ABSTRACT
Nosema ceranae is a new and emerging microsporidian parasite of European honey bees, Apis mellifera, that has been implicated in colony losses worldwide. RNA interference (RNAi), a posttranscriptional gene silencing mechanism, has emerged as a potent and specific strategy for controlling infections of parasites and pathogens in honey bees. While previous studies have focused on the silencing of parasite/pathogen virulence factors, we explore here the possibility of silencing a host factor as a mechanism for reducing parasite load. Specifically, we used an RNAi strategy to reduce the expression of a honey bee gene, naked cuticle (nkd), which is a negative regulator of host immune function. Our studies found that nkd mRNA levels in adult bees were upregulated by N. ceranae infection (and thus, the parasite may use this mechanism to suppress host immune function) and that ingestion of double-stranded RNA (dsRNA) specific to nkd efficiently silenced its expression. Furthermore, we found that RNAi-mediated knockdown of nkd transcripts in Nosema-infected bees resulted in upregulation of the expression of several immune genes (Abaecin, Apidaecin, Defensin-1, and PGRP-S2), reduction of Nosema spore loads, and extension of honey bee life span. The results of our studies clearly indicate that silencing the host nkd gene can activate honey bee immune responses, suppress the reproduction of N. ceranae, and improve the overall health of honey bees. This study represents a novel host-derived therapeutic for honey bee disease treatment that merits further exploration.
IMPORTANCE Given the critical role of honey bees in the pollination of agricultural crops, it is urgent to develop strategies to prevent the colony decline induced by the infection of parasites/pathogens. Targeting parasites and pathogens directly by RNAi has been proven to be useful for controlling infections in honey bees, but little is known about the disease impacts of RNAi silencing of host factors. Here, we demonstrate that knocking down the honey bee immune repressor-encoding nkd gene can suppress the reproduction of N. ceranae and improve the overall health of honey bees, which highlights the potential role of host-derived and RNAi-based therapeutics in controlling the infections in honey bees. The information obtained from this study will have positive implications for honey bee disease management practices.
INTRODUCTION
European honey bees, Apis mellifera, play a critical role in the pollination of important crops. However, honey bee populations have suffered high losses in much of the world (1), coincident with an increase in agricultural demand for honey bee pollination (2). Specifically, honey bee colony losses in the United States have been exacerbated since the report of colony collapse disorder (CCD), a syndrome that comprises large-scale, unexplained losses of managed honey bees (3–9). High levels of parasites and pathogens have been linked to the decline of honey bee colonies (10, 11).
Nosema is a genus of obligate, intracellular microsporidian parasites that infect many diverse animal species, including honey bees (12, 13). For years, Nosema disease of European honey bees was exclusively attributed to a single Nosema species, Nosema apis. Another species, Nosema ceranae, was originally detected in Asian honey bees, Apis cerana (14), and subsequently found to infect European honey bees, A. mellifera (15, 16). Since then, the infection of A. mellifera by N. ceranae has been reported worldwide (17–20), and nosemosis of A. mellifera caused by N. ceranae is now far more prevalent than that by its native sympatric congener N. apis (17, 21–25). Although there are no outward disease symptoms reported (12), N. ceranae infection can cause energetic stress and behavioral changes in worker bees (26–29), leading to a reduced life span of infected bees. As an emerging parasite, N. ceranae has often been linked to colony losses worldwide. A study based on Spanish honey bee populations showed that natural infection by N. ceranae could cause colony collapse (30, 31). New evidence has shown that N. ceranae interacts synergistically with pesticides, resulting in more complex and severe disease in honey bees (32–35). So far, the only registered treatment for nosemosis in North America is fumagillin. With prolonged use of fumagillin, the issues of drug resistance have arisen (36). As a result, novel parasite-specific and environmentally friendly therapeutic options are urgently needed for Nosema treatment.
RNA interference (RNAi), a posttranscriptional gene silencing mechanism, is an efficient and specific method of gene silencing which functions by inducing degradation of homologous mRNAs (37, 38). RNAi technology has been explored to protect honey bees from infection by pathogens and parasites (39). Three honey bee viruses, Israeli acute paralysis virus (IAPV), deformed wing virus (DWV), and Chinese sacbrood virus (CSBV), have been successfully inhibited by RNAi under laboratory conditions by feeding bees with virus-specific double-stranded RNAs (dsRNAs) or small interfering RNAs (siRNAs) (40–43). Moreover, a large-scale field application of IAPV dsRNA improved bee survival, colony size, and honey yield (44). RNAi has also been used to help control the parasitic mite Varroa destructor (45). In one previous study, N. ceranae ADP/ATP transporter genes were targeted and silenced by corresponding dsRNAs, resulting in the decline of spore loads and alleviation of disease in infected bees (46).
All applications of RNAi in treating honey bee diseases to date have targeted the genes of parasites or pathogens. Nevertheless, disease always involves interactions between hosts and parasites/pathogens, and it is also possible to mitigate infections from the host perspective, i.e., to use RNAi to manipulate host factors that interact with parasites or pathogens. In fact, Nosema infection dramatically alters honey bee transcriptional responses (47–49), providing potential targets for host-based RNAi manipulation.
The Wnt signaling pathway is an important regulator of immune function in mammals (50) and has recently been found to function in regulating immune pathways, specifically Toll pathways, in insects (Drosophila) (51). There are several genes that serve as antagonists of the Wnt pathway, and thus, upregulated expression of these genes should suppress immune function. One of these genes has been found in Drosophila—the naked cuticle gene (52). As suppressors of immune function, these genes may serve as excellent targets for parasite manipulation of the hosts' transcriptional pathways: if the parasite can upregulate these antagonists, it can suppress the host's immune response. In light of the fact that nkd is a negative regulator of the Wnt signaling pathway, we hypothesized that nkd expression might be regulated by Nosema infection and thereby could serve as a potential target of RNAi to mitigate Nosema infection in honey bees. In the work described here, we confirmed that nkd mRNA levels were upregulated when bees were infected by N. ceranae. Our RNAi experiments showed that silencing nkd led to the enhancement of immune responses through an increase in immune gene expression, reduction in parasite spore load, and improvement in the life expectancy of N. ceranae-infected bees. We provide unequivocal evidence that silencing the nkd gene is an efficient way to control N. ceranae infection and improve honey bee health.
MATERIALS AND METHODS
Honey bees.
The honey bees used in this study were collected from colonies of Apis mellifera ligustica maintained at the USDA-ARS Bee Research Laboratory, Beltsville, MD. Newly emerged bees were obtained by removing honeycomb frames with sealed brood from strong and healthy colonies that were identified as Nosema negative, placing the frames into mesh-walled cages individually, and maintaining the frames in an insect growth chamber at 34 ± 1°C and 55% ± 5% relative humidity (RH) overnight. Emerging adult worker bees were collected the next day (<24 h). In order to make sure that the experimental bees were free of N. ceranae infection before proceeding to the experimental inoculation, we confirmed the negative status of Nosema infection using a hemocytometer and light microscopy. Briefly, 30 abdomens of newly emerged bees were dissected and ground up thoroughly in 30 ml of deionized H2O. Ten microliters of the homogenate was loaded onto a hemocytometer, and the presence or absence of spores was determined under a light microscope following a previously described method (53).
Inoculum preparation.
N. ceranae spores were purified from foragers collected outside the entrance of identified N. ceranae-infected colonies. Midguts were pulled out and homogenized in sterile distilled water (dH2O). The purification of Nosema spores from the homogenate was performed as described by Fries et al. (54). The homogenate was filtered through a nylon mesh cloth (65-μm pore size) by centrifuging it for 5 min at 3,000 × g. The supernatant was discarded, and the pellet was resuspended in sterile water and centrifuged for 10 min at 5,000 × g; this step was repeated twice. Finally, the pellet was resuspended in dH2O and stored at room temperature for no more than 1 week. The inoculum was obtained by diluting spore solution with sucrose solution, with a final concentration of 2.0 × 107 spores/ml in 50% (mass/vol) sucrose solution.
Inoculation.
The newly emerged bees were collected and starved for 3 h in an incubator (32°C and 75% RH) before being inoculated with Nosema spores in solution. Individual feeding was performed for each bee by holding two wings of a bee on each side with one hand and feeding the bee with 5-μl inoculum (100,000 spores) with a pipette with the other hand. Thirty bees were then distributed into each cup cage, which is a plastic bee-rearing cup with a top-feeder design (55). A 3-ml syringe filled with 50% (mass/vol) sucrose solution was inserted in the top of the cage to feed the bees, and the solution was changed every 3 days. A small pollen patty was supplied in the bottom of the cage for 6 days. The same number of control bees (without spore inoculation) were transferred into a cup cage as well. There were 4 replicates for each group. All cup cages were maintained in an incubator (32°C and 75% RH). Five bees were sampled from each cage at days 6 (D6), 9 (D9), 12 (D12), 15 (D15), and 18 (D18) postinoculation. The collected bee samples were immediately frozen at −80°C and stored until processing.
Production of dsRNA.
Primers were designed from the sequence of the A. mellifera nkd gene (GenBank accession no. XM_001120899) by using the E-RNAi web service (56), and primers for GFP, which served as the control gene, were used from previous studies (57). All primer sequences were fused with the T7 promoter sequence (Table 1). PCRs were performed using different templates individually: the cDNA of an adult bee was used for the amplification of nkd, and the pGFP vector (Clontech) was used for the amplification of GFP. The 100-μl PCR mixture contained the following: 78 μl H2O, 10 μl 10× reaction buffer (Invitrogen), 3 μl MgCl2, 2 μl deoxynucleoside triphosphate (dNTP) mixture (10 mM; Invitrogen), 2 μl forward primer (20 μM), 2 μl reverse primer (20 μM), 1 μl Taq polymerase (Invitrogen), and 2 μl DNA template. The PCR program was 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 90 s, and then 72°C for 10 min. After each PCR amplification, the products were verified in 1.0% agarose gels, purified, and used as the templates for the in vitro transcription reaction. The production of dsRNAs was carried out by using the MEGAscript RNAi kit (Ambion). The transcription reaction mixtures were assembled according to the manufacturer's instruction, and the time of incubation at 37°C was extended to 15 h. The following steps, such as nuclease digestion, purification, and elution, were performed using the materials associated with the kit. The quality of the dsRNAs was tested using 1.0% agarose gels, and their concentrations were determined with a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Inc.). The products were diluted with sucrose solution to final concentrations of 10 μg/ml, 20 μg/ml, and 40 μg/ml dsRNA in 50% (mass/vol) sucrose solution. The final solutions were stored at −80°C until use.
TABLE 1.
Primer sequences used in this study
| Purpose | Gene | Accession no. | Primer | Sequence (5′→3′)a | Amplicon |
Reference or source | |
|---|---|---|---|---|---|---|---|
| Location | Length (bp) | ||||||
| RNAi | nkd | XM_001120899 | nkd-RNAi-F | TAATACGACTCACTATAGGGCGACGCGCTTATGTTCAACCTC | 1889–2122 | 234 | This study |
| nkd-RNAi-R | TAATACGACTCACTATAGGGCGAGGTCGCGTGTTTCAAATGAT | ||||||
| GFP | AF324407 | GFP-RNAi-F | TAATACGACTCACTATAGGGCGATTCCATGGCCAACACTTGTCA | 173–674 | 502 | 57 | |
| GFP-RNAi-R | TAATACGACTCACTATAGGGCGATCAAGAAGGACCATGTGGTC | ||||||
| qPCR | nkd | nkd-F | AGGATGACGGTGAAAATGCG | 1365–1540 | 176 | This study | |
| nkd-R | ATTAGTCGTGAGGAGAGGCG | ||||||
| β-actin | NM_001185145 | actin-F | TGCCAACACTGTCCTTTCTG | 1018–1173 | 156 | 73 | |
| actin-R | AGAATTGACCCACCAATCCA | ||||||
| Abaecin | NM_001011617 | Abaecin-F | AGATCTGCACACTCGAGGTCTG | 14–214 | 201 | 74 | |
| Abaecin-R | TCGGATTGAATGGTCCCTGA | ||||||
| Apidaecin | NM_001011613 | Apidaecin-F | TTTTGCCTTAGCAATTCTTGTTG | 58–137 | 80 | This study | |
| Apidaecin-R | GCAGGTCGAGTAGGCGGATCT | ||||||
| Defensin-1 | NM_001011616 | Defensin-1-F | TGTCGGCCTTCTCTTCATGG | 88–288 | 201 | This study | |
| Defensin-1-R | TGACCTCCAGCTTTACCCAAA | ||||||
| PGRP-S2 | NM_001163716 | PGRP-S2-F | TTGCACAAAATCCTCCGCC | 146–274 | 129 | This study | |
| PGRP-S2-R | CACCCCAACCCTTCTCATCT | ||||||
Underlining shows the T7 promoter sequence.
RNAi treatment.
Before the RNA interference (RNAi) treatment, the newly emerged bees were collected, inoculated with N. ceranae spores as described above, and then transferred into bee-rearing cages. In each cage, 20 bees were supplied with 1.5 ml of 50% sucrose solution containing nkd or GFP dsRNA in a 3-ml syringe and a small pollen patty in the bottom of the cage on the same day. Bees were fed with the dsRNA for 15 days; the dsRNA solution was changed daily. Pollen patties were supplied for the first 6 days and changed every 3 days. All cages were incubated at 32°C and 75% RH, and dead bees were removed every day.
To test the efficiency of gene knockdown, three different concentrations of nkd dsRNA solution, 10 μg/ml, 20 μg/ml, and 40 μg/ml, were applied to separate cages, and 20 μg/ml GFP dsRNA solution was used for control bees. There were three replicates for each treatment. The bees were sampled at D9 and D15 after the ingestion of dsRNA and stored at −80°C until use.
To study the biological responses to the knockdown of nkd, 20 μg/ml of nkd dsRNA and GFP dsRNA were fed to infected bees. Another control group was set up with bees that were not given any treatment and fed only 50% sucrose solution and pollen. Each group contained three replicates. The numbers of dead bees were recorded daily, and dead bees were then removed. All the bees were collected at D15 and stored at −80°C until use.
Spore counting.
To evaluate the Nosema infection levels in the RNAi-treated bees, the spores were counted in individual bees. First, the abdomens were separated, put into 1.5-ml Eppendorf tubes individually, and homogenized thoroughly in 1 ml dH2O using a pestle. Then, each homogenate was diluted 100 times. Ten microliters of the diluted solution was loaded onto a hemocytometer, and spores were counted under a light microscope as described by Cantwell (53).
RNA extraction and cDNA synthesis.
TRIzol reagent (Invitrogen) was used to extract total RNA from the abdomens of individual bees, following the manufacturer's protocols. Any genomic DNA contamination was removed by treatment with DNase I (DNA-free kit; Ambion). The purity and quantity of RNA samples were examined by using a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Inc.). All RNAs were stored at −80°C until use. First-strand cDNA was produced from a 20-μl reverse transcription reaction mixture that contained 2 μl total RNA (approximately 1 μg/μl), 1 μl dNTP mixture (10 mM), 1 μl random primers (0.15 μg/μl), 1 μl dithiothreitol (DTT) (0.1 M), 4 μl 5× first-strand buffer, 1 μl SuperScript III reverse transcriptase (200 U/μl, Invitrogen), and 10 μl nuclease-free water. The reaction program was as follows: 25°C for 5 min and 50°C for 45 min, followed by 70°C for 15 min. The cDNAs were stored at −20°C until use.
qPCR.
Quantitative PCRs (qPCRs) were run on a CFX384 Touch real-time PCR system (Bio-Rad, Hercules, CA), and SYBR green was selected as the detection signal. The primers used here were designed with Primer3 (Table 1) (58). β-actin served as the reference gene, and all primer pairs were validated as described in reference 59. Each 10-μl PCR mixture was assembled by mixing 5 μl 2× Brilliant III ultrafast SYBR green qPCR mix (Stratagene, La Jolla, CA), 0.25 μl forward primer (20 mM), 0.25 μl reverse primer (20 mM), 0.5 μl cDNA, and 4 μl nuclease-free water. Each reaction was run in duplicate. The PCR program was 95°C for 3 min, followed by 40 cycles of 95°C for 10 s and 60°C for 45 s. Melting curves and no-template-control (NTC) reactions were monitored to evaluate the quality and specificity of amplification. Only a single peak was seen in all melting curves, and no peaks showed for the NTCs. The PCR products were run on a 1% agarose gel to confirm the expected amplification sizes. The threshold cycle (CT) values were generated by using CFX Manager 3.1 (Bio-Rad). The relative quantification of gene expression was calculated with the comparative CT (ΔΔCT) method (60). For each gene, the average CT value of the target was normalized with the corresponding β-actin value using the formula ΔCT = average CT(target) − average CT(β-actin), and the group of bees with the lowest level of gene expression was chosen as a calibrator [ΔCT(calibrator)]. The ΔCT value of each group was subtracted from the ΔCT(calibrator) value to generate the ΔΔCT. The concentration of each target in each group was calculated using the formula 2−(ΔΔCT) and expressed as the n-fold difference relative to the calibrator value.
Bioinformatic and statistical analyses.
Multiple alignments of insect nkd protein sequences were carried out with ClustalX 2.0 (61). Protein domain identification and secondary structure prediction of A. mellifera nkd were performed with the InterProScan tool and EMBOSS: Garnier algorithm of Geneious version 9.1.3 (Biomatters), respectively.
The dynamics of nkd gene expression during the course of Nosema infection, the levels of immune gene expression, and the spore loads of infected bees after RNAi treatment were analyzed using independent-sample t tests. The mRNA levels of nkd after dsRNA treatment were analyzed by one-way analysis of variance (ANOVA) with all compared groups passing an equal variance test, and the post hoc effects were determined using the Tukey honestly significant difference (HSD) test. Survival analysis was performed using the Kaplan-Meier method, and log rank and Wilcoxon tests were computed to assess the overall homogeneity between the treatment strata. Pairwise comparisons were carried out using Wilcoxon tests. In all cases, a P value of <0.05 was taken to be significant. All analyses were carried out using PASW Statistics 18 (SPSS, Inc.).
RESULTS
Identification of a highly conserved nkd gene in honey bees.
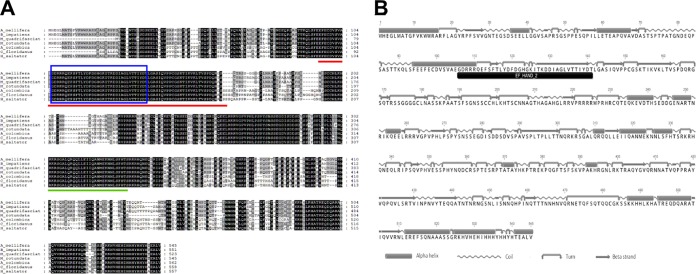
The identified honey bee nkd gene encodes a predicted protein of 545 amino acids (aa), which shares 92.9% and 89.2% sequence identity with the nkd-encoded proteins of the bumble bee Bombus impatiens and stingless bee Melipona quadrifasciata, respectively (Fig. 1). This protein is highly conserved in Hymenoptera, as remarkable sequence identity is also seen in non-Apidae species, ranging from 65.5% (Camponotus floridanus) to 85.8% (Megachile rotundata) (Fig. 1). A conserved EF-hand domain (InterPro accession number IPR002048), which is known to bind calcium, was identified in the nkd proteins (Fig. 1). Additional conserved regions included a region responsible for interaction with Dishevelled (Dsh), a component of the Wnt signaling pathway (62), and the nuclear localization motif that is required for nuclear localization and inhibition of Wnt signaling (Fig. 1) (63). Although the diversity of nkd protein sequences increases dramatically when the comparison is expanded to different insect orders, the sequence features mentioned above are still highly conserved (see Fig. S1 in the supplemental material).
FIG 1.
Sequence conservation and predicted secondary structure of A. mellifera nkd-encoded protein. (A) Multiple alignment of A. mellifera (XP_001120899), Bombus impatiens (XP_012249347), Megachile rotundata (XP_003702467), Melipona quadrifasciata (KOX70301), Atta colombica (KYM87061), Harpegnathos saltator (XP_011151892), and Camponotus floridanus (EFN66676) nkd protein sequences. Black and gray shadings indicate identity and a high degree of conservation of amino acids, respectively. The region responsible for interaction with dsh is indicated by the red bar. The nuclear localization motif is underlined by the green bar. The blue box highlights the EF-hand domain. (B) Protein domain identification and secondary structure prediction of A. mellifera nkd. The conserved domain is shown by a black bar.
N. ceranae infection upregulates the expression of the nkd gene.
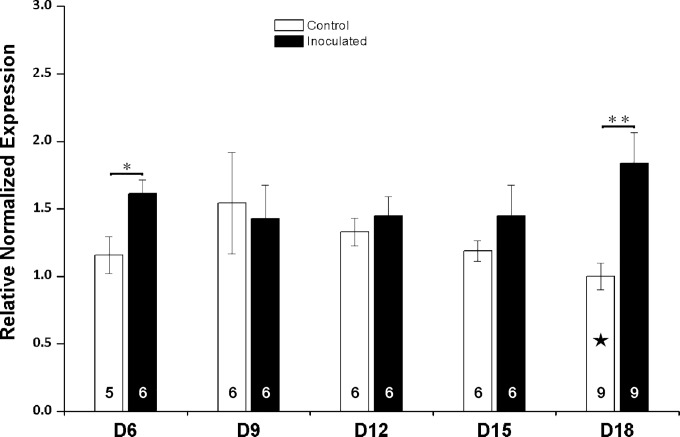
As shown by the results in Fig. 2, the dynamics of nkd gene expression was altered during the N. ceranae infection. The expression of nkd in the infected bees was significantly upregulated compared with that of control bees at day 6 (D6) and D18 postinoculation (for D6, t test, t = −2.774, df = 9, P = 0.022; and for D18, t test, t = −3.387, df = 10.860, P = 0.006) (Fig. 2). Collectively, N. ceranae infection upregulated the nkd gene expression (t test, t = −2.872, df = 63, P = 0.006).
FIG 2.
Expression profile of nkd during N. ceranae infection. The x axis indicates the days postinoculation with Nosema spores. The relative gene expression levels (y axis) are shown as mean values ± standard errors of the means (SEM). The sample size is shown in the bottom of each bar, and the solid five-pointed star indicates the calibrator used to normalize the gene expression. Data were analyzed by the independent-sample t test. Significant differences between groups are indicated by asterisks (*, P < 0.05; **, P < 0.01).
Ingestion of dsRNA silences the nkd gene expression in N. ceranae-infected bees.
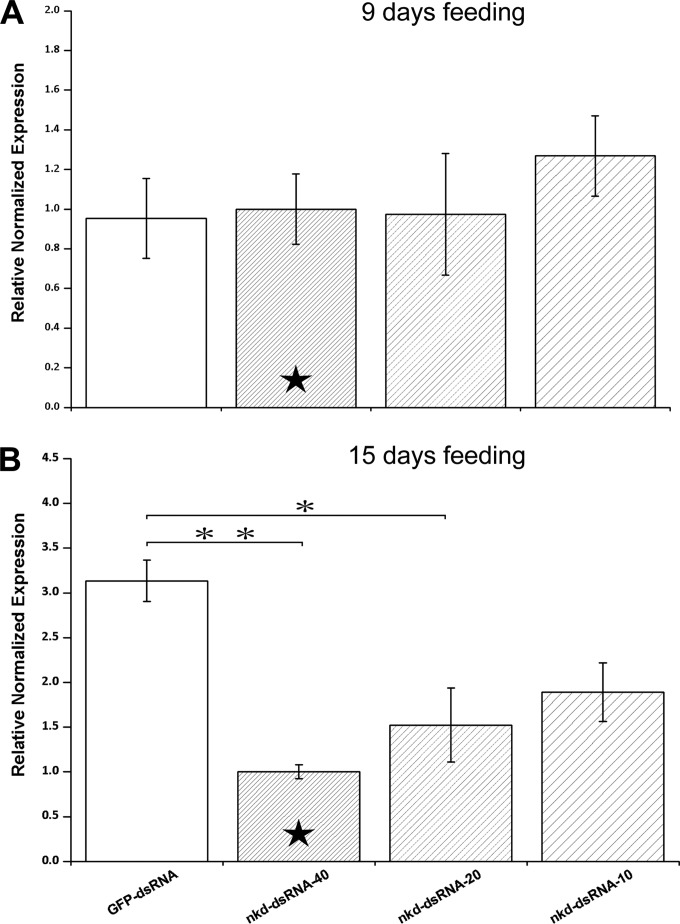
As shown by the results in Fig. 3, 9 days of feeding nkd dsRNA was insufficient to reduce the mRNA levels of the target gene in adult bees (ANOVA, F1,11 = 0.442, P = 0.742), but the gene expression could be significantly silenced by 15 days of ingesting the corresponding dsRNA (ANOVA, F1,11 = 9.1781, P = 0.005). Moreover, the knockdown of nkd was dosage dependent (Fig. 3): along with an increase in the dsRNA concentration, the effect of gene silencing increased. When 10 μg/ml of dsRNA was applied, a 40% knockdown of nkd mRNA was achieved, while larger amounts of 20 or 40 μg/ml dsRNA resulted in 50% and 70% gene silencing, and the differences compared to the nkd expression in controls became significant (P = 0.018 for 20 μg/ml and P = 0.004 for 40 μg/ml) (Fig. 3).
FIG 3.
Knockdown of nkd gene in adult bees by dsRNA ingestion. All groups of adult bees were inoculated with Nosema spores first and then fed with sucrose solution containing dsRNA for 15 days. The silencing effect was examined after 9 days (A) and 15 days (B) of feeding dsRNA. The control bees (GFP-dsRNA) were fed with the dsRNA derived from the GFP sequence. For the treatment groups, three different concentrations of nkd dsRNA were examined, as follows: 40 μg/ml (nkd-dsRNA-40), 20 μg/ml (nkd-dsRNA-20), and 10 μg/ml (nkd-dsRNA-10). The relative gene expression levels are shown as mean values ± SEM. All groups had the same sample size (n = 3). The calibrator used to normalize the gene expression is indicated by a solid five-pointed star inside the bar. One-way ANOVA was employed to analyze the differences between data, and post hoc effects were identified by Tukey HSD tests. Significant differences between groups are indicated by asterisks (*, P < 0.05; **, P < 0.01).
Silencing of the nkd gene upregulates immune gene expression in N. ceranae-infected bees.
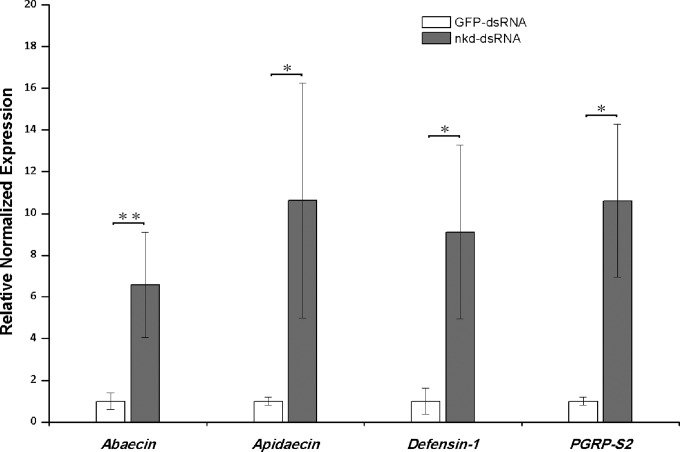
As shown by the results in Fig. 4, the nkd-silenced bees expressed significantly larger amounts of mRNA for three antimicrobial peptide (AMP) genes, Abaecin, Apidaecin, and Defensin-1, relative to the expression of these genes in the control bees (for Abaecin, t test, t = −3.689, df = 9, P = 0.005; for Apidaecin, t test, t = −3.047, df = 4.982, P = 0.029; and for Defensin-1, t test, t = −2.855, df = 9, P = 0.019). Moreover, the expression of a peptidoglycan recognition protein (PGRP) gene, PGRP-S2, was also significantly upregulated in nkd-silenced bees compared with its expression in the control bees (t test, t = −3.183, df = 5.043, P = 0.024) (Fig. 4).
FIG 4.
Effects of nkd gene silencing on immune gene expression in Nosema-infected bees. The relative mRNA levels of immune genes (x axis) were compared between control bees fed with 20 μg/ml GFP dsRNA (GFP-dsRNA; n = 6) and treatment bees fed with 20 μg/ml nkd dsRNA (nkd-dsRNA; n = 5). Both groups were inoculated with Nosema spores before dsRNA feeding. The expression levels are shown as mean values ± SEM. Data were analyzed by independent-sample t test. Significant differences between groups are indicated by asterisks (*, P < 0.05; **, P < 0.01).
Silencing of nkd gene reduces Nosema spore levels and extends the life span of N. ceranae-infected bees.
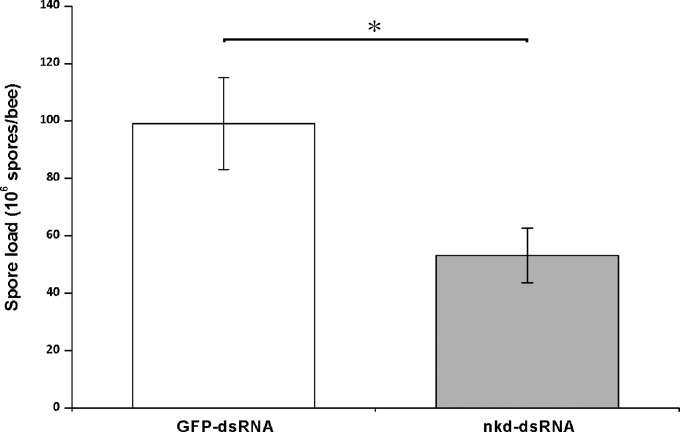
After feeding with nkd dsRNA, the spore load of infected bees was significantly (approximately 50%) lower than that in controls (fed with GFP dsRNA) (t test, t = 2.458, df = 15.485, P = 0.026) (Fig. 5), indicating that knockdown of nkd gene expression results in a decrease in Nosema infection levels. Survival analysis was performed to further examine the effect of silencing the nkd gene on the life span of honey bees after Nosema infection. The survival distributions for the groups tested were significantly different (log rank test, χ2 = 24.472, df = 2, P < 0.001; and Wilcoxon test, χ2 = 25.020, df = 2, P < 0.001) (Fig. 6). Nosema infection indeed induced higher mortality in the infected honey bees (no treatment versus GFP dsRNA treatment, Wilcoxon test, χ2 = 25.218, P < 0.001) (Fig. 6); however, silencing of nkd significantly reduced the incidence of death of adult bees (Fig. 6) (nkd dsRNA versus GFP dsRNA, Wilcoxon test, χ2 = 4.165, P = 0.041).
FIG 5.
Effect of nkd gene silencing on Nosema infection levels in adult bees. The Nosema infection levels were determined by spore counting. The spore loads of the two groups of bees, the control bees fed with 20 μg/ml GFP dsRNA (n = 11) and the treatment bees fed with 20 μg/ml nkd dsRNA (n = 8), were compared. The spore loads are expressed as mean values ± SEM. Significant difference between data was analyzed by independent-sample t test and is indicated with an asterisk (*, P < 0.05).
FIG 6.
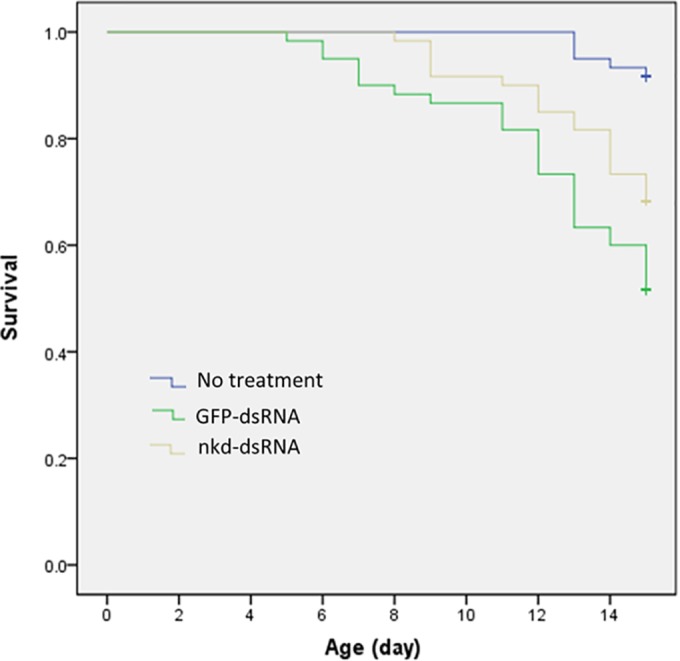
Effect of nkd gene silencing on the life span of honey bees infected by N. ceranae. Survival curves for bees inoculated with 100,000 N. ceranae spores at day 0 (i.e., within 24 h after adult emergence) and fed with 50% (mass/vol) sucrose solution containing 20 μg/ml nkd dsRNA (nkd-dsRNA; n = 60) or 20 μg/ml GFP dsRNA (GFP-dsRNA; n = 60) for 15 days and for the bees that did not receive any treatment and were only fed with 50% sucrose solution (no treatment; n = 60). Knockdown of nkd gene reduced the incidence of death (P = 0.041 by Wilcoxon test).
DISCUSSION
Significant progress has been made in exploring RNAi as a therapeutic strategy for controlling diseases in honey bees, with much attention focused on pathogen-specific virulence determinants. Nosema are obligate intracellular parasites and, therefore, require host cell proteins and pathways to support their replication and many phases of their life cycles (12, 13). In the present study, the identification and characterization of a host-based factor that is required for parasite pathogenesis in hosts allow us to obtain important insights into the host-parasite interactions and to propose a potential drug target against the parasite.
Wnt signaling is an evolutionarily conserved pathway that plays a critical role in embryogenesis and cell proliferation and differentiation (64). Additionally, recent studies found that activation of the Wnt signaling pathway regulates the immune response to certain pathogenic bacterial infections by upregulating the expression of anti-inflammatory factors and downregulating the expression of inflammatory factors (65–68). nkd is linked to Wnt signaling and was originally found to act as an inducible antagonist of Wnt signaling during embryonic development in Drosophila (52). The results of our study confirmed our hypothesis that nkd and Wnt signaling might be also involved in the honey bee responses to Nosema infections. Indeed, Nosema infection can result in dramatic host transcriptional responses (47–49), and the expression of the nkd gene is significantly upregulated during N. ceranae infection in honey bees, suggesting that nkd and Wnt signaling might be targeted by N. ceranae during the infection process to alter the defensive function of hosts.
Knockdown of nkd by feeding Nosema-infected bees with dsRNA specific to nkd led to several remarkable alternations, one of which was the modulation of host immune responses. Previous studies have revealed that N. ceranae infection induces immunosuppression in honey bees by downregulating several immune genes, such as the AMP genes Apidaecin, Abaecin, Defensin-1, and Hymenoptaecin (69, 70). Our results show that silencing the nkd gene in bees infected with N. ceranae can reverse immune suppression and enhance the host immune response. The exact mechanism of host immune induction by nkd silencing remains unclear. However, silencing of the nkd gene ultimately reduces the Nosema infection levels and extends the life span of infected adult bees.
Since infection always involves interaction between host and parasite/pathogen, infections can theoretically be controlled by targeting the host of the parasite/pathogen. All previous gene-based efforts to control honey bee parasites or pathogens have targeted these biotic threats directly. Here, by targeting the honey bee nkd gene with RNAi, we demonstrate that silencing a honey bee gene can suppress the reproduction of parasites/pathogens and improve the overall health of honey bees. Our results provide a novel host-derived strategy to mitigate honey bee disease. Similar studies have been reported in various species. For example, silencing of the Cactus gene, an inhibitor of the Toll pathway, reduces the extent of dengue virus infection in the midgut by fourfold in Aedes aegypti mosquitoes (71). Downregulation of scavenger receptor class B type 1 (SR-B1) expression by RNAi dramatically decreases the susceptibility of human hepatoma cells to hepatitis C virus (HCV) infection, resulting in the inhibition of this virus infection (72). These studies together indicate that targeting host factors by RNAi can potentially protect the hosts from infections of parasites/pathogens and promote the overall health of hosts.
RNAi technology has great potential for relieving the impacts of honey bee diseases. Other previous studies (46), combined with our efforts, demonstrate that silencing parasite/host genes by RNAi manipulation is efficient to suppress parasite development and improve honey bee health to some extent in the laboratory. It is likely that the combination of both strategies, meaning targeting both host and parasite genes in the same RNAi manipulation, will lead to better results for controlling Nosema infection. As for field application, more experiments are needed to determine the ideal treatment time, the suitable concentration of dsRNA, and other factors. The present study will help direct the application of RNAi to mitigate N. ceranae infection in honey bees.
In sum, these studies have identified a host factor required for Nosema infection and highlight the potency of host-derived RNAi-based therapeutics to inhibit not only microsporidian parasite infection but also potentially a wide range of pathogens and parasites that cause serious diseases in honey bees.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bart Smith, Sam Abban, and Andy Ulsamer for their laboratory and field assistance. We also thank the anonymous reviewers for their helpful comments.
Support for this study was provided by USDA-NIFA (grant 2014-67013-21784 to Y.P.C.), and C.R.-G. was supported by a Ph.D. INIA-FEDER Spanish grant (INIA-FEDER project RTA2012-00072-C02-01).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02105-16.
REFERENCES
- 1.vanEngelsdorp D, Meixner MD. 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol 103(Suppl):S80–S95. doi: 10.1016/j.jip.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Breeze TD, Vaissière BE, Bommarco R, Petanidou T, Seraphides N, Kozák L, Scheper J, Biesmeijer JC, Kleijn D, Gyldenkærne S, Moretti M, Holzschuh A, Steffan-Dewenter I, Stout JC, Pärtel M, Zobel M, Potts SG. 2014. Agricultural policies exacerbate honeybee pollination service supply-demand mismatches across Europe. PLoS One 9:e82996. doi: 10.1371/journal.pone.0082996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.vanEngelsdorp D, Hayes J, Underwood RM, Pettis JS. 2010. A survey of honey bee colony losses in the United States, fall 2008 to spring 2009. J Apic Res 49:7–14. doi: 10.3896/IBRA.1.49.1.03. [DOI] [Google Scholar]
- 4.vanEngelsdorp D, Hayes J, Underwood RM, Caron D, Pettis J. 2011. A survey of managed honey bee colony losses in the USA, fall 2009 to winter 2010. J Apic Res 50:1–10. doi: 10.3896/IBRA.1.50.1.01. [DOI] [Google Scholar]
- 5.vanEngelsdorp D, Caron D, Hayes J, Underwood R, Henson M, Rennich K, Spleen A, Andree M, Snyder R, Lee K, Roccasecca K, Wilson M, Wilkes J, Lengerich E, Pettis J. 2012. A national survey of managed honey bee 2010-11 winter colony losses in the USA: results from the Bee Informed Partnership. J Apic Res 51:115–124. doi: 10.3896/IBRA.1.51.1.14. [DOI] [Google Scholar]
- 6.Spleen AM, Lengerich EJ, Rennich K, Caron D, Rose R, Pettis JS, Henson M, Wilkes JT, Wilson M, Stitzinger J, Lee K, Andree M, Snyder R, vanEngelsdorp D. 2013. A national survey of managed honey bee 2011-12 winter colony losses in the United States: results from the Bee Informed Partnership. J Apic Res 52:44–53. doi: 10.3896/IBRA.1.52.2.07. [DOI] [Google Scholar]
- 7.Steinhauer NA, Rennich K, Wilson ME, Caron DM, Lengerich EJ, Pettis JS, Rose R, Skinner JA, Tarpy DR, Wilkes JT, vanEngelsdorp D. 2014. A national survey of managed honey bee 2012-2013 annual colony losses in the USA: results from the Bee Informed Partnership. J Apic Res 53:1–18. doi: 10.3896/IBRA.1.53.1.01. [DOI] [Google Scholar]
- 8.van Engelsdorp D, Hayes J Jr, Underwood RM, Pettis J. 2008. A survey of honey bee colony losses in the U.S., fall 2007 to spring 2008. PLoS One 3:e4071. doi: 10.1371/journal.pone.0004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.vanEnglesdorp D, Underwood R, Caron D, Hayes JJ. 2007. An estimate of managed colony losses in the winter of 2006-2007: a report commissioned by the Apiary Inspectors of America. Am Bee J 147:599–603. [Google Scholar]
- 10.Genersch E. 2010. Honey bee pathology: current threats to honey bees and beekeeping. Appl Microbiol Biotechnol 87:87–97. doi: 10.1007/s00253-010-2573-8. [DOI] [PubMed] [Google Scholar]
- 11.Ratnieks FLW, Carreck NL. 2010. Clarity on honey bee collapse? Science 327:152–153. doi: 10.1126/science.1185563. [DOI] [PubMed] [Google Scholar]
- 12.Fries I. 2010. Nosema ceranae in European honey bees (Apis mellifera). J Invertebr Pathol 103(Suppl):S73–S79. doi: 10.1016/j.jip.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 13.Holt HL, Grozinger CM. 2016. Approaches and challenges to managing Nosema (Microspora: Nosematidae) parasites in honey bee (Hymenoptera: Apidae) colonies. J Econ Entomol 109:1487–1503. doi: 10.1093/jee/tow103. [DOI] [PubMed] [Google Scholar]
- 14.Fries I, Feng F, da Silva A, Slemenda SB, Pieniazek NJ. 1996. Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur J Protistol 32:356–365. doi: 10.1016/S0932-4739(96)80059-9. [DOI] [Google Scholar]
- 15.Higes M, Martín R, Meana A. 2006. Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J Invertebr Pathol 92:93–95. doi: 10.1016/j.jip.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Huang W-F, Jiang J-H, Chen Y-W, Wang C-H. 2007. A Nosema ceranae isolate from the honeybee Apis mellifera. Apidologie 38:30–37. doi: 10.1051/apido:2006054. [DOI] [Google Scholar]
- 17.Chen Y, Evans JD, Smith IB, Pettis JS. 2008. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J Invertebr Pathol 97:186–188. doi: 10.1016/j.jip.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Giersch T, Berg T, Galea F, Hornitzky M. 2009. Nosema ceranae infects honey bees (Apis mellifera) and contaminates honey in Australia. Apidologie 40:117–123. doi: 10.1051/apido/2008065. [DOI] [Google Scholar]
- 19.Higes M, Martín-Hernández R, Garrido-Bailón E, Botías C, Meana A. 2009. The presence of Nosema ceranae (Microsporidia) in North African honey bees (Apis mellifera intermissa). J Apic Res 48:217–219. doi: 10.3896/IBRA.1.48.3.12. [DOI] [Google Scholar]
- 20.Invernizzi C, Abud C, Tomasco IH, Harriet J, Ramallo G, Campá J, Katz H, Gardiol G, Mendoza Y. 2009. Presence of Nosema ceranae in honeybees (Apis mellifera) in Uruguay. J Invertebr Pathol 101:150–153. doi: 10.1016/j.jip.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Martín-Hernández R, Meana A, Prieto L, Salvador AM, Garrido-Bailón E, Higes M. 2007. Outcome of colonization of Apis mellifera by Nosema ceranae. Appl Environ Microbiol 73:6331–6338. doi: 10.1128/AEM.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams GR, Shafer ABA, Rogers REL, Shutler D, Stewart DT. 2008. First detection of Nosema ceranae, a microsporidian parasite of European honey bees (Apis mellifera), in Canada and central USA. J Invertebr Pathol 97:189–192. doi: 10.1016/j.jip.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Traver BE, Fell RD. 2011. Prevalence and infection intensity of Nosema in honey bee (Apis mellifera L.) colonies in Virginia. J Invertebr Pathol 107:43–49. doi: 10.1016/j.jip.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Yoshiyama M, Kimura K. 2011. Distribution of Nosema ceranae in the European honeybee, Apis mellifera in Japan. J Invertebr Pathol 106:263–267. doi: 10.1016/j.jip.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Martínez J, Leal G, Conget P. 2012. Nosema ceranae an emergent pathogen of Apis mellifera in Chile. Parasitol Res 111:601–607. doi: 10.1007/s00436-012-2875-0. [DOI] [PubMed] [Google Scholar]
- 26.Goblirsch M, Huang ZY, Spivak M. 2013. Physiological and behavioral changes in honey bees (Apis mellifera) induced by Nosema ceranae infection. PLoS One 8:e58165. doi: 10.1371/journal.pone.0058165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higes M, García-Palencia P, Martín-Hernández R, Meana A. 2007. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J Invertebr Pathol 94:211–217. doi: 10.1016/j.jip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Mayack C, Naug D. 2009. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J Invertebr Pathol 100:185–188. doi: 10.1016/j.jip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Paxton RJ, Klee J, Korpela S, Fries I. 2007. Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 38:558–565. doi: 10.1051/apido:2007037. [DOI] [Google Scholar]
- 30.Higes M, Martín-Hernández R, Botías C, Bailón EG, González-Porto AV, Barrios L, del Nozal MJ, Bernal JL, Jiménez JJ, Palencia PG, Meana A. 2008. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ Microbiol 10:2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x. [DOI] [PubMed] [Google Scholar]
- 31.Higes M, Martín-Hernández R, Garrido-Bailón E, González-Porto AV, García-Palencia P, Meana A, Del Nozal MJ, Mayo R, Bernal JL. 2009. Honeybee colony collapse due to Nosema ceranae in professional apiaries. Environ Microbiol Rep 1:110–113. doi: 10.1111/j.1758-2229.2009.00014.x. [DOI] [PubMed] [Google Scholar]
- 32.Alaux C, Brunet J-L, Dussaubat C, Mondet F, Tchamitchan S, Cousin M, Brillard J, Baldy A, Belzunces LP, Le Conte Y. 2010. Interactions between Nosema microspores and a neonicotinoid weaken honeybees (Apis mellifera). Environ Microbiol 12:774–782. doi: 10.1111/j.1462-2920.2009.02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettis J, vanEngelsdorp D, Johnson J, Dively G. 2012. Pesticide exposure in honey bees results in increased levels of the gut pathogen Nosema. Naturwissenschaften 99:153–158. doi: 10.1007/s00114-011-0881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidau C, Diogon M, Aufauvre J, Fontbonne R, Viguès B, Brunet J-L, Texier C, Biron DG, Blot N, El Alaoui H, Belzunces LP, Delbac F. 2011. Exposure to sublethal doses of fipronil and thiacloprid highly increases mortality of honeybees previously infected by Nosema ceranae. PLoS One 6:e21550. doi: 10.1371/journal.pone.0021550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu JY, Smart MD, Anelli CM, Sheppard WS. 2012. Honey bees (Apis mellifera) reared in brood combs containing high levels of pesticide residues exhibit increased susceptibility to Nosema (Microsporidia) infection. J Invertebr Pathol 109:326–329. doi: 10.1016/j.jip.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Huang W-F, Solter LF, Yau PM, Imai BS. 2013. Nosema ceranae escapes fumagillin control in honey bees. PLoS Path 9:e1003185. doi: 10.1371/journal.ppat.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannon GJ. 2002. RNA interference. Nature 418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 38.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 39.Grozinger CM, Robinson GE. 2015. The power and promise of applying genomics to honey bee health. Curr Opin Insect Sci 10:124–132. doi: 10.1016/j.cois.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X, Zhang Y, Yan X, Han R. 2010. Prevention of Chinese sacbrood virus infection in Apis cerana using RNA interference. Curr Microbiol 61:422–428. doi: 10.1007/s00284-010-9633-2. [DOI] [PubMed] [Google Scholar]
- 41.Chen YP, Pettis JS, Corona M, Chen WP, Li CJ, Spivak M, Visscher PK, DeGrandi-Hoffman G, Boncristiani H, Zhao Y, vanEngelsdorp D, Delaplane K, Solter L, Drummond F, Kramer M, Lipkin WI, Palacios G, Hamilton MC, Smith B, Huang SK, Zheng HQ, Li JL, Zhang X, Zhou AF, Wu LY, Zhou JZ, Lee M-L, Teixeira EW, Li ZG, Evans JD. 2014. Israeli acute paralysis virus: epidemiology, pathogenesis and implications for honey bee health. PLoS Pathog 10:e1004261. doi: 10.1371/journal.ppat.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desai SD, Eu YJ, Whyard S, Currie RW. 2012. Reduction in deformed wing virus infection in larval and adult honey bees (Apis mellifera L.) by double-stranded RNA ingestion. Insect Mol Biol 21:446–455. doi: 10.1111/j.1365-2583.2012.01150.x. [DOI] [PubMed] [Google Scholar]
- 43.Maori E, Paldi N, Shafir S, Kalev H, Tsur E, Glick E, Sela I. 2009. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Mol Biol 18:55–60. doi: 10.1111/j.1365-2583.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 44.Hunter W, Ellis J, vanEngelsdorp D, Hayes J, Westervelt D, Glick E, Williams M, Sela I, Maori E, Pettis J, Cox-Foster D, Paldi N. 2010. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog 6:e1001160. doi: 10.1371/journal.ppat.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garbian Y, Maori E, Kalev H, Shafir S, Sela I. 2012. Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathog 8:e1003035. doi: 10.1371/journal.ppat.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paldi N, Glick E, Oliva M, Zilberberg Y, Aubin L, Pettis J, Chen Y, Evans JD. 2010. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl Environ Microbiol 76:5960–5964. doi: 10.1128/AEM.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aufauvre J, Misme-Aucouturier B, Viguès B, Texier C, Delbac F, Blot N. 2014. Transcriptome analyses of the honeybee response to Nosema ceranae and insecticides. PLoS One 9:e91686. doi: 10.1371/journal.pone.0091686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dussaubat C, Brunet J-L, Higes M, Colbourne JK, Lopez J, Choi J-H, Martín-Hernández R, Botías C, Cousin M, McDonnell C, Bonnet M, Belzunces LP, Moritz RFA, Le Conte Y, Alaux C. 2012. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS One 7:e37017. doi: 10.1371/journal.pone.0037017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holt H, Aronstein K, Grozinger C. 2013. Chronic parasitization by Nosema microsporidia causes global expression changes in core nutritional, metabolic and behavioral pathways in honey bee workers (Apis mellifera). BMC Genomics 14:799. doi: 10.1186/1471-2164-14-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staal FJT, Luis TC, Tiemessen MM. 2008. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 51.Gordon MD, Dionne MS, Schneider DS, Nusse R. 2005. WntD is a feedback inhibitor of Dorsal/NF-[kappa]B in Drosophila development and immunity. Nature 437:746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng W, Wharton KA, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP. 2000. naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403:789–795. doi: 10.1038/35001615. [DOI] [PubMed] [Google Scholar]
- 53.Cantwell GE. 1970. Standard methods for counting Nosema spores. Am Bee J 110:222–223. [Google Scholar]
- 54.Fries I, Chauzat MP, Chen YP, Doublet V, Genersch E, Gisder S, Higes M, McMahon DP, Martin-Hernandez R, Natsopoulou M, Paxton RJ, Tanner G, Webster TC, Williams GR. 2013. Standard methods for Nosema research. J Apic Res 52:1–28. doi: 10.3896/IBRA.1.52.1.08. [DOI] [Google Scholar]
- 55.Evans JD, Chen YP, di Prisco G, Pettis J, Williams V. 2009. Bee cups: single-use cages for honey bee experiments. J Apic Res 48:300–302. doi: 10.1080/00218839.2009.11101548. [DOI] [Google Scholar]
- 56.Horn T, Boutros M. 2010. E-RNAi: a web application for the multi-species design of RNAi reagents—2010 update. Nucleic Acids Res 38:W332–W339. doi: 10.1093/nar/gkq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amdam GV, Norberg K, Page JRE, Erber J, Scheiner R. 2006. Downregulation of vitellogenin gene activity increases the gustatory responsiveness of honey bee workers (Apis mellifera). Behav Brain Res 169:201–205. doi: 10.1016/j.bbr.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Huang ZY, Liu F, Li Z, Yan L, Zhang S, Chen S, Zhong B, Su S. 2013. Molecular cloning and characterization of juvenile hormone acid methyltransferase in the honey bee, Apis mellifera, and its differential expression during caste differentiation. PLoS One 8:e68544. doi: 10.1371/journal.pone.0068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 61.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 62.Rousset R, Mack JA, Wharton KA, Axelrod JD, Cadigan KM, Fish MP, Nusse R, Scott MP. 2001. naked cuticle targets dishevelled to antagonize Wnt signal transduction. Genes Dev 15:658–671. doi: 10.1101/gad.869201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waldrop S, Chan C-C, Cagatay T, Zhang S, Rousset R, Mack J, Zeng W, Fish M, Zhang M, Amanai M, Wharton KA. 2006. An unconventional nuclear localization motif is crucial for function of the Drosophila Wnt/Wingless antagonist naked cuticle. Genetics 174:331–348. doi: 10.1534/genetics.106.061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 65.Chen K, Yin L, Nie X, Deng Q, Wu Y, Zhu M, Li D, Li M, Wu M, Huang X. 2013. β-Catenin promotes host resistance against Pseudomonas aeruginosa keratitis. J Infect 67:584–594. doi: 10.1016/j.jinf.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 66.Liu X, Wu S, Xia Y, Li XE, Xia Y, Zhou ZD, Sun J. 2011. Wingless homolog Wnt11 suppresses bacterial invasion and inflammation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 301:G992–G1003. doi: 10.1152/ajpgi.00080.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silva-García O, Valdez-Alarcón JJ, Baizabal-Aguirre VM. 2014. The Wnt/β-catenin signaling pathway controls the inflammatory response in infections caused by pathogenic bacteria. Mediators Inflamm 2014:310183. doi: 10.1155/2014/310183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Lu R, Wu S, Zhang Y-G, Xia Y, Sartor RB, Sun J. 2012. Wnt2 inhibits enteric bacterial-induced inflammation in intestinal epithelial cells. Inflamm Bowel Dis 18:418–429. doi: 10.1002/ibd.21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antúnez K, Martín-Hernández R, Prieto L, Meana A, Zunino P, Higes M. 2009. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ Microbiol 11:2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x. [DOI] [PubMed] [Google Scholar]
- 70.Chaimanee V, Chantawannakul P, Chen Y, Evans JD, Pettis JS. 2012. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J Insect Physiol 58:1090–1095. doi: 10.1016/j.jinsphys.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 71.Xi Z, Ramirez JL, Dimopoulos G. 2008. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog 4:e1000098. doi: 10.1371/journal.ppat.1000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset F-L, Wakita T, Jaeck D, Doffoel M, Royer C, Soulier E, Schvoerer E, Schuster C, Stoll-Keller F, Bartenschlager R, Pietschmann T, Barth H, Baumert TF. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 73.Mutti NS, Dolezal AG, Wolschin F, Mutti JS, Gill KS, Amdam GV. 2011. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J Exp Biol 214:3977–3984. doi: 10.1242/jeb.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang X, Cox-Foster DL. 2005. Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc Natl Acad Sci U S A 102:7470–7475. doi: 10.1073/pnas.0501860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.