SUMMARY
Yeast cells are well suited to visualizing organelles by 4D confocal microscopy. Typically, one or more cellular compartments are labeled with a fluorescent protein or dye, and a stack of confocal sections spanning the entire cell volume is captured every few seconds. Under appropriate conditions, organelle dynamics can be observed for many minutes with only limited photobleaching. Images are captured at a relatively low signal-to-noise ratio and are subsequently processed to generate movies that can be analyzed and quantified. Here, we describe methods for acquiring and processing 4D data using conventional scanning confocal microscopy.
Keywords: yeast, confocal, 4D microscopy, photobleaching, deconvolution, ImageJ
1. INTRODUCTION
Live-cell fluorescence microscopy provides crucial information about organelle dynamics. Ideally, an entire 3D cell volume (Z-stack) is captured at each time point, yielding a 4D data set that allows intracellular structures to be tracked for many minutes. In the case of yeast cells, 4D imaging is facilitated by the small size of the cells and the relatively low copy numbers of some compartments. A variety of methods have been described for yeast 4D imaging, including widefield microscopy with a specialized deconvolution algorithm [1] and spinning-disk confocal microscopy with a custom high-sensitivity system [2]. Here, we describe our preferred method using conventional laser-scanning confocal microscopy. This approach uses readily available instruments and software.
The major challenge in 4D imaging is to minimize photobleaching and the accompanying photodamage to the cells. Yeast organelles are frequently tagged with fluorescent proteins such as GFP and mCherry. These fluorophores, as well as endocytic tracers such as the dye FM 4–64 [3], are prone to bleaching by confocal lasers. A single Z-stack of a yeast cell typically comprises at least 20 optical slices, and a 4D movie typically involves capturing a Z-stack every few seconds for 5–15 minutes, so the total acquisition can run to thousands of individual confocal sections. Preservation of fluorescence signals under these conditions requires careful attention to multiple parameters.
To minimize photobleaching, the laser power should be as low as possible. We find that photobleaching rates are nonlinear with respect to laser intensity. During multicolor imaging, a laser used to excite one fluorophore can also bleach a second fluorophore—e.g., a 488-nm laser used to excite GFP can also bleach mCherry—so reducing the laser power can have dramatic benefits. In addition, the dwell time of the laser should be low. To meet these criteria, we use the fastest available scan rates and then perform line averaging to obtain usable signals.
Pixel size as determined by the Zoom setting is a crucial parameter. To image a fluorescent structure in a manner that avoids information loss, the sampling interval should be no higher than the Nyquist limit, which is ~2.3 times smaller than the resolution of the optical system [4]. Yeast imaging generally employs a 1.4-NA oil immersion objective, giving a resolution on the order of 200 nm, so the pixel size should be <90 nm. In practice, oversampling down to a pixel size about half of the Nyquist limit tends to improve image quality, but this luxury is usually unavailable for 4D imaging because reducing the pixel size results in slower scans and more photobleaching. As a compromise, we use a pixel size of 65–70 nm, or ~85 nm for weak fluorescence signals.
Similarly, the Z-step interval between confocal sections must be small enough to avoid information loss, but large enough to keep photobleaching low and to enable rapid acquisition of a Z-stack. We use a Z-step interval of ~0.25 μm, or ~0.35 μm for weak fluorescence signals. Enough confocal sections should be used to ensure that fluorescent structures throughout the cell volume will be fully captured, even if the sample shows drift along the Z-axis during movie acquisition. We usually collect 20–30 optical slices per Z-stack.
The signal-to-noise ratio for fluorescent organelles can be enhanced in several ways. A simple tactic is to overexpress the tagged protein of interest. This approach is sometimes unavoidable, in which case control experiments can be performed to confirm that overexpression has not substantially changed the biological system [5]. Whenever possible, a protein should be expressed at endogenous levels by adding a tag through gene replacement at the chromosomal locus. For both overexpression and endogenous gene tagging, we routinely use cassettes encoding 3x or 6x tandem copies of a fluorescent protein [6]. The fluorescence signal scales with the copy number of the fluorescent protein [7]. In addition, we minimize the background noise from the cell by using completely nonfluorescent minimal media devoid of riboflavin and folic acid [8].
Even when the fluorescence signal is strong and clean as viewed by widefield microscopy, the confocal sections captured under conditions suitable for 4D imaging can look alarmingly noisy. The number of photons per pixel is usually very low, so labeled structures may not look smooth and may be embedded in a background that includes shot noise. It is important to accept relatively noisy images at the collection stage with the expectation that the noise can be reduced through image processing. A simple way to reduce shot noise and smooth the true signal is with a 3×3 hybrid median filter, which can be applied once or iteratively [9]. A more sophisticated approach is deconvolution with commercial software. Standard deconvolution methods are not ideal for noisy fluorescence images and may erase weak signals [1]. Until better deconvolution methods become available, we recommend that if the signal-to-noise ratio is low, the data should be preprocessed with a hybrid median filter before deconvolution. In general, empirical testing is needed to devise an image processing routine that adequately preserves the desired signal while removing most of the noise.
After filtering and/or deconvolution, it may be useful to correct for photobleaching. For most fluorophores, the bleaching rate approximates an exponential decay, and a 4D data set can be corrected for exponential photobleaching using an ImageJ plugin. This correction makes it easier to view and quantify the final movies. However, the information content of the Z-stacks diminishes with photobleaching, so if the photobleaching is severe at later time points, little or no signal will be recovered. Reducing the laser power may be beneficial because even though the early images will be of somewhat lower quality, the usable signal will persist for significantly longer.
A 4D movie can be viewed using a volume renderer such as the commercial Imaris or Volocity software, but a simpler and often sufficient alternative is to project each Z-stack to create a 2D image. The projections can then be assembled into a movie. Although maximum intensity projections are often used for convenience, they exaggerate weakly labeled structures, so average projections are preferable for quantitation [9]. A disadvantage of projections is that structures located at different depths in the cell volume may appear to be close together or merged. To address this issue, a labeled structure of interest can be tracked in the original 4D data set, and an edited movie can be generated by manually erasing the other fluorescence signals and projecting only the signal from the labeled structure [5, 10]. Such edited movies are ideal for quantifying the dynamics of individual labeled organelles.
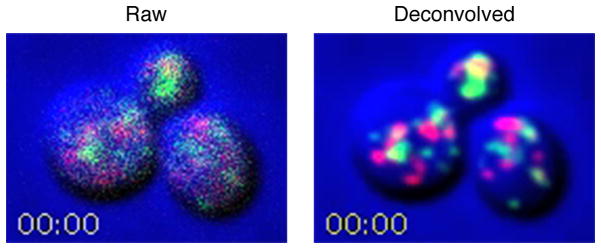
As an example, Fig. 1 shows the first frames from Video 1 and Video 2, which are 4D confocal movies of a yeast strain expressing a GFP-tagged early Golgi protein and a DsRed-tagged late Golgi protein [5]. These markers gave relatively strong signals. In the left panel of Fig. 1 and in Video 1, the raw data were average projected to illustrate the noise level in the images. In the right panel of Fig. 1 and in Video 2, the deconvolved data were average projected to illustrate how noisy images can be processed to obtain usable movies.
Fig. 1.
Illustration of the effect of image processing on 4D movies. Shown are the first frames from Video 1 and Video 2, which are 4D movies from the same data set before and after deconvolution, respectively. Yeast cells expressing GFP-Vrg4 as an early Golgi marker and Sec7-DsRed as a late Golgi marker were imaged by confocal microscopy, with cell images in the blue channel. Z-stacks of 24 optical sections each were collected every 2 sec. Where indicated, the red and green channels were deconvolved. The movie frames are average projections of the Z-stacks. In the deconvolved movie, maturation events can be observed when green Golgi cisternae turn red [5].
2. INSTRUMENTATION AND SOFTWARE
Microscope: Leica SP5 or comparable laser scanning confocal microscope with inverted optics, a fast scanner, and high sensitivity detectors.
Objective lens: Plan-Apo 100x or 63x lens with an NA of 1.4 or greater. The magnification is less important than the NA and lens quality.
Lasers: Multiple laser lines, minimally including a 488-nm laser for exciting green fluorophores and a 561-nm laser for exciting red fluorophores.
Stage adaptor: Standard adaptor for holding dishes with coverglass bottoms.
Z-step motor: Piezoelectric stepper motor for rapid and accurate collection of Z-stacks.
Image manipulation and analysis software: ImageJ freeware obtained from http://imagej.nih.gov/ij/. Custom ImageJ plugins for 4D data analysis can be found in the supplemental material for a recent paper [10], and an ImageJ plugin that corrects for photobleaching can be obtained from http://cmci.embl.de/downloads/bleach_corrector.
Deconvolution software: Huygens Essential (Scientific Volume Imaging).
Movie processing software: QuickTime Player 7 (Apple).
3. MATERIALS
Confocal imaging is performed with yeast strains that have been either engineered to express one or more fluorescently tagged marker proteins, or incubated with a fluorescent tracer dye. For expressing tagged proteins, we typically modify the endogenous chromosomal copy of a gene by clean gene replacement using the pop-in/pop-out method [11, 12]. If desired, a tagged gene can be overexpressed by integrating a vector containing a strong promoter [5]. These approaches have the advantage that the cells do not carry free plasmids, so the expression level within the culture is uniform and the strain is stable even when grown in nonselective medium. However, in some cases it is useful to express a tagged gene on a centromeric plasmid, because cell-to-cell variability in expression enables the choice of cells with an appropriate level of fluorescence.
Yeast cells are grown with shaking in baffled flasks either in SD dropout medium consisting of 0.67% yeast nitrogen base with ammonium sulfate, 2% glucose, and CSM complete supplement mixture or dropout mixture (Sunrise Science Products), or in a nonfluorescent SD medium (NSD) in which the yeast nitrogen base is replaced with a mixture of salts and vitamins lacking riboflavin and folic acid (see Note 1). For a strain that does not require selection to maintain a plasmid, a preculture is grown in rich YPD medium (1% yeast extract, 2% peptone, 2% glucose) and stored at 4°C, and this preculture is used to inoculate a culture in SD or NSD medium at a dilution of 1:1000 to 1:5000 for overnight growth to logarithmic phase (optical density at 600 nm of about 0.4 to 0.8).
Coverglass-bottom dishes with No. 1.5 thickness are obtained from Bioptechs or MatTek, optimally with high tolerance glass of 170 ± 5 μm.
Concanavalin A (Sigma-Aldrich) is dissolved in water to 2 mg/mL, and stored at 4°C for up to a week.
4. METHODS
4.1 Preparing the Cells
Grow one or more yeast strains overnight to logarithmic phase in 5 mL NSD medium in a 50-mL baffled flask.
Prepare a coverglass bottom dish: pipet 250 μL concanavalin A onto the dish, wait 15 min, wash thoroughly with ddH2O, and let dry.
Just before imaging, adhere cells to the dish: pipet 250 μL from the culture onto the dish, wait 10 min, and rinse gently several times with NSD by pipetting. Leave 2–3 mL of fresh NSD in the dish.
If treatment with a drug or dye is required, replace the NSD with fresh medium containing the drug or dye. If appropriate, remove the medium and replace it with fresh medium before imaging.
4.2 Confocal Imaging
-
1
Configure the microscope to use a 100x or 63x high-NA oil immersion lens. Ensure that the DIC prism is not in the light path, or the fluorescence images will be sheared and distored. In the case of a 100x objective with a Leica STED system, the quarter wave plate should be in place or the red and green signals will be offset along the Z-axis.
-
2
Set the frame size to approximately 256×128 (see Note 2).
-
3
Set the Zoom to a level that will give a pixel size of 65–70 nm, or up to ~85 nm if the signal is weak.
-
4
Set the scanner to operate at maximum speed. Use bidirectional mode, as long as the brightfield image confirms that the images from the two directions can be accurately aligned.
-
7
Set the pinhole to 1.2 Airy units (see Note 3).
-
8
Set the line averaging at 4 to 8 (see Note 4).
-
9
Choose a Z-step size of ~0.25 μm, or up to ~0.35 μm if the signal is weak. Note the directionality of Z-stack acquisition (toward or away from the coverslip). To avoid confusion, be consistent with this setting between movies.
-
10
Set an appropriate time interval between Z-stacks. An interval of 2 s is typically suitable, but shorter intervals may be needed to track very dynamic compartments, and somewhat longer intervals may be preferable if the signal is weak.
-
11
Set the lasers to the lowest levels that will generate workable signals (3–10% on the SP5) (see Note 5). For fluorescence data, use the highest sensitivity detectors (HyD on the SP5). For brightfield data, a less sensitive detector is adequate (PMT on the SP5). Set the detector gains to appropriate levels (400–500 for the HyD and 300–350 for the PMT on the SP5).
-
12
Choose emission windows that will maximize the information collected while minimizing bleedthrough between channels. For visualization of GFP that is excited with a 488-nm laser, the emission window is 495–550 nm, and for visualization of mCherry that is excited with a 561-nm laser, the emission window is 575–750 nm (see Note 6).
-
13
Store each confocal section as an 8-bit RGB image using the microscope vendor’s standard file format, with the brightfield view of the cells stored in the blue channel.
4.3 Filtering
For weak signals, deconvolution often erases the structures of interest, leading to flickering movies that are impossible to analyze. In this case, it may help considerably to pre-process the data with a hybrid median filter followed by a Gaussian blur.
In ImageJ, open the 4D data file as a TIFF hyperstack. Then use Image > Color > Split Channels to separate the channels. With the custom plugin called “Filter Hybrid Median”, filter the individual optical slices in the fluorescence channels using a single iteration of the 3D hybrid median filter. If this filter removes too much information, try a single iteration of the standard 2D hybrid median filter (see Note 7).
After hybrid median filtering, use Process > Filters > Gaussian Blur to do a 2D Gaussian blur with a 1-pixel radius for the optical slices in the fluorescence channels. Then use Image > Color > Merge Channels to merge the channels once again into a hyperstack.
4.4 Deconvolution and Bleach Correction
Although the relatively large pixels and Z-steps used for 4D imaging are not optimal for deconvolution, this processing method is useful for cleaning up noisy images and generating smooth objects that are suitable for downstream analysis. We use the Huygens algorithm, but other deconvolution algorithms should give similar results. As described above, when the signal is weak, the Huygens software may erase a structure unless the data are pre-processed by filtering. This effect can reportedly be attributed to the mathematical form of the deconvolution algorithm, which is not ideal for noisy fluorescence data [1].
If the 4D data file was produced directly by the confocal microscope software, the Huygens interface should provide an option for reading the imaging parameters directly. Alternatively, if the 4D data set was pre-processed in ImageJ to generate a TIFF file, these parameters will need to be entered manually. Ensure that the deconvolution software has the correct setting for the directionality of Z-stack acquisition. The refractive index of the embedding medium, which is a yeast cell, can be estimated as 1.35–1.40.
Deconvolve the fluorescence channels but not the brightfield channel. A signal-to-noise ratio of 10 is usually appropriate. Other parameters such as background subtraction can be varied in empirical tests until the output appears to be an accurate rendering of the original fluorescence signals.
Save the output from the deconvolution as an 8-bit TIFF image sequence.
In ImageJ, choose File > Import > Image Sequence to convert the image sequence to a stack. Then choose Image > Hyperstacks > Stack to Hyperstack and input the appropriate order for the confocal sections (e.g., “xyzct”), number of channels, slices per stack, and frames.
To correct for photobleaching (see Note 8), first use Image > Color > Split Channels to separate the channels. For each of the fluorescence channels, use Plugins > EMBLtools > Bleach Correction, and choose “Exponential Fit (Frame-wise)”. Make sure the plot shows a good curve fit, then close the plot window and log window and original channel window, leaving the new corrected channel window. Finally, use Image > Color > Merge channels to regenerate a hyperstack with the bleach-corrected fluorescence data.
4.5 Editing 4D Data Sets with Custom ImageJ Plugins
These procedures are used when the goal is to create an edited 4D data set that includes only one or a few structures. The edited data set can be analyzed to quantify the time course of the fluorescence signals.
Examine a projected movie that was generated as described in Section 4.6 from a non-edited 4D data set. Identify candidate structures that can potentially be tracked for an extended period without interference from other nearby structures, keeping in mind that the 4D data set may permit resolution of structures that occasionally overlap in a projection.
Identify the 8-bit TIFF hyperstack that was used to generate the non-edited projected movie. With the custom plugin called “Make Montage Series”, open this hyperstack, scale the images, and create the montage series.
With the custom plugin called “Edit Montage Series”, choose a single structure that will be tracked through part or all of the movie. Delete the fluorescence signals for all of the other structures at each time point.
With the custom plugin called “Montage Series to Hyperstack”, generate a hyperstack for the edited 4D data set. The images in this hyperstack will normally be magnified due to scaling that occurred during creation of the montage series.
With the custom plugin called “Analyze Edited Movie”, open the hyperstack for the edited 4D data set and specify the time interval between Z-stacks. The “Red” and “Green” columns in the output file represent the total fluorescence signals from the structure of interest at each time point. Save this file, and open it in Microsoft Excel or another data analysis program to plot the fluorescence signals as a function of time.
4.6 Processing 4D Data Sets for Presentation
After performing any or all of the filtering, deconvolution, bleach correction, and editing steps, a 4D data set needs to be converted from a hyperstack to a form that is suitable for presentation and publication.
The first step is to convert the data to 16-bit, to prevent information loss during the subsequent average projection. In ImageJ, choose Image > Color > Split Channels. For each of the resulting three windows, choose Image > Type > 16-bit. Then choose Image > Color > Merge Channels to restore the hyperstack. Finally, choose Process > Math > Multiply, and use a multiplication factor of 256 (see Note 9).
If desired, merge the fluorescence from two 4D movies using the custom ImageJ plugin called “Merge Two Hyperstacks”. This plugin allows two edited 4D movies to be merged, or alternatively allows one 4D movie to be placed above the other—e.g., an original movie above the corresponding edited movie.
The next step is to create an average projection. In ImageJ, choose Image > Stacks > Z Project. If desired, select a subset of each Z-stack by choosing the Start and Stop slices. Choose the “Average Intensity” option.
Adjust the brightness and contrast of the individual channels as follows. Choose Image > Adjust > Brightness/Contrast. For each channel, press the “Auto” button. These settings can be fine tuned using the sliders or the “Set” button.
To add a time stamp, use Image > Stacks > Label. Use the format 00:00, with the appropriate time interval in seconds. To place the label in the lower left corner, set the Y location to be 1 less than the Y value of the lowest row of pixels in the image.
The final step is to make a movie. In ImageJ, choose File > Save As > AVI. Choose PNG compression. A frame rate of 10 fps is generally suitable. If desired, open the resulting AVI file with QuickTime Player 7 and then export a MOV file, which will often have a much smaller file size with no loss of image quality.
Supplementary Material
Footnotes
Nonfluorescent glucose medium (NSD) can be made by assembling the components of normal SD medium except for riboflavin and folic acid. To make 100 mL of a 500x vitamin stock solution, add 100 mg calcium pantothenate, 500 mg myo-inositol, 20 mg niacin, 10 mg p-aminobenzoic acid, 20 mg pyridoxine hydrochloride, 20 mg thiamine hydrochloride, and 20 mg biotin, and store at 4°C. To make a 500x stock solution of cobalt chloride, add 0.1g of cobalt chloride hexahydrate per liter, and store at 4°C. To make 1 liter of NSD, add 20 g glucose, 5 g ammonium sulfate, 5 g potassium phosphate monobasic, 1 g magnesium sulfate heptahydrate, 0.5 g sodium chloride, and 0.1 g calcium chloride dihydrate. Add 2 mL of the vitamin stock solution and 2 mL of the cobalt chloride stock solution. Add YNB trace elements (ANACHEM) from a concentrated stock solution, which is stored at 4°C. If desired, add a CSM complete supplement mixture or dropout mixture. Adjust the pH to 5.5 with NaOH.
Frame height is the main determinant of the total scan time. If desired, a wider frame can be used to capture more data.
The typical recommendation is to use 1.0 Airy unit. However, that setting assumes optimal conditions with no refractive index mismatch between the sample and the oil/glass, whereas the actual refractive index of the cell is lower than that of the oil/glass. Empirically, we find that increasing the pinhole to 1.2 Airy units results in the capture of significantly more light with a negligible decrease in resolution.
Fast scans with line averaging tend to cause less photobleaching than slower scans without line averaging. Image quality is dramatically improved by line averaging, but at the expense of increasing the acquisition time. A line averaging setting of 4–8 is typically a good compromise.
Each laser should be used at a power that yields a decent signal while keeping the photobleaching rate acceptably low. During collection, the signal often is barely visible and is accompanied by significant noise, but such data sets can be processed to obtain usable movies. Set the laser power to 5% or even lower if possible. Photobleaching increases nonlinearly with laser intensity, so a reduction in laser power may yield a signal that is weaker at first but persists for much longer.
Ensure that the confocal microscope is configured with notch filters to suppress background signals from the excitation lasers.
Hybrid median filters work optimally when the image collection settings are near the Nyquist limit. For imaging with a high-NA objective, the pixel size should be in the range of 60–90 nm and the Z-stack interval should be in the range of 0.2–0.4 μm. Filtering before deconvolution is not generally encouraged by makers of deconvolution software because it alters the characteristics of the data, but empirically, this approach sometimes improves the movies by preserving biologically meaningful signals.
Unless the signal is quite strong, photobleaching is usually significant. The correction procedure involves fitting the signal intensity decay to an exponential curve and then multiplying each Z-stack by an appropriate factor to compensate. This procedure is effective up to a point, but as the original signal becomes fainter, the weaker signals in the data set will be progressively lost and will not be recovered after the correction for photobleaching.
After conversion to 16-bit and multiplication by 256, the image display may not be scaled appropriately, but the data are still suitable for further processing.
References
- 1.Arigovindan M, et al. High-resolution restoration of 3D structures from widefield images with extreme low signal-to-noise-ratio. Proc Natl Acad Sci USA. 2013;110:17344–17349. doi: 10.1073/pnas.1315675110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurokawa K, et al. Live cell visualization of Golgi membrane dynamics by super-resolution confocal live imaging microscopy. Methods Cell Biol. 2013;118:235–242. doi: 10.1016/B978-0-12-417164-0.00014-8. [DOI] [PubMed] [Google Scholar]
- 3.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawley JB. Handbook of Biological Confocal Microscopy. 3. Springer; 2006. p. 985. [Google Scholar]
- 5.Losev E, et al. Golgi maturation visualized in living yeast. Nature. 2006;22:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- 6.Genove G, Glick BS, Barth AL. Brighter reporter genes from multimerized fluorescent proteins. BioTechniques. 2005;39:814–822. doi: 10.2144/000112056. [DOI] [PubMed] [Google Scholar]
- 7.Connerly PL, et al. Sec16 is a determinant of transitional ER organization. Curr Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 8.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 9.Hammond AT, Glick BS. Raising the speed limits for 4D fluorescence microscopy. Traffic. 2000;1:935–940. [PubMed] [Google Scholar]
- 10.Papanikou E, et al. COPI selectively drives maturation of the early Golgi. doi: 10.7554/eLife.13232. Submitted, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossanese OW, et al. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.