Abstract
Background
Epidemiological studies and secondary analyses of randomized trials supported the hypothesis that selenium and vitamin E lower prostate cancer risk. However, the Selenium and Vitamin E Cancer Prevention Trial (SELECT) showed no benefit of either supplement. Genetic variants involved in selenium or vitamin E metabolism or transport may underlie the complex associations of selenium and vitamin E.
Methods
We undertook a case-cohort study of SELECT participants randomized to placebo, selenium or vitamin E. The subcohort included 1,434 men; our primary outcome was high-grade prostate cancer (N=278 cases, Gleason 7 or higher cancer). We used weighted Cox regression to examine the association between SNPs and high-grade prostate cancer risk. To assess effect modification, we created interaction terms between randomization arm and genotype and calculated log likelihood statistics.
Results
We noted statistically significant (p<0.05) interactions between selenium assignment, SNPs in CAT, SOD2, PRDX6, SOD3, and TXNRD2 and high-grade prostate cancer risk. Statistically significant SNPs that modified the association of vitamin E assignment and high-grade prostate cancer included SEC14L2, SOD1, and TTPA. In the placebo arm, several SNPs, hypothesized to interact with supplement assignment and risk of high-grade prostate cancer, were also directly associated with outcome.
Conclusion
Variants in selenium and vitamin E metabolism/transport genes may influence risk of overall and high-grade prostate cancer, and may modify an individual man’s response to vitamin E or selenium supplementation with regards to these risks.
Impact
The effect of selenium or vitamin E supplementation on high-grade prostate cancer risk may vary by genotype.
Keywords: Prostate cancer, antioxidant genes, single nucleotide polymorphisms, selenium, vitamin E
INTRODUCTION
Primary prevention of prostate cancer holds promise to reduce the burden of this disease, yet specific preventive factors remain elusive. In the 1990s, secondary analyses of two randomized clinical trials – the Alpha-Tocopherol & Beta Carotene Cancer Prevention Trial (ATBC) and the Nutritional Prevention of Cancer Trial – yielded provocative results suggesting that supplementation with selenium or vitamin E might markedly reduce the risk of clinically significant prostate cancer.(1-3) Moreover, there was corroborating epidemiological evidence suggesting that higher endogenous levels of vitamin E or selenium might be associated with lower risk of prostate cancer.(4-10)
These data supported the development and implementation of the Selenium and Vitamin E Cancer Prevention Trial (SELECT) in which 35,533 men were randomized to supplementation with 200 μg/d selenium (L-selenomethionine) alone, 400 IU/d vitamin E (alpha-tocopheryl acetate) alone, both, or placebo. The men were cancer-free at baseline and were followed prospectively for prostate cancer incidence. The trial was stopped early due to lack of efficacy of either supplement, and subsequent reports have indicated that men assigned to the vitamin E arm had a 17% greater risk of overall prostate cancer (hazard ratio (HR) 1.17, 99% Confidence Interval (CI) 1.004-1.36, P=.008).(11) Furthermore, men with higher baseline selenium or alpha-tocopherol levels assigned to selenium supplementation had greater risk of high-grade prostate cancer, while men assigned to vitamin E supplements who had low baseline selenium levels were at increased risk of prostate cancer.(12, 13)
The SELECT results clearly do not support the use of supplemental selenium or vitamin E in adult life for primary prevention of prostate cancer. However, there is intriguing data that variation in genes associated with selenium or vitamin E metabolism or transport may underlie the complex associations and unexpected results among the clinical trials. (14-18)} We leveraged the unique study design of SELECT and evaluated variation across 21 genes that were hypothesized a priori to be related to selenium or vitamin E metabolism or transport (Supplementary Table 1) and the risk of overall and high-grade prostate cancer. We specifically hypothesized that variation in these genes may influence prostate cancer risk as a function of randomization to vitamin E or selenium supplementation (compared to placebo) and the risk of prostate cancer, particularly for risk of high-grade prostate cancer.
MATERIALS AND METHODS
Study Population
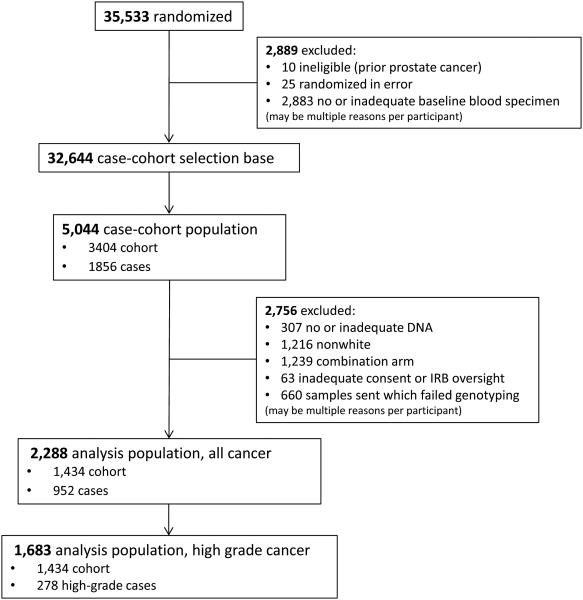
SELECT recruited 35,533 men from sites in the United States, Canada and Puerto Rico. Details on the eligibility and enrollment methods can be found in Lippman et al, 2009.(11) To control for population stratification, we limited the study to Caucasian men with available germline DNA samples, who consented to use the sample, and who were randomized to placebo, selenium alone or vitamin E alone. We did not include the combination arm of vitamin E and selenium given the apparent interaction between the two supplements and prostate cancer risk.(11) We used a case-cohort design and sampled the subcohort stratified by age group (55-59, 60-64, 65-69, ≥ 70 years). Figure 1 presents an overview of the case-cohort sampling for this study. The subcohort included 1,434 men, of whom 98 had been diagnosed with prostate cancer, including 29 with high-grade disease (defined as Gleason 7 or higher). We further included all remaining 854 prostate cancer cases, for a total of 952 cases of whom 278 had high-grade disease.
Figure 1.
Consort diagram summarizing the case-cohort design, including selection of the cohort and cases.
Genotyping
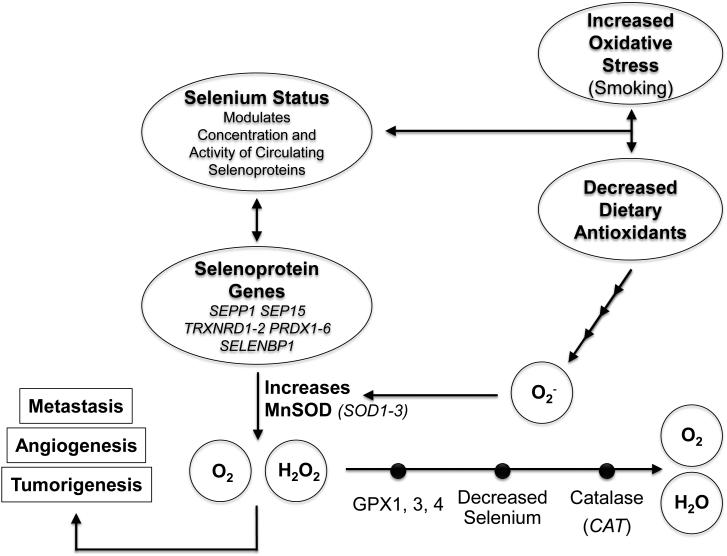
We selected 21 genes that had previously been reported to interact either with selenium or vitamin E levels, metabolism, or transport, in relation to prostate cancer risk (Supplemental Table 1). These included 18 genes with putative selenium-related antioxidant properties (CAT, GPX1, GPX3, GPX4, PRDX1-6; SELENBP1, SEP15, SEPP1, SOD1, SOD2, SOD3, TXNRD1, TXNRD2 Figure 2) (15, 17, 19-24); 2 genes involved in vitamin E transport (SEC14L2, TTPA) (18); and a DNA repair gene that interacted with vitamin E and prostate cancer in multiple reports (XRCC1) (25-29). We focused on single nucleotide polymorphisms (SNPs) to capture variation across these genes. The inclusion of the peroxiredoxin genes (PRDX 1-6) was more exploratory, based on limited data suggesting their (potentially selenium-dependent (30)) antioxidant properties, and corroborative studies indicating that somatic expression of PRDX influences androgen pathways in prostate cancer. (30-40) While chosen primarily for their putative interaction with selenium, for completeness, we also examined the interaction of SOD1, SOD3, and SOD3 with vitamin E assignment, based on a prior report of an interaction for prostate cancer. (41)
Figure 2.
Schematic overview of the potential role of variants in genes in the antioxidant pathways to modify the effect of selenium on risk of aggressive prostate cancer.
Using the HapMap3 R28 database, we undertook a haplotype tagging approach to capture genetic variation with an R2>0.80 across each of the 21 genes, as well as 5 kilobase pairs up- and downstream using pairwise tagging. Selection was restricted to SNPs with a minor allele frequency >5% in the International HapMap CEPH samples. We tagged 135 SNPs. Genotyping was performed on DNA extracted from buffy coat using the Sequenom iPLEX platform assay at the Genotyping Core Facility at Children’s Hospital, Boston. On each 96-well plate, we included 4% quality control specimens. 130 SNPs had high genotyping success (>90%) (Supplementary Table 1); the 5 SNPs that failed genotyping (rs1001179, rs35741824, rs1799895, rs548649, and rs5993853) were excluded from future analyses. We further excluded 6 SNPs with a minor allele frequency in our study population <5% (GPX3 rs8177425, PRDX2 rs35866106, PRDX4 rs6653694, SOD1 rs17885303, TXNRD2 rs4485648, XRCC1 rs25489). In addition, we excluded data from 318 participants because of low genotyping quality (<85%). The sample size total varies by SNP, as genotyping a particular SNP may have failed for some participants.
Outcome and statistical analyses
Our primary outcome was time to diagnosis of high-grade prostate cancer, defined as a Gleason 7 or greater tumor. We additionally examined the risk of prostate cancer overall as a secondary outcome.
We used Cox proportional hazards models to examine the association between each SNP and risk of high-grade prostate cancer, as well as overall prostate cancer risk. Models were stratified by the four age groups to account for the case-cohort design, and weighted based on the fraction of men selected to the cohort from each stratum compared to the total trial analysis population (Caucasian, 3 treatment arms). A second type of weight was used to construct the pseudolikelihood function, where all cohort members were weighted equally regardless of future prostate cancer diagnosis, and cases outside of the cohort were weighted only at the time of diagnosis as described by Prentice(42). The sampling and case-cohort weights were used to calculate hazard ratios (HR) and 95% confidence intervals (CI) of the association between each SNP and prostate cancer risk. For analyses of high-grade prostate cancer, participants in the subcohort diagnosed with low-grade prostate cancer were censored at time of diagnosis.
For the associations of the genotypes and prostate cancer risk, we calculated hazard ratios and 95% CIs using a co-dominant genetic model, and estimated p-values of linear trend across the genotypes using an additive model. For homozygous rare genotypes with a frequency less than 5%, we modeled SNPs using a dominant genetic model. To assess effect modification, we created interaction terms between randomization arm and each genotype assuming an additive model and calculated log likelihood statistics. Supplementary Table 1 presents the minor allele frequencies (MAF) of the investigated 130 SNPs in the 21 antioxidant-related genes of interest and also summarizes the specific evaluation of SNP-treatment interactions which were restricted depending on the gene function and its hypothesized role in either selenium or vitamin E. Five SNPs violated Hardy-Weinberg Equilibrium (p<0.001) after sample filtering based on Pearson’s goodness of fit, but these were retained in the analyses.
Analyses were performed with SAS statistical software versions 9.3 and 9.4 (SAS Institute, Cary, NC) and P values are two-sided. As we undertook a pathway-based approach to test specific a priori hypotheses, we did not control for multiple comparisons; P<0.05 was considered statistically significant.
RESULTS
Table 1 compares baseline demographic, lifestyle and clinical factors among men in the subcohort as well as with high-grade prostate cancer, separately in the placebo arm, selenium arm and vitamin E arm. The median age of men in the subcohort was 63 years; among the men with high-grade disease, the median age was 64-65 years.
Table 1.
Distribution of Baseline Demographic and Health-Related Characteristics by Treatment Arm, Analysis Population (n=1,683)a
| Placebo Arm | Selenium Arm | Vitamin E Arm | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Characteristic | Cohortb
(n = 481) N (%) |
High- Gradec (n = 78) N (%) |
Cohortb
(n = 476) N (%) |
High-Gradec
(n = 97) N (%) |
Cohortb
(n = 477) N (%) |
High-Gradec
(n = 103) N (%) |
| Age, years | ||||||
| Median (IQR) | 63 (59,68) | 65 (60,69) | 63 (59,69) | 64 (60,69) | 63 (59,68) | 65 (61,69) |
| <60 | 126 (26.2%) | 19 (24.4%) | 131 (27.5%) | 22 (22.7%) | 120 (25.2%) | 14 (13.6%) |
| 60-64 | 150 (31.2%) | 20 (25.6%) | 135 (28.4%) | 29 (29.9%) | 146 (30.6%) | 33 (32.0%) |
| 65-69 | 116 (24.1%) | 22 (28.2%) | 121 (25.4%) | 26 (26.8%) | 131 (27.5%) | 33 (32.0%) |
| ≥70 | 89 (18.5%) | 17 (21.8%) | 89 (18.7%) | 20 (20.6%) | 80 (16.8%) | 23 (22.3%) |
| Family history of prostate cancer |
||||||
| No | 406 (84.4%) | 56 (71.8%) | 403 (84.7%) | 71 (73.2%) | 396 (83.0%) | 78 (75.7%) |
| Yes | 74 (15.4%) | 22 (28.2%) | 73 (15.3%) | 26 (26.8%) | 81 (17.0%) | 25 (24.3%) |
| Unknown | 1 (0.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Body mass index, kg/m2 |
||||||
| Median (IQR) | 28.1 (25.6,31.4) |
28.5 (26.1,30.5) |
27.7 (25.6,30.8) |
28 (25.5,31.1) |
28.1 (25.8,31) |
27.8 (25.2,31.7) |
| <25 | 93 (19.3%) | 14 (17.9%) | 89 (18.7%) | 18 (18.6%) | 83 (17.4%) | 24 (23.3%) |
| 25-<30 | 223 (46.4%) | 40 (51.3%) | 243 (51.1%) | 47 (48.5%) | 243 (50.9%) | 46 (44.7%) |
| ≥30 | 164 (34.1%) | 24 (30.8%) | 143 (30.0%) | 31 (32.0%) | 150 (31.4%) | 33 (32.0%) |
| Unknown | 1 (0.2%) | 0 (0.0%) | 1 (0.2%) | 1 (1.0%) | 1 (0.2%) | 0 (0.0%) |
| Diabetes | ||||||
| No | 441 (91.7%) | 73 (93.6%) | 437 (91.8%) | 88 (90.7%) | 430 (90.1%) | 95 (92.2%) |
| Yes | 40 (8.3%) | 5 (6.4%) | 39 (8.2%) | 9 (9.3%) | 47 (9.9%) | 8 (7.8%) |
| Prostate Specific Antigen (ng/mL)d |
||||||
| Median (IQR) | 1.1 (0.7,1.8) | 2.2 (1.5,3.1) | 1.2 (0.7,2) | 2.5 (1.8,3.2) | 1.1 (0.6,1.9) | 2.4 (1.7,3.1) |
| <1.0 | 211 (43.9%) | 6 (7.7%) | 184 (38.7%) | 7 (7.2%) | 200 (41.9%) | 9 (8.7%) |
| 1.01-1.99 | 165 (34.3%) | 24 (30.8%) | 166 (34.9%) | 26 (26.8%) | 161 (33.8%) | 27 (26.2%) |
| 2.00-2.99 | 61 (12.7%) | 25 (32.1%) | 77 (16.2%) | 30 (30.9%) | 75 (15.7%) | 37 (35.9%) |
| 3.00-3.99 | 44 (9.1%) | 23 (29.5%) | 49 (10.3%) | 34 (35.1%) | 40 (8.4%) | 30 (29.1%) |
| ≥4.00 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0%) | 0 (0%) |
| Smoking Status | ||||||
| Never | 201 (41.8%) | 34 (43.6%) | 206 (43.3%) | 48 (49.5%) | 200 (41.9%) | 50 (48.5%) |
| Former | 252 (52.4%) | 39 (50.0%) | 241 (50.6%) | 45 (46.4%) | 256 (53.7%) | 49 (47.6%) |
| Current | 25 (5.2%) | 3 (3.8%) | 29 (6.1%) | 4 (4.1%) | 20 (4.2%) | 4 (3.9%) |
| Unknown | 3 (0.6%) | 2 (2.6%) | 0 (0.0%) | 0 (0.0%) | 1 (0.2%) | 0 (0.0%) |
| Highest level of education completed |
||||||
| High school or less |
96 (20.0%) | 10 (12.8%) | 91 (19.1%) | 22 (22.7%) | 81 (17.0%) | 23 (22.3%) |
| Some college or vocational school |
113 (23.5%) | 20 (25.6%) | 135 (28.4%) | 23 (23.7%) | 129 (27.0%) | 22 (21.4%) |
| College graduate or higher |
268 (55.7%) | 46 (59.0%) | 249 (52.3%) | 52 (53.6%) | 266 (55.8%) | 58 (56.3%) |
| Unknown | 4 (0.8%) | 2 (2.6%) | 1 (0.2%) | 0 (0.0%) | 1 (0.2%) | 0 (0.0%) |
| Marital status | ||||||
| Currently married/ cohabitating |
421 (87.5%) | 66 (84.6%) | 391 (82.1%) | 82 (84.5%) | 414 (86.8%) | 86 (83.5%) |
| Previously married |
45 (9.4%) | 7 (9.0%) | 62 (13.0%) | 13 (13.4%) | 46 (9.6%) | 15 (14.6%) |
| Never married | 12 (2.5%) | 3 (3.8%) | 21 (4.4%) | 2 (2.1%) | 15 (3.1%) | 2 (1.9%) |
| Unknown | 3 (0.6%) | 2 (2.6%) | 2 (0.4%) | 0 (0.0%) | 2 (0.4%) | 0 (0.0%) |
| % of annual PSA tests done |
||||||
| <25% | 47 (9.8%) | 2 (2.6%) | 46 (9.7%) | 3 (3.1%) | 36 (7.5%) | 2 (1.9%) |
| 25-<50% | 47 (9.8%) | 7 (9.0%) | 54 (11.3%) | 5 (5.2%) | 56 (11.7%) | 5 (4.9%) |
| 50-<75% | 185 (38.5%) | 30 (38.5%) | 167 (35.1%) | 35 (36.1%) | 182 (38.2%) | 38 (36.9%) |
| 75-<100% | 154 (32.0%) | 28 (35.9%) | 155 (32.6%) | 23 (23.7%) | 164 (34.4%) | 25 (24.3%) |
| 100% | 48 (10.0%) | 11 (14.1%) | 54 (11.3%) | 31 (32.0%) | 39 (8.2%) | 33 (32.0%) |
| Supplemental Vitamin E (IU/dy) |
||||||
| Median (IQR) | 15 (10,20) | 15 (11,20) | 15 (10,22) | 16 (11,23) | 15 (10,23) | 15 (11,22) |
| <25 | 400 (83.2%) | 64 (82.1%) | 390 (81.9%) | 77 (79.4%) | 379 (79.5%) | 85 (82.5%) |
| 25-<50 | 68 (14.1%) | 12 (15.4%) | 74 (15.5%) | 19 (19.6%) | 89 (18.7%) | 15 (14.6%) |
| 50-<75 | 11 (2.3%) | 2 (2.6%) | 9 (1.9%) | 1 (1.0%) | 8 (1.7%) | 3 (2.9%) |
| 75-<100 | 0 (0.0%) | 0 (0.0%) | 3 (0.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| ≥100 | 2 (0.4%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.2%) | 0 (0.0%) |
| Supplemental Selenium (μg/dy) |
||||||
| Median (IQR) | 134 (101,172) | 136 (97,183) | 131 (94,174) | 148 (113,189) |
133 (98,170) | 132 (96,175) |
| 0 | 2 (0.4%) | 2 (2.6%) | 1 (0.2%) | 0 (0.0%) | 1 (0.2%) | 0 (0.0%) |
| <50 | 14 (2.9%) | 1 (1.3%) | 17 (3.6%) | 1 (1.0%) | 9 (1.9%) | 1 (1.0%) |
| 50-<100 | 103 (21.4%) | 18 (23.1%) | 119 (25.0%) | 16 (16.5%) | 116 (24.3%) | 28 (27.2%) |
| 100-<150 | 170 (35.3%) | 27 (34.6%) | 159 (33.4%) | 35 (36.1%) | 175 (36.7%) | 34 (33.0%) |
| ≥150 | 192 (39.9%) | 30 (38.5%) | 180 (37.8%) | 45 (46.4%) | 176 (36.9%) | 40 (38.8%) |
Analysis population was taken from the pre-existing SELECT case-cohort, restricted to Caucasian only, and to the Placebo, Selenium, and Vitamin E treatment arms only.
The cohort includes high-grade cases as follows: Placebo arm, 9; Selenium Arm, 14; Vitamin E arm, 6.
High-grade cases are those with Gleason scores available for the diagnostic biopsy, with Gleason score 7 or higher.
One patient in the cohort had a baseline PSA of 10.6, although the study eligibility criteria required PSA <= 4.00. This participant was retained in analyses.
IQR: inter-quartile range; PSA: Prostate Specific Antigen
Table 2 presents statistically significant (p<0.05) results for effect modification between antioxidant SNPs, selenium assignment and risk of high-grade prostate cancer. This analysis included 1109 participants randomized to selenium alone or placebo, including 934 in the subcohort and 175 high-grade cases. The identified SNPs included several in CAT (rs10836233, rs533425, rs7944397), 1 in SOD2 (rs7855), 1 in PRDX6 (rs11580117), and multiple SNPs in SOD3 (rs699473, rs8192287) and TXNRD2 (rs3804047, rs8141691). Full results for all analyzed SNPs and interactions with selenium assignment, for both high-grade prostate cancer and overall disease, can be found in Supplementary Table 2.
Table 2.
Statistically significant (p < 0.05)a interactions between antioxidant SNPs and selenium supplementation for risk of high-grade prostate cancer (N=1,109)
| Genotype frequency | Hazard ratio (95% CI) high-grade cancer |
|||||
|---|---|---|---|---|---|---|
| Gene | Genotype | N (%) cases | N (%) controls |
Placebo arm | Selenium arm | p-valueb |
| CAT | rs10836233 | |||||
| GG | 145 (83.3%) | 750 (80.6%) | 1.0 | 1.02 (0.71,1.46) | 0.005 | |
| Any A rs533425 |
29 (16.7%) | 180 (19.4%) | 0.38 (0.18,0.83) | 1.56 (0.90,2.71) | ||
| GG | 58 (33.3%) | 351 (37.7%) | 1.0 | 2.58 (1.42,4.68) | 0.003 | |
| AG | 91 (52.3%) | 443 (47.6%) | 2.31 (1.29,4.13) | 2.28 (1.28,4.07) | ||
| AA rs7944397 | 25 (14.4%) | 136 (14.6%) | 2.35 (1.13,4.87) | 1.51 (0.67,3.37) | ||
| AA | 143 (82.7%) | 676 (72.8%) | 1.0 | 1.02 (0.71,1.46) | 0.02 | |
| Any G | 30 (17.3%) | 253 (27.2%) | 0.30 (0.14,0.62) | 0.93 (0.55,1.60) | ||
| PRDX6 | rs11580117 | |||||
| AA | 157 (89.7%) | 843 (90.3%) | 1.0 | 1.08 (0.75,1.54) | 0.05 | |
| any G | 18 (10.3%) | 91 (9.7%) | 0.72 (0.36,1.42) | 1.84 (1.09,3.13) | ||
| SOD2 | rs7855 | |||||
| AA | 157 (89.7%) | 843 (90.3%) | 1.0 | 1.46 (1.04,2.06) | 0.02 | |
| Any G | 18 (10.3%) | 91 (9.7%) | 2.14 (1.06,4.31) | 0.77 (0.31,1.88) | ||
| SOD3 | rs699473 | |||||
| TT | 73 (46.2%) | 379 (44.1%) | 1.0 | 2.18 (1.29,3.69) | 0.04 | |
| CT | 37 (23.4%) | 248 (28.8%) | 1.40 (0.75,2.63) | 1.11 (0.57,2.17) | ||
| CC rs8192287 | 48 (30.4%) | 233 (27.1%) | 1.68 (0.92,3.07) | 1.66 (0.88,3.11) | ||
| GG | 153 (87.4%) | 826 (88.5%) | 1.0 | 1.44 (1.02,2.03) | 0.04 | |
| any T | 22 (12.6%) | 107 (11.5%) | 1.69 (0.89,3.23) | 0.85 (0.39,1.87) | ||
| TXNRD2 | rs3804047 | |||||
| AA | 87 (50.9%) | 483 (52.8%) | 1.0 | 1.78 (1.12,2.84) | 0.03 | |
| AG | 72 (42.1%) | 355 (38.8%) | 1.48 (0.89,2.48) | 1.51 (0.91,2.50) | ||
| GG rs8141691 | 12 (7.0%) | 77 (8.4%) | 1.63 (0.67,3.94) | 0.91 (0.33,2.50) | ||
| GG | 65 (37.8%) | 398 (43.1%) | 1.0 | 1.64 (0.97,2.77) | 0.05 | |
| AG | 77 (44.8%) | 388 (42.0%) | 1.34 (0.78,2.31) | 1.76 (1.06,2.94) | ||
| AA | 30 (17.4%) | 138 (14.9%) | 2.18 (1.11,4.25) | 1.29 (0.65,2.56) | ||
Full results for all analyzed SNP x selenium assignment interactions are presented in Supplementary Table 2
P-value for test for interaction between SNP and selenium assignment for the outcome of risk of high-grade prostate cancer
Table 3 provides statistically significant SNPs that appeared to modify the association of vitamin E assignment and risk of high-grade prostate cancer. This analysis included 1,124 participants (943 controls and 181 high-grade prostate cancer cases) in the vitamin E alone and placebo arms. The significant SNPs included SEC14L2 (rs5753106) and TTPA (rs12679996, rs4606052). Full results for all analyzed SNPs and interactions with vitamin E assignment, for both high-grade prostate cancer and overall disease, can be found in Supplementary Table 3.
Table 3.
Statistically significant (p<0.05) interactionsa between antioxidant & vitamin E transport SNPs and vitamin E supplementation interactions (p < 0.05) for risk of high-grade prostate cancer (N=1,124)
| Genotype frequency | Hazard ratio (95% CI) high- grade cancer |
|||||
|---|---|---|---|---|---|---|
| Gene | Genotype | N (%) cases | N (%) controls |
Placebo arm | Vitamin E arm | p-valueb |
| SEC14L2 | rs5753106 | |||||
| AA | 106 (59.2%) | 543 (57.6%) | 1.0 | 0.98 (0.64,1.49) | 0.008 | |
| AG | 65 (36.3%) | 347 (36.8%) | 0.74 (0.44,1.24) | 1.25 (0.80,1.97) | ||
| GG | 8 (4.5%) | 52 (5.5%) | 0.14 (0.02,0.99) | 1.57 (0.63,3.91) | ||
| TTPA | rs12679996 | |||||
| CC | 69 (38.5%) | 361 (38.9%) | 1.0 | 2.29 (1.32,3.97) | 0.001 | |
| CT | 77 (43.0%) | 426 (46.0%) | 1.34 (0.74,2.40) | 1.70 (0.98,2.94) | ||
| TT rs4606052 | 33 (18.4%) | 140 (15.1%) | 2.76 (1.44,5.29) | 1.33 (0.60,2.97) | ||
| CC | 62 (35.4%) | 271 (29.9%) | 1.0 | 2.25 (1.26,4.03) | 0.007 | |
| CT | 68 (38.9%) | 434 (48.0%) | 0.97 (0.53,1.79) | 1.27 (0.71,2.25) | ||
| TT | 45 (25.7%) | 200 (22.1%) | 2.02 (1.06,3.82) | 1.38 (0.69,2.75) | ||
Full results for all analyzed SNP x vitamin E assignment interactions are presented in Supplementary Table 3
P-value for test for interaction between SNP and selenium assignment for the outcome of risk of high-grade prostate cancer
We also identified several SNPs that were nominally statistically significant (p<0.05) for overall prostate cancer risk in the placebo arm only (Table 4), several of which were also associated with high-grade prostate cancer. Of note, SNPs in CAT (rs10836233, rs533425, rs7944397), SEC14L2 (rs5753106), SOD2 (rs2070424), TTPA (rs12679996, rs4606052), and TXRND2 (rs8141691) that were significantly associated with high-grade risk overall were also identified as modifiers of randomized supplement assignment. Full results for the association between each of the individual SNPs and risk of overall and high-grade prostate cancer are in Supplementary Table 4.
Table 4.
Statistically significant associationsa between antioxidant metabolism and transport SNPs and risk of high-grade prostate cancer, in the placebo group of SELECT (N=550)
| Genotype Frequency | High grade prostate cancer |
||||
|---|---|---|---|---|---|
| Gene | Genotype | N (%) cases | N (%) controls |
HR (95% CI) | p-value |
| CAT | rs10836233 | ||||
| GG | 69 (89.6%) | 363 (77.4%) | 1.0 | 0.02 | |
| Any A rs533425 | 8 (10.4%) | 102 (21.7%) | 0.39 (0.18,0.84) | ||
| GG | 18 (23.1%) | 190 (40.3%) | 1.0 | 0.005 | |
| AG | 45 (57.7%) | 215 (45.6%) | 2.29 (1.28,4.09) | ||
| AA rs7944397 | 15 (19.2%) | 66 (14.0%) | 2.31 (1.12,4.76) | ||
| AA | 68 (88.3%) | 325 (69.3%) | 1.0 | 0.001 | |
| Any G | 9 (11.7%) | 144 (30.7%) | 0.30 (0.14,0.62) | ||
| GPX1 | rs17650792 | ||||
| AA | 30 (39.0%) | 139 (29.9%) | 1.0 | 0.04 | |
| AG | 37 (48.1%) | 231 (49.7%) | 0.71 (0.42,1.18) | ||
| GG | 10 (13.0%) | 95 (20.4%) | 0.48 (0.23,1.03) | ||
| SEC14L2 | rs5753106 | ||||
| AA | 52 (67.5%) | 263 (55.7%) | 1.0 | 0.01 | |
| AG | 24 (31.2%) | 178 (37.7%) | 0.71 (0.43,1.19) | ||
| GG | 1 (1.3%) | 31 (6.6%) | 0.14 (0.02,1.00) | ||
| SELENBP1 | rs2769264 | ||||
| TT | 46 (59.0%) | 334 (71.2%) | 1.0 | 0.05 | |
| any G | 32 (41.0%) | 121 (25.8%) | 1.65 (1.01,2.69) | ||
| SOD1 | rs2070424 | ||||
| AA | 74 (94.9%) | 400 (85.8%) | 1.0 | 0.04 | |
| Any G | 4 (5.1%) | 62 (13.3%) | 0.33 (0.12,0.95) | ||
| SOD2 | rs7855 | ||||
| AA | 66 (84.6%) | 433 (91.7%) | 1.0 | 0.03 | |
| any G | 12 (15.4%) | 39 (8.3%) | 2.16 (1.08,4.33) | ||
| TTPA | rs12679996 | ||||
| CC | 22 (28.9%) | 189 (41.1%) | 1.0 | 0.004 | |
| CT | 31 (40.8%) | 196 (42.6%) | 1.29 (0.72,2.31) | ||
| TT rs4606052 | 23 (30.3%) | 75 (16.3%) | 2.82 (1.48,5.38) | ||
| CC | 20 (26.7%) | 142 (31.5%) | 1.0 | 0.04 | |
| CT | 28 (37.3%) | 209 (46.3%) | 0.96 (0.52,1.76) | ||
| TT | 27 (36.0%) | 100 (22.2%) | 2.03 (1.07,3.85) | ||
| TXNRD2 | rs8141691 | ||||
| GG | 28 (36.4%) | 217 (46.4%) | 1.0 | 0.03 | |
| AG | 33 (42.9%) | 194 (41.5%) | 1.37 (0.80,2.37) | ||
| AA | 16 (20.8%) | 57 (12.2%) | 2.19 (1.12,4.28) | ||
Full results for all SNP * supplement interactions presented in Supplementary Table 3
DISCUSSION
In this large case-cohort study nested within SELECT, we found that genetic variants in several key antioxidant genes were nominally associated with risk of high-grade prostate cancer, including SNPs in CAT, GPX1, SOD1, SOD2, SOD3, TXNRD2, SEC14L2, and TTPA. Moreover, the associations of several of these genetic variants and high-grade prostate cancer differed as a function of selenium or vitamin E supplementation. For example, we observed significant effect modification of three SNPs in CAT by selenium supplementation. For rs7944397, we found an inverse association of the rare variant allele with high-grade disease among men in the placebo arm, whereas there was no association among men in the selenium arm. Similarly, for rs533425, we observed a significantly increased risk with the rare variant allele in the placebo arm, and no association in the supplementation arm. Given the high compliance of men in SELECT, these data suggest that the effect of these genetic variants on high-grade prostate cancer depends on endogenous levels of selenium.
It is noteworthy that none of the SNPs examined in SEP15, GPX3, GPX4, SEPP1, or XRCC1 were associated with high-grade prostate cancer in SELECT, either individually or through an interaction with selenium or vitamin E supplement assignment, whereas at least one prior report had indicated a potential direct association or interaction between these genes and vitamin E or selenium intake or levels, and risk of prostate cancer. (14-17, 20, 21, 29, 43-45)
SOD2, GPX1, and CAT have been researched most commonly in relation to various human diseases, including asthma, cardiovascular disease, diabetes, and cancer, including prostate cancer.(46) Of these, SOD2 has been investigated the most with regards to prostate cancer, and several,(47-49) but not all(50) meta-analyses have reported significant associations between genetic variants in SOD2 and risk of prostate cancer, particularly for more aggressive disease. We and others have previously reported on potential interaction effects between SNPS in SOD1, SOD2, selenoprotein or selenobinding proteins, and selenium status, and risk of aggressive prostate cancer.(15, 17, 19, 20, 23, 45, 51-53) SNPs in TXNRD1 and TXNRD2 have been reported to modify the association of circulating selenium and risk of aggressive prostate cancer;(54) and variants in TXNRD1 and GPX4 have been associated with prostate cancer-specific mortality, though results for the latter were not statistically significant after consideration for multiple comparisons.(21) Lower CAT activity measured in blood has been associated with higher Gleason grade in one small study.(55) The exact function of all the SNPs noted to interact potentially with selenium assignment for risk of high-grade prostate cancer, is not known. However, rs7855 is in the 3’ UTR of SOD2 and could be influencing splicing or acting as an enhancer. Also, rs10836233 in CAT is in LD (r2 = 0.93) with rs11032717 in ELF5, which is an ETS transcription factor gene that has been implicated in androgen sensitivity and aggressiveness of prostate cancer cell lines (56-58); and rs533425 in CAT is in moderate linkage disequilibrium (R2=0.42) with the functional 262 C/T SNP (rs1001179) that has been associated with advanced stage prostate cancer risk. (59)
Data from the ATBC trial indicated there is potential effect modification between a variant in SEC14L2 (rs2299829), vitamin E assignment, and risk of prostate cancer; and between variants in SEC14L2 (rs2299825, rs2299826), dietary intake of alpha-tocopherol, and risk of advanced prostate cancer.(18) This is noteworthy given the strong linkage disequilibrium (r2 = 0.86) between rs2299825 with rs5753106 that we identified in the current study. We previously reported on potential interaction effects between GPX4, gamma-tocopherol, and risk of lethal prostate cancer.(60) Our observation of potential interaction between a variant in SEC14L2 (rs5753106), vitamin E assignment, and high-grade prostate cancer was somewhat consistent with the prior report from the ATBC trial, as rs2299825 is in strong linkage disequilibrium with rs5753106. Additionally, rs5753106 in SEC14L2 is strong LD with the 3’- and 5’-UTR regions for several other genes, including: SF3A1, CCDC157, and RNF215. Further, we identified a potential interaction between vitamin E assignment and rs4606052 in TTPA and risk of high-grade prostate cancer. While rs4606052 is intronic, it is in strong LD (r2=0.99) with rs4587328 in the 3’-UTR of TTPA, which encodes instructions for making alpha-tocopherol transport protein that controls the delivery and distribution of vitamin E from food throughout the body.
While our data are consistent with prior reports indicating potential interactions between SOD2, SOD3, and TXNRD2, and selenium status and prostate cancer risk,(45, 51-54) the specific SNPs previously implicated in each of these genes in our study genes were not statistically significantly related to the outcomes of interest in the current study and not the same as the SNPs we identified. (61) The differences across studies may in part be due to these genes having multiple roles at different time points in prostate cancer progression, and each study addressed a slightly different question based on their study populations and outcome definitions. Moreover, the SNPs studied to date may be tagging to varying degree the true causal SNP within each of these genes. Further research is warranted to understand the downstream functional effects of these individual SNPS to confirm and elucidate the role of these genes on selenium metabolism and prostate cancer.
There are strengths and limitations to consider in assessing the impact of these findings. This is the first study to examine potential interactions between selenium-related genes and selenium supplementation and risk of aggressive prostate cancer using a randomized design. Leveraging the randomized design of SELECT reduces potential confounding in the gene-antioxidant interactions. The study includes a large number of high-grade prostate cancers, and comprehensively assesses genetic variation across 21 unique selenium- or vitamin E-related genes. Nonetheless, the results must be interpreted with caution given the large number of potential effects evaluated. We focused on pre-specified hypotheses and did not adjust for multiple testing in our analyses. It is noteworthy, however, that many of the SNPs that were nominally associated with high-grade prostate cancer were also the SNPs that were significant in the interaction analyses. We also did not have sufficient numbers to examine any minority populations individually, and results presented are for Caucasian participants only. Additionally, SELECT assigned participants to higher doses of selenium and vitamin E than would usually be consumed by diet alone, and at least for selenium, data suggest that the biological relationship may be U-shaped (i.e., highest and lowest levels confer adverse health effects, whereas a middle level is considered optimal).(62-64) Thus, caution is warranted in generalizing these results to comment on the potential interaction effects between these gene variants and dietary intakes of selenium and vitamin E, and prostate cancer risk.
In conclusion, this report on more than 130 SNPs in 21 genes provides support for the hypothesis that genetic variation in selenium and vitamin E metabolism/transport genes may influence risk of overall and high-grade prostate cancer, and may modify an individual man’s response to vitamin E or selenium supplementation with regards to these risks.
Supplementary Material
Acknowledgements
We are grateful to the participants in SELECT.
Financial support: The trial was funded by NIH R01 CA106947-01A1 (J.M. Chan). This work was funded in part by Public Health Service grants U10 CA37429 (A.K. Darke/E.A. Klein/I.M. Thompson/C.M. Tangen/J.M. Rae/P.J. Goodman) and UM1 CA182883 (I.M. Thompson/C.M. Tangen/A.K. Darke/P.J. Goodman) from the National Cancer Institute.
Footnotes
Trial registration ID (for SELECT): NCT00006392
Conflict of Interest Statement: There are no conflicts to disclose.
REFERENCES
- 1.Clark LC, Dalkin B, Krongrad A, Combs GF, Turnbull BW, Slate EH, et al. Decreased incidence of prostate cancer with selenium supplementation: results of a double-blind cancer prevention trial. Br J Urol. 1998;81:730–4. doi: 10.1046/j.1464-410x.1998.00630.x. [DOI] [PubMed] [Google Scholar]
- 2.Heinonen OP, Albanes D, Virtamo J, Taylor PR, Huttunen JK, Harman AM, et al. Prostate cancer and supplementation with alpha-tocopherol and beta-carotene: Incidence and mortality in a controlled trial. J Natl Cancer Inst. 1998;90:440–6. doi: 10.1093/jnci/90.6.440. [DOI] [PubMed] [Google Scholar]
- 3.Clark LC, Combs GF, Jr., Turnbull BW, Slate EH, Chalker DK, Chow J, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. Jama. 1996;276:1957–63. [PubMed] [Google Scholar]
- 4.Helzlsouer KJ, Huang HY, Alberg AJ, Hoffman S, Burke A, Norkus EP, et al. Association between alpha-tocopherol, gamma-tocopherol, selenium, and subsequent prostate cancer. J Natl Cancer Inst. 2000;92:2018–23. doi: 10.1093/jnci/92.24.2018. [DOI] [PubMed] [Google Scholar]
- 5.Nomura AM, Lee J, Stemmermann GN, Combs GF. Serum selenium and subsequent risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:883–7. [PubMed] [Google Scholar]
- 6.Brooks JD, Metter EJ, Chan DW, Sokoll LJ, Landis P, Nelson WG, et al. Plasma selenium level before diagnosis and the risk of prostate cancer development. J Urol. 2001;166:2034–8. [PubMed] [Google Scholar]
- 7.Helzlsouer KJ, Huang HY, Alberg AJ, Hoffman S, Burke A, Norkus EP, et al. Association Between alpha-Tocopherol, gamma-Tocopherol, Selenium, and Subsequent Prostate Cancer. J Natl Cancer Inst. 2000;92:2018–23. doi: 10.1093/jnci/92.24.2018. [DOI] [PubMed] [Google Scholar]
- 8.van den Brandt PA, Zeegers MP, Bode P, Goldbohm RA. Toenail selenium levels and the subsequent risk of prostate cancer: a prospective cohort study. Cancer Epidemiol Biomarkers Prev. 2003;12:866–71. [PubMed] [Google Scholar]
- 9.Weinstein SJ, Wright ME, Pietinen P, King I, Tan C, Taylor PR, et al. Serum alpha-tocopherol and gamma-tocopherol in relation to prostate cancer risk in a prospective study. J Natl Cancer Inst. 2005;97:396–9. doi: 10.1093/jnci/dji045. [DOI] [PubMed] [Google Scholar]
- 10.Yoshizawa K, Willett WC, Morris SJ, Stampfer MJ, Spiegelman D, Rimm EB, et al. Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J Natl Cancer Inst. 1998;90:1219–24. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- 11.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA : the journal of the American Medical Association. 2009;301:39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albanes D, Till C, Klein EA, Goodman PJ, Mondul AM, Weinstein SJ, et al. Plasma Tocopherols and Risk of Prostate Cancer in the Selenium and Vitamin E Cancer Prevention Trial (SELECT) Cancer Prev Res (Phila) 2014;7:886–95. doi: 10.1158/1940-6207.CAPR-14-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristal AR, Darke AK, Morris JS, Tangen CM, Goodman PJ, Thompson IM, et al. Baseline selenium status and effects of selenium and vitamin e supplementation on prostate cancer risk. J Natl Cancer Inst. 2014;106:djt456. doi: 10.1093/jnci/djt456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer SR, Richman EL, Sosa E, Weinberg V, Song X, Witte JS, et al. Antioxidant and vitamin E transport genes and risk of high-grade prostate cancer and prostate cancer recurrence. The Prostate. 2013;73:1786–95. doi: 10.1002/pros.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penney KL, Li H, Mucci LA, Loda M, Sesso HD, Stampfer MJ, et al. Selenoprotein P genetic variants and mrna expression, circulating selenium, and prostate cancer risk and survival. Prostate. 2013;73:700–5. doi: 10.1002/pros.22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Platz EA. Is prostate cancer prevention with selenium all in the genes? Cancer Prev Res (Phila) 2010;3:576–8. doi: 10.1158/1940-6207.CAPR-10-0072. [DOI] [PubMed] [Google Scholar]
- 17.Penney KL, Schumacher FR, Li H, Kraft P, Morris JS, Kurth T, et al. A large prospective study of SEP15 genetic variation, interaction with plasma selenium levels, and prostate cancer risk and survival. Cancer Prev Res (Phila) 2010;3:604–10. doi: 10.1158/1940-6207.CAPR-09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright ME, Peters U, Gunter MJ, Moore SC, Lawson KA, Yeager M, et al. Association of variants in two vitamin e transport genes with circulating vitamin e concentrations and prostate cancer risk. Cancer Res. 2009;69:1429–38. doi: 10.1158/0008-5472.CAN-08-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinbrecher A, Meplan C, Hesketh J, Schomburg L, Endermann T, Jansen E, et al. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev. 2010;19:2958–68. doi: 10.1158/1055-9965.EPI-10-0364. [DOI] [PubMed] [Google Scholar]
- 20.Geybels MS, van den Brandt PA, Schouten LJ, van Schooten FJ, van Breda SG, Rayman MP, et al. Selenoprotein gene variants, toenail selenium levels, and risk for advanced prostate cancer. J Natl Cancer Inst. 2014;106:dju003. doi: 10.1093/jnci/dju003. [DOI] [PubMed] [Google Scholar]
- 21.Geybels MS, Hutter CM, Kwon EM, Ostrander EA, Fu R, Feng Z, et al. Variation in selenoenzyme genes and prostate cancer risk and survival. Prostate. 2013;73:734–42. doi: 10.1002/pros.22617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundmark B, Zethelius B, Garmo H, Holmberg L. Serum levels of selenium and smoking habits at age 50 influence long term prostate cancer risk; a 34 year ULSAM follow-up. BMC Cancer. 2011;11:431. doi: 10.1186/1471-2407-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper ML, Adami HO, Gronberg H, Wiklund F, Green FR, Rayman MP. Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Res. 2008;68:10171–7. doi: 10.1158/0008-5472.CAN-08-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerome-Morais A, Wright ME, Liu R, Yang W, Jackson MI, Combs GF, Jr., et al. Inverse association between glutathione peroxidase activity and both selenium-binding protein 1 levels and Gleason score in human prostate tissue. Prostate. 2012;72:1006–12. doi: 10.1002/pros.21506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–30. [PubMed] [Google Scholar]
- 26.Rybicki BA, Conti DV, Moreira A, Cicek M, Casey G, Witte JS. DNA repair gene XRCC1 and XPD polymorphisms and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:23–9. doi: 10.1158/1055-9965.epi-03-0053. [DOI] [PubMed] [Google Scholar]
- 27.van Gils CH, Bostick RM, Stern MC, Taylor JA. Differences in base excision repair capacity may modulate the effect of dietary antioxidant intake on prostate cancer risk: an example of polymorphisms in the XRCC1 gene. Cancer Epidemiol Biomarkers Prev. 2002;11:1279–84. [PubMed] [Google Scholar]
- 28.Zhang J, Dhakal IB, Greene G, Lang NP, Kadlubar FF. Polymorphisms in hOGG1 and XRCC1 and risk of prostate cancer: effects modified by plasma antioxidants. Urology. 2010;75:779–85. doi: 10.1016/j.urology.2009.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JY, Huang Y, Sellers TA. Single nucleotide polymorphisms in DNA repair genes and prostate cancer risk. Methods Mol Biol. 2009;471:361–85. doi: 10.1007/978-1-59745-416-2_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YJ, Baek SH, Bogner PN, Clement IP, Rustum YM, Fakih MG, et al. Targeting the Nrf2-Prx1 Pathway with Selenium to Enhance the Efficacy and Selectivity of Cancer Therapy. J Cancer Molecules. 2007;3:37–43. [Google Scholar]
- 31.Raatikainen S, Aaaltomaa S, Karja V, Soini Y. Increased Peroxiredoxin 6 Expression Predicts Biochemical Recurrence in Prostate Cancer Patients After Radical Prostatectomy. Anticancer Res. 2015;35:6465–70. [PubMed] [Google Scholar]
- 32.Whitaker HC, Patel D, Howat WJ, Warren AY, Kay JD, Sangan T, et al. Peroxiredoxin-3 is overexpressed in prostate cancer and promotes cancer cell survival by protecting cells from oxidative stress. Br J Cancer. 2013;109:983–93. doi: 10.1038/bjc.2013.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ummanni R, Barreto F, Venz S, Scharf C, Barett C, Mannsperger HA, et al. Peroxiredoxins 3 and 4 are overexpressed in prostate cancer tissue and affect the proliferation of prostate cancer cells in vitro. J Proteome Res. 2012;11:2452–66. doi: 10.1021/pr201172n. [DOI] [PubMed] [Google Scholar]
- 34.Shiota M, Yokomizo A, Kashiwagi E, Takeuchi A, Fujimoto N, Uchiumi T, et al. Peroxiredoxin 2 in the nucleus and cytoplasm distinctly regulates androgen receptor activity in prostate cancer cells. Free Radic Biol Med. 2011;51:78–87. doi: 10.1016/j.freeradbiomed.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Basu A, Banerjee H, Rojas H, Martinez SR, Roy S, Jia Z, et al. Differential expression of peroxiredoxins in prostate cancer: consistent upregulation of PRDX3 and PRDX4. Prostate. 2011;71:755–65. doi: 10.1002/pros.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz MA, Abdel-Mageed AB, Mondal D. The nrf1 and nrf2 balance in oxidative stress regulation and androgen signaling in prostate cancer cells. Cancers (Basel) 2010;2:1354–78. doi: 10.3390/cancers2021354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chhipa RR, Lee KS, Onate S, Wu Y, Ip C. Prx1 enhances androgen receptor function in prostate cancer cells by increasing receptor affinity to dihydrotestosterone. Molecular cancer research : MCR. 2009;7:1543–52. doi: 10.1158/1541-7786.MCR-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SY, Yu X, Ip C, Mohler JL, Bogner PN, Park YM. Peroxiredoxin 1 interacts with androgen receptor and enhances its transactivation. Cancer Res. 2007;67:9294–303. doi: 10.1158/0008-5472.CAN-07-0651. [DOI] [PubMed] [Google Scholar]
- 39.Shen C, Nathan C. Nonredundant antioxidant defense by multiple two-cysteine peroxiredoxins in human prostate cancer cells. Mol Med. 2002;8:95–102. [PMC free article] [PubMed] [Google Scholar]
- 40.Arner ES, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–6. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Kang D, Lee KM, Park SK, Berndt SI, Peters U, Reding D, et al. Functional variant of manganese superoxide dismutase (SOD2 V16A) polymorphism is associated with prostate cancer risk in the prostate, lung, colorectal, and ovarian cancer study. Cancer Epidemiol Biomarkers Prev. 2007;16:1581–6. doi: 10.1158/1055-9965.EPI-07-0160. [DOI] [PubMed] [Google Scholar]
- 42.Prentice RL. A Case-Cohort Design for Epidemiologic Cohort Studies and Disease Prevention Trials. Biometrika. 1986;73:1–11. [Google Scholar]
- 43.Major JM, Yu K, Weinstein SJ, Berndt SI, Hyland PL, Yeager M, et al. Genetic variants reflecting higher vitamin e status in men are associated with reduced risk of prostate cancer. J Nutr. 2014;144:729–33. doi: 10.3945/jn.113.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Gils CH, Bostick RM, Stern MC, Taylor JA. Differences in base excision repair capacity may modulate the effect of dietary antioxidant intake on prostate cancer risk: an example of polymorphisms in the XRCC1 gene. Cancer Epidemiol Biomarkers Prev. 2002;11:1279–84. [PubMed] [Google Scholar]
- 45.Gerstenberger JP, Bauer SR, Van Blarigan EL, Sosa E, Song X, Witte JS, et al. Selenoprotein and antioxidant genes and the risk of high-grade prostate cancer and prostate cancer recurrence. Prostate. 2015;75:60–9. doi: 10.1002/pros.22892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crawford A, Fassett RG, Geraghty DP, Kunde DA, Ball MJ, Robertson IK, et al. Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene. 2012;501:89–103. doi: 10.1016/j.gene.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Sun GG, Wang YD, Lu YF, Hu WN. Different association of manganese superoxide dismutase gene polymorphisms with risk of prostate, esophageal, and lung cancers: evidence from a meta-analysis of 20,025 subjects. Asian Pac J Cancer Prev. 2013;14:1937–43. doi: 10.7314/apjcp.2013.14.3.1937. [DOI] [PubMed] [Google Scholar]
- 48.Mao C, Qiu LX, Zhan P, Xue K, Ding H, Du FB, et al. MnSOD Val16Ala polymorphism and prostate cancer susceptibility: a meta-analysis involving 8,962 subjects. J Cancer Res Clin Oncol. 2010;136:975–9. doi: 10.1007/s00432-009-0742-x. [DOI] [PubMed] [Google Scholar]
- 49.Wang S, Wang F, Shi X, Dai J, Peng Y, Guo X, et al. Association between manganese superoxide dismutase (MnSOD) Val-9Ala polymorphism and cancer risk - A meta-analysis. Eur J Cancer. 2009;45:2874–81. doi: 10.1016/j.ejca.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 50.Liwei L, Chunyu L, Ruifa H. Association between manganese superoxide dismutase gene polymorphism and risk of prostate cancer: a meta-analysis. Urology. 2009;74:884–8. doi: 10.1016/j.urology.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Kantoff PW, Giovannucci E, Leitzmann MF, Gaziano JM, Stampfer MJ, et al. Manganese superoxide dismutase polymorphism, prediagnostic antioxidant status, and risk of clinical significant prostate cancer. Cancer Res. 2005;65:2498–504. doi: 10.1158/0008-5472.CAN-04-3535. [DOI] [PubMed] [Google Scholar]
- 52.Abe M, Xie W, Regan MM, King IB, Stampfer MJ, Kantoff PW, et al. Single-nucleotide polymorphisms within the antioxidant defence system and associations with aggressive prostate cancer. BJU Int. 2011;107:126–34. doi: 10.1111/j.1464-410X.2010.09344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan JM, Oh WK, Xie W, Regan MM, Stampfer MJ, King IB, et al. Plasma selenium, manganese superoxide dismutase, and intermediate- or high-risk prostate cancer. J Clin Oncol. 2009;27:3577–83. doi: 10.1200/JCO.2008.18.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meplan C, Rohrmann S, Steinbrecher A, Schomburg L, Jansen E, Linseisen J, et al. Polymorphisms in thioredoxin reductase and selenoprotein K genes and selenium status modulate risk of prostate cancer. PLoS One. 2012;7:e48709. doi: 10.1371/journal.pone.0048709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Battisti V, Maders LD, Bagatini MD, Reetz LG, Chiesa J, Battisti IE, et al. Oxidative stress and antioxidant status in prostate cancer patients: relation to Gleason score, treatment and bone metastasis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2011;65:516–24. doi: 10.1016/j.biopha.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Koyama K, Takahara K, Inamoto T, Ibuki N, Minami K, Uehara H, et al. E74-like factor inhibition induces reacquisition of hormone sensitiveness decreasing period circadian protein homolog 1 expression in prostate cancer cells. Prostate Int. 2015;3:16–21. doi: 10.1016/j.prnil.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yao B, Zhao J, Li Y, Li H, Hu Z, Pan P, et al. Elf5 inhibits TGF-beta-driven epithelial-mesenchymal transition in prostate cancer by repressing SMAD3 activation. Prostate. 2015;75:872–82. doi: 10.1002/pros.22970. [DOI] [PubMed] [Google Scholar]
- 58.Xie BX, Zhang H, Wang J, Pang B, Wu RQ, Qian XL, et al. Analysis of differentially expressed genes in LNCaP prostate cancer progression model. J Androl. 2011;32:170–82. doi: 10.2164/jandrol.109.008748. [DOI] [PubMed] [Google Scholar]
- 59.Geybels MS, van den Brandt PA, van Schooten FJ, Verhage BA. Oxidative stress-related genetic variants, pro- and antioxidant intake and status, and advanced prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2015;24:178–86. doi: 10.1158/1055-9965.EPI-14-0968. [DOI] [PubMed] [Google Scholar]
- 60.Van Blarigan EL, Ma J, Kenfield SA, Stampfer MJ, Sesso HD, Giovannucci EL, et al. Plasma antioxidants, genetic variation in SOD2, CAT, GPX1, GPX4, and prostate cancer survival. Cancer Epidemiol Biomarkers Prev. 2014;23:1037–46. doi: 10.1158/1055-9965.EPI-13-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amin Al Olama A, Dadaev T, Hazelett DJ, Li Q, Leongamornlert D, Saunders EJ, et al. Multiple novel prostate cancer susceptibility signals identified by fine-mapping of known risk loci among Europeans. Hum Mol Genet. 2015;24:5589–602. doi: 10.1093/hmg/ddv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hurst R, Hooper L, Norat T, Lau R, Aune D, Greenwood DC, et al. Selenium and prostate cancer: systematic review and meta-analysis. Am J Clin Nutr. 2012;96:111–22. doi: 10.3945/ajcn.111.033373. [DOI] [PubMed] [Google Scholar]
- 63.Kenfield SA, Van Blarigan EL, DuPre N, Stampfer MJ, Chan JM. Selenium supplementation and prostate cancer mortality. J Natl Cancer Inst. 2015;107:360. doi: 10.1093/jnci/dju360. E LG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richman EL, Chan JM. Selenium and prostate cancer: the puzzle isn't finished yet. The American journal of clinical nutrition. 2012;96:1–2. doi: 10.3945/ajcn.112.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.