ABSTRACT
Studying the host-associated butyrate-producing bacterial community is important, because butyrate is essential for colonic homeostasis and gut health. Previous research has identified the butyryl coenzyme A (CoA):acetate-CoA transferase (EC 2.3.8.3) as a gene of primary importance for butyrate production in intestinal ecosystems; however, this gene family (but) remains poorly defined. We developed tools for the analysis of butyrate-producing bacteria based on 12 putative but genes identified in the genomes of nine butyrate-producing bacteria obtained from the swine intestinal tract. Functional analyses revealed that eight of these genes had strong But enzyme activity. When but paralogues were found within a genome, only one gene per genome encoded strong activity, with the exception of one strain in which no gene encoded strong But activity. Degenerate primers were designed to amplify the functional but genes and were tested by amplifying environmental but sequences from DNA and RNA extracted from swine colonic contents. The results show diverse but sequences from swine-associated butyrate-producing bacteria, most of which clustered near functionally confirmed sequences. Here, we describe tools and a framework that allow the bacterial butyrate-producing community to be profiled in the context of animal health and disease.
IMPORTANCE Butyrate is a compound produced by the microbiota in the intestinal tracts of animals. This compound is of critical importance for intestinal health, and yet studying its production by diverse intestinal bacteria is technically challenging. Here, we present an additional way to study the butyrate-producing community of bacteria using one degenerate primer set that selectively targets genes experimentally demonstrated to encode butyrate production. This work will enable researchers to more easily study this very important bacterial function that has implications for host health and resistance to disease.
INTRODUCTION
Short-chain fatty acids (SCFAs) play a central role in the maintenance of colonic homeostasis, which is the delicate balance between the host, its immune system, and the gastrointestinal microbial partners (1). Butyrate in particular has potent effects on host tissues. As with other SCFAs, butyrate is consumed by the host as an energy source; however, unlike the other common SCFAs, such as propionate and acetate, butyrate is the preferred energy source for colonocytes (2) and is rapidly absorbed and used by the colonic epithelium. This rapid oxidation of butyrate reduces local oxygen concentrations, causing the epithelia to become hypoxic and thus limiting the growth of facultative aerobic pathogens, such as Salmonella species (3, 4). In addition, butyrate alters host gene expression to promote immune tolerance to the colonic microbiota and to improve the barrier function of the colonic epithelium. For example, butyrate has been shown to increase the secretion of antimicrobial peptides and mucus as well as the expression of tight junction proteins, thickening and strengthening the barrier while making it less hospitable to invasive microbes (5–7). Most of butyrate's immunomodulatory activities result in anti-inflammatory effects, including the production of extrathymic T-regulatory (T-reg) cells (8), the limitation of proinflammatory CD4+ T cell activity (9), the stimulation of epithelial cells to produce retinoic acid (10), and the desensitization of colonic epithelial cells to gamma interferon (IFN-γ) (11). Although the maintenance of immune tolerance is complex and requires a balance among many regulatory factors, butyrate is a major signal for the host immune system leading to the inhibition of proinflammatory responses and to toleration of microbes that are present (12).
Because of butyrate's importance in maintaining colonic homeostasis and host health, characterizing and manipulating the bacterial populations responsible for its production are of great interest. Butyrate-producing bacteria do not form a monophyletic group, and at least four different fermentation pathways lead to butyrate production (13). The most common pathway for butyrate production in colonic environments entails the condensation of two molecules of acetyl coenzyme A (acetyl-CoA), followed by reduction to butyryl CoA. After butyryl CoA has been generated, two different enzymes are responsible for the final conversion to butyrate: butyrate kinase (Buk) and butyryl-CoA:acetate-CoA transferase (But) (14), with the But protein being the most common in the colonic environment (13). This enzyme takes the CoA group from butyryl-CoA and transfers it to acetate, yielding acetyl-CoA, thus regenerating a substrate of the main butyrate production pathway. This transferase is thought to be especially advantageous in the colonic ecosystem due to the high levels of acetate, allowing butyrate producers to take up and use a waste product of other microbes (15). Many studies have suggested that the majority of butyrate production in hindgut fermenters is the product of But enzyme activity, including in swine (13, 14, 16–18). It should be noted that previous work has suggested that not all enzymes capable of But activity are homologous. Previous work demonstrated that some bacteria from clostridial cluster XVI isolated from a chicken cecum had But activity despite lacking genes with significant homology to the but gene family (19). The authors identified genes similar to those encoding other known propionyl-CoA transferases in these genomes and suggested that these genes were responsible for the observed But activity. This work focuses only on genes encoding But enzymes commonly found in the Ruminococcaceae and Lachnospiraceae families (previously known as clostridial clusters IV and XIVa, respectively) and is not applicable to But-active enzymes with different evolutionary origins.
The sequence variation for the but gene family is poorly defined and currently includes closely related transferases that have differing substrate preferences (16, 17, 20). The FunGene But protein database is a large repository of But-like protein sequences and is an excellent resource; however, it contains But proteins and similar transferases that have distinct substrate specificities. Furthermore, few But proteins in this database have been functionally confirmed. Here, we have analyzed the but gene from previously identified butyrate-producing bacteria from swine (18), defined the functional diversity of the but sequences, developed degenerate but primers for PCR, and investigated the butyrate-producing bacterial community in the swine colonic environment. The results show that the degenerate but primers preferentially amplify genes encoding functional But enzymes over their paralogues, and that diverse but genes are transcribed in the swine colon.
MATERIALS AND METHODS
Identifying potential but-encoding sequences.
Previous work identified nine strains of swine-associated intestinal bacteria as butyrate producers, as determined by gas chromatography. Additionally, these strains were also found to exhibit But activity, although the active genes could not be identified in all cases (18). These strains were subjected to shotgun genomic sequencing to identify the genes encoding their But activity. Genomic DNAs were isolated using a previously described protocol (21). Sequencing was performed using a HiSeq 2500 sequencer (2 × 150 bp, rapid mode; Illumina, San Diego, CA) or a MiSeq (2 × 300 bp) at the Iowa State University Office of Biotechnology (DNA facility, Ames, IA), a Pacific Biosciences sequencer (P6-C4 chemistry; PacBio, Menlo Park, CA) at the Yale Center for Genome Analysis (New Haven, CT), and Roche FLX-Titanium chemistry (Roche Diagnostics, Branford, CT, USA). Libraries were prepared according to the manufacturer's directions. The resulting data included some combination of PacBio reads, Roche FLX 2.3-kb mate-pair library reads, and Illumina 7.9-kb mate-pair library reads. These were assembled using the MIRA assembler in a de novo hybrid assembly (22). Potential but genes were identified in the genomes by performing a BLAST search with the amino acid sequence of the butyryl-CoA:acetate-CoA transferase gene from Roseburia intestinalis L1-82 (GenBank accession no. EEV00989).
Testing for butyrate transferase activity.
Candidate genes were cloned into the pET-TOPO-101 vector (Invitrogen, Carlsbad, CA) and transformed into TOP10 Escherichia coli chemically competent cells, according to the manufacturer's instructions (primers used for cloning are listed in Table S1 in the supplemental material). Plasmid DNAs were isolated using the MinElute miniprep kit (Qiagen, Valencia, CA), and positive clones were confirmed to have full-length gene inserts by sequencing on an Applied Biosystems 3730xl DNA analyzer. Cloned DNAs were additionally transformed into E. coli BL21 Star competent cells for protein expression, in accordance with the kit protocol. Cultures (100 ml) were grown for 12 h in LB containing 50 μg/ml carbenicillin. Expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After an additional 6 h of growth, cultures were harvested by centrifugation, washed, and resuspended in 10 ml of sterile phosphate-buffered saline (PBS). Cells were lysed by two passages through a French press (Aminco, Silver Spring, MD). Lysates were centrifuged at 19,000 × g for 10 min to remove remaining unlysed cells. Protein expression was confirmed, and the amount of recombinant protein in each lysate was estimated by running 15 μg of total protein (determined by the Bradford assay [23]) in a 15% SDS-PAGE gel, staining with Coomassie blue, and comparing the 49-kDa band to all bands in the sample using a densitometry analysis in the ImageJ software package (24). Activities were normalized to the amount of protein present in the 49-kDa band.
Butyryl-CoA transferase (EC 2.8.3.8) activity was tested using the citrate synthase assay, as described previously (16), and activity was measured with acetate and butyryl-CoA as substrates (Sigma). The acetyl-CoA generated by butyrate transferase is condensed with oxaloacetate, liberating CoA, which reacts with 5,5′-dithio-bis-(2-nitrobenzoate) to form a yellow thiophenolate anion. The reaction rates were measured by monitoring the absorbance at 412 nm at 39°C on a Beckman (Indianapolis, IN) DU-650 spectrophotometer. Crude cell lysates were diluted with sterile water as necessary to achieve the linear range for the rate of the reaction. The reaction was repeated in the absence of acetate to confirm that the measured rate was not due to CoA-hydrolase activity.
Designing and validating conserved primers to but.
All full-length functionally validated but-like genes were aligned using CLC Genomics Workbench (Aarhus, Denmark), and conserved regions were identified. Degenerate primers (funbut-FWD, 5′-CARYTIGGIATYGGIGGIATSCC; funbut-REV, 5′-TGTCCGCCIGYICCRSWRAT) were designed to preferentially amplify those but genes with confirmed activity.
Full-length genes were downloaded from the FunGene but database on 21 March 2016, including only those sequences with a score of 275 or higher (25), resulting in 1,144 full-length sequences after removing redundant entries. The number of mismatches between the funbut primers and each gene in this data set was calculated with a Python script utilizing the Biopython libraries (26) (see Table S1 in the supplemental material). Previously published primer sets from Vital et al. (17) and Louis and Flint (20) were also analyzed for comparison. This script yielded a table listing the number of mismatches to each primer set for each gene entry. FastTree (27) was used to generate a phylogenetic tree from full-length amino acid sequences, and the R packages APE (28) and ggtree (29) were used to generate primer coverage figures. Sequences considered likely to amplify were those with two or fewer total mismatches to the primers.
To investigate potential amplification biases, the funbut primers were used in qPCR assays to determine which genes are preferentially amplified. Full-length gene amplicons were generated for each gene included in this study (see Table 1 for primer sequences). The amplicons were evaluated via NanoDrop (30) and diluted in 2 μg/ml sheared salmon sperm DNA to 107 copies/μl. The quantitative PCRs (qPCRs) were conducted with the Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA) containing each of the funbut primers at 500 nM and 1 μl of but gene amplicon DNA in 20 μl on a Stratagene 3005P thermocycler (San Diego, CA). The cycling conditions were as follows: 95°C for 30 s, 53°C for 30 s, and 72° for 30 s, for 40 cycles in total. Inferences about amplification preference were made by comparing threshold cycle (CT) values for each gene. As each reaction mixture contained the exact same primer concentrations and numbers of target molecules, any difference in the CT values among the different targets is due to amplification preferences.
TABLE 1.
Primers used in cloning each but-like gene as well as for generating full-length gene amplicons for use in qPCR amplification preference assaya
| Gene | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Annealing temp (°C) | PCR product length (bp) |
|---|---|---|---|---|
| Butyricicoccus #1 | CACCATGAGTATTTTTACAGAATACAGAAGCAAACTGCG | CAATATCGTTTGTTAGAGCGCCGC | 53 | 1,338 |
| Butyricicoccus #2 | CACCATGGATTATCAGGCGCTCTATCAG | TTATCTGCGGTTGGAGGTTCTCCA | 55 | 1,344 |
| Megasphaera #1 | CACCATGTACAAACAGAAACTGATGA | CCGTTTATTGCTATATCTCCAGATTTTCAT | 50 | 1,323 |
| Megasphaera #2 | CACCATGTACAAACTTTCACAAATTCC | TTACATGGAAATCTTGCTGGTCTT | 50 | 1,347 |
| 499 | CACCATGGATTTTAATCAGGAATACCAGC | TTATTTATTGCTTCTTCTCCAAATATGCAT | 53 | 1,341 |
| 35-6-1 | CACCATGGATTATTCTAAAGAATATCAA | TTATCTCTTATTAGTTCTTCTCCAAATATTC | 48 | 1,344 |
| 494a | CACCATGTCATTTACTCAAGAATATCAGA | TTATTTGTTAGATCTTCTCCAGATGTTAGC | 53 | 1,341 |
| 831b | CACCATGGATTTTTCAACTGAATACAAAC | TTATTTGTTGCTTCTTCTCCAGATGTGCAT | 53 | 1,341 |
| 992a | CACCATGAGTTACGCAAACGAATATCAA | TTACTTATTGCTGCGTCTCCAGATATGAG | 54 | 1,341 |
| 27-5-10 | CACCATGGAGCAAACAGAACTTTACCGG | TTACTTTCGGTTGCTTCTGCGCC | 55 | 1,344 |
| 68-3-10 #1 | CACCATGAGTAAGGATTTTGGATTGT | TTATTTCATCACCTTGCTGCTCG | 50 | 1,383 |
| 68-3-10 #2 | CACCATGTACAAGGTTTCGACTTTAG | CTAGAACGATGTTTTACTGGTGTATTTCC | 50 | 1,359 |
See Fig. 3.
MiSeq but amplicon library prep.
Swine proximal colon contents (10 cm distal from the cecum) were immediately placed in RNAlater and quickly homogenized to preserve the integrity of nucleic acids. Samples were subsequently frozen at −80°C until extraction (within 1 month). DNAs and RNAs were extracted using the PowerClean DNA and RNA extraction kits (MO BIO Laboratories, Carlsbad, CA) from proximal colon contents from six pigs fed a standard diet and associated with a different study (51). The iScript Select kit (Bio-Rad, Hercules, CA) was used to generate cDNA from the RNA using random hexamer primers. Amplicon sequencing libraries were prepared according to Illumina's 16S metagenomic sequencing library preparation (part 15044223 revision B), substituting the funbut primers for the 16S primers. This protocol uses a 2-step PCR procedure: the first step generates the amplicons from environmental samples, and the second step adds the indices and the sequencing adapters. In the first step, the funbut primers were used to amplify a 359-bp fragment using the AccuPrime Taq high-fidelity PCR system (Invitrogen). Due to the inclusion of multiple inosine bases, we were unable to produce a PCR product using a proofreading polymerase alone (31, 32), necessitating the use of a procedure that included Taq as well. The first-step PCR mixtures contained each primer at 500 nM and 100 ng of template and used an annealing temperature of 53°C for 35 cycles. The second PCR step was performed in accordance with the protocol using Kapa HiFi polymerase (Kapa Biosystems, Wilmington, MA) and the Nextera XT version 2 indices (Illumina). This library was sequenced on a MiSeq using a 2 × 300 version 3 reagent kit to generate 300-bp paired-end reads.
Sequence analysis.
Sequences were processed using mothur (33) according to a modified version of the MiSeq standard operating procedure (SOP) (http://www.mothur.org/wiki/MiSeq_SOP). Paired-end reads were joined, quality screened, and aligned to full-length high-quality but genes downloaded from FunGene, as previously mentioned. Sequences passing quality filters were clustered at a 97% similarity cutoff, and representative sequences were obtained for each operational taxonomic unit (OTU). These representatives were used in a BLASTn search against a database comprising full-length sequences from the FunGene but gene data set plus genes from the current work to determine the closest matching published sequence (% identity). Communities were subsampled to 1,556 sequences per sample prior to further analysis. Sequences used as references in the phylogenetic trees have had their activities confirmed either in this work or in previously published work. Genes from human strains have been cloned and their activities confirmed in previous works, such as those of Charrier et al. (16) and Louis et al. (34). Other work has tested crude cell lysates for But enzyme activity, such as the study by Duncan et al. (15), and inferred but gene presence by measuring butyrate production, acetate consumption, and the presence of a gene homologous to confirmed but gene sequences. Additionally, reviews, such as that by Louis et al. (35), identified isolates with confirmed But enzyme activity. These reference sequences were trimmed to the length of the representative sequences for the OTUs and used to generate a maximum likelihood tree using RAxML (36).
Accession number(s).
Bacterial genomes, butyrate transferase sequences, and amplicon sequencing data were deposited in GenBank under BioProject PRJNA341691. The code and data used to generate the figures in this paper are available at https://github.com/Jtrachsel/AEM-funbuts.
RESULTS
Activity encoded by the but gene is associated with an amino acid sequence motif.
Putative But-encoding genes were identified in the genomic sequence data from the butyrate-producing bacteria isolated from swine. All of the nine genomes analyzed yielded at least one potential but gene, and three genomes were predicted to contain two (those of Megasphaera, Butyricicoccus, and Eubacterium spp.), resulting in a total of 12 putative But-encoding genes. Functional analyses revealed that eight genomes had strong But activity ranging from 7,004 μM · mg−1 · min−1 (strain 27-5-10) to 27,819 μM · mg−1 · min−1 (strain 831b; Table 2). Only one but gene per genome showed appreciable activity, with the exception of the Eubacterium (strain 68-5-10), in which neither putative But protein was highly active (Table 2). This lack of appreciable But activity in strain 68-5-10 is consistent with previous work with this strain using native whole-cell lysate in the same assay (18). All genes that showed strong activity also exhibited similar activity when propionyl-CoA was used as a substrate (Table 2). These results are in agreement with previous characterizations of this gene family (16) and demonstrate that these sequences encode But activity.
TABLE 2.
But enzyme kinetics and amino acid analysesa
| Strain of origin | Closest related species (% 16S BLAST identity) | Gene name | Length (aa)b | Crude activity (μM/min · mg) |
Similarity to Roseburia query sequence (aa)c |
||
|---|---|---|---|---|---|---|---|
| Butyryl-CoA | Propionyl-CoA | % ID | % POS | ||||
| 68-3-10 | Eubacterium nodatum (93) | 68-3-10 #1 | 460 | 77.3 | 406.4 | 47 | 66 |
| 68-3-10 | E. nodatum (93) | 68-3-10 #2 | 452 | 218.1 | 335.4 | 48 | 69 |
| 1161 | Megasphaera elsdenii LC-1 (99) | Megasphaera #1 | 441 | 655.5 | 637.2 | 55 | 69 |
| 1161 | M. elsdenii LC-1 (99) | Megasphaera #2 | 448 | 23,515.6 | 20,747.3 | 49 | 69 |
| BB10 | Butyricicoccus pullicaecorum (94) | Butyricicoccus #1 | 445 | 231.2 | 785.4 | 48 | 67 |
| BB10 | B. pullicaecorum (94) | Butyricicoccus #2 | 447 | 13,367.9 | 12,665.3 | 73 | 85 |
| 35-6-1 | Peptoniphilus grossensis (97) | 36-5-1 | 447 | 18,343.5 | 13,495.2 | 62 | 76 |
| 27-5-10 | Intestinimonas butyriciproducens (99) | 27-5-10 | 447 | 7,004.6 | 5,725.3 | 72 | 85 |
| 494a | Anaerostipes butyraticus (96) | 494a | 446 | 13,692.3 | 12,767.3 | 74 | 85 |
| 992a | Anaerostipes hadrus (95) | 992a | 446 | 16,152.8 | 12,193.9 | 77 | 85 |
| 499 | Roseburia hominis (96) | 499 | 446 | 8,276.8 | 7,946.9 | 83 | 91 |
| 831b | R. hominis (97) | 831b | 446 | 27,819.8 | 27,892.9 | 86 | 91 |
Sequences with strong activity are shown in bold.
aa, amino acids.
ID, identity; POS, positive.
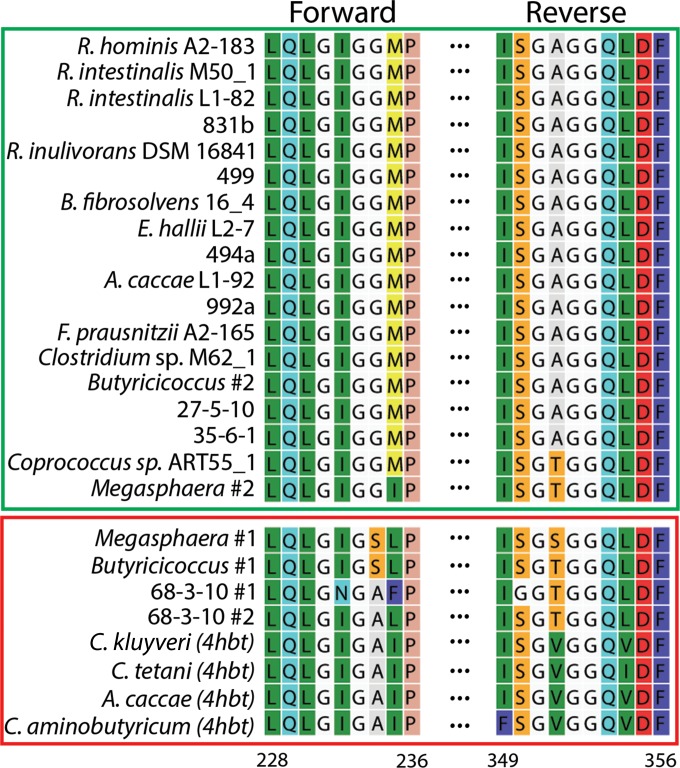
To determine sequence motifs associated with active But proteins, an amino acid alignment of all 12 putative but genes was generated. The alignment yielded several differences that demarked those with activity from those without. These differences occurred in a conserved region containing the amino acid motif LQLGIGG (Fig. 1). This motif was identical in all highly active But proteins; however, the proteins with low But activity contained at least one substitution in this motif. These data suggested that primers annealing to this site could be designed to preferentially amplify genes similar to those with high But activity, thus distinguishing genes with potential But activity from nonfunctional paralogues. The reverse primer-binding site was nondiscriminating, since we designed it to a gene region where But and But-like proteins shared similar amino acid residues. Unfortunately, no suitable alternative reverse primer-binding site would preferentially amplify all functional genes and still allow a primer of reasonable degeneracy for amplification.
FIG 1.
An amino acid alignment of the primer-binding regions. Residue numbering is based on the full-length But protein sequence from Roseburia intestinalis L1-82 GenBank accession no. EEV00989. Functionally confirmed sequences occupy the top green-bordered box. Sequences with little activity are bordered by the red box. The glycine at position 234 of this alignment is conserved in all highly active sequences. Amino acid residues are colored according to the RasMol convention. 4hbt, 4-hydroxybutyryl-CoA:acetate-CoA transferase (outgroup).
Degenerate primers preferentially target function-associated But protein-coding sequences and amplify diverse swine-associated but genes.
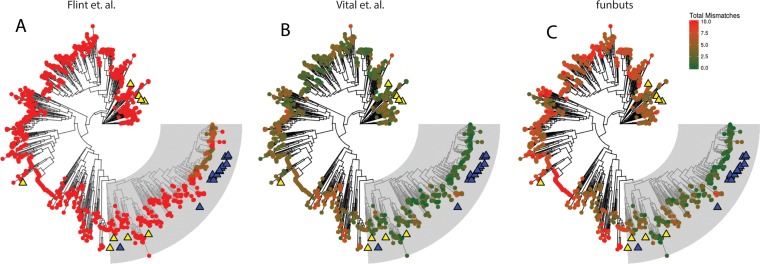
An in silico analysis compared the primer coverage of currently available but-targeting primer sets to the funbut primer pair and revealed that the funbut primers preferentially cover the clade containing all functionally confirmed sequences while having little coverage outside this clade (Fig. 2). The funbut primers are likely to amplify (two or fewer mismatches) 194 sequences, with 95% of these (184 sequences) in the main functional clade of interest. In contrast, the primers published by Vital et al. (17) are likely to amplify 517 sequences, 228 of these being in the clade of interest (44%). Finally, the primers published by Louis and Flint (20) are likely to amplify 5 sequences, all of which are in the main functional clade. It should be noted that the estimates for the number of sequences likely to be amplified are based only on the number of mismatches; amplification conditions also play a large role. Each of these primer sets could amplify more or less diversity than our estimate suggests depending on the exact conditions of the PCR. These results show that the funbut primers are more specific to diverse genes encoding functional But enzymes than are previously published but primers.
FIG 2.
Primer coverage for the three available but-targeting primer sets. (A) Flint et al. (2). (B) Vital et al. (3). (C) funbut primers. Maximum likelihood trees of full-length protein coding sequences from the FunGene but database, with the tips of each branch colored to reflect the total number of mismatches each primer set has to that particular sequence (red to green). The clade containing all verified but genes is shaded gray. Sequences with confirmed activity are marked with a blue triangle, and but paralogues are marked with yellow triangles.
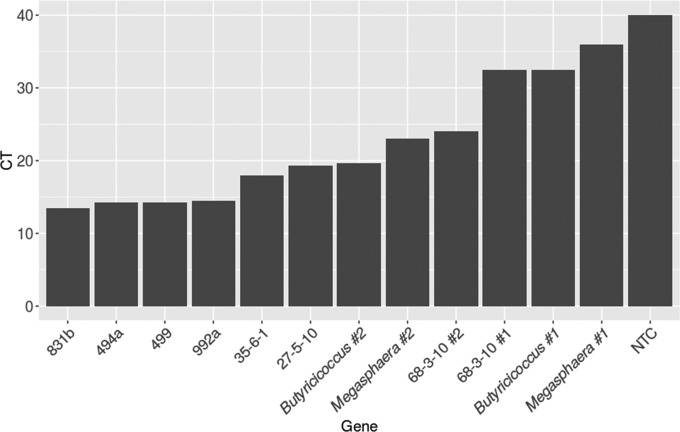
The in silico validation of the funbut primers suggests that they will preferentially amplify functional but genes over their paralogues, and to verify these findings, we investigated the amplification preference of our primers for the functionally validated But-encoding genes in this study. The funbut primers preferentially amplified sequences associated with But enzyme function over sequences associated with little or no But enzyme activity (Fig. 3). However, some but genes were amplified in fewer qPCR cycles and therefore more readily than others, revealing amplification biases even among the functionally confirmed genes (Fig. 3).
FIG 3.
qPCR CT values using the funbut primers against each gene identified in this study at 107 target molecules per reaction. Genes with lower CT values are preferentially amplified over those with higher CT values. NTC, no-template control.
A large unexplored diversity of but genes exists in the swine hindgut.
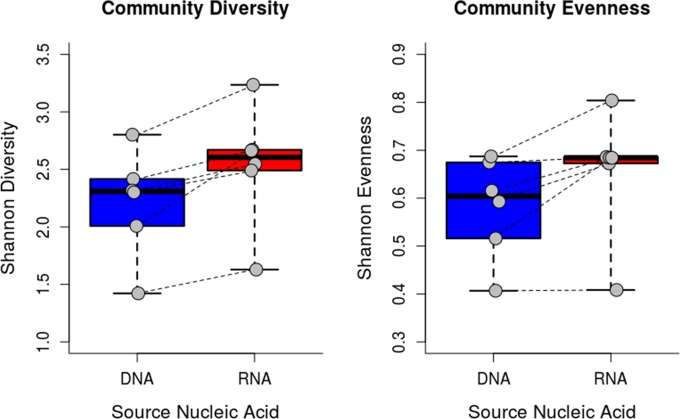
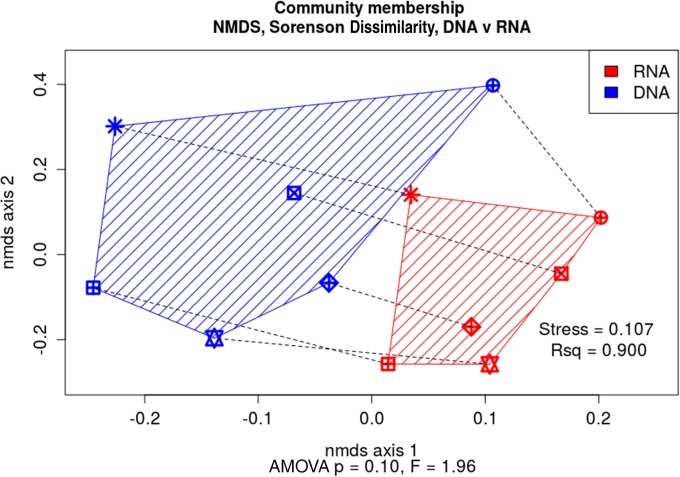
The funbut primers were applied to nucleic acids from a gut microbial community to evaluate but gene detection in this ecosystem. Diverse but genes were amplified from total DNA and RNA from swine proximal colonic contents. The colonic contents of six pigs yielded 90 OTUs from total DNA and 86 OTUs from total RNA (92 total unique OTUs, 97% similarity). Several OTUs were present in all animals, but these OTUs differed depending on which nucleic acid was used to profile the community. Fourteen OTUs were detected in RNA libraries from every pig, four OTUs were detected in all DNA libraries, and three OTUs were detected in every library regardless of nucleic acid type (OTU4, OTU14, and OTU23). The RNA-based communities harbored a greater diversity of but sequences, and these communities were more even than the DNA-based communities from the same sample (Fig. 4; Shannon diversity index with Wilcoxon paired test, P = 0.03; Shannon evenness with Wilcoxon paired test, P = 0.03). Similarly, community membership tended to differ between the RNA- and DNA-based communities (Fig. 5), suggesting that the most active butyrate producers may not be the most abundant.
FIG 4.
Shannon diversity and evenness indices of DNA and RNA but gene sequences, with communities from the same pig joined by a dotted line. The horizontal line within the box indicates the median, the boundaries of the box indicate the 25th and 75th percentiles, the whiskers extend to the minimum and maximum values, and individual data points are represented by gray circles.
FIG 5.
Nonmetric multidimensional scaling plot of Sorenson dissimilarity distances (membership) of the but gene sequence communities from six swine colons. Communities from the same animal are joined with a dotted line. AMOVA, analysis of molecular variance; nmds, nonmetric multidimensional scaling; Rsq, R2.
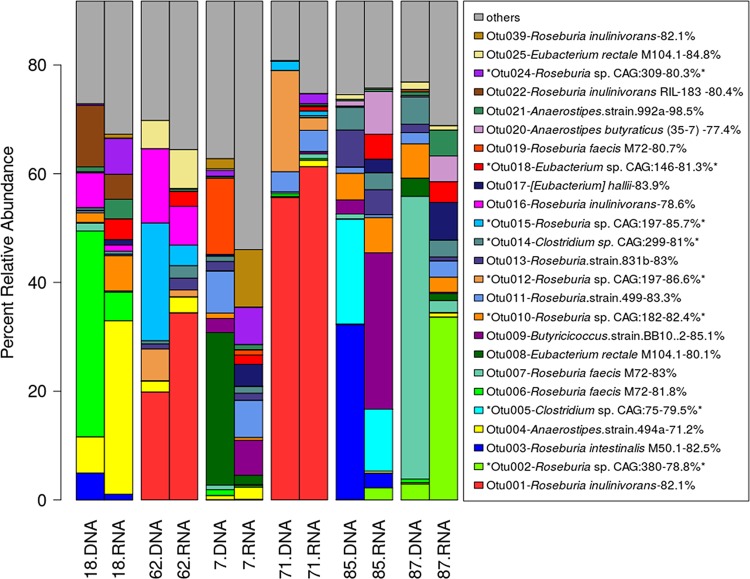
Representative sequences of many of the OTUs differed greatly from previously known cultured butyrate producers, and some showed more similarity to but genes from organisms detected only in metagenomes (Fig. 6). In total, 33 out of 92 OTUs were represented by sequences that most closely matched organisms detected only in metagenomic data sets. Similarly, many OTUs were represented by sequences with relatively low identity to any previously detected but gene. The maximum identity detected was 100%, and the minimum identity was 71.2% (OTU4). Representative sequences from 82 OTUs showed <90% identity, and 23 showed <80% identity to previously detected genes in the reference databases, whether they were of metagenomic origin or not. These findings suggest that many as-yet-uncultured butyrate producers exist in the swine gut and that this community is underrepresented in databases. Furthermore, the abundances of many OTUs from the same animal differed greatly depending on whether gene copy abundance (DNA) or transcript copy abundance (RNA) was considered (Fig. 6), supporting the idea that the transcriptionally active population is distinct from the most abundant.
FIG 6.
Twenty-five most abundant OTUs, clustered at 97% similarity at the DNA level. Listed are the names of the genomes containing the top homologues of representative sequences for each cluster, followed by percent identity (top BLAST hit). Asterisks denote genomes assembled from fecal metagenomes. Target sequences for this BLAST search were from the FunGene but gene database as well as the genes identified in this work.
Predicting function from phylogenetic analysis of But protein sequences.
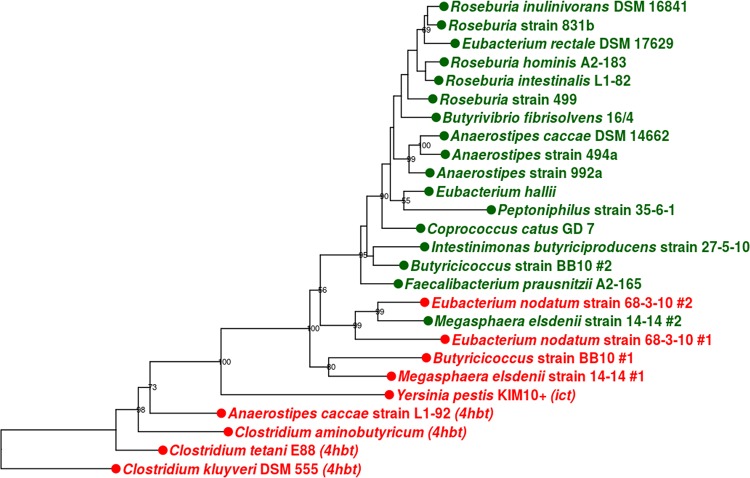
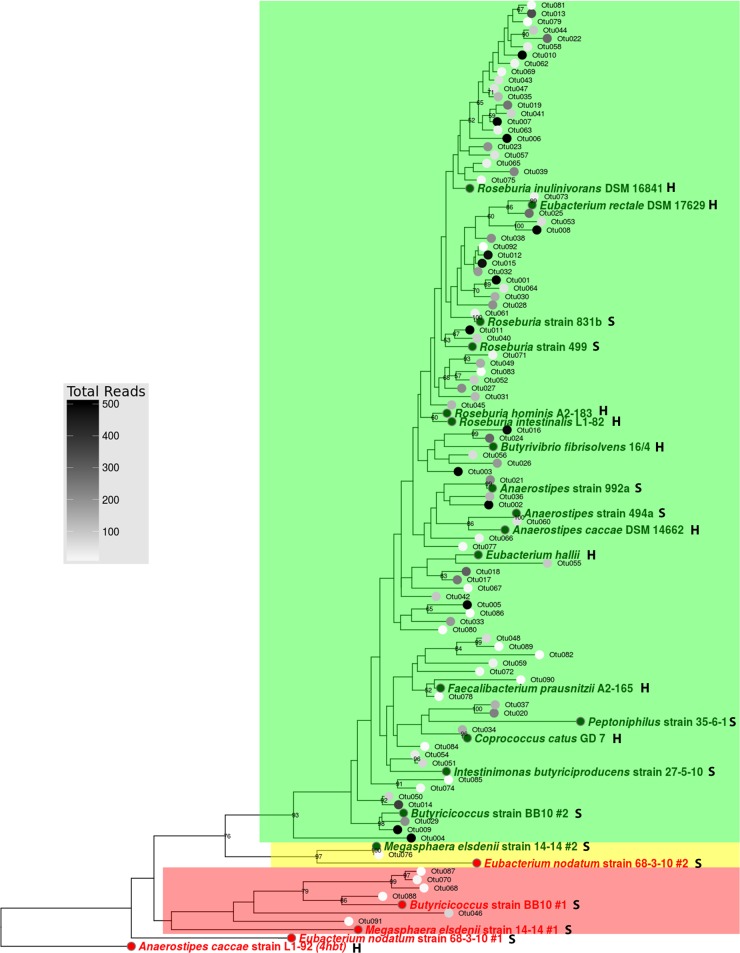
The genes encoding highly functional But proteins are phylogenetically separated from potential paralogues, but this separation is not perfect (Fig. 7), with some verified But enzymes and potential paralogues occupying the same clades on the tree. An example of this is the Megasphaera #2 gene, which encoded stronger But enzyme activity than the paralogue Megasphaera #1 gene from the same genome, but it was more divergent from the large functional clade than other sequences with confirmed activity. Phylogenetic placement was used to determine whether the OTUs detected by the funbut primer set encoded highly functional But enzymes or paralogues with lower activity. The representative DNA sequence from each OTU was aligned with confirmed but DNA sequences from the literature and this work (trimmed to the amplicon length), and a maximum likelihood phylogenetic tree was constructed from this alignment (Fig. 8). Similar to the tree constructed with full-length sequences, most of the confirmed reference sequences grouped together in one main clade apart from potential but paralogues, with the Megasphaera #2 gene being the exception. The vast majority of the OTUs detected by the funbut primers clustered more closely with but sequences encoding highly active enzymes than with sequences encoding low activity. Out of 18,672 total sequences, 18,559 sequences (99.4%) were contained within OTUs in the main functional clade, while only 113 sequences (0.6%) were outside this clade near potential but paralogues. The results suggest that phylogenetic relatedness is predictive of function for the majority of but gene sequences and that the funbut primers preferentially amplify functional but genes. However, function is more difficult to predict for distantly related deeply branching sequences within the but gene family.
FIG 7.
A phylogenetic tree (maximum likelihood) of full-length protein coding but sequences. Sequences with confirmed function are shown in green, and potential paralogues are shown in red. Note how functional But-encoding sequences are interspersed with paralogues toward the root of the tree. 4hbt, 4-hydroxybutyryl-CoA:acetate-CoA transferase; ict, itaconate-CoA transferase.
FIG 8.
A phylogenetic tree (maximum likelihood) of representative sequences from OTUs from this study, and previously studied But-like sequences identified in Vital et al. (1). Sequences were generated with the funbut primers and clustered at 97% similarity. Sequences with confirmed But enzyme activity are shown in green, and sequences that have failed to demonstrate activity in functional assays are shown in red. Numbers at the nodes are bootstrap values after 1,000 resamplings. The main clade containing all functionally confirmed reference sequences is shaded green, the clade containing both confirmed But-encoding genes and potential paralogues is shaded yellow, and the clade containing only potential paralogues with low activity is shaded red. Reference sequences from human-associated bacterial isolates are marked with an H, and those from swine-associated bacterial isolates are marked with an S.
DISCUSSION
Butyrate is centrally important to colonic homeostasis and is present in every vertebrate gut system studied to date (37). The butyrate-producing microbiota has been implicated in host health in many different disease models (35, 38–47), resulting in increased interest in studying this community. It is therefore important to have a reliable tool to identify butyrate-producing microbes and to detect their activity. The but gene is an excellent candidate for this probe due to its ubiquity in colonic environments, and we show the rational design of a primer set that detects but genes associated with butyrate transferase function. However, any analysis of the butyrate-producing community that examines only the but gene cannot be considered exhaustive. The bacterial butyrate synthesis pathway can be completed by other proteins, such as Buk and Ato, as well as nonhomologous enzymes capable of But activity (13, 19, 48). Characterizing a wide variety of functional genes is a critical step in designing targeted primers and probes. These data are additionally valuable when conducting comparative analyses of amplicon and metagenomic data sets from gut bacterial communities under different conditions.
Comparison of currently available but primer sets.
Several primer sets targeting the but gene are currently available. The first was described by Louis and Flint in 2007 (20). This primer set was designed to be used in qPCR assays to estimate the total number of but gene copies in complex environments, such as feces. Much care was taken to avoid any amplification of nontarget, closely related transferases, resulting in a conservative primer set. These primers are unlikely to amplify paralogues of the but gene but also miss much of the full diversity of functionally verified gene products. Due to the specificity of these primers, very little spurious amplification is observed when used on complex samples, such as feces.
Conversely, Vital et al. recently described a more promiscuous set of but primers (17). These primers were designed to amplify a wide range of but-like sequences, acknowledging that they would amplify closely related non-but genes. The primers were used to elucidate the diversity of but gene sequences in humans and many other vertebrate species via Roche's 454 pyrosequencing (17, 37). The in silico processing pipeline they describe attempts to remove some of the non-but sequences by eliminating those that closely match reference sequences that reside outside the phylogenetic cluster formed by functionally confirmed But-encoding genes. This method, while useful, is imperfect; the functional validation of but gene family members presented here has revealed that the phylogenetic separation of verified But-encoding genes and their paralogues is not absolute (Fig. 7 and 8). Additionally, because of the degeneracy of this primer set, spurious off-target amplification and incorrectly sized PCR products regularly occur with these primers, necessitating the inclusion of a gel extraction step in sequencing library prep (17, 37).
The development of the funbut primer set built upon these two approaches by amplifying functionally verified yet diverse but gene sequences. The funbut primer set amplifies a greater diversity of but genes than the primers described by Louis and Flint (20) and fewer non-but paralogues than the primers described by Vital et al. (17). However, as is the case with most primer sets, these will likely misrepresent or underrepresent some important groups. For example, although we detected some but sequences similar to those of Faecalibacterium spp., these appeared at a much lower abundance than would be expected from 16S rRNA gene sequence-based studies of the swine gut. Importantly, due to the increased specificity of the funbut primers compared to the primers in the study by Vital et al. (17), no incorrectly sized PCR product has been observed when amplifying from complex fecal or mucosal samples, even at annealing temperatures as low as 45°C. This allows for the omission of the gel extraction step when preparing sequencing libraries and for the possibility of using these primers in SYBR-based qPCR assays.
Active members of the butyrate-producing community differ from the abundant members.
The funbut but primers detected striking differences in community composition from the same starting material (proximal colon contents) depending on whether DNA or RNA was used as the template. Sequences detected in DNA-derived libraries are not necessarily being transcribed and translated into proteins and may be representative of microbes simply passing through the intestinal tract, producing butyrate through alternative pathways, or utilizing metabolisms not involving butyrate production. At best, DNA-based but libraries represent the functional potential in the ecosystem. In contrast, sequences detected in RNA-based libraries represent microbes that are actively transcribing but genes. These active microbes represent a subset of the total but-containing community; however, we detected a greater diversity of but genes in the RNA-based libraries than in the DNA-based libraries. Other studies have identified similar differences between the metagenome and metatranscriptome, such as the observation that functional genes for methanogenesis in the human gut were far more abundant in the metatranscriptome than in the metagenome (49). This reinforces the idea that the gut ecosystem contains microbes that may be in low abundance but are highly active, and that DNA-based studies may overlook their importance. When profiling the butyrate-producing community, RNA may be a more appropriate source molecule than DNA.
Phylogenetic relatedness of But protein sequences informs potential function.
Within the currently defined But protein family, there are genes encoding functional But enzymes and very similar paralogues. These two groups are more similar to each other than to the next most similar gene family, the 4-hydroxybutyrate transferases. One large clade harbored the vast majority of functionally confirmed but genes as well as the majority of all but-related OTUs detected, supporting the use of phylogenetic analyses to predict function for this family of But proteins. Outside this clade, functionally confirmed and unconfirmed But enzymes were interspersed (Fig. 7 and 8). However, this work does not rule out the possibility that paralogues also exist in the main functional clade as well. Because But enzyme activity is advantageous in colonic ecosystems, it is possible that enzymes specializing in this function have evolved multiple times from different ancestor proteins. Work by Eeckhaut et al. identified But enzyme activity from bacteria that lacked genes similar to but genes or their paralogues (19). They proposed that genes most similar to propionyl-CoA transferases were responsible for But enzyme activity in these organisms. Additional discovery and analysis of butyrate-producing organisms are required to delineate the full functional sequence diversity of deeply branching But protein sequences and to identify other protein families capable of But enzyme activity.
Further emphasizing the need to more fully characterize this family, and in agreement with previous gene-targeted studies (17, 37), we detected many OTUs with low identity to both confirmed sequences and cultured organisms. This work reveals many gaps in our knowledge of the but gene family. Due to the importance of this bacterial function in nearly all colonic ecosystems, better characterization of this community is necessary. It follows that culturing novel butyrate producers and identifying their functional genes remain important steps to improve but data sets.
Potential identities of some but gene paralogues.
Many similar fatty acid-CoA transferases are easily confused for but genes. The 4-hydroxybutyryl-CoA:acetate-CoA transferases (4-hbt) are known to be similar; however, several researchers have been investigating genes that are more similar to but genes than 4-hbt genes and are required for full pathogenicity in Yersinia pestis and Salmonella species (50). These genes have been proposed to be itaconate-CoA transferases. They transfer a CoA group from succinyl-CoA onto itaconate, thus activating it and enabling its degradation into acetyl-CoA and pyruvate. Indeed, many entries in the FunGene database for but-like genes are from Salmonella and Yersinia genomes. These genes cluster more closely with functionally confirmed but genes than to the 4-hbt genes. It is likely that many other genes that closely resemble but genes act to move CoA moieties among various fatty acids.
Analyses of butyryl-CoA transferases in the animal intestinal ecosystem enable the study of a functional aspect of the gut microbiota and how it relates to health and disease. This research provides a tool to investigate functional butyrate transferases in the swine gut microbiota and could also be applied to other animals or other environmental samples, or it could be used to generate but amplicon data sets from metagenomic samples. This advances the analyses of the host-associated butyrate-producing community for enhancing swine health and improving food safety.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sam Humphrey, Stephanie Jones, Joel Nott, Thad Stanton, Lisa Lai, Jennifer Jones, Tom Casey, and David Alt for technical support and helpful advice. We appreciate the caretakers in the Animal Resources Unit for managing the animals.
Julian Trachsel was supported by a fellowship from the Office of Biotechnology, Iowa State University. The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S Department of Agriculture. The USDA is an equal opportunity provider and employer.
Funding Statement
This work was funded by the USDA | Agricultural Research Service (ARS).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02307-16.
REFERENCES
- 1.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donohoe DR, Wali A, Brylawski BP, Bultman SJ. 2012. Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLoS One 7:e46589. doi: 10.1371/journal.pone.0046589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Baumler AJ. 2016. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell Y, Fantacone ML, Gombart AF. 2012. Regulation of antimicrobial peptide gene expression by nutrients and by-products of microbial metabolism. Eur J Nutr 51:899–907. doi: 10.1007/s00394-012-0415-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatayama H, Iwashita J, Kuwajima A, Abe T. 2007. The short chain fatty acid, butyrate, stimulates MUC2 mucin production in the human colon cancer cell line, LS174T. Biochem Biophys Res Commun 356:599–603. doi: 10.1016/j.bbrc.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 7.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. [DOI] [PubMed] [Google Scholar]
- 8.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontenelle B, Gilbert KM. 2012. n-Butyrate anergized effector CD4+ T cells independent of regulatory T cell generation or activity. Scand J Immunol 76:457–463. doi: 10.1111/j.1365-3083.2012.02740.x. [DOI] [PubMed] [Google Scholar]
- 10.Schilderink R, Verseijden C, Seppen J, Muncan V, van den Brink GR, Lambers TT, van Tol EA, de Jonge WJ. 2016. The SCFA butyrate stimulates the epithelial production of retinoic acid via inhibition of epithelial HDAC. Am J Physiol Gastrointest Liver Physiol 310:G1138–G1146. doi: 10.1152/ajpgi.00411.2015. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman MA, Singh N, Martin PM, Thangaraju M, Ganapathy V, Waller JL, Shi H, Robertson KD, Munn DH, Liu K. 2012. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am J Physiol Gastrointest Liver Physiol 302:G1405–G1415. doi: 10.1152/ajpgi.00543.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishio J, Honda K. 2012. Immunoregulation by the gut microbiota. Cell Mol Life Sci 69:3635–3650. doi: 10.1007/s00018-012-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5(2):e00889-14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol 186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 68:5186–5190. doi: 10.1128/AEM.68.10.5186-5190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charrier C, Duncan GJ, Reid MD, Rucklidge GJ, Henderson D, Young P, Russell VJ, Aminov RI, Flint HJ, Louis P. 2006. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology 152:179–185. doi: 10.1099/mic.0.28412-0. [DOI] [PubMed] [Google Scholar]
- 17.Vital M, Penton CR, Wang Q, Young VB, Antonopoulos DA, Sogin ML, Morrison HG, Raffals L, Chang EB, Huffnagle GB, Schmidt TM, Cole JR, Tiedje JM. 2013. A gene-targeted approach to investigate the intestinal butyrate-producing bacterial community. Microbiome 1:8. doi: 10.1186/2049-2618-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine UY, Looft T, Allen HK, Stanton TB. 2013. Butyrate-producing bacteria, including mucin degraders, from the swine intestinal tract. Appl Environ Microbiol 79:3879–3881. doi: 10.1128/AEM.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eeckhaut V, Van Immerseel F, Croubels S, De Baere S, Haesebrouck F, Ducatelle R, Louis P, Vandamme P. 2011. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb Biotechnol 4:503–512. doi: 10.1111/j.1751-7915.2010.00244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louis P, Flint HJ. 2007. Development of a semiquantitative degenerate real-time PCR-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol 73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson K. 1987. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol Chapter 2:Unit 2.4. [DOI] [PubMed] [Google Scholar]
- 22.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56. In Computer science and biology. Proceedings of the German Conference on Bioinformatics, GCB '99. GCB, Hannover, Germany. [Google Scholar]
- 23.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR. 2013. FunGene: the functional gene pipeline and repository. Front Microbiol 4:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 29.Yu G, Smith D, Zhu H, Guan Y, Lam TT-Y. 22 September 2016. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 30.Desjardins P, Conklin D. 2010. NanoDrop microvolume quantitation of nucleic acids. J Vis Exp 2565:2565. doi: 10.3791/2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiwara H, Fujiwara K, Hashimoto K. 1995. PCR with deoxyinosine-containing primers using DNA polymerases with proofreading activity. PCR Methods Appl 4:239–240. doi: 10.1101/gr.4.4.239. [DOI] [PubMed] [Google Scholar]
- 32.Knittel T, Picard D. 1993. PCR with degenerate primers containing deoxyinosine fails with Pfu DNA polymerase. PCR Methods Appl 2:346–347. doi: 10.1101/gr.2.4.346. [DOI] [PubMed] [Google Scholar]
- 33.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louis P, McCrae SI, Charrier C, Flint HJ. 2007. Organization of butyrate synthetic genes in human colonic bacteria: phylogenetic conservation and horizontal gene transfer. FEMS Microbiol Lett 269:240–247. doi: 10.1111/j.1574-6968.2006.00629.x. [DOI] [PubMed] [Google Scholar]
- 35.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 36.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vital M, Gao J, Rizzo M, Harrison T, Tiedje JM. 2015. Diet is a major factor governing the fecal butyrate-producing community structure across Mammalia, Aves and Reptilia. ISME J 9:832–843. doi: 10.1038/ismej.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerisuelo A, Marin C, Sanchez-Vizcaino F, Gomez EA, de la Fuente JM, Duran R, Fernandez C. 2014. The impact of a specific blend of essential oil components and sodium butyrate in feed on growth performance and Salmonella counts in experimentally challenged broilers. Poult Sci 93:599–606. doi: 10.3382/ps.2013-03528. [DOI] [PubMed] [Google Scholar]
- 39.Metzler-Zebeli BU, Hooda S, Pieper R, Zijlstra RT, van Kessel AG, Mosenthin R, Ganzle MG. 2010. Nonstarch polysaccharides modulate bacterial microbiota, pathways for butyrate production, and abundance of pathogenic Escherichia coli in the pig gastrointestinal tract. Appl Environ Microbiol 76:3692–3701. doi: 10.1128/AEM.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khan S, Jena GB. 2014. Protective role of sodium butyrate, a HDAC inhibitor on beta-cell proliferation, function and glucose homeostasis through modulation of p38/ERK MAPK and apoptotic pathways: study in juvenile diabetic rat. Chem Biol Interact 213:1–12. doi: 10.1016/j.cbi.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Kumar A, Alrefai WA, Borthakur A, Dudeja PK. 2015. Lactobacillus acidophilus counteracts enteropathogenic E. coli-induced inhibition of butyrate uptake in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 309:G602–G607. doi: 10.1152/ajpgi.00186.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolho K-L, Korpela K, Jaakkola T, Pichai MVA, Zoetendal EG, Salonen A, de Vos WM. 2015. Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol 110:921–930. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 43.Chiba M, Tsuji T, Nakane K, Komatsu M. 2015. High amount of dietary fiber not harmful but favorable for Crohn disease. Perm J 19:58–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cushing K, Alvarado DM, Ciorba MA. 2015. Butyrate and mucosal inflammation: new scientific evidence supports clinical observation. Clin Transl Gastroenterol 6:e108. doi: 10.1038/ctg.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chassard C, Dapoigny M, Scott KP, Crouzet L, Del'homme C, Marquet P, Martin JC, Pickering G, Ardid D, Eschalier A, Dubray C, Flint HJ, Bernalier-Donadille A. 2012. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther 35:828–838. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 46.Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, Duncan G, Johnstone AM, Lobley GE, Wallace RJ, Duthie GG, Flint HJ. 2011. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr 93:1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- 47.Stoll ML, Kumar R, Morrow CD, Lefkowitz E, Cui X, Genin A, Cron RQ, Elson CO. 2014. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther 16:486. doi: 10.1186/s13075-014-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bui TP, Ritari J, Boeren S, de Waard P, Plugge CM, de Vos WM. 2015. Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat Commun 6:10062. doi: 10.1038/ncomms10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J, Garrett WS, Chan AT, Huttenhower C. 2014. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A 111:E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasikaran J, Ziemski M, Zadora PK, Fleig A, Berg IA. 2014. Bacterial itaconate degradation promotes pathogenicity. Nat Chem Biol 10:371–377. doi: 10.1038/nchembio.1482. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Bass BE, Bandrick M, Loving CL, Brockmeier SL, Looft T, Trachsel J, Madson DM, Thomas M, Casey TA, Frank JW, Stanton TB, Allen HK. Fermentation products as feed additives mitigate some ill-effects of heat stress in pigs. J Anim Sci, in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.