on TSA plates, respectively, and 2.32 ± 0.14 and 3.19 ± 0.22 log10 cycles on VRBGA plates, respectively.
Finally, MT6.5 displayed PbolA-gfp and PdnaK-gfp expression levels similar to those of its parental MT6 strain (data not shown), suggesting that the increased recovery speed of MT6.5 was not mediated by improved RpoS or RpoH activity.
DISCUSSION
Since E. coli O157:H7 is a notorious foodborne pathogen with a low infectious dose and the capacity to cause hemorrhagic colitis and hemolytic uremic syndrome in humans (12), its potential for resistance development in the context of mild food processing needs to be thoroughly documented and investigated. In this context, our observations reveal that E. coli O157:H7 strain ATCC 43888 can straightforwardly and reproducibly acquire significant resistance against mild heat after a limited number of exposures. In fact, one of the most heat-resistant ATCC 43888 mutants isolated in this study (i.e., MT6) was ca. 106-fold more resistant than its parent after a 15-min treatment at 58.0°C and displayed less than 1,000-fold inactivation after a 15-min exposure to 59.0°C. Moreover, the acquisition of heat resistance coincided with cross-resistance against HHP intensities that are relevant for the food industry (26) and, as such, compromises the potential impact of this nonthermal mild hurdle.
Nevertheless, the heat-resistant mutants isolated in this study did not survive exposures of 60°C or higher, underscoring the limits of their genomic evolvability in this respect. It should be underscored, however, that in these laboratory-directed evolution experiments, evolvability is typically restricted to the available genomic repertoire, while in more natural settings, lateral gene transfer can be an important driver of adaptation. In this context, it was recently shown that the ability of extremely heat-resistant E. coli strains, which were isolated from a slaughterhouse plant using steam-based carcass decontamination, to survive exposures up to 71°C stemmed from the lateral acquisition of a genomic island termed the locus of heat resistance (27, 28).
Closer analysis of a few of the most heat-resistant mutant strains identified in this study (i.e., MT3, MT6, and MT9) revealed that they each displayed upregulated basal RpoS activity. Improved RpoS activity tends to increase general stress resistance (13, 17), which likely explains at least part of the observed cross-resistance to HHP. Furthermore, MT3 and MT9 also displayed improved RpoH activity. While improved RpoH activity can logically increase heat resistance, upregulation of the heat-shock response was previously correlated with improved HHP resistance (17, 20). Nevertheless, intrinsic heat and HHP resistance are not necessarily correlated with each other, as the most heat-resistant E. coli strains are not per se the most HHP resistant, and vice versa (13, 29). Development of heat resistance through mutational upregulation of the activity of global regulators, such as RpoS and RpoH, seems to comply with the expected pleiotropic systemic impact of heat stress on the cell, although identification of the most important downstream effectors of these extensive stress regulons might help us to better delineate the exact nature of incurred cellular injury.
Given the extensive regulatory network determining RpoS and RpoH activities inside the cell (30, 31), it can be expected that many different evolutionary pathways would result in an equivalent modulation of their activity, although those mutations coinciding with a growth defect are likely to become counterselected during the growth stages in between repetitive exposures to heat. Such different evolutionary routes are, in fact, evident in the comparison of MT3, MT6, and MT9. Indeed, while MT3 and MT9 display upregulation of RpoS and RpoH activities, MT6 displays only upregulated RpoS activity. In addition, while MT3 and MT6 display similar upregulation of RpoS activity, their corresponding heat resistance was not similarly affected by RssB-mediated quenching of this activity, further suggesting that there are different mechanisms of upregulation. Future whole-genome sequencing and genetic reconstruction efforts, however, should reveal and sort out the mutations and evolutionary routes that contribute to the constitutive upregulation of RpoH and/or RpoS activity. Furthermore, it remains to be addressed whether other E. coli O157:H7 isolates or other foodborne pathogenic bacteria would evolve along the same routes in response to heat stress.
Interestingly, mutant E. coli ATCC 43888 strains with a shorter resuscitation time were easily selected after repeated exposures to the same heat treatment. More specifically, since MT6 already displayed a strong heat resistance that could hardly be improved, strains with shorter resuscitation times were likely to be enriched during the imposed isothermal selection regimen. As such, our data underscore that, aside from absolute resistance, resuscitation speed itself is an evolvable trait. Previous work showed that E. coli can also improve its resuscitation time in response to recurring freeze-thaw cycles (32, 33), suggesting that selection for faster recovery is a more general phenomenon that can be triggered by several stresses. Interestingly, most recent evidence seems to suggest that E. coli can evolve to increase the time it needs or takes before growth resumption to avoid being killed by antibiotics (34). Together, these observations reveal the very flexible nature of resuscitation and lag times.
In this report, we document the rapid evolvability of E. coli O157:H7 ATCC 43888 in terms of resistance and speed of recovery under heat stress selection. The resulting increased survival and earlier outgrowth are both important concerns for food safety and deserve further attention.
ACKNOWLEDGMENTS
This work was supported by doctoral (1135116N; to A.C.) and postdoctoral (12P9815N; to E.G.) fellowships from the Research Foundation–Flanders (FWO-Vlaanderen) and a postdoctoral fellowship (F+/13/040; to E.G.) and grants (STRT1/10/036 and DBOF/12/035) from the KU Leuven Research Fund.
We thank Kristof Vanoirbeek and Nele Rutten for technical assistance.
REFERENCES
- 1.Leistner L. 2000. Basic aspects of food preservation by hurdle technology. Int J Food Microbiol 55:181–186. doi: 10.1016/S0168-1605(00)00161-6. [DOI] [PubMed] [Google Scholar]
- 2.Ross AIV, Griffiths MW, Mittal GS, Deeth HC. 2003. Combining nonthermal technologies to control foodborne microorganisms. Int J Food Microbiol 89:125–138. doi: 10.1016/S0168-1605(03)00161-2. [DOI] [PubMed] [Google Scholar]
- 3.Raso J, Barbosa-Canovas GV. 2003. Nonthermal preservation of foods using combined processing techniques. Crit Rev Food Sci Nutr 43:265–285. doi: 10.1080/10408690390826527. [DOI] [PubMed] [Google Scholar]
- 4.Feyaerts J, Rogiers G, Corthouts J, Michiels CW. 2015. Thiol-reactive natural antimicrobials and high pressure treatment synergistically enhance bacterial inactivation. Innov Food Sci Emerg Technol 27:26–34. doi: 10.1016/j.ifset.2014.12.005. [DOI] [Google Scholar]
- 5.Abee T, Koomen J, Metselaar KI, Zwietering MH, den Besten HMW. 2016. Impact of pathogen population heterogeneity and stress-resistant variants on food safety. Annu Rev Food Sci Technol 7:439–456. doi: 10.1146/annurev-food-041715-033128. [DOI] [PubMed] [Google Scholar]
- 6.Dragosits M, Mozhayskiy V, Quinones-Soto S, Park J, Tagkopoulos I. 2013. Evolutionary potential, cross-stress behavior and the genetic basis of acquired stress resistance in Escherichia coli. Mol Syst Biol 9:643. doi: 10.1038/msb.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris DR, Pollock SV, Wood EA, Goiffon RJ, Klingele AJ, Cabot EL, Schackwitz W, Martin J, Eggington J, Durfee TJ, Middle CM, Norton JE, Popelars MC, Li H, Klugman SA, Hamilton LL, Bane LB, Pennacchio LA, Albert TJ, Perna NT, Cox MM, Battista JR. 2009. Directed evolution of ionizing radiation resistance in Escherichia coli. J Bacteriol 191:5240–5252. doi: 10.1128/JB.00502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajkovic A, Smigic N, Uyttendaele M, Medic H, de Zutter L, Devlieghere F. 2009. Resistance of Listeria monocytogenes, Escherichia coli O157:H7 and Campylobacter jejuni after exposure to repetitive cycles of mild bactericidal treatments. Food Microbiol 26:889–895. doi: 10.1016/j.fm.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Sagarzazu N, Cebrián G, Pagán R, Condón S, Mañas P. 2013. Emergence of pulsed electric fields resistance in Salmonella enterica serovar Typhimurium SL1344. Int J Food Microbiol 166:219–225. doi: 10.1016/j.ijfoodmicro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Vanlint D, Mitchell R, Bailey E, Meersman F, McMillan PF, Michiels CW, Aertsen A. 2011. Rapid acquisition of gigapascal-high-pressure resistance by Escherichia coli. mBio 2:e00130-10. doi: 10.1128/mBio.00130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanlint D, Rutten N, Michiels CW, Aertsen A. 2012. Emergence and stability of high-pressure resistance in different food-borne pathogens. Appl Environ Microbiol 78:3234–3241. doi: 10.1128/AEM.00030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Loughlin EV, Robins-Browne RM. 2001. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes Infect 3:493–507. doi: 10.1016/S1286-4579(01)01405-8. [DOI] [PubMed] [Google Scholar]
- 13.Álvarez-Ordóñez A, Alvseike O, Omer MK, Heir E, Axelsson L, Holck A, Prieto M. 2013. Heterogeneity in resistance to food-related stresses and biofilm formation ability among verocytotoxigenic Escherichia coli strains. Int J Food Microbiol 161:220–230. doi: 10.1016/j.ijfoodmicro.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Carter MQ, Louie JW, Huynh S, Parker CT. 2014. Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157:H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol 44:108–118. doi: 10.1016/j.fm.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Robey M, Benito A, Hutson RH, Pascual C, Park SF, Mackey BM. 2001. Variation in resistance to high hydrostatic pressure and rpoS heterogeneity in natural isolates of Escherichia coli O157:H7. Appl Environ Microbiol 67:4901–4907. doi: 10.1128/AEM.67.10.4901-4907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong T, Chiang SM, Joyce C, Yu R, Schellhorn HE. 2009. Polymorphism and selection of rpoS in pathogenic Escherichia coli. BMC Microbiol 9:118. doi: 10.1186/1471-2180-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanlint D, Rutten N, Govers SK, Michiels CW, Aertsen A. 2013. Exposure to high hydrostatic pressure rapidly selects for increased RpoS activity and general stress-resistance in Escherichia coli O157:H7. Int J Food Microbiol 163:28–33. doi: 10.1016/j.ijfoodmicro.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 18.Ferenci T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol 57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- 19.Vanlint D, Pype BJY, Rutten N, Vanoirbeek KGA, Michiels CW, Aertsen A. 2013. Loss of cAMP/CRP regulation confers extreme high hydrostatic pressure resistance in Escherichia coli O157:H7. Int J Food Microbiol 166:65–71. doi: 10.1016/j.ijfoodmicro.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Aertsen A, Vanoirbeek K, De Spiegeleer P, Sermon J, Hauben K, Farewell A, Nyström T, Michiels CW. 2004. Heat shock protein-mediated resistance to high hydrostatic pressure in Escherichia coli. Appl Environ Microbiol 70:2660–2666. doi: 10.1128/AEM.70.5.2660-2666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose RE. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res 16:355. doi: 10.1093/nar/16.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdivia RH, Falkow S. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol 22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 23.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. doi: 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 24.Van Dyk TK, Majarian WR, Konstantinov KB, Young RM, Dhurjati PS, LaRossa RA. 1994. Rapid and sensitive pollutant detection by induction of heat shock gene-bioluminescence gene fusions. Appl Environ Microbiol 60:1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos JM, Freire P, Vicente M, Arraiano CM. 1999. The stationary-phase morphogene bolA from Escherichia coli is induced by stress during early stages of growth. Mol Microbiol 32:789–798. doi: 10.1046/j.1365-2958.1999.01397.x. [DOI] [PubMed] [Google Scholar]
- 26.Balasubramaniam VM, Martinez-Monteagudo SI, Gupta R. 2015. Principles and application of high pressure-based technologies in the food industry. Annu Rev Food Sci Technol 6:435–462. doi: 10.1146/annurev-food-022814-015539. [DOI] [PubMed] [Google Scholar]
- 27.Dlusskaya EA, McMullen LM, Gänzle MG. 2011. Characterization of an extremely heat-resistant Escherichia coli obtained from a beef processing facility. J Appl Microbiol 110:840–849. doi: 10.1111/j.1365-2672.2011.04943.x. [DOI] [PubMed] [Google Scholar]
- 28.Mercer RG, Zheng J, Garcia-Hernandez R, Ruan L, Gänzle MG, McMullen LM. 2015. Genetic determinants of heat resistance in Escherichia coli. Front Microbiol 6:932. doi: 10.3389/fmicb.2015.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Hernandez R, McMullen L, Gänzle MG. 2015. Development and validation of a surrogate strain cocktail to evaluate bactericidal effects of pressure on verotoxigenic Escherichia coli. Int J Food Microbiol 205:16–22. doi: 10.1016/j.ijfoodmicro.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 30.Arsène F, Tomoyasu T, Bukau B. 2000. The heat shock response of Escherichia coli. Int J Food Microbiol 55:3–9. doi: 10.1016/S0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 31.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sleight SC, Orlic C, Schneider D, Lenski RE. 2008. Genetic basis of evolutionary adaptation by Escherichia coli to stressful cycles of freezing, thawing and growth. Genetics 180:431–443. doi: 10.1534/genetics.108.091330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sleight SC, Lenski RE. 2007. Evolutionary adaptation to freeze-thaw-growth cycles in Escherichia coli. Physiol Biochem Zool 80:370–385. doi: 10.1086/518013. [DOI] [PubMed] [Google Scholar]
- 34.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]