ABSTRACT
Microbially influenced corrosion (MIC) is a major cause of damage to steel infrastructure in the marine environment. Despite their ability to grow directly on Fe(II) released from steel, comparatively little is known about the role played by neutrophilic iron-oxidizing bacteria (FeOB). Recent work has shown that FeOB grow readily on mild steel (1018 MS) incubated in situ or as a substrate for pure cultures in vitro; however, details of how they colonize steel surfaces are unknown yet are important for understanding their effects. In this study, we combine a novel continuously upwelling microcosm with confocal laser scanning microscopy (CLSM) to determine the degree of colonization of 1018 MS by the marine FeOB strain DIS-1. 1018 MS coupons were incubated with sterile seawater (pH 8) inoculated with strain DIS-1. Incubations were performed both under oxic conditions and in an anoxic-to-oxic gradient. Following incubations of 1 to 10 days, the slides were removed from the microcosms and stained to visualize both cells and stalk structures. Stained coupons were visualized by CLSM after being mounted in a custom frame to preserve the three-dimensional structure of the biofilm. The incubation of 1018 MS coupons with strain DIS-1 under oxic conditions resulted in initial attachment of cells within 2 days and nearly total coverage of the coupon with an ochre film within 5 days. CLSM imaging revealed a nonadherent biofilm composed primarily of the Fe-oxide stalks characteristic of strain DIS-1. When incubated with elevated concentrations of Fe(II), DIS-1 colonization of 1018 MS was inhibited.
IMPORTANCE These experiments describe the growth of a marine FeOB in a continuous culture system and represent direct visualizations of steel colonization by FeOB. We anticipate that these experiments will lay the groundwork for studying the mechanisms by which FeOB colonize steel and help to elucidate the role played by marine FeOB in MIC. These observations of the interaction between an FeOB, strain DIS-1, and steel suggest that this experimental system will provide a useful model for studying the interactions between microbes and solid substrates.
INTRODUCTION
Microbially influenced corrosion (MIC) is a major contributor to the degradation of structural steel in the marine environment, and the resultant damages are estimated to be in the billions of dollars per year (1). Recent studies have demonstrated that microbial activity can result in corrosion rates up to 3-fold higher than would otherwise be expected (2, 3). An especially pervasive form of marine corrosion is accelerated low-water corrosion (ALWC), which occurs close to the tidal low water mark on permanent structures like piers and bridges (2). Microbial involvement is known to be a factor in ALWC. The result of these processes is the need for more frequent maintenance of structures, such as piers, bridges, and pipelines, as well as an increased risk of catastrophic failure. The majority of the existing literature suggests that MIC is primarily a result of surface colonization by sulfate-reducing bacteria (SRB) (2–26), and while it is clear that the SRB play a major role in MIC, recent reports (2, 3, 15, 17, 27, 28) have demonstrated the presence of lithotrophic Fe-oxidizing bacteria (FeOB) on steel surfaces impacted by MIC. FeOB utilize Fe(II) released from the steel surface as their primary energy source and are autotrophic, requiring only a few micronutrients in addition to Fe(II) to grow (29). As a result, they are well suited to be early colonizers of steel. Several studies have shown that during in situ incubations of mild steel, FeOB are capable of early colonization, and this pioneering colonization is followed by an increase in diversity of the surface community, including development of SRB populations (3, 17, 27). The FeOB that naturally colonize either mineral or steel surfaces are known to produce porous mats comprised of interwoven stalks of polysaccharides and poorly crystalline iron oxyhydroxides (17, 23, 28, 30, 31). For example, a single, micrometer-sized cell of the marine FeOB Mariprofundus ferrooxydans can make at least 30 μm of stalk per cell division (30); thus, there is the potential for these cells to produce a substantial amount of external structure as a result of their growth. It has been speculated that the mats created by the FeOB on steel surfaces serve as substrates for further colonization by diverse communities, including anaerobes such as sulfate-reducing bacteria, that result in accelerated MIC microbial communities (3, 17, 27).
The dynamics of biocorrosion in the environment presents a number of challenges to studying MIC. In situ studies are of great value in understanding processes that occur in nature; however, it is difficult to constrain, or even identify, all of the potential variables affecting the process, as well as to identify what role particular groups of microbes are playing. Conversely, static incubations in the laboratory may fail to fully address the continual addition of nutrients and removal of waste products, as seen under natural conditions. The development of laboratory microcosms or continuous culture systems is an effective way to study processes under controlled but more environmentally relevant conditions. Such systems have been used effectively for studying processes related to anaerobic MIC in the marine environment; for example, Angell et al. used a multiple-chemostat system to continually supply fresh cultures of Thiobacillus ferrooxydans, Desulfovibrio vulgaris, and Pseudomonas aeruginosa to a potentiostatically controlled electrochemical cell (32). Continuous culture or microcosm-based systems have not been used for studying the role of FeOB in corrosion. Such a system would be useful for understanding attachment and colonization of steel surfaces, as well as looking at the effects of redox gradients on the process.
In this report, we describe the design of a continuous culture system developed to assess colonization and growth of FeOB on steel surfaces. We used a novel isolate, Mariprofundus sp. strain DIS-1, that was isolated from a steel coupon incubated in situ in the ocean to study the colonization of the steel surface under different conditions. Further, we report on the development of a novel method for three-dimensional (3D), 2-color visualization of steel colonization by FeOB via confocal microscopy.
MATERIALS AND METHODS
Isolation of Mariprofundus sp. strain DIS-1.
Mariprofundus sp. strain DIS-1 was isolated from 1018 mild steel (MS) coupons incubated in West Boothbay Harbor (lat 43.84443, long −69.64095) at a depth of 5 to 7 m using polyvinyl chloride (PVC) pipe samplers as described by McBeth et al. (17). The coupons were incubated in situ for 2 weeks in January 2012 at a temperature of 4.5°C. Following recovery of the samplers, 100-μl aliquots were used to inoculate petri plates containing ca. 60 mg of autoclaved 200-mesh Fe(0) powder (Alfa Aesar, Ward Hill, MA) and artificial seawater (ASW) as described by Emerson and Floyd (33). Plates were incubated at room temperature in a sealed acrylic jar with a BBL CampyPak plus microaerophilic system envelope (Becton, Dickinson, and Co., NJ). Following incubation, orange-colored floc was observed. Phase-contrast microscopy revealed these flocs were composed largely of the Fe-oxyhydroxide-coated stalks that are characteristic of some FeOB. Isolation of a pure culture from these plates was carried out by following the protocol reported by Krepski et al. (14). A single particle was selected from the enrichment culture using a sterile Pasteur pipet, washed five times in sterile ASW, and serially diluted 1:10 seven times, giving a final dilution of 1 × 10−7. Following an incubation of 2 to 5 days, these dilutions were examined microscopically, and a single particle containing stalks was selected using an Eppendorf Transferman NK2 micromanipulator (Eppendorf, Hamburg, Germany) fitted to a Zeiss Axio Observer inverted microscope (Zeiss, Jena, Germany). The selected particle was then diluted and transferred onto new plates containing Fe(0) and ASW. This process was repeated 6 times; after the 5th transfer, the culture was streaked onto ASW-R2A solid medium to rule out the continued presence of heterotrophic bacteria in the culture.
Following incubation of the isolated organism, strain DIS-1, DNA was extracted and the complete 16S rRNA gene was sequenced. Sequencing revealed that the DIS-1 16S rRNA gene sequence was identical to that of Zetaproteobacterium sp. clone Dock D2b-C6 (GenBank accession number HQ206656), first reported by McBeth et al. (17) as a clone from DNA extracted from a steel coupon incubated in situ in Boothbay Harbor.
Microcosm design and construction.
A schematic of the microcosms is depicted in Fig. 1, showing the flow path from a reservoir of autoclaved seawater, through a peristaltic pump, through a column packed with zero-valent iron (ZVI), and into the microcosm. Microcosms were constructed from 50-ml conical centrifuge tubes (VWR) which had been modified as follows.
FIG 1.
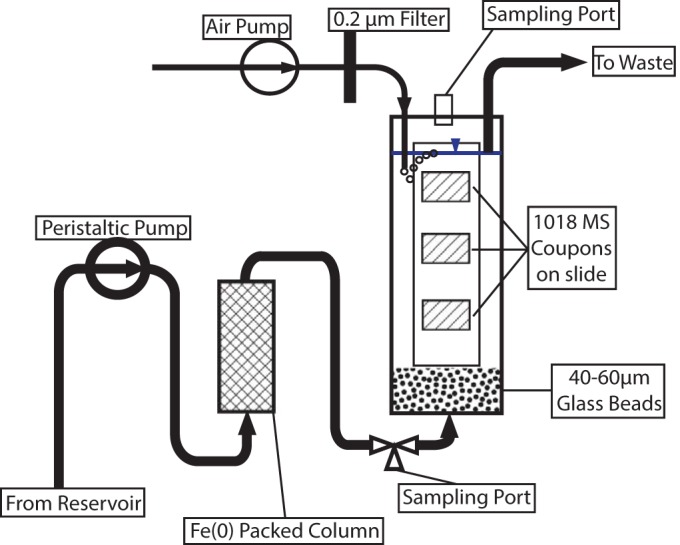
Schematic of microcosm setup showing flow direction and valves.
A 3.175-mm hole was drilled through the bottom of the tube, and the barbed end of a polypropylene Luer fitting with a 3.175-mm hose barb was inserted. The conical bottom of the tube up to the tip of the barb was then filled with epoxy (Devcon) to seal the fitting to the tube. In addition, the fitting was heat welded to the exterior of the centrifuge tube. A thin layer of glass wool was placed in the bottom of the tube, after which 15 g of acid-washed 40- to 60-μm glass beads (Sigma) was added.
The lid of the 50-ml centrifuge tube was modified as follows. Three holes were drilled, one of 3.175 mm, one of 1.5875 mm, and one to accommodate the threaded portion of a 2-ml screw-cap microcentrifuge tube. A male Luer-Lok fitting with a 3.175-mm hose barb was inserted into the 3.175-mm hole, a male Luer-Lok fitting with a 1.5875-mm hose barb was inserted into the 1.5875-mm hole, and the top 2.5 mm of a 2-ml cryogenic storage tube (including the cap) was inserted for use as a sampling port. Following insertion, these fittings were heat welded to the cap. The end of the 3.175-mm barb protruded approximately 1 mm down into the top of the microcosm, and this was used to maintain the medium at this level below the top of the microcosm. Two cm of 0.7938-mm-inner-diameter (ID) Tygon tubing (Cole Parmer) was attached to the 1.5875-mm barb and used to bubble filtered air into the microcosm.
Air was supplied to the microcosm from an aquarium pump at approximately 30 ml min−1 into the 1.5875-mm fitting and was filtered through a 0.22-μm, 13-mm Pall syringe filter with a 0.22-μm-nominal-pore-size nylon membrane (Pall, Port Washington, NY).
Medium was pumped from a 20-liter reservoir using an Ismatec 24 channel peristaltic pump (Cole-Parmer). Medium was supplied to the microcosms using 2.06-mm-ID PVC pump tubing. The use of this tubing with a setting of 30 on the pump allowed for a medium input rate of 1 ml min−1. A stable medium level in the microcosm was maintained by using 2.79-mm-ID PVC pump tubing, which ensured that the rate of removal was in excess of the rate of addition; thus, the level would not exceed the outlet port. Medium was supplied to and removed from the reactor using 3.175-mm-ID Tygon tubing (Cole-Parmer) and Luer fittings throughout. A sampling port was provided by installing a 4-way valve in the line between the inlet of the microcosm and the outlet of the ZVI column.
After a 24-h period to ensure that the epoxy had fully cured, the microcosms were assembled, wrapped in foil, and autoclaved for 30 min prior to use. All tubing, fittings, and reservoirs were autoclaved prior to use.
ZVI columns.
ZVI columns were prepared in 5-ml Luer-Lok syringes (Becton, Dickinson, and Company, Franklin Lakes, NY) as follows. A plug of glass wool approximately 1 cm thick was inserted into the bottom of the syringe. Fifteen g of coarse iron filings (Aqua Solutions, Deer Park, TX) was then added to the syringe, followed by an additional 1 cm of glass wool. The syringe was sealed with a 20-mm butyl rubber stopper and aluminum crimp seal.
Coupon preparation.
1018 MS coupons (12 mm by 15.0625 mm by 3.175 mm) were progressively sanded with 200-, 400-, and 600-grit sandpaper as described in McBeth et al. (17). Following sanding, the coupons were affixed to standard 75-mm by 25-mm glass microscope slides using silicone glue. Three coupons were mounted on each slide, with center points at 16, 38, and 60 mm as shown in Fig. 2. The mounted coupons were rinsed with 95% ethanol and acetone prior to UV sterilization as described by McBeth et al. (17). Final assembly of the microcosms and slides was performed under sterile conditions in a laminar-flow hood.
FIG 2.

Mild steel coupons shown mounted on slide, with staining frame and coverslip in place. The staining frame and coverslip are added after incubation.
Measurements of iron.
Iron was assayed using the ferrozine method (34) adapted for reading on a Mikroscan 96-well plate reader. Concentrations of Fe(II) and total iron were obtained, with Fe(III) concentrations calculated by subtracting the Fe(II) concentration from the total Fe concentration. Total aqueous Fe concentrations were measured with ferrozine following reduction with hydroxylamine hydrochloride. For the −Fe experimental series, 900-μl samples were obtained and immediately acidified with 100 μl of 0.01N HCl to preserve speciation, and for the +Fe experimental series, 100-μl samples were diluted with 900 μl of 0.01N HCl.
Experimental conditions.
The medium used in these experiments consisted of sand-filtered Damariscotta River seawater. This seawater was sterilized by autoclaving for 5 h in 20-liter polypropylene carboys. Following autoclaving and cooling, the pH was adjusted to 8 (±0.2) by bubbling with sterile filtered CO2 prior to use. Following sterilization and assembly, the microcosms were filled and allowed to equilibrate for 1 h prior to inoculation with 1 ml of DIS-1 culture grown on Fe(0) as described by McBeth et al. (17). The microcosms were inoculated by adding 1 ml of DIS-1 culture to the microcosm through the top sampling port with a sterile micropipette. One hour after inoculation, a 1-ml sample was removed for Fe assay from both the inlet and outlet sampling ports.
Two sets of experiments were performed in order to assess the growth of Mariprofundus sp. strain DIS-1. For each experimental series, 5 microcosms were prepared in parallel. The first set omitted the ZVI column in order to assess the attachment and growth of DIS-1 to a steel surface under dynamic, aerobic conditions that simulate in situ colonization of an exposed steel structure. For this experimental series, one microcosm was sacrificed each day for 5 days. All microcosms were sampled daily for Fe speciation and concentration at the inlet and outlet, while redox and dissolved oxygen (DO) profiles were taken only on the microcosm to be sacrificed in order to minimize disturbance and the potential for contamination of the remaining microcosms. The second set of microcosms included the ZVI column and assessed the attachment and growth of DIS-1 on steel when subjected to a gradient of both dissolved oxygen and Fe(II), as might occur in structural steel at the sediment-water interface. As in the first experimental series, these microcosms were sampled daily at the inlet and outlet for iron speciation and concentration. This experimental series was run for 10 days, with microcosms sacrificed for imaging on days 2, 4, 6, 8, and 10.
Microcosm redox and dissolved oxygen profiling.
DO profiles were obtained using a FireSting optode-based oxygen meter (Pyro Science GmbH, Aachen, Germany) fitted with a Pyro Science OXR230 probe attached to a micromanipulator. The FireSting instrument was calibrated and operated according to the manufacturer's instructions. To obtain the profile, the oxygen probe was inserted through the top sampling port in the microcosm and moved through the microcosm at 5-mm intervals. At each interval, DO measurements were taken for 30 s at 5-s intervals. When measurements indicated the presence of an oxic/anoxic transition zone, measurements were taken at 2.5-mm intervals to more closely constrain the location of the transition zone.
Redox profiles were obtained with a Sentex redox probe attached to an Oakton pH/oxidation-reduction potential (ORP) meter, which was calibrated using the Orion ORP standard. The redox probe was mounted on a micromanipulator and inserted through the sampling port at the top of the microcosm. Measurements were taken through the microcosm at 5-mm intervals, and the probe/meter was allowed to stabilize at each interval before the measurement was recorded.
After inlet/outlet samples, redox profiles, and DO profiles were obtained, the coupon slides were removed from the microcosms, digital photographs were immediately recorded with a Sony DSC-W50 digital camera, and the coupons were prepared for confocal imaging without allowing them to dry.
Staining and imaging of coupons.
Following incubation, each coupon was overlaid with a staining and embedding solution consisting of 1.5 μl 2 mg/ml rhodamine-conjugated Ricinus communis agglutinin I (Vector Laboratories, Burlingame, CA) and 5 μl SYTO13 (Life Technologies, Grand Island, NY) in 144.5 μl 0.5% low-melting-point agarose in MilliQ (Millipore, Billerica, MA) reagent-grade water. Following the application of the staining solution, a 3.430-mm-thick nylon frame with outer dimensions of 25 by 75 mm and inner dimensions of 19 by 69 mm with a 24- by 60-mm coverslip was placed over the slide (Fig. 2). This frame allowed for approximately 150 μm of clearance between the top of the coupon and the underside of the coverslip. These frames were produced to the authors' specifications using stereolithography by Ponoko, Inc. After the frames were applied, the slides were incubated for 1 h at 4°C prior to imaging with a Zeiss LSM700 confocal scanning laser microscope controlled by Zeiss ZEN software. At least 5 image stacks (0.5-μm slice thickness, 639.5 by 639.5 μm) at ×200 magnification were obtained per coupon, with the laser intensity and gain settings optimized for each image stack. These image stacks were imported into the FIJI implementation of ImageJ (35, 36), and the number of cells was calculated using the three-dimensional (3D) object counter with an intensity cutoff of 35 and a size cutoff of 25 to 200 voxels. Cell counts were normalized to square millimeters.
Genomics of DIS-1.
For genome sequencing, 100 ml of culture was grown on ZVI plates, described above, and harvested in the late log phase of growth. The cell pellet was extracted using a MoBio PowerSoil DNA extraction kit, and approximately 1 μg of DNA was recovered. Preparation of DNA for sequencing was done as described by Field et al. (37), and the sequencing of 150- by 2-bp paired-end reads was done on an Illumina Nextgen instrument at the Single Cell Genomic Center at Bigelow Laboratory. The assembled sequence had 57 contigs with an N50 of 101 kb.
Accession number(s).
The DIS-1 genome is publically available from the Joint Genome Institutes, Integrated Microbial Genomes (IMG), under genome identifier 2571042359.
RESULTS
Strain DIS-1 grew as an obligate Fe oxidizer. It consistently produced a stalk. It did not grow on heterotrophic medium or on H2, and it did not grow unless Fe(II) was available. Its doubling time has not been determined precisely but is comparable to those of other FeOB.
It took several iterations and test runs of the system to develop a microcosm that was reproducible in terms of controlling flow rates, pH, Fe(II), and O2 levels, as well as having the capability for removal of coupons with delicate but intact biofilms for microscopic visualization and analysis. For experimental runs, the microcosms were operated for 5 days in the case of the series without supplemental iron and for 10 days for the series with supplemented iron. Under all conditions, the flow rate of 1 ml min−1 was constant, and the pH was maintained at 8 (±0.2) U. A photograph of the five parallel microcosms is shown in Fig. S1 in the supplemental material, with the critical components of the system highlighted.
Iron concentrations.
In the −Fe series, the microcosm inlet Fe(II) and Fe(III) concentrations were below the detection limit (∼1 μM) throughout the experiment, as expected (Fig. 3A). At the outlet, Fe(III) was at its highest level (12 μM) at the beginning of the experiment, decreasing below the detection limit by the end of the experimental period. The outlet Fe(II) concentration was also at its peak (9 μM) at the beginning of the experiment, decreasing to near the detection limit by the second day, after which it was not detected (Fig. 3A).
FIG 3.
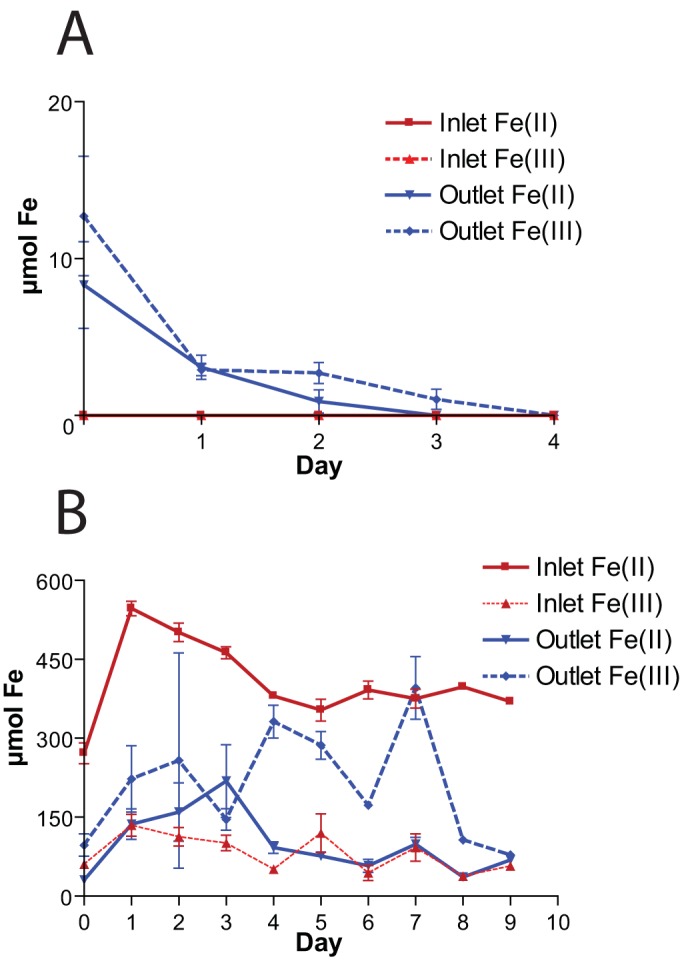
Fe(II) and Fe(III) concentrations over time in the −Fe (A) and +Fe (B) experiments.
In the +Fe series (Fig. 3B), the inlet Fe(II) peaked at ∼500 μM at day 1 and decreased to approximately 400 μM by day 4, and it was stable for the remainder of the experiment. The inlet Fe(III) concentration ranged from ∼75 to ∼125 μM over the course of the experiment, averaging ∼100 μM. The outlet Fe(III) concentrations were highly variable (Fig. 3B) due to the unavoidable entrainment of various amounts of flocculent iron oxyhydroxides during sampling. Outlet Fe(II) concentrations peaked at approximately ∼200 μM at the beginning of the experiment, with an average Fe(II) concentration of approximately 100 μM.
In the −Fe series, the DO and redox profile measurements indicated that oxic and oxidizing conditions prevailed throughout the column. Little to no change in either the oxidation reduction potential or the DO concentration was observed across the depth of the column (data not shown).
In the +Fe experiment, a sharp transition from oxic to anoxic and from oxidizing to reducing was observed (Fig. 4). The transition from oxidizing conditions occurred across approximately 10 mm, between 40 and 50 mm below the liquid surface in the microcosm. The location of this transition zone ensured that at least one of the coupons was always in the anoxic and reducing portion of the microcosm (Fig. 4).
FIG 4.
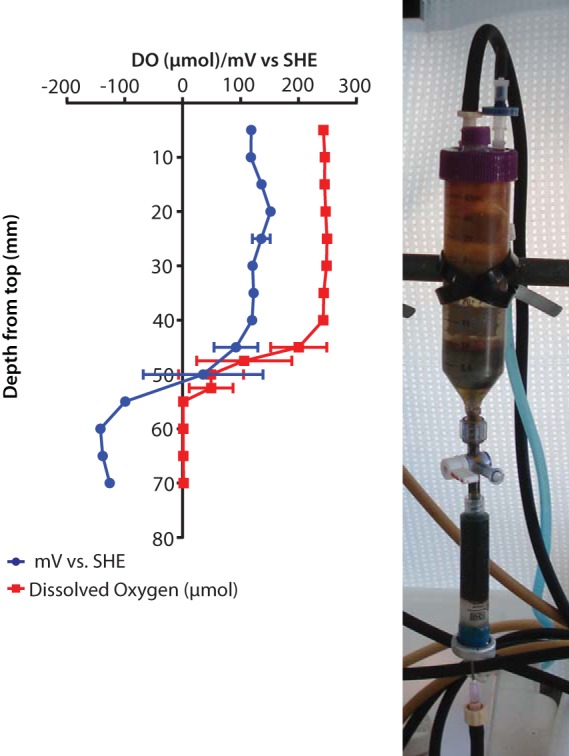
Redox and dissolved oxygen (DO) profile from the 6th day of incubation with supplemental Fe. The oxic/anoxic transition occurred between 40 and 50 mm below the surface of the microcosm, as can also be seen in the transition from clear to reddish in the photograph. SHE, standard hydrogen electrode.
Colonization of 1018 mild steel coupons.
The coupons were colonized much more rapidly and completely in the −Fe series (Fig. 5). Complete coverage of all coupons was observed by day 4 of the experiment. The uppermost coupon (15 mm) had the most variable coverage; this was most likely the result of turbulence caused by air bubbles from the sparger. Confocal imaging (Fig. 5A) shows that the ochre-colored material covering the coupon is comprised primarily of the stalks characteristic of DIS-1, with many cells attached to the stalks. Confocal imaging also clearly shows that the mat covering the coupon surface is a highly porous, three-dimensional structure and not an adherent biofilm (see Movie S1 in the supplemental material). The absolute thickness of the mature mat could not be measured using the confocal laser scanning microscope. This was because, with time, the uppermost layer of the mat reached a density that effectively blocked laser penetration and prevented imaging of the coupon surface. Based upon images from earlier time points, when both the coupon surface and mat surface could be visualized, analysis of the confocal image stacks indicated that the mat reached a thickness of at least 50 μm by day 4 (data not shown).
FIG 5.
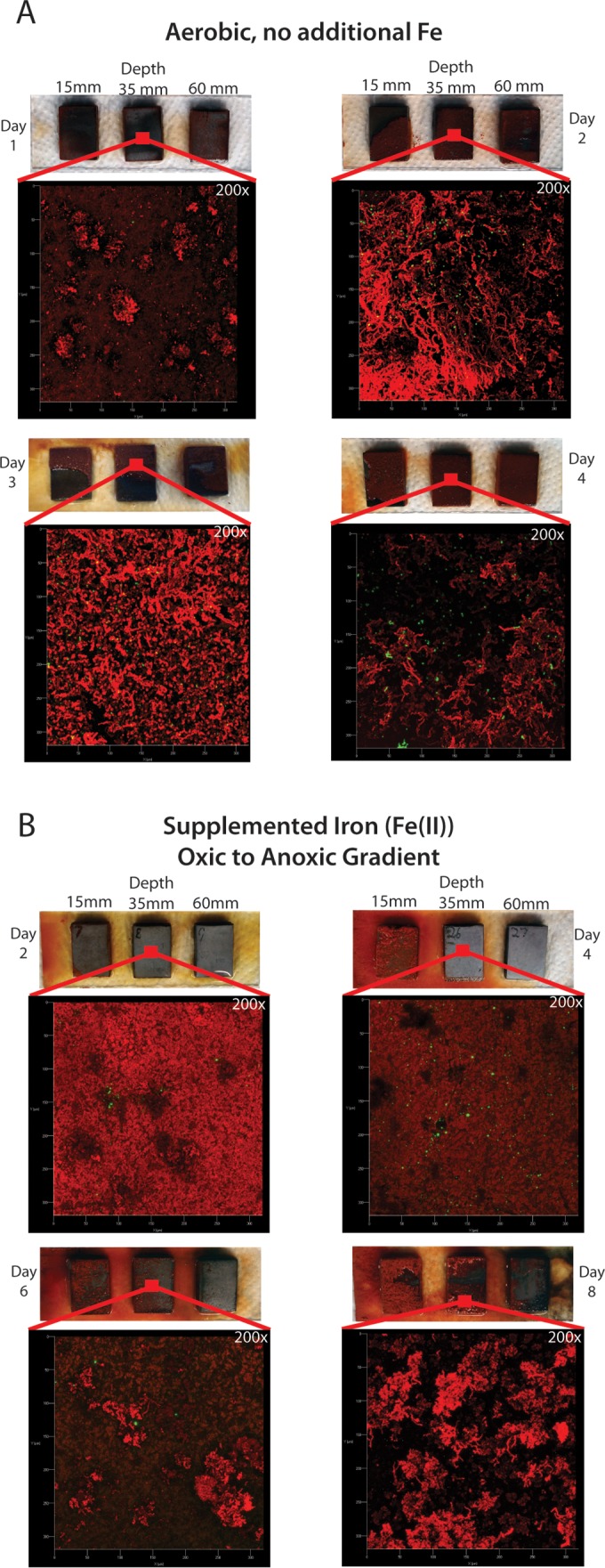
Photographs and confocal micrographs demonstrating progress of colonization over 4 days under aerobic conditions with no supplemented iron (A) and over 9 days in an oxic/anoxic gradient with supplemented iron (B). All confocal micrographs are at ×200 magnification, showing a 300-μm by 300-μm area of the slide incubated at 35-mm depth.
Over the course of the +Fe experiment, coupons incubated in the O2/Fe(II) gradient showed clear differences in colonization based on their location in the oxic/anoxic and redox gradient (Fig. 5B). While colonization of the uppermost (15-mm-depth) coupon in the microcosm followed a trend similar to that seen in the −Fe series, the lowest (60-mm-depth) coupon remained uncolonized throughout the course of the experiment, consistent with it being in the anoxic zone. Colonization was not observed on the intermediate (35-mm-depth) coupon until the 6th day of incubation, at which point individual clusters of stalks could be seen. We observed that the stalks on the 15-mm coupon were much more heavily mineralized than those seen in the −Fe microcosms. Presumably the much higher concentration of Fe(II) in the aqueous phase resulted in increased auto-oxidation and buildup of Fe-oxyhydroxides on the stalks. While a mat was beginning to develop on the 35-mm coupon by day 8, it is notable that this mat is much less developed than that present on the 15-mm coupon. Images were not obtained for the upper coupon on day 10, as the mat had become too thick for the nominal 150-μm clearance provided by the slide frame, resulting in destruction of the mat when the frame was applied to the slide.
Cell counts.
Cell counts increased for the first 3 days of the −Fe series and then decreased between the 3rd and 4th days (Fig. 6A). The decrease in cells on day 4 is attributed to the mat being disrupted due to direct contact with the coverslip, indicating that the mat had filled the approximately 150 μm of space between the surface of the coupon and the coverslip. This was the case for all three coupons. As a result of this, we report the cell counts at day 3, the last day which could be reliably imaged. At day 3, the average cell counts were 4,037 ± 3,677 cells/mm2 on the 15-mm-depth coupon, 1,415 ± 821 cells/mm2 on the 35-mm-depth coupon, and 2,867 ± 1,416 cells/mm2 on the 60-mm-depth coupon. This large error associated with these counts is attributed both to the somewhat patchy nature of the mat and to the uneven distribution of cells within the mat.
FIG 6.
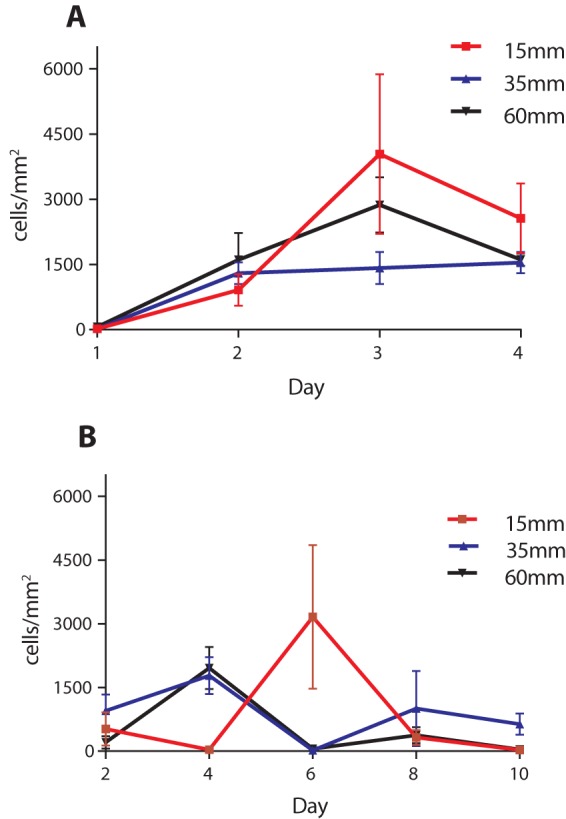
Cell counts from −Fe (A) and +Fe (B) experiments. Error bars represent standard deviations calculated from 5 image stacks.
Cell counts were highly variable in the +Fe microcosms (Fig. 6B), reflecting the patchy distribution of mats and cells across the coupon surfaces in the 15-mm and 35-mm coupons (Fig. 5B). While a large number of cells was observed on day 4, these did not cooccur with the presence of stalks and may be due to weakly adherent cells remaining from the initial inoculation. After 10 days of incubation, a limited number of cells and stalks were observed, with cell counts found to be 27 ± 25 cells/mm2 on the coupon at 15 mm, 634 ± 503 cells/mm2 on the coupon at 35 mm, and 43 ± 29 cells/mm2 on the coupon at 60 mm. During this experiment, the mat thickness never exceeded the approximately 150 μm of clearance between the coupon surface and the coverslip, allowing for all coupons to be imaged and counted at all time points.
Genomic analysis.
Genomic sequencing of DIS-1 resulted in the recovery of 2,945,093 total base pairs of DNA sequence contained in 57 scaffolds. The genome has a GC content of 48.58%. Annotation with IMG (38) found 2,934 open reading frames, including 47 tRNA genes and two 16S rRNA genes. The two 16S genes shared 100% identity with one another and 95.3% identity with the 16S gene of Mariprofundus ferrooxydans PV-1, and the average nucleotide identity (ANI) with other pure-culture genomes of zetaproteobacteria ranged between 70 and 73.6% (Table 1). Together, these results confirmed that DIS-1 was a member of the zetaproteobacteria but could be a new species in the genus Mariprofundus. More details about the genome will be published elsewhere; however, salient aspects are considered here. A gene encoding a homolog of the cyc-2PV-1 protein was present (see Table S1 in the supplemental material); this protein has been proposed to be an important component of the pathway for energy conservation from growth on Fe(II) (39). The genome did not reveal any other obvious metabolic pathways for energy generation from substrates alternative to iron. Consistent with a lithoautotrophic lifestyle, DIS contained genes for the Calvin-Benson-Bassham pathway for fixation of CO2. This included key genes for both the form I and form II ribulose 1,5-bisphosphate carboxylase (RubisCO) genes (see Table S1). Because DIS-1 is capable of growing on the surface of steel coupons in fully oxygenated seawater, we were particularly curious about its capacity to tolerate high O2 concentrations and the presence of reactive oxygen species (ROS). As shown in Table 1, DIS-1 is unusual compared to other marine FeOB in the number and variety of different genes it has with the potential to defend against ROS; this is discussed further below.
TABLE 1.
Comparison of genome sizes, ANI values for different FeOB genomes, and abundance of genes involved in ROS protection
| Parameter | Value for: |
|||||
|---|---|---|---|---|---|---|
| DIS-1 | PV-1 | JV-1 | M34 | EKF-M39 | TAG1 | |
| Genome size (Mb) | 2.94 | 2.86 | 2.85 | 2.74 | 2.72 | 2.16 |
| ANI (%) | 100 | 73.6 | 73.6 | 73.4 | 73.2 | 70 |
| No. of genes | ||||||
| Catalase | 2 | 0 | 0 | 0 | 0 | 0 |
| Catalase-peroxidase | 2 | 0 | 0 | 0 | 0 | 1 |
| Cytochrome c peroxidase | 3 | 2 | 2 | 2 | 4 | 2 |
| Glutathione peroxidase | 1 | 0 | 0 | 0 | 0 | 0 |
| Fe-dependent peroxidase | 1 | 1 | 1 | 1 | 1 | 0 |
| Superoxide dismutase | 1 | 1 | 1 | 1 | 1 | 0 |
| Peroxiredoxins | 5 | 5 | 5 | 6 | 6 | 4 |
| Total | 15 | 9 | 9 | 10 | 12 | 7 |
DISCUSSION
The work presented here uses a novel, continuously flowing seawater microcosm to show that a pure culture of an FeOB can rapidly colonize the surface of mild steel. The use of a custom-made slide holder coupled with lectin staining of the stalk material produced by DIS-1 allowed us to study the adherent microbial mat produced by the FeOB and gain insight into the colonization process. The first important observation is that DIS-1 cells did not make a tightly adherent biofilm. Typically, microbial biofilms produce adherent layers by sticking to the surfaces they colonize and then expanding to cover the surface with a layer that may become several cell layers thick (40). This clearly was not the case for DIS-1. These cells attached initially to surfaces of the steel coupons, but following attachment their growth and stalk production resulted in translocation away from the surface. This growth strategy is somewhat surprising, especially for cells growing in the −Fe conditions where the only source of Fe(II) is what is released from the steel surface. Translocation away from the steel surface moves the cells further from the Fe(II) source they require for growth.
The result was a loosely adherent mat composed primarily of stalk material that formed a porous matrix. Physically this mat provides a large surface in the form of Fe-oxyhydroxide-encrusted stalks that extend above the steel surface. It is likely this growth strategy reflects a response to DIS-1's natural growth conditions rather than a steel surface that is not naturally occurring. It is notable that Mariprofundus sp. strain DIS-1 is closely related to environmental clones of zetaproteobacteria that come from the surface of steel coupons incubated in full-strength seawater (3). The diversity of zetaproteobacteria in those enrichments was low, in some cases consisting of a single clone. This suggests that DIS-1 is a good representative of natural FeOB capable of colonizing steel surfaces. The behavior of DIS-1 growing on the coupon surface is also reminiscent of stalk-forming FeOB that grow at hydrothermal vents and are related to DIS-1 (41). In this case, zetaproteobacteria also produce loosely adherent mats on the surfaces of basaltic lava substrata around the vents (11). This matrix of biogenic iron oxides can become colonized by other bacteria as well as be a substrate for continued auto-oxidation of Fe(II) and the adsorption of other minerals and elements, like silica and phosphorus. We have also observed small (approximately 1-mm-diameter) tufts of iron oxides produced by stalk-forming FeOB growing around the openings of worm burrows in coastal marine sediments, where they are presumably growing on Fe(II) that is fluxing from within the sediment (D. Emerson, unpublished observations). It is likely that this is the natural habitat of DIS-1. A recent study that captured intact vent mats showed stalk-forming FeOB were early colonizers and the primary architect of these mats (41). Stalk formation occurred in a highly parallel fashion, such that the mat grew with a uniform directionality perpendicular to the rock face and showed remarkable temporal and spatial coordination in the formation of the intact microbial mat (41). While we were unable to quantify the orientation of DIS-1 growing on coupons, 3D imaging of their growth did not indicate stalks formed in a highly parallel way (Fig. 5; also see Movie S1 in the supplemental material). Instead, the stalks were more randomly oriented, although there was a bias toward growing away from the steel surface, i.e., we did not observe stalks growing directly along the surface. This suggests the cells were not responding to strong external gradients, for example, oxygen, that might result in more directional growth.
The comparison of treatments with and without added Fe(II) proved informative. In the +Fe treatment there was a relatively steep redox gradient through the microcosm column, from anoxic at the bottom to fully oxic in the top 1 to 2 cm. There were also much higher concentrations of Fe(II) in the bulk water, ranging from 100 to 500 μM Fe(II), compared to the −Fe treatment, where Fe(II) was at the lower detection limit. Somewhat counterintuitively, this resulted in slower colonization and growth on the surface of the coupons, as seen both visually, comparing day 2 or day 4 coupons between +Fe and −Fe microcosms (Fig. 5), and in cell counts where −Fe coupons were more rapidly and consistently colonized (Fig. 6). A potential explanation for this is that with abundant Fe(II) available throughout the microcosm, the cells did not need to attach to the steel surface and may have preferred to attach to other surfaces in the microcosm, either plastic or glass. Consistent with this, we observed significant wall growth in the +Fe microcosm, while such growth was virtually absent from the −Fe microcosm (Fig. 4). In the −Fe microcosm the primary source of Fe(II) to support growth was only what was released from the steel surface, thus forcing the cell to remain close to the steel to acquire Fe(II) for growth.
The other major difference between the two microcosms was the O2 concentration and redox gradient. In the −Fe treatment, coupons at all levels were well oxygenated, and approximately equivalent levels of colonization were observed throughout the microcosm. Based on our understanding of the physiology of zetaproteobacteria as being microaerophiles, we had originally hypothesized that aerobic conditions would inhibit the growth of FeOB and that colonization and growth under fully oxygenated conditions would be limited; however, this did not appear to be the case for DIS-1. In the +Fe microcosm, as expected, we found little to no colonization of the coupon incubated in the anoxic, reducing zone of the microcosm. Somewhat surprisingly, however, there was little difference in colonization between the middle (low O2) and the top coupon, which was exposed to the highest O2 concentration, indicating fully oxygenated conditions did not inhibit DIS-1.
We examined the genome of DIS-1 to better understand possible adaptations that allowed it to tolerate high O2 levels. Compared to other zetaproteobacteria, DIS-1 has all four of the major complexes of the electron transport chain. It possesses two different gene operons that encode a ccb3-type terminal oxidase (complex IV) that has a high affinity for O2, but there was no evidence for a low-affinity cytochrome c oxidase (cox) in the genome (see Table S1 in the supplemental material). This suggests DIS-1 is adapted for microaerobic conditions, although it is clearly capable of tolerating fully oxygenated environments. Consistent with DIS-1 being adapted to higher O2 levels is the fact that it has both type I and type II RubisCO. Type I RubisCO is less O2 sensitive and has a higher affinity for CO2 than type II (42). Most of the other zetaproteobacterial genomes only have type II RubisCO (37).
It has previously been noted that among oxygen-dependent FeOB there is a lack of the most well-recognized genes involved in defense against ROS (37, 43). For example, to date, the gene for catalase, the most commonly occurring enzyme that breaks down hydrogen peroxide (H2O2) (44), has not been found in the genomes of any of the cultured zetaproteobacteria. This is surprising, since these bacteria grow in environments with high concentrations of Fe(II) that also have oxygen and H2O2 present, resulting in conditions that are conducive to the formation of destructive hydroxyl radicals via Fenton chemistry (45). As shown in Table 1, DIS-1 is an exception to this trend, with a number of genes devoted to protection against ROS. There are two copies of the canonical catalase gene that is found in the cytoplasm of most aerobic bacteria but absent from the other FeOB. There are two additional genes (these are paralogs) annotated as catalase-peroxidase genes (see Table S1 in the supplemental material); the corresponding proteins have signal peptides and transmembrane sequences, suggesting they are membrane-bound proteins. Again, most of the other zetaproteobacterial genomes lack this gene(s). Interestingly, there are three cytochrome c peroxidase (CCP) genes in DIS-1, two of which are paralogs. This enzyme is common in Gram-negative bacteria, although it is often in a single copy in the chromosome (46). CCP is an excreted enzyme, located in the periplasm, and is thought to help protect against exogenous sources of H2O2. It can also interact with the electron transport chain, such that H2O2 is used as a terminal electron acceptor. Compared to the other cultured zetaproteobacterial genomes, EKF-M39, an isolate from an iron mat at Loihi Seamount, has four copies of CCP, while the other strains of zetaproteobacteria each have two copies (Table 1). This suggests a general trend among marine FeOB that CCP serves an important function in protection against ROS. Peroxiredoxins that utilize a cysteine-based thiol redox mechanism to break down H2O were represented by 4 to 6 gene copies in the different zetaproteobacterial genomes, thus providing another mechanism for dealing with ROS (47, 48). In addition to these peroxidases, DIS-1 has a glutathione peroxidase and an Fe-dependent peroxidase belonging to the DyP family of peroxidases. All the zetaproteobacteria, except TAG-1, a new isolate from the Mid-Atlantic Ridge, have a single copy of a superoxide dismutase gene of the Fe-Mn type (Table 1). Thus, in total, DIS-1 has a robust repertoire of genes that can help protect it against ROS. This may help explain its ability to colonize steel surfaces in fully oxygenated seawater.
Taking the results together, this work demonstrates how iron-oxidizing zetaproteobacteria colonize steel surfaces under dynamic, aerobic conditions. While the zetaproteobacteria have not been directly implicated in accelerated MIC, these experiments demonstrate how the mats created by these organisms under aerobic conditions could create anaerobic microenvironments suitable for colonization by other microbes, including sulfate-reducing bacteria (SRB), in a successional process as recently reported by McBeth and Emerson (3). The rapid development of an FeOB mat, followed by SRB colonization, would also have the potential to give rise to the conditions required for accelerated low-water corrosion as reported by Marty et al. (2). Further work, including the coculturing of sulfate-reducing bacteria with iron-oxidizing bacteria within the microcosms described here, will serve to further this hypothesis.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the Bigelow Single Cell Genomics Center for their work in sequencing the genome of DIS-1. We appreciate the assistance of Kim Dempsey, an undergraduate from Bowdoin College, for help in establishing the microcosm system, and especially thank Joyce McBeth for her assistance in isolating DIS-1 and Anna Leavitt for isolating the DNA from DIS-1 used for sequencing. We thank Andreas Kappler for providing the opportunity for I.J.A. to participate in this project. We are grateful for many discussions with, and assistance from, Erin Field, Jarrod Scott, and Emily Fleming.
This work was funded by a grant from the Office of Naval Research, ONR N00014-08-1-0334, and I.J.A. was supported by the European Science Foundation through their Earth Sciences Networking Program.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01990-16.
REFERENCES
- 1.Koch GH, Brongers MPH, Thompson NG, Virmani YP, Payer JH. 2002. Corrosion costs and preventative strategies in the United States. Summary of US FHWA publication no. FHWA-RD-01-156 NACE International, Houston, TX: https://www.nace.org/uploadedFiles/Publications/ccsupp.pdf. [Google Scholar]
- 2.Marty F, Gueuné H, Malard E, Sánchez-Amaya JM, Sjögren L, Abbas B, Quillet L, van Loosdrecht MCM, Muyzer G. 2014. Identification of key factors in accelerated low water corrosion through experimental simulation of tidal conditions: influence of stimulated indigenous microbiota. Biofouling 30:281–297. doi: 10.1080/08927014.2013.864758. [DOI] [PubMed] [Google Scholar]
- 3.McBeth JM, Emerson D. 2016. In situ microbial community succession on mild steel in estuarine and marine environments: exploring the role of iron-oxidizing bacteria. Front Microbiol 7:767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anonymous. 2004. Further readings in geomicrobiology. Geomicrobiol J 21:431–432. doi: 10.1080/01490450490489940. [DOI] [Google Scholar]
- 5.Dearaujojorge TC, Coutinho C, Deaguiar LEV. 1992. Sulfate-reducing bacterial associated with biocorrosion–a review. Mem Inst Oswaldo Cruz 87:329–337. doi: 10.1590/S0074-02761992000300001. [DOI] [Google Scholar]
- 6.Dinh HT, Kuever J, Muszmann M, Hassel AW, Stratmann M, Widdel F. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829–832. doi: 10.1038/nature02321. [DOI] [PubMed] [Google Scholar]
- 7.El Mendili Y, Abdelouas A, Bardeau JF. 2013. Insight into the mechanism of carbon steel corrosion under aerobic and anaerobic conditions. Phys Chem Chem Phys 15:9197–9204. doi: 10.1039/c3cp50853f. [DOI] [PubMed] [Google Scholar]
- 8.Enning D, Garrelfs J. 2014. Corrosion of iron by sulfate-reducing bacteria: new views of an old problem. Appl Environ Microbiol 80:1226–1236. doi: 10.1128/AEM.02848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enning D, Venzlaff H, Garrelfs J, Dinh HT, Meyer V, Mayrhofer K, Hassel AW, Stratmann M, Widdel F. 2012. Marine sulfate-reducing bacteria cause serious corrosion of iron under electroconductive biogenic mineral crust. Environ Microbiol 14:1772–1787. doi: 10.1111/j.1462-2920.2012.02778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin M, White DC, Little B, Ray R, Pope R. 2000. The role of bacteria in pit propagation of carbon steel. Biofouling 15:13–23. doi: 10.1080/08927010009386294. [DOI] [PubMed] [Google Scholar]
- 11.Henri PA, Rommevaux-Jestin C, Lesongeur F, Mumford A, Emerson D, Godfroy A, Ménez B. 2015. Structural iron (II) of basaltic glass as an energy source for zetaproteobacteria in an abyssal plain environment, off the Mid Atlantic Ridge. Front Microbiol 6:1518–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernández Gayosso MJ, Zavala Olivares G, Ruiz Ordaz N, Juárez Ramirez C, García Esquivel R, Padilla Viveros A. 2004. Microbial consortium influence upon steel corrosion rate, using polarisation resistance and electrochemical noise techniques. Electrochim Acta 49:4295–4301. doi: 10.1016/j.electacta.2004.03.038. [DOI] [Google Scholar]
- 13.Hubert C, Nemati M, Jenneman G, Voordouw G. 2005. Corrosion risk associated with microbial souring control using nitrate or nitrite. Appl Microbiol Biotechnol 68:272–282. doi: 10.1007/s00253-005-1897-2. [DOI] [PubMed] [Google Scholar]
- 14.Krepski ST, Hanson TE, Chan CS. 2012. Isolation and characterization of a novel biomineral stalk-forming iron-oxidizing bacterium from a circumneutral groundwater seep. Environ Microbiol 14:1671–1680. doi: 10.1111/j.1462-2920.2011.02652.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee AK, Newman DK. 2003. Microbial iron respiration: impacts on corrosion processes. Appl Microbiol Biotechnol 62:134–139. doi: 10.1007/s00253-003-1314-7. [DOI] [PubMed] [Google Scholar]
- 16.Little B, Wagner P, Mansfeld F. 1992. An overview of microbiologically influenced corrosion. Electrochim Acta 37:2185–2194. doi: 10.1016/0013-4686(92)85110-7. [DOI] [Google Scholar]
- 17.McBeth JM, Little BJ, Ray RI, Farrar KM, Emerson D. 2011. Neutrophilic iron-oxidizing “zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl Environ Microbiol 77:1405–1412. doi: 10.1128/AEM.02095-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehanna M, Basséguy R, Délia M-L, Bergel A. 2009. Effect of Geobacter sulfurreducens on the microbial corrosion of mild steel, ferritic and austenitic stainless steels. Corrosion Sci 51:2596–2604. doi: 10.1016/j.corsci.2009.06.041. [DOI] [Google Scholar]
- 19.Neria-González I, Wang ET, Ramírez F, Romero JM, Hernández-Rodríguez C. 2006. Characterization of bacterial community associated to biofilms of corroded oil pipelines from the southeast of Mexico. Anaerobe 12:122–133. doi: 10.1016/j.anaerobe.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen A, Hermansson M. 1991. Bacterial corrosion of iron in seawater in situ, and in aerobic and anaerobic model systems. FEMS Microbiol Lett 86:139–147. [Google Scholar]
- 21.Potekhina JS, Sherisheva NG, Povetkina LP, Pospelov AP, Rakitina TA, Warnecke F, Gottschalk G. 1999. Role of microorganisms in corrosion inhibition of metals in aquatic habitats. Appl Microbiol Biotechnol 52:639–646. doi: 10.1007/s002530051571. [DOI] [Google Scholar]
- 22.Rao TS, Sairam TN, Viswanathan B, Nair KVK. 2000. Carbon steel corrosion by iron oxidising and sulphate reducing bacteria in a freshwater cooling system. Corrosion Sci 42:1417–1431. doi: 10.1016/S0010-938X(99)00141-9. [DOI] [Google Scholar]
- 23.Sung EH, Han JS, Ahn CM, Seo HJ, Kim CG. 2011. Biological metal corrosion in saline systems by sulfur-reducing and iron-oxidizing bacteria. Water Qual Res J Canada 46:321–331. doi: 10.2166/wqrjc.2011.009. [DOI] [Google Scholar]
- 24.Venzlaff H, Enning D, Srinivasan J, Mayrhofer KJJ, Hassel AW, Widdel F, Stratmann M. 2013. Accelerated cathodic reaction in microbial corrosion of iron due to direct electron uptake by sulfate-reducing bacteria. Corrosion Sci 66:88–96. doi: 10.1016/j.corsci.2012.09.006. [DOI] [Google Scholar]
- 25.Xu C, Zhang Y, Cheng G, Zhu W. 2007. Localized corrosion behavior of 316L stainless steel in the presence of sulfate-reducing and iron-oxidizing bacteria. Mater Sci Eng A 443:235–241. doi: 10.1016/j.msea.2006.08.110. [DOI] [Google Scholar]
- 26.Xu CM, Zhang YH, Cheng GX, Zhu WS. 2008. Pitting corrosion behavior of 316L stainless steel in the media of sulphate-reducing and iron-oxidizing bacteria. Mater Charact 59:245–255. doi: 10.1016/j.matchar.2007.01.001. [DOI] [Google Scholar]
- 27.Dang H, Chen R, Wang L, Shao S, Dai L, Ye Y, Guo L, Huang G, Klotz MG. 2011. Molecular characterization of putative biocorroding microbiota with a novel niche detection of epsilon- and zetaproteobacteria in Pacific Ocean coastal seawaters. Environ Microbiol 13:3059–3074. doi: 10.1111/j.1462-2920.2011.02583.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee JS, McBeth JM, Ray RI, Little BJ, Emerson D. 2013. Iron cycling at corroding carbon steel surfaces. Biofouling 29:1243–1252. doi: 10.1080/08927014.2013.836184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emerson D, Fleming EJ, McBeth JM. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 30.Chan CS, Fakra SC, Emerson D, Fleming EJ, Edwards KJ. 2011. Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J 5:717–727. doi: 10.1038/ismej.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C, Moyer CL. 2007. A novel lineage of proteobacteria involved in formation of marine fe-oxidizing microbial mat communities. PLoS One 2:667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angell P, Machowski WJ, Paul PP, Wall CM, Lyle FF Jr. 1997. A multiple chemostat system for consortia studies on microbially influenced corrosion. J Microbiol Methods 30:173–178. doi: 10.1016/S0167-7012(97)00057-2. [DOI] [Google Scholar]
- 33.Emerson D, Floyd M. 2005. Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol 397:112–123. doi: 10.1016/S0076-6879(05)97006-7. [DOI] [PubMed] [Google Scholar]
- 34.Stookey LL. 1970. Ferrozine- a new spectrophotometric reagent for iron. Anal Chem 42:779–781. doi: 10.1021/ac60289a016. [DOI] [Google Scholar]
- 35.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Field EK, Sczyrba A, Lyman AE, Harris CC, Woyke T, Stepanauskas R, Emerson D. 2015. Genomic insights into the uncultivated marine zetaproteobacteria at Loihi Seamount. ISME J 9:857–870. doi: 10.1038/ismej.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barco RA, Emerson D, Sylvan JB, Orcutt BN, Jacobson Meyers ME, Ramírez GA, Zhong JD, Edwards KJ. 2015. New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl Environ Microbiol 81:5927–5937. doi: 10.1128/AEM.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang H, Lovell CR. 2016. Microbial surface colonization and biofilm development in marine environments. Microbiol Mol Biol Rev 80:91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. 2016. The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol 7:796–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badger MR, Bek EJ. 2008. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot 59:1525–1541. [DOI] [PubMed] [Google Scholar]
- 43.Emerson D, Field E, Chertkov O, Davenport K, Goodwin L, Munk C, Nolan M, Woyke T. 2013. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol 4:254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White D. 2007. The physiology and biochemistry of prokaryotes. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 45.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atack JM, Kelly DJ. 2006. Structure, mechanism and physiological roles of bacterial cytochrome c peroxidases. Adv Microb Physiol 52:73–106. doi: 10.1016/S0065-2911(06)52002-8. [DOI] [PubMed] [Google Scholar]
- 47.Rhee SG. 2016. Overview on peroxiredoxin. Mol Cells 39:1–5. doi: 10.14348/molcells.2016.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rhee SG, Kang SW, Chang T-S, Jeong W, Kim K. 2001. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52:35–41. doi: 10.1080/15216540252774748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


